Abstract
Chlamydia pneumoniaeencodes a functional arginine decarboxylase (ArgDC), AaxB, that activates upon self-cleavage and converts L-arginine to agmatine. In contrast, most Chlamydia trachomatis serovars carry a missense or nonsense mutation in aaxB abrogating activity. The G115R missense mutation was not predicted to impact AaxB functionality, making it unclear if AaxB variations in other Chlamydia species also result in enzyme inactivation. To address the impact of gene polymorphism on functionality, we investigated the activity and production of the Chlamydia AaxB variants. Since ArgDC plays a critical role in the Escherichia coli acid stress response, we studied the ability of these Chlamydia variants to complement an E. coli ArgDC mutant in an acid shock assay. Active AaxB was detected in four additional species: C. caviae, C. pecorum, C. psittaci, and C. muridarum. Of the C. trachomatis serovars, only E appears to encode active enzyme. To determine when functional enzyme is present during the chlamydial developmental cycle, we utilized an anti-AaxB antibody to detect both uncleaved and cleaved enzyme throughout infection. Uncleaved enzyme production peaked around 20 hours post-infection, with optimal cleavage around 44 hours. While the role ArgDC plays in Chlamydia survival or virulence is unclear, our data suggest a niche-specific function.
Keywords: Chlamydia evolution, arginine decarboxylase, inactivating mutation
Introduction
Infection with Chlamydia, a genus of Gram-negative obligate intracellular bacteria, may result in ocular, genital, or pneumonic disease, depending on route of entry and bacterial species/serovar. While the majority of Chlamydia species are zoonotic, infecting a wide range of mammalian and avian hosts, the Chlamydia trachomatis serovars are human-specific pathogens (Rohde et al., 2010; Carlson et al., 2005). All species undergo a unique biphasic developmental cycle transitioning between the extracellular, infectious elementary body (EB) and the intracellular, replicative form known as the reticulate body (RB) (AbdelRahman and Belland, 2005).
Arginine decarboxylases (ArgDC), which catalyze the conversion of arginine into agmatine, are conserved in bacteria and play dual roles in acid resistance and the metabolism of polyamines such as putrescine (Lin et al., 1995; Tabor and Tabor, 1984). In bacteria such as Yersinia, functional ArgDC is required to produce biofilms, making this enzyme essential for virulence (Patel et al., 2006). Two ArgDC are encoded by E. coli: the acid-inducible adiA and a constitutive speA that functions in polyamine biosynthesis (Stim and Bennett, 1993). In Chlamydia, the only known ArgDC is encoded by aaxB, which resides in an operon between the putative porin aaxA and the characterized arginine-agmatine antiporter, aaxC (Giles and Graham, 2007) (Figure 1A). Although AaxB is functionally equivalent to E. coli AdiA, the enzyme itself is actually a member of the pyruvoyl-dependent ArgDC (PvlArgDC), and shares more similarities with ArgDC from organisms such as Methanococcus jannaschii (Graham et al., 2002).
Figure 1. Gene organization and protein alignment.
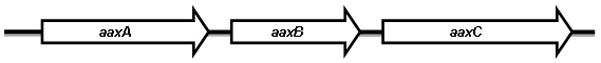
(a) aaxB is transcribed in an operon with aaxA, an outer membrane protein of unknown function, and aaxC, a characterized arginine-agmatine antiporter (Smith and Graham, 2008). (b) ClustalW alignment of different unique Chlamydia AaxB proteins, representative of all fully annotated strains and high quality draft genomes from clinical isolates found on the NCBI server as of June 23, 2012. The conserved Thr52Ser53autocleavage site essential for activity is in brackets. * indicates the nonsense mutation in C. trachomatis serovar L2 that results in early truncation of the protein. A circle indicates the glycine to arginine mutation that abrogates cleavage and therefore activity; this mutation is conserved in all C. trachomatis A, B, D, and G lineages. Rabbit polyclonal antibody was raised against the highly conserved 18 amino acid peptide (outlined in the box) present in all AaxB sequences except the truncated C. trachomatis serovar L2.
