Abstract
Circulating adiponectin has been associated with both clinical and subclinical cardiovascular disease (CVD). Variants of the adiponectin gene (ADIPOQ) are associated with clinical CVD, but little is known about associations with subclinical CVD. We studied the association of 11 ADIPOQ SNPs with common and internal carotid intima media thickness (cIMT), presence of coronary artery calcification (CAC), and CAC scores (in those with CAC) in 2847 participants in the Multi-Ethnic Study of Atherosclerosis (MESA). Participants were Caucasian (n=712), African-American (n=712), Chinese (n=718), and Hispanic (n=705). All models were adjusted for age, sex, and field site, and stratified by race/ethnic group. African-Americans with genotypes AG/GG of rs2241767 had 36% greater (95% CI (16%, 59%), p=0.0001) CAC prevalence; they also had a larger common cIMT (p=0.0043). Also in African-Americans, genotypes AG/AA of rs1063537 were associated with a 35% (95% CI (14%, 59%), p=0.0005) greater CAC prevalence. Hispanics with the AA genotype of rs11711353 had a 37% (95% CI (14%, 66%), p=0.0011), greater CAC prevalence compared to those with the GG genotype. Additional adjustment for ancestry in African-American and Hispanic participants did not change the results. No single SNP was associated with subclinical CVD phenotypes in Chinese or Caucasian participants. There appears to be an association between ADIPOQ SNPs and subclinical CVD in African-American and Hispanics. Replication as well as assessment of other ADIPOQ SNPs appears warranted.
Introduction
Subclinical cardiovascular disease (CVD) measures, including carotid intima media thickness (cIMT) and coronary artery calcium (CAC), reflect overall atherosclerotic burden and can be used to non-invasively assess the presence, amount, and severity of atherosclerosis(1,2). Many studies have shown CAC and cIMT are associated with clinical CVD outcomes and also improve prediction of risk for future CVD events(1,2) even in asymptomatic individuals(3,4). A number of studies have evaluated the association between circulating adiponectin and coronary heart disease (CHD) or CVD outcomes with some showing higher levels of adiponectin associated with decreased risk(5,6) while other studies show an increased risk(7,8). However, associations of adiponectin with CAC and common and internal cIMT have shown more consistency, with higher adiponectin generally associated with thinner cIMT and less CAC(9,10,11).
The adiponectin gene (ADIPOQ) encodes the circulating protein adiponectin and is expressed primarily in the adipose tissue of various organs, but it is expressed in vascular tissue as well(12). Only a small proportion of all ADIPOQ polymorphisms have been investigated in relation to circulating adiponectin, and CHD or CVD and it is likely that many functional variants responsible for these observed associations have yet to be studied(13). A number of studies have examined the association of single nucleotide polymorphisms (SNPs) and in ADIPOQ with circulating adiponectin levels and/or CHD, but these studies have had mixed results and have concentrated mostly on just a few SNPs, usually in just one ethnic group(14,15,16,17,18). Few studies have evaluated the association of SNPs in ADIPOQ with subclinical CVD measures and those that exist have again used only a few SNPs in examining these associations, examined the associations in one ethnic group, or had small sample sizes(19,20,21,22).
Thus, given the evidence of the roles of both circulating adiponectin and ADIPOQ in clinical CVD and possibly subclinical CVD measures, this study examined the association of ADIPOQ SNPs chosen by a tagSNP method with the presence and amount of CAC and cIMT in four racial/ethnic groups in the Multi-Ethnic Study of Atherosclerosis (MESA).
Methods
Participants
MESA participants were recruited from six field sites in the United States – Forsyth County, NC, Northern Manhattan/Bronx, NY, Baltimore/Baltimore County, MD, St. Paul, MN, Chicago, IL, and Los Angeles County, CA. The MESA cohort comprises 6,814 men and women of diverse ethnic background who were 45 to 84 years old at the baseline exam and free of clinically overt cardiovascular disease. The cohort was 53% women with a racial/ethnic composition of approximately 38% white, 28% African American, 23% Hispanic and 11% Asian, primarily of Chinese descent. Further details of the objectives and design of MESA were published elsewhere(23).
A subsample of 2847 MESA subjects was selected for a candidate gene study from participants who gave informed consent for DNA extraction. ADIPOQ SNPs were genotyped on this subsample. The candidate gene study included 712 African American, 705 Hispanic, 718 Chinese, and 712 Caucasian participants and approximately equal numbers of men and women. Further details have been previously published on the selection of this same participant subsample (24) and are also available in the supplemental methods.
DNA extraction and SNP selection and genotyping
DNA was extracted by use of a commercially available DNA isolation kit (Puregene; Gentra Systems, Minneapolis, MN) from peripheral leukocytes isolated from packed EDTA and citrate cells of anticoagulated blood frozen at –70 degrees C. Tag SNPs were selected in based on compatibility with the Illumina GoldenGate technology and had to have a minor allele frequency (MAF)>0.05. Further details for SNP selectioncan be found in the supplemental methods and have been published elsewhere(24). A total of 12 ADIPOQ SNPs were chosen using this method.
Illumina Genotyping Services (Illumina Inc., San Diego, CA) performed genotyping using the GoldenGate assay. Illumina performed initial quality control to identify samples and SNPs that failed genotyping. After removal of failed SNPs and samples, the genotype call rate was 99.93%. Further details on quality control can be found elsewhere(24).
