Abstract
Summary: Enterococci are common, commensal members of gut communities in mammals and birds, yet they are also opportunistic pathogens that cause millions of human and animal infections annually. Because they are shed in human and animal feces, are readily culturable, and predict human health risks from exposure to polluted recreational waters, they are used as surrogates for waterborne pathogens and as fecal indicator bacteria (FIB) in research and in water quality testing throughout the world. Evidence from several decades of research demonstrates, however, that enterococci may be present in high densities in the absence of obvious fecal sources and that environmental reservoirs of these FIB are important sources and sinks, with the potential to impact water quality. This review focuses on the distribution and microbial ecology of enterococci in environmental (secondary) habitats, including the effect of environmental stressors; an outline of their known and apparent sources, sinks, and fluxes; and an overview of the use of enterococci as FIB. Finally, the significance of emerging methodologies, such as microbial source tracking (MST) and empirical predictive models, as tools in water quality monitoring is addressed. The mounting evidence for widespread extraenteric sources and reservoirs of enterococci demonstrates the versatility of the genus Enterococcus and argues for the necessity of a better understanding of their ecology in natural environments, as well as their roles as opportunistic pathogens and indicators of human pathogens.
INTRODUCTION
Enterococci are important members of gut communities in many animals (e.g., see references 82, 86, 87, 101, 105, 144, 195, 202, and 240) and opportunistic pathogens that cause millions of infections annually (231, 233). Their abundance in human and animal feces, the ease with which they are cultured, and their correlation with human health outcomes in fresh and marine waters have led to their widespread use as tools for assessing recreational water quality worldwide (333, 335, 345–347). The enterococci are most frequently used as fecal indicator bacteria (FIB), or general indicators of fecal contamination, but they are also used as surrogates for pathogens and/or health effects in risk assessment and other modeling applications (61, 214, 285, 303, 329, 346). Research spanning more than 3 decades, however, has shown that these bacteria are widely distributed in a variety of environmental habitats, even when there is little or no input from human and/or animal fecal sources. These extraenteric habitats include soil and sediments, beach sand, aquatic and terrestrial vegetation, and ambient waters (rivers, streams, and creeks); they may also be considered heterothermic habitats, in which temperatures are variable, in contrast to the gastrointestinal tract of warm-blooded animals, where the temperature is relatively constant.
The overall goal of this review of enterococci is to present the reader with an understanding of (i) the taxonomy and phylogeny, (ii) the microbial ecology (occurrence, persistence, and survival in nonenteric habitats), and (iii) the use of these bacteria in protecting human health from waterborne illnesses. In this review, unless otherwise stated, we define “environmental enterococci” as those bacteria found in a variety of extraenteric habitats, such as ambient waters, aquatic and terrestrial vegetation, beach sand, soil, and sediments.
ENTEROCOCCI AND THE GENUS ENTEROCOCCUS
Previously classified in the genus Streptococcus, the enterococci were proposed to be a division comprised of bacteria that generally grow at temperatures of between 10°C and 45°C in 6.5% NaCl at pH 9.6 and to survive at 60°C for 30 min (66, 68, 218, 293). This classification scheme, proposed previously by Sherman (293), correlated with a serological scheme developed by Lancefield in the 1930s, wherein the enterococci reacted with group D antisera whereas nonenterococcal streptococci reacted with antiserum group A, B, C, E, F, or G (198). In 1984, Enterococcus was proposed as a unique genus, separate from Streptococcus, when DNA-DNA and DNA-rRNA hybridization revealed that species such as Streptococcus faecalis and S. faecium (now Enterococcus faecalis and E. faecium, respectively) were relatively distantly related to nonenterococcal streptococci such as Streptococcus bovis (67, 283). Presently, there are 36 known Enterococcus species, classified into five groups (Table 1).
Table 1.
Species of the genus Enterococcus and their currently known habitats
| Group | Species | Known habitat(s) | Human pathogen | Reference(s) |
|---|---|---|---|---|
| E. faecalis | E. faecalis | Human, animal (multiple), plant, insect | Yes | 70, 203, 235, 283 |
| E. haemoperoxidus | Surface water | 320 | ||
| E. moraviensis | Surface water | 320 | ||
| E. silesiacus | Drinking water | 323 | ||
| E. termitis | Animal (termite) | 323 | ||
| E. caccae | Human | 54 | ||
| E. faecium | E. faecium | Human, animal (multiple), plant, insect | Yes | 70, 192, 203, 235, 283 |
| E. durans | Human, animal (multiple), insect | Yes | 67, 70, 203 | |
| E. hirae | Animal (multiple), plant | 101, 203, 235 | ||
| E. mundtii | Soil, plant | Yes | 66 | |
| E. villorum | Animal (hog) | 339 | ||
| E. canis | Animal (dog) | 82 | ||
| E. ratti | Animal (rat) | 324 | ||
| E. asini | Animal (donkey) | 86 | ||
| E. phoeniculicola | Animal (bird) | 202 | ||
| E. canintestini | Animal (dog) | 243 | ||
| E. thailandicus | Human, animal (cattle) | 55, 295, 316 | ||
| E. avium | E. avium | Human, animal (multiple) | Yes | 67, 121, 203, 257 |
| E. pseudoavium | Human | 65 | ||
| E. malodoratus | Animal (cattle) | 67 | ||
| E. raffinosus | Human | Yes | 65, 241 | |
| E. gilvus | Human | 330 | ||
| E. pallens | Human | 330 | ||
| E. hermanniensis | Animal (dog) | 195 | ||
| E. devriesei | Animal (cattle) | 322 | ||
| E. viikkiensis | Animal (broiler plant) | 269 | ||
| E. gallinarum | E. gallinarum | Human, animal (multiple), insect | Yes | 67, 70, 203 |
| E. casseliflavus | Plant, soil, human, animal (multiple) | Yes | 67, 203, 239, 241 | |
| E. cecorum | E. cecorum | Animal (chickens) | 88, 360 | |
| E. columbae | Animal (pigeon) | 87 | ||
| Ungrouped | E. saccharolyticus | Animal (cattle), sewage | 203, 270 | |
| E. aquimarinus | Seawater | 321 | ||
| E. sulfureus | Plant | 218 | ||
| E. dispar | Human | 68 | ||
| E. italicus | Animal (cattle) | 107 | ||
| E. camelliae | Plant | 316 |
We have adopted the following conventions of nomenclature and terminology. When referring to a confirmed member of the genus Enterococcus, the technical genus and/or species (e.g., E. faecalis) or the inclusive equivalent (Enterococcus sp. or spp.) is used. When referring to organisms that are identified only by isolation and the correct phenotype on selective-differential medium, the generic term “enterococci” is employed. Finally, in older publications where designations such as “fecal streptococci” or previous species names (e.g., S. faecalis or S. faecium) were used, we employ that terminology with the understanding that the terms, especially fecal streptococci, are largely synonymous with enterococci. The term “intestinal enterococci,” used by the European Union to describe the FIB group used for water quality assessments, is largely interchangeable with enterococci (363) but has been defined by biochemical characteristics set by the International Organization for Standardization (168).
It is noteworthy that the early classification system proposed by Sherman (293) is occasionally still used today to differentiate enterococci from nonfecal streptococci as well as to identify enterococci based upon reactions to group D and, in some cases, group Q antisera (241, 257). Enterococci are spherical or ovoid cells arranged in pairs or chains (144, 241). The enterococci are Gram positive, non-spore-forming, obligately fermentative chemoorganotrophs. They are catalase negative, although some species produce pseudocatalase, and they are usually homofermentative, producing lactic acid (144, 192, 241). Motility differs among species; e.g., E. gallinarum and E. casseliflavus are motile, and E. asini and E. phoeniculicola are not (66, 86, 202). Pigmentation also differs among species; i.e., yellow-pigmented species include E. sulfureus, E. casseliflavus, and E. mundtii (66, 218), and pigmented species are commonly found among plants (1). Enterococci are also found in the gut of insects (e.g., Drosophila) (70). The known habitats of various Enterococcus spp. are catalogued in Table 1, but it is important to note that as more environmental habitats are explored, and as methods for the identification of organisms to the species level become less labor-intensive and more standardized, the list of known habitats for members of the genus Enterococcus will doubtless increase.
In general, enterococci are commensal bacteria, potentially helping in digestion and other gut metabolic pathways. Some Enterococcus spp., such as E. faecium and E. faecalis, are used in probiotics to treat diarrhea and improve host immunity (108). While most species of Enterococcus are commensal organisms, some species are opportunistic human pathogens. E. faecalis and E. faecium have become particularly important etiological agents of nosocomial infections (231, 233), including urinary tract infections, endocarditis, bacteremia, neonatal infections, central nervous system (CNS) infections, and abdominal and pelvic infections (231, 241). Of particular concern is the intrinsic antibiotic resistance among certain species, particularly resistance to aminoglycosides and cephalosporins, or acquired resistance to many others, most prominently vancomycin (233, 325). While E. faecalis is the species most commonly implicated in nosocomial infections, E. faecium has shown resistance to the widest array of antibiotics (233, 325), and E. avium, E. casseliflavus, E. durans, E. gallinarum, and E. raffinosus have been isolated from patients diagnosed with enterococcal infections (241). Because of their near-ubiquitous distribution in the feces of animals, including humans, they are commonly used as FIB, or surrogates for pathogens, in water quality analyses (see Use of Enterococci as Fecal Indicator Bacteria).
Several genotyping techniques have been employed with this group to obtain correct identification to the species level and further discrimination at the subspecies level for clinical, environmental, and food-related issues, including ribotyping (232), repetitive extragenic palindromic PCR (REP-PCR) or BOX-PCR (16, 244), pulsed-field gel electrophoresis (PFGE) (128, 326), 16S rRNA gene sequencing (62, 244), and multilocus sequence typing (MLST) (276). REP-PCR has been used to target repetitive genetic sequences and, based on the genome size and the location of the repetitive elements, to generate unique banding patterns (or “fingerprints”) to differentiate strains. While one of the major criticisms of this technique has been the lack of a consensus on interpretations of the resulting fingerprints (216), horizontal, fluorophore-enhanced REP-PCR (HFERP) improves alignments between multiple gels and reduces within-gel groupings in the resulting dendrograms (185). HFERP can identify isolates with 77% agreement with 16S rRNA genetic sequencing and superior discrimination among environmental isolates (244).
The current standard in the clinical identification of Enterococcus spp. and strain typing is PFGE (163, 242). Whereas ribotyping techniques accurately discriminate among species (140) and are less expensive than PFGE, the latter method has better discrimination among closely related strains (128). MLST has proven to be useful in epidemiological studies of E. faecalis and E. faecium (56, 91, 276), and studies have shown an accuracy equivalent to that of PFGE for the identification of organisms to the subspecies level (163, 242).
ECOLOGY
Responses to Environmental Stressors
When enterococci are released from the gastrointestinal tract of warm-blooded animals into secondary habitats such as environmental waters, aquatic vegetation, or sediment, they are subjected to a host of biotic and abiotic stressors that generally lead to a decline in the population over time.
Sunlight.
Sunlight has been a suspected stressor of bacteria since at least 1877 (95). Major mechanisms of sunlight damage to microorganisms include the direct absorption of UV light by DNA or the indirect effect of the formation of endogenous and exogenous reactive oxygen species. The ability of DNA to absorb UV light was discovered in 1929 (117), leading to studies of the mechanism of UV damage to DNA and microbial inactivation (33, 97, 189). Many mesocosm studies have noted the germicidal effect (defined as a loss of culturability) of sunlight on enterococci (77, 112, 187, 255, 286, 306–308) (Table 2); however, the reported time required to achieve a 90% reduction in the concentration (T90) (equivalent to a 1-log reduction) varies widely according to geographic and seasonal factors and the experimental design (e.g., the source of the inoculum or physicochemical properties of the water). For example, in marine and estuarine waters inoculated with sewage, the reported T90 values range from 2 h (112) to 35 h (187). Generally, mesocosm studies of saline waters conducted in warmer climates (112, 306–308), during summer months (255, 306, 307), and in waters with relatively low turbidity (112, 187) reported lower T90 values (more rapid die-off) than those reported for colder climates (187), winter months (255, 306–308), and turbid waters (187). A similar trend was observed in freshwater mesocosm studies, where the reduction in levels of enterococci was enhanced at higher temperatures (i.e., during summer months and in warmer climates) (180, 255, 308). Mesocosm studies comparing sunlight inactivation in fresh versus marine waters (112, 255, 308) have generally found inactivation to be more pronounced in the latter.
Table 2.
Environmental stressors that negatively impact survival of enterococci
| Stressor | Type of stress | Source(s) of enterococci | References |
|---|---|---|---|
| Sunlight | Ambient and simulated sunlight | Environmental strains; sewage | 30, 59, 60, 71, 73, 77–81, 94, 103, 112, 113, 180, 187, 215, 217, 255, 280, 286, 306–308, 344 |
| Salinity | Estuarine and marine waters | Environmental strains; sewage | 10, 53, 75, 94, 187, 229, 308 |
| Disinfection | Chlorine/UV/peracetic acid | Sewage; pure cultures of E. faecalis | 25, 52, 57, 58, 152, 156, 182, 194, 222, 272, 273, 328 |
| Starvation | Oligotrophic conditions, glucose deficiency | Pure cultures of E. faecium, E. durans, E. flavescens, E. avium, E. pseudoavium, E. malodoratus, E. raffinosus, E. mundtii, E. faecalis, E. hirae, E. gallinarum, and E. casseliflavus | 160, 210–212, 302 |
| Predation | Bacterivorous protozoa | Environmental strains; pure cultures of E. faecalis | 75, 126, 147, 166, 169, 170, 229, 317 |
While sunlight inactivation of microorganisms is a natural, low-cost process for the treatment of contaminated water, its efficacy depends on numerous environmental factors, including the chemical composition of the water (e.g., dissolved oxygen and turbidity) and site characteristics (e.g., depth). The depth (77–79, 112, 180, 215) and turbidity (60, 78, 113, 187) of irradiated water are inversely proportional to the effectiveness of sunlight disinfection. Both factors are positively correlated with the absorbance, which is the difference between the amount of light energy (measured at a specific wavelength) that enters a sample and the amount that passes through it (338). Once absorbed, UV light loses its germicidal properties; thus, nonspecific absorption (by substances other than the intended target) hinders the efficiency of UV light disinfection. The sunlight-mediated inactivation of FIB in waste stabilization ponds (WSPs) has been shown to increase with dissolved oxygen (DO) concentrations in a wide variety of systems, including anaerobic, secondary facultative, and maturation systems and algal ponds (64, 67, 70–72, 267). The proposed mechanism for the observed synergistic action between DO concentrations and sunlight inactivation postulates that endogenous chemicals (e.g., porphyrin derivatives, flavins, and menaquinone) can act as “sensitizers” when they absorb light; reactions between excited sensitizer molecules and oxygen lead to the formation of reactive oxygen species (singlet oxygen, superoxide, hydrogen peroxide, and hydroxyl radicals), resulting in photo-oxidative damage to the organism (73, 80, 81).
Although mesocosm studies are helpful for isolating factors that may contribute to the variability in the survival of enterococci, they cannot reflect all of the complex biotic and abiotic interactions that occur in aquatic environments. Furthermore, the use of single laboratory-grown strains (as opposed to wastewater isolates) as a mesocosm inoculum may elevate inactivation rates for enterococci (103), thus further compounding the issue. In general, based on mesocosm studies, the time required to achieve a decrease in the concentration of enterococci of 3 orders of magnitude (i.e., 99.9%) ranges from 0.9 to 52 h (76, 83, 103, 255, 308). Sassoubre et al. (280) investigated field-relevant dark inactivation and photoinactivation rates by sampling two sites in San Pedro Creek, CA, hourly over 25 h. Between 6 a.m. and 6 p.m. (time points before and after sunlight exposure), concentrations of enterococci decreased by approximately 1 order of magnitude (280). Comparable results were observed in a study in Hawaii, where 22 streams were sampled during the time before the sun rose (a.m.) and at noon (p.m.); the concentrations of enterococci were approximately 0.5 logs lower in the afternoon (344). Boehm et al. (30) conducted a 72-h experiment to investigate the diurnal variation in microorganism concentrations at Avalon Beach, CA, and noted that concentrations of enterococci during the day were approximately 0.5 logs lower than those during the night (30). A similar study conducted under dry weather conditions in a tidally influenced salt marsh found a fairly wide range of concentrations of enterococci (spanning up to 3 orders of magnitude), with the early-morning concentrations generally being 1 to 2 orders of magnitude higher than those in the late afternoon (94). Hourly sampling in South Korea resulted in similar conclusions: under dry weather conditions, sunlight inactivation of enterococci was responsible for a reduction of 1 to 2 orders of magnitude (59). A recent study showed that exceedances of current regulatory standards for enterococci at marine beaches (i.e., 104 CFU/100 ml) are more frequent during the night and late-afternoon hours (from approximately 6 p.m. to 8 a.m.) than during the morning and early afternoon, suggesting that sampling at different times of the day can significantly influence beach management decisions (100). Interestingly, a recent study described a possible mechanism for the extended survival of some enterococcal species exposed to sunlight: carotenoid pigment quenching of reactive oxygen species in certain strains appears to confer a competitive advantage against sunlight-induced inactivation (over nonpigmented isolates) (217).
Salinity.
The ability of enterococci to grow in the presence of salt (6.5% NaCl) is one of the distinguishing characteristics of the genus (see Enterococci and the Genus Enterococcus). The greater salt tolerance of enterococci than of fecal coliforms and Escherichia coli probably contributes to their better performance as indicators of human health risk in marine recreational waters than members of the coliform group (see Use of Enterococci as Fecal Indicator Bacteria).
Many field (53, 94, 344) and mesocosm (10, 75, 187, 308) studies reported an inverse relationship between salinity and the detection/survival of enterococci (Table 2). A field study conducted in tidally influenced Hawaiian streams (salinity range of 0.60‰ to 37.3‰) found a negative relationship between concentrations of enterococci and physicochemical water parameters (temperature, salinity, and dissolved oxygen content) (344). Similar findings were reported for coastal Mississippi waters (salinity range of 0.00‰ to 26.4‰) and a salt marsh in California (salinity range of 29.4‰ to 30.5‰) (53, 94).
In a mesocosm study conducted in England using raw sewage as a source of enterococci and ambient water of various salinities (6.00‰ to 40.3‰), researchers observed an inverse relationship between salinity and the time required to achieve a 90% reduction in concentrations of enterococci (i.e., less time was required at higher salinities) (187). In a New Zealand mesocosm study utilizing freshwater and marine waters inoculated with either raw sewage or an inoculum from a waste stabilization pond (WSP), enterococci persisted longer in freshwater than in marine waters; interestingly, enterococci from raw sewage were more sensitive than WSP enterococci to salinity (308). The decay of enterococci in freshwater and marine subtropical environments seems to follow the same pattern. Anderson et al. (10) investigated the persistence of FIB from dog feces, wastewater, and soil known to be contaminated with feces in mesocosms filled with either river or Gulf of Mexico waters and sediments (10). Overall, decay rates of enterococci in the marine mesocosms were at least 2-fold higher than in the freshwater mesocosms, regardless of the location (i.e., water column or sediments) (10). Similar results were recorded in a mesocosm study conducted in Australia, where researchers investigated the decay of indigenous sediment enterococci from freshwater (salinity range of 1.6‰ to 3.3‰) and marine (salinity range of 33.8‰ to 35.4‰) environments (75). Over a period of 60 days, concentrations of enterococci from marine sediments decreased by up to 2 orders of magnitude, while freshwater sediments maintained nearly intact concentrations for the duration of the experiment (75) (see also “Environmental Reservoirs and Extraenteric Habitats”).