The AaxB proteins of Chlamydia pneumoniae and C. trachomatis serovars D and L2 were previously characterized (Giles and Graham, 2007; Giles et al., 2009). All sequenced C. pneumoniae encode a 25 kDa proenzyme, which requires autocleavage between the conserved Thr52 Ser53 residues to produce 16 kDa α and 9 kDa β subunits. The cleaved subunits are then free to assemble into the active (αβ)3 complex.
In contrast, C. trachomatis serovars D and L2 have inactivated AaxB through one of two independent mutations (Giles et al., 2009). The Gly115Arg substitution mutation in serovar D (also present and predicted to inactivate AaxB from B/D/G and F) disrupts the necessary auto-cleavage event; in serovar L2, a nonsense mutation midway through the gene results in early truncation. The Gly115Arg mutation present in strains of D was not predicted to result in enzyme inactivation based on sequence analysis alone, making it unclear if AaxB sequence variations seen in other Chlamydia alter AaxB activity.
To further our understanding of this enzyme and determine if inactivation of AaxB is restricted to the human-specific C. trachomatis serovars, we completed an activity panel using variant Chlamydia AaxB proteins in a surrogate E. coli acid shock assay. A pan-chlamydial anti-AaxB antibody was used to detect enzyme production and processing during the developmental cycle using a cell culture infection model. Collectively, our data indicate that non-C. trachomatis species (and a single C. trachomatis serovar: E) produce active AaxB.
Methods and Materials
Strains
Chlamydia strains used in this study include: Chlamydia muridarum strain Nigg, Chlamydia trachomatis serovar D strain UW-3/CX, Chlamydia psittaci strain 6BC, Chlamydia caviae strain SP6 (Binet et al., 2010), and Chlamydia trachomatis serovar E strain UW-5/CX. C. pecorum strain E58 DNA was provided by Patrik Bavoil (University of Maryland). The previously unreported aaxB sequences for C. caviae SP6 and C. trachomatis E strain UW-5/CX were deposited in Genbank under accession numbers JX287368 and JX287367, respectively. E. coli strain MG1655 was used for the acid resistance complementation assays, while E. coli Rosetta-gami2 (DE3) (Novagen) was used for AaxB expression and purification.
Cloning of aaxB
A pBAD/HisA vector (modified during cloning to remove the histidine tag coding region) (Invitrogen) carrying aaxB from C. pneumoniae strain Kajaani 6 or adiA from E. coli strain MG1655 was provided by David Graham (Oak Ridge National Laboratory). Primers used to amplify the different aaxB variants are listed in Table 1. PCR-amplified products were digested and ligated into the NcoI and HindIII sites on the pBAD/HisA vector (without the Histidine tag). Constructs were then electroporated into ΔadiA E. coli strain MG1655. The aaxB gene from C. caviae also was PCR-amplified (primers listed in Table 1) for cloning into a pET-19b expression vector (Invitrogen). PCR-amplified products were digested and ligated into the NdeI and BamHI sites on pET-19b, then electroporated into E. coli strain Rosetta-gami2 (DE3). All constructs were sequence verified at the Biomedical Instrumentation Center at the Uniformed Services University.
Deletion of E. coli adiA
The adiA gene was deleted from E. coli strain MG1655 using the lambda red method of linear recombination with the primers listed in Table 1 (Datsenko and Wanner, 2000). After PCR verification of the constructed ΔadiA::kan mutation, the allele was moved into a clean E. coli MG1655 background via P1L4 transduction (Miller, 1972). Transductants were selected on LB agar containing 100 μg mL−1 kanamycin, verified by PCR, and checked for their acid resistance phenotype.
Acid resistance assay
The acid resistance assay of Castanie-Cornet et al. (Castanie-Cornet et al., 1999), as modified by David Graham (Giles and Graham, 2007), was utilized. Acid resistance was expressed as the percentage of viable bacteria remaining after one hour acid shock compared to the number of viable bacteria determined immediately following acid shock.
Production of AaxB antibody
The highly conserved chlamydial AaxB peptide 137HAKMWLKKSLQHELDLRS154 (part of the α subunit) was commercially synthesized by Pierce Custom Antibody production service and used to raise polyclonal rabbit antibodies using the standard 90-day protocol.