Outcomes
Both common and internal cIMT were measured by high-resolution B-mode ultrasonography on a Logiq 700 ultrasound machine (GE Medical Systems, Waukesha, WA) at the baseline examination and calculation of these values was performed at the central MESA ultrasound reading center (Tufts-New England Medical Center, Boston, MA). Overall common and internal cIMT were calculated as the mean of all available maximum wall thicknesses across all scans, for both left and right sides, and for the near and far walls. Coronary artery calcium (CAC) scanning was performed by either ECG-gated electron beam (EBCT) or multidetector (MDCT) computed tomography per the MESA protocol(25). CAC was quantified using the Agatston method. Scans were read centrally at the Los Angeles Biomedical Research Institute at Harbor-UCLA. Two CT scans were performed for each participant, and the results were averaged after phantom attenuation adjustment. Further details have been published elsewhere(25).
Covariates
Information on age, sex, and race/ethnicity, smoking, alcohol use, education and income was obtained via baseline interview and questionnaire. Waist circumference, systolic and diastolic blood pressure, trigylcerides, HDL cholesterol, LDL cholesterol, fasting glucose, and C-reactive protein were all measured using standard methods and assays. Further details on methods and specific variable definitions can be found in the supplementary methods.
Statistical Analysis
For SNPs, pairwise linkage disequilibrium (LD) was calculated (r2) and LD plots were generated using Haploview V4.0(26). Hardy-Weinberg equilibrium (HWE) tests were conducted within each racial/ethnic group using the exact test. Baseline characteristics for each racial/ethnic group were compared across groups by ANOVA, chi-square or Kruskal-Wallis as appropriate.
Outcomes included presence or absence of CAC (CAC>0 vs. CAC=0), and CAC score as a continuous outcome among those with values > 0, as well as common and internal cIMT measures. In the MESA candidate gene substudy, the prevalence of CAC was high, so log-binomial models were used to assess associations of each SNP with CAC presence, as odds ratios from logistic regression will over-estimate the prevalence ratios in this case. In the event that the log binomial model did not converge, a Gaussian error distribution was used and robust standard errors were estimated. As common and internal cIMT and CAC score > 0 were skewed, these were natural log-transformed. Linear regression was used to assess the association of the SNPs with these outcomes and geometric means are presented.
All analyses were stratified by self-reported race/ethnicity to control for population stratification. For significant SNPs in African American and Hispanic models, additional control for population stratification was achieved by adjusting for percent European ancestry for African Americans and Hispanics, and additionally for percent Native American ancestry for Hispanics. Ancestry was estimated separately in each racial/ethnic group from 199 ancestry informative markers using STRUCTURE V2.2, software that employs a Bayesian Markov Chain Monte Carlo simulation approach(27).
Univariate associations of each SNP with each outcome were evaluated first by 2 degree of freedom tests in models adjusting for age, sex, and field center. Individual SNPs with a 2 degree of freedom p<0.02 were examined further, adding ancestry, and then cardiovascular risk factors, to each model. Multiple comparisons were controlled for using a Bonferroni correction for each racial/ethnic group and outcome, α = 0.05/11 = 0.0045. Analyses were conducted using SAS V9.1 (SAS Institute, Cary, NC).
Results
Supplementary Table 1 shows the minor allele frequencies and HWE p-values for ADIPOQ SNPs by racial/ethnic group. SNPs rs9877202 and rs1403697 were not polymorphic among the Chinese; these SNPs also had only one Caucasian participant with the heterozygous genotype and none with the rare homozygous genotype. Minor allele frequencies for most SNPs differed somewhat between the ethnic groups, but the differences were not substantial. However, two SNPs did show a different minor allele for some of the racial/ethnic groups. There were no substantial deviations from HWE (p<0.001) for anySNP.
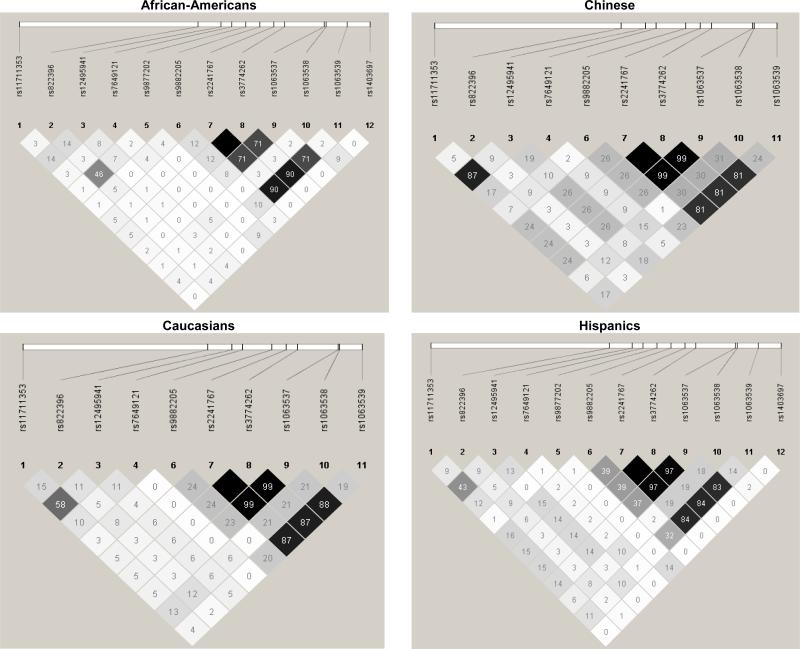
The LD plots show that rs2241767 and rs3774262 were perfectly correlated (r2=1) within every racial/ethnic group, so analyses proceeded with rs2241767 but not rs3774262 (Figure 1). Thus, the Bonferroni-corrected p-value threshold used to determine significance for single SNP associations within each racial/ethnic group was α = 0.05/11 = 0.0045. Among African Americans, rs2241767 was in high LD with rs1063539 (Figure 1). In Chinese, rs11711353 and rs1249541 were in high LD; rs1063537 and rs2241767 were in high LD among Chinese, Caucasians, and Hispanics (Figure 1).