Conflicting results were reported in a field study conducted on the River Seine and the Belgian coast of the North Sea, where the differences in concentrations of enterococci between two different water types were minimal (229). The authors of that study attributed the decrease in concentrations of enterococci to the actions of predatory protozoa and stipulated that salinity may play a more important role in culturability than in mortality rates, which were assessed by the loss of [3H]thymidine-labeled strains (229).
Disinfection.
Disinfection of wastewater is a barrier that is intended to prevent the contamination of receiving waters with FIB and pathogens. The abilities of microorganisms to survive disinfection vary both with the organism and with the disinfection method. Although fecal coliforms or total coliforms are generally used to assess the efficacy of disinfection (152), some studies suggested that the survival of enterococci against disinfection is a better predictor of the fate of viruses than are coliforms (84, 368). Because ineffective wastewater treatment can allow enterococci and pathogens to enter environmental waters, the responses of enterococci to disinfection methods are discussed here.
The most common disinfection strategy in the United States is the utilization of chlorine (or chlorine derivatives such as chloramines) followed by UV light irradiation. In a study examining the efficacy of chlorine disinfection by adding various concentrations of chlorine (11.8 mg/liter to 23.2 mg/liter in the form of sodium hypochlorite) to filter-sterilized wastewater effluent from the primary treatment stage, concentrations of enterococci decreased by more than 5 orders of magnitude after 15 min of contact time (25). Similar results were reported almost 3 decades later, when comparable concentrations of sodium hypochlorite (8.0 to 30.0 mg/liter) were added to wastewater effluent from the primary treatment stage seeded with pure cultures of selected organisms (including E. faecalis) (328). In both instances, enterococci exhibited first-order decay: a rapid decrease of culturable enterococci measuring approximately five orders of magnitude was observed after 5 to 15 minutes of contact time, depending on the chlorine concentration (328). Concurring results were reported in a more realistic scenario, where concentrations of enterococci in wastewater influent and disinfected effluent were compared for six wastewater reclamation facilities (five of which used chlorine disinfection) (152). Like fecal coliforms, enterococci were highly susceptible to disinfection, as both FIB were found in only 27% of disinfected effluent samples; however, the decrease in the concentration of fecal coliforms through wastewater treatment was higher than that of enterococci. A higher percentage of disinfected effluent samples contained other types of indicator organisms, including total coliforms, Clostridium perfringens, F-specific (F+) coliphage, and somatic coliphage (152). While chlorine appears to be an effective disinfectant against enterococci (Table 2) (and other non-spore-forming bacteria), the potential for the formation of harmful by-products has led to the exploration of other modes of disinfection, specifically UV light and ozonation.
A review detailing the effectiveness of UV disinfection against enterococci (and other organisms) in wastewater and drinking water systems was recently reported (162). The reported reductions in levels of enterococci are somewhat variable (spanning approximately 2 to 5 orders of magnitude), depending on the treatment processes prior to UV exposure (e.g., sedimentation or coagulation) and the type and intensity of the UV source (155, 162, 174, 194, 301), with higher reductions observed with more extensive downstream processes and stronger UV dosages. Comparisons of UV disinfection efficacies on a seeded laboratory strain of E. faecalis versus environmental isolates indicated that the former is more resistant (57, 156, 222). Meta-analyses of the existing literature indicated that chlorine disinfection considerably outperforms UV radiation in reducing concentrations of enterococci by as much as 2 orders of magnitude (58, 152, 182, 272, 273) (Table 2).
Starvation.
The transition from the animal gastrointestinal tract, a nutrient-rich environment, to oligotrophic environmental waters exposes enterococci to nutrient starvation, one of the abiotic factors detrimental to their survival. One of the first reports on the survival of enterococci under nutrient starvation conditions indicated that E. faecalis survived for extended periods in sterilized sewage (presumably due to the availability of organic nutrients) but declined rapidly in sterile lake water and phosphate buffer, indicating that oligotrophic conditions (exemplified by the sterile lake water and phosphate buffer) were deleterious to the survival of enterococci (302).
Previous studies have identified at least 42 proteins in E. faecalis that are induced under starvation conditions (e.g., glucose depletion or incubation under oligotrophic conditions) (122, 124, 146). Furthermore, carbohydrate starvation can enhance resistance to multiple stressors, including heat, oxidative stress, acid, ethanol, and sodium hypochlorite (122, 124, 146, 199). One protein in particular (gls24), belonging to the class A starvation proteins in E. faecalis (synthesized in both growing and resting cells but differentially expressed during starvation) (123), was overexpressed under both starvation conditions mentioned above (122, 123). An additional analysis of the protein (and the corresponding gls24 gene) revealed that the gene is under the control of a stress-inducible operon and that a mutation in gls24 has a pleiotropic effect on cellular morphology (the formation of shorter chains of cocci), stress sensitivity (reduced growth in the presence of bile salts), and the expressions of several genes involved in pyruvate metabolism (124).
Sigma factors are regulatory proteins that modify stress responses in bacteria by controlling the initiation of transcription and are well characterized for E. coli (σS) and Bacillus subtilis (σB) (158, 159, 161); however, their counterpart in the genome of Enterococcus spp. has not been fully described. Benachour et al. (24) identified two genes (sigV and rsiV) in E. faecalis that are under the control of the same operon and are predicted to encode sigma and anti-sigma factors. Further analysis indicated the differential expression of the operon in response to exposure to various stresses; notably, it was overexpressed under conditions of glucose starvation and complete starvation, suggesting that it plays an important role in the response of enterococci to nutrient depletion (24). Another E. faecium regulatory protein (σ54) was suggested to be a potential virulence factor capable of influencing the rate of autolysis (and, by extension, the nature and composition of the biofilm matrix [173]) and governing sensitivity to certain bacteriocins (51, 74).
The viable-but-nonculturable (VBNC) phenomenon describes a state in which bacteria that can normally be cultured under a defined set of conditions lose that ability while retaining viability, as assessed by measurements of membrane potential, infectivity, mRNA expression, the ability to reproduce, or cell envelope integrity (160, 188, 259, 341). A series of studies explored the starvation-induced existence of the VBNC state in Enterococcus spp. (160, 210–212), in which viability assessments included the presence of mRNA, cell envelope integrity, and the ability to reproduce (assessed by Kogure direct viable counts). The authors of those studies found marked differences in the time to the loss of culturability for various Enterococcus spp. and evidence supporting the existence of the VBNC state for E. faecalis and E. hirae. It is particularly important to improve our understanding of the existence and nature of the VBNC state in enterococci, because VBNC cells would be recognized by molecular methods, such as quantitative PCR (qPCR), but not counted by culture methods, which could well contribute to the large differences in quantities estimated by conventional and emerging methods (318, 341).
Predation.
Grazing by bacterivorous protozoa, bacteriophage infection followed by virus-mediated lysis, and predation by some bacteria are among the biotic effects that control the abundance of prokaryotic organisms in the environment. Predation by bacteria has been well described for Vibrio spp., most notably Vibrio parahaemolyticus, where infection by predatory Bdellovibrio spp. plays a role in the population dynamics of these species (230, 319). Bacteriophage infection affects a much wider range of bacteria, and viral infection was suggested to be a mechanism responsible for the elimination of up to 50% of autochthonous bacteria from aquatic habitats (109, 266, 327). Bacteriophages that infect various Enterococcus spp. (“enterophage”) from different sources (i.e., raw sewage, cow manure, and environmental waters) were recently described (31, 223, 268, 279). However, the effect of enterophage on bacterial survival was not tested directly, since the main objective of these works was to examine the utility of enterophage as a microbial source tracking marker. Nonetheless, the relatively high concentrations of enterophage that specifically infects E. casseliflavus, E. mundtii, or E. gallinarum from cow fecal slurry (104 to 105 PFU/100 ml) and E. faecalis or E. faecium (∼103 PFU/100 ml) from raw sewage (268) indicate that, at least in these instances, lysis by enterophage can be a predatory factor on populations of enterococci.
Protozoan grazing is an important top-down control of bacterial populations in aquatic environments (e.g., see references 18, 75, 125, 224, and 229), including allochthonous bacteria such as enterococci (20, 263, 289). Some estimates suggest that protozoan grazing is responsible for up to 90% of the overall mortality of both autochthonous and allochthonous microorganisms from freshwater and marine environments (8, 229). Factors that affect predation rates include temperature and characteristics of prey populations. Digestion rates of both flagellated and ciliated protozoa increased exponentially at temperatures between 12°C and 22°C (294), and a direct correlation between rates of predation and temperature was found in a variety of environments, with more vigorous grazing and an increase in protozoan concentrations at higher temperatures (7, 9, 19, 225, 294). Prey characteristics such as cell wall morphology and the physiological state may also influence the magnitude and efficiency of protozoan grazing (23, 127, 220, 300, 343). Notably, lower rates of grazing were observed for Gram-positive organisms (including E. faecalis) than for E. coli (75, 126, 169, 170, 253). Nonetheless, several experiments conducted in mesocosms and environmental chambers documented decreases in concentrations of enterococci in marine (29, 75, 146, 229) and freshwater (75, 229, 317) environments in the presence of protozoa (Table 2).
The apparent predilection of protozoa for Gram-negative organisms may be explained by the physiological state of the enterococci and the preferences of different types of protozoa for particular prey. Hartke et al. (147) showed a more active grazing of zooflagellate protozoa on E. faecalis cells harvested from the exponential growth phase than on glucose-starved cells, while nanoflagellates did not appear to exhibit a preference (147). Similarly, it was shown that while E. faecalis concentrations decreased by more than an order of magnitude over 72 h in coculture with Acanthamoeba polyphaga, amoeba levels were ∼80% lower than those of the negative controls that were grown in the absence of enterococci (166). That same study found increases in amoeba numbers when cocultured with E. coli, Bacillus cereus, and Salmonella enterica serovar Typhimurium, indicating that enterococci are not a good food source for A. polyphaga (166).
Environmental Reservoirs and Extraenteric Habitats
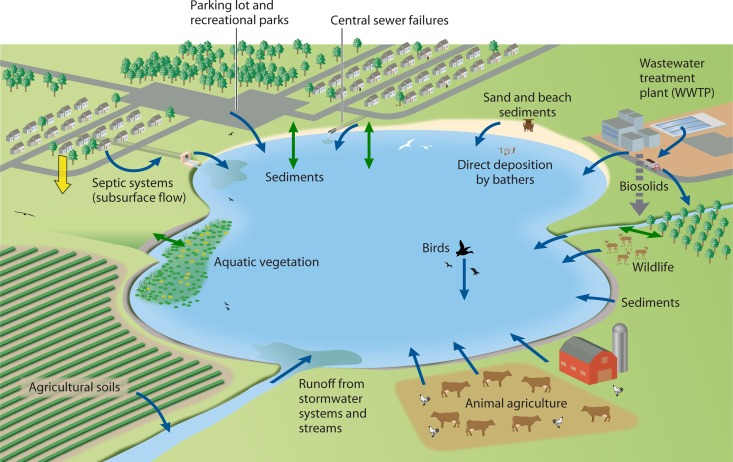
In contrast to the initial conception of Enterococcus spp. as inhabitants of the gastrointestinal tract, we are gaining an understanding of the extent to which environmental habitats can serve as sources and sinks of this group. Although the environmental stressors discussed above negatively impact the survival of enterococci, many studies have clearly demonstrated the persistent nature of some Enterococcus spp. and strains in extraenteric habitats. Figure 1 illustrates our current understanding of the major sources and sinks of enterococci in environmental habitats. The concepts presented in this graphical illustration are discussed in Use of Enterococci as Fecal Indicator Bacteria. Table 3 provides an overview of studies and findings on the occurrence, persistence, growth, and population genetics of enterococci in extraenteric habitats.
Fig 1.
Sources of enterococci in water bodies (blue arrows) as well as sinks where enterococci are immobilized (yellow arrow) and areas of flux, in which enterococci can transition from a reservoir to the water column and vice versa (green arrows). Fluxes act as secondary sources or sinks depending upon the conditions.
Table 3.
Occurrence, persistence, growth, and population genetics of enterococci in extraenteric habitats
| Habitat for enterococci | Occurrence (reference[s]) | Persistence/survival (reference[s]) | Growth (reference[s]) | Population genetics (reference[s]) |
|---|---|---|---|---|
| Soil | Recovered in tropical and temperate soils (36, 40, 85, 110, 145, 237) | Persist longer than E. coli; survive longer than other FIB (37, 305) | May grow in soil under certain conditions (40, 85) | Different Enterococcus spp. have been recovered in tropical soils (40); metabolically diverse strains of Enterococcus spp. are found in soil (40, 111) |
| Sediment | Enterococci are found in both freshwater and marine water sediments (102, 116, 207, 252, 258) | Survive longer in sediments than in water (10, 282); found mostly in surficial layers, with no seasonal trends in distribution (258); hydrometeorological events influence the bacterial flux between sediments and water (116, 134, 282); differential survival observed for freshwater and marine water sediments (75) | May grow in sediments under certain conditions (85, 102, 116) | |
| Beach sand | Found in both freshwater and marine beaches (44, 120, 142, 264, 278, 363, 366); bacterial distribution is patchy over space and time (44, 142) | Persists longer in moist beach sand and in nearshore and backshore areas (44) | In situ growth has been suggested for persistent populations of enterococci in sand (148, 205, 367) | Numerous Enterococcus spp., including E. faecalis, E. faecium, E. casseliflavus, E. mundtii, and E. hirae, have been recovered from beach sand/intertidal sediments (44, 102) |
| Vegetation | Found in both aquatic and terrestrial vegetation, including algae (358), beach wrack (11, 131, 167), submerged vegetation (14, 15), flowering plants (237), and forage crops (235, 261) | Few days to months for submerged vegetation (14, 15), dried algae (358) | High bacterial densities in vegetation have been attributed to growth (14, 42, 358) | Enterococcus spp. have been recovered from the rhizosphere and the phyllosphere (235, 261); evidence for clonal populations (14) |
| Freshwater | Found in both tropical and temperate freshwaters (110, 114, 136, 145, 181, 193) | Differential survival of enterococci compared to other FIB has been observed (10, 308) | Sporadic growth in nutrient-rich waters has been recorded in the presence of algae (42) | Strains of environmental origin found in catchments (3, 15); diverse Enterococcus spp. and strains observed (14, 244) |
| Marine water | Found in marine waters (27, 297) | Survive longer than E. coli (208) | Prolonged survival and potential growth are most likely in moist sand/sediments and associated vegetation (102, 131, 167, 178, 179, 367) | Diverse Enterococcus strains have been observed (14) |
Aquatic and terrestrial vegetation.
Cladophora, a macrophytic green alga, is found in both fresh and marine waters. Until recently, the impact of Cladophora on beach water quality was unexamined, but in a seminal work, Whitman et al. (358) showed that the algal mats collected along shorelines of southern and northern Lake Michigan in the Great Lakes were a significant source of FIB (E. coli and enterococci), with densities often exceeding 100,000 CFU/g (dry weight). These findings have been confirmed by a number of studies of Lake Michigan and elsewhere (342). Aside from FIB, enteric pathogens, such as Shiga toxin-producing E. coli (STEC), Shigella, Salmonella, and Campylobacter, have also been isolated from these mats, indicating that Cladophora may serve as an environmental source of these pathogens in recreational water (41). Enterococci survived in sun-dried algal mats stored at 4°C for over 6 months and displayed the ability to grow to high concentrations (∼108 CFU/g) upon rehydration (358).
The high densities of FIB, including enterococci, in fresh Cladophora mats have been attributed to in situ growth (42). For instance, enterococci grew over 100-fold in undiluted algal leachate at 35°C in 24 h, suggesting that Cladophora provides enough nutrients to sustain these bacteria, which are known to have fastidious growth requirements, as evident by the commercial media used for these bacteria (6). Cladophora is perennial in nature, and in temperate waters (e.g., Great Lakes), it overwinters, leaving behind scattered basal stumps; however, there have been no reports of residual enterococci or other FIB surviving in these stumps under wintery conditions.
Other aquatic macrophytes that have been identified as sources of enterococci include decaying seaweed (11, 131, 167). In New Zealand, Anderson et al. (11) observed that the densities of enterococci in drifting seaweed exceeded those in seawater by 2 to 4 orders of magnitude; Grant et al. (131) found densities of enterococci as high as a 450,000 most probable number (MPN)/100 g in a marsh in southern California. In addition to seaweed, recent studies have expanded the occurrence of enterococci to submerged aquatic vegetation (SAV) (mostly Hydrilla verticillata) (15, 16). In laboratory studies, Badgley et al. (15) found that enterococci survived longer and at much higher densities in mesocosms containing SAV than in those without SAV. Furthermore, the recovery of a dominant E. casseliflavus strain indicated that this genotype was likely adapted to or naturalized on this vegetation. Aside from macrophytic plants, enterococci have also been associated with planktonic communities and macroinvertebrates (221, 299). Data reported by Signoretto et al. (299) suggested that attachment and the shift to a VBNC state contribute to the prolonged survival of enterococci in marine waters. Furthermore, the time of survival of enterococci is longer in sediments than in water (see “Sediments” below) and in the presence of aquatic vegetation (14, 358).
Some of the earliest findings for the association of enterococci with terrestrial vegetation were demonstrated by Mundt (236), who recovered these bacteria from flowers and buds of different plant species. Recent studies have expanded these findings to forage and crop species (235, 261). Mundt (237) initially suggested that the occurrence of enterococci in plants was seasonal, with maximum recovery in late summer (September), and that these bacteria were transient populations most likely introduced by insects and wind (237). Shortly thereafter, Mundt et al. demonstrated the ability of E. faecalis to grow on plants (238), and many other studies have argued for the existence of epiphytic enterococci (204, 235, 261). Furthermore, the finding that certain strains of Enterococcus spp. may have environmental adaptations, for instance, E. casseliflavus in submerged aquatic vegetation (15) and E. casseliflavus, E. faecalis, E. faecium, E. hirae, E. mundtii, E. sulfureus, and many other strains resembling E. faecalis from forage crops (50, 235, 261), strongly supports the existence of plant-associated enterococci.
Beach sand.
The sanitary quality of beach sand, i.e., the extent of contamination by FIB, has been a subject of public health concern in recent years (reviewed in reference 142). Numerous investigations across marine and freshwater beaches have repeatedly shown that enterococci and other FIB and pathogens (e.g., E. coli, Salmonella, and Campylobacter) are common microbial contaminants in beach sand, with potential implications for shoreline water quality (120, 278, 290, 355, 363). Whether enterococci are part of the natural resident or transient microflora of beach sand remains unknown; different Enterococcus spp. have been recovered in sand from freshwater and marine beaches, e.g., E. faecium, E. casseliflavus, E. durans, and many unidentified species from Lake Michigan (44), and E. faecalis, E. faecium, E. hirae, E. casseliflavus, and E. mundtii from marine sediments (16, 102). However, the recent demonstration of biofilm-associated enterococci in beach sands (265) argues that some of these populations are resident.