Purification of C. caviae AaxB
E. coli Rosetta-gami2 (DE3) was transformed with pET-19b carrying aaxB from C. caviae and grown in LB containing 100 μg mL−1 ampicillin to an OD600 of 0.6. AaxB expression was induced with 1 mM IPTG for 23 hours at 20° C, and bacteria were collected by centrifugation. Bacteria were resuspended in equilibration buffer (50 mM monobasic sodium phosphate, 300 mM sodium chloride, and 10 mM imidazole, pH adjusted to 7.4) with 1x protease inhibitor (Roche) and 1x phosphatase inhibitors 2 and 3 (Sigma). Bacteria were lysed via sonication, centrifuged to remove debris, and the supernatant passed through a 0.45 μm filter (Millipore). HisPur™ cobalt resin (Thermo Scientific) was applied to the supernatant and the batch method of purification was carried out as per manufacturer’s instructions. Purified protein samples were eluted (50 mM sodium phosphate, 300 mM sodium chloride, 500 mM imidazole, pH adjusted to 7.4), then applied to a 3K Amicon filter (Millipore) for concentration. Samples were resuspended in 50 mM Bis-Tris buffer (pH 6.0), and quantified by the Bio-Rad Protein Assay (Bio-Rad). Protein identity and purity was assessed using SDS-PAGE followed by Coomassie Brilliant Blue Staining or Western blotting with the anti-AaxB antibody (at a 1:250 dilution).
Detection of chlamydial AaxB in EBs and during infection
Chlamydia were grown in and harvested from mouse fibroblast L2 cells. EBs were titered using an infection forming unit assay (IFU) and stored at −80° C in Sucrose Phosphate Glutamic acid buffer (SPG) (7.5% w/v sucrose, 17 mM Na2HPO4, 3 mM NaH2PO4, 5 mM L-glutamic acid, pH 7.4) until use (Binet et al., 2010). For time course experiments, L2 cells were infected at an MOI of 5 (10 hr samples), an MOI of 1 (20, 30, and 44 hr samples), or mock infected (Giles et al., 2009). Samples were disrupted directly in Laemmli buffer and run on 12% SDS-PAGE gels for Western blot analysis with either anti-AaxB antibodies or anti-Hsp60 antibodies (provided by Dan Rockey, Oregon State University; Yuan et al., 1992). Detection of Hsp60 (60 kDa heat shock protein, GroEL) was used to demonstrate successful infection and equal loading of protein. To detect AaxB in EBs, bacteria were disrupted in Laemmli buffer and 1 × 107 IFU were used per SDS-PAGE gel lane.
Results
Alignment of Chlamydia AaxB
The AaxB sequences from the available Chlamydia genome projects were aligned to assess amino acid variability. All Chlamydia species have at least one sequenced strain available in the Genbank database except C. suis, a porcine pathogen. As many strains within the same species or serovar had identical protein sequences, duplicates were discarded and only unique AaxB sequences are shown in Figure 1B.
Despite differences in amino acid sequence, all AaxB variants carried the highly conserved Thr52 Ser53 cleavage site. C. trachomatis serovars A/B/D/F and G carry a missense mutation, a glycine to arginine substitution (Gly115Arg) that was shown to abrogate cleavage of the protein and therefore activity in the serovar D variant (Giles et al., 2009). In C. trachomatis serovar L2, an ochre codon at position 128 truncates the gene in mid-open reading frame. This truncated protein lacks activity (Giles et al., 2009). Both inactivating mutations are present in high quality draft genomes of clinical isolates, suggesting that these mutations did not arise from laboratory adaptation. Neither C. trachomatis serovar E, nor any of the remaining Chlamydia species, carry either of the known mutations that have been shown to inactivate AaxB. However, there are variations in the amino acid sequence of these proteins compared to the amino acid sequence of the active C. pneumoniae AaxB. As the missense mutation in C. trachomatis serovars A/B/D/F and G was not indicative of protein inactivation, we measured the activity of the remaining variants.