Figure 1.
Pairwise linkage disequilibrium (r2) for adiponectin gene SNPs
Figure 1 shows the pairwise linkage disequilibrium (r2) by racial/ethnic group for single nucleotide polymorphisms in the adiponectin gene.
Table 1 shows baseline characteristics for the MESA candidate gene study by racial/ethnic group. There were some significant differences in cIMT between racial/ethnic groups. Caucasians had the highest prevalence of CAC while African-Americans had the largest common cIMT. No participants were missing CAC measures, and only 31 were missing common cIMT and 66 were missing internal cIMT.
Table 1.
Baseline characteristics among the n=2847 MESA participants from the candidate gene substudy
| Characteristic | Caucasian n=712 | African-American n=712 | Chinese n=718 | Hispanic n=705 | p-valuea |
|---|---|---|---|---|---|
| Age, years | 61.5±10.4 | 61.5±9.9 | 62.3±10.3 | 61.2±10.1 | 0.18 |
| Gender | |||||
| Male | 332 (46.6) | 321 (45.1) | 348 (48.5) | 324 (46.0) | 0.63 |
| Female | 380 (53.4) | 391 (54.9) | 370 (51.5) | 381 (54.0) | |
| Field Center | |||||
| Wake Forest | 183 (25.7) | 179 (25.1) | 0 (0) | 1 (0.14) | <0.001 |
| Columbia | 59 (8.3) | 153 (21.5) | 2 (0.28) | 244 (34.6) | |
| Johns Hopkins | 115 (16.2) | 215 (30.2) | 0 (0) | 0 (0) | |
| Minnesota | 170 (23.9) | 0 (0) | 0 (0) | 202 (28.7) | |
| Northwestern | 133 (18.7) | 104 (14.6) | 267 (37.2) | 0 (0) | |
| UC-Los Angeles | 52 (7.3) | 61 (8.6) | 449 (62.5) | 258 (36.6) | |
| Education ≥ college | 552 (77.5) | 492 (69.1) | 423 (58.9) | 239 (33.9) | <0.001 |
| Income ≥$40,000/year | 463 (65.1) | 328 (46.1) | 249 (34.7) | 172 (24.4) | <0.001 |
| Current smoker | 112 (15.7) | 135 (19.0) | 38 (5.3) | 106 (15.0) | <0.001 |
| Hypertension | 277 (38.9) | 398 (55.9) | 269 (37.5) | 295 (41.8) | <0.001 |
| Diabetes | 32 (4.5) | 97 (13.6) | 71 (9.9) | 95 (13.5) | <0.001 |
| Chronic kidney disease | 89 (12.5) | 47 (6.6) | 72 (10.0) | 56 (7.9) | 0.001 |
| Systolic BP, mmHg | 124.2±21.1 | 129.6±20.9 | 124.7±21.7 | 126.0±21.8 | <0.001 |
| Diastolic BP, mmHg | 70.6±9.9 | 73.9±9.8 | 71.9±10.2 | 71.3±10.3 | <0.001 |
| Fasting glucose, mg/dL | 97.2±19.7 | 107.0±33.2 | 105.7±27.4 | 110.4±40.5 | <0.001 |
| HDL cholesterol, mg/dL | 52.6±16.0 | 52.9±15.4 | 49.4±12.4 | 47.5±12.6 | <0.001 |
| LDL cholesterol, mg/dL | 117.3±31.0 | 116.3±33.0 | 115.3±28.3 | 119.9±32.4 | 0.03 |
| Triglycerides, mg/dLb | 113.0 (87.0) | 91.0 (57.0) | 124.0 (86.0) | 137.0 (88.5) | <0.001 |
| Body mass index, kg/m2 | 27.7±5.3 | 30.1±5.8 | 24.0±3.3 | 29.5±5.1 | <0.001 |
| Waist circumference, cm | 97.8±15.1 | 101.0±14.5 | 87.2±9.9 | 100.4±12.8 | <0.001 |
| C-reactive protein, mg/Lb | 1.79 (3.63) | 2.59 (4.54) | 0.87 (1.32) | 2.43 (3.69) | <0.001 |
| Common carotid IMT, mm | 0.87±0.21 | 0.91±0.19 | 0.83±0.18 | 0.86±0.18 | <0.001 |
| Internal carotid IMT, mm | 1.09±0.59 | 1.10±0.61 | 0.88±0.47 | 1.02±0.56 | <0.001 |
| Coronary artery calcium >0 | 393 (55.2) | 301 (42.3) | 359 (50.0) | 317 (45.0) | <0.001 |
| Coronary artery calcium score (>0)b | 107.7 (335.3) | 77.7 (310.4) | 70.6 (181.6) | 73.6 (238.4) | 0.18 |
| Lipid-lowering med use | 112 (15.7) | 122 (17.1) | 101 (14.1) | 103 (14.6) | 0.37 |
| Anti-hypertension med use | 230 (32.3) | 351 (49.3) | 207 (28.8) | 231 (32.8) | <0.001 |
p-value by ANOVA, chi-square or Kruskal-Wallis as appropriate
Median (Interquartile range)
Results of the 2 degree of freedom demographic adjusted single SNP association tests for all racial/ethnic groups and outcomes are presented in Supplemental Figure 1. In some cases, SNPs with very rare homozygote genotypes had to be combined with the heterozygote genotype. Two SNPs reached the Bonferroni-corrected p=0.0045 significance level, both in the African American group (Figure 1). rs2241767 was strongly associated with CAC presence (p=0.0029) in African Americans. Similar significant results were observed for rs1063539 (a SNP highly correlated with rs2241767, r2=0.90) and CAC presence. Results that nearly reached the Bonferroni-corrected significance level for African Americans included rs1063537 and CAC presence (p=0.0063), rs2241767 and common cIMT (p=0.0071). rs1063538 was nominally associated with CAC score > 0, p=0.0148. Among the Hispanics, rs11711353 was nominally associated with CAC presence (p=0.0121), while rs7649121 was nominally associated with common cIMT (p=0.0174) (Figure 1). No associations among Caucasians or Chinese had p<0.02 (Figure 1).