In recent years, efforts have been directed toward an understanding of the sources and contributions of FIB and their potential interactions in the “beachshed” ecosystem, which comprises the various sources of FIB, and their influences, within the beach and its related watershed, as described by Whitman et al. (356) (Fig. 1). Shoreline birds, particularly geese and gulls, have received significant attention because of their abundance and potential influence on water quality (105, 136, 240). Other potential contributors may include beach visitors themselves as carriers of sand-borne bacteria during recreational activities (99, 262); an individual bather might contribute as many as 6.0 × 105 CFU of enterococci through sand particles adhered to the skin (99). Hydrological processes, such as overflows, runoff, and wave surges (27, 136, 271), are among the contributors to diffuse nonpoint sources of enterococci that influence water quality at recreational beaches. A growing body of data suggests that in situ bacterial growth may be an alternative explanation for high levels of enterococci in beach sand (148, 367). Studies conducted in sand-filled columns found that transient growth of enterococci occurred after intermittent wetting of sand (367). At the same time, the replication of enterococci under natural conditions is likely to be limited because of desiccation and other environmental stresses. Despite these limitations, the widespread occurrence of enterococci and other FIB in beach sand (44, 120, 355, 363) can be attributed to repeated seeding from birds and other sources (105, 240) as well as residual surviving populations, especially in moist, subsurface sand in both foreshore and backshore areas (44).
Previous studies have shown that the densities of enterococci in sand vary within and between locations (44, 264, 366). Densities of enterococci as high as 7,200 CFU/100 g were reported in a study conducted in coastal California (366). Lower densities of enterococci in beach sand have been reported in other studies; e.g., densities in samples of wet sand at Avalon Beach, CA, averaged a 310 MPN/100 g (ranging from nondetected to a 4,200 MPN/100 g) (143). Likewise, in a 13-month study at two Great Lakes beaches along southern Lake Michigan, Dunbar, and West Beach, the densities of enterococci (log MPN ± standard error [SE]) in moist subsurface sand near the water table averaged 1.1 (±0.15) and 1.1 (±0.08) MPN/100 g (44). Such variations in densities of enterococci probably reflect contaminant sources, the locations of sampling (e.g., foreshore and backshore areas) (44), and, importantly, methods of sample collection and analysis (28). Currently, there is no established or approved method(s) for FIB analysis in beach sand/sediment substrates, and a patchy distribution of these bacteria in sand environments is highly probable, as observed for other substrates (e.g., soil) (36, 40).
Despite concerted efforts, the management of beach sanitary quality has proven difficult. Various management strategies, such as sand replacement (355), beach grooming (190, 191), and bird harassment using trained dogs, have been attempted, with various degrees of success. The total eradication of FIB and other microbial contaminants might be difficult or simply impractical. For instance, enterococci can survive and persist in moist subsurface sand (44), countering the effects of surface treatments. Other beach management practices, such as beach grooming—a tool routinely applied for esthetic purposes—might help reduce the problem but can be counterproductive when the particulate-associated FIB get dispersed and are deposited deeper, further protecting them and prolonging their survival in sand environments (191). In these situations, the sand can serve as a continuous source or reservoir of FIB and associated pathogens to nearshore waters.
Sediments.
Numerous studies have shown that both freshwater and marine sediments are significant sources or reservoirs of enterococci (116, 119, 258, 296), with (sediment) bacterial densities being typically several orders of magnitude higher than those in the overlying water on a per-mass basis (10, 72, 102, 129, 252). High bacterial densities in sediments have been attributed to better resistance to environmental stressors, in particular predation, solar inactivation, starvation, possible regrowth, vegetation, and related factors (10, 141, 167, 179) (see also “Responses to Environmental Stressors”). The prolonged survival of enterococci has similarly been observed in freshwater (10, 15, 141) and estuarine (10, 179) sediments.
Whether enterococci can grow under most environmental conditions remains speculative; however, Mundt et al. (238) demonstrated the growth of E. faecalis on germinating seeds and plants. High bacterial densities in sediments (85, 102, 205), in aquatic vegetation (15, 42, 358), and in detritus and planktonic communities (234) suggest that enterococci grow in these nonenteric habitats under certain conditions. Growth of enterococci has been observed in several mesocosm studies: in beach sand (367), algal washings (42), rehydrated algal (Cladophora) mats (358), and aquatic vegetation (15). These findings collectively support their growth capabilities in the environment; however, more studies are needed to better understand this ecological process.
Because of their close interaction with surface water, sediments play a major role in influencing shoreline water quality through the resuspension of the particle-bound bacteria in the water column. While the quantification of bacterial loads from sediments by conventional methods might be difficult, alternative techniques such as hydrodynamic or empirical modeling have increasingly been used in recent years to better understand this process (118, 130). In large bodies of water, such as the upper Chesapeake Bay, more than 80% of indicator organisms, including fecal streptococci and fecal coliforms, were found to be associated with suspended sediments (282). Such processes are often mediated by mechanical disturbances during recreational activities (7), hydrometeorological events (including high-flow, wind, and erosional conditions [116, 175, 274, 312] and river outfalls [98, 249]), as well as dredging operations (134, 135). Collectively, such events can increase FIB densities in the water even in the absence of any significant human inputs.
Soil.
Some of the earliest research on the survival of enterococci in soils was conducted with experimental plots by van Donsel et al. (340), who observed that the rates of survival of Streptococcus faecalis were higher than those of fecal coliforms during spring and winter, and while there was no difference in the survival patterns in the autumn, fecal coliforms survived longer than S. faecalis during summer months. Interestingly, many of the early investigations of the survival and persistence of enterococci/fecal streptococci in soil environments focused on watersheds impacted by anthropogenic activities, particularly cattle grazing and field lot operations (92, 165, 176). Recent studies confirmed that populations of FIB (E. coli and enterococci) are equally abundant in relatively less-impacted soils (46, 85, 110, 200). For instance, a survey of soils on the island of Oahu, HI, showed that enterococci were nearly ubiquitous compared to E. coli (98% and 54% frequencies in surveyed soils, respectively), with enterococcal counts often exceeding a 1,000 MPN/g soil (40).
High densities of enterococci in soils may be attributed, in part, to the greater survival abilities of Gram-positive bacteria (e.g., enterococci and staphylococci) than of Gram-negative bacteria (e.g., E. coli, Pseudomonas spp., and Rhizobium spp.) in the face of environmental stresses, particularly cellular injury and desiccation (17, 245). In one mesocosm study, densities of seeded E. faecalis remained nearly constant (∼6.0 log CFU/g dry soil) for 8 days when the moist soil (35% moisture, corresponding to a 60% water-holding capacity) was allowed to desiccate (12% moisture) under laboratory conditions (25°C). E. coli densities, on the other hand, declined drastically from 6.0 log CFU/g to <1 CFU/g in 4 days but returned to the original levels upon rehydration (37). Similarly, enterococci survive longer than other enteric bacteria under certain field conditions: in cow feces, a 90% inactivation of enterococci occurred after 56 days, followed by E. coli (48 days), Salmonella enterica (38 days), nonenterococcal fecal streptococci (35 days), and Campylobacter jejuni (6.2 days) (305).
It has been argued that soil environments provide the necessary niche for populations of FIB to survive, adapt, and grow in these heterothermic habitats (115, 171, 361). While the growth requirements of E. coli are relatively simple because of its ability to synthesize cellular macromolecules from glucose and minerals (12), enterococci require complex nutrients (e.g., growth factors), even when grown under laboratory conditions on commercial media such as m-Enterococcus agar and mEI (6). Although enterococci are relatively common in some tropical soils (40, 110, 145), studies of growth characteristics in these environments are rather limited. In one study, enterococci grew only marginally in the presence of full competition from the native microbiota; however, in the presence of nutrients (peptone) and reduced competition (achieved by the addition of sodium azide), enterococci grew more than 100,000-fold over 13 days (38).
The paucity of available nutrients may thus limit the growth of FIB in soil environments, yet a likely habitat that provides conditions for spurts of growth is the plant rhizosphere region, where microbial activity is known to be severalfold higher than in the adjacent bulk soil (310). The various compounds released by plant roots as exudates into the surrounding soil are highly diverse and complex, including amino acids, growth-promoting and growth-inhibiting substances, low-molecular-weight sugars, organic acids, polysaccharides, and proteins (139, 310, 349). Additional studies are needed to better understand pathogen and FIB ecology in the rhizosphere.
Populations of enterococci represent only a small part of the soil microflora. For instance, in six soil samples collected on the campus of the University of Hawaii, culturable heterotrophic bacteria were about 10,000- to 10,000,000-fold more numerous than enterococci (37). Furthermore, the widespread range of these bacteria in soils throughout the island of Oahu (40), comprised of at least six different species of Enterococcus (40, 111), strongly supports the hypothesis of environmentally adapted or autochthonous populations in soil environments.
While the original source of populations of enterococci in soil is debatable in some cases, potential sources include human and animal (including wildlife) waste (Table 1 and Fig. 1), and over time, a subset of the original population may have adapted to the soil environment. In summary, aquatic and terrestrial vegetation, beach sand, freshwater and marine water sediments, and soil have been identified as some of the major environmental sources of enterococci and other FIB. FIB derived from these sources can potentially impact the water quality of associated beaches and watersheds, and thus, there is a need for a better understanding of their fate in these ecosystems.
USE OF ENTEROCOCCI AS FECAL INDICATOR BACTERIA
For over a century, FIB have been used to assess water quality and protect humans from the myriad of enteric pathogens that are transmitted by the waterborne route by acting as fecal indicators (reviewed in references 277 and 362). FIB are generally commensal inhabitants of the gastrointestinal tracts of many warm-blooded animals and are shed in feces at high densities; thus, they are easily detected in contaminated waters. Ostrolenk et al. (260) were among the first to suggest that the enterococci might be more appropriate FIB than E. coli (260), and studies conducted in the 1970s confirmed this suggestion for marine waters (49, 96). More recently, multiple studies have shown a correlation between elevated concentrations of enterococci and the risks of humans contracting gastroenteritis during recreational water use, particularly when point source contamination is present (186, 267, 333, 335).
The use of enterococci as FIB has been criticized almost since their adoption as a regulatory tool (64), because the epidemiology studies on which the standards were based were focused solely on waters contaminated by point source (particularly human sewage) pollution (47, 333). Little was known about the relationship of enterococci and other FIB to human health in recreational waters contaminated by nonpoint sources when the regulations were promulgated (115). Recently, some studies found an association between densities of enterococci and illness rates at beaches impacted by nonpoint sources of contamination (104, 304). While there was an increased incidence of gastrointestinal illness, respiratory illness, and skin illness in bathers in one study (104), the only health effect with a dose-response relationship to concentrations of indicator bacteria in both studies was skin illnesses (104, 304). Furthermore, in a comparison of analytical methods, the dose-response relationship for skin illness was seen only with samples analyzed by membrane filtration (304). Enterococci are currently the only FIB recommended by the U.S. Environmental Protection Agency (EPA) for brackish and marine waters, since they correlate better with human health outcomes than other FIB, such as fecal coliforms or Escherichia coli (346–348). Several epidemiological studies have also shown a correlation between concentrations of enterococci in beach sands and gastrointestinal illness in bathers (31, 157).
Characteristics associated with “ideal” FIB include a lack of virulence; the existence of a simple, rapid methodology for enumeration; survival characteristics that are similar to those of pathogens in extraenteric environments; and a strong association with the presence of pathogens (49). In contrast to this ideal, studies have shown that populations of enterococci may be endogenous in sediments and soils and not exclusively of fecal origin, which may confound accurate water quality assessments (37, 85). Furthermore, many domestic and wild animals can contribute enterococci to water bodies (Table 1), which complicates the FIB-pathogen relationship since the suite of pathogens associated with various animal gastrointestinal tracts and the risk associated with fecal contamination are highly variable (309, 364). Figure 1 depicts some of the many possible sources (blue arrows) of enterococci in environmental waters, which include human sources, such as sewage and its many derived products, e.g., biosolids, and fecal shedding from recreational water users. Other important sources are agricultural contributions, which may come directly from animals, e.g., cattle or swine defecating in and near water bodies, or indirectly from activities such as the spreading of manure or poultry litter on fields (334). Wildlife (e.g., birds, deer, feral hogs, and raccoons) (Table 1) can be sources of enterococci in urban and rural environments, either via direct deposition (represented by the gull depicted mid-lake in Fig. 1) or in runoff. The particulate matter in storm water contributes to the transport of enterococci in receiving waters and eventual deposition into sediments (90). Enterococci may also attach to aquatic vegetation and detritus (14–16, 234). When sediment is disturbed by high flow, waves, or the activity of humans or animals, enterococci can recontaminate the water column in what can be considered a flux (Fig. 1, green arrows). Here, we define a flux as a transport pathway for enterococci that begins with the primary source (e.g., feces), followed by deposition to a sink, in which enterococci are temporarily sequestered (e.g., sediments). In the case of a flux, the sink is temporary and eventually becomes a secondary source when organisms reenter the water column following a disturbance (183, 207). Likewise, fluxes of enterococci from aquatic vegetation to the water column or runoff from a field to a stream can constitute a secondary source. Permanent sinks, in which enterococci are deposited into an area from which they have very little probability of being transported to water, are less common (Fig. 1, yellow arrow); appropriate examples would be properly functioning on-site wastewater disposal systems such as septic systems and pit toilets.
Several studies have reported difficulty in finding media that can effectively enumerate the broad range of Enterococcus spp. without sacrificing specificity to the genus (204, 288), and the identification of isolates of enterococci from environmental matrices (e.g., plants, soil, sediments, sand, and water) remains challenging (14, 43, 89, 151, 235). Upon the initial introduction into an extraenteric environment, enterococci may become rapidly inactivated (see also “Responses to Environmental Stressors”), which could potentially result in false-negative results when enterococci are used as pathogen surrogates (226, 281). Conversely, the underlying sediments and aquatic vegetation can act as reservoirs for enterococci (see also “Environmental Reservoirs and Extraenteric Habitats”) (14–16, 85, 205), which may lead to overestimates of health risks when pathogens are not similarly persistent.
Several methods for the detection and enumeration of enterococci have been successfully used and are prescribed by regulatory agencies to predict health risks. The epidemiological studies conducted in the 1970s that were used to set recreational water quality criteria (48, 49) concentrated enterococci by membrane filtration and cultured them on mEI medium; consequently, membrane filtration methods are the current “gold standard” for water quality assessments (336). In addition to standard methods using membrane filtration, numerous monitoring laboratories have also relied on alternative culturing techniques. A comparison of membrane filtration with multiple-tube fermentation and chromogenic substrate methods, i.e., Enterolert (34), showed that results did not vary significantly by method (132) and were being used interchangeably to manage beaches across a large portion of Southern California. With the increasing frequency and number of beaches being monitored for enterococci since the passage of the BEACH Act (22), many locations are using either chromogenic substrate or membrane filtration analytical techniques, with results being used interchangeably across jurisdictions for beach management (246).
The benefits of both culture-based methodologies discussed above are that the techniques are easily learned and the methods are not costly (254). Furthermore, concentrations of enterococci obtained by using culture-dependent methods have shown significant correlations with human health risks in estuarine and marine waters (47, 333). Despite the demonstrated advantages, the drawback of these culture-based methods is that they have a lengthy time lag (18 to 24 h) before results are obtained (201). This lag results in the postponement of decisions on risk management for recreational water use, potentially exposing humans to health threats between the sample collection time and the reporting of results, as FIB concentrations can vary widely across small spatial and temporal scales (32, 201, 250, 254, 354). In other words, by the time the testing results are reported, the contamination that caused the elevated FIB concentrations may have dissipated, leaving the water body safe for use by the time a warning is eventually posted.
EMERGING TECHNOLOGIES FOR DETERMINING CONTAMINATION SOURCES, ASSESSING WATER QUALITY, AND DETERMINING HUMAN HEALTH RISKS
Microbial Source Tracking
While the presence of enterococci in the feces of a wide range of animals is a useful characteristic for a general indicator of fecal contamination, no information on the contamination source is provided by the quantification of the group as a whole. Knowledge of fecal contamination sources is useful or required in many scenarios, e.g., for total maximum daily load (TMDL) assessment, risk assessment for water use, and remediation of polluted water bodies (137, 153, 337). Microbial source tracking (MST) methods, which target host-specific microorganisms as identifiers of fecal or sewage sources in water bodies, have repeatedly addressed the lack of specificity of conventional FIB (recently reviewed in references 137, 153, and 275).
The enterococci have been the focus of the development of several MST methodologies (reviewed in reference 315). Library-dependent methods require a large database of FIB from the feces of host species; FIB are isolated from feces and genotyped or phenotyped (149) to identify specific characteristics or traits for discrimination among strains. Once the accuracy of the library categorization of isolates by host source is ascertained, isolates from water or other matrices are then compared to library isolates for assignment to source categories. Although field studies that used enterococci as source identifiers for library-dependent MST methods initially indicated promise for use in a regulatory context (138, 154, 359), the expense, difficulty in the interpretation of results, and uncertain accuracy of such methods (315) have discouraged their general use. The potential for the extended persistence and possible growth of enterococci in extraenteric habitats (Fig. 1) further complicates the interpretation of results from library-dependent MST methods. Instead, the focus of MST has turned to library-independent methods, which generally rely on PCR to identify gene fragments (markers) specific for microorganisms that are host associated (153).
The esp gene of E. faecium (espfm) is strongly human associated (5, 287, 357), although a low frequency of cross-reactivity with nonhuman feces has been noted, and it is not readily detected in some sewage sources, such as on-site (septic) systems (4, 357). The occurrence of espfm was correlated with human polyomaviruses in polluted surface waters in Florida (227) and with fecal coliforms in another study (197). It has also been used in field studies in Florida, the Great Lakes, and Australia (5, 39, 196, 209). A quantitative PCR (qPCR) method for espfm has been developed and used in field studies in Australia (4). Interestingly, the presence of the esp gene was found to affect the transport of E. faecium in saturated quartz sands by lowering bacterial mobility through increased attachment to sand particles (184).
A novel approach for identifying MST markers of enterococci associated with various hosts was proposed by Soule et al. (311), who used DNA microarrays to identify candidate host-specific Enterococcus species and associated genes. The use of bacteriophages specific to certain strains of Enterococcus spp. was also recently explored (32, 268) (see also “Responses to Environmental Stressors”). Many other MST methods rely on microbial groups other than enterococci (recently reviewed in references 275 and 365), including anaerobes such as the Bacteroidales (e.g., see references 26, 291, and 292) and Methanobrevibacter smithii (331) and viruses (106, 227, 228). The correlation of MST marker detection or concentrations with concentrations of enterococci has varied across studies: Harwood et al. (150) found no correlation between concentrations of enterococci and levels of human sewage markers in untreated sewage; however, enterococci and the human Lachno2 marker were strongly correlated in a freshwater harbor that received combined sewer overflows (251). As is the case for currently recognized FIB such as enterococci, the usefulness of MST for water quality assessment is ultimately predicated on the correlation of human health risk and pathogen presence with host-specific markers; however, there are many data gaps remaining in this growing area of research.