Activity of Chlamydia AaxB
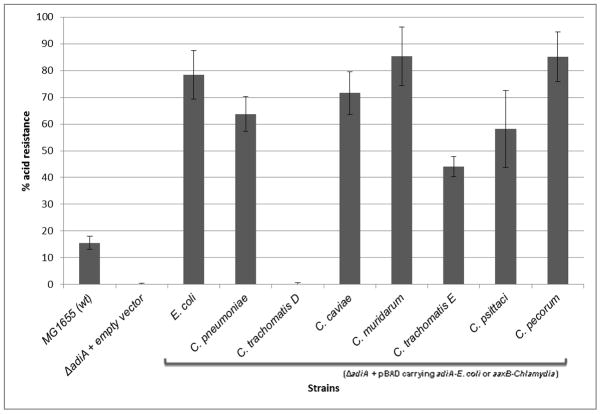
Previously, Giles and Graham demonstrated that expression of functional AaxB from C. pneumoniae can rescue an E. coli ΔadiA mutant from acid shock, demonstrating activity of the Chlamydia enzyme in a surrogate system (Giles and Graham, 2007). To test the remaining Chlamydia variants, an ΔadiA knockout of E. coli MG1655 was constructed and transformed with wild type E. coli adiA or Chlamydia aaxB genes cloned into a vector under control of an arabinose-inducible promoter. The different AaxB variants from C. caviae, C. muridarum, C. trachomatis serovar E, C. psittaci, and C. pecorum were tested in the acid resistance assay, with AaxB variants from C. pneumoniae and C. trachomatis serovar D serving as positive and negative controls, respectively (Figure 2A). All Chlamydia AaxB tested restored acid shock survival in the E. coli ΔadiA mutant, suggesting that C. caviae, C. muridarum, C. trachomatis serovar E, C. psittaci, and C. pecorum all encode active enzyme.
Figure 2. Activity analysis of AaxB proteins.
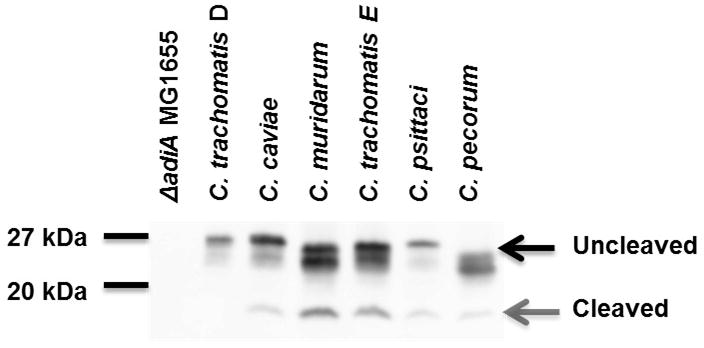
(a) Strains were grown for 22 hours at 37° C in the presence of 0.2% arabinose. ~3 × 107 cfu was exposed to acid shock buffer containing 0.4% arginine (pH 2.5) for one hour, and percent survival determined by viable plate counts. Strains were tested in three independent experiments performed in triplicate. Wild type MG1655 was significantly acid resistant compared to the ΔadiA E. coli empty vector control using a standard t-test with unequal variance (p<0.005); all other strains, with the exception of C. trachomatis serovar D, were able to complement the ΔadiA E. coli mutant over wild type. The difference in acid resistance between the ΔadiA E. coli strain expressing AaxB from C. trachomatis serovar D and the empty vector control was not statistically significant (p>0.1). (b) AaxB protein production and cleavage was measured using Western blotting with the anti-AaxB antibody. Molecular weight markers are shown to the left of the blot, and the proenzyme and α fragments (if present) are indicated with black and gray arrows, respectively.
Protein expression and cleavage of the AaxB variants was measured via Western blotting with anti-AaxB antibody (Figure 2B). All constructs used in the acid shock experiments expressed uncleaved AaxB protein, and each active AaxB variant was capable of auto-cleavage as evidenced by detection of the α fragment (Figure 2B); i.e. the cleavage profile correlates with acid resistance. The deviation in protein size between the AaxB variants may be due to variation in molecular weight and isoelectric point; the predicted pI fluctuates within a range of approximately 0.3 pH and the predicted molecular weight fluctuates within a range of approximately 500 Da.
Time course expression of AaxB in Chlamydia
The timing of AaxB protein production and cleavage, and therefore activity, during infection is unknown. To determine when active enzyme is present during the chlamydial developmental cycle, the highly Chlamydia-conserved peptide 137HAKMWLKKSLQHELDLRS154 was used to produce rabbit polyclonal antibodies. This antibody recognizes both the inactive, uncleaved proenzyme form of AaxB, as well as the activated α subunit, and therefore cleavage of this protein can be directly measured during infection.
