Abstract
The Esophageal cancer related gene-4 (Ecrg4) is a candidate tumor suppressor gene whose secreted protein product has been implicated in the development and progression of epithelial cancers, neuroprogenitor cell activation after central nervous system injury, cell senescence in neurodegeneration, and the survival of hematopoietic stem cells. Here, we investigated the temporal and spatial localization of Ecrg4 expression in healthy and injured mouse skin, and evaluated the biological activity of Ecrg4 using viral-mediated gene delivery in cutaneous wound healing models. Using in situ hybridization and immunohistochemistry, we found both Ecrg4 mRNA and its protein product localized to the epidermis, dermis, and hair follicles of healthy mouse skin. Upon cutaneous injury, Ecrg4 redistributed to the wound margins where gene microarray and quantitative RT-PCR showed an increased gene expression 5–10 days post injury as a late phase injury response gene. Ecrg4 over-expression inhibited the directional migration of fibroblasts in modified Boyden chambers in vitro, but had no effect on rates of fibroblast proliferation. Ecrg4 over-expression in vivo at the wound margins delayed the rate of wound closure at one and two days after full-thickness punch injury. These findings point to the candidate tumor suppressor gene Ecrg4 as a novel, biologically active, constituent of skin and skin injury. The possibility that Ecrg4 serves as a wound termination factor during wound resolution is discussed.
Keywords: Ecrg4, tumor suppressor gene, skin, wound healing, wound closure
INTRODUCTION
Cutaneous wound healing is a well-coordinated process that transitions across distinct inflammatory, secretory, and remodeling phases in a temporal sequence designed to restore homeostasis to the wounded area after injury. The response to cutaneous injury involves a complex interplay of soluble and insoluble factors that are induced, suppressed, and up-regulated at the site of injury [4]. Numerous trophic signaling factors, including growth factors, chemokines, low-molecular weight compounds and their respective receptors, have been implicated in the initiation and up-regulation of the cutaneous injury response [39]. Similarly, there exists numerous extracellular ligands in skin that represent candidate wound resolution factors that participate late after injury [14,15,21,22,5] that have yet to be identified.
Since many growth factors are analogous to the proto-oncogene ligands that participate in early repair and regeneration following injury, many believe the opposing biology of wound resolution and cancer progression [30,9,32] implicates a physiological role for tumor suppressor genes (TSG) in the later stages of wound repair. Due to the importance of TSGs in cancer [9,30,32], we became interested in the possibility that there might exist novel, and currently unrecognized, TSGs in skin that influence cutaneous injury. Presumably down-regulated by the initial injury response, these growth inhibitory genes would be predicted to be up-regulated during the last stages of wound resolution when cell growth is least active. The proposal that down-regulated, intra-cellular TSGs may play an important physiological role in wound healing [17,2] lead us in search of previously uncharacterized TSGs present in skin.
In the course of mining public bioinformatic databases like GENSAT and GEOprofiles, we recognized the potential of one particular gene, Ecrg4 (also called human C2orf40 or mouse 1500015O10Rik). Originally detected in the cDNA libraries of expressed genes [36], Ecrg4 was found to be differentially down-regulated in a host of human malignancies with an expression profile that was related to the increased migration, proliferation and metastatic capabilities of tumor cells [13,19,20,25,40]. We specifically focused on Ecrg4 because of the unique features of this candidate TSG: its product is secreted, ligand-like, tethered to the cell surface, and has been implicated in organ specific injury responses [24,29,12,1]. As opposed to intracellular TSGs like p53 and Rb94 [2,17], we hypothesized Ecrg4 may act on nearby proliferating target cells to help resolve the cutaneous injury response.
In this study, we identified Ecrg4 mRNA and its encoded peptide product in the epidermis, dermis, and hair follicles of healthy mouse skin. Following cutaneous injury, we found Ecrg4 gene expression increased as a late phase injury response gene and localized to the adjacent wound margins. Next, we identified a biological role for Ecrg4 in wound healing as viral-mediated Ecrg4 over-expression inhibited fibroblast migration in vitro and delayed the kinetics of wound closure at one and two days post-injury. Together, these findings identify the candidate TSG, Ecrg4, as a novel constituent of skin that we propose may, by virtue of its late phase expression kinetics and inhibitory biological activity, function as a wound termination factor during wound resolution.
MATERIALS AND METHODS
Ethics Statement
All animal experiments were performed with oversight of the UCSD IACUC to ensure animal welfare and the humane treatment of research animals.
Tissues
Tissue studies were performed using skin, spleen, whole blood, heart, and lung tissues harvested from 8–12 wk-old Balb/c mice (Jackson Labs, Bar Harbor, ME, USA) after anesthesia and cervical dislocation.
Adenovirus (AD)
AD vectors containing Ecrg4 (AD-Ecrg4) were prepared according to manufacturer's instructions using the AdEasy XL Adenoviral Vector System (Agilent Technologies, Santa Clara, CA, USA). Authenticity of the final products was demonstrated by DNA sequencing and immunoblotting after transduction of HEK, PC3, MEF, BHK and Caco cell lysates ([29] and unpublished data). Adenovirus vector containing green fluorescent protein (AD-GFP) was purchased from Vector BioLabs (Philadelphia, PA, USA).
