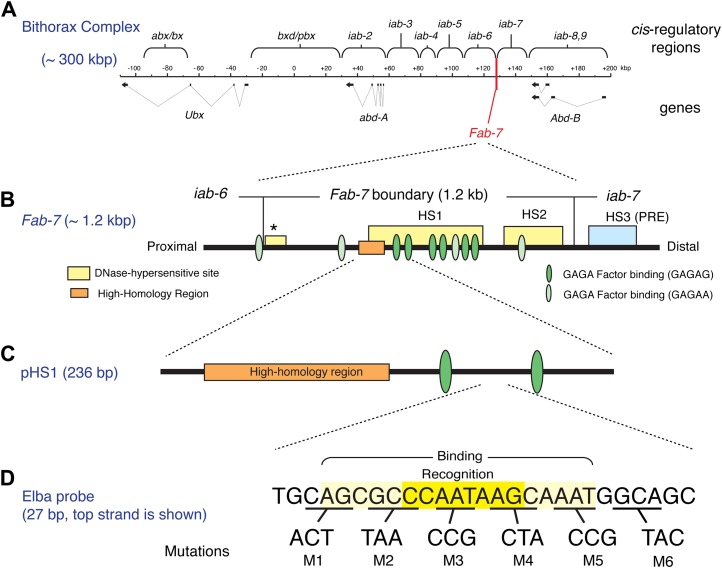
Figure 1. The Bithorax Complex, the Fab-7 boundary and the Elba recognition element.
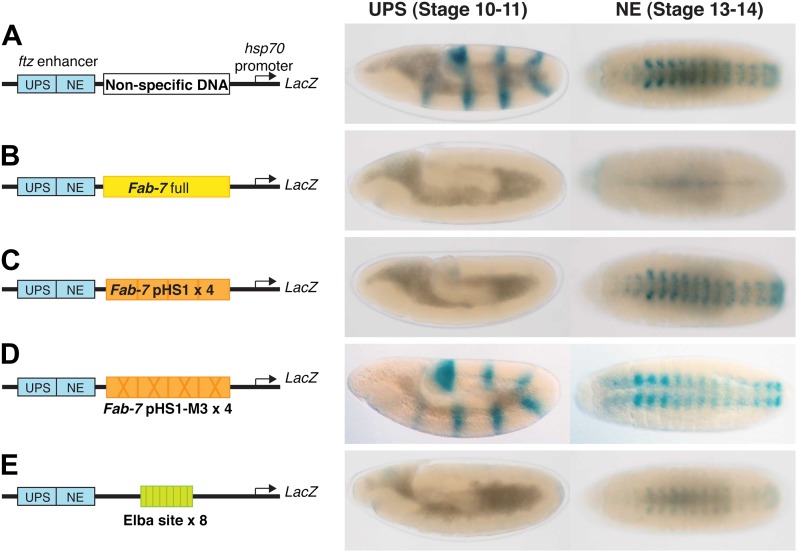
(A) Drosophila Bithorax complex (BX-C). BX-C spans ∼300 kb and includes three Hox-family genes Ultrabithorax, abdominal-A and Abdominal-B. Parasegment specific expression of these three homeotic genes is generated by a series of functionally autonomous cis-regulatory domains: abx/bx, bxd/pbx and iab-2-iab8, 9. Functionally autonomy depends upon boundary elements that lie between each cis-regulatory domain (Maeda and Karch, 2010). One of these boundary elements is Fab-7, which is located in between the iab-6 and iab-7 cis-regulatory domains. Both in the context of BX-C and in transgene assays, the Fab-7 boundary can block the action of enhancers/silencers at all stages of development, apparently irrespective of tissue or cell type (Galloni et al., 1993; Hagstrom et al., 1996; Mihaly et al., 1997; Schweinsberg et al., 2004). (B) The Fab-7 boundary spans a sequence of 1.2 kb and consists of two prominent and one minor (*) chromatin specific nuclease hypersensitive regions (shown as yellow boxes). There is a third prominent nuclease hypersensitive region (blue) just distal to the boundary, which corresponds to a Polycomb Response Element (PRE) for the iab-7 cis-regulatory domain (Maeda and Karch, 2010). The orange box is a ∼100 base pair (bp) high-homology region which is conserved among Drosophila species (>90%) (Aoki et al., 2008). The ovals are binding sites for Trithorax-like (GAGA factor). (C) pHS1 is a 236-bp fragment from the proximal side of HS1 which has enhancer-blocking activity only in early embryos (Schweinsberg and Schedl, 2004). pHS1 includes the high-homology region and two GAGA-binding sites. These two GAGA sites are important for the early boundary activity of Fab-7, while GAGA sites elsewhere in Fab-7 are needed later in development (Schweinsberg et al., 2004). In addition to the GAGA sites, the enhancer-blocking activity of pHS1 in early embryos also depends upon an 8-bp sequence, CCAATAAG, called Elba (Early boundary activity). Mutations in this sequence compromise the blocking activity of a 4×pHS1 multimer, while multimerization of a 27-bp oligo spanning the Elba sequence (8×Elba) [see (D)] is sufficient to confer early blocking activity. The Elba sequence is recognized by the stage-specific Elba DNA-binding factor. Elba factor binding is detected in 0–6 hr nuclear extracts, but it is absent in 6–12 hr (and 6–18 hr) nuclear extracts (Aoki et al., 2008). (D) Sequence of the 27-bp oligo used as the Elba probe in the EMSA experiments shown in Figures 3A, 4, 5B, and 6. The Elba factor in 0–6 hr nuclear extracts recognizes the 8-bp Elba sequence (shaded by yellow) and requires an additional 5 bp both upstream and downstream for full binding activity (shaded by light yellow). The bases underlined were altered as indicated in the mutant oligos, M1–M6. These mutant oligos were used as cold competitors in Figures 4C and 6C as indicated. For the DNA affinity beads, a 27-bp oligo containing the mutation M3 was used as the mutant Elba sequence.