Abstract
In this study, we describe the production of buffalo parthenogenetic blastocysts and subsequent isolation of parthenogenetic embryonic stem cell (PGESC)-like cells. PGESC colonies exhibited dome-shaped morphology and were clearly distinguishable from the feeder layer cells. Different stages of development of parthenogenetic embryos and derived embryonic stem cell (ESC)-like cells expressed key ESC-specific markers, including OCT-4, NANOG, SOX-2, FOXD3, REX-1, STAT-3, TELOMERASE, NUCLEOSTEMIN, and cMYC. Immunofluorescence-based studies revealed that the PGESCs were positive for surface-based pluripotent markers, viz., SSEA-3, SSEA-4, TRA 1-80, TRA 1-60, CD-9, and CD-90 and exhibited high alkaline phosphatase (ALP) activity. PGEC cell-like cells formed embryoid body (EB)-like structures in hanging drop cultures and when cultured for extended period of time spontaneously differentiated into derivatives of three embryonic germ layers as confirmed by RT-PCR for ectodermal (CYTOKERATIN8, NF-68), mesodermal (MSX1, BMP-4, ASA), and endodermal markers (AFP, HNF-4, GATA-4). Differentiation of PGESCs toward the neuronal lineage was successfully directed by supplementation of serum-containing media with retinoic acid. Our results indicate that the isolated ESC-like cells from parthenogenetic blastocyst hold properties of ESCs and express markers of pluripotency. The pluripotency markers were also expressed by early cleavage-stage of buffalo embryos.
Introduction
Embryonic stem cells (ESCs) isolated from the inner cell mass (ICM) of the blastocyst have the ability to proliferate indefinitely, and under controlled conditions they differentiate into specialized cell types (Smith et al., 2001). ESC lines have been isolated from several species, including mouse, human, pig, bovine, equine, sheep, and buffalo (Anand et al. 2011; Evans and Kaufman et al., 1981; Martin, 1981; Mitalipova et al., 2001; Notarianni et al., 1991; Saito et al., 2002; Sharma et al., 2011; Thomson et al., 1998; Verma et al., 2007; Wheeler et al., 1994). ESCs have also been isolated from blastocysts produced by in vivo fertilization, in vitro fertilization (IVF), and parthenogenetic activation (Mitalipova et al., 2001; Nakatasuji et al., 2002; Notarianni et al., 1991; Saito et al., 2002; Thomson et al., 1996; Verma et al., 2007). ESCs offer a convenient approach in reproductive biotechnology for genetic engineering and developing disease models. This technology holds great potential for domestic animals as well (Wilmut et al., 1994). Establishment of ESC lines in domestic animals, especially from cow, buffalo, sheep, and goat, would be useful for production of pharmacological products for treatment of human and animal diseases. Parthenogenetically produced embryos carry only maternal chromosomes and can be generated by inducing oocytes to resume meiosis without fertilization. The first cell lines derived from parthenogenetic embryos [parthenogenetic embryonic stem cells (PGESCs)] were established from mice (Kaufman et al., 1983). After that pioneering work, several studies have been carried out by various workers to establish cell lines in nonhuman primates, monkey, rabbit, human, rat (Cibelli et al., 2002; Fang et al., 2006; Iannaccone et al., 1994; Revazova et al., 2007; Vrana et al., 2003; Wang et al., 2006), pig (Wheeler et al., 1994), equine (Saito et al., 2002), sheep (Notarianni et al., 1991), bovine, and buffalo (Mayam et al., 2010; Sritanaudomchai et al., 2007; Strelchenko et al., 1996).
The buffalo is a fundamental livestock species in many developing South East Asian countries and provides milk, meat, and draft power. In vitro embryo production in buffalo has been poor in terms of blastocyst development, 10–20% in comparison to 30–40% in cattle (Gasparrini et al., 2003; Liang et al., 2007; Nandi et al., 2002; Palta and Chauhan, 1998; Yang et al., 1998) due to poor oocyte yield and quality obtained from slaughterhouse ovaries. Therefore, parthenogenetically produced blastocysts could be one of the possible alternatives to isolate and culture ESCs from buffalo. In ICMs isolated from bovine parthenogenetic embryos, developmental defects have been observed and these ICMs do not proliferate to maintain an ICM population and start differentiating (Wang et al., 2005). In buffalo, there are two studies that report the isolation and characterization of parthenogenetic ESCs (Muzaffar et al. 2012; Sritanaudomchai et al., 2007). In the present study, we tried to determine the expression of pluripotency markers in buffalo parthenogenetic embryos and putative PGESCs and examine their spontaneous and directed differentiation.
Materials and Methods
All chemicals and media were purchased from Sigma Chemical Company (St. Louis, MO, USA). All plasticware was from Nunc (Roskilde, Denmark) unless otherwise indicated.
In vitro production of parthenogenetic embryos
Follicular (2–8 mm in diameter) oocytes were aspirated from buffalo ovaries (obtained from an abattoir) with an 18-gauge needle in aspiration medium [M-199 containing 0.3% bovine serum albumin (BSA)]. The oocytes were washed four to six times with the washing medium, which consisted of M-199, 10% fetal bovine serum (FBS), 0.8 mM sodium pyruvate, 2 mM L-glutamine, and 50 μg mL−1 gentamicin. Only those cumulus–oocytes complexes (COCs) that had a compact and unexpanded cumulus mass with more than two to three layers of cumulus cells and with homogeneous evenly granular ooplasm were used for in vitro maturation (IVM).
The oocytes were washed three times with the IVM medium [M-199 supplemented with 10% FBS, 5 μg mL−1 porcine follicle-stimulating hormone (pFSH), 2 mM L-glutamine, 0.81 mM sodium pyruvate, and 50 μg mL−1 gentamicin]. Groups of 15–20 COCs were cultured in 100-μL droplets of the IVM medium, overlaid with sterile mineral oil in 35-mm petri dishes, and kept for 24 h in a CO2 incubator (5% CO2, 90–95% relative humidity) at 38.5°C. The mature oocytes were subjected to parthenogenetic activation (24 h post IVM), as described earlier (Gasparrini et al., 2004), with slight modification. Briefly, after being cultured in maturation medium, buffalo oocytes were denuded of their cumulus cells by incubation in 0.2% hyaluronidase in Dulbecco's phosphate-buffered saline (DPBS) for 2 min. The denuded oocytes with a prominent polar body were parthenogenetically activated by exposure to 7% ethanol for 7 min, followed by incubation with 2 mM 6-dimethyl aminopurine (6-DMAP) in mCR2aa medium for 4 h in a CO2 incubator (5% CO2, 90–95% relative humidity) at 38.5°C. The presumptive parthenotes were cultured for 8 days in mCR2aa medium containing 0.6% BSA and 10% FBS for production of embryos at different developmental stages.