L2 cells were infected with C. caviae, and the expression and cleavage of AaxB into active subunits over the course of infection was studied (Figure 3A). A unique band of ~20 kDa representing uncleaved proenzyme was initially detected at 20 hours post-infection, with very little cleaved protein (<20 kDa) appearing. Over the next 24 hours, this ratio slowly shifted, and by 44 hours post-infection, the majority of protein was in the cleaved, active state.
Figure 3. Time course of C. caviae AaxB production.
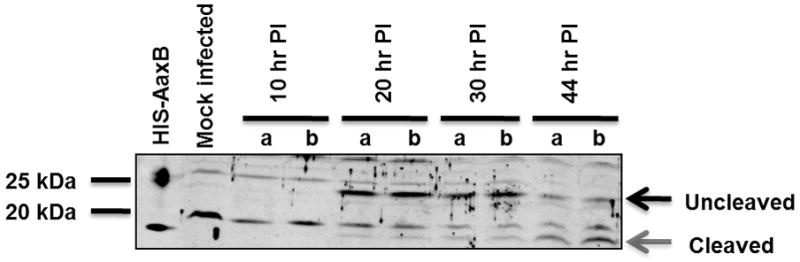

L2 cells were infected at an MOI of 5 (10 hr samples), an MOI of 1 (20, 30, and 44 hr samples), or mock infected. Western blotting was performed with either anti-AaxB antibodies (a) or anti-Hsp60 antibodies as a control to demonstrate infection and for semi-quantitative comparison of chlamydial protein (b). Purified his-tagged AaxB is shown in the first lane. Two parallel-run infection samples are shown for each time point. Proenzyme and the α fragment are indicated by black and gray arrows to the right of the blot. Protein molecular weight marker positions are shown to the left of each blot.
Interestingly, this pattern did not necessarily hold true across all the Chlamydia species (Figure 4A). In C. muridarum, while the majority of uncleaved protein also appeared at 20 hours post-infection, cleaved protein production likewise peaked at this time, then waned at subsequent time points. C. psittaci produced very little detectable cleaved protein.
Figure 4. Time course of AaxB production by different Chlamydia spp.
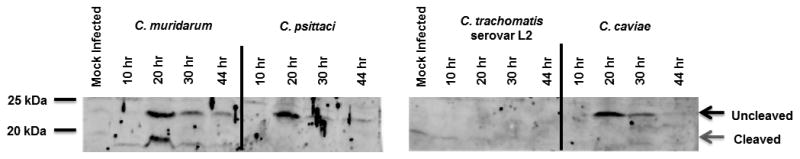

L2 cells were infected at an MOI of 10 (10 hr samples), an MOI of 1 (20, 30, and 44 hr samples), or mock infected. At the indicated times, samples were disrupted directly in Laemmli buffer and run on 12% SDS-PAGE gels. Protein was detected by Western blotting with either anti-AaxB antibodies (a) or anti-Hsp60 antibodies as a control to demonstrate infection and for semi-quantitative comparison of chlamydial protein (b). Full length AaxB is indicated by the black arrows and processed AaxB is indicated by the grey arrows. Protein molecular weight marker positions are shown to the left of each blot.
Cleavage of AaxB also was assessed in EBs in comparison to samples from cells infected for 20 hours when full-length protein appears to be the predominant species (Supplementary Figure 1A). The cleaved form predominates in EBs, and very little, if any, detectable proenzyme remains. Despite equal loading of bacteria, AaxB was undetectable in C. trachomatis serovar D.
Discussion
Previously, a functional arginine decarboxylase enzyme, AaxB, was identified and characterized in C. pneumoniae (Giles and Graham, 2007). In this study, we demonstrate that several additional Chlamydia species, including C. caviae, C. muridarum, C. psittaci, and C. pecorum, encode functional AaxB. Although previous publications established that the majority of the C. trachomatis serovars encode nonfunctional AaxB due to one of two inactivating mutations (Graham et al., 2009), we now show that the AaxB variant of C. trachomatis serovar E is capable of cleavage and activity.