Cell Culture and in vitro experiments
Primary mouse embryonic fibroblasts (MEFs) (Millipore, Billerica, MA) and normal human dermal fibroblasts (NHDF) (Lonza, Hopkinton, MA) were propagated as recommended by the manufacturer. MEFs were plated onto cover slips and grown for 24–48 hours until they reached 70–85% confluence. For the purposes of over-expressing Ecrg4 via gene delivery in MEFs [29], Ad-Ecrg4 (MOI 50–100) or control virus Ad-GFP was added to cells and incubated for 72 hours. To conduct modified Boyden chamber haptotaxis assays [16,10], 50,000 MEFs were plated onto the top well of trans-well culture inserts containing 8 μm pores (Corning Incorporated) that were previously coated on the bottom surface with 10 μg/ml rat tail collagen type I (Upstate Biotech). After a 6-hour incubation, MEFs migrating to the bottom surface were stained with 1% crystal violet, imaged with an inverted microscope (N=3) and quantified by a double-blinded observer. The experiment was repeated 3 times. For the cell proliferation assays, 20,000 NHDF cells/ml were plated in 24 well plates, incubated with AD-Ecrg4 or GFP (MOI 100) for 48 hours, and evaluated for cell count. Cells were numerated from triplicate samples on the third day and the counts confirmed using the Countess Automated Cell Counter (Life Technologies, Grand Island, NY).
Antibody Generation and Immunohistochemistry
Polyclonal antibodies were raised in chickens by immunization with a recombinant fusion protein expressing the amino acid sequence 71–148 of the full length predicted product of the Ecrg4 open reading frame (Genway, San Diego, CA). Antibodies were immunoaffinity purified with the antigen, where pre-immune IgY was used for controls. Specificity was validated by immunoblotting recombinant Ecrg4 and Ecrg4-transduced cells. For immunohistochemistry, tissue samples were fixed in 4% paraformaldehyde overnight at 4° C and rinsed in 30% sucrose in PBS at 4° C. Skin was embedded in OCT and 10mm cryosections were mounted on super-frost plus slides. Tissue sections were blocked for 20 minutes at room temperature with 15% normal donkey serum diluted in Tris-buffered saline (TBS) containing 0.5% Tween and 2% bovine serum albumin (BSA). Sections were then incubated with primary antibody (1:2000) overnight at 4°C, washed, and then stained with biotin-conjugated donkey anti-chicken antibody for 30 minutes at room temperature (Jackson ImmunoResearch). Sections were quenched in 0.3% H2O2 in methanol. After three further washings in TBS, sections were processed with an Avidin Biotin Complex (ABC) kit (Vectastatin, Burlingame, Ca) and detected with diaminobenzine (Vector) substrate. Sections were finally counterstained with hematoxylin, dehydrated and mounted.
Immunoblotting
To determine the molecular weight of the Ecrg4 peptide product in mouse tissues, samples were harvested and immediately frozen on dry ice and kept at −80°C until analyzed. Total protein was extracted from mouse tissue by homogenization and sonication in 4× reducing lithium dodecyl sulfate (LDS) buffer (Invitrogen). Protein was size-fractionated by polyacrylamide gel electrophoresis (PAGE) on a 4–12% bis-tris gradient gel run in 2-(N-morpholino) ethanesulfonic acid (MES) buffer and transferred to a polyvinylidene fluoride membrane. Affinity purified chicken anti-Ecrg4 antibody (1:20,000) or pre-immune IgY was used to detect Ecrg4. Horseradish peroxidase (HRP) conjugated goat anti-chicken antibody (BioRad) was used as the secondary antibody in Western blots. An IVIS® Lumina imaging system (Caliper Life Sciences, Hopkinton, MA, USA) was used to detect the chemi-luminescent signal.
In situ Hybridization
Restriction enzyme linearized (Sal I) plasmid pCMV-Sport 6 expressing the mouse Ecrg4 cDNA (RIKEN cDNA 1500015O10Rik) was originally obtained from Origene (MC200116) and used as template for the generation of digoxigenin-labeled RNA probe using T7 polymerase as indicated by manufacturer's recommendations (Roche, Indianapolis, IN). Control RNA probes were generated from pSPT18-Neo plasmid while labeling efficiency was verified by dot blot comparison of RNA probes with pre-labeled standards provided in the kit. Tissue samples were fixed in 4% paraformaldehyde overnight at 4° C and rinsed in 30% sucrose in PBS at 4° C. Skin was embedded in OCT and 10mm cryosections were mounted on super-frost plus slides. Sections were fixed on slides in 4% paraformaldehyde, washed in PBS, followed by 2×SSC, and incubated in pre-hybridization buffer using hybridization granules reconstituted according to manufacturer's directions (Roche). Digoxigenin-labeled complementary RNA probe was added to sections following a 2 minute denaturation at 95° C and incubated at 45° C overnight. Non-specific binding was removed by washes in 2× SSC followed by 60% formamide in 0.2× SSC. Detection of the digoxigenin probe was performed with a horseradish peroxidase conjugated sheep anti-digoxigenin primary antibody (Roche) and detected with a DAB substrate. Images were acquired with an Olympus FSX100 microscope.
Punch Biopsy Injury Model
8–12 week-old Balb/c mice (n=3) were anesthetized with isoflurane, the surgical site shaved, and prepared in a routine aseptic fashion. After verifying adequate anesthesia, a full-thickness 2 mm diameter wound was performed on the right dorsum using a biopsy punch. Animals were housed in separate cages with a 12 hour light/dark cycle and given access to feed and water. Tissues were harvested 5 days after injury. For the purposes of examining the biological effects of Ecrg4 over-expression in mouse skin, mice (N=4) were given two daily 5 μl intra-dermal injections of 1011 MOI/ml of either AD-GFP or AD-Ecrg4 starting 48 hours prior to punch injury. Another 15 μL of AD-GFP or AD-Ecrg4 was administered directly onto the wound bed immediately after injury. Wound size was measured daily with calipers. To assess the efficiency of skin transduction with adenovirus, Balb/c mice were injected intradermally with 1011 MOI/ml AD-GFP and 48 hours later, imaged in a non-invasive fluorescence imager [33,28] to observe and quantify GFP gene expression in skin.