Assessment of blastocyst quality
For examining the health of the embryos, the total cell number of trophectoderm (TE) and ICM of day-7 blastocysts was determined by differential staining, as described by Thouas et al. (2001). With little modifications, briefly the blastocysts were washed with DPBS for 10–15 sec and transferred into 500 μL of solution I (5 μg/mL Hoechst 33342) and incubated for 40 min at 37°C. Then blastocysts were washed with DPBS and immediately transferred into 500 μL of solution II (0.04% Triton X-100) for 1 min. Blastocysts were washed again with DPBS, followed by incubation in 25 μg/mL propidium iodide (PI) for 40 sec. The number of nuclei was counted using an inverted microscope (Nikon Diaphot) fitted with an ultraviolet (UV) lamp and excitation filters (excitation wavelength, 330–380 nm; barrier filter, 420 nm). ICM cells stained blue whereas TE cells stained red.
Isolation and culture of ICM
The hatched and early/expanded parthenogenetically generated blastocysts obtained on days 7 and 8 were used for isolation of ICMs that were seeded on a mitomycin C–treated buffalo fetal fibroblast feeder layer (Verma et al., 2007) to isolate ESCs. For hatched blastocysts, the ICMs were isolated mechanically with the help of two fine glass needles under a zoom stereomicroscope. The procedure for isolation of ICM from early/expanded blastocysts was described earlier (George et al., 2011). Briefly, the zona pellucida was removed by 1.0% pronase treatment in PBS (wt/vol). The zona-free parthenogenetic blastocysts were transferred into 0.25% trypsin–EDTA solution and observed under a zoom stereomicroscope until the trophectodermal cells became loose and were shed from the ICM by gentle pipetting. Isolated ICMs were then seeded on feeder layer, and cultured in knockout Dulbecco's modified Eagle medium (KO-DMEM) supplemented with 20% FBS, 1000 IU/mL murine leukemia inhibitory factor (mLIF), 5 ng mL−1 fibroblast growth factor-2 (FGF-2), 1% nonessential amino acids, 0.1 mM β-mercaptoethanol, and 2 mM l-glutamine. Three to four 100-μL droplets of the stem cell medium were prepared in a 35-mm dish and then cell culture–tested mineral oil (Sigma Chemical Company, St. Louis, MO, USA) was added to these dishes to cover all of the drops and keep them intact. The feeder cells were cultured in 25-cm2 tissue culture flasks, and treated with mitomycin C for 3–4 h. Before seeding feeder cells in the 100-μL droplets (described above) for preparing feeder layers, cells were washed many times with DPBS and trypsinized. Mitomycin C–treated cells were then seeded in these 100-μL droplets at the required concentration for preparing feeder layers. Clumps of stem cell colonies or blastocysts were directly introduced into these drops using a glass Pasteur pipette. The culture medium was changed every 24 h, and proliferation of colony cells was observed routinely.
Cryopreservation of BuPGESCs
ESC cryopreservation was performed as described earlier (George et. al., 2011; Verma et al., 2007) with slight modifications. Colonies of ESC-like cells, at different passages were vitrified in groups of five to seven in French ministraws (IMV, L'Aigle, Cedex, France). Vitrification solutions were based on holding medium (HM) consisting of DMEM and 20% FBS. Colonies were incubated in vitrification solution 1 (VS1), consisting of 10% dimethyl sulfoxide (DMSO) and 10% ethylene glycol, for 1 min, followed by incubation in VS2, consisting of 20% DMSO, 20% ethylene glycol (EG), and 0.5 M sucrose, for 20–30 sec and finally loaded into the straws. Just after sealing the open end of the straws with polyvinyl alcohol, the straws were immediately plunged into liquid nitrogen. For thawing, straws were plunged into water at 37°C for 30 sec; the sealed end was cut to allow colonies to enter and remain in the HM containing 0.2 M sucrose, for 1 min, followed by 5 min of incubation in HM containing 0.1 M sucrose. The colonies were further incubated twice (5 min each) in HM before seeding on mitomycin C–treated feeder layer.
Expression pattern of pluripotent markers
Gene expression analysis of transcription based -pluripotent markers like REX-1, OCT-4, NANOG, SOX-2, NUCLEOSTEMIN, FOXD-3, TELOMERASE, STAT-3, and c-MYC genes was performed in different embryonic developmental stages (immature oocytes, mature oocytes, two-cell, four-cell, eight- to 16-cell, morula, and blastocyst), TE cells, ESCs, embryoid body (EB) cells, and fetal and adult fibroblast cells. A two-step reverse transcriptase PCR (RT-PCR) was carried out using Cells-to-cDNA Kit-II (Ambion, Austin, TX, USA), as recommended by manufacturer. Briefly, five oocytes/embryos from each group of developmental stages were used for RT-PCR. Initially the cells were washed with ice-cold PBS and transferred to 50 μL of cold cell lysis buffer and incubated in a thermal cycler at 75°C for 10 min. The cell lysate was treated with DNase I at 37°C for 30 min to degrade genomic DNA and then heated at 75°C for 5 min to inactivate the DNase I. A total of 10 μL of cell lysate was used to reverse transcribe total RNA to cDNA. The list of primers and PCR conditions used are listed in Table S1 (See Supplementary Data at www.liebertpub/cell/).
The expression of surface markers, like SSEA-1, SSEA-3, SSEA-4, TRA-1-60, TRA-1-81, CD-9, and CD-90, was examined by carrying out immunofluorescence (IF) staining. Alkaline phosphatase (ALP) staining was performed according to the manufacturer (Sigma Aldrich). For IF staining, parthenogenetically generated buffalo ESCs were fixed in 4% paraformaldehyde in DPBS for 30 min, washed three times with DPBS, and then permeabilized by treatment with 0.1% Triton X-100 in DPBS for 30 min. After thorough washing with DPBS, the ESCs were incubated in the blocking solution (4% normal goat serum) for 30 min at room temperature and then with the primary antibody at a dilution of 1:20 for 1 h at room temperature. After washing three times with DPBS, the ESCs were incubated with appropriate fluorescein isothiocyanate (FITC)-labeled secondary antibody [anti-rat immunoglobin M (IgM) or anti-mouse IgG or IgM, diluted to 1:200] for 2 h at room temperature and then examined under a fluorescence microscope. The addition of primary antibody was omitted for respective controls. Chromosomal integrity of buffalo ESCs were analyzed by performing karyotyping at regular intervals.
EB formation and directed differentiation into neuron cells
For evaluation of in vitro differentiation capacity of buffalo PGESCs, cells at different passages were disaggregated by pipetting into small clumps. The clumps were cultured for 2 days in hanging drops (20 μL of ESC culture medium without LIF) followed by culturing in noncoated dishes. These EBs were plated on coated tissue culture dishes for adhesion and growth. The cells were fed every other day by tilting the plate, allowing the cells to settle, and carefully replacing the medium. For characterization of three germ layers, the expression of lineage-specific markers, i.e., GATA-4, AFP, and HNF-4 (endoderm), MSX-1, BMP-4 and ASA (mesoderm), and NF-68 and CYTOKARATIN (ectoderm), was studied using RT-PCR. After EBs formed, they were transferred onto gelatin-coated dishes in DMEM supplemented with 10% FBS and 10−6 M all-trans retinoic acid for formation of neuron-like cells. The neuron cells were characterized by RT-PCR with mRNA expression of NF-68. The morphology of the cells at the periphery of the EBs changed, demonstrating that these cells had differentiated to several types of cells.