AaxB undergoes maximal autocleavage during the mid to late Chlamydia developmental cycle, with slight variations on timing between the different species. At the extremes, optimal cleavage of C. muridarum AaxB occurs around 20 hours post-infection, with C. caviae AaxB cleaving around 44 hours. Although cellular conditions for autocleavage are not yet clear, timing of cleavage may be influenced by differences in amino acid composition between variants or post-translational modification.
We were unable to detect AaxB from C. trachomatis serovar D. Since this enzyme appears to be nonfunctional, production of AaxB would squander bacterial energy resources. While a transcriptome analysis by Belland et al. suggests that this gene is still transcribed, it is probable that expression of the inactive aaxB was maintained due to the necessity of transcribing the remaining operon genes (Belland et al., 2003; Giles et al., 2009). Due to the early truncation of the C. trachomatis serovar L2 AaxB, the anti-AaxB antibody, which was developed against a conserved peptide after the truncation, would not recognize this serovar if truncated protein is produced. However, previous data using an E. coli surrogate expression system indicates that this protein may not be produced (Giles et al., 2009).
The total protein level of AaxB in C. trachomatis serovar E also appeared to be lower than the non-C. trachomatis variants (possibly indicating decreased expression levels), and the acid resistance phenotype of the serovar E AaxB producing strain was the weakest of the complementing strains. As the only C. trachomatis serovar expressing active AaxB, it is possible that the serovar E strains represent an intermediate phenotype between isolates that have maintained or lost enzyme functionality. Several studies suggest that there is no association between infections with C. trachomatis serovar E and presence or absence of clinical infection or specific symptoms, although this serovar is one of the most prevalent worldwide (Morré et al., 2000; review, Byrne, 2010; van der Laar et al., 1996). As the other genital serovars (D, F-K) occupy the same niche, it is unlikely that serovar E requires active AaxB when the other serovars have lost functionality. This, coupled with the low AaxB levels detected during in vitro infection, suggest that although C. trachomatis serovar E currently retains active AaxB, this serovar may be in the process of inactivating this enzyme.
While C. pneumoniae and many of the non-C. trachomatis serovars retain an active ArgDC, the function of this enzyme in Chlamydia remains obscure. Although ArgDCs in other bacteria play roles in acid resistance and/or polyamine metabolism, neither function appears relevant to Chlamydia. The Chlamydia inclusion remains at neutral pH throughout infection, so encounters with acidic environments are unlikely (Schramm et al., 1996; Al-Younes et al., 1999; Grieshaber et al., 2002). Additionally, there are no known Chlamydia enzymes able to metabolize agmatine, such as the agmatine ureohydrolase, and therefore AaxB cannot contribute to polyamine synthesis. Finally, in certain cell lines, addition of exogenous agmatine alone may provide protection against cellular apoptosis (Arndt et al., 2009), but investigation in our laboratory suggests that this is likely not a factor during Chlamydia infection (data not shown).
As Giles and colleagues have speculated previously, the most likely function for the arginine decarboxylase system during Chlamydia infection is depletion of host cell arginine reserves (Giles and Graham, 2007). L-arginine, a substrate for the nitric oxide synthase (iNOS) in macrophages and granulocytes, catalyzes the production of the reactive nitrogen intermediate nitric oxide (NO), which may play a role in decreasing bacterial infectivity and/or inhibiting the dissemination of Chlamydia to systemic sites (Xie et al., 1992; Mayer et al., 1993; Igietseme et al., 1998). Removal of the substrate L-arginine (which would be degraded to agmatine and pumped back into the cytosol in counter-exchange for arginine by AaxC) could therefore promote Chlamydia survival and/or fitness, particularly in strains that are known to infect and replicate within these specialized host cells, such as C. pneumoniae and C. psittaci (Redecke et al., 1998; Wyrick and Brownridge, 1978). The timing of cleavage, and presumably corresponding activity, of AaxB in these strains may correlate with optimal iNOS activation in infected macrophages, and ultimately allow Chlamydia to avoid the detrimental consequences of NO production prior to bacterial exit from the host cell. Alternatively, the presence of processed AaxB in EBs may indicate that EBs are ‘pre-loaded’ with functional AaxB that is used to protect against NO production during the immediate-early stage of infection.