Integra™ grafting model
8–12 week old Balb/c mice (n=3) were subjected to full-thickness injury and grafted with Integra™, as previously described in Shaterian et al. [33]. Briefly, one full-thickness (1.5 cm diameter, marked by template) circular wound was excised from the right side of the dorsum. The excisions were deep to the panniculus carnosus, removing epidermal, dermal, subcutaneous, and fascia layers. Integra grafts (Integra LifeSciences Corporation, Plainsboro, NJ, USA) 1.5 cm in diameter were secured with seven 6–0 silk interrupted sutures (Sherwood Davis & Geck, St. Louis, MO, USA), equidistant from each other. Tissues were harvested 5 days after injury.
Gene Array and qPCR
To analyze Ecrg4 mRNA in a 1 mm skin punch injury model, skin immediately adjacent to the wound site was collected over a time course of 6 hours to 10 days and analyzed (n=3) as we have previously described in Chen et al [4]. To determine gene expression levels of Ecrg4, total RNA was extracted using TriZol (Invitrogen, Carlsbad, CA, USA) and treated with DNase I (Invitrogen). Reverse transcription was performed using the RETROscript kit (Ambion Inc., Austin, TX, USA). All qPCR runs were performed in duplicate using StepOnePlus Real Time PCR system (Applied Biosystems, Carlsbad, California, USA), and the amplification cycle threshold (Ct) for Ecrg4 was normalized to glyceraldehyde-3-phosphate (GAPDH) using SYBR green detection (Applied Biosystems). The delta-delta Ct (2e-ΔΔCt) method was used to calculate fold-change relative to normal skin. Efficiencies for both primer sets were 95–100%. Primer sequences for Ecrg4 and GAPDH were:
Ecrg4-forward 5'-AAGCGTGCCAAACGACAGCTGTGGGAC-3';
Ecrg4-reverse 5'-TTAATAGTCATCATAGTTGACACTGGC-3';
GAPDH-forward 5'- TCACCACCATGGAGAAGGC-3';
GAPDH-reverse 5'- GCTAAGCAGTTGGTGGTGCA-3'.
RESULTS
Ecrg4 mRNA and encoded peptide product are expressed in skin
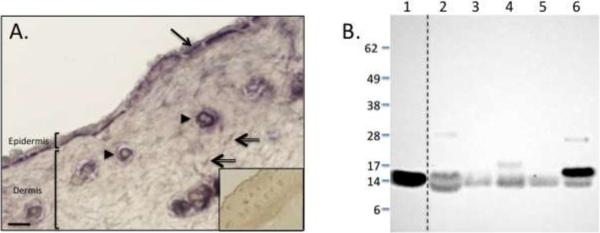
To assess for the presence of Ecrg4 and confirm the expression of this putative TSG in skin, we analyzed mouse skin sections for Ecrg4 mRNA and its encoded peptide product. We subjected mouse skin tissue sections to in situ hybridization to detect Ecrg4 mRNA (Figure 1A). Additional skin sections were incubated with a non-Ecrg4 coding digoxigenin mRNA probe as a control for specificity (Figure 1A, inset). We observed a localization of Ecrg4 mRNA in the epithelium (arrow), hair follicles (arrowhead), and occasional dermal fibroblast-like cells (double-lined arrow). These dermal cells potentially represent a fibroblast population based on morphological features. Previous reports have suggested that multiple isoforms of Ecrg4 can be detected in transfected cultured cell models [27], thus we used an anti-Ecrg4 antibody to detect endogenous Ecrg4 protein in mouse skin. A single 14 kDa immunoreactive band was detected (Figure 1B3), that corresponds to the molecular weight of the predicted product, amino acids 31–148, of Ecrg4. Although the Ecrg4 protein sequence is highly conserved across species [12], the tissue distribution of endogenous Ecrg4 protein has not been determined. Therefore, we subjected whole cell lysates of mouse blood, skin, spleen, lung and heart to immunoblotting with an anti-Ecrg4 antibody (Figure 1B). Here, we observed that all of the tissues expressed Ecrg4 with a molecular weight that co-migrated with the recombinant Ecrg4 (31–148) standard (Figure 1B, lane 1). We noted that blood, spleen and heart expressed additional 17 and 28 kDa Ecrg4 products of unknown function that could represent secondary Ecrg4 isoform multimers or post-translational modifications. Taken together, these data demonstrate that although other tissues expressed significant levels of Ecrg4 (i.e. blood and heart), Ecrg4 mRNA was present in healthy mouse skin and the molecular weight of the encoded protein was consistent with the predicted molecular weight of a 14 kDa Ecrg4 (31–148).
Figure 1. Ecrg4 mRNA and protein detected in healthy mouse skin.
A: In situ hybridization of Ecrg4 in healthy mouse skin. The constitutive nature of Ecrg4 expression was confirmed by in situ hybridization with antisense RNA to Ecrg4 mRNA. Ecrg4 was expressed in epidermis (arrow), hair follicles (arrowhead), and dermal cells (double-lined arrow). No staining was detected in control sections treated with sense strand of RNA (inset). Magnification bar=100mm
B: Immunoblotting of Ecrg4 protein in different mouse tissues. Lanes: (1) Recombinant fusion protein of amino acid sequences 31–148 of the full-length predicted product of Ecrg4; (2) Whole blood; (3) Skin; (4) Spleen; (5) Lung; (6) Heart. Dotted line indicates different parts of the same gel. Molecular weight standards (kDaltons) are indicated.