Results and Discussion
Here we report the isolation and characterization of buffalo ESCs from ICMs of blastocysts produced through parthenogenetic activation.
Production of buffalo parthenotes and derivation of embryo ESCs
Oocytes collected from slaughtered buffalo ovaries were matured in vitro and subjected to parthenogenetic activation to produce blastocysts (Fig. 1A). A total of 606 oocytes were taken for in vitro maturation, out of which 372 cleaved (61.39%) and 87 blastocysts (23.38%) were formed, which were then used for deriving ESC colonies (n=47), differential staining (n=10) of the blastocysts, and gene expression studies (n=30). In vitro embryo production in buffalo through parthenogenetic activation was higher than that for IVF. The quality of frozen semen, poor oocyte yield, and quality of oocytes obtained from slaughterhouse and improper capacitation, besides some yet unknown culture factors, might be possible reasons for low embryo development through IVF in buffalo (Chauhan et al., 1998; Mishra et al., 2008; Nandi et al., 2001). The quality assessment of blastocysts produced through parthenogenetic activation was done on the basis of total cell number and the ICM/TE ratio using differential staining (Fig. 1B). The total cell number of the blastocysts was observed to be 174±3.03 whereas the ICM/TE ratio was found to be 0.11±0.008, which is in agreement with other reports (Muzaffar et al., 2012).
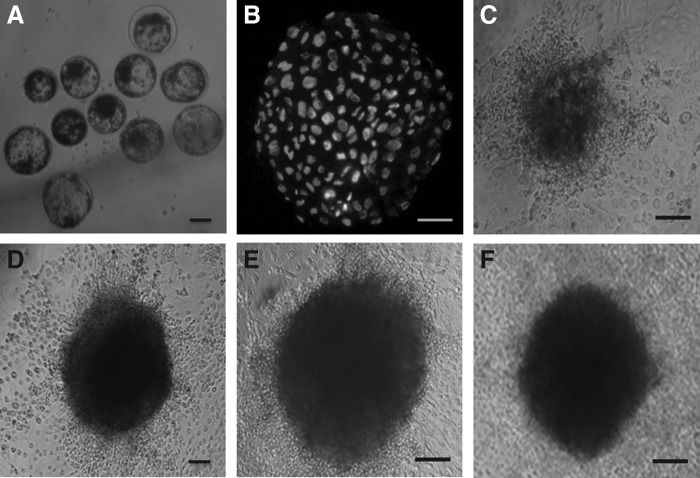
FIG. 1.
(A) Blastocysts produced parthenogenetically. (B) Differential staining of the blastocyst. (C) Primary colony of PGESCs. (D) Colony of PGESCs at passage 10. (E) Colony of PGESCs at passage 20. (F) Colony of PGESCs at passage 30. Scale bar, 200 μm, except for A and D, which is 100 μm.
Mechanically isolated ICMs from hatched blastocysts, when cultured on feeder layers, resulted in primary colonies formation after 5–8 days (Fig. 1C). Buffalo PGESCs were found to be morphologically similar to ESCs derived from IVF blastocysts. Earlier, it was reported that mouse ESCs cells formed dome-shaped colonies (Evans et al., 1981; Martin, 1981), but human and monkey ESCs formed flat colonies (Thomson et al., 1998). Morphologically, our colonies were compact and dome shaped with well-defined boundaries (Fig. 1D, E). These primary colonies were mechanically dissected in 1:2 or 1:3 split ratios, depending on the size of colony, and reseeded onto fresh feeder layers, with discarding TE cells to the maximum possible extent. The clumps of ESC-like cells were seeded individually in a 100-μL droplet containing ESC culture medium on the fresh feeder layers, requiring an average of 3–6 days between subsequent passages. The colonies were mechanically passaged and subcultured up to 30 passages (150 days) (Fig. 1F).
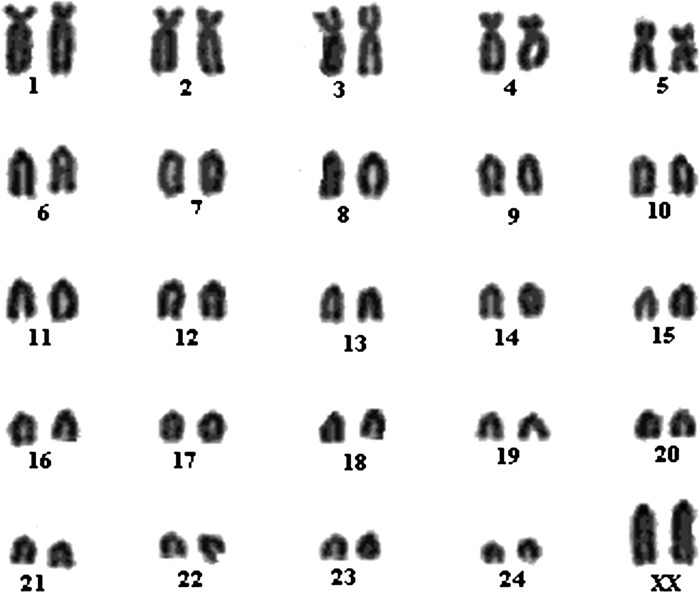
In addition, to confirming the viability and pluripotencey of cultured ESC-like cells, we also vitrified the clumps of ESC-like cells at regular passages. When vitrified clumps were recovered after thawing and cultured on a mitomycin C–treated feeder layer, they were found to proliferate after 3–4 days. ALP activity and pluripotency markers expression was detected in all the vitrified-thawed ESCs. Moreover, the ESCs were found to contain normal diploid set of chromosomes after karyotyping (Fig. 2).
FIG. 2.
Karyotype of parthenogenetic embryonic stem cells after 15 passages.
Expression of pluripotency markers in buffalo embryos and putative ESCs
To our knowledge, there is no report on expression of pluripotent markers in different developmental stages of bovine or bubaline embryos, produced parthenogenetically. Here we report for the first time in buffalo the expression of OCT-4, NANOG, SOX-2, REX-1, FOXD-3, TELOMERASE, NUCLEOSTEMIN, and STAT-3 genes in different stages of embryos produced through parthenogenetic activation. All the genes were expressed in immature and mature oocytes, two-cell, four-cell, eight- to 16-cell, morula, and blastocyst stages (Fig. 3 and Table 1). Magnani and Cabot (2008) reported the expression of OCT-4, NANOG, and SOX-2 in all the developmental stages of porcine embryos generated through parthenogenesis and revealed that OCT-4 transcript levels were highest at the two-cell stage whereas NANOG and SOX-2 were highest at four-cell stage embryos. Lee et al. (2006) reported that OCT-4 transcripts were high in the oocyte followed by reduction at the four- to eight-cell stage, and then leading to an increase at the blastocyst stage in both IVF and cloned embryos of pig. Van Eijk et al. (1999) and Daniels et al. (2000) studied the expression of OCT-4 in bovine IVF embryos and reported its expression in all the stages, starting from oocyte to blastocyst stage. The OCT-4 and NANOG transcript are expressed in the pig oocytes and remain visible throughout early cleavage and blastocyst formation (Brevini et al., 2007). Magnani and Cabot (2008) reported that SOX-2 expression was positive in pig oocytes, two-cell, four-cell, eight-cell, and blastocyst, derived from IVF, parthenogenetic activation (PA), and somatic cell nuclear transfer (SCNT).