The AG/GG genotypes of rs2241767 were associated with a 36% greater CAC prevalence among African Americans as compared to the AA genotype in demographic adjusted models, p=0.0002 (Table 2). Further adjustment for percent European ancestry did not change this association. Similar results were observed for the rs1063537-CAC presence association, with the AG/AA genotypes associated with a 35% greater CAC prevalence as compared to those with the GG genotype in fully adjusted models, p=0.0005 (Table 2). Although it did not quite meet the multiple comparisons p-value threshold, those with the GG genotype of rs1063538 had an average CAC score of 120.1, which was higher when compared to those with the AA genotype (average CAC score 54.9), p=0.0056. Further adjustment for European ancestry only slightly attenuated this association. Common cIMT differed by genotype for rs2241767 in African Americans, with those with the AG/GG genotypes having larger common cIMT (p=0.0071) in demographic adjusted models (Table 2). Adjustment for European ancestry strengthened this association slightly and the p-value criteria for multiple comparisons was met (p=0.0043). Further adjustment for cardiovascular risk factors, including smoking, alcohol use, education, income, diabetes, chronic kidney disease, hypertension, anti-hypertensive and lipid lowering medication use, body mass index, waist circumference, systolic and diastolic blood pressure, trigylcerides, HDL cholesterol, LDL cholesterol, fasting glucose, C-reactive protein, did not substantially alter any of the above associations. No SNP-SNP interactions were present for any ethnic group for SNPs with p<0.05 by 2 df test.
Table 2.
Association of ADIPOQ SNPs with subclinical CVD in African Americans
| SNP-outcome | Demographic Adjusted PR or Means (95% CI)a | p-value | Ancestry Adjusted PR or Means (95% CI)b | p-value | Fully Adjusted PR or Means (95% CI)c | p-value |
|---|---|---|---|---|---|---|
| rs2241767-CAC presence | ||||||
| AA (n=648) | ref | -- | ref | -- | ref | -- |
| AG/GG (n=64)d | 1.36 (1.16, 1.62) | 0.0002 | 1.36 (1.16, 1.59) | 0.0001 | 1.37(1.13, 1.66) | 0.0013 |
| rs1063537-CAC presence | ||||||
| GG (n=666) | ref | -- | ref | -- | ref | -- |
| AG/AA (n=46)d | 1.39 (1.16, 1.66) | 0.0004 | 1.35 (1.14, 1.59) | 0.0005 | 1.36(1.13, 1.64) | 0.0013 |
| rs2241767-Common | ||||||
| 0.89 mm (0.88, 0.90) | ref | 0.89 mm (0.88, 0.90) | ref | 0.89 mm (0.87, 0.90) | ref | |
| cIMT | ||||||
| 0.95 mm (0.91, 0.99) | 0.0071 | 0.95 mm (0.91, 1.00) | 0.0043 | 0.97 mm (0.92, 1.01) | 0.0007 | |
| AA (n=634) | ||||||
| AG/GG(n=63)d |
PR=prevalence ratio for CAC presence outcome; Means=geometric means for common cIMT outcome; models adjusted for age, sex, and field center
Models adjusted for age, sex, field center and percent European ancestry
Models adjusted for age, sex, field center, percent European ancestry, current smoking, alcohol use, education, income, diabetes, chronic kidney disease, hypertension, anti-hypertensive and lipid lowering medication use, body mass index, waist circumference, systolic and diastolic blood pressure, trigylcerides, HDL cholesterol, LDL cholesterol, fasting glucose, C-reactive protein
Heterozygous and rare homozygous genotypes combined due to only one African-American participant with the rare homozygous genotype
Among Hispanics, rs11711353 was associated with presence of CAC, with those with the AA genotype having a 37% greater CAC prevalence compared to the GG genotype (p=0.0011) (Table 3). Further adjustment for European ancestry and Native American ancestry did not change the strength of the association. Common cIMT differed somewhat between the AA and TT genotypes of rs7649121; however, the difference was not significant, p=0.02. Further adjustment for cardiovascular risk factors did not alter any of the above associations, and there were no SNP-SNP interactions present.
Table 3.