Ecrg4 protein localization in mouse skin
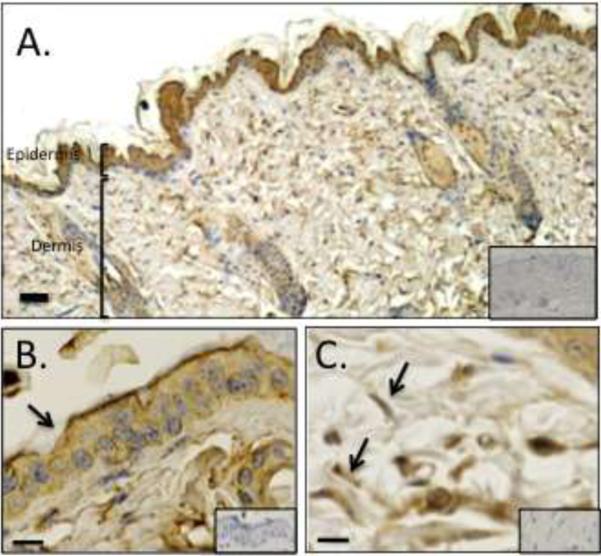
To determine the localization of Ecrg4 protein in skin, we subjected tissue sections to immunostaining with an anti-Ecrg4 antibody as described previously [29]. While Ecrg4 immunoreactivity localized to various cell types in skin (Figure 2A), we found punctuate staining in the epidermis (Figure 2B) and dermis (Figure 2C), with lower levels in hair follicles (data not shown). We also noted that the immunoreactive dermal cells have a characteristic fibroblast-like morphology. Immunostaining with isotype-matched control antibodies was performed to demonstrate specificity (Figure 2, insets). While in situ hybridization revealed a specific Ecrg4 mRNA expression profile in the skin, Ecrg4 protein was more widely expressed in tissues, consistent with previous studies suggesting that Ecrg4 protein can be secreted from the cell surface.
Figure 2. Ecrg4 protein localization in healthy mouse skin.
A) Low-power photomicrographs of healthy mouse skin stained with an anti-Ecrg4 antibody. Size bar = 100 μm. High-power photomicrographs show Ecrg4 immunolocalization within the B) epidermis, and C) dermal fibroblast-like cells. Control isotype antibody immunostaining is shown in the insets. Size bar=10μm.
Ecrg4 is a late injury response gene
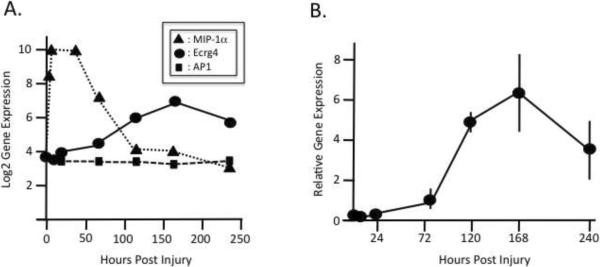
To examine the role of Ecrg4 in cutaneous injury, we mined an Affymetrix™ gene chip microarray of gene expression [4] and analyzed the kinetics of global gene expression following 1 mm cutaneous punch injury. We compared Ecrg4 mRNA expression levels to a known early response gene, macrophage inflammatory protein-1α (MIP-1α), as well as to a constitutively expressed gene involved in intracellular vesicle transport, AP1 complex subunit-2 (AP1) (Figure 3A). As expected [26], a 64× fold increase in MIP-1α was observed as early as the first 6 hour time point with gene expression levels returning to baseline by 120 hours. In contrast, Ecrg4 mRNA expression began increasing 24 hours after injury until achieving a maximal 8× fold expression 170 hours post injury. Gene expression levels of AP1, a constitutively expressed gene, remained relatively unchanged throughout the time course of the analyses (Figure 3A). To confirm the results of the Affymetrix microarray, we performed qRT-PCR and normalized Ecrg4 expression to levels of the reference gene GAPDH (Figure 3B). As with the microarray data, a late peak in Ecrg4 expression was observed 170 hours post-injury. Therefore, in addition to being expressed at constitutive baseline levels in healthy skin (Figure 1), Ecrg4 was induced upon injury and remained up-regulated for 5–10 days after injury. From these findings, we concluded that Ecrg4 is a late phase injury-response gene and hypothesized that Ecrg4 may have a potential role in cutaneous wound resolution.
Figure 3. Ecrg4 expression kinetics following cutaneous injury.
A: An Affymetrix GeneChip Mouse Genome chip was used for analysis of a 1 mm full-thickness punch wound in mice (n=3) and examined for changes in gene expression of Ecrg4 (sold circles), AP1 complex subunit-2 (solid squares) and macrophage inflammatory protein-1 (solid triangles).
B: qRT-PCR of Ecrg4 gene expression in skin after injury. Ecrg4 mRNA was processed from margins of 1 mm punch-injured tissue (n=3) and cDNAs evaluated by qPCR for Ecrg4 gene expression and data normalized to GAPDH expression.
Ecrg4 expression in the skin post-injury
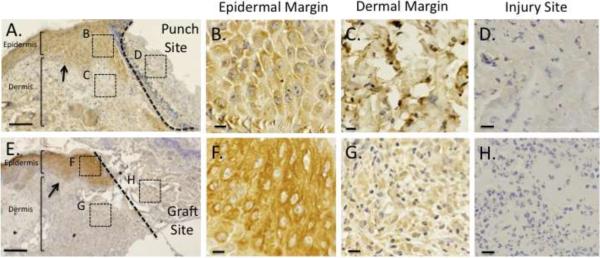
To determine the effects of tissue injury on Ecrg4 protein localization, we evaluated several models of cutaneous injury in mouse by immunohistochemistry. First, we examined the distribution of Ecrg4 protein in mouse skin following a 2 mm full-thickness punch wound, and observed Ecrg4 immunostaining within the epidermal and dermal cell margins adjacent to the site of injury (Figure 4, panels A–C). The presenting neoepithelium revealed Ecrg4 immunoreactivity across multiple epithelial cells layers (Figure 4A, arrows) and extended for approximately 20–60 cells lateral to the wound margin when evaluated at high magnification (data not shown). In contrast, the granulation tissue of the injury site lacked significant Ecrg4 immunoreactivity (Figure 4D). Next, we examined Ecrg4 protein localization in mouse skin that was subjected to a full-thickness excisional wound grafted with Integra™, a biosynthetic dermal regeneration matrix commonly used in burn patients. We have previously described the utility of this model in examining wound resolution kinetics following cutaneous excisional injury [33]. Similar to the punch model, Ecrg4 immunoreactivity was observed in the epidermal and dermal wound margins (Figure 4, Panels E–G), but absent from the granulation tissue underlying the Integra™ graft (Figure 4H). In combination with the Ecrg4 expression kinetics, the contrast between robust Ecrg4 expression at the wound margins with the lack of Ecrg4 expression at the wound site suggests that Ecrg4 is a late injury response gene that localizes to the adjacent wound margins after injury.
