Abstract
Summary: Pneumococcal infections cause up to 2 million deaths annually and raise a large economic burden and thus constitute an important threat to mankind. Because of the increase in the antibiotic resistance of Streptococcus pneumoniae clinical isolates, there is an urgent need to find new antimicrobial approaches to triumph over pneumococcal infections. Toxin-antitoxin (TA) systems (TAS), which are present in most living bacteria but not in eukaryotes, have been proposed as an effective strategy to combat bacterial infections. Type II TAS comprise a stable toxin and a labile antitoxin that form an innocuous TA complex under normal conditions. Under stress conditions, TA synthesis will be triggered, resulting in the degradation of the labile antitoxin and the release of the toxin protein, which would poison the host cells. The three functional chromosomal TAS from S. pneumoniae that have been studied as well as their molecular characteristics are discussed in detail in this review. Furthermore, a meticulous bioinformatics search has been performed for 48 pneumococcal genomes that are found in public databases, and more putative TAS, homologous to well-characterized ones, have been revealed. Strikingly, several unusual putative TAS, in terms of components and genetic organizations previously not envisaged, have been discovered and are further discussed. Previously, we reported a novel finding in which a unique pneumococcal DNA signature, the BOX element, affected the regulation of the pneumococcal yefM-yoeB TAS. This BOX element has also been found in some of the other pneumococcal TAS. In this review, we also discuss possible relationships between some of the pneumococcal TAS with pathogenicity, competence, biofilm formation, persistence, and an interesting phenomenon called bistability.
INTRODUCTION
Among diseases caused by bacterial pathogens, those originating from Gram-positive (G+) bacteria, particularly Streptococcus pneumoniae, Staphylococcus aureus, and Enterococcus faecalis, pose a major threat to the human population because of their high rates of morbidity and mortality, economic impact, role as reservoirs of antibiotic resistance (3), and virulence genes that are horizontally spread (6, 143). These three bacteria cause many nosocomial diseases, show elevated levels of resistance to antibiotics, and play a key role in the spread of resistance (76).
S. pneumoniae (the pneumococcus) is the causal agent of pneumococcal pneumonia, of more than 50% of meningitis cases, and also of sepsis, otitis media, and other less serious infections. Pneumococcal infections are a major cause of mortality worldwide, with recent estimates that pneumonia kills approximately 1.4 million children under the age of 5 years annually, more than AIDS, malaria, and tuberculosis combined (http://www.who.int/mediacentre/factsheets/fs331/en/index.html) (100). In the United States and Europe, the annual incidence of invasive pneumococcal disease ranges from 10 to 100 cases per 100,000 population (http://www.who.int/ith/diseases/pneumococcal/en/index.html). Infections by S. pneumoniae in these countries are the most common causes of community-acquired bacterial pneumonia in adults. The rate of carriage of the bacteria varies between 5% and 70% depending upon age, geographic area, and socioeconomic conditions (14). In general, newly acquired pneumococcal colonization of the nasopharynx of healthy individuals is not associated with the development of disease. However, it is a source of the constant presence of the bacteria within the human population, which in turn would result in continued transmission. It is when the host immunological system is compromised or when a more virulent variant is acquired that mild to serious diseases would appear (112).
Understandably enough, there is an urgent need to find novel targets to tackle pneumococcal infections. Several strategies, not mutually exclusive, have been proposed for dealing with pneumococcal infections. First, the development of new antibiotics (http://www.who.int/bulletin/volumes/89/2/11-030211/en/index.html) or the employment of rotation schemes for the use of known antibiotics has been proposed as a measure to prevent the spread of resistance, the idea being that the alleviation of the selective pressure for a particular antibiotic will lead to the elimination of antibiotic-resistant bacteria from the environment (4). However, studies on the fitness of antibiotic-resistant bacteria indicated that when the numbers of resistant cells increase above a threshold level, they are likely to remain there for a long time (5). In addition, the pneumococcus is naturally transformable with exogenous DNA, and horizontal DNA transfer among closely related streptococci is evident from the mosaic structure of certain virulence factors, such as the choline- and penicillin-binding proteins (106). Thus, the therapy of pneumococcal diseases is hampered by the increasing prevalence of antibiotic-resistant strains and the suboptimal clinical efficacy of the available vaccines. Vaccination has greatly reduced the number of pneumococcal infections, but it has also led to the selection of serotypes for which there are no available vaccines (serotype replacement). In addition to that, the numbers of clinical isolates of S. pneumoniae that exhibit antibiotic resistances have also increased during the past decade (55). Other proposed strategies include the targeting of key virulence factors (101), the inhibition of gene spread by the discovery of conjugation inhibitors (50), bacteriophage therapy, and the use of genomic approaches to identify genes that are essential for microbial survival or virulence. Now that the sequences of a large number of pneumococcal genomes are completed or are under way, it will be possible to search for genes conserved among these bacterial isolates and relate them to other bacteria that can survive in similar sites within the human microbiome: these might represent potential targets for antimicrobials. Furthermore, the profiling of a large cohort of patients based on their metagenome will facilitate the findings of the functional compositions of microbiomes within the human populations and, in turn, will help to find associations between healthy and diseased individuals, to find patterns in host-microbe interactions and immune system responses.
The use of naturally occurring peptide antibiotics or peptide-based inhibitors as design templates for the synthesis of compounds with similar physiochemical properties is also an approach used in pharmaceutical research (7). Finally, the employment of molecules designed to take advantage of the choline-containing cell wall of pneumococcal cells has been used to develop pneumococcus-specific inhibitors of cell growth by mimicking the specific feature of the presentation of choline residues in the cell wall. Molecules termed dendrimers have been successfully employed to develop inhibitors of pneumococcal cell wall hydrolysis, yielding very interesting antipneumococcal molecules (71).
The use of the toxin proteins from the toxin-antitoxin (TA) systems (TAS) as potential targets for the development of new antibacterials has also been proposed (4, 114), since TAS are found in a large number of prokaryotes but not in eukaryotes (90, 122). TA pairs are usually organized as a single operon of two genes: the toxin gene encodes a relatively stable protein, whereas the antitoxin gene either encodes a labile antitoxin protein or is transcribed but remains untranslated as RNA (59, 61). Under steady-state conditions, the toxin is neutralized by the antitoxin, generating a harmless complex which autoregulates their synthesis. However, under certain circumstances, such as nutritional stress, one or more TA operons will be triggered, in which the antitoxin, which is more labile, will be degraded more rapidly, leaving the toxin protein to act on its cellular target. Genes encoding chromosomal TA pairs are also potential targets for the development of drugs that would interfere with the ability of pneumococci to adapt to those stresses (150).
In this review, we focus on the TA genes of S. pneumoniae because they constitute a fairly unexplored world: only three pneumococcal TA pairs, namely, RelBE2, PezAT, and YefM-YoeB, have been studied so far, and only the PezAT TA pair has been characterized in terms of structure and function (21, 82, 113, 117). Surprisingly enough, an in-depth study of possible TAS present in the genomes of pneumococcal strains that have been sequenced showed that their number is far larger than envisaged (90), and our results presented here show that their number can be even larger. Furthermore, the highly recombinogenic nature of this bacterium has apparently led to novel putative TAS with genetic organizations not previously reported (see below).
BACTERIAL TAS: WHAT ARE THEY REALLY FOR?
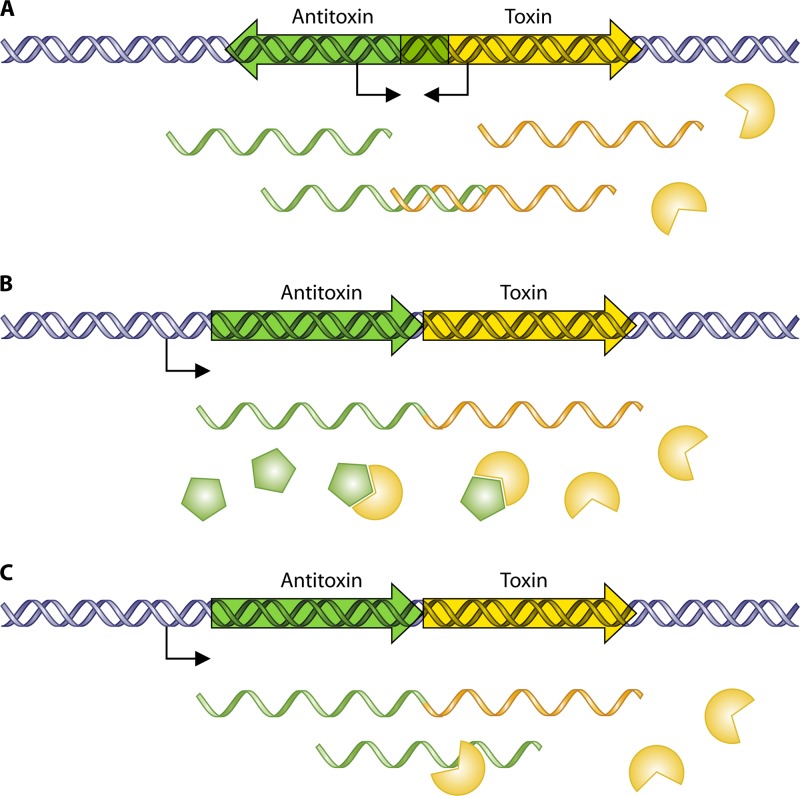
TAS can be broadly classified into three types (schematically depicted in Fig. 1). In type I systems, the antitoxin is an antisense RNA that prevents the translation of the toxin by binding to its complementary toxin mRNA (Fig. 1A). Different variations on the theme can also exist, as shown by recent reviews (52, 61). In type II (proteic) systems, both toxin and antitoxin proteins will be translated, and the antitoxin protein neutralizes the toxin by forming a tight innocuous complex with the toxin protein (Fig. 1B) (68). A type III TA was recently described, whereby the antitoxin RNA inactivates the toxin protein by direct RNA-protein interactions (Fig. 1C) (51).
Fig 1.
Classification of TAS. (A) Type I TAS. Several genetic organizations of this type have been reported (52, 61), and one of the examples is shown here: the two genes encoding the antitoxin and the toxin are organized in opposite orientations, and they are transcribed individually from their own promoters. The antitoxin is an antisense mRNA which prevents the translation of the toxin (a protein). (B) Type II (proteic) TAS. Both TA genes are arranged as an operon, with the antitoxin gene usually preceding the toxin gene. The two genes encoding the TAS are cotranscribed as a single mRNA transcript from the promoter(s), which is placed upstream of the antitoxin gene and subsequently translated. The antitoxins counteract the toxic effect of the cognate toxins by binding avidly to the toxin protein, thus neutralizing the toxic effect. (C) Type III TAS. The organization of both TA genes is the same as that of type II TAS, but instead of protein-protein interactions, the toxin protein is inactivated by the antitoxin mRNA. Green indicates the antitoxin components, whereas beige indicates the toxin components.
The last decade has seen a tremendous increase in publications and citations of TAS, as these systems have been associated with many important bacterial processes. TAS were initially reported to exist on plasmids, where they function to mediate plasmid maintenance via postsegregational killing (15, 16, 38, 60), although more subtle processes, like the coupling of plasmid replication and maintenance, have been shown for the plasmid R1-harbored kis-kid TAS (97). However, when TA genes are chromosomally carried, their function seems to be varied and debatable. Chromosomal TAS were shown, at least, to be involved (i) in the stress response (27), (ii) in programmed cell death (44), (iii) in persistence and antibiotic tolerance (92, 108), (iv) as antiaddiction modules (131), (v) as phage abortive infection systems (51), and (vi) in the maintenance of integrative conjugative elements (152), a function which mirrors that of their plasmid-borne counterparts. Perhaps, the function of the chromosomal TA genes cannot be generalized, as these would likely depend on where the locus resides in the genome, its mode of action, and whether it imparts a selective advantage to its host.