Association of rs11711353 with CAC presence in Hispanics
| Demographic Adjusted | p-value | Ancestry Adjusted | p-value | Fully Adjusted | p-value | |
|---|---|---|---|---|---|---|
| PR (95% CI)a | PR (95% CI)b | PR (95% CI)c | ||||
| GG (n=337) | ref | -- | ref | -- | ref | -- |
| AG (n=298) | 1.03 (0.91, 1.17) | 0.63 | 1.02 (0.90, 1.16) | 0.74 | 1.03 (0.90, 1.18) | 0.64 |
| AA (n=69) | 1.37 (1.14, 1.66) | 0.0011 | 1.37 (1.14, 1.65) | 0.0008 | 1.37 (1.14, 1.65) | 0.0007 |
PR=prevalence ratio; model adjusted for age, sex, and field center
Model adjusted for age, sex, field center, percent European ancestry, and percent Native American ancestry
Model adjusted for age, sex, field center, percent European ancestry, percent Native American ancestry, current smoking, alcohol use, education, income, diabetes, chronic kidney disease, hypertension, anti-hypertensive and lipid lowering medication use, body mass index, waist circumference, systolic and diastolic blood pressure, trigylcerides, HDL cholesterol, LDL cholesterol, fasting glucose, C-reactive protein
We calculated detectable effect sizes given the set paramters of 80% power, allele frequencies observed, and a sample size of ~700 for each racial/ethnic group. For a SNP such as rs2241767, modeled as a dominant SNP (1 degree of freedom) with the heterozygous/rare homozygous combined group having a frequency of ~9%, we have 80% power to detect an RR of ~1.65 for CAC presence, and a beta coefficient of ~0.04 mm for common cIMT among African-Americans with a type 1 error rate of 0.005. For a SNP such as rs11711353, modeled as 2 degree of freedom SNP, with a minor allele frequency of ~1%, we have 80% power to detect an RR of ~3.50 for CAC presence among Hispanics with a type 1 error rate of 0.005.
Discussion
Among African Americans, rs2241767 in the adiponectin gene appears to be an important SNP in explaining variation in subclinical CVD as it was significantly associated with CAC presence and common cIMT in models adjusted for demographics and European ancestry, as well as additional adjustment for cardiovascular risk factors. rs1063537 and rs1063539 were also associated with CAC presence, although these SNPs were in high LD with each other in African Americans, r2=0.90. In demographic, ancestry, and CVD risk factor adjusted models, rs11711353 was associated with CAC presence among Hispanics.
Few previous studies have examined the effects of ADIPOQ SNPs with subclinical measures of CVD. rs2241766 and rs1501299 have been the most frequently studied variants of ADIPOQ in relation to subclinical CVD, but the results have been inconsistent overall. Among pre-hypertensive participants in the Mexico City Diabetes study, the G allele of rs2241766 (+45G/T) SNP was associated with increased common cIMT (RR 1.45 95% CI (1.04-2.01) for the top quintile versus lowest quintile)(19). Mackevics et al(21) found no association of rs2241766 or rs1501299 (+276G/T) SNPs with cIMT in the Salzburg Atherosclerosis Prevention program in subjects at High Individual Risk (SAPHIR) cohort. Patel et al(22), in 1306 Caucasian men and women from the Relationship between Insulin Sensitivity and Cardiovascular disease (RISC) study, found rs266729 (-11377C/G) significantly associated with common IMT in fully adjusted models; those with the G allele had significantly higher cIMT than those with the CC genotype (601 versus 590 μm, p=0.011). Other ADIPOQ SNPs, including rs2241766, were not associated with common cIMT in that study. In a study of 708 Korean participants with type 2 diabetes, Kim et al(20) found the GG genotype of rs2241766 was associated with higher odds of coronary artery plaques compared to the T allele carriers. That study found no association of rs2241766 or rs1501299 with common cIMT. All of the above studies were in one ethnic group, used just a few ADIPOQ SNPs, were small, or were not stringent about adjusting for multiple comparisons. By contrast, we examined the association of ADIPOQ and subclinical CVD in four ethnic groups of ~700 participants each, with 11 variants different from those described above, and adjusted for multiple comparisons. Unfortunately, none of the MESA SNPs overlapped with the above studies, so direct SNP comparisons are not possible.
In the earliest stages of atherosclerosis, circulating adiponectin stimulates production of nitric oxide in endothelial cells(28) and has been shown to suppress expression of cellular adhesion molecules by inhibiting endothelial NF-κB signaling through the activation of cAMP protein kinase C(29,30,31,32). At the later stages of atherosclerosis, adiponectin inhibits the proliferation and migration of smooth muscle cells by binding to platelet derived growth factor(32,33). Adiponectin also promotes plaque stability in the more advanced stages of atherosclerosis by increasing the expression of a tissue inhibitor of matrix metalloproteinases in macrophages(33). A study by Matsuura et al(34) suggests that adiponectin may mediate lipid production and protect against atherosclerosis by increasing protein and mRNA levels of ABCA1 and apolipoprotein A-1 which in turn increases HDL assembly.
Circulating adiponectin promotes adipocyte differentiation and enhanced insulin sensitivity. Low levels of circulating adiponectin have also been linked to obesity, particularly central adiposity, and insulin resistance(35,36,37). Experimental evidence has shown that adipocytes that over-express adiponectin lead to more adipocyte differentiation and increased insulin-responsive glucose transport activity(35). It has also been hypothesized that adiponectin may increase insulin sensitivity and lower glucose levels by decreasing hepatic production, increasing fatty acid oxidation, and lowering intramyocellular lipid content(36).