FIG. 3.
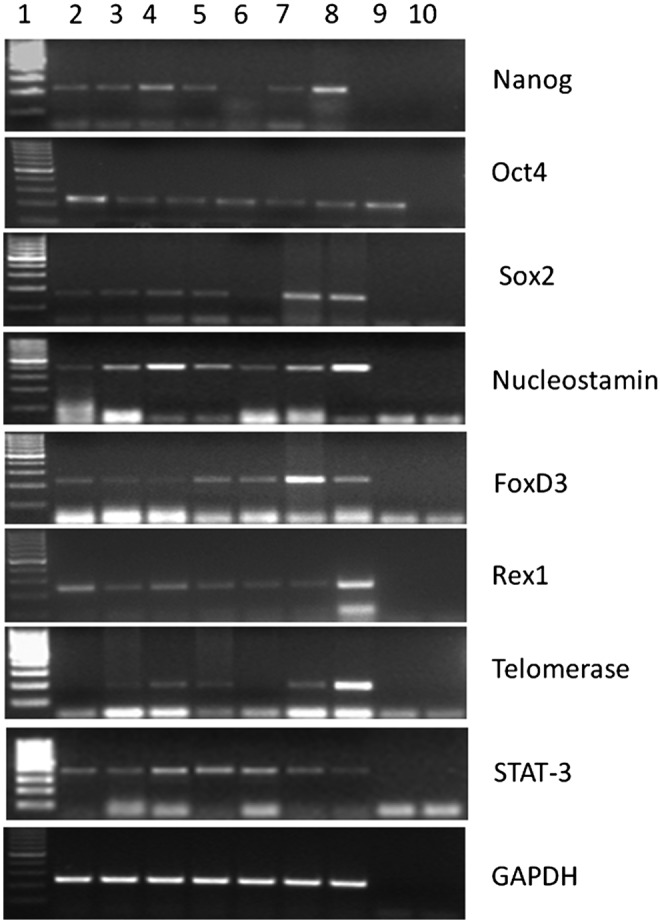
Expression pattern of pluripotent markers in developmental stages of parthenotes. Lane 1, 100-bp marker; lane 2, immature oocytes; lane 3, mature oocytes; lane 4, two-cell; lane 5, four-cell; lane 6, eight- to 16-cell; lane 7, morula; lane 8, blastocyst; lane 9, negative RT; lane 10, negative PCR. A negative RT control was included in the reaction, which consisted of all of the components of the cDNA synthesis excluding the reverse transcriptase enzyme given in the kit. The negative PCR included all the components for PCR reaction except the template. GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Table 1.
Summary of Marker Expression in Different Developmental Stages, Buffalo PGESCs, EB Cells, Fetal and Adult Fibroblast Cells, and TE Cells
| Markers | IO | MO | 2C | 4C | 8–16C | MO | BL | ES | EB | BFF | BAF | TE |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OCT-4 | + | + | + | + | + | + | + | + | + | − | − | + |
| NANOG | + | + | + | + | + | + | + | + | + | + | + | − |
| SOX-2 | + | + | + | + | + | + | + | + | + | − | − | − |
| REX-1 | + | + | + | + | + | + | + | + | − | + | + | + |
| FOXD-3 | + | + | + | + | + | + | + | + | + | + | + | − |
| TERT | + | + | + | + | + | + | + | + | − | + | + | − |
| NUCL | + | + | + | + | + | + | + | + | + | + | + | − |
| STAT-3 | + | + | + | + | + | + | + | + | + | + | + | − |
IO, immmature oocytes; MO, mature oocytes; 2C, two-cell stage; 4C, four-cell stage; 8–16C, eight to 16-cell stage; MO, morula; BL, blastocyst; ES, embryonic stem cells; EB, embryoid body; BFF, buffalo fetal fibroblast; BAF, buffalo adult fibroblast; TE, trophectoderm cells; TERT, TELOMERASE; NUCL, NUCLEOSTEMIN.
The characterization of PGESCs with the help of pluripotent markers was carried out at different passages to confirm the pluripotency of ESCs. In ESCs, RT-PCR analysis revealed mRNA expression of OCT-4, NANOG, SOX-2, REX-1, FOXD-3, NUCLEOSTEMIN, TELOMERASE, STAT-3, and c-MYC (Fig. 4 and Table 1). The expression of pluripotency markers was also studied in TE cells to determine the markers that are exclusive to the ICM. It was reflected from our results that OCT-4 and REX-1 expression was not confined to the ICM alone and was also observed in TE cells (Fig. 4 and Table 1). Moreover, expression of NANOG, SOX-2, FOXD-3, NUCLEOSTEMIN, and STAT-3 was exclusive to ICM and absent from TE cells (Fig. 4 and Table 1). Kurosaka et al. (2004) and Lee et al. (2006) also reported OCT-4 expression in TE cells of bovine and pig embryos, respectively. Munoz et al. (2008) reported the expression of NANOG protein in both ICM and TE of bovine blastocysts. Rogers et al. (1991) reported REX-1 expression in blastocyst, ICM, and TE cells of mouse. Although we did not find FOXD-3 expression in buffalo TE cells, Tompers et al. (2005) reported expression of FOXD-3 in mouse TE cells.
FIG. 4.