A number of cellular targets for the type II toxins have been elucidated, and intriguingly, most of these toxins function as translational inhibitors, with the majority acting as endoribonucleases, and are termed mRNA interferases (69, 154). The TAS mRNA interferases can be classified into two groups: (i) the ribosome-dependent mRNA interferases, which cleave mRNAs at the ribosomal A site and are typified by the RelE toxin (125), and (ii) the ribosome-independent mRNA interferases, which are typified by the MazF toxin (110, 155, 159). Other toxins, such as Doc and RatA, inhibit translation through binding with the 30S and 50S ribosomal subunits, respectively (96, 158), whereas HipA was suggested to inhibit translation through the phosphorylation of elongation factor Tu (134), and the VapC toxin specifically cleaves within the stem-loop region of the initiator tRNAfMet (151). Other targets of type II toxins include DNA gyrase (12, 31, 34, 145), topoisomerase I (73), cytoskeletal proteins (139), and cell wall assembly via peptidoglycan synthesis (113).
Bioinformatics approaches indicated that TAS are abundant in both bacterial and archaeal genomes. Some bacteria, like Mycobacterium tuberculosis, harbor at least 80 TAS in their genomes (126), while Escherichia coli K-12 harbors at least 36, at the last count (155). The numbers of type II TAS were also found to vary between isolates from the same species, indicating that they are likely to move from one genome to another via horizontal gene transfer (90, 122). The presence of so many TAS in a single bacterium is ambiguous, and whether they cross talk remains to be elucidated, although recent reports suggested the possibility of a coordinated regulation of a network of TAS under various stress conditions (155, 156, 160).
The last few years have seen an explosion in the number of TAS that have been discovered. Six years ago, type II TA pairs could be classified into eight toxin families (58), and later on, the number of toxin families was increased to 10 (146). A more recent thorough bioinformatics search resulted in the discovery of many more novel toxins and antitoxins, a few of which were experimentally validated by using a simple killing-rescue assay; i.e., the expression of the putative toxins led to growth inhibition, while the coexpression of the putative cognate antitoxins restored normal growth (90). In a more recent paper, type II toxins were classified into 12 toxin superfamilies, whereas antitoxins were grouped into 20 superfamilies. These toxin and antitoxin families were observed to have originated from distinct ancestors and have likely been assembled multiple times during the course of evolution, indicating that they might be selfish DNA sequences, as suggested by Leplae et al. (90).
Chromosomal TAS of Gram-negative (G−) bacteria, particularly those of E. coli, have been well studied. In the E. coli K-12 genome, there are at least 11 type II TA loci that have been functionally characterized, namely, relBE (64), dinJ-yafQ (107), yefM-yoeB (23), prlF-yhaV (133), mazEF (2), chpSB (103), hicAB (77), hipBA (86), yafNO, higBA (ygjNM), and ygiUT (mqsR-mqsA) (28). Even though TAS of G+ bacteria are also widely distributed and have been found in both chromosomes and plasmids, they are comparatively less well characterized. For instance, some of the well-studied TA genes of G− bacteria have also been discovered in G+ bacteria, such as mazEF in S. aureus (40) and relBE and yefM-yoeB in S. pneumoniae (117). Nevertheless, there are cases in which specific TAS seem to play different roles in G+ and in G− bacteria: the epsilon-zeta TA cassette stabilizes plasmids in E. coli less efficiently than in Bacillus subtilis. Moreover, the expression of the Zeta toxin was shown to be bactericidal for B. subtilis but bacteriostatic for E. coli (161).
TAS IN S. PNEUMONIAE: SO FEW
The number of TAS reported to exist in S. pneumoniae was relatively small, with at most five in strain R6 and six in virulent strain TIGR4, when they were first reported in 2005 to 2006 (117, 122). About 4 years later, the number was increased to up to eight TAS in the genome of S. pneumoniae (118). However, as we discuss below, these figures are an underestimation of the number and, most importantly, the complexity of the pneumococcal TAS. Data mining of the increasing number of pneumococcal genomes available has shown that the number of type II TAS in S. pneumoniae is indeed large. In addition to the well-studied two-component (toxin and antitoxin) TAS, namely, relBE, yefM-yoeB, and pezAT, we have found here an intriguing number of the so-called three-component TAS and even more complex organizations (see below). Furthermore, in the case of S. pneumoniae, we propose that at least some of its chromosomal TA genes cannot be considered “selfish,” but rather, they might play an important role in the pneumococcal life-style (see below). To date, only three chromosomal pneumococcal TA operons have been studied in some detail, namely, relBE2 (GI 15903147 and 15903146 in the R6 genome), pezAT (GI 15902995 and 15902996 in the R6 genome), and yefM-yoeB (GI 15903628 and 15903627 in the R6 genome).
In the case of the pneumococcal relBE genes, analyses performed on bacterial genomes showed the existence of two putative pneumococcal toxin genes that shared homologies to the E. coli relE gene; the toxin genes were preceded by putative antitoxin (relB) counterparts, and the TA loci were termed relBE1 and relBE2 (122). The percent similarities observed for the RelE toxins were relatively high: 43% (E. coli RelE and pneumococcal RelE1), 51% (E. coli RelE and pneumococcal RelE2), and 50% (pneumococcal RelE1 and RelE2). The putative antitoxins also exhibited significant sequence similarities: 52% (E. coli RelB and pneumococcal RelB1), 34% (E. coli RelB and pneumococcal RelB2), and 39% (pneumococcal RelB antitoxins). The sequence similarities of the antitoxins presently found by us support the keeping of the name of the pneumococcal relBE TA genes (4, 39, 118), in spite of some reluctance by others (90). The pneumococcal relBE2 genes were shown to be functional (117), but this was reportedly not the case for relBE1 (GI 15902296 and 15902297 in the R6 genome). The latter TA pair was shown to be unable to cleave mRNA after the induction of transcription of the pneumococcal relE1 gene in E. coli (26); however, we still cannot rule out the functionally of the pneumococcal relBE1 genes, as an assessment of the cell growth profile after the overexpression of the relE1 gene was not conducted (26, 117). The chromosomal pezAT (pneumococcal epsilon-zeta antitoxin-toxin) genes were named after their homologue, the epsilon-zeta TAS, which was discovered in plasmid pSM19035 of Streptococcus pyogenes (18, 20, 82). Finally, the pneumococcal yefM-yoeB TAS showed homology to the E. coli counterpart (116, 122) as well as to the axe-txe TAS of plasmid pRUM of Enterococcus faecium (65, 116). These three functional pneumococcal TA pairs (relBE2, pezAT, and yefM-yoeB) are reviewed below in terms of their genetic organizations, their transcriptional regulation, their functional activities, and, finally, the available details on their structures.
GENETIC ORGANIZATION AND TRANSCRIPTIONAL REGULATION
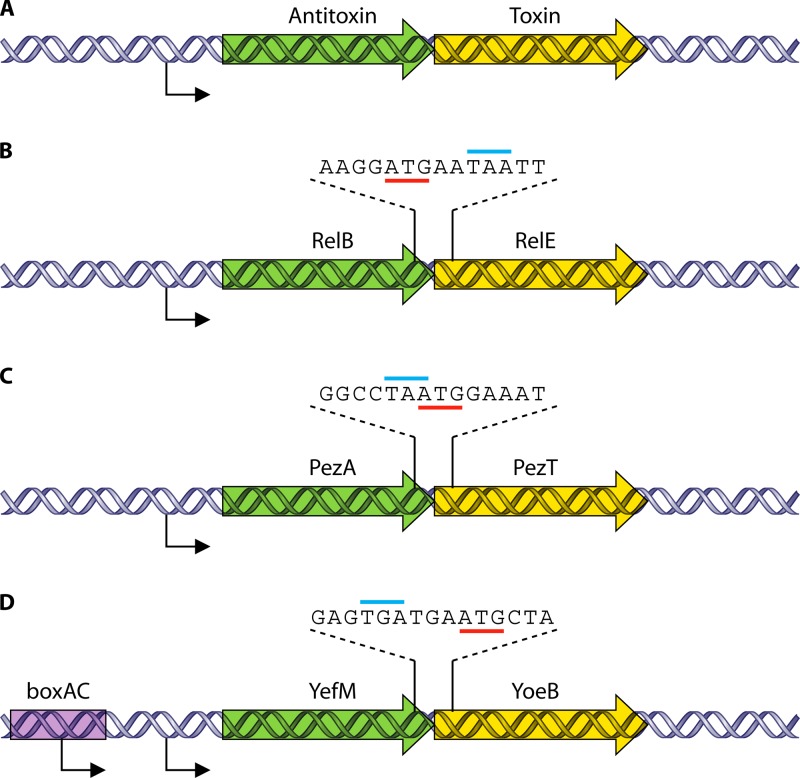
The three pneumococcal TAS, as is usual in most (but not in all) systems, are organized as operons of two genes in which the antitoxin gene precedes the toxin gene (Fig. 2A). Sequence analyses of the region between the antitoxin and the toxin genes in the three pneumococcal TA pairs showed an organization indicative of translational coupling (Fig. 2). In relBE2, the relB2 antitoxin gene overlaps the relE2 toxin gene by 8 nucleotides (nt) (Fig. 2B), and in pezAT, the TAA stop codon of the pezA antitoxin gene overlaps the ATG initiation codon of the pezT toxin gene (Fig. 2C). In the case of yefM-yoeB, there are two stop codons of the yefM antitoxin gene followed by the ATG start codon of the yoeB toxin gene (Fig. 2D).
Fig 2.
Genetic organization of pneumococcal type II TAS. (A) Typical organization of type II TAS, in which the antitoxin gene precedes the toxin gene and both genes constitute an operon. Both genes usually overlap by 1 to 4 nt and are cotranscribed from a single promoter located upstream of the antitoxin gene. The transcription of the operon is negatively autoregulated by the TA protein complex. (B to D) The organization of the three studied pneumococcal TAS is similar to that of the typical type II TAS: the genes overlapped by 8 nt (relBE2) (B) or by 1 nt (pezAT TA genes) (C) or were separated by 3 nt (yefM-yoeB TA genes) (D). The yefM-yoeB TAS (D), however, is somewhat dissimilar to the other two pneumococcal TAS, as this operon is transcribed from two promoters: PyefM2 is regulated by the YefM and YoeB proteins, and PyefM1, which is located upstream of PyefM2, is 15-fold weaker than PyefM2, but it is a constitutive, unregulated promoter. In addition, a boxAC element (purple rectangle) is found to overlap the −35 sequence of PyefM1. The antitoxin and toxin genes are indicated in green and beige, respectively. The lines indicate the start (red) and stop (cyan) codons.
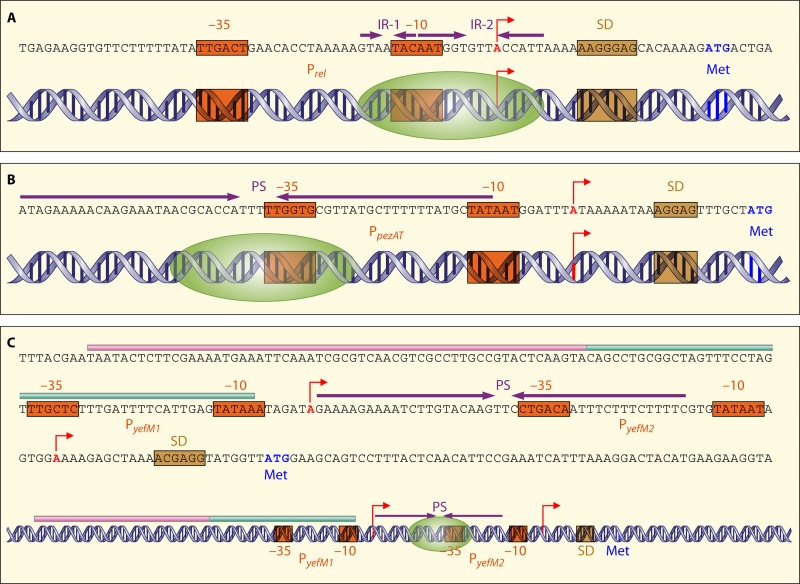
The relBE2 promoter region (Fig. 3A) contains two short inverted repeats (IR-1 and IR-2) that span nucleotides −16 to +6 (with position +1 being the nucleotide where transcription initiates). The right and left arms of IR-1 and IR-2, respectively, overlap the −10 region of the single promoter Prel (117). Electrophoretic mobility shift assays showed that the antitoxin RelB2 and the RelB2-RelE2 protein complex bind to a DNA fragment containing this region, which is indicative of the existence of an operator that has not been characterized further so far (105). A putative Shine-Dalgarno (SD) sequence, which complies with the reported consensus sequence for S. pneumoniae (Chang Bioscience), was located at position +11.