The adiponectin gene, ADIPOQ, encodes the circulating protein adiponectin. A number of studies have shown that variants of the gene are associated with circulating adiponectin levels(14,15,17,18) and one hypothesis is that ADIPOQ affects adiponectin levels, which in turn could affect cIMT and calcification. Recently, however, it has been shown that ADIPOQ is also expressed in vascular tissue, not just in adipose tissue(12). Treatment of patients with type 2 diabetes with quinapril increased adiponectin gene expression in the vascular tissue by two times(12). In addition, another study found that ADIPOQ expression in epicardial adipose tissue was increased in patients with coronary artery disease compared to those without, and this expression increased with the number of injured arteries(38). This evidence possibly supports a more direct role of ADIPOQ in the various stages of the atherosclerotic process (rather than just through affecting circulating adiponectin levels), which then could lead to thicker IMT and increased calcification.
The most interesting SNPs within ADIPOQ in our study were in the intron 2 (rs2241767), the 3’ untranslated region (UTR) (rs1063537, rs1063539), or in the 5’UTR of ADIPOQ (rs11711353). Introns are non-coding regions of a gene; however, there is evidence that introns of the protein-coding gene transcripts can affect gene expression by repressing translation or cleaving RNA transcripts (39). It is possible that a SNP in the intron could be in high LD with a causal variant in the exon. Menzaghi et al(13), in a meta-analysis and review of ADIPOQ variants and associations with serum adiponectin, CVD, and type 2 diabetes, further notes that the 3’ UTR of the gene, where some variants of ADIPOQ have been found that influence serum adiponectin levels, is shown to control gene expression through binding to proteins that regulate mRNA translation and degradation. Additionally, variants in the promoter region or 5’UTR of adiponectin affect circulating adiponectin levels, enhancing ADIPOQ promoter activity(13). Areas upstream of a gene are also thought to regulate mRNA translation(40).
This study has several important strengths but also some limitations. Due to the varied ethnicity of the MESA cohort, we could examine the SNP-subclinical CVD association in Caucasian, African American, Chinese and Hispanic populations; many other studies have concentrated only on one ethnic group. Using the tagSNP approach to choose SNPs enables the capturing of the variation across the adiponectin gene rather than just choosing a few SNPs from the adiponectin gene as previous studies have done. Furthermore, tagSNPs were chosen from both HapMap and Seattle SNPs with an initial minor allele frequency of >0.05. No previous studies of the adiponectin gene and subclinical CVD have used a tagSNP approach to examine associations or have examined these associations in a variety of ethnicities. Many steps were taken to minimize measurement error and to ensure the quality of the cIMT and CAC measures in the MESA. Many genetic association studies do not control for population stratification within racial/ethnic groups; in this study, there were well-characterized ancestry measurements for the admixed groups (African Americans and Hispanics). It is possible that there are SNPs with minor allele frequencies of less than 0.05 that are important. There were some ADIPOQ SNPs reported as important in previous studies, particularly rs2241766 and rs1501299, which were not genotyped in MESA. Due to the complexity of the algorithm for choosing the ADIPOQ SNPs in MESA, some of the tagSNPs tag each other, i.e. they have an r2>0.80, and some previously studied SNPs such as rs266729, rs2241766, and rs1501299 were not chosen as tagSNPs. Furthermore, 2 SNPs chosen (rs9877202, rs1403697) were not polymorphic for the Chinese or Caucasian groups. Given that associations of 11 ADIPOQ SNPs and subclinical CVD were examined for four outcomes and four ethnic groups, multiple testing is a limitation. This study did not have serum adiponectin levels available in order to test whether ADIPOQ SNPs were associated with subclinical CVD independent of serum adiponectin. Additionally, this study focused on subclinical CVD measures in participants with no known clinical CVD; thus the full range of CVD severity was not examined.
We found variants of ADIPOQ, many of which have not previously been studied, significantly associated with subclinical CVD in both Hispanics and African Americans in MESA. However, further, larger studies are warranted to replicate these findings. There are still many ADIPOQ SNPs that have not been examined; thus far only a small proportion of them have been studied(13). Further study will also be needed to determine which SNPs may be functional variants. Information such as this could eventually be used in developing better risk profiles to predict cardiovascular disease. With new genetics initiatives and consortia currently in progress, large studies with consistent phenotyping and genotyping methods will be possible, and may help in addressing some of these remaining questions.
Supplementary Material
Acknowledgements
CLW was supported by the NIH training grant in cardiovascular genetic epidemiology (T32 HL097972) and cardiovascular epidemiology and prevention (T32HL07779) during this research. This research was supported by contracts N01-HC-95159 through N01-HC-95169 and grants R01HL071051, R01HL071205, R01HL071250, RO1HL071251, R01HL071252, R01HL071258, and R01HL071259 from the National Heart, Lung, and Blood Institute. The authors thank the other investigators, the staff, and the participants of the MESA study and of the MESA Family Study for their valuable contributions. A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org.
Footnotes
Disclosures
None.