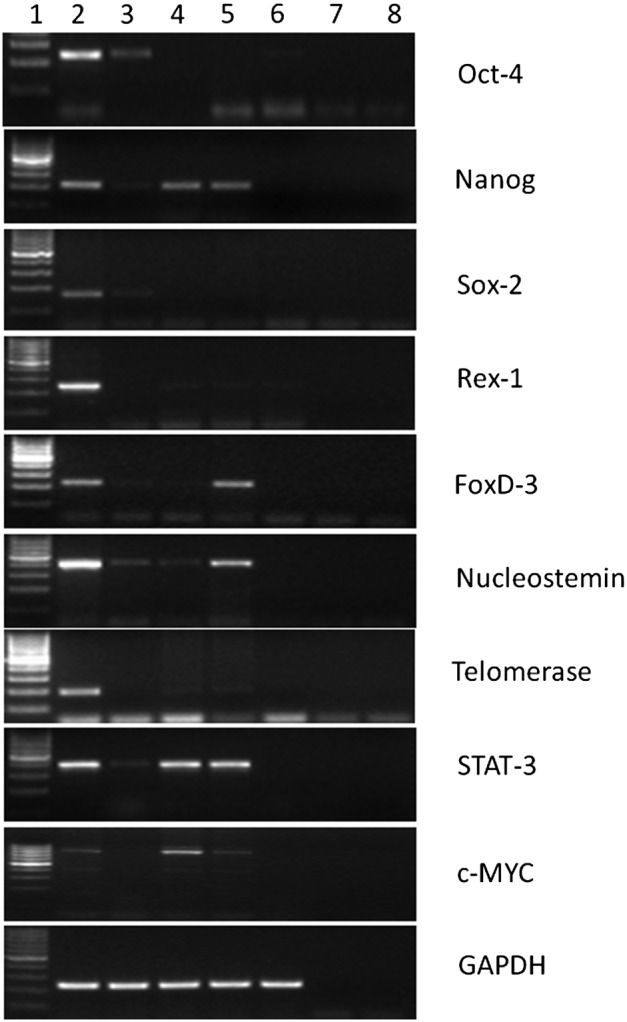
Expression pattern of pluripotent markers in PGESCs and other cells. Lane 1, 100-bp marker; lane 2, PGESCs; lane 3, EB cells; lane 4, fetal fibroblast cells; lane 5, adult fibroblast cells; lane 6, TE cells; lane 7, negative RT; lane 8, negative PCR. GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Buffalo fetal and adult fibroblast cells were used as negative controls in the expression study, and expression of NANOG, REX-1, TELOMERASE, NUCLEOSTAMIN, c-MYC, and STAT-3 was observed whereas OCT-4 and SOX-2 were found to be negative (Fig. 4 and Table 1). Brevini et al. (2005, 2007) reported expression of NANOG in somatic tissues like ovary, heart, and muscle in pig. Huang et al. (2008) reported the presence of NANOG in buffalo ESCs cells but its absence in fibroblast cells. Page et al. (2009) reported the presence of NANOG and SOX-2 in human fibroblast cells without induction. Carlin et al. (2006) reported that porcine fibroblasts cells and umbilical cord matrix cells expressed OCT-4, SOX-2, and NANOG. Hoei-Hansen et al. (2005) also found the expression of NANOG in human testis. We have also studied the expression of pluripotent markers in EB cells. These cells expressed all of the markers except REX-1 and TELOMERASE (Fig. 4 and Table 1).
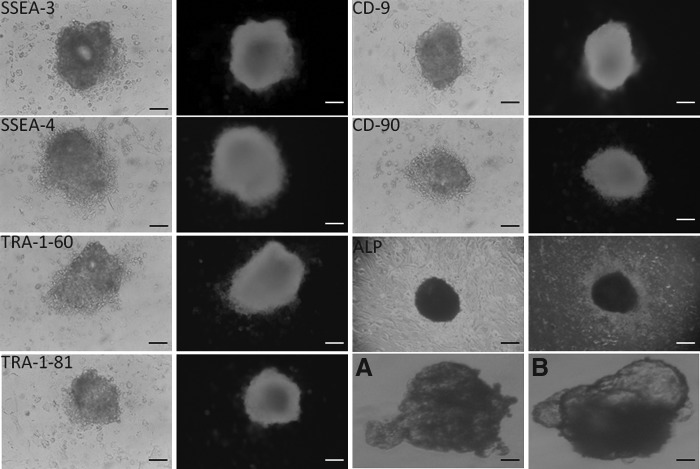
The ESCs derived from buffalo parthenotes were also characterized by studying expression of ALP and surface-based markers like SSEA-1, SSEA-3, SSEA-4, TRA-1-80, TRA-1-60, CD-9, and CD-90. Parthenogenetic ESCs exhibited high ALP activity (Fig. 5). The expression of all the surface markers was positive (Fig. 5), except for SSEA-1, which was negative in parthenogenetic ESCs. All of the surface markers were found to be negative in the fetal fibroblast cells that were used as negative controls.
FIG. 5.
Immunoflurences staining of buffalo PGESC SSEA-3, SSA-4, TRA-1-60, TRA1- 81, CD-9, CD-90, and EBs produced from PGESCs. (A) Simple EBs. (B) Cystic EBs. Scale bar, 100 μm.
Previously ESC-like cells have also been isolated from parthenogenetically derived buffalo embryos (Sritanaudomchai et al., 2007), which were positive for SSEA-1, SSEA-4, TRA-1-60, and TRA-1-81, but negative for SSEA-3. Huang et al. (2008) reported the expression of surface markers SSEA-1, SSEA-3, and SSEA-4 in buffalo ESCs generated through IVF embryos. Recently, we also reported the expression of SSEA-4, TRA-1-60, and TRA-1-81 in IVF buffalo embryos and ESCs (Anand et al., 2011), but we could not detect SSEA-1 and SSEA-3 in both embryos and ES-like cells. Wang et al. (2005) reported that embryonic stem cell (ntES) cells derived from cloned bovine blastocysts expressed SSEA-4, but were found to be negative for SSEA-1 and TRA-1. Munoz et al. (2008) also reported the expression of SSEA-4, TRA-1-81, and TRA-1-60 in ESC-like cells.
Spontaneous and directed differentiation of PEGSCs
To test the differentiation potential of buffalo PGESCs, colonies were mechanically dissected into small clumps and cultured in hanging drops of 20 μL of DMEM supplemented with 15% knockout serum replacement (KSR) in low-adherence dishes without growth factors or feeder cells for EB formation. After 2–4 days of culture in hanging drops, the simple EBs were formed (Fig. 5A). From the third day onward, the EBs were transferred to noncoated dishes for further culture. Compact or cystic EBs were formed within 4–7 days (Fig. 5B). When these EBs were cultured for a longer time, the size and number of cysts in cystic EBs increased. Expression of the germ layer–specific markers, i.e., GATA-4, alpha fetoprotein (AFP) and hepatocyte nuclear factor-4 (HNF-4) (endoderm), MSH homeobox-1 (MSX-1), bone morphogenic protein (BMP-4), α-skeletal actin (ASA) (mesoderm), neurofilament-68 (NF-68), CYTOKARATIN (ectoderm), were observed using RT-PCR (Fig. 6). We have also checked the expression of the markers of the three germ layers in the differentiated ESCs found on the periphery of colonies of ESCs (Fig. 6). Upon culturing EBs for 7–11 days with and without retinoic acid, the periphery of EBs proliferated to give neuron-like cells (Fig. 7A) and epithelial-like cells (Fig. 7E), because these cells exhibited NF-68 mRNA (Fig 7B) and NESTIN protein (Fig. 7C, D) expression for neuron and CYTOKARATIN 18 for epithelial cells, respectively (Fig. 7F).
FIG. 6.