Fig 3.
Schematic representation of the DNA double helix at the promoter regions of the pneumococcal relBE2 (A), pezAT (B), and yefM-yoeB (C) TAS as well as the (putative) binding sites of their respective proteins. The inverted repeats (IR) or palindrome sequences (PS) are indicated by purple arrows, the transcriptional start sites are indicated by red arrows, and the boxA and boxC subelements are indicated by pink and green lines, respectively. The −10 and −35 promoter sequences (orange), the Shine-Dalgarno (SD) sequences (brown), as well as the start codon “Met” (navy blue) are also indicated. The proposed binding sites of the TA complexes (green ovals indicate the TA proteins) are also shown.
The pezA and pezT genes are cotranscribed from a single promoter located upstream of pezA (Fig. 3B). Their homologues, epsilon-zeta of plasmid pSM19035 together with its upstream gene omega, are a three-component TAS in which the transcription of the whole operon is regulated by the Omega repressor protein (36). Neither the Epsilon antitoxin nor the Zeta toxin is involved in the regulation of this operon, although there is a weak promoter detected upstream of the epsilon gene (20, 35). Unlike the Epsilon antitoxin, PezA functions as a repressor that binds to a 56-bp-long palindrome sequence located upstream of the pezA gene. This imperfect palindrome encompasses nearly the entire promoter of the operon. Although no detailed analyses have been performed on the contacts between the PezA protein and its DNA target, all or part of the palindrome could be the target of the antitoxin, whereas PezT serves as a corepressor in a PezA-PezT complex to further repress the pezAT promoter (82). The Epsilon-Zeta genetic module was initially presumed to be restricted to G+ bacteria (104), but homologues of the Epsilon-Zeta systems are now known to be widely distributed among bacterial phyla (90). Close homologues of PezT are chromosomally encoded and associated with PezA-like antitoxins, with no homologues of the Omega repressor to be found elsewhere in their respective genomes, and therefore, they constitute a classical two-component TAS. Likewise, nearly all homologues that are more closely related to the pSM19035-encoded Zeta toxin are associated with Epsilon-like antitoxins along with Omega-like repressors and thus form a three-component Omega-Epsilon-Zeta TAS. A third group of PezT/Zeta-like proteins is both chromosomally and plasmid encoded and shares between 30 and 40% sequence identity with Zeta as well as PezT. Besides a lack of an apparent antitoxin homologue, another distinctive feature of this third group of Zeta-like proteins is their size, which, at more than 500 amino acid residues, is around twice the size of Zeta (287 residues) and PezT (253 residues). The functionality of these apparently solo Zeta-like proteins has yet to be determined.
The pneumococcal yefM-yoeB TA pair was initially proposed to be designated relBE3 due to its low-level but significant similarity to the E. coli relBE system (58); however, we preferred to name it yefM-yoeB in previous work (21, 116) and in this review. In addition to the low level of similarity between the gene products (38% between the E. coli RelB and the pneumococcal YefM proteins and 33% for the E. coli RelE and the pneumococcal YoeB proteins), differences also exist in the promoter regions. This is reflected mostly by the pneumococcal yefM-yoeB operon, in which a pneumococcal BOX element (32, 102) was inserted upstream of the “natural” promoter, generating a second promoter which is not regulated by the TA protein complex (Fig. 3C). Compared to other TA operons, the regulation of the pneumococcal yefM-yoeB operon is atypical because of the insertion of this putative mobile sequence, designated the boxAC element, upstream of its main promoter (Fig. 3C). BOX elements are abundant repeated sequences that are placed within the intergenic regions of the genome of S. pneumoniae, and the presence of these BOX elements was proposed to affect the expressions of neighboring genes (102). Without this element, the regulation of yefM-yoeB appears to be similar to those of other TA pairs in which the YefM antitoxin serves as a repressor of this operon by binding to a palindrome sequence that overlaps the −35 sequence of the promoter PyefM2, whereas the YoeB toxin serves as a corepressor to further repress transcription (21). The incorporation of the boxAC element upstream of PyefM2 created an additional promoter, designated PyefM1, which is about 15-fold weaker than PyefM2 but not regulated by YefM and YoeB (Fig. 3C). Instead, transcriptional fusion assays indicated that the presence of boxAC (PyefM1) together with PyefM2 and the yefM reading frame in cis led to transcriptional activation in E. coli. It was suggested that activation could be due to still unknown host binding factors and/or other cis-acting elements (21). The presence of this boxAC element is universal, as it is found in all sequenced pneumococcal strains that harbor an intact yefM-yoeB locus but is absent in other yefM-yoeB homologues of different bacteria. The conservation of the boxAC-yefM-yoeB arrangement in pneumococcal genomes indicated that the insertion of boxAC was not recent and is likely advantageous to its host. By increasing the overall basal transcription level of the yefM-yoeB locus, the boxAC element could possibly enable the pneumococcal cells to better adapt to and survive unfavorable conditions.
FUNCTIONALITY
The production of the pneumococcal RelE2 toxin in E. coli yielded a pattern similar to that of E. coli RelE, with cleavages at the transfer-messenger RNA (tmRNA) stop and UUA codons, suggesting that the pneumococcal RelE2 toxin inhibits translation by mRNA cleavage (27). The transcription of the relE2 gene was toxic to both S. pneumoniae and E. coli; the overproduction of RelE2 in the latter host resulted in cell growth arrest, which was rescued by the induction of the cognate relB2 gene. However, the prolonged exposure of E. coli cells to the pneumococcal RelE2 toxin led to cells that were unable to form colonies, which is indicative of the antitoxin being effective only within a certain time frame (117). This contrasted with the response of E. coli cells to their cognate RelBE TAS, since in this case, cells could be rescued after antitoxin overproduction, even after long periods of exposure to the toxin (124). This phenomenon could reflect differences in the activities of the pneumococcal and the E. coli RelE toxins, but this hypothesis has not been investigated further.
The overproduction of the pneumococcal PezT toxin in E. coli resulted in growth inhibition for the first 3 h, but a restoration of cellular growth was observed subsequently, without the need for a concomitant synthesis of the PezA antitoxin (82). This growth profile is similar to the profile reported for E. coli cells overproducing the Zeta toxin (161). The toxicity of PezT was abolished with the coexpression of the pezA gene. In another study, the production of a truncated PezT protein (lacking the last 11 amino acids) after half an hour in E. coli yielded cells forming bulges following membrane permeabilization and lysis, whereas the intact cells displayed small and ovoid morphologies (113). This toxicity is more severe in fast-dividing cells than in slow-growing or persister cells. PezT/Zeta was subsequently shown to inhibit bacterial cell wall synthesis by phosphorylating UDP-N-acetylglucosamine (UNAG) to UDP-N-acetylglucosamine-3′-phosphate (UNAG-3P) in the presence of ATP and Mg2+. UNAG-3P inhibits MurA, the catalytic enzyme in the initial stage of peptidoglycan biosynthesis, and also competes with the synthesis of other glycoconjugates and subsequently provokes cell autolysis (113). UNAG is found in all kingdoms of life (42) and thus explains the toxicity of the S. pyogenes-encoded Zeta protein in eukaryotes such as Saccharomyces cerevisiae (162).
The overproduction of the pneumococcal YoeB toxin in E. coli led to the inhibition of cell growth and a reduction in the number of cells that were able to form colonies. The reductions were about 8 logarithmic units for the B strain (BL21) and 4 to 5 logarithmic units for the other two K-12 strains (MG1655 and TOP10) after 6 h of exposure to YoeB (116). On the other hand, the induction of the entire yefM-yoeB locus was nontoxic to the cells. The reduction in the number of cells that were able to form colonies following yoeB transcription could be alleviated by the later transcription of yefM: the numbers of CFU were reduced to 60% after 2 h and only 4% after 4 h of yoeB transcription, compared to 0.4% without YefM production, signifying that there is a window period for cell resuscitation (116). Moreover, the expression of yoeB in a Lon-deficient strain led to 85% of cells being able to form colonies after 1 h of yoeB overexpression, compared to only 25% of the colonies formed by the wild-type strain (116), indicating that Lon protease could play a role in YoeB-mediated toxicity. The target of pneumococcal YoeB has not been determined so far, but E. coli YoeB was shown to inhibit translation initiation by causing the cleavage of mRNAs at 3 bases downstream of the initiation codons in vivo (157). E. coli YoeB alone does not have RNase activity but is found to be associated with the 50S ribosomal subunit in 70S ribosomes and interacts with the ribosomal A site, leading to the cleavage of the mRNA (157). However, in another report, the E. coli YoeB toxin was shown to have in vitro RNase activity that preferentially cleaves at the 3′ end of purine ribonucleotides (79). Besides overexpression assays, the pneumococcal yefM-yoeB TAS seemed to be able to increase the stability of a segregationally unstable mini-F replicon in E. coli, although no further results were sought (116).
STRUCTURAL INFORMATION
Analytical ultracentrifugation assays showed that the pneumococcal RelB2 antitoxin behaved as a dimer in solution, whereas the TA complex was, in its vast majority, a heterohexamer (105). This finding contrasted with the known structures of two RelBE protein complexes of archaeal origins, both of them being heterotetramers (53, 138). In the case of the E. coli RelBE complex, two possibilities have been reported: a heterotetramer (93) or a heterotrimer in solution (120). Further structural data might solve this apparent discrepancy. Circular dichroism experiments showed that the pneumococcal RelB antitoxin has a relatively high average number of secondary structures (around 35% of α-helices), which would increase after the formation of the RelBE protein complex, in agreement with previous results for the E. coli counterpart, so that both antitoxins would not be totally unfolded proteins (24, 105).
Concerning the predicted structure of the pneumococcal antitoxin RelB2, molecular modeling suggested that the N-terminal region of each monomer could fold during dimerization, with perhaps complete folding and dimerization being part of the same process. Thus, the dimer would have a ribbon-helix-helix (RHH) motif, by which it would bind to its target DNA, similar to the Arc-CopG-Omega family of transcriptional repressors (37, 62, 111, 121); in fact, the 28 N-terminal amino acids of the pneumococcal RelB protein share 64% similarity with the 45-residue CopG transcriptional repressor encoded by streptococcal plasmid pMV158 (37, 105). The RHH DNA-binding motif also seems to be present in the N-terminal regions of the RelB and YefM proteins from E. coli (79, 93, 119) and in the pneumococcal RelB2 protein (105) but not in the archaeal RelB proteins (53, 138) or in the pneumococcal YefM protein (our unpublished observations).