Figure 4. Expression of Ecrg4 protein post-injury.
Low-power photomicrograph of the wound bed/ wound margin interface showing Ecrg4 protein localization in mouse skin five days after A) 2 mm full-thickness punch biopsy and E) 1.5cm full-thickness excisional wound grafted with Integra. Dotted line delineates the wound bed and arrows depict the neoepithelium. Size Bar = 100 μm. High-power photomicrographs show absence of immunoreactivity at the injury sites (Panels D, H,) while maintaining localization within the epidermis and dermis of wound margins (Panels B, C, F, G). Size Bar=10μm.
Ecrg4 over-expression inhibits fibroblast migration and wound closure
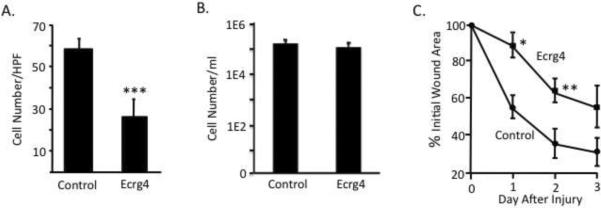
The later stages of tissue repair involve fibroblast proliferation/migration, granulation tissue invasion, and wound contraction [23,35,37]. As such, we investigated the effects of Ecrg4 as a late phase injury response gene on representative wound healing processes in vitro. As shown in figure 5, cultured fibroblasts were transduced with adenovirus expressing Ecrg4 or a control GFP gene, and then evaluated for changes in the rate of migration and proliferation in vitro. When Ecrg4- transduced fibroblasts were evaluated in collagen-coated modified Boyden chambers [16,10], a significant decrease in the rate of directional migration in vitro was observed (Figure 5A, P<0.05). In contrast, when we evaluated the effects of Ecrg4 over-expression on fibroblast proliferation in culture [6], we found similar rates of proliferation compared to controls (Figure 5B). Together, these data suggest that Ecrg4 gene expression preferentially inhibits fibroblasts migration vs. proliferation in vitro.
Figure 5. Ecrg4 over-expression interferes with fibroblast migration and wound closure.
A: Delivery of Ecrg4 inhibits fibroblast migration in vitro. Fibroblasts were transduced with ADEcrg4 or control ADgfp, and directional migration evaluated in modified Boyden chambers (N=4). Significance (p <0.01) was established by Student's T-test and ANOVA.
B: Delivery of Ecrg4 did not have a significant affect on fibroblast proliferation in vitro. Fibroblasts were transduced with ADEcrg4 or control ADgfp, incubated for 48hrs, and evaluated for cell count on day 3.
C: Delivery of exogenous Ecrg4 in skin delays the rate of wound closure at 1 and 2 days after 2 mm full thickness punch biopsy wound (p<0.05). ADEcrg4 (solid squares) or control ADgfp (solid circles) was administered intra-dermally 48hrs and 24hrs prior to injury, and again directly onto the wound bed immediately after injury, to maximize Ecrg4 gene expression. Results are presented as percentages of the initial wound area.
Since fibroblasts are key mediators of wound contraction and wound closure [37,3], we used viral-mediated intra-dermal gene delivery to examine the effects of Ecrg4 over-expression on cutaneous wound closure following a 2 mm full-thickness punch biopsy injury. Adenovirus-expressing either Ecrg4 or control GFP was administered intra-dermally at 48 and 24 hours prior to injury, and once again onto the wound bed immediately after injury, to maximize local gene expression. Daily measurements of the punch site diameter revealed a significant delay in the rate of wound closure in AD-Ecrg4 vs. control AD-GFP-injected animals at one and two days post-injury (Figure 5C, P< 0.05). This Ecrg4-mediated reduction in wound closure was transient however, possibly due to short-term adenovirus expression kinetics. Taken together, these data ascribe a novel biological activity to Ecrg4 in skin and suggest a role for Ecrg4 in fibroblast migration and wound closure.
Discussion
In this study, we investigated the temporal and spatial expression of Ecrg4 in normal vs. injured skin, and examined several models of viral-mediated Ecrg4 over-expression to gain insight into its biological activity in skin. In healthy skin, Ecrg4 mRNA and protein product localized to the epidermis, dermis, and hair follicles. Following cutaneous injury, Ecrg4 gene expression increased as a late phase injury response gene suggesting that Ecrg4 may function in the resolution phases of wound healing. Immunohistochemical analyses of Ecrg4 protein expression after cutaneous injury demonstrated that Ecrg4 protein localized to the cell margins adjacent to the site of injury, and were in sharp contrast to its absence in the injury site itself. Next, we found Ecrg4 over-expression inhibited the directional migration of fibroblasts in vitro, and decreased the rate of wound closure at one and two days after cutaneous injury in vivo. Taken together, these data suggest Ecrg4 as a novel, biologically active constituent of skin that may be induced in the later stages of injury as a wound termination factor.