References
- 1.Greenland P, Bonow RO, Brundage BH, et al. ACCF/AHA 2007 clinical expert consensus document on coronary artery calcium scoring by computed tomography in global cardiovascular risk assessment and in evaluation of patients with chest pain: a report of the American College of Cardiology Foundation Clinical Expert Consensus Task Force (ACCF/AHA Writing Committee to Update the 2000 Expert Consensus Document on Electron Beam Computed Tomography). Circulation. 2007;115:402–426. doi: 10.1161/CIRCULATIONAHA..107.181425. [DOI] [PubMed] [Google Scholar]
- 2.Stein JH, Korcarz CE, Hurst RT, et al. Use of carotid ultrasound to identify subclinical vascular disease and evaluate cardiovascular disease risk: a consensus statement from the American Society of Echocardiography Carotid Intima-Media Thickness Task Force. Endorsed by the Society for Vascular Medicine. J Am Soc Echocardiogr. 2008;21:93–111. doi: 10.1016/j.echo.2007.11.011. quiz 189-90. [DOI] [PubMed] [Google Scholar]
- 3.Greenland P, LaBree L, Azen SP, et al. Coronary artery calcium score combined with Framingham score for risk prediction in asymptomatic individuals. JAMA. 2004;291:210–215. doi: 10.1001/jama.291.2.210. [DOI] [PubMed] [Google Scholar]
- 4.van der Meer IM, Bots ML, Hofman A, et al. Predictive value of noninvasive measures of atherosclerosis for incident myocardial infarction: the Rotterdam Study. Circulation. 2004;109:1089–1094. doi: 10.1161/01.CIR.0000120708.59903.1B. [DOI] [PubMed] [Google Scholar]
- 5.Pischon T, Girman CJ, Hotamisligil GS, et al. Plasma adiponectin levels and risk of myocardial infarction in men. JAMA. 2004;291:1730–1737. doi: 10.1001/jama.291.14.1730. [DOI] [PubMed] [Google Scholar]
- 6.Sattar N, Wannamethee G, Sarwar N, et al. Adiponectin and coronary heart disease: a prospective study and meta-analysis. Circulation. 2006;114:623–629. doi: 10.1161/CIRCULATIONAHA.106.618918. [DOI] [PubMed] [Google Scholar]
- 7.Kanaya AM, Wassel Fyr C, Vittinghoff E, et al. Serum adiponectin and coronary heart disease risk in older Black and White Americans. J Clin Endocrinol Metab. 2006;91:5044–5050. doi: 10.1210/jc.2006-0107. [DOI] [PubMed] [Google Scholar]
- 8.Laughlin GA, Barrett-Connor E, May S, et al. Association of adiponectin with coronary heart disease and mortality: the Rancho Bernardo study. Am J Epidemiol. 2007;165:164–174. doi: 10.1093/aje/kwk001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maahs DM, Ogden LG, Kinney GL, et al. Low plasma adiponectin levels predict progression of coronary artery calcification. Circulation. 2005;111:747–753. doi: 10.1161/01.CIR.0000155251.03724.A5. [DOI] [PubMed] [Google Scholar]
- 10.Nilsson PM, Engstrom G, Hedblad B, et al. Plasma adiponectin levels in relation to carotid intima media thickness and markers of insulin resistance. Arterioscler Thromb Vasc Biol. 2006;26:2758–2762. doi: 10.1161/01.ATV.0000249638.01416.4b. [DOI] [PubMed] [Google Scholar]
- 11.Iglseder B, Mackevics V, Stadlmayer A, et al. Plasma adiponectin levels and sonographic phenotypes of subclinical carotid artery atherosclerosis: data from the SAPHIR Study. Stroke. 2005;36:2577–2582. doi: 10.1161/01.STR.0000190834.00284.fd. [DOI] [PubMed] [Google Scholar]
- 12.Hermann TS, Li W, Dominguez H, et al. Quinapril treatment increases insulin-stimulated endothelial function and adiponectin gene expression in patients with type 2 diabetes. J Clin Endocrinol Metab. 2006;91:1001–1008. doi: 10.1210/jc.2005-1231. [DOI] [PubMed] [Google Scholar]
- 13.Menzaghi C, Trischitta V, Doria A. Genetic Influences of Adiponectin on Insulin Resistance, Type 2 Diabetes, and Cardiovascular Disease. Diabetes. 2007 doi: 10.2337/db06-0506. [DOI] [PubMed] [Google Scholar]
- 14.Filippi E, Sentinelli F, Romeo S, et al. The adiponectin gene SNP+276G>T associates with early-onset coronary artery disease and with lower levels of adiponectin in younger coronary artery disease patients (age <or=50 years). J Mol Med. 2005;83:711–719. doi: 10.1007/s00109-005-0667-z. [DOI] [PubMed] [Google Scholar]
- 15.Hoefle G, Muendlein A, Saely CH, et al. The -11377 C > G promoter variant of the adiponectin gene, prevalence of coronary atherosclerosis, and incidence of vascular events in men. Thromb Haemost. 2007;97:451–457. [PubMed] [Google Scholar]
- 16.Pischon T, Pai JK, Manson JE, et al. Single nucleotide polymorphisms at the adiponectin locus and risk of coronary heart disease in men and women. Obesity (Silver Spring) 2007;15:2051–2060. doi: 10.1038/oby.2007.244. [DOI] [PubMed] [Google Scholar]
- 17.Qi L, Li T, Rimm E, et al. The +276 polymorphism of the APM1 gene, plasma adiponectin concentration, and cardiovascular risk in diabetic men. Diabetes. 2005;54:1607–1610. doi: 10.2337/diabetes.54.5.1607. [DOI] [PubMed] [Google Scholar]
- 18.Vasseur F, Helbecque N, Dina C, et al. Single-nucleotide polymorphism haplotypes in the both proximal promoter and exon 3 of the APM1 gene modulate adipocyte-secreted adiponectin hormone levels and contribute to the genetic risk for type 2 diabetes in French Caucasians. Hum Mol Genet. 2002;11:2607–2614. doi: 10.1093/hmg/11.21.2607. [DOI] [PubMed] [Google Scholar]