In the case of the PezA antitoxin, its C-terminal region shares homology to the C-terminal region of the plasmid-encoded Epsilon antitoxin. However, differences are found at their N-terminal moieties: PezA shows a helix-turn-helix (HTH) motif, which shares 30 to 40% sequence identity with the Xre and CI/Cro families of transcriptional repressors. This HTH motif is absent in the N-terminal moiety of the Epsilon antitoxin as well as in the Omega repressor of the epsilon-zeta TAS (82). In the case of Omega, this protein has an RHH motif (149) and belongs to the Arc-CopG family of proteins (62). The three-dimensional structures of the PezT toxin and the C terminus of PezA are similar to the structures of the Zeta toxin and Epsilon antitoxin, respectively. Mutations in equivalent residues of PezT that are involved in Zeta toxicity also abolished PezT toxicity (82). The similarities of the structures of Epsilon-Zeta and PezA-PezT are such that a degree of cross-reactivity has been observed between them: Epsilon was able to interact with PezT and could partly alleviate PezT toxicity, and conversely, the toxicity of Zeta could also be alleviated by PezA (C. C. Yeo and C. K. Lim, unpublished data). PezT/Zeta toxins have a characteristic Walker A motif for ATP/GTP binding along with a phosphoryltransferase active site. The antitoxins PezA and Epsilon function to negate the toxicity by occluding the ATP/GTP-binding site through tight binding with their respective toxins (82, 104). Nonetheless, toxin release is impeded in both E. coli and B. subtilis due to proteolytic resistance when both the PezA and PezT proteins are bound together (18, 94, 115), which was distinct from their Epsilon-Zeta homologues, in which a Lon- and ClpXP-dependent degradation of Epsilon occurred on a time scale of minutes in the presence of Zeta when the translation of this operon was inhibited in B. subtilis. The PezAT complex was also shown to have an increased thermodynamic stability and an enhanced electrostatic potential compared to both individual proteins. The femtomolar affinity of PezA and PezT is the strongest reported among all TAS, and even Epsilon-Zeta has only micromolar affinity (115).
Circular dichroism analyses showed that about 55% of the pneumococcal YefM antitoxin was unstructured at 4°C, but it did not have complete unfolding, even at elevated temperatures (116), which was in contrast to the results obtained for the E. coli YefM antitoxin (23). In the thermal stability assays, the melting temperature of the pneumococcal YefM antitoxin (about 45°C) was much lower than that of the pneumococcal YefM-YoeB complex (about 70°C), indicating that YefM may lack a significant hydrophobic core, a feature perhaps important to keep the antitoxin sensitive to proteolysis. Axe-Txe of E. faecium and the E. coli-encoded YefM-YoeB TAS could complement each other, but this was not the case for pneumococcal YefM-YoeB and E. coli YefM-YoeB (116). Based on molecular modeling, a positively charged moiety defined by an R72 residue in E. coli YefM, which is responsible for the interaction with the negatively charged pocket on the interface of E. coli YoeB, was observed for Axe but not pneumococcal YefM, thus implying the possible reason for the failure of pneumococcal YefM to neutralize the E. coli YoeB toxin. The pneumococcal YoeB structural model also showed a larger exposed surface than those of the E. coli YoeB and Txe toxins, which perhaps increases the potential interaction surface, leading to the ineffectiveness of E. coli YefM in counteracting the pneumococcal toxin (116).
TAS IN THE GENOME OF S. PNEUMONIAE: AND YET SO MANY
Bioinformatic searches for TAS in the sequenced S. pneumoniae genomes that are available in the NCBI database were performed. The search was based on data expanded from previous studies (99, 122, 136). The BLASTP program (cutoff point of 10−4) was used to find homologues of the TAS families, namely, CcdAB, HicAB, HipBA, MazEF, PemIK, ParDE, Phd-Doc, RelBE, HigBA (also known as RelE-Xre [136]), YefM-YoeB, VapBC, MosAT, YeeUV, PezAT, Xre-COG2856, and Xre-Bro. Only complete TA pairs were included in this study, whereas solo toxins or solo antitoxins were excluded. A total of 352 TA pairs were found in 48 pneumococcal strains with completely sequenced genomes or contigs (from whole-genome shotgun sequences). The TBLASTN program (cutoff point of 10−4) was then used to find homologues of the TA pairs that could have been missed due to the discrepancy of the annotation (Table 1). Indeed, we found another 17 additional TA pairs in all the genomes studied. Among the TAS that we searched, only HicAB, Phd-Doc, RelBE, HigBA, YefM-YoeB, PezAT, Xre-COG2856, and Xre-Bro were present in the genomes of S. pneumoniae. Others, like MazEF and VapBC, which are highly abundant in bacteria and archaea, were absent. The total number of TA pairs ranged from 4 to 10 in individual strains, with the average number of TA pairs being 7 (Table 1). These numbers could still be underestimated.
Table 1.
TA homologues in S. pneumoniae strains
| S. pneumoniae strain | No. of TA homologues |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HicBA | Phd-Doc | RelBE1 | RelBE2 | HigBA | YefM-YoeB | PezAT | Xre-COG2856CA | Xre-COG2856B | Xre-Bro1 | Bro2-Xre | Total | |
| AP200 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 | ||||
| 70585 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 | |||
| 670-6B | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 | |||
| ATCC 700669 | 1 | 1 | 1 | 2 | 2 | 1 | 2 | 10 | ||||
| BS397 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 9 | ||
| BS455 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 9 | ||
| BS457 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 9 | ||
| BS458 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 9 | ||
| CCRI 1974 | 1 | 1 | 1 | 1 | 1 | 5 | ||||||
| CCRI 1974M2 | 1 | 1 | 1 | 1 | 1 | 5 | ||||||
| CDC0288-04 | 1 | 1 | 1 | 1 | 1 | 1 | 6 | |||||
| CDC1087-00 | 1 | 1 | 1 | 1 | 1 | 1 | 6 | |||||
| CDC1873-00 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 10 | ||
| CDC3059-06 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 1 | 10 | |||
| CGSP14 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 8 | ||||
| D39 | 1 | 1 | 1 | 1 | 1 | 1 | 6 | |||||
| G54 | 1 | 1 | 1 | 1 | 1 | 5 | ||||||
| GA04375 | 1 | 1 | 1 | 1 | 4 | |||||||
| GA17545 | 1 | 1 | 1 | 1 | 1 | 5 | ||||||
| GA17570 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 | ||||
| GA41301 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 9 | |||
| GA41317 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 | ||||
| GA47368 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 10 | ||
| GA47901 | 1 | 1 | 1 | 1 | 1 | 5 | ||||||
| Hungary 19A-6 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 10 | |
| INV104 | 1 | 1 | 1 | 1 | 1 | 1 | 6 | |||||
| INV200 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 | ||||
| JJA | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 9 | |||
| MLV-016 | 1 | 1 | 1 | 1 | 1 | 1 | 6 | |||||
| OXC141 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 | |||
| P1031 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 10 | |||
| R6 | 1 | 1 | 1 | 1 | 1 | 1 | 6 | |||||
| SP11-BS70 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 | |||
| SP14-BS292 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 9 | ||
| SP14-BS69 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 | |||
| SP18-BS74 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 | ||||
| SP195 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 9 | |||
| SP19-BS75 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 | ||||
| SP23-BS72 | 1 | 1 | 1 | 1 | 1 | 1 | 6 | |||||
| SP3-BS71 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 | |||
| SP6-BS73 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 | ||||
| SP9-BS68 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 | ||||
| SP-BS293 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 9 | ||
| Canada MDR_19A | 1 | 1 | 1 | 1 | 1 | 5 | ||||||
| Canada MDR_19F | 1 | 1 | 1 | 1 | 1 | 5 | ||||||
| Taiwan 19F-14 | 1 | 1 | 1 | 1 | 1 | 5 | ||||||
| TCH8431/19A | 1 | 1 | 1 | 1 | 1 | 5 | ||||||
| TIGR4 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 | |||
| Total | 48 | 43 | 47 | 55 | 1 | 29 | 34 | 45 | 28 | 2 | 20 | 352 |
Of all these TAS, HicBA and functional RelBE2 were present in all the strains that were analyzed in this review, whereas Phd-Doc, RelBE1, and Xre-COG2856CA (see below) were present in nearly all the strains. The other two functionally characterized TAS, pezAT and yefM-yoeB, were also found in more than half of the strains. In certain strains, RelBE2 (7 out of 48 strains), PezAT (3 out of 31 strains), and Xre-COG2856B (3 out of 25 strains) (see below) were present as two copies (Table 1). Although the duplicates within a strain shared high percent similarities with each other (72% for RelE2, 96% for PezT, and 72% for COG2856B), their neighboring genes were very dissimilar and not clustered together. Interestingly, the second copy of RelBE2 is always associated with Xre-COG2856B (see below), and Bro2-Xre along with an integrase were also found in the neighboring sequences. We have chosen a few TAS that we think are interesting for further discussion and also as a wealth of information for future research: most of these pneumococcal TAS are putative, and no experimental information is available so far.
RelBE1 and RelBE2
As mentioned above, two different RelBE TAS exhibiting low levels of sequence similarity between them were found. The pneumococcal RelBE1 TAS was present in all the strains examined except GA04375. The putative RelE1 toxin was mostly conserved in all the strains except for strains SP19-BS75 and SP23-BS72, in which half of the relE1 sequences were truncated. Sequence comparisons of the putative RelE1 pneumococcal toxin with Pyrococcus horikoshii RelE (79, 80, 118) showed that some amino acid residues that are likely involved in the toxic activity were absent in RelE1: two of the amino acid residues in P. horikoshii RelE (R65 and R85) that are responsible for the protein synthesis-inhibitory activity were not found in pneumococcal RelE1, but the other three important amino acid residues (R40, L48, and R58) of P. horikoshii RelE were conserved in pneumococcal RelE1 (corresponding to R43, L49, and R59). In the case of pneumococcal RelE2, all these five residues were conserved, which explained the endoribonuclease activity observed for RelE2 but not for RelE1. In spite of this, the presence of an intact relBE1 locus in nearly all the pneumococcal strains examined is indicative to us that the functionality of this TAS has to be reassessed.