Supplementary Material
L2 cells were infected at an MOI of 1 or mock infected for 20 hrs (a). 1×107 purified EBs were loaded per lane in (b). Samples were disrupted directly in Laemmli buffer prior to 12% SDS-PAGE/Western blotting analysis with anti-AaxB antibodies. Proenzyme and the α fragment are indicated by black and gray arrows to the right of the blot. Protein molecular weight marker positions are shown to the left of each blot.
Acknowledgments
This study was supported by grants AI44033 from the National Institute of Allergy and Infectious Diseases (Maurelli), 1F32AI078655-01 from the National Institute of Allergy and Infectious Diseases (Fisher), and the USUHS Graduate Education Office (Bliven). The opinions or assertions contained herein are the private ones of the authors and are not to be construed as official or as reflecting the views of the Department of Defense or the Uniformed Services University.
References
- AbdelRahman YM, Belland RJ. The chlamydial developmental cycle. FEMS Microbiol Rev. 2005;29:949–59. doi: 10.1016/j.femsre.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Al-Younes HM, Rudel T, Meyer TF. Characterization and intracellular trafficking pattern of vacuoles containing Chlamydia pneumoniae in human epithelial cells. Cell Microbiol. 1999;1:237–47. doi: 10.1046/j.1462-5822.1999.00024.x. [DOI] [PubMed] [Google Scholar]
- Arndt MA, Battaglia V, Parisi E, et al. The arginine metabolite agmatine protects mitochondrial function and confers resistance to cellular apoptosis. Am J Physiol Cell Physiol. 2009;296:C1411–9. doi: 10.1152/ajpcell.00529.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belland RJ, Zhong G, Crane DD, Hogan D, Sturdevant D, Sharma J, Beatty WL, Caldwell HD. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. P Natl Acad Sci USA. 2003;100:8478–83. doi: 10.1073/pnas.1331135100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binet R, Bowlin AK, Maurelli AT, Rank RG. Impact of azithromycin resistance mutations on the virulence and fitness of Chlamydia caviae in guinea pigs. Antimicrob Agents Chemother. 2010;54:1094–101. doi: 10.1128/AAC.01321-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne GI. Chlamydia trachomatis strains and virulence: rethinking links to infection prevalence and disease severity. J Infect Dis. 2010;201:S126–33. doi: 10.1086/652398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson JH, Porcella SF, McClarty G, Caldwell HD. Comparative genomic analysis of Chlamydia trachomatis oculotropic and genitotropic strains. Infect Immun. 2005;73:6407–18. doi: 10.1128/IAI.73.10.6407-6418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanie-Cornet M, Penfound TA, Smith D, Elliott JF, Foster JW. Control of acid resistance in Escherichia coli. J Bacteriol. 1999;181:3525–35. doi: 10.1128/jb.181.11.3525-3535.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. P Natl Acad Sci USA. 2000;97:6640–5. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles TN, Graham DE. Characterization of an acid-dependent arginine decarboxylase enzyme from Chlamydophila pneumoniae. J Bacteriol. 2007;189:7376–83. doi: 10.1128/JB.00772-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles TN, Fisher DJ, Graham DE. Independent inactivation of arginine decarboxylase genes by nonsense and missense mutations led to pseudogene formation in Chlamydia trachomatis serovar L2 and D strains. BMC Evol Biol. 2009;9:166. doi: 10.1186/1471-2148-9-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham DE, Xu H, White RH. Methanococcus jannaschii uses a pyruvoyl-dependent arginine decarboxylase in polyamine biosynthesis. J Biol Chem. 2002;277:23500–07. doi: 10.1074/jbc.M203467200. [DOI] [PubMed] [Google Scholar]
- Grieshaber S, Swanson JA, Hackstadt T. Determination of the physical environment within the Chlamydia trachomatis inclusion using ion-selective ratiometric probes. Cell Microbiol. 2002;4:273–83. doi: 10.1046/j.1462-5822.2002.00191.x. [DOI] [PubMed] [Google Scholar]