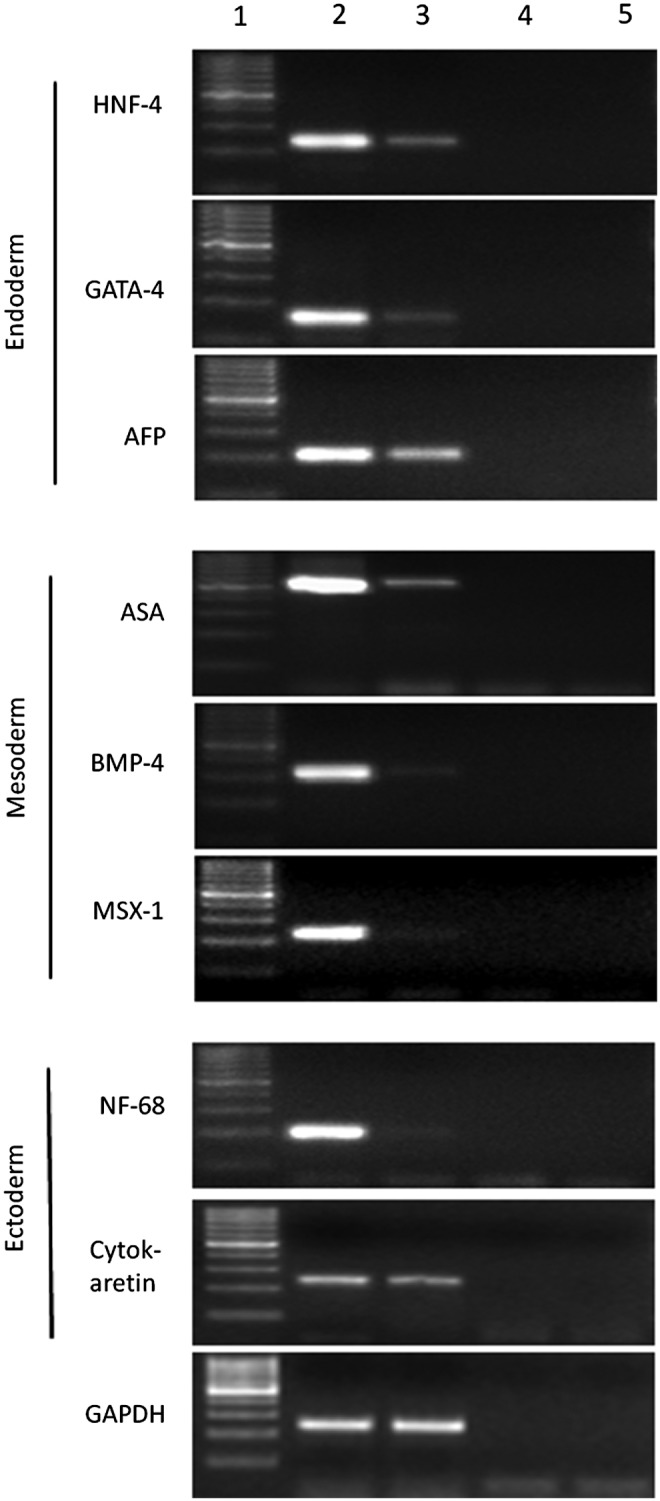
Lineage-specific markers expression in EB. Lane 1, 100-bp marker; lane 2, EB cells; lane 3, differentiated PGESCs; lane 4, negative RT; lane 5, negative PCR. GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
FIG. 7.
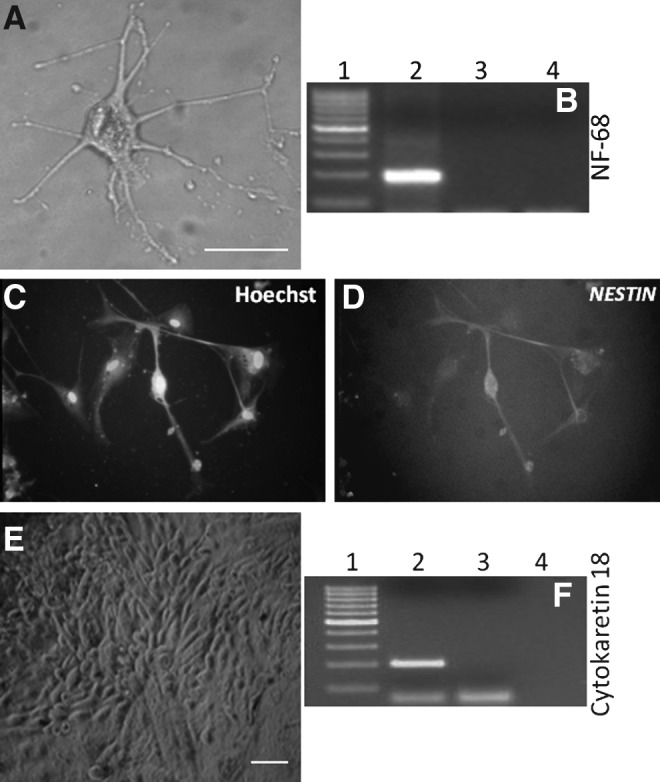
In vitro differentiation of PGESCs. (A) Neuron-like cells. (B) Characterization of neuron-like cells by NF-68 gene expression. Lane 1, 100-bp marker; lane 2, neuron-like cells; lane 3, negative RT; lane 4, negative PCR. (C) Hoechst staining of neuron cells. (D) Immunostaining of neuron cells by NESTIN. (E) Epithelial-like cells. (F) Characterization of epithelial-like cells by CYTOKARATIN 18 gene expression. Lane 1, 100-bp marker; lane 2, epithelial-like cells; lane 3, negative RT; lane 4, negative PCR. Scale bar, 50 μm.
Multilineage differentiation of buffalo ESCs was obtained via EB formation and spontaneous differentiation. We were able to detect the markers of all the three lineages in our EBs, similar to results reported by Sritanaudomchai et al. (2007). Upon directed differentiation of our ESC-like cells toward the neuronal lineage, we could observe neurons that expressed the neuron-specific markers NF-68 and NESTIN. Upon spontaneous differentiation, mainly epithelial-like cells were identified after characterization.
PGESCs are an efficient alternate for IVF-derived cell lines and can serve as a valuable in vitro model to study effects of imprinting on cell differentiation. Moreover, in buffalo, the blastocyst formation rate of parthenogenetic embryos is much more than IVF embryos and as such they provide a more valuable source for ESC derivation. There are many differences in blastocysts derived from IVF and parthenogenesis, but whether these differences are manifested in the respective derived ESC lines is still an open question. Among the many different alternative ways to pluripotency, artificial parthenogenesis represents an interesting method for deriving pluripotent cell lines that could also be considered ethically acceptable. PGESC lines are a unique model for studies addressed to a better understanding of mechanisms driving early embryonic development as well as genetic makeup of these cells.
In conclusion, here we report for the first time the expression of OCT-4, NANOG, SOX-2, REX-1, FOXD-3, TELOMERASE, NUCLEOSTEMIN, and STAT-3 genes in different stages of buffalo embryos produced through parthenogenetic activation, immature oocytes, and mature oocytes. Only OCT-4 and REX-1 were expressed in TE cells. The fetal and adult fibroblast cells also expressed the markers of pluripotency and only OCT-4 and SOX-2 were negative. It has been shown that the pluripotent ESCs can be generated in large animal models, and these cells express markers of pluripotency and can give rise to the derivatives of the three germ layers in vitro. Our results suggest that the cell line generated from parthenogenetically produced blastocysts maintained the properties of ESCs and can be used as an in vitro model to study the effects of imprinting on cell differentiation and as an invaluable material for extensive molecular studies on imprinted genes.