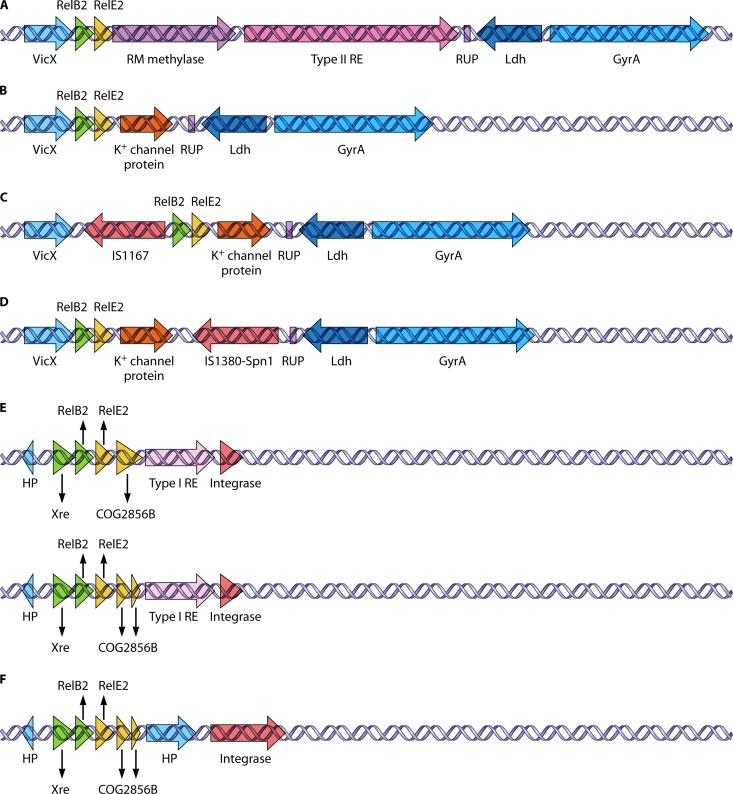
Conversely, RelBE2 has been proven as a bona fide TAS in S. pneumoniae and E. coli (117). We found that the relBE2 locus is present in all the strains analyzed here, and in 7 out of 48 strains, two copies of the locus were found. When the gene organizations of relBE2 in all the strains were carefully examined, we discovered that the strains that harbored the second copy of relBE2 (Fig. 4E and F) had a different gene orientation than that of the first copy of relBE2 (Fig. 4A to D). For the first copy of relBE2 (which is present in all strains), four different groups of gene organizations were found, but in general, this relBE2 operon was located downstream of the vicX gene (metal-dependent hydrolase), and the genes downstream of this operon were comprised of another kind of enigmatic pneumococcal repeat, the 107-nt-long repeat unit of pneumococcus or RUP unit (32), followed by ldh (lactate dehydrogenase) as well as gyrA (the A subunit of DNA gyrase) (Fig. 4). For group 1, genes encoding a putative type II restriction-modification system were found between relE2 and the RUP unit (Fig. 4A); in group 2, the restriction-modification genes were replaced by a gene encoding a putative K+/cation channel protein (Fig. 4B). Group 3 showed a genetic structure similar to that of group 2 but with IS1167 inserted upstream of the relBE2 operon (Fig. 4C). Group 4 is also similar to group 2 but with an IS1380-Spn1 insertion element inserted downstream of the gene encoding the putative K+/cation channel protein (Fig. 4D). On the other hand, for the second copy of the relBE2 operon, the locus was flanked by another putative TAS, the xre-COG2856B genes (see below), and downstream of these two sets of TAS, a gene encoding a type I restriction endonuclease was found, followed by a transposase, and this organization was designated group 5. There are cases in which a stop codon was found within the COG2856B gene that separated this gene in two, but we still considered them to have a group 5 organization (Fig. 4E). Last but not least, in Group 6, which was similar to Group 5, the gene encoding the type I restriction endonuclease was replaced by an unknown gene (Fig. 4F). From the presence of insertion sequence (IS)-integrases and perhaps some phage-carried genes in neighboring sequences, we deduced that this second copy of the relBE2 operon together with its neighboring genes were acquired foreign pieces of DNA. Moreover, when the sequences of all these relE2 homologues were compared, the sequences were highly conserved, but the first and second copies of relE2 were distinct from each other (72% similarity). However, the five amino acid residues that were likely involved in the toxic activity were conserved in both copies of RelE2 in all the strains.
Fig 4.
Polymorphisms associated with the genetic organization of relBE2 and its neighboring genes found in S. pneumoniae clinical isolates (118) and NCBI databases. In general, the relBE2 operon is flanked by vicX (metal-dependent hydrolase) at its upstream region, whereas a RUP (repeat unit of pneumococcus) element, ldh (lactate dehydrogenase), as well as gyrA (the A subunit of DNA gyrase) are located downstream. (A to D) Variations are found, as follows. (A) Genes that encode the putative restriction-modification (RM) methylase and type II restriction endonuclease (RE) are found immediately downstream of relE2; (B) a gene encoding a K+/cation channel protein is located immediately downstream of relE2 instead; (C) same as in panel B but with a gene encoding an IS1167 insertion sequence located immediately upstream of relB2 in the opposite orientation; (D) same as panel B but with an IS1380-SpnI element found downstream of the gene encoding the K+/cation channel protein and in the opposite orientation while keeping the RUP element. (E and F) On the other hand, we found strains which harbor a second set of relBE2 genes, which seems to be an acquired foreign DNA. They have totally different organization patterns from the types described above. (E) The relBE2 genes are flanked by another putative set of TA genes, xre-COG2856B. Besides, an unknown hypothetical protein (HP) is found further upstream, and a type I restriction endonuclease and an integrase are found downstream of the COG2856B gene; in certain strains, a stop codon was found within COG2856B, which leads to the truncation of the gene. (F) Similar to panel E, but instead of a type I restriction endonuclease, an unknown hypothetical protein is found downstream of the COG2856B gene.
HigBA or RelE-Xre (136) also shares significant similarity with the pneumococcal RelBE family. The HigB putative toxin shares 44% similarity with RelE1 and 32% similarity with RelE2; whereas the HigA putative antitoxin shares 46% similarity with RelB1 and 38% similarity with RelB2. These percent similarities are high enough for us to propose it as a bona fide TA pair. However, the HigBA TA pair has a different gene organization from the RelBE family, in which the putative antitoxin higA is located downstream of the putative toxin higB. Nonetheless, this putative higAB locus was found in only one strain in this study, TIGR4.
The Peculiar Xre-COG2856 TAS
COG2856 (Clusters of Orthologous Genes) belongs to the family of metzincin Zn-dependent proteases, and it was suggested to be a potential toxin (99, 137). The proteins of COG2856 are usually associated with the HTH domain-containing protein of the Xre family. The functions of these pneumococcal proteins are still unclear. After scrutinizing all the COG2856 protein sequences found in various strains of S. pneumoniae as suggested previously by Makarova et al. (99), we found that these proteins could be further subdivided into three groups due to the low percent sequence similarities among each other. We thus propose to rename them COG2856A (GI 15903793 in the R6 genome), COG2856B (GI 169832735 in the Hungary 19A-6 genome), and COG2856C (GI 15903794 in the R6 genome). The percent similarities among them were 42% for COG2856A and COG2856B, 30% for COG2856A and COG2856C, and 26% for COG2856B and COG2856C. In all the strains that harbor COG2856A (i.e., 45 of 48 strains), COG2856C was located upstream of COG2856A (Fig. 5A). In addition, both putative toxins share a putative antitoxin xre gene (which may also function as the transcriptional regulator of the system) that overlaps the 5′ end of the gene of COG2856C. Thus, we renamed this putative TA Xre-COG2856CA. Although the sequences of these genes are conserved in general, the annotations vary among strains: the xre (∼162 nt) and COG2856C (∼207 nt) genes mostly overlap by 14 nt, whereas in strain R6, both genes overlap by 50 nt (Table 2). In general, most TA genes overlap by 1 nt (in which the stop codon of the antitoxin gene overlaps the start codon of the toxin gene) or 4 nt (122) so that both genes are cotranscribed, and the proteins are also coexpressed by translational coupling (45). However, there are also cases in which both TA genes are separated by 3 nt (like the pneumococcal yefM-yoeB genes) or overlap by 8 nt (like the pneumococcal relBE2 genes). Hence, the annotation of the xre gene and the gene of COG2856C as overlapping by 14 nt sounds plausible here. On the other hand, in most strains, the gene of COG2856C is either 72 nt or 50 nt apart from the gene of COG2856A (∼333 nt). Besides, there is a strain (AP200) in which both genes were annotated as being separated by 157 nt (Table 2). The analysis of the sequences of the intergenic region led us to identify a possible promoter upstream of the gene of COG2856A. However, the distance between the putative SD sequence and the possible start codon TTG is 18 nt, which is perhaps a bit too far from the norm, and thus, if the gene of COG2856A is expressed, the translation could be less efficient. Interestingly, in strain SP6-BS73, there is a point mutation at the COG2856C stop codon TAA→CAA, which leads to the combination of both COG2856C and COG2856A as a single open reading frame (ORF) that could potentially encode a 298-amino-acid-residue protein. Having two toxin genes sharing an antitoxin counterpart is not common, as the antitoxin protein should be in excess of or at least equal to the toxin protein in order to neutralize the toxic effect of the toxin protein on cells under normal conditions. However, the stoichiometry of the TA complex could be 1 antitoxin to 2 toxins, as seen for E. coli yefM-yoeB (25), making it possible that the two toxin genes (COG2856C and COG2856A) would share the single xre antitoxin gene (Fig. 5A).
Fig 5.
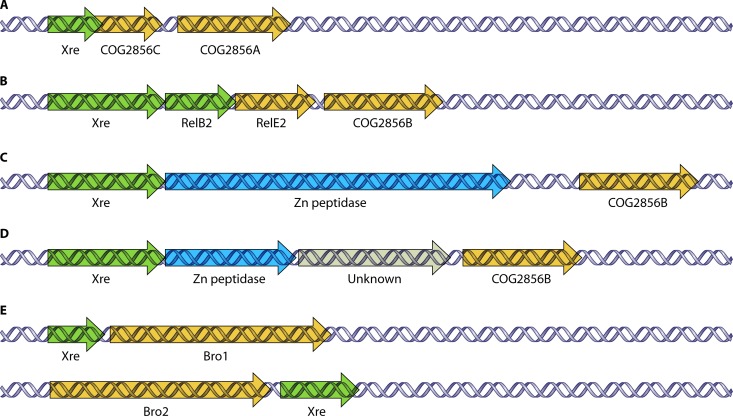
Genetic organization of novel putative pneumococcal TAS found in this study. (A) Three-component Xre-COG2856CA TAS. The gene for the COG2856C toxin is located upstream of the COG2856A toxin gene, with both toxins possibly sharing a common Xre antitoxin. The xre reading frame overlapped the upstream portion of the COG2856C reading frame. (B to D) For Xre-COG2856B, besides the typical gene organization in which the Xre antitoxin precedes the COG2856B toxin, relBE2 (B), a Zn-peptidase (C), as well as a Zn-peptidase and an unknown gene (D) are found between Xre and COG2856B. (E) For the putative Xre-Bro TAS family, two contrasting gene organizations were evident: the xre antitoxin gene preceded the Bro1 toxin gene, whereas the Bro2 toxin gene preceded the xre antitoxin gene.
Table 2.
Numbers of nucleotides in which the N-terminal-encoding portion of COG2856C overlaps with xre as well as the distance between COG2856C and COG2856A in various sequenced genomes of S. pneumoniae
| No. of S. pneumoniae strainsa | No. of overlapping nt of xre-COG2856C | Distance between COG2856C and COG2856A (nt) |
|---|---|---|
| 12 | 14 | 72 |
| 5 | 14 | 50 |
| 1 | 50 | 50 |
| 1 | 14 | 157 |
| 1 | 14 | Both genes combined |
| 22 | COG2856C is not annotated | |
| 3 | No annotationb |
Each strain harbors only one copy of the xre-COG2856AC genes.
Although no ORF is annotated, homologues of COG2856 were found in these sequences.
On the other hand, the Xre-COG2856B pair is less prevalent, since it is present in only 25 out of 48 strains, but it is present as two copies instead of one copy in strains ATCC 700669, CDC3059-06, and SP195 (Table 1). The Xre-COG2856B pair has diverse gene arrangements. First, in most of the strains, the putative xre antitoxin precedes COG2856B, and both genes are either 12 nt (14 out of 28 cases) or 21 nt (3 out of 28 cases) apart. Second, the relBE2 genes are located between the xre and COG2856B genes in the same orientation, with xre overlapping relB2 by 1 nt and relB2 overlapping relE2 by 1 nt, whereas relE2 is 12 nt apart from COG2856B (in 7 out of 28 cases) (Fig. 5B). This operon-like organization gives a hint that both TAS could be interrelated. Interactions between TAS were previously reported; e.g., the overexpression of the Doc toxin (from the Phd-Doc TAS) was shown to activate RelE-mediated mRNA cleavage in E. coli (56), but they do not belong to the same operon. However, the pneumococcal xre-relB2-relE2-COG2856B organization would be the first instance of two different TAS within a single operon. Third, a Zn2+-dependent peptidase-like protein appeared to be located between Xre and COG2856B (in 3 out of 28 cases) (Fig. 5C). Finally, a Zn2+ peptidase-like protein and an ORF of unknown function were located between Xre and COG2856B (in 1 out of 28 cases) (Fig. 5D). The diversity of the gene arrangements hints at the highly recombinogenic nature of this region of the pneumococcal genome. The functionality of these putative TAS, their link to the relBE2 system, and their possible role(s) in S. pneumoniae biology are new and exciting avenues waiting to be explored.