Quantitative Microbial Risk Assessment
The term quantitative microbial risk assessment (QMRA) refers to a risk analysis framework and process for defining the type(s) of microbial hazard that is likely to be encountered in a given situation and the magnitude of the probable harm (risk), usually to some human population (177). Over the past decade, QMRA has increasingly been applied to hazard estimations for recreational water quality, and enterococci are frequently employed in these models (13, 329). Schoen et al. (285) found that measurements of levels of enterococci by culture methods are likely to underestimate the risk of gastroenteritis caused by enteric viruses in recreational waters where contamination is from mixed sources; in contrast, qPCR estimates of densities of enterococci were more reliable predictors of norovirus and human health risk. QMRA has been used to estimate the relative risk from contamination by human sewage versus animal sources in models that use the U.S. EPA's recreational water quality criterion for enterococci (35 CFU/100 ml) as one reference point (284, 309). Among the sources examined, gull fecal contamination carried the least human health risk, and cattle contamination carried the greatest (309). Another study estimated that rain events and storm water runoff increase health risks to surfers (329). QMRA has been recommended as an important component of a “holistic” approach to recreational water quality assessment (13), which includes extensive knowledge of the watershed(s), including potential pathogen sources and transport pathways. An important caveat in all risk assessment models that use FIB as surrogates is that the ratio of FIB to pathogens is highly variable in contaminating fecal material and in water samples (13); therefore, users must be cognizant of the limitations of such models.
Rapid Testing Methods
While epidemiological studies conducted at sewage-impacted beaches continue to support the association between concentrations of enterococci and rates of swimming-related illnesses (345, 346), the time lapse between sample collection and the availability of results severely compromises their usefulness in making appropriate decisions regarding the opening or closing of beaches. Efforts to overcome these shortcomings have included the use of rapid enumeration methods such as qPCR (155), alternative indicators that are more specific to the contaminant sources (e.g., human-associated Bacteroides, Catellicoccus gull fecal markers, and Brevibacterium poultry fecal markers) (26, 213, 351, 352), direct monitoring for potential pathogens and QMRA (see above) (13), and predictive modeling (164, 249), but cost, ease of use, and sustainability as a monitoring program must all be considered in optimizing the application of any newer method or monitoring technology. More information on the sensitivities and specificities of individual tests are provided in other sections.
Alternate methods for the enumeration of enterococci in surface waters that do not rely on bacterial growth and therefore are more rapid and have the potential to become “real-time” tools for water quality assessment have been developed in recent years. Among these methods, qPCR (e.g., see reference 155) has been the most widely tested method and is currently under consideration for application in beach programs. While membrane filtration relies on the detection of living and culturable enterococci, qPCR quantifies DNA from both living and dead cells, a difference with potential implications for regulatory and management decisions. Some studies identified a correlation between the two endpoints in side-by-side comparisons with culture-dependent methods (45, 201, 298). Direct comparisons of the results of the two tests have been discouraged due to differences in variation along a concentration gradient and fluctuations in outcomes due to the original source of the enterococci (201, 353). Inhibition during qPCR analysis has been a significant issue, and efforts to refine the technique have led to numerous modifications of the original protocol, including purification kits, additional filtration steps, and smaller sample volumes (256). The lack of universal standards, calibrators, and methods complicates the use of this test as a monitoring standard. Recent method validation studies have begun to address these concerns (93, 201, 350).
Another emerging technology that has been widely tested is immunomagnetic separation-ATP (IMS-ATP) (206). In this analysis, target enterococci or other FIB are concentrated and separated through the use of specific, antibody-coated immunomagnetic beads; the cells are then quantified by measuring the bioluminescence response from the bacterium's ATP (206). Unlike qPCR, IMS-ATP targets only metabolically active cells; however, the use of ATP as the target can result in the detection of organisms that may not be culturable by using standard culture methods (35). Because IMS-ATP depends on active cellular metabolism, it may underestimate target concentrations compared to methods that measure total cells, such as qPCR. In a comparison study of numerous test methods, IMS-ATP analysis and culturable enterococci showed a strong correlation between the two results, with the exception of one location (35). Further comparisons indicated that IMS-ATP suffered from a large number of false-positive results (133). The cost of the equipment and analytical reagents and the need for technical expertise/personnel may limit its application for routine monitoring.
Further analytical approaches have sought to target multiple potential indicators and pathogens simultaneously by using molecular techniques. For example, the Luminex (Luminex Corporation, Austin, TX) detection system has been developed to test multiple targets through the detection of DNA, RNA, or proteins; this technique has been used to analyze FIB (e.g., E. coli and Enterococcus spp.) and pathogenic bacteria such as Shigella spp. in a multiplex format (21). In brief, the extracted DNA is marked with probes, and a detection system determines the overall concentration of the target microbes. Experiments using natural waters found that the methodology worked best on river water samples. The targets were not as concentrated in beach water and sand; therefore, group-specific primers were developed to optimize the technique for these natural waters (21). Initial results indicated that the system could detect Enterococcus spp., but quantities were often quite different from those obtained by culture methods (21). Further method validation indicated that the Luminex system had the highest specificity and sensitivity for Enterococcus over those of IMS-ATP or any of the currently used DNA-based methods, such as PCR and qPCR, with no false-positive results for the negative controls (133). That study cautioned that the system has yet to be fully developed for use with natural water samples; as with many molecular methods, natural water samples introduce inhibition and other obstacles to accurate detection (313).
Aside from comparisons among analytical methods, the issue of primary importance is the usefulness of a given method for predicting health risk. Thus, any new application with a weak or no clear relationship between the measured parameter and human health would be less useful for the management of recreational water quality. Overall, qPCR results for enterococci have generally correlated well with illness rates at sites impacted by point sources (345, 346, 348). However, if the contaminants are from nonpoint sources (e.g., storm water), evidence for effects on health is thus far conflicting; i.e., several studies did not find a correlation between qPCR for enterococci and health effects (2, 64, 104, 304), but one did (63).
Predictive Modeling of Levels of FIB
In addition to advancing molecular technologies to develop rapid tests for enterococci, efforts have been made to improve reporting accuracy by predicting concentrations of enterococci in situ by using statistical models. Predictive models have also been encouraged by the U.S. EPA (332), and as such, they have been used in numerous locations in the United States. Unlike many of the rapid analytical tests developed, predictive models are not hindered by interference from other materials suspended in natural beach water. Typically, beach monitoring data for FIB are collected in concert with data for hydrometeorological variables, such as wave height, solar insolation, and wind direction, and the combination of parameters that best predicts the concentration of enterococci is determined through statistical modeling (246). Statistical models have included regression (246, 298), Bayesian analysis (69), and neural networks (219); the complexity of the type of model that is tested is determined by need and application: more simple models can be used for daily predictions, while complex models integrate numerous parameters and may be used for determining contamination sources and pathways in order to develop mitigation plans. Simple models, such as rainfall threshold, allow for immediate management decisions (314), while more complex models, incorporating multiple predictive variables, require additional technology and expertise (246). Predictive models have met with uneven success; the source of contamination and characteristics of the beach structure generally influence the predictability of concentrations of FIB (247, 298). Common predictors can be linked directly to the physical persistence of enterococci in water: solar insolation affects bacterial die-off, wave height influences the resuspension of settled particle-attached bacteria, and wind direction influences the advection of bacterium-containing plumes from point sources such as rivers and streams (246). Predictive models can be developed which provide results in a fraction of the time currently required for FIB culturing techniques and even rapid molecular methods. Public health improvements with the use of a predictive model have not been adequately assessed, but one study indicated an improvement in overall health protection with the use of a model over standard, culture-dependent techniques (248).
CONCLUSIONS
Major advances in the understanding of the phylogeny of the enterococci and the ecology of the group in secondary habitats have been made over the past several decades. While these advances aid in the protection of human, animal, and environmental health, many gaps remain. From our perspective, among the most important areas for further research is the gaining of a better understanding of the relationship between environmental enterococci and a range of human pathogens (e.g., Campylobacter, Salmonella, enterotoxigenic E. coli, Vibrio spp., and Listeria monocytogenes) commonly transmitted through contaminated water and food networks. Additional epidemiological studies of waters impacted by nonpoint source pollution that include the identification of enterococci to the species level and the quantification of pathogens are also needed to better understand the human health risks associated with elevated levels of FIB. Measurements of nutrient levels (e.g., total organic carbon, nitrogen, phosphorous, and micronutrients) in conjunction with survival studies that are realistic simulations of aquatic environments will provide a further understanding of the drivers of the fate of enterococci in extraintestinal habitats. The possible ecological roles of enterococci in extraintestinal habitats, e.g., the decomposition of organic matter, competition with other members of the microbial community, and the protection of plants from pathogens, warrant further investigation. When used in conjunction with the quantification and species-level identification of enterococci, next-generation sequencing technologies may well revolutionize our understanding of the ecology of these organisms and their continued usefulness as FIB for recreational waters worldwide. A coordinated, international effort that focuses on the issues outlined above and that involves academic and regulatory research scientists could produce a comprehensive data set that allows us to define the impact of ecological factors on the survival of enterococci and the relationships among enterococci, pathogens, and human health in environmental settings.
ACKNOWLEDGMENTS
Any use of trade, product, or firm names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
The U.S. Environmental Protection Agency, through its Office of Research and Development, partially funded and collaborated in the research described here; it has been subjected to agency review and approved for publication. This work was funded in part by the U.S. Geological Survey Ocean Research Priorities Plan.
We express our gratitude to Teresa Ruby, senior graphic designer (SRA International, Inc.) for drafting Fig. 1.
Biographies

Muruleedhara N. Byappanahalli, Ph.D., is a Research Microbiologist with the U.S. Geological Survey Great Lakes Science Center. His research interests include microbial ecology, soil microbiology, and water quality and public health microbiology. Dr. Byappanahalli has published extensively on the ecology of indicator bacteria (E. coli and enterococci) in natural environments, such as soils and sediments, beach sand, and aquatic vegetation. His research was among the first to document the natural occurrence, persistence, and growth of indicator bacteria in nonenteric habitats. Dr. Byappanahalli's other research interests include exploring gene-based (molecular) techniques for identifying sewage markers in environmental waters. Dr. Byappanahalli is a member of the American Society for Microbiology and the Society for Applied Microbiology. He serves on the editorial board of the journal Applied and Environmental Microbiology. He has adjunct faculty positions at Purdue University and Indiana University.

Meredith B. Nevers is a research ecologist with the U.S. Geological Survey Great Lakes Science Center in Porter, IN, where she has worked for the past 15 years. Her research interests include microbiological contamination of beaches, improving public health protection through beach monitoring, impact assessments on biological communities, and aquatic ecology. Ms. Nevers has published extensively on beach health and improving monitoring accuracy through predictive modeling as well as in the ecology and natural occurrence of fecal indicator bacteria and is frequently invited to present her research at national and international professional meetings. She is the team leader for predictive modeling in her research group. Ms. Nevers serves on the board of the Great Lakes Beach Association and is an active member of the American Society for Microbiology and the International Water Association.

Asja Korajkic, Ph.D., is a postdoctoral fellow in the National Exposure Research Laboratory at the U.S. Environmental Protection Agency. She received her doctoral degree in Environmental and Ecological Microbiology at the University of South Florida in Tampa under the guidance of Dr. Valerie J. Harwood. Her research has been published in several peer-reviewed journals and presented at numerous international conferences. Her current research is focused on the fate and transport of fecal indicator bacteria, microbial source tracking (MST) markers, and pathogens in aquatic habitats.

Zachery R. Staley is a Ph.D. student in the Department of Integrative Biology at the University of South Florida in Tampa. He has published two peer-reviewed manuscripts focusing on the impact of agrochemicals on the fate and survival of fecal indicator bacteria. He has also participated in studies utilizing microbial source tracking (MST) to determine and remediate sources of fecal contamination in local water bodies.

Valerie J. Harwood, Ph.D., is a Professor in the Department of Integrative Biology at the University of South Florida in Tampa. She has published over 55 peer-reviewed manuscripts on subjects ranging from the biochemistry of hyperthermophiles and the ecology of Vibrio spp. to the persistence of fecal indicator bacteria in aquatic habitats and their relationship to pathogens. She was among the first to investigate the potential of microbial source tracking (MST) to determine animal and human contributions to fecal pollution in water bodies and is coeditor of the 2011 book Microbial Source Tracking: Methods, Applications, and Case Studies. She is a member of the Editorial Board of Applied and Environmental Microbiology.
Footnotes
This paper is contribution 1706 of the U.S. Geological Survey Great Lakes Science Center.
REFERENCES
- 1. Aarestrup FM, Butaye P, Witte W. 2002. Nonhuman reservoirs of enterococci, p 55–99. In Gilmore MS. (ed), The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC [Google Scholar]
- 2. Abdelzaher AM, et al. 2010. Presence of pathogens and indicator microbes at a non-point source subtropical recreational marine beach. Appl. Environ. Microbiol. 76: 724–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ahmed W, Katouli M. 2008. Phenotypic variations of enterococci in surface waters: analysis of biochemical fingerprinting data from multi-catchments. J. Appl. Microbiol. 105: 452–458 [DOI] [PubMed] [Google Scholar]
- 4. Ahmed W, Stewart J, Gardner T, Powell D. 2008. A real-time polymerase chain reaction assay for quantitative detection of the human-specific enterococci surface protein marker in sewage and environmental waters. Environ. Microbiol. 10: 3255–3264 [DOI] [PubMed] [Google Scholar]
- 5. Ahmed W, Stewart J, Powell D, Gardner T. 2008. Evaluation of the host-specificity and prevalence of enterococci surface protein (esp) marker in sewage and its application for sourcing human fecal pollution. J. Environ. Qual. 23: 1583–1588 [DOI] [PubMed] [Google Scholar]
- 6. American Public Health Association 2005. Standard methods for the examination of water and wastewater, 21st ed. American Public Health Association, Washington, DC [Google Scholar]
- 7. An YJ, Kampbell DH, Breidenbach GP. 2002. Escherichia coli and total coliforms in water and sediments at lake marinas. Environ. Pollut. 120: 771–778 [DOI] [PubMed] [Google Scholar]
- 8. Anderson A, Larsson U, Hagstrom A. 1986. Size selective grazing by a microflagellate on pelagic bacteria. Mar. Ecol. Prog. Ser. 33: 51–57 [Google Scholar]
- 9. Anderson IC, Rhodes MW, Kator HI. 1983. Seasonal variation in survival of Escherichia coli exposed in situ in membrane diffusion chambers containing filtered and nonfiltered estuarine water. Appl. Environ. Microbiol. 45: 1877–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Anderson KL, Whitlock JE, Harwood VJ. 2005. Persistence and differential survival of fecal indicator bacteria in subtropical waters and sediments. Appl. Environ. Microbiol. 71: 3041–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Anderson SA, Turner SJ, Lewis GD. 1997. Enterococci in the New Zealand environment: implications for water quality monitoring. Water Sci. Technol. 35(11-12): 325–331 [Google Scholar]
- 12. Andrews JH. 1991. Comparative ecology of microorganisms and macroorganisms. Springer-Verlag, New York, NY [Google Scholar]
- 13. Ashbolt NJ, Schoen ME, Soller JA, Roser DJ. 2010. Predicting pathogen risks to aid beach management: the real value of quantitative microbial risk assessment (QMRA). Water Res. 44: 4692–4703 [DOI] [PubMed] [Google Scholar]
- 14. Badgley BD, Nayak BS, Harwood VJ. 2010. The importance of sediment and submerged aquatic vegetation as potential habitats for persistent strains of enterococci in a subtropical watershed. Water Res. 44: 5857–5866 [DOI] [PubMed] [Google Scholar]
- 15. Badgley BD, Thomas FIM, Harwood VJ. 2010. The effects of submerged aquatic vegetation on the persistence of environmental populations of Enterococcus spp. Environ. Microbiol. 42: 1271–1281 [DOI] [PubMed] [Google Scholar]
- 16. Badgley BD, Thomas FIM, Harwood VJ. 2011. Quantifying environmental reservoirs of fecal indicator bacteria associated with sediment and submerged aquatic vegetation. Environ. Microbiol. 3: 932–942 [DOI] [PubMed] [Google Scholar]
- 17. Bale MN, Bennett PM, Beringer JE, Hinton M. 1993. The survival of bacteria exposed to desiccation on surfaces associated with farm buildings. J. Appl. Bacteriol. 75: 519–528 [DOI] [PubMed] [Google Scholar]
- 18. Barcina I, Ayo B, Muela A, Egea L, Iriberri J. 1991. Predation rates of flagellate and ciliated protozoa on bacterioplankton in a river. FEMS Microbiol. Ecol. 85: 141–149 [Google Scholar]
- 19. Barcina I, Gonzalez JM, Iriberri J, Egea L. 1991. Role of protozoa in the regulation of enteric bacteria populations in seawater. Mar. Microb. Food Webs 5: 179–188 [Google Scholar]
- 20. Barcina I, Lebaron P, VivesRego J. 1997. Survival of allochthonous bacteria in aquatic systems: a biological approach. FEMS Microbiol. Ecol. 23: 1–9 [Google Scholar]
- 21. Baums IB, et al. 2007. Luminex detection of fecal indicators in river samples, marine recreational water, and beach sand. Mar. Pollut. Bull. 54: 521–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Beaches Environmental Assessment and Coastal Health Act 2000. Public law 106-284, H.R. 99-106th Congress. http://www.govtrack.us/congress/bills/106/hr99 Accessed 20 September 2012
- 23. Beardsley C, Pernthaler J, Wosniok W, Amann R. 2003. Are readily culturable bacteria in coastal North Sea waters suppressed by selective grazing mortality? Appl. Environ. Microbiol. 69: 2624–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Benachour A, et al. 2005. The Enterococcus faecalis SigV protein is an extracytoplasmic function sigma factor contributing to survival following heat, acid, and ethanol treatments. J. Bacteriol. 187: 1022–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Berg G, Dahling DR, Brown GA, Berman D. 1978. Validity of fecal coliforms, total coliforms, and fecal streptococci as indicators of viruses in chlorinated primary sewage effluents. Appl. Environ. Microbiol. 36: 880–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bernhard AE, Field KG. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 66: 4571–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boehm AB, et al. 2002. Decadal and shorter period variability of surf zone water quality at Huntington Beach, California. Environ. Sci. Technol. 36: 3885–3892 [DOI] [PubMed] [Google Scholar]