Our studies have focused on the characterization of Ecrg4, a candidate TSG, as a mediator of the wound healing response in skin. TSGs with biological activity have been previously reported in skin. Examples include TAK1, a TSG up-regulated in wound healing found to coordinate cell migration and keratinocyte proliferation [38]. The TSG thrombospondin-1, also described in cutaneous wound repair, is involved in granulation tissue formation and angiogenesis [37]. Mdm2, also a TSG, has been shown to regulate epidermal stem cell senescence and premature aging [11], while the TSG p63 has been found to mediate morphogenesis of skin epidermis [18]. The insights taken from TSG studies in wound healing have established novel mechanisms regulating cell growth, migration, and differentiation, suggesting that in combination with the underlying cell biology of Ecrg4 and its relevance to skin injury, TSGs function in normal skin tissues mediating homeostasis and injury responses.
The response to cutaneous injury is a dynamic process that transitions across biologically distinct phases that are each regulated in part by transcriptional gene regulation at the wound site [31,7]. In work by Chen, DiPietro and colleagues [4], the temporal regulation of injury response genes were examined and clusters of “early” or “late” wound response genes were identified [7,4]. Microarray analyses of injured skin identified early response genes as those originating in the wound and inflammatory responses and included various cytokines, growth and chemotactic factors, and their respective receptors. In contrast, late response genes related to extracellular matrix remodeling, and hence represented collagens, structural proteins, ECM-receptor proteins and enzymes [4]. The characterization of Ecrg4 in this study extends these observations to include a previously unrecognized trophic factor that is induced with injury and may, by inference of its localization, expression kinetics, and biological activity, contribute to normal cutaneous homeostasis and the resolution stages of wound healing.
Wound resolution plays an important role in wound repair and is necessary for the return of functionality to the skin [34]. Accordingly, the ability to down-regulate the early and active cellular responses during injury is needed to achieve optimal healing [34,8]. To this end, we suggest Ecrg4 may play a role in this “stop phase” of wound healing as a wound termination factor. Up-regulated in the later phases of injury, this candidate TSG product demonstrates inhibitory biological activity on processes such as fibroblast migration and wound closure that must cease to prevent over-exuberant wound healing. In this model, the expression of Ecrg4 at the wound margin may serve to down regulate the early cellular processes prior to final wound resolution. Similarly, the absence of Ecrg4 at the injury site itself may foster robust and active cellular responses characteristic of granulation tissue. To this end, the up-regulation of Ecrg4 five to nine days after injury may help signal the beginning of wound resolution and help terminate specific cellular responses active in early wound healing. As such, Ecrg4 could join other epigenetically regulated genes in wound margin cells that have been associated with down-regulating the active wound healing response during resolution [34].
Our findings have also shown that Ecrg4 is expressed at baseline in healthy skin. This suggests that Ecrg4 may also play a constitutive role in maintaining the normal homeostatic environment of healthy skin. In this model, a TSG such as Ecrg4, participates in maintaining the homeostatic environment of skin, that upon cutaneous injury, becomes regionally dysinhibited until returning to quiescence after the final phases of wound healing when Ecrg4 expression levels return to baseline. The widespread expression of Ecrg4 in various other healthy quiescent tissues has lead us to speculate that Ecrg4 may serve a similar role in maintaining homeostasis, and regulating injury responses, in other mammalian tissues.
Acknowledgements
Research supported by National Institutes of Health grants GM078421 (AB), EY018479 (AB), 1TL1RR03197 (AS) and HL73396 (BE), the CDMRP BC073891 (AB), GM078426 (LDP), the BBSRC grant BB/C50466X/1 (AMG), and the expert assistance of Ana Maria Gonzalez, Ph.D., Jasmine Chukwueke, Sofia Peeva, Xitong Dang, Ph.D., Sonia Podvin, Ph.D., Emelie Amburn, Alexandra Borboa, Shuman Sun, and Paul Wolf, M.D.
Footnotes
Conflict of Interest: The authors state no conflict of interest.