The Paradoxical Xre-Bro TAS
Two relatively different Bro (baculovirus repeat ORF) sequences, renamed here Bro1 (GI 169833276 in the Hungary 19A-6 genome) and Bro2 (GI 169834001 in the Hungary 19A-6 genome) (34% similarity), as well as their respective putative antitoxin counterparts, which are Xre homologues, were found in a search of S. pneumoniae strains performed by Makarova et al. (99). Bro proteins are a family of DNA binding proteins encoding transcription regulators of DNA viruses that were also proposed to exist in bacterial TAS (74, 99). The Xre-Bro TAS is similar to phage repressor-antirepressor proteins that determine the state of the phage by regulating the expression of lytic genes (70, 99). The Ant (antirepressor) domains found in the C-terminal region of Bro1 were suggested to be toxic to bacteria (70, 99). Xre-Bro1 was present in only two strains (Hungary 19A-6 and SP18-BS74), and its organization is that of a typical TAS, in which the putative xre antitoxin gene precedes the putative Bro1 toxin. Both genes are separated by 1 nt, indicating that both genes are likely cotranscribed and translationally coupled. Conversely, Bro2-Xre is present in 20 strains (Table 1), and strikingly, it has an orientation opposite that of its homologue Xre-Bro1, in which the putative Bro2 toxin is located upstream of the Xre antitoxin (Fig. 5E). Both the Bro2 and Xre genes are annotated to be between 11 and 13 nt apart (Table 3). Whether Xre-COG2856 and Xre-Bro are truly functional TAS awaits future investigations.
Table 3.
Distance (number of nucleotides) between the N-terminal-encoding portion of Bro2 and xre in various sequenced genomes of S. pneumoniae
Each strain harbors only one copy of the xre-Bro2 genes.
Although no ORF is annotated, homologues of xre-Bro were found in these sequences.
The Enigmatic BOX Elements
BOX elements are considered to be mobile sequences found exclusively in pneumococci and closely related species (102). They are distributed randomly and are numerous in intergenic regions in the genomes of strains TIGR4 (127 copies [109, 141]) and R6 (115 copies [72]). The BOX elements consist of three different modules: boxA (59 nt), boxB (45 nt), and boxC (50 nt) (102). BoxB is located between boxA and boxC and is present at between zero and eight copies. BOX elements which contain boxA and boxC modules have the potential to form a stable stem-loop structure, which could affect the expression levels of neighboring genes (84). The secondary structure seems to be more important than the base changes within the consensus sequence, as in certain cases, the BOX elements differ from the consensus, but the secondary structure remains the same (102). The BOX elements might enhance gene expression by either stabilizing mRNAs (e.g., increasing the half-life of the mRNA) or serving as DNA-binding sites for regulatory proteins (33, 84). They are also probably involved in the regulation of virulence, genetic competence (84), and phase variation (132).
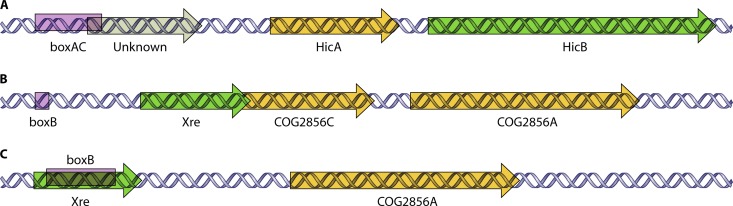
The influence of the BOX subelement boxAC on the expression of the yefM-yoeB TAS of S. pneumoniae was reported recently by our laboratory (21); this boxAC is highly conserved in all the S. pneumoniae strains that harbor the yefM-yoeB TAS. In the strains that we studied here, BOX elements were annotated in only two instances, R6 and ATCC 700669. In strain R6, besides yefM-yoeB, a boxAC element was also detected 264 nt upstream of the putative hicAB TAS. However, there is an ORF encoding an unknown protein of 59 amino acid residues annotated 103 nt upstream of the hicA gene. The ORF overlaps the 3′ portion of boxAC by 25 nt (Fig. 6A). Whether this unknown protein is translated and whether the boxAC element (no prominent promoter sequence was observed) would affect the regulation of the hicAB TAS have yet to be determined. Nevertheless, a boxB subelement was also found to be associated with Xre-COG2856CA. In strain R6, a 21-nt boxB was located 138 nt upstream of Xre (Fig. 6B). However, a different annotation was assigned for strain ATCC 700669, whereby a 138-nt boxB was annotated within the Xre reading frame, and COG2856C was not annotated (Fig. 6C), even when both strains share similar sequences. Further research is needed to verify the discrepancy of these annotations.
Fig 6.
Putative pneumococcal TAS associated with BOX elements. (A) A boxAC element is found overlapping a gene upstream of hicA in the hicAB locus of strain R6. (B) A small boxB subelement was identified upstream of xre of the Xre-COG2856CA TAS in strain R6. (C) A different annotation in which a boxB subelement was found within the xre gene of the Xre-COG2856CA TAS (which has a similar sequence in strain R6) in strain ATCC 700669.
DO PNEUMOCOCCAL TAS PLAY A ROLE IN THE BACTERIAL LIFE-STYLE?
The natural niche of S. pneumoniae is the mucosal surface of the upper respiratory tract in healthy individuals. Pneumococcal colonization of the upper respiratory tract seems to be asymptomatic but represents the environment in which S. pneumoniae exists most commonly and from which the bacteria spread to colonize new tissues (especially when the immune system is compromised) or new hosts. Thus, factors that contribute to bacterial persistence during colonization might increase its virulence (78). The triggering of TAS as a response to stress has been related to bacterial persistence, allowing the bacteria to survive under conditions of nutrient limitation (98). In this sense, chromosomal TAS may contribute to the survival of bacteria in their hosts, either as mediators of the so-called “altruistic” response to bacterial programmed cell death (44) or as modulators of bacterial cell growth that can lead to tolerance to antibiotics (118), perhaps under conditions that resemble the situation found in biofilms. In addition to the analyses described above, analyses of the genomes of 12 dangerous pandemic bacteria (including S. pneumoniae) showed that they possess not only a larger number of TAS than controls but also a smaller genome, implying that TAS could be preferably retained in pathogenic bacteria (57).
Interestingly, an in-depth analysis of the pneumococcal relBE2 operon, which was performed for nearly 100 strains (70 from Spanish and Polish clinical isolates and 30 from genomes available in databases) showed that the operon was functionally present in all of them (118). In addition to several point mutations that did not seem to affect the functionality of the operon, several rearrangements were detected in the chromosomal region surrounding the relBE2 operon, and these rearrangements were classified into three major groups, as shown in Fig. 4A to C (118). These three groups (118) have been enlarged here to up to six different arrangements (Fig. 4). Such a panoply of arrangements found in this region of the pneumococcal chromosome indicated that most of the clinical strains have been subjected to processes of accelerated evolution, probably due to the high selective pressure posed by the antibiotic treatments to which the patients were subjected. The advantage of keeping a functional relBE2 cassette is presently unknown. Furthermore, the three earlier classes of polymorphisms corresponded well with the multiple-locus sequence types (MLST) rather than with the serotypes of the strains, making attractive the proposal that serotypes should not be the main criterion for pneumococcal typing but rather MLSTs (118). A similar proposal was recently made as a result of a large study (of 4,257 isolates) of Salmonella enterica (1).
The other two pneumococcal TAS, pezAT and yoeB-yefM, were not much conserved, and the former was found to be absent in several clinical isolates of S. pneumoniae (82). Intriguingly, the pezAT locus is located within pneumococcal pathogenicity island 1 (PPI1), and PezT had been linked with the virulence of S. pneumoniae when a mutant strain with a disruption in pezT showed significantly attenuated infection progression in mice but without any general growth defects compared to wild-type strains (17, 82). Furthermore, when S. pneumoniae serotype 1 isolates from native Australian patients were recently compared, the pezAT locus was found within the PPI-1v recombination hot spot in all hypervirulent isolates but was absent in noninvasive and intermediate-virulent strains (67). This finding is not surprising considering the high rate of recombination exhibited by S. pneumoniae (66), so rather, the unexpected finding was the conservation of the relBE2 locus in many clinical isolates (118). PezT was postulated to contribute to pneumococcal virulence by triggering cell lysis, which is a requisite for the release of pneumolysin, one of the major pneumococcal virulence factors which causes host cellular damage and accelerates the progress of infection (115). Partial autolysis and the general inhibition of capsular polysaccharide synthesis by PezT would in turn favor the formation of biofilms. On the other hand, slowly dividing cells or persisters will survive the PezT release, and these cells will eventually recover by the production of PezA in the absence of stress conditions (115), which would in turn point to a behavior linked to bistability (see below). The postulated role of the PezT toxin in pneumococcal virulence was extensively discussed in a recent review (114).
Concerning the pneumococcal yefM-yoeB operon, preliminary data from our laboratory indicate that it might be involved in pneumococcal biofilm formation (I. Moreno-Córdoba, M. Moscoso, W. T. Chan, E. García, C. Nieto, and M. Espinosa, unpublished data), a role for TAS which was shown previously for E. coli (83). However, the presence of the BOX element upstream of the pneumococcal operon suggested to us that it might be involved in processes of bistability, although at present, it is only a matter of attractive speculation based on the ascribed role of the BOX element (see below). We further discuss below the various possible general roles of pneumococcal TAS in important aspects of the bacterial life-style, in particular persistence and bistable behavior.
Persistence
Bacterial persistence is the ability of individual cells to randomly enter a period of dormancy, during which the cells are protected against antibiotics and other harmful substances, so that they can survive harsh conditions and enhance their chances of survival (48). Bacterial persistence was discovered by Bigger in the 1940s, when he treated S. aureus cultures with a high concentration of penicillin and found that a small fraction of cells survived in the cultures (13). Dormancy is also a mechanism by which pathogens can escape the immune system of the host that they colonize, guaranteeing a successful invasion. The persisters constitute a small fraction of cells in an otherwise homogeneous population, and they play an important role in optimizing the decision-making process in bacterial populations (43). Persisters are genetically identical to the rest of the nontolerant cells, and they are able to exit dormancy by mechanisms not fully understood (47), perhaps by the stochastic appearance of “explorer cells,” which would check whether the environmental conditions are favorable (47). These explorers would determine whether the entire bacterial population should continue to be dormant (and the explorers would be killed) or the population can resume growth.
In E. coli, the deletion of 10 individual TAS encoding mRNases or the deletion of up to four of the TA loci did not significantly affect bacterial persistence. However, the successive deletion of the 10 TA mRNases gradually reduced the formation of persisters (98). In addition, it was shown that the formation of persisters also depended on the Lon protease-mediated degradation of the antitoxins, whereby antitoxin degradation activated the mRNase activity of the toxin, leading to the inhibition of global translation and the induction of dormancy and persistence (98). On the other hand, the overproduction of the RelE (49, 81), HipA (49), or YgiT (135) toxin in E. coli increased the production of persister cells in response to different antibiotics, which coincided with recent findings by Maisonneuve et al. (98). However, it appears that the RelE toxin-mediated induction of persistence would take place under conditions of high cell density, which would resemble more the situation present in biofilms (140). It is worth noting that the overproduction of DnaJ in E. coli and PmrC in Salmonella enterica serovar Typhimurium, which are genes unrelated to TAS, also led to an increase in the number of persister cells (147). In fact, E. coli as well as other microbial pathogens have been shown to possess redundant pathways for the formation of persisters (92).