- 28. Boehm AB, et al. 2009. Faecal indicator bacteria enumeration in beach sand: a comparison study of extraction methods in medium to coarse sands. J. Appl. Microbiol. 107: 1740–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Boehm AB, Keymer DP, Shellenbarger GG. 2005. An analytical model of enterococci inactivation, grazing, and transport in the surf zone of a marine beach. Water Res. 39: 3565–3578 [DOI] [PubMed] [Google Scholar]
- 30. Boehm AB, et al. 2009. Covariation and photoinactivation of traditional and novel indicator organisms and human viruses at a sewage-impacted marine beach. Environ. Sci. Technol. 43: 8046–8052 [DOI] [PubMed] [Google Scholar]
- 31. Bonilla N, et al. 2010. Enterophages, a group of phages infecting Enterococcus faecalis, and their potential as alternate indicators of human faecal contamination. Water Sci. Technol. 61(2): 293–300 [DOI] [PubMed] [Google Scholar]
- 32. Bonilla TD, et al. 2007. Prevalence and distribution of fecal indicator organisms in South Florida beach sand and preliminary assessment of health effects associated with beach sand exposure. Mar. Pollut. Bull. 54: 1472–1482 [DOI] [PubMed] [Google Scholar]
- 33. Brandt CL, Giese AC. 1956. Photoreversal of nuclear and cytoplasmic effects of short ultraviolet radiation on Paramecium caudatum. J. Gen. Physiol. 39: 35–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Budnick GE, Howard RT, Mayo DR. 1996. Evaluation of Enterolert for enumeration of enterococci in recreational waters. Appl. Environ. Microbiol. 62: 3881–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bushon RN, Likirdopulos CA, Brady AMG. 2009. Comparison of immunomagnetic separation/adenosine triphosphate rapid method to traditional culture-based method for E. coli and enterococci enumeration in wastewater. Water Res. 43: 4940–4946 [DOI] [PubMed] [Google Scholar]
- 36. Byappanahalli M, Fowler M, Shively D, Whitman R. 2003. Ubiquity and persistence of Escherichia coli in a Midwestern stream. Appl. Environ. Microbiol. 69: 4549–4555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Byappanahalli M, Fujioka R. 2004. Indigenous soil bacteria and low moisture may limit but allow faecal bacteria to multiply and become a minor population in tropical soils. Water Sci. Technol. 50(1): 27–32 [PubMed] [Google Scholar]
- 38. Byappanahalli MN. 2000. Assessing the persistence and multiplication of fecal indicator bacteria in Hawaii's soil environment. Ph.D. thesis. University of Hawaii at Manoa, Honolulu, HI [Google Scholar]
- 39. Byappanahalli MN, Przybyla-Kelly K, Shively DA, Whitman RL. 2008. Environmental occurrence of the enterococcal surface protein (esp) gene is an unreliable indicator of human fecal contamination. Environ. Sci. Technol. 42: 8014–8020 [DOI] [PubMed] [Google Scholar]
- 40. Byappanahalli MN, Roll BM, Fujioka RS. 2012. Evidence for occurrence, persistence, and growth of Escherichia coli and enterococci in Hawaii's soil environments. Microbes Environ. 27: 164–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Byappanahalli MN, et al. 2009. Seasonal stability of Cladophora-associated Salmonella in Lake Michigan watersheds. Water Res. 43: 806–814 [DOI] [PubMed] [Google Scholar]
- 42. Byappanahalli MN, Shively DA, Nevers MB, Sadowsky MJ, Whitman RL. 2003. Growth and survival of Escherichia coli and enterococci populations in the macro-alga Cladophora (Chlorophyta). FEMS Microbiol. Ecol. 46: 203–211 [DOI] [PubMed] [Google Scholar]
- 43. Byappanahalli MN, Whitman RL. 2009. Clostridium botulinum type E occurs and grows in the green alga Cladophora glomerata. Can. J. Fish. Aquat. Sci. 66: 879–882 [Google Scholar]
- 44. Byappanahalli MN, et al. 2006. Seasonal persistence and population characteristics of Escherichia coli and enterococci in deep backshore sand of two freshwater beaches. J. Water Health 4: 313–320 [DOI] [PubMed] [Google Scholar]
- 45. Byappanahalli MN, Whitman RL, Shively DA, Nevers MB. 2010. Linking non-culturable (qPCR) and culturable enterococci densities with hydrometeorological conditions. Sci. Total Environ. 408: 3096–3101 [DOI] [PubMed] [Google Scholar]
- 46. Byappanahalli MN, et al. 2012. The population structure of Escherichia coli isolated from subtropical and temperate soils. Sci. Total Environ. 417-418: 273–279 [DOI] [PubMed] [Google Scholar]
- 47. Cabelli VJ. 1983. Health effects criteria for marine recreational waters. EPA-600/1-80-031. US Environmental Protection Agency, Research Triangle Park, NC [Google Scholar]
- 48. Cabelli VJ. 1978. New standards for enteric bacteria, p 2333. In Mitchel R. (ed), Water pollution microbiology. Wiley, New York, NY [Google Scholar]
- 49. Cabelli VJ, Dufour AP, Levin MA, McCabe LJ, Haberman PW. 1979. Relationship of microbial indicators to health effects at marine bathing beaches. Am. J. Public Health 69: 690–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cai Y. 1999. Identification and characterization of Enterococcus species isolated from forage crops and their influence on silage fermentation. J. Dairy Sci. 82: 2466–2471 [DOI] [PubMed] [Google Scholar]
- 51. Calvez S, Rince A, Auffray Y, Prevost H, Drider D. 2007. Identification of new genes associated with intermediate resistance of Enterococcus faecalis to divercin V41, a pediocin-like bacteriocin. Microbiology 153: 1609–1618 [DOI] [PubMed] [Google Scholar]
- 52. Caretti C, Lubello C. 2003. Wastewater disinfection with PAA and UV combined treatment: a pilot plant study. Water Res. 37: 2365–2371 [DOI] [PubMed] [Google Scholar]
- 53. Carr MR, Wang SY, McLean TI, Flood CJ, Ellender RD. 2010. Salmonella rarely detected in Mississippi coastal waters and sediment. J. Appl. Microbiol. 109: 2191–2199 [DOI] [PubMed] [Google Scholar]
- 54. Carvalho MGS, et al. 2006. Enterococcus caccae sp. nov., isolated from human stools. Int. J. Syst. Evol. Microbiol. 56: 1505–1508 [DOI] [PubMed] [Google Scholar]
- 55. Carvalho MGS, et al. 2008. Designation of the provisional new Enterococcus species CDC PNS-E2 as Enterococcus sanguinicola sp. nov., isolated from human blood, and identification of a strain previously named Enterococcus CDC PNS-E1 as Enterococcus italicus Fortina, Ricci, Mora, and Manachini 2004. J. Clin. Microbiol. 46: 3473–3476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cha JO, Jung YH, Lee HR, Yoo JI, Lee YS. 26 April 2012. Comparison of genetic epidemiology of vancomycin-resistant Enterococcus faecium isolates from humans and poultry. J. Med. Microbiol. doi:10.1099/jmm.0.037549-0. [DOI] [PubMed]
- 57. Chang JC, et al. 1985. UV inactivation of pathogenic and indicator microorganisms. Appl. Environ. Microbiol. 49: 1361–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chauret C, Springthorpe S, Sattar S. 1999. Fate of Cryptosporidium oocysts, Giardia cysts, and microbial indicators during wastewater treatment and anaerobic sludge digestion. Can. J. Microbiol. 45: 257–262 [PubMed] [Google Scholar]
- 59. Cho KH, et al. 2010. Meteorological effects on the levels of fecal indicator bacteria in an urban stream: a modeling approach. Water Res. 44: 2189–2202 [DOI] [PubMed] [Google Scholar]
- 60. Christensen J, Linden KG. 2003. How particles affect UV light in the UV disinfection of unfiltered drinking water. J. Am. Water Works Assoc. 95: 179–189 [Google Scholar]
- 61. Cizek AR, et al. 2008. Comparing the partitioning behavior of Giardia and Cryptosporidium with that of indicator organisms in stormwater runoff. Water Res. 42: 4421–4438 [DOI] [PubMed] [Google Scholar]
- 62. Clarridge JE., III 2004. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 17: 840–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Colford JM, et al. 2012. Using rapid indicators for Enterococcus to assess the risk of illness after exposure to urban runoff contaminated marine water. Water Res. 46: 2176–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Colford JM, Jr, et al. 2007. Water quality indicators and the risk of illness at beaches with nonpoint sources of fecal contamination. Epidemology 18: 27–35 [DOI] [PubMed] [Google Scholar]
- 65. Collins MD, Facklam RR, Farrow JA, Williamson R. 1989. Enterococcus raffinosus sp. nov., Enterococcus solitarius sp. nov., and Enterococcus pseudoavium sp. nov. FEMS Microbiol. Lett. 48: 283–288 [DOI] [PubMed] [Google Scholar]
- 66. Collins MD, Farrow JAE, Jones D. 1986. Enterococcus mundtii sp. nov. Int. J. Syst. Bacteriol. 36: 8–12 [Google Scholar]
- 67. Collins MD, Jones D, Farrow JAE, Kilpper-Bälz R, Schleifer KH. 1984. Enterococcus avium nom.rev., comb. nov.; E. casseliflavus nom. rev., comb. nov.; E. durans nom. rev., comb. nov.; E. gallinarum comb. nov.; and E. malodoratus sp. nov. Int. J. Syst. Bacteriol. 34: 220–223 [Google Scholar]
- 68. Collins MD, Rodrigues UM, Pigott NE, Facklam RR. 1991. Enterococcus dispar sp. nov.: a new Enterococcus species from human sources. Lett. Appl. Microbiol. 12: 95–98 [DOI] [PubMed] [Google Scholar]
- 69. Coulliette AD, Money ES, Serre ML, Noble RT. 2009. Space/time analysis of fecal pollution and rainfall in an eastern North Carolina estuary. Environ. Sci. Technol. 43: 3728–3735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cox CR, Gilmore MS. 2007. Native microbial colonization of Drosophila melanogaster and its use as a model of Enterococcus faecalis pathogenesis. Infect. Immun. 75: 1565–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Craggs RJ, Sukias JP, Tanner CT, Davies-Colley RJ. 2004. Advanced pond system for dairy-farm effluent treatment. N. Z. J. Agric. Res. 47: 449–460 [Google Scholar]
- 72. Craig DL, Fallowfield HJ, Cromar NJ. 2002. Enumeration of faecal coliforms from recreational coastal sites: evaluation of techniques for the separation of bacteria from sediments. J. Appl. Microbiol. 93: 557–565 [DOI] [PubMed] [Google Scholar]
- 73. Curtis TP, Mara DD, Silva SA. 1992. Influence of pH, oxygen, and humic substances on ability of sunlight to damage fecal coliforms in waste stabilization pond water. Appl. Environ. Microbiol. 58: 1335–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Dalet K, Briand C, Cenatiempo Y, Hechard Y. 2000. The rpoN gene of Enterococcus faecalis directs sensitivity to subclass IIa bacteriocins. Curr. Microbiol. 41: 441–443 [DOI] [PubMed] [Google Scholar]
- 75. Davies CM, Long JA, Donald M, Ashbolt NJ. 1995. Survival of fecal microorganisms in marine and freshwater sediments. Appl. Environ. Microbiol. 61: 1888–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Davies CM, Roser DJ, Feitz AJ, Ashbolt NJ. 2009. Solar radiation disinfection of drinking water at temperate latitudes: inactivation rates for an optimised reactor configuration. Water Res. 43: 643–652 [DOI] [PubMed] [Google Scholar]
- 77. Davies-Colley RJ, Bell RG, Donnison AM. 1994. Sunlight inactivation of enterococci and fecal coliforms in sewage effluent diluted in seawater. Appl. Environ. Microbiol. 60: 2049–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Davies-Colley RJ, Craggs RJ, Park J, Nagels JW. 2005. Optical characteristics of waste stabilization ponds: recommendations for monitoring. Water Sci. Technol. 51(12): 153–161 [PubMed] [Google Scholar]
- 79. Davies-Colley RJ, Craggs RJ, Nagels JW. 2003. Disinfection in a pilot-scale “advanced” pond system (APS) for domestic sewage treatment in New Zealand. Water Sci. Technol. 48(2): 81–87 [PubMed] [Google Scholar]
- 80. Davies-Colley RJ, Donnison AM, Speed DJ. 1997. Sunlight wavelengths inactivating faecal indicator microorganisms in waste stabilisation ponds. Water Sci. Technol. 35(11-12): 219–225 [Google Scholar]
- 81. Davies-Colley RJ, Donnison AM, Speed DJ, Ross CM, Nagels JW. 1999. Inactivation of faecal indicator microorganisms in waste stabilisation ponds: interactions of environmental factors with sunlight. Water Res. 33: 1220–1230 [Google Scholar]
- 82. de Graef EM, et al. 2003. Description of Enterococcus canis sp. nov. from dogs and reclassification of Enterococcus porcinus Teixeira et al. 2001 as a junior synonym of Enterococcus villorum Vancanneyt et al. Int. J. Syst. Evol. Microbiol. 53: 1069–1074 [DOI] [PubMed] [Google Scholar]
- 83. Deller S, Mascher F, Platzer S, Reinthaler FF, Marth E. 2006. Effect of solar radiation on survival of indicator bacteria in bathing waters. Cent. Eur. J. Public Health 14: 133–137 [DOI] [PubMed] [Google Scholar]
- 84. De Luca G, Sacchetti R, Zanetti F, Leoni E. 2008. Comparative study on the efficiency of peracetic acid and chlorine dioxide at low doses in the disinfection of urban wastewaters. Ann. Agric. Environ. Med. 15: 217–224 [PubMed] [Google Scholar]
- 85. Desmarais TR, Solo-Gabriele HM, Palmer CJ. 2002. Influence of soil on fecal indicator organisms in a tidally influenced subtropical environment. Appl. Environ. Microbiol. 68: 1165–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. de Vaux A, Laguerre G, Diviès C, Prévost H. 1998. Enterococcus asini sp. nov. isolated from the caecum of donkeys (Equus asinus). Int. J. Syst. Bacteriol. 48: 383–387 [DOI] [PubMed] [Google Scholar]
- 87. Devriese LA, Ceyssens K, Rodrigues UM, Collins MD. 1990. Enterococcus columbae, a species from pigeon intestines. FEMS Microbiol. Lett. 71: 247–251 [DOI] [PubMed] [Google Scholar]
- 88. Devriese LA, Dutta GN, Farrow JAE, Van De Kerckhove A, Phillips BA. 1983. Streptococcus cecorum, a new species isolated from chickens. Int. J. Syst. Bacteriol. 33: 772–776 [Google Scholar]
- 89. Devriese LA, Pot B, Collins MD. 1993. Phenotypic identification of the genus Enterococcus and differentiation of phylogenetically distinct enterococcal species and species groups. J. Appl. Bacteriol. 75: 399–408 [DOI] [PubMed] [Google Scholar]
- 90. Dickenson JA, Sansalone JJ. 24 December 2011. Distribution and disinfection of bacterial loadings associated with particulate matter fractions transported in urban wet weather flows. Water Res. [Epub ahead of print.] http://dx.doi.org/10.1016/j.watres.2011.12.039 [DOI] [PubMed]
- 91. Djahmi N, et al. 8 May 2012. Molecular epidemiology of Enterococcus sp. isolated in a university hospital in Algeria. Scand. J. Infect. Dis. doi:10.3109/00365548.2012.673232. [DOI] [PubMed]
- 92. Doran JE, Linn DM. 1979. Bacteriological quality of runoff water from pastureland. Appl. Environ. Microbiol. 37: 985–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Dorevitch S, et al. 2010. Knowledge and gaps in developing microbial criteria for inland recreational waters. Environ. Health Perspect. 118: 871–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Dorsey JH, Carter PM, Bergquist S, Sagarin R. 2010. Reduction of fecal indicator bacteria (FIB) in the Ballona Wetlands saltwater marsh (Los Angeles County, California, USA) with implications for restoration actions. Water Res. 44: 4630–4642 [DOI] [PubMed] [Google Scholar]
- 95. Downes A, Blunt TP. 1877. Researchers on the effect of light upon bacteria and other organisms. Proc. R. Soc. Lond. B Biol. Sci. 26: 488–500 [Google Scholar]
- 96. Dufour AP. 1984. Health effects criteria for fresh recreational waters. EPA report EPA-600/1-84-004. US Environmental Protection Agency, Cincinnati, OH [Google Scholar]
- 97. Dulbecco R. 1950. Experiments on photoreactivation of bacteriophages inactivated with ultraviolet radiation. J. Bacteriol. 59: 329–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Dwight RH, Baker DB, Semenza JC. 2002. Association of urban runoff with coastal water quality in Orange County, California. Water Environ. Res. 74: 82–90 [DOI] [PubMed] [Google Scholar]
- 99. Elmir SM, et al. 2007. Quantitative evaluation of bacteria released by bathers in a marine water. Water Res. 41: 3–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Enns AA, et al. 2012. Spatial and temporal variation in indicator microbe sampling is influential in beach management decisions. Water Res. 46: 2237–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Farrow JAE, Collins MD. 1985. Enterococcus hirae, a new species that includes amino acid assay strain NCDO 1258 and strains causing growth depression in young chickens. Int. J. Syst. Bacteriol. 35: 73–75 [Google Scholar]
- 102. Ferguson DM, Moore DF, Getrich MA, Zhowandai MH. 2005. Enumeration and speciation of enterococci found in marine and intertidal sediments and coastal water in southern California. J. Appl. Microbiol. 99: 598–608 [DOI] [PubMed] [Google Scholar]
- 103. Fisher MB, Iriarte M, Nelson KL. 2012. Solar water disinfection (SODIS) of Escherichia coli, Enterococcus spp., and MS2 coliphage: effects of additives and alternative container materials. Water Res. 46: 1745–1754 [DOI] [PubMed] [Google Scholar]
- 104. Fleisher JM, et al. 2010. The BEACHES Study: health effects and exposures from non-point source microbial contaminants in subtropical recreational marine waters. Int. J. Epidemiol. 39: 1291–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Fogarty LR, Haack SK, Wolcott MJ, Whitman RL. 2003. Abundance and characteristics of the recreational water quality indicator bacteria Escherichia coli and enterococci in gull faeces. J. Appl. Microbiol. 94: 865–878 [DOI] [PubMed] [Google Scholar]
- 106. Fong T, Griffin D, Lipp E. 2005. Molecular assays for targeting human and bovine enteric viruses in coastal waters and their application for library-independent source tracking. Appl. Environ. Microbiol. 71: 2070–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Fortina MG, Ricci G, Mora D, Manachini PL. 2004. Molecular analysis of artisanal Italian cheeses reveals Enterococcus italicus sp. nov. Int. J. Syst. Evol. Microbiol. 54: 1717–1721 [DOI] [PubMed] [Google Scholar]
- 108. Franz CMAP, Huch M, Abriouel H, Holzapfel W, Galvez A. 2011. Enterococci as probiotics and their implications in food safety. Int. J. Food Microbiol. 151: 125–140 [DOI] [PubMed] [Google Scholar]
- 109. Fuhrman JA, Noble RT. 1995. Viruses and protists cause similar bacterial mortality in coastal seawater. Limnol. Oceanogr. 40: 1236–1242 [Google Scholar]
- 110. Fujioka R, Sian-Denton C, Borja M, Castro J, Morphew K. 1999. Soil: the environmental source of Escherichia coli and enterococci in Guam's streams. J. Appl. Microbiol. 85( Suppl 1): 83S–89S doi:10.1111/j1365-2672.1998.tb05286.x. [DOI] [PubMed] [Google Scholar]
- 111. Fujioka RS, Byappanahalli MN. 2001. Microbial ecology controls the establishment of fecal bacteria in tropical soil environment, p 273–283. In Matsuo T, Hanaki K, Takizawa S, Satoh H. (ed), Advances in water and wastewater treatment technology: molecular technology, nutrient removal, sludge reduction, and environmental health. Elsevier Science, Amsterdam, Netherlands [Google Scholar]
- 112. Fujioka RS, Hashimoto HH, Siwak EB, Young RH. 1981. Effect of sunlight on survival of indicator bacteria in seawater. Appl. Environ. Microbiol. 41: 690–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Fujioka RS, Narikawa OT. 1982. Effect of sunlight on enumeration of indicator bacteria under field conditions. Appl. Environ. Microbiol. 44: 395–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Fujioka RS, Tenno K, Kansako S. 1988. Naturally occurring fecal coliforms and fecal streptococci in Hawaii's freshwater streams. Tox. Assess. 3: 613–630 [Google Scholar]
- 115. Fujioka RSB, Byappanahalli MN. (ed). 2003. Proceedings and report: tropical water quality indicator workshop SR-2004–01. University of Hawaii Water Resources Research Center, Honolulu, HI: http://www.wrrc.hawaii.edu/tropindworkshop.html. [Google Scholar]
- 116. Gary HL, Adams JC. 1985. Indicator bacteria in water and stream sediments near the snowy range in southern Wyoming. Water Air Soil Pollut. 25: 133–144 [Google Scholar]
- 117. Gates FL. 1929. A study of the bactericidal action of ultra violet light. I. The reaction to monochromatic radiations. J. Gen. Physiol. 13: 231–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Ge Z, Whitman RL, Nevers MB, Phanikumar MS, Byappanahalli MN. 2012. Evaluating the role of an embayed beach as a reservoir and a net source of fecal contamination. Limnol. Oceanogr. 57: 362–381 [Google Scholar]
- 119. Gerba CP, McLeod JS. 1976. Effect of sediments on the survival of Escherichia coli in marine waters. Appl. Environ. Microbiol. 32: 114–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Ghinsberg RC, Bar Dov L, Rogol M, Sheinberg Y, Nitzan Y. 1994. Monitoring of selected bacteria and fungi in sand and sea water along the Tel Aviv coast. Microbios 77: 29–40 [PubMed] [Google Scholar]
- 121. Ghosh A, KuKanich K, Brown CE, Zurek L. 21 February 2012. Resident cats in small animal veterinary hospitals carry multi-drug resistant enterococci and are likely involved in cross-contamination of the hospital environment. Front. Microbiol. doi:10.3389/fmicb.2012.00062. [DOI] [PMC free article] [PubMed]
- 122. Giard JC, et al. 1996. Starvation-induced multiresistance in Enterococcus faecalis JH2-2. Curr. Microbiol. 32: 264–271 [DOI] [PubMed] [Google Scholar]
- 123. Giard JC, Hartke A, Flahaut S, Boutibonnes P, Auffray Y. 1997. Glucose starvation response in Enterococcus faecalis JH2-2: survival and protein analysis. Res. Microbiol. 148: 27–35 [DOI] [PubMed] [Google Scholar]
- 124. Giard JC, Rince A, Capiaux H, Auffray Y, Hartke A. 2000. Inactivation of the stress- and starvation-inducible gls24 operon has a pleiotrophic effect on cell morphology, stress sensitivity, and gene expression in Enterococcus faecalis. J. Bacteriol. 182: 4512–4520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Gonzalez JM, Iriberri J, Egea L, Barcina I. 1992. Characterization of culturability, protistan grazing, and death of enteric bacteria in aquatic ecosystems. Appl. Environ. Microbiol. 58: 998–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Gonzalez JM, Iriberri J, Egea L, Barcina I. 1990. Differential rates of digestion of bacteria by freshwater and marine phagotrophic protozoa. Appl. Environ. Microbiol. 56: 1851–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Gonzalez JM, Sherr EB, Sherr BF. 1990. Size-selective grazing on bacteria by natural assemblages of estuarine flagellates and ciliates. Appl. Environ. Microbiol. 56: 583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Gordillo ME, Singh KV, Murray BE. 1993. Comparison of ribotyping and pulsed-field gel electrophoresis for subspecies differentiation of strains of Enterococcus faecalis. J. Clin. Microbiol. 31: 1570–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Goyal SM, Gerba CP, Melnick JL. 1977. Occurrence and distribution of bacterial indicators and pathogens in canal communities along the Texas coast. Appl. Environ. Microbiol. 34: 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Grant SB, Litton-Mueller RM, Ahn JH. 13 May 2011. Measuring and modeling the flux of fecal bacteria across the sediment-water interface in a turbulent stream. Water Resour. Res. doi:10.1029/2010WR009460.