- Igietseme JU, Perry LL, Ananaba GA, Uriri IM, Ojior OO, Kumar SN, Caldwell HD. Chlamydial infection in inducible nitric oxide synthase knockout mice. Infect Immun. 1998;66:1282–6. doi: 10.1128/iai.66.4.1282-1286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Lee IS, Frey J, Slonczewski JL, Foster JW. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J Bacteriol. 1995;177:4097–104. doi: 10.1128/jb.177.14.4097-4104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer J, Woods ML, Vavrin Z, Hibbs JB. Gamma interferon-induced nitric oxide production reduces Chlamydia trachomatis infectivity in McCoy cells. Infect Immun. 1993;61:491–7. doi: 10.1128/iai.61.2.491-497.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press; Plainview, NY: 1972. [Google Scholar]
- Morré SA, Rozendaal L, van Valkengoed IGM, Boeke AJP, van Voorst Vader PC, Schirm J, de Blok S, van der hoek JAR, van Doornum GJJ, Meijer CJLM, van der Brule AJC. Urogenital Chlamydia trachomatis serovars in men and women with a symptomatic or asymptomatic infection: an association with clinical manifestations? J Clin Microbiol. 2000;38:2292–6. doi: 10.1128/jcm.38.6.2292-2296.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel CN, Wortham BW, Lines JL, Fetherston JD, Perry RD, Oliveira MA. Polyamines are essential for the formation of plague biofilm. J Bacteriol. 2006;188:2355–63. doi: 10.1128/JB.188.7.2355-2363.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redecke V, Dalhoff K, Bohnet S, Braun J, Maass M. Interaction of Chlamydia pneumoniae and human alveolar macrophages: infection and inflammatory response. Am J Respir Cell Mol Biol. 1998;19:721–7. doi: 10.1165/ajrcmb.19.5.3072. [DOI] [PubMed] [Google Scholar]
- Rohde G, Straube D, Essig A, Reinhold P, Sachse K. Chlamydial zoonoses. Dtsch Arztebl Int. 2010;107:174–180. doi: 10.3238/arztebl.2010.0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm N, Bagnell CR, Wyrick PB. Vesicles containing Chlamydia trachomatis serovar L2 remain above pH 6.0 within HEC-1B cells. Infect Immun. 1996;64:1208–14. doi: 10.1128/iai.64.4.1208-1214.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CB, Graham DE. Outer and inner membrane proteins compose an arginine-agmatine exchange system in Chlamydophila pneumoniae. J Bacteriol. 2008;190:7431–40. doi: 10.1128/JB.00652-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stim KP, Bennett GN. Nucleotide sequence of the adi gene, which encodes the biodegradative acid-induced arginine decarboxylase of Escherichia coli. J Bacteriol. 1993;175:1221–34. doi: 10.1128/jb.175.5.1221-1234.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor CW, Tabor H. Polyamines. Annual Rev Biochem. 1984;53:749–90. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- Van der Laar MJW, Lan J, van Duynhoven YTHP, Fennema JSA, Ossewaarde JM, van den Brule AJC, van Doornum GJJ, Coutinho RA, van den Hoek JAR. Differences in clinical manifestations of genital chlamydial infections related to serovars. Genitourin Med. 1996;72:261–5. [PMC free article] [PubMed] [Google Scholar]
- Wyrick PB, Brownridge EA. Growth of Chlamydia psittaci in macrophages. Infect Immun. 1978;19:1054–60. doi: 10.1128/iai.19.3.1054-1060.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie QW, Cho HJ, Calaycay J, Mumford RA, Swiderek KM, Lee TD, Ding A, Troso T, Nathan C. Cloning and characterization of inducible nitric oxide synthase from mouse macrophages. Science. 1992;256:225–8. doi: 10.1126/science.1373522. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Lyng K, Zhang YX, Rockey DD, Morrison RP. Monoclonal antibodies define genus-specific, species-specific, and cross-reactive epitopes of the chlamydial 60-kilodalton heat shock protein (hsp60): specific immunodetection and purification of chlamydial hsp60. Infect Immun. 1992;60:2288–96. doi: 10.1128/iai.60.6.2288-2296.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
L2 cells were infected at an MOI of 1 or mock infected for 20 hrs (a). 1×107 purified EBs were loaded per lane in (b). Samples were disrupted directly in Laemmli buffer prior to 12% SDS-PAGE/Western blotting analysis with anti-AaxB antibodies. Proenzyme and the α fragment are indicated by black and gray arrows to the right of the blot. Protein molecular weight marker positions are shown to the left of each blot.