Supplementary Material
Acknowledgments
Authors are thankful to Dr. B. Prakash (Cytogenetics Lab, NBAGR) for help with karyotyping. This work was supported in part by National Agriculture Innovative Project grant (C4-C2067&75) to MSC.
Author Disclosure Statement
The authors declare that there are no competing interests.
References
- Anand T. Kumar D. Singh M.K., et al. Buffalo (Bubalus bubalis) embryonic stem cell-like cells and preimplantation embryos exhibit comparable expression of pluripotency-related antigens. Reprod. Domest. Anim. 2011;46:50–58. doi: 10.1111/j.1439-0531.2009.01564.x. [DOI] [PubMed] [Google Scholar]
- Brevini T.A.L. Cillo F. Antonini S., et al. Expression pattern of nanog gene in porcine tissue and parthenogenetic embryos. Reprod. Domest. Anim. 2005;40:384. [Google Scholar]
- Brevini T.A.L. Tostti V. Crestan M., et al. Derivation and characterization of pluripotent cell lines from pig embryos of different origins. Theriogenology. 2007;67:54–63. doi: 10.1016/j.theriogenology.2006.09.019. [DOI] [PubMed] [Google Scholar]
- Carlin R. Davis D. Weiss M., et al. Expression of early transcription factors OCT-4, SOX-2 and NANOG by porcine umbilical cord (PUC) matrix cells. Reprod. Biol. Endocrinol. 2006;4:8. doi: 10.1186/1477-7827-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan M.S. Singla S.K. Palta P., et al. In vitro maturation and fertilization, and subsequent development of buffalo (Bubalus bubalis) embryos, effects of oocyte quality and type of serum. Reprod. Fertil. Dev. 1998;10:173–177. doi: 10.1071/r97080. [DOI] [PubMed] [Google Scholar]
- Cibelli J.B. Grant K.A. Chapman K.B., et al. Parthenogenetic stem cells in nonhuman primates. Science. 2002;295:819. doi: 10.1126/science.1065637. [DOI] [PubMed] [Google Scholar]
- Daniels R. Hall V. Trounson A.O., et al. Analysis of gene transcription in bovine nuclear transfer embryos reconstructed with granulose cell nuclei. Biol. Reprod. 2000;63:1034–1040. doi: 10.1095/biolreprod63.4.1034. [DOI] [PubMed] [Google Scholar]
- Evans M.J. Kaufman M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Fang Z.F. Gai H. Huang Y.Z., et al. Rabbit embryonic stem cell lines derived from fertilized, parthenogenetic or somatic cell nuclear transfer embryos. Exp. Cell Res. 2006;312:3669–3682. doi: 10.1016/j.yexcr.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Gasparrini B. Sayoud H. Neglia G., et al. Glutathione synthesis during in vitro maturation of buffalo (Bubalus bubalis) oocytes: Effects of cysteamine on embryo development. Theriogenology. 2003;60:943–952. doi: 10.1016/s0093-691x(03)00098-0. [DOI] [PubMed] [Google Scholar]
- Gasparrini B. Boccia L. Rosa A.D., et al. Chemical activation of buffalo (Bubalus bubalis) oocytes by different methods: Effect of aging on post parthenogenetic development. Theriogenology. 2004;62:1627–1637. doi: 10.1016/j.theriogenology.2004.03.005. [DOI] [PubMed] [Google Scholar]
- George A. Sharma R. Singh K.P., et al. Production of cloned and transgenic embryos using buffalo (Bubalus bubalis) embryonic stem cell like cells isolated from in vitro fertilized and cloned blastocysts. Cell. Reprogram. 2011;13:263–272. doi: 10.1089/cell.2010.0094. [DOI] [PubMed] [Google Scholar]
- Hoei-Hansen C.E. Almstrup K. Nielsen J.E., et al. Stem cell pluripotency factor NANOG is expressed in human fetal gonocytes, testicular carcinoma in situ and germ cell tumours. Histopathology. 2005;47:48–56. doi: 10.1111/j.1365-2559.2005.02182.x. [DOI] [PubMed] [Google Scholar]
- Huang B. Li T. Wang X.L., et al. Generation and characterization of embryonic stem cell lines derived from in vitro fertilization buffalo (Bubalus bubalis) embryos. Reprod. Domest. Anim. 2008;45:122–128. doi: 10.1111/j.1439-0531.2008.01268.x. [DOI] [PubMed] [Google Scholar]
- Iannaccone P.M. Taborn G.U. Garton R.L., et al. Pluripotent embryonic stem cells from the rat are capable of producing chimeras. Dev. Biol. 1994;163:288–292. doi: 10.1006/dbio.1994.1146. [DOI] [PubMed] [Google Scholar]
- Kaufman M.H. Robertson E.J. Handyside A.H., et al. Establishment of pluripotential cell lines from haploid mouse embryos. J. Embryol. Exp. Morphol. 1983;73:249–261. [PubMed] [Google Scholar]
- Kurosaka S. Eckardt S. Maclaughlin K.J., et al. Pluripotent lineage definition in bovine embryos by Oct-4 transcription localization. Biol. Reprod. 2004;71:1578–1582. doi: 10.1095/biolreprod.104.029322. [DOI] [PubMed] [Google Scholar]
- Lee E. Lee S.H. Kim S., et al. Analysis of nuclear reprogramming in cloned miniature pig embryos by expression of Oct-4 and Oct-4 related genes. Biochem. Biophys. Res. Commun. 2006;348:1419–1428. doi: 10.1016/j.bbrc.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Liang X. Zhang X. Yang B., et al. Pregnancy and calving rates following transfer of in vitro produced river and F1 (river x swamp) buffalo (Bubalus bubalis) embryos in recipients on natural oestrus or synchronized for ovulation. Reprod. Fertil. Dev. 2007;19:670–676. doi: 10.1071/rd07048. [DOI] [PubMed] [Google Scholar]
- Magnani L. Cabot R.A. In vitro and in vivo derived porcine embryos possess similar, but not identical, patterns of Oct4, Nanog and Sox2 mRNA expression during cleavage development. Mol. Reprod. Dev. 2008;75:1726–1735. doi: 10.1002/mrd.20915. [DOI] [PubMed] [Google Scholar]
- Martin G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayam P.A. Khodadadi K. Holland M.K., et al. The efficient generation of cell lines from bovine parthenotes. Cell. Reprogram. 2010;12:571–579. doi: 10.1089/cell.2009.0118. [DOI] [PubMed] [Google Scholar]
- Mishra V. Misra A.K. Sharma R. A comparative study of parthenogenic activation and in vitro fertilization of bubaline oocytes. Anim. Reprod. Sci. 2008;103:249–259. doi: 10.1016/j.anireprosci.2006.12.019. [DOI] [PubMed] [Google Scholar]