Persistence in G− bacteria is better studied than in G+ bacteria. Besides S. aureus, persistence was also reported to exist in streptococcal species such as Staphylococcus epidermidis, Streptococcus mutans, and S. pneumoniae. For pneumococci, it was reported that the relBE2 locus was associated with antibiotic tolerance, because a mutation that abolished the synthesis of RelE2 led to cells that were more tolerant to treatment with antibiotics like erythromycin, although long-term persistence in biofilms was not analyzed (118). Nevertheless, a recent report showed that the overexpression of the RelBE and MazEF systems in S. mutans led to a 1-log-fold increase in the number of persister cells (91). However, that same study showed that a ΔmazEF ΔrelBE double deletion mutant did not exhibit any effect on persister formation, strongly indicating an apparent redundancy of persister genes in S. mutans, as had been found for E. coli and other bacteria. Indeed, a subsequent screening of an expression library led to the discovery of several other candidate persister genes, including some of those involved in transcription and replication, sugar and energy metabolism, and cell wall synthesis (91). Interestingly, that same study also showed that an intraspecies S. mutans quorum-sensing system, the competence-stimulating peptide (CSP)-ComDE regulatory circuit, was directly involved in persister development (91).
The modes of action of the TAS may provide a clue as to how the persisters are formed, such as TA-encoding mRNases reducing the global translation levels or PezT inhibiting cell wall synthesis. However, the determination of which cell is the fortunate one that could escape from the adverse condition and survive as a persister still remains puzzling, or perhaps it is due merely to stochasticity, as was found for E. coli persistence mediated by hipBA (130). It is interesting that the decision to follow a vegetative life or to sporulate in the G+ bacterium B. subtilis seems to be determined by a “molecular race” between differentiation programs competing with each other in time and not by complex cross-regulatory mechanisms (88). Nonetheless, we can postulate that persistence, just like sporulation, is likely a bistable process that constitutes a smart yet optimal strategy to survive environmental changes for the benefit of the bacterium as a whole and that TAS may play an important role in this benefit (see below).
Bistability
In certain circumstances, such as stressful conditions, a population of genetically identical bacteria that grow under nearly homogenous conditions will separate stochastically into two or more distinct subpopulations, a phenomenon termed bi- or multistability, respectively (75). The bistable behavior would represent the possibility of switching between two semistable states, coexisting simultaneously in the same population and environment and in which the possession of a panoply of regulatory feedback (positive or negative) loops would be beneficial for the population as a whole (128). Bistability has been observed for a number of bacterial processes, such as the formation of persisters (see above), genetic competence, escape from sporulation, and biofilm formation (29, 30, 41). A mixture of cells expressing different sets of genes within a population coexisting in the same environment could improve the chances of survival of part of the population, so that stochastic switching could be the best strategy for optimal survival, i.e., a bet-hedging strategy. Although population heterogeneity can incur a fitness cost, since part of the population would always be maladaptive in a given environment, theoretical studies indicated that under conditions of environmental fluctuations, stochastic switching led to a net fitness gain for the population (54, 142).
Reports of bistable processes in S. pneumoniae and closely related bacteria include at least phase variation (132), persistence (91), competence (30, 41), and the expression of type I pilus genes (11), with most of the repertoire of genes participating in these processes also being involved in virulence. An interesting example was provided by a global analysis of transcription during the onset of competence in pneumococci. It showed the transcriptional activation of two choline-binding proteins, and a functional role of one of them in competence was demonstrated (129). Furthermore, autolysis and competence have been shown for S. pneumoniae (30), and a suggestion that at least one of these choline-binding proteins could play a role in autolysis or in changes in the cell wall during competence was made (129). These findings, obtained well before the role of PezAT in cell wall integrity was demonstrated (113), could be used by us to argue in favor of a connection between competence and this specific TA, all within a bistable behavior.
The discovery of a BOX element associated with yefM-yoeB and the putative hicAB and xre-COG2856CA TAS in S. pneumoniae raised the question of whether these TAS have any other role that is related to other cell processes. There have been reports showing that most BOX elements are located in the immediate vicinity of genes that are related to competence in the genome of S. pneumoniae, such as the comA, hexB, and, possibly, mmsA genes (22, 102). Besides competence, BOX elements are also found close to genes such as neuA, which encodes the pneumococcal virulence factor neuraminidase (19), and ply, which encodes another virulence factor, termed pneumolysin (148). The location of these BOX elements in the immediate vicinity of genes related to competence and virulence raises the intriguing possibility of BOX elements being involved in coordinating the expression and modulation of these genes, perhaps being dependent on the copy numbers of the boxB subelement and its orientation, as suggested by Knutsen et al. (84). It is worth noting that competence and virulence (both likely bistable) are also viewed as global responses of S. pneumoniae upon stress, just like the activation of TAS. In another report, it was shown that a BOX element, although not necessary for the expression of opacity, increased the frequency of (bistable) phase variation and made the observation of opaque variants more likely (132). It would thus appear that the hyperrecombinogenic nature of S. pneumoniae provides the bacterium with selective advantages when subjected to stressful conditions, such as the incorporation of an integrative piece, like the BOX element, into an operative genome piece, such as the comA, neuA, and ply genes and the yefM-yoeB operon (8, 9). This “molecular attunement” of the cells to their changing environment may have led to a better fitness of the bacteria, although strains totally devoid of all BOX elements have not been constructed so far (32). Interestingly, a recent in silico evolutionary simulation showed that in fluctuating environments, the emergence and maintenance of bistability occur only in the presence of noise, and the emergence of bistability enables a faster adaptation to the fluctuating environment (89). Selective forces that lead to higher/lower noise might also enhance/inhibit the evolution of bistability and nonlinearity in gene regulation (89). The conservation of BOX elements in pneumococcal genomes is indicative of their utility to the bacterial cell. The integration of the BOX element upstream of the yefM-yoeB locus led to the creation of an additional promoter that was not regulated by either YefM or the YefM-YoeB complex (21). It could thus be hypothesized that the additional promoter afforded by the BOX element enabled higher noise in the expression of yefM-yoeB, thereby enhancing its bistable behavior and enabling a faster adaptation to changing environments. Whether BOX elements play similar roles in other pneumococcal genes with which they are associated awaits experimental validation.
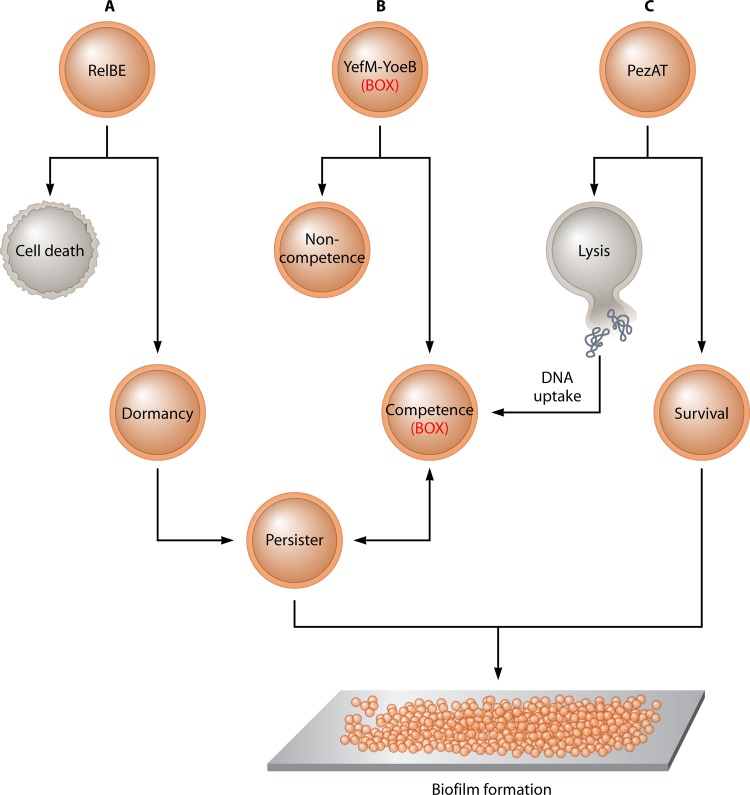
How would S. pneumoniae benefit from the genetic inducement brought about by the acquisition of the, at least, three TAS that we have analyzed here? Any events that could disrupt the interaction of the TA complex would release the toxin to exert its lethal effect. This may affect a subpopulation (bistable) of bacteria in which a cell death program would be triggered after an environmental stress (46, 85). Stressful conditions that trigger the RelBE TAS could lead to the RelE-mediated cell death of a part of the population, whereas the other part will cease growing (118), which in turn may lead to the development of persisters (Fig. 7A). In the case of yefM-yoeB, the presence of BOX may prompt pneumococci to processes that could belong to an evolutionary schedule, where a fraction of the population starts a program that can lead to entry into a genetic competence state for transformation (30) (Fig. 7B). In both cases, we have preliminary evidence that the deletion of either yefM-yoeB or relBE results in a slight, but significant, decrease in the frequency of transformation, which, in addition, was cumulative in a mutant deleted in both systems (Moreno-Córdoba et al., unpublished). In the case of PezAT, it was shown that the activation of the PezT toxin led to cell lysis (113). In this situation (Fig. 7C), part of the population will survive, while lysis would lead to the release of DNA, which would be used by other competent bacteria: the killing of sister cells (fratricide) is common in S. pneumoniae (29). Thus, the triggering of one or all of the toxins would result in different bistable (or even, perhaps, multistable) situations, which would lead to persister development and in turn would facilitate biofilm formation (Fig. 7), and we have preliminary evidence that deletions of the relBE and yefM-yoeB operons lead to a substantial reduction in the ability of the cells to generate biofilms (Moreno-Córdoba et al., unpublished).
Fig 7.
Model of the possible bistable behavior of S. pneumoniae cells subjected to stress. (A) Antibiotic stress may lead to RelE2-mediated cell death for part of the population, but part of the cells would undergo dormancy instead (i.e., persister cells) and are thus tolerant toward the antibiotics. (B) Preliminary results indicate that the deletion of the yefM-yoeB TA locus led to decreased competence in S. pneumoniae (Moreno-Córdoba et al., unpublished). Under stressful conditions, a fraction of cells would undergo a genetic competent state, whereas the rest would remain noncompetent. yefM-yoeB and some genes involved in competence were related to the pneumococcal BOX element, raising speculation on the role of BOX. (C) PezT was demonstrated to be involved in cell lysis under harsh environmental conditions, thus releasing DNA that could be taken up by the competent sibling cells, whereas the cells that survive, which also include persister cells, could in turn facilitate biofilm formation. All these processes would result in S. pneumoniae cells displaying bistable behavior under conditions of stress that perhaps involve, directly or indirectly, the three known pneumococcal TAS.
All in all, we propose that the triggering of TAS leads S. pneumoniae to a situation in which evolutionary processes would be at work and leads to situations which might prompt bacteria to competence and persister development. These processes, as well as those reported to occur when B. subtilis cells escape from the sporulation developmental program (63), are mediated by the killing of part of the sister cells through cannibalism (B. subtilis) or fratricide (S. pneumoniae) and belong to a bistable variation within a bacterial population (41). However, the mechanisms leading to their response (altruism, persistence, cannibalism, or fratricide) might not be related to but rather pertain to global evolutionary processes. Indeed, it was proposed previously that a general response to stressful conditions triggers several regulons involved in genetic competence (30). The presence of BOX elements upstream of genes involved in pneumococcal competence (84) and the presence of the boxAC element upstream of the yefM-yoeB operon (21) make it tempting to speculate that the responses to stress mediated by at least the three specific TAS belong to a bistable variation and that the BOX elements play a role in enhancing the evolution of bistability in these systems and, thus, the adaptability of the bacteria to fluctuating environments.