- 131. Grant SB, et al. 2001. Generation of enterococci bacteria in a costal saltwater marsh and its impact on surf zone water quality. Environ. Sci. Technol. 35: 2407–2416 [DOI] [PubMed] [Google Scholar]
- 132. Griffith JF, et al. 2006. Comparison and verification of bacterial water quality indicator measurement methods using ambient coastal water samples. Environ. Monit. Assess. 116: 335–344 [DOI] [PubMed] [Google Scholar]
- 133. Griffith JF, Cao Y, McGee CD, Weisberg SB. 2009. Evaluation of rapid methods and novel indicators for assessing microbiological beach water quality. Water Res. 43: 4900–4907 [DOI] [PubMed] [Google Scholar]
- 134. Grimes DJ. 1980. Bacteriological water quality effects of hydraulically dredging contaminated upper Mississippi River bottom sediment. Appl. Environ. Microbiol. 39: 782–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Grimes DJ. 1975. Release of sediment-bound fecal coliforms by dredging. Appl. Microbiol. 29: 109–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Haack SK, Fogarty LR, Wright C. 2003. Escherichia coli and enterococci at beaches in the Grand Traverse Bay, Lake Michigan: sources, characteristics, and environmental pathways. Environ. Sci. Technol. 37: 3275–3282 [DOI] [PubMed] [Google Scholar]
- 137. Hagedorn C, Blanch AR, Harwood VJ. (ed). 2011. Microbial source tracking: methods, applications, and case studies. Springer, New York, NY [Google Scholar]
- 138. Hagedorn C, et al. 1999. Determining sources of fecal pollution in a rural Virginia watershed with antibiotic resistance patterns in fecal streptococci. Appl. Environ. Microbiol. 65: 5522–5531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Hale HG, Moore LD, Griffin CJ. 1978. Root exudates and exudation, p 163–203.In Dommergues YR, Krupa SV. (ed), Interactions between non-pathogenic soil microorganisms and plants. Elsevier Scientific Publishing, New York, NY [Google Scholar]
- 140. Hall LMC, Duke B, Guiney M, Williams R. 1992. Typing of Enterococcus species by DNA restriction fragment analysis. J. Clin. Microbiol. 30: 915–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Haller L, Amedegnato E, Poté J, Wildi W. 2009. Influence of freshwater sediment characteristics on persistence of fecal indicator bacteria. Water Air Soil Pollut. 203: 217–227 [Google Scholar]
- 142. Halliday E, Gast RJ. 2011. Bacteria in beach sands: an emerging challenge in protecting coastal water quality and bather health. Environ. Sci. Technol. 45: 370–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Halliday E, Griffith JF, Gast RJ. 2010. Use of an exogenous plasmid standard and quantitative PCR to monitor spatial and temporal distribution of Enterococcus spp. in beach sands. Limnol. Oceanogr. Methods 8: 146–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Hardie JM, Whiley RA. 1997. Classification and overview of the genera Streptococcus and Enterococcus, p 1S–11S. In Andrew PW, Mitchell TJ. (ed), The biology of streptococci and enterococci. Blackwell Science, Oxford, United Kingdom [Google Scholar]
- 145. Hardina CM, Fujioka RS. 1991. Soil: the environmental source of Escherichia coli and enterococci in Hawaii's streams. Environ. Toxicol. Water Qual. 6: 185–195 [Google Scholar]
- 146. Hartke A, Giard JC, Laplace JM, Auffray Y. 1998. Survival of Enterococcus faecalis in an oligotrophic microcosm: changes in morphology, development of general stress resistance, and analysis of protein synthesis. Appl. Environ. Microbiol. 64: 4238–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Hartke A, Lemarinier S, Pichereau V, Auffray Y. 2002. Survival of Enterococcus faecalis in seawater microcosms is limited in the presence of bacterivorous zooflagellates. Curr. Microbiol. 44: 329–335 [DOI] [PubMed] [Google Scholar]
- 148. Hartz A, et al. 2008. Survival potential of Escherichia coli and enterococci in subtropical beach sand: implications for water quality managers. J. Environ. Qual. 37: 898–905 [DOI] [PubMed] [Google Scholar]
- 149. Harwood VJ. 2007. Assumptions and limitations of microbial source tracking methods, p 33–64.In Santo Domingo JW, Sadowsky MJ. (ed), Microbial source tracking. ASM Press, Washington, DC [Google Scholar]
- 150. Harwood VJ, et al. 2009. Validation and field testing of library-independent microbial source tracking methods in the Gulf of Mexico. Water Res. 43: 4812–4819 [DOI] [PubMed] [Google Scholar]
- 151. Harwood VJ, et al. 2004. Molecular confirmation of Enterococcus faecalis and E. faecium from clinical, faecal and environmental sources. Lett. Appl. Microbiol. 38: 476–482 [DOI] [PubMed] [Google Scholar]
- 152. Harwood VJ, et al. 2005. Validity of the indicator organism paradigm for pathogen reduction in reclaimed water and public health protection. Appl. Environ. Microbiol. 71: 3163–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Harwood VJ, Ryu H, Santo Domingo JW. 2010. Microbial source tracking, p 189–216.In Sadowsky MJ, Whitman RL. (ed), The fecal bacteria. ASM Press, Washington, DC [Google Scholar]
- 154. Harwood VJ, Whitlock JE, Whitington V. 2000. Classification of antibiotic resistance patterns of indicator bacteria by discriminant analysis: use in predicting the source of fecal contamination in subtropical waters. Appl. Environ. Microbiol. 66: 3698–3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Haugland RA, Siefring SC, Wymer LJ, Brenner KP, Dufour AP. 2005. Comparison of Enterococcus measurements in freshwater at two recreational beaches by quantitative polymerase chain reaction and membrane filter culture analysis. Water Res. 39: 559–568 [DOI] [PubMed] [Google Scholar]
- 156. Havelaar AH, Pot-Hogeboom WM. 1988. F-specific RNA-bacteriophages as model viruses in water hygiene: ecological aspects. Water Sci. Technol. 20(11-12): 399–407 [Google Scholar]
- 157. Heaney CD, et al. 2012. Fecal indicators in sand, sand contact, and risk of enteric illness among beachgoers. Epidemiology 23: 95–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Hecker M, Volker U. 2001. General stress response of Bacillus subtilis and other bacteria. Adv. Microb. Physiol. 44: 35–91 [DOI] [PubMed] [Google Scholar]
- 159. Hecker M, Volker U. 1998. Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the sigmaB regulon. Mol. Microbiol. 29: 1129–1136 [DOI] [PubMed] [Google Scholar]
- 160. Heim S, Lleo MM, Bonato B, Guzman CA, Canepari P. 2002. The viable but nonculturable state and starvation are different stress responses of Enterococcus faecalis, as determined by proteome analysis. J. Bacteriol. 184: 6739–6745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Hengge-Aronis R. 2000. The general stress response in Escherichia coli, p 161–178.In Storz G, Hengge-Aronis R. (ed), Bacterial stress response. ASM Press, Washington, DC [Google Scholar]
- 162. Hijnen WA, Beerendonk EF, Medema GJ. 2006. Inactivation credit of UV radiation for viruses, bacteria and protozoan (oo)cysts in water: a review. Water Res. 40: 3–22 [DOI] [PubMed] [Google Scholar]
- 163. Homan WL, et al. 2002. Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 40: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Hou D, Rabinovici SJM, Boehm AB. 2006. Enterococci predictions from partial least squares regression models in conjunction with a single-sample standard improve the efficacy of beach management advisories. Environ. Sci. Technol. 40: 1737–1743 [DOI] [PubMed] [Google Scholar]
- 165. Howell JM, Coyne MS, Cornelius PL. 1995. Fecal bacteria in agricultural waters of the bluegrass region of Kentucky. J. Environ. Qual. 24: 411–419 [Google Scholar]
- 166. Huws SA, Morley RJ, Jones MV, Brown MR, Smith AW. 2008. Interactions of some common pathogenic bacteria with Acanthamoeba polyphaga. FEMS Microbiol. Lett. 282: 258–265 [DOI] [PubMed] [Google Scholar]
- 167. Imamura GJ, Thompson RS, Boehm AB, Jay JA. 2011. Wrack promotes the persistence of fecal indicator bacteria in marine sands and seawater. FEMS Microbiol. Ecol. 77: 40–49 [DOI] [PubMed] [Google Scholar]
- 168. International Organization for Standardization 1998. Water quality—detection and enumeration of intestinal enterococci in surface and waste water. Part 1: miniaturized method (most probable number) by inoculation in liquid medium. International Organization for Standardization, Geneva, Switzerland [Google Scholar]
- 169. Iriberri J, Ayo B, Artolozaga I, Barcina I, Gea LE. 1994. Grazing on allochthonous vs autochthonous bacteria in river water. Lett. Appl. Microbiol. 18: 12–14 [Google Scholar]
- 170. Iriberri J, Azua I, Labiruaiturburu A, Artolozaga I, Barcina I. 1994. Differential elimination of enteric bacteria by protists in a fresh-water system. J. Appl. Bacteriol. 77: 476–483 [DOI] [PubMed] [Google Scholar]
- 171. Ishii S, Sadowsky MJ. 2008. Escherichia coli in the environment: implications for water quality and human health. Microbes Environ. 23: 101–108 [DOI] [PubMed] [Google Scholar]
- 172. Ishii S, et al. 2006. Cladophora (Chlorophyta) spp. harbor human bacterial pathogens in nearshore water of Lake Michigan. Appl. Environ. Microbiol. 72: 4545–4553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Iyer VS, Hancock LE. 2012. Deletion of σ(54) (rpoN) alters the rate of autolysis and biofilm formation in Enterococcus faecalis. J. Bacteriol. 194: 368–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Jacangelo JG, Loughran P, Petrik B, Simpson D, McIlroy C. 2003. Removal of enteric viruses and selected microbial indicators by UV irradiation of secondary effluent. Water Sci. Technol. 47(9): 193–198 [PubMed] [Google Scholar]
- 175. Jamieson RC, Gordon RJ, Tattrie SC, Stratton GW. 2003. Sources and persistence of fecal coliform bacteria in a rural watershed. Water Qual. Res. J. Can. 38: 33–47 [Google Scholar]
- 176. Jawson MD, Elliott LF, Saxton KE, Fortier DH. 1982. The effect of cattle grazing on indicator bacteria in runoff from a Pacific Northwest watershed. J. Environ. Qual. 11: 621–627 [Google Scholar]
- 177. Jaykus LA. 1996. The application of quantitative risk assessment to microbial food safety risks. Crit. Rev. Microbiol. 22: 279–293 [DOI] [PubMed] [Google Scholar]
- 178. Jeng HC, England AJ, Bradford HB. 2005. Indicator organisms associated with stormwater suspended particles and estuarine sediment. J. Environ. Sci. Health A Toxic Hazard. Subst. Environ. Eng. 40: 779–791 [DOI] [PubMed] [Google Scholar]
- 179. Jeng HC, Sinclair R, Daniels R, Englande AJ. 2005. Survival of Enterococci faecalis in estuarine sediments. Int. J. Environ. Stud. 62: 283–291 [Google Scholar]
- 180. Jenkins MB, Fisher DS, Endale DM, Adams P. 2011. Comparative die-off of Escherichia coli O157:H7 and fecal indicator bacteria in pond water. Environ. Sci. Technol. 45: 1853–1858 [DOI] [PubMed] [Google Scholar]
- 181. Jenkins TM, Scott TM, Morgan MR, Rose JB. 2005. Occurrence of alternative fecal indicators and enteric viruses in Michigan rivers. J. Great Lakes Res. 31: 22–31 [Google Scholar]
- 182. Jimenez B, Chavez A, Maya C, Jardines L. 2001. Removal of microorganisms in different stages of wastewater treatment for Mexico City. Water Sci. Technol. 43(10): 155–162 [PubMed] [Google Scholar]
- 183. Jin G, Englande AJ, Bradford H, Jeng HW. 2004. Comparison of E. coli, enterococci, and fecal coliform as indicators for brackish water quality assessment. Water Environ. Res. 76: 245–255 [DOI] [PubMed] [Google Scholar]
- 184. Johanson JJ, Feriancikova L, Xu S. 2012. Influence of enterococcal surface protein (esp) on the transport of Enterococcus faecium within saturated quartz sands. Environ. Sci. Technol. 46: 1511–1518 [DOI] [PubMed] [Google Scholar]
- 185. Johnson LK, et al. 2004. Sample size, library composition, and genotypic diversity among natural populations of Escherichia coli from different animals influence accuracy of determining sources of fecal pollution. Appl. Environ. Microbiol. 70: 4478–4485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Kay D, et al. 1994. Predicting likelihood of gastroenteritis from sea bathing: results from randomised exposure. Lancet 344: 905–909 [DOI] [PubMed] [Google Scholar]
- 187. Kay D, et al. 2005. Decay of intestinal enterococci concentrations in high-energy estuarine and coastal waters: towards real-time T90 values for modelling faecal indicators in recreational waters. Water Res. 39: 655–667 [DOI] [PubMed] [Google Scholar]
- 188. Kell DB, Kaprelyants AS, Weichart DH, Harwood CR, Barer MR. 1998. Viability and activity in readily culturable bacteria: a review and discussion of the practical issues. Antonie Van Leeuwenhoek 73: 169–187 [DOI] [PubMed] [Google Scholar]
- 189. Kelner A. 1950. Light-induced recovery of microorganisms from ultraviolet radiation injury, with special reference to Escherichia coli. Bull. N. Y. Acad. Med. 26: 189–199 [PMC free article] [PubMed] [Google Scholar]
- 190. Kinzelman J, Pond K, Longmaid K, Bagley R. 2004. The effect of two mechanical beach grooming strategies on Escherichia coli density in beach sand at a southwestern Lake Michigan beach. Aquat. Ecosyst. Health Manage. 7: 425–432 [Google Scholar]
- 191. Kinzelman JL, Whitman RL, Byappanahalli M, Jackson E, Bagley RC. 2003. Evaluation of beach grooming techniques on Escherichia coli density in foreshore sand at North Beach, Racine, WI. Lake Reserv. Manage. 19: 349–354 [Google Scholar]
- 192. Klein G. 2003. Taxonomy, ecology, and antibiotic resistance of enterococci from food and the gastro-intestinal tact. Int. J. Food Microbiol. 88: 123–131 [DOI] [PubMed] [Google Scholar]
- 193. Knee KL, Layton BA, Street JH, Boehm AB, Paytan A. 2008. Sources of nutrients and fecal indicator bacteria to nearshore waters on the north shore of Kaua'i (Hawai'i, USA). Estuar. Coast 31: 607–622 [Google Scholar]
- 194. Koivunen J, Heinonen-Tanski H. 2005. Inactivation of enteric microorganisms with chemical disinfectants, UV irradiation and combined chemical/UV treatments. Water Res. 39: 1519–1526 [DOI] [PubMed] [Google Scholar]
- 195. Koort J, Coenye T, Vandamme P, Sukura A, Björkroth J. 2004. Enterococcus hermanniensis sp. nov., from modified-atmosphere-packaged broiler meat and canine tonsils. Int. J. Syst. Evol. Microbiol. 54: 1823–1827 [DOI] [PubMed] [Google Scholar]
- 196. Korajkic A, Badgley BD, Brownell MJ, Harwood VJ. 2009. Application of microbial source tracking methods in a Gulf of Mexico field setting. J. Appl. Microbiol. 107: 1518–1527 [DOI] [PubMed] [Google Scholar]
- 197. Korajkic A, Brownell MJ, Harwood VJ. 2011. Investigation of human sewage pollution and pathogen analysis at Florida Gulf Coast beaches. J. Appl. Microbiol. 110: 174–183 [DOI] [PubMed] [Google Scholar]
- 198. Lancefield RC. 1933. A serological differentiation of human and other groups of hemolytic streptococci. J. Exp. Med. 57: 571–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Laplace JM, Thuault M, Hartke A, Boutibonnes P, Auffray Y. 1997. Sodium hypochlorite stress in Enterococcus faecalis: influence of antecedent growth conditions and induced proteins. Curr. Microbiol. 34: 284–289 [DOI] [PubMed] [Google Scholar]
- 200. Lasalde C, Rodriguez R, Toranzos GA, Smith HH. 2005. Heterogeneity of uidA gene in environmental Escherichia coli populations. J. Water Health 3: 297–304 [DOI] [PubMed] [Google Scholar]
- 201. Lavender JS, Kinzelman JL. 2009. A cross comparison of qPCR to agar-based or defined substrate test methods for the determination of Escherichia coli and enterococci in municipal water quality monitoring programs. Water Res. 43: 4967–4979 [DOI] [PubMed] [Google Scholar]
- 202. Law-Brown J, Meyers PR. 2003. Enterococcus phoeniculicola sp. nov., a novel member of the enterococci isolated from the uropygial gland of the red-billed woodhoopoe, Phoeniculus purpureus. Int. J. Syst. Evol. Microbiol. 53: 683–685 [DOI] [PubMed] [Google Scholar]
- 203. Layton BA, Walters SP, Lam LH, Boehm AB. 2010. Enterococcus species distribution among human and animal hosts using multiplex PCR. J. Appl. Microbiol. 109: 539–547 [DOI] [PubMed] [Google Scholar]
- 204. Leclerc H, Devriese LA, Mossel DAA. 1996. Taxonomical changes in intestinal (faecal) enterococci and streptococci: consequences on their use as indicators of faecal contamination in drinking water. J. Appl. Bacteriol. 81: 459–466 [DOI] [PubMed] [Google Scholar]
- 205. Lee CM, et al. 2006. Persistence of fecal indicator bacteria in Santa Monica Bay beach sediments. Water Res. 40: 2593–2602 [DOI] [PubMed] [Google Scholar]
- 206. Lee J, Deininger RA. 2004. Detection of E. coli in beach water within 1 hour using immunomagnetic separation and ATP bioluminescence. Luminescence 19: 31–36 [DOI] [PubMed] [Google Scholar]
- 207. Le Fevre NM, Lewis GD. 2003. The role of resuspension in enterococci distribution in water at an urban beach. Water Sci. Technol. 47(3): 205–210 [PubMed] [Google Scholar]
- 208. Lessard EJ, Sieburth JM. 1983. Survival of natural sewage populations of enteric bacteria in diffusion and batch chambers in the marine environment. Appl. Environ. Microbiol. 45: 950–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Liu L, et al. 2006. Modeling the transport and inactivation of E. coli and enterococci in the near-shore region of Lake Michigan. Environ. Sci. Technol. 40: 5022–5028 [DOI] [PubMed] [Google Scholar]