LITERATURE CITED
- 1.Baird A, Coimbra R, Dang X, Lopez N, Lee J, Krzyzaniak M, Winfield R, Potenza B, Eliceiri BP. Cell surface localization and release of the candidate tumor suppressor Ecrg4 from polymorphonuclear cells and monocytes activate macrophages. J Leukoc Biol. 2012;91(5):773–781. doi: 10.1189/jlb.1011503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartkova J, Gron B, Dabelsteen E, Bartek J. Cell-cycle regulatory proteins in human wound healing. Arch Oral Biol. 2003;48(2):125–132. doi: 10.1016/s0003-9969(02)00202-9. [DOI] [PubMed] [Google Scholar]
- 3.Baum CL, Arpey CJ. Normal cutaneous wound healing: clinical correlation with cellular and molecular events. Dermatol Surg. 2005;31(6):674–686. doi: 10.1111/j.1524-4725.2005.31612. discussion 686. [DOI] [PubMed] [Google Scholar]
- 4.Chen L, Arbieva ZH, Guo S, Marucha PT, Mustoe TA, DiPietro LA. Positional differences in the wound transcriptome of skin and oral mucosa. BMC Genomics. 2010;11:471. doi: 10.1186/1471-2164-11-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Croce CM. Oncogenes and cancer. N Engl J Med. 2008;358(5):502–511. doi: 10.1056/NEJMra072367. [DOI] [PubMed] [Google Scholar]
- 6.Demaria M, Misale S, Giorgi C, Miano V, Camporeale A, Campisi J, Pinton P, Poli V. STAT3 can serve as a hit in the process of malignant transformation of primary cells. Cell Death Differ. 2012 doi: 10.1038/cdd.2012.20. doi:10.1038/cdd.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deonarine K, Panelli MC, Stashower ME, Jin P, Smith K, Slade HB, Norwood C, Wang E, Marincola FM, Stroncek DF. Gene expression profiling of cutaneous wound healing. J Transl Med. 2007;5:11. doi: 10.1186/1479-5876-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doshi BM, Perdrizet GA, Hightower LE. Wound healing from a cellular stress response perspective. Cell Stress Chaperones. 2008;13(4):393–399. doi: 10.1007/s12192-008-0059-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med. 1986;315(26):1650–1659. doi: 10.1056/NEJM198612253152606. [DOI] [PubMed] [Google Scholar]
- 10.Eliceiri BP, Klemke R, Stromblad S, Cheresh DA. Integrin alphavbeta3 requirement for sustained mitogen-activated protein kinase activity during angiogenesis. The Journal of cell biology. 1998;140(5):1255–1263. doi: 10.1083/jcb.140.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gannon HS, Donehower LA, Lyle S, Jones SN. Mdm2-p53 signaling regulates epidermal stem cell senescence and premature aging phenotypes in mouse skin. Dev Biol. 2011;353(1):1–9. doi: 10.1016/j.ydbio.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalez AM, Podvin S, Lin SY, Miller MC, Botfield H, Leadbeater WE, Roberton A, Dang X, Knowling SE, Cardenas-Galindo E, Donahue JE, Stopa EG, Johanson CE, Coimbra R, Eliceiri BP, Baird A. Ecrg4 expression and its product augurin in the choroid plexus: impact on fetal brain development, cerebrospinal fluid homeostasis and neuroprogenitor cell response to CNS injury. Fluids Barriers CNS. 2011;8(1):6. doi: 10.1186/2045-8118-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gotze S, Feldhaus V, Traska T, Wolter M, Reifenberger G, Tannapfel A, Kuhnen C, Martin D, Muller O, Sievers S. ECRG4 is a candidate tumor suppressor gene frequently hypermethylated in colorectal carcinoma and glioma. BMC Cancer. 2009;9:447. doi: 10.1186/1471-2407-9-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo S, Dipietro LA. Factors affecting wound healing. J Dent Res. 2010;89(3):219–229. doi: 10.1177/0022034509359125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gurtner GC, Werner S, Barrandon Y, Longaker MT. Wound repair and regeneration. Nature. 2008;453(7193):314–321. doi: 10.1038/nature07039. [DOI] [PubMed] [Google Scholar]
- 16.Kamiya K, Ryer E, Sakakibara K, Zohlman A, Kent KC, Liu B. Protein kinase C delta activated adhesion regulates vascular smooth muscle cell migration. The Journal of surgical research. 2007;141(1):91–96. doi: 10.1016/j.jss.2007.02.025. [DOI] [PubMed] [Google Scholar]
- 17.Kane CD, Greenhalgh DG. Expression and localization of p53 and bcl-2 in healing wounds in diabetic and nondiabetic mice. Wound Repair Regen. 2000;8(1):45–58. doi: 10.1046/j.1524-475x.2000.00045.x. [DOI] [PubMed] [Google Scholar]
- 18.King KE, Weinberg WC. p63: defining roles in morphogenesis, homeostasis, and neoplasia of the epidermis. Mol Carcinog. 2007;46(8):716–724. doi: 10.1002/mc.20337. [DOI] [PubMed] [Google Scholar]
- 19.Li L, Zhang C, Li X, Lu S, Zhou Y. The candidate tumor suppressor gene ECRG4 inhibits cancer cells migration and invasion in esophageal carcinoma. J Exp Clin Cancer Res. 2010;29:133. doi: 10.1186/1756-9966-29-133. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Li W, Liu X, Zhang B, Qi D, Zhang L, Jin Y, Yang H. Overexpression of candidate tumor suppressor ECRG4 inhibits glioma proliferation and invasion. J Exp Clin Cancer Res. 2010;29:89. doi: 10.1186/1756-9966-29-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X, Mohan S, Gu W, Baylink DJ. Analysis of gene expression in the wound repair/regeneration process. Mamm Genome. 2001;12(1):52–59. doi: 10.1007/s003350010230. [DOI] [PubMed] [Google Scholar]
- 22.Lindblad WJ. The future of human wound repair--regeneration science. Wound Repair Regen. 2004;12(3):261. doi: 10.1111/j.1067-1927.2004.012301.x. [DOI] [PubMed] [Google Scholar]
- 23.Martin P. Wound healing--aiming for perfect skin regeneration. Science. 1997;276(5309):75–81. doi: 10.1126/science.276.5309.75. [DOI] [PubMed] [Google Scholar]