- Mitalipova M. Beyhan Z. First N.L. Pluripotency of bovine embryonic cell line derived from precompacting embryos. Cloning. 2001;3:59–67. doi: 10.1089/15204550152475563. [DOI] [PubMed] [Google Scholar]
- Munoz M. Rodriguez A. Frutos C.D., et al. Conventional pluripotency markers are unspecified for bovine embryonic derived cell-lines. Theriogenology. 2008;69:1159–1164. doi: 10.1016/j.theriogenology.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Muzaffar M. Selokar N.L. Singh K.P, et al. Equivalency of buffalo (Bubalus bubalis) embryonic stem cells derived from fertilized, parthenogenetic and handmade cloned embryos. Cell. Reprogram. 2012;14:267–279. doi: 10.1089/cell.2011.0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuji N. Suemori H. Embryonic stem cell of nonhuman primates. Sci. World J. 2002;26:1762–1773. doi: 10.1100/tsw.2002.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi S. Chauhan M.S. Palta P. Effect of environmental temperature on quality and developmental competence in vitro of buffalo oocytes. Vet. Rec. 2001;148:278–279. doi: 10.1136/vr.148.9.278. [DOI] [PubMed] [Google Scholar]
- Nandi S. Raghu H.M. Ravindranatha V.M., et al. Production of buffalo (Bubalus bubalis) embryos in vitro: Premises and Promises. Reprod. Domest. Anim. 2002;37:65–74. doi: 10.1046/j.1439-0531.2002.00340.x. [DOI] [PubMed] [Google Scholar]
- Notarianni E. Galli C. Laurie S., et al. Derivation of pluripotent, embryonic cell lines from pig and sheep. J. Reprod. Fertil. 1991;43:255–260. [PubMed] [Google Scholar]
- Page R.L. Ambady S. Holmes W.F., et al. Induction of stem cell gene expression in adult human fibroblast without transgenes. Cloning Stem Cells. 2009;1:417–425. doi: 10.1089/clo.2009.0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palta P. Chauhan M.S. Laboratory production of buffalo (Bubalus bubalis) embryos. Reprod. Fert. Dev. 1998;10:379–391. doi: 10.1071/rd98085. [DOI] [PubMed] [Google Scholar]
- Revazova E.S. Turovets N.A. Kochetkova O.D., et al. Patient-specific stem cell lines derived from human parthenogenetic blastocysts. Cloning Stem Cells. 2007;9:432–449. doi: 10.1089/clo.2007.0033. [DOI] [PubMed] [Google Scholar]
- Rogers M.B. Hosler B.A. Gudas L.Z. Specific expression of a retinoic acid regulated zinc-finger gene REX-1 in preimplantation embryos trophoblast and spermatocytes. Development. 1991;113:815–824. doi: 10.1242/dev.113.3.815. [DOI] [PubMed] [Google Scholar]
- Saito S. Ugai H. Sawai K., et al. Isolation of embryonic stem-like cells from equine blastocysts and their differentiation in vitro. FEBS Lett. 2002;531:389–396. doi: 10.1016/s0014-5793(02)03550-0. [DOI] [PubMed] [Google Scholar]
- Sharma R. George A. Kamble N.M., et al. Optimization of culture condition to support long term self renewal of buffalo (Bubalus bubalis) embryonic stem cell like cells. Cell. Reprogram. 2011;13:539–549. doi: 10.1089/cell.2011.0041. [DOI] [PubMed] [Google Scholar]
- Smith AG. Embryo-derived stem cells: Of mice and men. Cell Dev. Biol. 2001;17:433–462. doi: 10.1146/annurev.cellbio.17.1.435. [DOI] [PubMed] [Google Scholar]
- Sritanaudomchai H. Pavasuthipaisit K. Kitiyanant Y., et al. Characterization and multilineage differentiation of embryonic stem cells derived from a buffalo parthenogenetic embryos. Mol. Reprod. Dev. 2007;74:1295–1302. doi: 10.1002/mrd.20592. [DOI] [PubMed] [Google Scholar]
- Strelchenko N. Bovine pluripotent stem cells. Theriogenology. 1996;45:131–140. [Google Scholar]
- Thomson J.A. Kalishman J. Golos T.G., et al. Pluripotent cell lines derived from common marmoset (Callithrix jacchus) blastocysts. Biol. Reprod. 1996;55:254–259. doi: 10.1095/biolreprod55.2.254. [DOI] [PubMed] [Google Scholar]
- Thomson J.A. Itskovitzeldor J. Shapiro S.S., et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Tompers D.M. Foreman R.K. Wang Q., et al. FOXD3 is required in the trophoblast progenitor cell lineage of the mouse embryos. Dev. Biol. 2005;285:126–137. doi: 10.1016/j.ydbio.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Thouas G.A. Korfiatis N.A. French A.J., et al. Simplified technique for differential staining of inner cell mass and trophectoderm cells of mouse and bovine blastocysts. Reprod. Biomed. Online. 2001;3:25–29. doi: 10.1016/s1472-6483(10)61960-8. [DOI] [PubMed] [Google Scholar]
- Van Eijk M.J.T. Rooijen V.M.A. Modina S., et al. Molecular cloning, genetic mapping and developmental expression of bovine POU5F1. Biol. Reprod. 1999;60:1093–1103. doi: 10.1095/biolreprod60.5.1093. [DOI] [PubMed] [Google Scholar]
- Verma V. Gautam S.K. Singh B., et al. Isolation and characterization of embryonic stem cell- like cells in vitro - produced buffalo (Bubalus bubalis) embryos. Mol. Reprod. Dev. 2007;74:520–529. doi: 10.1002/mrd.20645. [DOI] [PubMed] [Google Scholar]
- Vrana K.E. Jason D.H. Ashley M.G., et al. Nonhuman primate parthenogenetic stem cells. Proc. Natl. Acad. Sci. USA. 2003;100:11911–11916. doi: 10.1073/pnas.2034195100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. Duan E. Sung L.Y., et al. Generation and characterization of pluripotent stem cells from cloned bovine embryos. Biol. Reprod. 2005;73:149–155. doi: 10.1095/biolreprod.104.037150. [DOI] [PubMed] [Google Scholar]
- Wang S. Tang X. Niu Y., et al. Generation and characterization of rabbit embryonic stem cells. Stem Cells. 2006;25:481–489. doi: 10.1634/stemcells.2006-0226. [DOI] [PubMed] [Google Scholar]
- Wheeler M.B. Development and validation of swine embryonic stem cells: A review. Reprod. Fertil. Dev. 1994;6:563–568. doi: 10.1071/rd9940563. [DOI] [PubMed] [Google Scholar]
- Wilmut I. Whitlaw C.B. Strategies for production of pharmaceutical proteins in milk. Reprod. Fertil. Dev. 1994;6:625–630. doi: 10.1071/rd9940625. [DOI] [PubMed] [Google Scholar]
- Yang X. Kubota C. Suzuki H., et al. Control of oocyte maturation in cows- biological factors. Theriogenology. 1998;49:471–482. doi: 10.1016/s0093-691x(97)00419-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.