- 210. Lleo MM, Bonato B, Benedetti D, Canepari P. 2005. Survival of enterococcal species in aquatic environments. FEMS Microbiol. Ecol. 54: 189–196 [DOI] [PubMed] [Google Scholar]
- 211. Lleo MM, et al. 2001. Resuscitation rate in different enterococcal species in the viable but non-culturable state. J. Appl. Microbiol. 91: 1095–1102 [DOI] [PubMed] [Google Scholar]
- 212. Lleo MM, Tafi MC, Canepari P. 1998. Nonculturable Enterococcus faecalis cells are metabolically active and capable of resuming active growth. Syst. Appl. Microbiol. 21: 333–339 [DOI] [PubMed] [Google Scholar]
- 213. Lu J, Santo Domingo JW, Lamendella R, Edge T, Hill S. 2008. Phylogenetic diversity and molecular detection of bacteria in gull feces. Appl. Environ. Microbiol. 74: 3969–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Ma L, Kornacki JL, Zhang G, Lin CM, Doyle MP. 2007. Development of thermal surrogate microorganisms in ground beef for in-plant critical control point validation studies. J. Food Prot. 70: 952–957 [DOI] [PubMed] [Google Scholar]
- 215. Maiga Y, Wethe J, Denyigba K, Ouattara AS. 2009. The impact of pond depth and environmental conditions on sunlight inactivation of Escherichia coli and enterococci in wastewater in a warm climate. Can. J. Microbiol. 55: 1364–1374 [DOI] [PubMed] [Google Scholar]
- 216. Malathum K, Singh KV, Weinstock GM, Murray BE. 1998. Repetitive sequence-based PCR versus pulsed-field gel electrophoresis for typing of Enterococcus faecalis at the subspecies level. J. Clin. Microbiol. 36: 211–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Maraccini PA, Ferguson DM, Boehm AB. 2012. Diurnal variation in Enterococcus species composition in polluted ocean water and a potential role for the enterococcal carotenoid in protection against photoinactivation. Appl. Environ. Microbiol. 78: 305–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Martinez-Murcia AJ, Collins MD. 1991. Enterococcus sulfureus, a new yellow-pigmented Enterococcus species. FEMS Microbiol. Lett. 80: 69–73 [DOI] [PubMed] [Google Scholar]
- 219. Mas DML, Ahlfeld DP. 2007. Comparing artificial neural networks and regression models for predicting faecal coliform concentrations. Hydrol. Sci. J. 52: 713–731 [Google Scholar]
- 220. Matz C, Boenigk J, Arndt H, Jurgens K. 2002. Role of bacterial phenotypic traits in selective feeding of the heterotrophic nanoflagellate Spumella sp. Aquat. Microb. Ecol. 27: 137–148 [Google Scholar]
- 221. Maugen TL, Carbone M, Fera MT, Irrera GP, Guliandolo C. 2004. Distribution of potentially pathogenic bacteria as free living and plankton associated in a marine coastal zone. J. Appl. Microbiol. 97: 354–361 [DOI] [PubMed] [Google Scholar]
- 222. Maya C, Beltran N, Jimenez B, Bonilla P. 2003. Evaluation of the UV disinfection process in bacteria and amphizoic amoeba inactivation. Water Sci. Technol. Water Supply 3: 285–291 [Google Scholar]
- 223. Mazaheri Nezhad Fard R, Barton MD, Heuzenroeder MW. 2011. Bacteriophage-mediated transduction of antibiotic resistance in enterococci. Lett. Appl. Microbiol. 52: 559–564 [DOI] [PubMed] [Google Scholar]
- 224. McCambridge J, McMeekin TA. 1980. Effect of temperature on activity of predators of Salmonella typhimurium and Escherichia coli in estuarine water. Aust. J. Mar. Freshw. Res. 31: 851–855 [Google Scholar]
- 225. McCambridge J, McMeekin TA. 1980. Relative effects of bacterial and protozoan predators on survival of Escherichia coli in estuarine water samples. Appl. Environ. Microbiol. 40: 907–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. McFeters GA, Bissonnette GK, Jezeski JJ, Thomson CA, Stuart DG. 1974. Comparative survival of indicator bacteria and enteric pathogens in well water. Appl. Microbiol. 27: 823–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. McQuaig SM, Scott TM, Harwood VJ, Farrah SR, Lukasik JO. 2006. Detection of human-derived fecal pollution in environmental waters by use of a PCR-based human polyomavirus assay. Appl. Environ. Microbiol. 72: 7567–7574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. McQuaig SM, Scott TM, Lukasik JO, Paul JH, Harwood VJ. 2009. Quantification of human polyomaviruses JC virus and BK virus by TaqMan quantitative PCR and comparison to other water quality indicators in water and fecal samples. Appl. Environ. Microbiol. 75: 3379–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Menon P, Billen G, Servais P. 2003. Mortality rates of autochthonous and fecal bacteria in natural aquatic ecosystems. Water Res. 37: 4151–4158 [DOI] [PubMed] [Google Scholar]
- 230. Mitchell R. 1971. Role of predators in the reversal of imbalances in microbial ecosystems. Nature 230: 257–258 [DOI] [PubMed] [Google Scholar]
- 231. Moellering RC., Jr 1992. Emergence of Enterococcus as a significant pathogen. Clin. Infect. Dis. 15: 58–62 [DOI] [PubMed] [Google Scholar]
- 232. Moore DF, et al. 2005. Evaluation of antibiotic resistance analysis and ribotyping for identification of fecal pollution sources in an urban watershed. J. Appl. Microbiol. 99: 618–628 [DOI] [PubMed] [Google Scholar]
- 233. Morrison D, Woodford N, Cookson B. 1997. Enterococci as emerging pathogens of humans, p 89S–99S.In Andrew PW, Mitchell TJ. (ed), The biology of streptococci and enterococci. Blackwell Science, Oxford, United Kingdom [Google Scholar]
- 234. Mote BL, Turner JW, Lipp EK. 2012. Persistence and growth of the fecal indicator bacteria enterococci in detritus and natural estuarine plankton communities. Appl. Environ. Microbiol. 78: 2569–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Muller T, Ulrich A, Ott EM, Muller M. 2001. Identification of plant-associated enterococci. J. Appl. Microbiol. 91: 268–278 [DOI] [PubMed] [Google Scholar]
- 236. Mundt JO. 1963. Occurrence of enterococci on plants in a wild environment. Appl. Microbiol. 11: 141–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Mundt JO. 1961. Occurrence of enterococci: bud, blossom, and soil studies. Appl. Microbiol. 9: 541–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Mundt JO, Coggins JH, Johnson LF. 1962. Growth of Streptococcus faecalis var. liquefaciens on plants. Appl. Microbiol. 10: 552–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Mundt JO, Graham WF. 1968. Streptococcus faecium var. casseliflavus, nov. var. J. Bacteriol. 95: 2005–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Muniesa M, Jofre J, Lucena F. 1999. Occurrence and numbers of bacteriophages and bacterial indicators in faeces of yellow-legged seagull (Larus cachinnans). Lett. Appl. Microbiol. 29: 421–423 [DOI] [PubMed] [Google Scholar]
- 241. Murray BE. 1990. The life and times of the Enterococcus. Clin. Microbiol. Rev. 3: 46–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Nallapareddy SR, Duh R-W, Singh KV, Murray BE. 2002. Molecular typing of selected Enterococcus faecalis isolates: pilot study using multilocus sequence typing and pulsed-field gel electrophoresis. J. Clin. Microbiol. 40: 868–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Naser SM, et al. 2005. Enterococcus canintestini sp. nov., from faecal samples of healthy dogs. Int. J. Syst. Evol. Microbiol. 55: 2177–2182 [DOI] [PubMed] [Google Scholar]
- 244. Nayak BS, Badgley B, Harwood VJ. 2011. Comparison of genotypic and phylogenetic relationships of environmental Enterococcus isolates by BOX-PCR typing and 16S rRNA gene sequencing. Appl. Environ. Microbiol. 77: 5050–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Neely AN, Maley MP. 2000. Survival of enterococci and staphylococci on hospital fabrics and plastics. J. Clin. Microbiol. 38: 724–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Nevers MB, Boehm AB. 2010. Modeling fate and transport of fecal bacteria in surface water, p 165–188.In Sadowsky MJ, Whitman RL. (ed), The fecal bacteria. ASM Press, Washington, DC [Google Scholar]
- 247. Nevers MB, et al. 2009. Geographic relatedness and predictability of Escherichia coli along a peninsular beach complex of Lake Michigan. J. Environ. Qual. 38: 2357–2364 [DOI] [PubMed] [Google Scholar]
- 248. Nevers MB, Whitman RL. 2011. Efficacy of monitoring and empirical predictive modeling at improving public health protection at Chicago beaches. Water Res. 45: 1659–1668 [DOI] [PubMed] [Google Scholar]
- 249. Nevers MB, Whitman RL. 2005. Nowcast modeling of Escherichia coli concentrations at multiple urban beaches of southern Lake Michigan. Water Res. 39: 5250–5260 [DOI] [PubMed] [Google Scholar]
- 250. Nevers MB, Whitman RL. 2010. Policies and practices of beach monitoring in the Great Lakes, USA: a critical review. J. Environ. Monit. 12: 581–590 [DOI] [PubMed] [Google Scholar]
- 251. Newton RJ, Vandewalle J, Borchardt MA, Gorelick MH, McLellan SL. 2011. Lachnospiraceae and Bacteroidales alternative fecal indicators reveal chronic human sewage contamination in an urban harbor. Appl. Environ. Microbiol. 77: 6972–6981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Niewolak S. 1998. Total viable count and concentration of enteric bacteria in bottom sediments from the Czarna Hancza River, Northeast Poland. Pol. J. Environ. Stud. 7: 295–306 [Google Scholar]
- 253. Nilsson JR. 1987. Structural aspects of digestion of Escherichia coli in Tetrahymena. J. Protozool. 34: 1–6 [Google Scholar]
- 254. Noble RT, Blackwood AD, Griffith JF, McGee CD, Weisberg SB. 2010. Comparison of rapid quantitative PCR-based and conventional culture-based methods for enumeration of Enterococcus spp. and Escherichia coli in recreational waters. Appl. Environ. Microbiol. 76: 7437–7443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Noble RT, Lee IM, Schiff KC. 2004. Inactivation of indicator micro-organisms from various sources of faecal contamination in seawater and freshwater. J. Appl. Microbiol. 96: 464–472 [DOI] [PubMed] [Google Scholar]
- 256. Noble RT, Weisberg SB. 2005. A review of technologies for rapid detection of bacteria in recreational waters. J. Water Health 3: 381–392 [DOI] [PubMed] [Google Scholar]
- 257. Nowlan SS, Deibel RH. 1967. Group Q streptococci. I. Ecology, serology, physiology, and relationship to established enterococci. J. Bacteriol. 94: 291–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Obiri-Danso K, Jones K. 2000. Intertidal sediments as reservoirs for hippurate negative campylobacters, salmonellae and faecal indicators in three EU recognised bathing waters in north west England. Water Res. 34: 519–527 [Google Scholar]
- 259. Oliver JD. 2010. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol. Rev. 34: 415–425 [DOI] [PubMed] [Google Scholar]
- 260. Ostrolenk M, Kramer N, Cleverdon RC. 1947. Comparative studies of enterococci and Escherichia coli as indices of pollution. J. Bacteriol. 53: 197–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Ott EM, et al. 2001. Population dynamics and antagonistic potential of enterococci colonizing the phyllosphere of grasses. J. Appl. Microbiol. 91: 54–66 [DOI] [PubMed] [Google Scholar]
- 262. Papadakis JA, Mavridou A, Richardson SC, Lampiri M, Marcelou U. 1997. Bather-related microbial and yeast populations in sand and seawater. Water Res. 31: 799–804 [Google Scholar]
- 263. Pernthaler J. 2005. Predation on prokaryotes in the water column and its ecological implications. Nat. Rev. Microbiol. 3: 537–546 [DOI] [PubMed] [Google Scholar]
- 264. Phillips MC, Solo-Gabriele HM, Piggot AM, Klaus JS, Zhang YJ. 2011. Relationships between sand and water quality at recreational beaches. Water Res. 45: 6763–6769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Piggot AM, Klaus JS, Johnson S, Phillips M, Solo-Gabriele HM. 15 June 2012. Enterococci levels are related to sediment biofilms at recreational beaches in South Florida. Appl. Environ. Microbiol. doi:10.1128/AEM.00603-12. [DOI] [PMC free article] [PubMed]
- 266. Proctor LM, Fuhrman JA. 1990. Viral mortality of marine-bacteria and cyanobacteria. Nature 343: 60–62 [Google Scholar]
- 267. Prüss A. 1998. Review of epidemiological studies on health effects from exposure to recreational water. Int. J. Epidemiol. 27: 1–9 [DOI] [PubMed] [Google Scholar]
- 268. Purnell SE, Ebdon JE, Taylor HD. 2011. Bacteriophage lysis of Enterococcus host strains: a tool for microbial source tracking? Environ. Sci. Technol. 45: 10699–10705 [DOI] [PubMed] [Google Scholar]
- 269. Rahkila R, Johansson P, Säde E, Björkroth J. 2011. Identification of enterococci from broiler products and a broiler processing plant and description of Enterococcus viikiensis sp. nov. Appl. Environ. Microbiol. 77: 196–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Rodrigues U, Collins MD. 1990. Phylogenetic analysis of Streptococcus saccharolyticus based on 16S rRNA sequencing. FEMS Microbiol. Lett. 71: 231–234 [DOI] [PubMed] [Google Scholar]
- 271. Roll BM, Fujioka RS. 1997. Sources of faecal indicator bacteria in a brackish, tropical stream and their impact on recreational water quality. Water Sci. Technol. 35(11-12): 179–186 [Google Scholar]
- 272. Rose JB, Dickson L, Farrah S, Carnahan R. 1996. Removal of pathogenic and indicator microorganisms by a full-scale water reclamation facility. Water Res. 30: 2785–2797 [Google Scholar]
- 273. Rose JB, et al. 2001. Reduction of enteric microorganisms at the Upper Occoquan Sewage Authority Water Reclamation Plant. Water Environ. Res. 73: 711–720 [DOI] [PubMed] [Google Scholar]
- 274. Roslev P, Bastholm S, Iverson N. 2008. Relationship between fecal indicators in sediment and recreational waters in a Danish estuary. Water Air Soil Pollut. 194: 13–21 [Google Scholar]
- 275. Roslev P, Bukh AS. 2011. State of the art molecular markers for fecal pollution source tracking in water. Appl. Microbiol. Biotechnol. 89: 1341–1355 [DOI] [PubMed] [Google Scholar]
- 276. Ruiz-Garbajosa P, et al. 2006. Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J. Clin. Microbiol. 44: 2220–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277. Sadowsky MJ, Whitman RL. (ed). 2010. The fecal bacteria. ASM Press, Washington, DC [Google Scholar]
- 278. Sanchez PS, Agudo EG, Castro FG, Alves MN, Martins MT. 1986. Evaluation of the sanitary quality of marine recreational waters and sands from beaches of the Sao Paulo State, Brazil. Water Sci. Technol. 18(10): 61–72 [Google Scholar]
- 279. Santiago-Rodríguez TM, et al. 2010. Characterization of Enterococcus faecalis-infecting phages (enterophages) as markers of human fecal pollution in recreational waters. Water Res. 44: 4716–4725 [DOI] [PubMed] [Google Scholar]
- 280. Sassoubre LM, Walters SP, Russell TL, Boehm AB. 2011. Sources and fate of Salmonella and fecal indicator bacteria in an urban creek. J. Environ. Monit. 13: 2206–2212 [DOI] [PubMed] [Google Scholar]
- 281. Savichtcheva O, Okabe S. 2006. Alternative indicators of fecal pollution: relations with pathogens and conventional indicators, current methodologies for direct pathogen monitoring and future application perspectives. Water Res. 40: 2463–2476 [DOI] [PubMed] [Google Scholar]
- 282. Sayler GS, Nelson J, Justice A, Colwell RR. 1975. Distribution and significance of fecal indicator organisms in the Upper Chesapeake Bay. Appl. Microbiol. 30: 625–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283. Schleifer KH, Kilpper-Bälz R. 1984. Transfer of Streptococcus faecalis and Streptococcus faecium to the genus Enterococcus nom. rev. as Enterococcus faecalis comb. nov. and Enterococcus faecium comb. nov. Int. J. Syst. Bacteriol. 34: 31–34 [Google Scholar]
- 284. Schoen ME, Ashbolt NJ. 2010. Assessing pathogen risk to swimmers at non-sewage impacted recreational beaches. Environ. Sci. Technol. 44: 2286–2291 [DOI] [PubMed] [Google Scholar]
- 285. Schoen ME, Soller JA, Ashbolt NJ. 2011. Evaluating the importance of faecal sources in human-impacted waters. Water Res. 45: 2670–2680 [DOI] [PubMed] [Google Scholar]
- 286. Schultz-Fademrecht C, Wichern M, Horn H. 2008. The impact of sunlight on inactivation of indicator microorganisms both in river water and benthic biofilms. Water Res. 42: 4771–4779 [DOI] [PubMed] [Google Scholar]
- 287. Scott TM, Jenkins TM, Lukasik J, Rose JB. 2005. Potential use of a host associated molecular marker in Enterococcus faecium as an index of human fecal pollution. Environ. Sci. Technol. 39: 283–287 [PubMed] [Google Scholar]
- 288. Scott TM, Rose JB, Jenkins TM, Farrah SR, Lukasik J. 2002. Microbial source tracking: current methodology and future directions. Appl. Environ. Microbiol. 68: 5796–5803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289. Servais P, Garcia-Armisen T, George I, Billen G. 2007. Fecal bacteria in the rivers of the Seine drainage network (France): sources, fate and modeling. Sci. Total Environ. 375: 152–167 [DOI] [PubMed] [Google Scholar]
- 290. Shah AH, et al. 2011. Indicator microbes correlate with pathogenic bacteria, yeasts and helminthes in sand at a subtropical recreational beach site. J. Appl. Microbiol. 110: 1571–1583 [DOI] [PubMed] [Google Scholar]