- 19.Femia R, Kozakova M, Nannipieri M, et al. Carotid Intima-Media Thickness in Confirmed Prehypertensive Subjects. Predictors and Progression. Arterioscler Thromb Vasc Biol. 2007 doi: 10.1161/ATVBAHA.107.149641. [DOI] [PubMed] [Google Scholar]
- 20.Kim SH, Kang ES, Hur KY, et al. Adiponectin gene polymorphism 45T>G is associated with carotid artery plaques in patients with type 2 diabetes mellitus. Metabolism. 2008;57:274–279. doi: 10.1016/j.metabol.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 21.Mackevics V, Heid IM, Wagner SA, et al. The adiponectin gene is associated with adiponectin levels but not with characteristics of the insulin resistance syndrome in healthy Caucasians. Eur J Hum Genet. 2006;14:349–356. doi: 10.1038/sj.ejhg.5201552. [DOI] [PubMed] [Google Scholar]
- 22.Patel S, Flyvbjerg A, Kozakova M, et al. Variation in the ADIPOQ gene promoter is associated with carotid intima media thickness independent of plasma adiponectin levels in healthy subjects. Eur Heart J. 2008;29:386–393. doi: 10.1093/eurheartj/ehm526. [DOI] [PubMed] [Google Scholar]
- 23.Bild DE, Bluemke DA, Burke GL, et al. Multi-ethnic study of atherosclerosis: objectives and design. Am J Epidemiol. 2002;156:871–881. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- 24.Bielinski SJ, Pankow JS, Li N, et al. ICAM1 and VCAM1 polymorphisms, coronary artery calcium, and circulating levels of soluble ICAM-1: the multi-ethnic study of atherosclerosis (MESA). Atherosclerosis. 2008;201:339–344. doi: 10.1016/j.atherosclerosis.2008.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carr JJ, Nelson JC, Wong ND, et al. Calcified coronary artery plaque measurement with cardiac CT in population-based studies: standardized protocol of Multi-Ethnic Study of Atherosclerosis (MESA) and Coronary Artery Risk Development in Young Adults (CARDIA) study. Radiology. 2005;234:35–43. doi: 10.1148/radiol.2341040439. [DOI] [PubMed] [Google Scholar]
- 26.Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 27.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H, Montagnani M, Funahashi T, et al. Adiponectin stimulates production of nitric oxide in vascular endothelial cells. J Biol Chem. 2003;278:45021–45026. doi: 10.1074/jbc.M307878200. [DOI] [PubMed] [Google Scholar]
- 29.Ekmekci H, Ekmekci OB. The role of adiponectin in atherosclerosis and thrombosis. Clin Appl Thromb Hemost. 2006;12:163–168. doi: 10.1177/107602960601200203. [DOI] [PubMed] [Google Scholar]
- 30.Karaduman M, Sengul A, Oktenli C, et al. Tissue levels of adiponectin, tumour necrosis factor-alpha, soluble intercellular adhesion molecule-1 and heart-type fatty acid-binding protein in human coronary atherosclerotic plaques. Clin Endocrinol (Oxf) 2006;64:196–202. doi: 10.1111/j.1365-2265.2006.02448.x. [DOI] [PubMed] [Google Scholar]
- 31.Li CJ, Sun HW, Zhu FL, et al. Local adiponectin treatment reduces atherosclerotic plaque size in rabbits. J Endocrinol. 2007;193:137–145. doi: 10.1677/JOE-06-0173. [DOI] [PubMed] [Google Scholar]
- 32.Shimada K, Miyazaki T, Daida H. Adiponectin and atherosclerotic disease. Clin Chim Acta. 2004;344:1–12. doi: 10.1016/j.cccn.2004.02.020. [DOI] [PubMed] [Google Scholar]
- 33.Szmitko PE, Teoh H, Stewart DJ, et al. ADIPONECTIN AND CARDIOVASCULAR DISEASE. Am J Physiol Heart Circ Physiol. 2006 doi: 10.1152/ajpheart.01072.2006. [DOI] [PubMed] [Google Scholar]
- 34.Matsuura F, Oku H, Koseki M, et al. Adiponectin accelerates reverse cholesterol transport by increasing high density lipoprotein assembly in the liver. Biochem Biophys Res Commun. 2007;358:1091–1095. doi: 10.1016/j.bbrc.2007.05.040. [DOI] [PubMed] [Google Scholar]
- 35.Lara-Castro C, Fu Y, Chung BH, et al. Adiponectin and the metabolic syndrome: mechanisms mediating risk for metabolic and cardiovascular disease. Curr Opin Lipidol. 2007;18:263–270. doi: 10.1097/MOL.0b013e32814a645f. [DOI] [PubMed] [Google Scholar]
- 36.Havel PJ. Control of energy homeostasis and insulin action by adipocyte hormones: leptin, acylation stimulating protein, and adiponectin. Curr Opin Lipidol. 2002;13:51–59. doi: 10.1097/00041433-200202000-00008. [DOI] [PubMed] [Google Scholar]
- 37.Ahima RS. Metabolic actions of adipocyte hormones: focus on adiponectin. Obesity (Silver Spring) 2006;14(Suppl 1):9S–15S. doi: 10.1038/oby.2006.276. [DOI] [PubMed] [Google Scholar]
- 38.Eiras S, Teijeira-Fernandez E, Shamagian LG, et al. Extension of coronary artery disease is associated with increased IL-6 and decreased adiponectin gene expression in epicardial adipose tissue. Cytokine. 2008 doi: 10.1016/j.cyto.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 39.Ying SY, Lin SL. Intron-derived microRNAs--fine tuning of gene functions. Gene. 2004;342:25–28. doi: 10.1016/j.gene.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 40.Morris DR, Geballe AP. Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol. 2000;20:8635–8642. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.