- 24.Mirabeau O, Perlas E, Severini C, Audero E, Gascuel O, Possenti R, Birney E, Rosenthal N, Gross C. Identification of novel peptide hormones in the human proteome by hidden Markov model screening. Genome Res. 2007;17(3):320–327. doi: 10.1101/gr.5755407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mori Y, Ishiguro H, Kuwabara Y, Kimura M, Mitsui A, Kurehara H, Mori R, Tomoda K, Ogawa R, Katada T, Harata K, Fujii Y. Expression of ECRG4 is an independent prognostic factor for poor survival in patients with esophageal squamous cell carcinoma. Oncol Rep. 2007;18(4):981–985. [PubMed] [Google Scholar]
- 26.O'Grady NP, Tropea M, Preas HL, 2nd, Reda D, Vandivier RW, Banks SM, Suffredini AF. Detection of macrophage inflammatory protein (MIP)-1alpha and MIP-1beta during experimental endotoxemia and human sepsis. J Infect Dis. 1999;179(1):136–141. doi: 10.1086/314559. [DOI] [PubMed] [Google Scholar]
- 27.Ozawa A, Lick AN, Lindberg I. Processing of proaugurin is required to suppress proliferation of tumor cell lines. Mol Endocrinol. 2011;25(5):776–784. doi: 10.1210/me.2010-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterson CY, Shaterian A, Borboa AK, Gonzalez AM, Potenza BM, Coimbra R, Eliceiri BP, Baird A. The noninvasive, quantitative, in vivo assessment of adenoviral-mediated gene delivery in skin wound biomaterials. Biomaterials. 2009;30(35):6788–6793. doi: 10.1016/j.biomaterials.2009.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Podvin S, Gonzalez AM, Miller MC, Dang X, Botfield H, Donahue JE, Kurabi A, Boissaud-Cooke M, Rossi R, Leadbeater WE, Johanson CE, Coimbra R, Stopa EG, Eliceiri BP, Baird A. Esophageal cancer related gene-4 is a choroid plexus-derived injury response gene: evidence for a biphasic response in early and late brain injury. PLoS One. 2011;6(9):e24609. doi: 10.1371/journal.pone.0024609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riss J, Khanna C, Koo S, Chandramouli GV, Yang HH, Hu Y, Kleiner DE, Rosenwald A, Schaefer CF, Ben-Sasson SA, Yang L, Powell J, Kane DW, Star RA, Aprelikova O, Bauer K, Vasselli JR, Maranchie JK, Kohn KW, Buetow KH, Linehan WM, Weinstein JN, Lee MP, Klausner RD, Barrett JC. Cancers as wounds that do not heal: differences and similarities between renal regeneration/repair and renal cell carcinoma. Cancer Res. 2006;66(14):7216–7224. doi: 10.1158/0008-5472.CAN-06-0040. [DOI] [PubMed] [Google Scholar]
- 31.Schafer M, Werner S. Transcriptional control of wound repair. Annu Rev Cell Dev Biol. 2007;23:69–92. doi: 10.1146/annurev.cellbio.23.090506.123609. [DOI] [PubMed] [Google Scholar]
- 32.Schafer M, Werner S. Cancer as an overhealing wound: an old hypothesis revisited. Nat Rev Mol Cell Biol. 2008;9(8):628–638. doi: 10.1038/nrm2455. [DOI] [PubMed] [Google Scholar]
- 33.Shaterian A, Borboa A, Sawada R, Costantini T, Potenza B, Coimbra R, Baird A, Eliceiri BP. Real-time analysis of the kinetics of angiogenesis and vascular permeability in an animal model of wound healing. Burns. 2009;35(6):811–817. doi: 10.1016/j.burns.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaw TJ, Martin P. Wound repair at a glance. J Cell Sci. 2009;122(Pt 18):3209–3213. doi: 10.1242/jcs.031187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stadelmann WK, Digenis AG, Tobin GR. Physiology and healing dynamics of chronic cutaneous wounds. Am J Surg. 1998;176(2A Suppl):26S–38S. doi: 10.1016/s0002-9610(98)00183-4. [DOI] [PubMed] [Google Scholar]
- 36.Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF, Zeeberg B, Buetow KH, Schaefer CF, Bhat NK, Hopkins RF, Jordan H, Moore T, Max SI, Wang J, Hsieh F, Diatchenko L, Marusina K, Farmer AA, Rubin GM, Hong L, Stapleton M, Soares MB, Bonaldo MF, Casavant TL, Scheetz TE, Brownstein MJ, Usdin TB, Toshiyuki S, Carninci P, Prange C, Raha SS, Loquellano NA, Peters GJ, Abramson RD, Mullahy SJ, Bosak SA, McEwan PJ, McKernan KJ, Malek JA, Gunaratne PH, Richards S, Worley KC, Hale S, Garcia AM, Gay LJ, Hulyk SW, Villalon DK, Muzny DM, Sodergren EJ, Lu X, Gibbs RA, Fahey J, Helton E, Ketteman M, Madan A, Rodrigues S, Sanchez A, Whiting M, Young AC, Shevchenko Y, Bouffard GG, Blakesley RW, Touchman JW, Green ED, Dickson MC, Rodriguez AC, Grimwood J, Schmutz J, Myers RM, Butterfield YS, Krzywinski MI, Skalska U, Smailus DE, Schnerch A, Schein JE, Jones SJ, Marra MA. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A. 2002;99(26):16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Streit M, Velasco P, Riccardi L, Spencer L, Brown LF, Janes L, Lange-Asschenfeldt B, Yano K, Hawighorst T, Iruela-Arispe L, Detmar M. Thrombospondin-1 suppresses wound healing and granulation tissue formation in the skin of transgenic mice. Embo J. 2000;19(13):3272–3282. doi: 10.1093/emboj/19.13.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan SH, Pal M, Tan MJ, Wong MH, Tam FU, Teo JW, Chong HC, Tan CK, Goh YY, Tang MB, Cheung PC, Tan NS. Regulation of cell proliferation and migration by TAK1 via transcriptional control of von Hippel-Lindau tumor suppressor. The Journal of biological chemistry. 2009;284(27):18047–18058. doi: 10.1074/jbc.M109.002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Werner S, Grose R. Regulation of wound healing by growth factors and cytokines. Physiological reviews. 2003;83(3):835–870. doi: 10.1152/physrev.2003.83.3.835. [DOI] [PubMed] [Google Scholar]
- 40.Yue CM, Deng DJ, Bi MX, Guo LP, Lu SH. Expression of ECRG4, a novel esophageal cancer-related gene, downregulated by CpG island hypermethylation in human esophageal squamous cell carcinoma. World J Gastroenterol. 2003;9(6):1174–1178. doi: 10.3748/wjg.v9.i6.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]