- 291. Shanks OC, et al. 2008. Quantitative PCR for detection and enumeration of genetic markers of bovine fecal pollution. Appl. Environ. Microbiol. 74: 745–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Shanks OC, Kelty CA, Sivaganesan M, Varma M, Haugland RA. 2009. Quantitative PCR for genetic markers of human fecal pollution. Appl. Environ. Microbiol. 75: 5507–5513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293. Sherman JM. 1937. The streptococci. Bacteriol. Rev. 1: 3–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294. Sherr BF, Sherr EB, Rassoulzadegan F. 1988. Rates of digestion of bacteria by marine phagotrophic protozoa: temperature dependence. Appl. Environ. Microbiol. 54: 1091–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. Shewmaker PL, et al. 2011. Reevaluation of the taxonomic status of recently described species of Enterococcus: evidence that E. thailandicus is a senior subjective synonym of “E. sanguinicola” and confirmation of E. caccae as a species distinct from E. silesiacus. J. Clin. Microbiol. 49: 2676–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296. Shiaris MP, et al. 1987. Distribution of indicator bacteria and Vibrio parahaemolyticus in sewage-polluted intertidal sediments. Appl. Environ. Microbiol. 53: 1756–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297. Shibata T, Solo-Gabriele HM, Fleming LE, Elmir S. 2004. Monitoring marine recreational water quality using multiple microbial indicators in an urban tropical environment. Water Res. 38: 3119–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298. Shibata T, et al. 2010. Evaluation of conventional and alternative monitoring methods for a recreational marine beach with nonpoint source of fecal contamination. Environ. Sci. Technol. 44: 8175–8181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299. Signoretto C, et al. 2004. Adhesion of Enterococcus faecalis in the nonculturable state to plankton is the main mechanism responsible for persistence of this bacterium in both lake and seawater. Appl. Environ. Microbiol. 70: 6892–6896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300. Simek K, Vrba J, Hartman P. 1994. Size-selective feeding by Cyclidium sp on bacterioplankton and various sizes of cultured bacteria. FEMS Microbiol. Ecol. 14: 157–167 [Google Scholar]
- 301. Simpson D, Jacangelo J, Loughran P, McIlroy C. 2003. Investigation of potential surrogate organisms and public health risk in UV irradiated secondary effluent. Water Sci. Technol. 47(9): 37–43 [PubMed] [Google Scholar]
- 302. Sinclair JL, Alexander M. 1984. Role of resistance to starvation in bacterial survival in sewage and lake water. Appl. Environ. Microbiol. 48: 410–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303. Sinclair RG, Rose JB, Hasham SA, Gerba CP, Haas CN. 2012. Criteria for selection of surrogates used to study the fate and control of pathogens in the environment. Appl. Environ. Microbiol. 78: 1969–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304. Sinigalliano CD, et al. 2010. Traditional and molecular analyses for fecal indicator bacteria in nonpoint source subtropical recreational marine waters. Water Res. 44: 3763–3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305. Sinton LW, Braithwaite RR, Hall CH, Mackenzie ML. 2007. Survival of indicator and pathogenic bacteria in bovine feces on pasture. Appl. Environ. Microbiol. 73: 7917–7925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306. Sinton LW, Davies-Colley RJ, Bell RG. 1994. Inactivation of enterococci and fecal coliforms from sewage and meatworks effluents in seawater chambers. Appl. Environ. Microbiol. 60: 2040–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307. Sinton LW, Finlay RK, Lynch PA. 1999. Sunlight inactivation of fecal bacteriophages and bacteria in sewage-polluted seawater. Appl. Environ. Microbiol. 65: 3605–3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308. Sinton LW, Hall CH, Lynch PA, Davies-Colley RJ. 2002. Sunlight inactivation of fecal indicator bacteria and bacteriophages from waste stabilization pond effluent in fresh and saline waters. Appl. Environ. Microbiol. 68: 1122–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309. Soller JA, Schoen ME, Bartrand T, Ravenscroft JE, Ashbolt NJ. 2010. Estimated human health risks from exposure to recreational waters impacted by human and non-human sources of faecal contamination. Water Res. 44: 4674–4691 [DOI] [PubMed] [Google Scholar]
- 310. Sorensen J. 1997. The rhizosphere as a habitat for soil microorganisms, p 21–45.In van Elsas JD, Trevors JT, Wellington EMH. (ed), Modern soil microbiology. Marcel Dekker, Inc, New York, NY [Google Scholar]
- 311. Soule M, Kuhn E, Loge F, Gay J, Call DR. 2006. Using DNA microarrays to identify library-independent markers for bacterial source tracking. Appl. Environ. Microbiol. 72: 1843–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312. Steets BM, Holden PA. 2003. A mechanistic model of runoff-associated fecal coliform fate and transport through a coastal lagoon. Water Res. 37: 589–608 [DOI] [PubMed] [Google Scholar]
- 313. Stewart JR, et al. 2008. The coastal environment and human health: microbial indicators, pathogens, sentinels and reservoirs. Environ. Health 7(Suppl 2): S3 doi:10.1186/1476-069x-7-S2-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314. Stidson RT, Gray CA, McPhail CD. 2012. Development and use of modelling techniques for real-time bathing water quality predictions. Water Environ. J. 26: 7–18 [Google Scholar]
- 315. Stoeckel DM, Harwood VJ. 2007. Performance, design, and analysis in microbial source tracking studies. Appl. Environ. Microbiol. 73: 2405–2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316. Sukontasing S, Tanasupawat S, Moonmangmee S, Lee J-S, Suzuki K-I. 2007. Enterococcus camelliae sp. nov., isolated from fermented tea leaves in Thailand. Int. J. Syst. Evol. Microbiol. 57: 2151–2154 [DOI] [PubMed] [Google Scholar]
- 317. Surbeck CQ, Jiang SC, Grant SB. 2010. Ecological control of fecal indicator bacteria in an urban stream. Environ. Sci. Technol. 44: 631–637 [DOI] [PubMed] [Google Scholar]
- 318. Süß J, Volz S, Obst U, Schwartz T. 2009. Application of a molecular biology concept for the detection of DNA damage and repair during UV disinfection. Water Res. 43: 3705–3716 [DOI] [PubMed] [Google Scholar]
- 319. Sutton DC, Besant PJ. 1994. Ecology and characteristics of bdellovibrios from 3 tropical marine habitats. Mar. Biol. 119: 313–320 [Google Scholar]
- 320. Švec P, et al. 2001. Enterococcus haemoperoxidus sp. nov. and Enterococcus moraviensis sp. nov. isolated from water. Int. J. Syst. Evol. Microbiol. 51: 1567–1574 [DOI] [PubMed] [Google Scholar]
- 321. Švec P, et al. 2005. Enterococcus aquimarinus sp. nov., isolated from sea water. Int. J. Syst. Evol. Microbiol. 55: 2183–2187 [DOI] [PubMed] [Google Scholar]
- 322. Švec P, et al. 2005. Enterococcus devriesei sp. nov., associated with animal sources. Int. J. Syst. Evol. Microbiol. 55: 2479–2484 [DOI] [PubMed] [Google Scholar]
- 323. Švec P, et al. 2006. Enterococcus silesiacus sp. nov. and Enterococcus termitis sp. nov. Int. J. Syst. Evol. Microbiol. 56: 577–581 [DOI] [PubMed] [Google Scholar]
- 324. Teixeira LM, et al. 2001. Enterococcus porcinus sp. nov. and Enterococcus ratti sp. nov., associated with enteric disorders in animals. Int. J. Syst. Evol. Microbiol. 51: 1737–1743 [DOI] [PubMed] [Google Scholar]
- 325. Tendolkar PM, Baghdayan AS, Shankar N. 2003. Pathogenic enterococci: new developments in the 21st century. Cell. Mol. Life Sci. 60: 2622–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326. Tenover FC, Arbeit RD, Goering RV. 1997. How to select and interpret molecular strain typing methods for epidemiological studies of bacterial infections: a review for healthcare epidemiologists. Infect. Control Hosp. Epidemiol. 18: 426–439 [DOI] [PubMed] [Google Scholar]
- 327. Thingstad TF. 2000. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol. Oceanogr. 45: 1320–1328 [Google Scholar]
- 328. Tree JA, Adams MR, Lees DN. 2003. Chlorination of indicator bacteria and viruses in primary sewage effluent. Appl. Environ. Microbiol. 69: 2038–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329. Tseng LY, Jiang SC. 2012. Comparison of recreational health risks associated with surfing and swimming in dry weather and post-storm conditions at southern California beaches using quantitative microbial risk assessment (QMRA). Mar. Pollut. Bull. 64: 912–918 [DOI] [PubMed] [Google Scholar]
- 330. Tyrrell GJ, et al. 2002. Enterococcus gilvus sp. nov. and Enterococcus pallens sp. nov. isolated from human clinical specimens. J. Clin. Microbiol. 40: 1140–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331. Ufnar JA, Wang S, Ufnar D, Ellender RD. 2007. Methanobrevibacter ruminantium as an indicator of domesticated-ruminant fecal pollution in surface waters. Appl. Environ. Microbiol. 73: 7118–7121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332. US Environmental Protection Agency 1999. Action plan for beaches and recreational waters. EPA/600/R-98/079. US Environmental Protection Agency, Washington, DC [Google Scholar]
- 333. US Environmental Protection Agency 1986. Ambient water quality criteria for bacteria 1986. EPA 440/5-84-002 Office of Water Regulations and Standards, US Environmental Protection Agency, Washington, DC [Google Scholar]
- 334. US Environmental Protection Agency 2005. Detecting and mitigating environmental impact of fecal pathogens originating from confined animal feeding operations: rev EPA/600/R-06/ 021 US Environmental Protection Agency, Cincinnati, OH [Google Scholar]
- 335. US Environmental Protection Agency 2004. Implementation guidance for ambient water quality criteria for bacteria. EPA-823-B-04-002. US Environmental Protection Agency, Washington, DC [Google Scholar]
- 336. US Environmental Protection Agency 2002. Method 1600—enterococci in water by membrane filtration using membrane-Enterococcus indoxyl-B-D-glucoside agar (mEI). EPA-821-R-02-022. US Environmental Protection Agency, Washington, DC [Google Scholar]
- 337. US Environmental Protection Agency 2005. Microbial source tracking guide document. EPA/600/R-05/064. US Environmental Protection Agency, Washington, DC [Google Scholar]
- 338. US Environmental Protection Agency 2006. Ultraviolet disinfection guidance manual for the final long term 2 enhanced surface water treatment rule. EPA 815-R-06-007. US Environmental Protection Agency, Washington, DC [Google Scholar]
- 339. Vancanneyt M, et al. 2001. Enterococcus villorum sp. nov., an enteroadherent bacterium associated with diarrhoea in piglets. Int. J. Syst. Evol. Microbiol. 51: 393–400 [DOI] [PubMed] [Google Scholar]
- 340. van Donsel DJ, Geldreich EE, Clarke NA. 1967. Seasonal variations in survival of indicator bacteria in soil and their contribution to storm-water pollution. Appl. Microbiol. 15: 1362–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341. Venieri D, Chatzisymeon E, Gonzalo MS, Rosal R, Mantzavinos D. 2011. Inactivation of Enterococcus faecalis by TiO2-mediated UV and solar irradiation in water and wastewater: culture techniques never say the whole truth. Photochem. Photobiol. Sci. 10: 1744–1750 [DOI] [PubMed] [Google Scholar]
- 342. Verhougstraete MP, Byappanahalli MN, Whitman RL, Rose JB. 2010. Cladophora in the Great Lakes: impacts on beach water quality and human health. Water Sci. Technol. 62(1): 68–76 [DOI] [PubMed] [Google Scholar]
- 343. Verity PG. 1991. Feeding in planktonic protozoans—evidence for nonrandom acquisition of prey. J. Protozool. 38: 69–76 [Google Scholar]
- 344. Viau EJ, et al. 2011. Bacterial pathogens in Hawaiian coastal streams—associations with fecal indicators, land cover, and water quality. Water Res. 45: 3279–3290 [DOI] [PubMed] [Google Scholar]
- 345. Wade TJ, et al. 2008. High sensitivity of children to swimming-associated gastrointestinal illness: results using a rapid assay of recreational water quality. Epidemiology 19: 375–383 [DOI] [PubMed] [Google Scholar]
- 346. Wade TJ, et al. 2006. Rapidly measured indicators of recreational water quality are predictive of swimming-associated gastrointestinal illness. Environ. Health Perspect. 114: 24–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347. Wade TJ, Pai N, Eisenberg JN, Colford JM. 2003. Do US Environmental Protection Agency water quality guidelines for recreational waters prevent gastrointestinal illness? A systematic review and meta-analysis. Environ. Health Perspect. 111: 1102–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348. Wade TJ, et al. 2010. Rapidly measured indicators of recreational water quality and swimming-associated illness at marine beaches: a prospective cohort study. Environ. Health 9: 66 doi:10.1186/1476-069x-9-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349. Walker TS, Bais HP, Grotewold E, Vivanco JM. 2003. Root exudation and rhizosphere biology. Plant Physiol. 132: 44–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350. Water Environment Research Foundation 2009. Experts scientific workshop on critical research and science needs for the development of recreational water quality criteria in inland waters, Dallas-Ft Worth, TX, 18 to 20 February 2009. Water Environment Research Foundation, Alexandria, VA [Google Scholar]
- 351. Weidhaas JL, Macbeth TW, Olsen RL, Harwood VJ. 2011. Correlation of quantitative PCR for a poultry-specific Brevibacterium marker gene with bacterial and chemical indicators of water pollution in a watershed impacted by land application of poultry litter. Appl. Environ. Microbiol. 77: 2094–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352. Weidhaas JL, et al. 2010. Identification of a Brevibacterium marker gene specific to poultry litter and development of a quantitative PCR assay. J. Appl. Microbiol. 109: 334–347 [DOI] [PubMed] [Google Scholar]
- 353. Whitman RL, et al. 2010. Relationship and variation of qPCR and culturable enterococci in ambient surface waters are predictable. Environ. Sci. Technol. 44: 5049–5054 [DOI] [PubMed] [Google Scholar]
- 354. Whitman RL, Nevers MB. 2004. Escherichia coli sampling reliability at a frequently closed Chicago beach: monitoring and management implications. Environ. Sci. Technol. 38: 4241–4246 [DOI] [PubMed] [Google Scholar]
- 355. Whitman RL, Nevers MB. 2003. Foreshore sand as a source of Escherichia coli in nearshore water of a Lake Michigan beach. Appl. Environ. Microbiol. 69: 5555–5562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356. Whitman RL, Nevers MB, Byappanahalli MN. 2006. Watershed-wide distribution of Escherichia coli along southern Lake Michigan: an integrated approach. Appl. Environ. Microbiol. 72: 7301–7310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357. Whitman RL, Przybyla-Kelly K, Shively DA, Byappanahalli MN. 2007. Incidence of the enterococcal surface protein gene in human and animal fecal sources. Environ. Sci. Technol. 41: 6090–6095 [DOI] [PubMed] [Google Scholar]
- 358. Whitman RL, Shively DA, Pawlik H, Nevers MB, Byappanahalli MN. 2003. Occurrence of Escherichia coli and enterococci in Cladophora (Chlorophyta) in nearshore water and beach sand of Lake Michigan. Appl. Environ. Microbiol. 69: 4714–4719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359. Wiggins BA. 1996. Discriminant analysis of antibiotic resistance patterns in fecal streptococci, a method to differentiate human and animal sources of fecal pollution in natural waters. Appl. Environ. Microbiol. 62: 3997–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360. Williams AM, Farrow JAE, Collins MD. 1989. Reverse transcriptase sequencing of 16S ribosomal RNA from Streptococcus cecorum. Lett. Appl. Microbiol. 8: 185–189 [Google Scholar]
- 361. Winfield MD, Groisman EA. 2003. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl. Environ. Microbiol. 69: 3687–3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362. Wolf HW. 1972. The coliform count as a measure of water quality, p 333–345.In Mitchell R. (ed), Water pollution microbiology. Wiley Interscience, New York, NY [Google Scholar]
- 363. World Health Organization 2003. Microbial aspects of beach sand quality, p 118–126.In World Health Organization (ed), Guidelines for safe recreational water environments, vol 1. Coastal and fresh waters World Health Organization, Geneva, Switzerland: http://whqlibdoc.who.int/publications/2003/9241545801.pdf. [Google Scholar]
- 364. Wright ME, Solo-Gabriele HM, Elmir S, Fleming LE. 2009. Microbial load from animal feces at a recreational beach. Mar. Pollut. Bull. 58: 1649–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365. Wuertz S, Wang D, Reischer GH, Farnleitner AH. 2011. Library-independent bacterial source tracking methods, p 61–112.In Hagedorn C, Blanch AR, Harwood VJ. (ed), Microbial source tracking: methods, applications, and case studies. Springer, New York, NY [Google Scholar]
- 366. Yamahara KM, Layton BA, Santoro AE, Boehm AB. 2007. Beach sands along the California coast are diffuse sources of fecal bacteria to coastal waters. Environ. Sci. Technol. 41: 4515–4521 [DOI] [PubMed] [Google Scholar]
- 367. Yamahara KM, Walters SP, Boehm AB. 2009. Growth of enterococci in unaltered, unseeded sands subjected to tidal wetting. Appl. Environ. Microbiol. 75: 1517–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368. Zanetti F, De Luca G, Sacchetti R, Stampi S. 2007. Disinfection efficiency of peracetic acid (PAA): inactivation of coliphages and bacterial indicators in a municipal wastewater plant. Environ. Technol. 28: 1265–1271 [DOI] [PubMed] [Google Scholar]