CONCLUSIONS AND PERSPECTIVES: USE OF TAS AS ANTIMICROBIALS
In the pathogenesis of disease and survival inside the host, bacteria are subjected to stress. A promising global solution to the potential lack of anti-infectives against pneumococci could be to unbalance the regulated synthesis of macromolecules. This could provide a target for anti-infective interventions that would assist existing therapy and the natural defenses of the host. One possibility would be to use the toxin of a given TA pair as a bactericidal or bacteriostatic compound. To achieve this, compounds that could disrupt the interactions between a toxin and its cognate antitoxin could be very useful as antibacterials: an unbalanced synthesis of each independent TAS triggers cell growth arrest or, for prolonged periods of time, cell death, with a survival rate of 1 × 10−6 to 10−7 per each TA pair (127). Thus, it is conceivable that the employment of two independent toxins would potentiate such cell death, and indeed, TAS have been used in the design of tools for the biological containment of strains designed for bioremediation and other industrial purposes (87, 123, 127).
However, even if this approach is feasible, its usefulness might be hampered by the finding that a disruption of the interaction between PezA and PezT, to activate PezT for potential antimicrobial therapy, would be difficult because of the strong binding of PezA and PezT (114). To monitor the possibility of such a disruption, several approaches were proposed (4), and specifically, one of them, bioluminescence resonance energy transfer (BRET) (153), was shown to be effective for the detection of interactions between the pneumococcal RelB antitoxin and RelE toxin in vivo (117). The adaptation of BRET assays to high-throughput formats allowed the screening of peptide libraries (nearly 50 million individual peptides), and two peptide mixtures were shown to inhibit, albeit slightly, the interactions between the Epsilon antitoxin and the Zeta toxin, opening interesting possibilities for the development of novel antibacterial compounds (95). The use of the artificial activation of the toxins of TAS as an antibacterial strategy was recently reviewed (150).
A most exciting approach has been made by the finding that small synthetic peptides derived from the ParE toxin from E. coli (included as a member of the RelE toxin superfamily) as well as peptides derived from the CcdB toxin were able to inhibit topoisomerases (from bacteria to humans) in vitro, a discovery which no doubt will open new uses for the toxicity of the toxins (10, 144). Whether this approach can be used to design small RNAs that encode the desired peptides and, if so, whether they can be used as potential antibacterials remain to be seen.
The acquisition of antibiotic resistances by pathogens is only a matter of time due to genetic evolvability and horizontal gene transfer, which consequently establish a diverse pool of resistance and related genes, the resistome. The discovery of novel alternatives that can extend the capabilities of therapy is critical, because a number of processes are at work: horizontal gene transfer, serotype replacements in populations subjected to pneumococcal vaccination, and the selection of antibiotic-resistant strains due to the indiscriminant use of these antimicrobials, to name a few. Findings like those discussed here, and the possibility of a stimulation of TAS synthesis as means of developing antimicrobials (150), open new avenues to explore the exploitation of the potential toxic activity of the pneumococcal toxins as a means to develop novel anti-infectives. Although there is a long way to go before effective drugs can be derived from these toxins, there is still a wealth of unexplored possibilities in pneumococcal genomes: novel genetic organizations, duplications of TAS in pairs, and the solving of enigmas like the (possible) role of BOX elements and the novel genetic organization of TAS in pneumococcal genomes will be matters for future experimentation.
ACKNOWLEDGMENTS
We thank Kenn Gerdes, the two anonymous reviewers, and members of the INTERMODS Consortium for critical reading and suggestions on the manuscript; present and former members of the Espinosa laboratory; and numerous colleagues from the Spanish Network of Extrachromosmomal Elements (REDEEX) for many fruitful discussions. C.C.Y. thanks A. Meinhart, J. A. Harikrishna, and past members of his laboratory for the productive work and discussions as well as the camaraderie.
While this review was written, our laboratories were supported by grants from the Spanish Ministry of Economy and Competitivity (grants CSD-2008-00013, INTERMODS, and BFU2010-19597 to M.E.), the European Union (grant EU-CP223111, CAREPNEUMO, to M.E.), and the Malaysian Ministry of Science, Technology and Innovation (grants 09-99-06-0104-EA001, 02-02-05-SF0019, and 02-02-05-SF0026 and SAGA grant M18 to C.C.Y.).
REFERENCES
- 1. Achtman M, et al. 2012. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog. 8: e1002776 doi:10.1371/journal.ppat.1002776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aizenman E, Engelberg-Kulka H, Glaser G. 1996. An Escherichia coli chromosomal “addiction module” regulated by guanosine 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. U. S. A. 93: 6059–6063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alekshun MN, Levy SB. 2007. Molecular mechanisms of antibacterial multidrug resistance. Cell 128: 1037–1050 [DOI] [PubMed] [Google Scholar]
- 4. Alonso JC, et al. 2007. Bacterial toxin-antitoxin systems as targets for the development of novel antibiotics, p 313–329 In Bonomo RA, Tolmasky ME. (ed), Enzyme-mediated resistance to antibiotics: mechanisms, dissemination, and prospects for inhibition. ASM Press, Washington, DC [Google Scholar]
- 5. Anderson DI. 2003. Persistence of antibiotic resistant bacteria. Curr. Opin. Microbiol. 6: 452–456 [DOI] [PubMed] [Google Scholar]
- 6. Anderson MT, Seifert HS. 2011. Opportunity and means: horizontal gene transfer from the human host to a bacterial pathogen. mBio 2(1): e00005–11 doi:10.1128/mBio.00005-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Arkin MR, Wells JA. 2004. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat. Rev. Drug Discov. 3: 301–317 [DOI] [PubMed] [Google Scholar]
- 8. Baquero F. 2009. Environmental stress and evolvability in microbial systems. Clin. Microbiol. Infect. 15: 5–10 [DOI] [PubMed] [Google Scholar]
- 9. Baquero F. 2004. From pieces to patterns: evolutionary engineering in bacterial pathogens. Nat. Rev. Microbiol. 2: 510–518 [DOI] [PubMed] [Google Scholar]
- 10. Barbosa LCB, et al. 2012. Design and synthesis of peptides from bacterial ParE toxin as inhibitors of topoisomerases. Eur. J. Med. Chem. 54: 591–596 [DOI] [PubMed] [Google Scholar]
- 11. Basset A, et al. 2012. An epigenetic switch mediates bistable expression of the type 1 pilus genes in Streptococcus pneumoniae. J. Bacteriol. 194: 1088–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bernard P, Couturier M. 1992. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J. Mol. Biol. 226: 735–745 [DOI] [PubMed] [Google Scholar]
- 13. Bigger JW. 1944. Treatment of staphylococcal infections with penicillin by intermittent sterilization. Lancet 244: 497–500 [Google Scholar]
- 14. Bogaert D, et al. 2004. Colonisation by Streptococcus pneumoniae and Staphylococcus aureus in healthy children. Lancet 363: 1871–1872 [DOI] [PubMed] [Google Scholar]
- 15. Bravo A, de Torrontegui G, Díaz R. 1987. Identification of components of a new stability system of plasmid R1, Par D, that is close to the origin of replication of this plasmid. Mol. Gen. Genet. 210: 101–110 [DOI] [PubMed] [Google Scholar]
- 16. Bravo A, Ortega S, de Torrontegui G, Díaz R. 1988. Killing of Escherichia coli cells modulated by components of the stability system ParD of plasmid R1. Mol. Gen. Genet. 215: 146–151 [DOI] [PubMed] [Google Scholar]
- 17. Brown JS, Gilliland SM, Spratt BG, Holden DW. 2004. A locus contained within a variable region of pneumococcal pathogenicity island 1 contributes to virulence in mice. Infect. Immun. 72: 1587–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Camacho AG, et al. 2002. In vitro and in vivo stability of the epsilon2zeta2 protein complex of the broad host-range Streptococcus pyogenes pSM19035 addiction system. Biol. Chem. 383: 1701–1713 [DOI] [PubMed] [Google Scholar]
- 19. Camara M, Mitchell TJ, Andrew PW, Boulnois GJ. 1991. Streptococcus pneumoniae produces at least two distinct enzymes with neuraminidase activity: cloning and expression of a second neuraminidase gene in Escherichia coli. Infect. Immun. 59: 2856–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ceglowski P, Boitsov A, Chai S, Alonso JC. 1993. Analysis of the stabilization system of pSM19035-derived plasmid pBT233 in Bacillus subtilis. Gene 136: 1–12 [DOI] [PubMed] [Google Scholar]
- 21. Chan WT, et al. 2011. Genetic regulation of the yefM-yoeBSpn toxin-antitoxin locus of Streptococcus pneumoniae. J. Bacteriol. 193: 4612–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chandler MS, Morrison DA. 1988. Identification of two proteins encoded by com, a competence control locus of Streptococcus pneumoniae. J. Bacteriol. 170: 3136–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cherny I, Gazit E. 2004. The YefM antitoxin defines a family of natively unfolded proteins: implications as a novel antibacterial target. J. Biol. Chem. 279: 8252–8261 [DOI] [PubMed] [Google Scholar]
- 24. Cherny I, et al. 2007. Structural and thermodynamic characterization of the Escherichia coli RelBE toxin-antitoxin system: indication for a functional role of differential stability. Biochemistry 46: 12152–12163 [DOI] [PubMed] [Google Scholar]
- 25. Cherny I, Rockah L, Gazit E. 2005. The YoeB toxin is a folded protein that forms a physical complex with the unfolded YefM antitoxin. J. Biol. Chem. 280: 30063–30072 [DOI] [PubMed] [Google Scholar]
- 26. Christensen SK, Gerdes K. 2003. RelE toxins from bacteria and archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol. Microbiol. 48: 1389–1400 [DOI] [PubMed] [Google Scholar]
- 27. Christensen SK, Pedersen K, Hansen FG, Gerdes K. 2003. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 332: 809–819 [DOI] [PubMed] [Google Scholar]
- 28. Christensen-Dalsgaard M, Jørgensen MG, Gerdes K. 2010. Three new RelE-homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses. Mol. Microbiol. 75: 333–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Claverys JP, Havarstein LS. 2007. Cannibalism and fratricide: mechanisms and raisons d'être. Nat. Rev. Microbiol. 5: 219–229 [DOI] [PubMed] [Google Scholar]
- 30. Claverys JP, Prudhomme M, Martin B. 2006. Induction of competence regulons as general stress responses in Gram-positive bacteria. Annu. Rev. Microbiol. 60: 451–475 [DOI] [PubMed] [Google Scholar]
- 31. Critchlow SE, et al. 1997. The interaction of the F plasmid killer protein, CcdB, with DNA gyrase: induction of DNA cleavage and blocking of transcription. J. Mol. Biol. 273: 826–839 [DOI] [PubMed] [Google Scholar]
- 32. Croucher NJ, Vernikos GS, Parkhill J, Bentley SD. 2011. Identification, variation and transcription of pneumococcal repeat sequences. BMC Genomics 12: 120 doi:10.1186/1471-2164-12-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Croucher NJ, et al. 2009. Role of conjugative elements in the evolution of the multidrug-resistant pandemic clone Streptococcus pneumoniae Spain23F ST81. J. Bacteriol. 191: 1480–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dao-Thi MH, et al. 2005. Molecular basis of gyrase poisoning by the addiction toxin CcdB. J. Mol. Biol. 348: 1091–1102 [DOI] [PubMed] [Google Scholar]
- 35. de la Hoz AB, et al. 2000. Plasmid copy-number control and better-than-random segregation genes of pSM19035 share a common regulator. Proc. Natl. Acad. Sci. U. S. A. 97: 728–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. de la Hoz AB, et al. 2004. Recognition of DNA by Omega protein from the broad-host range Streptococcus pyogenes plasmid pSM19035: analysis of binding to operator DNA with one to four heptad repeats. Nucleic Acids Res. 32: 3136–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. del Solar G, Hernández-Arriaga AM, Gomis-Rüth FX, Coll M, Espinosa M. 2002. A genetically economical family of plasmid-encoded transcriptional repressors in control of plasmid copy number. J. Bacteriol. 184: 4943–4951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Diago-Navarro E, et al. 2010. parD toxin-antitoxin system of plasmid R1—basic contributions, biotechnological applications and relationships with closely-related toxin-antitoxin systems. FEBS J. 277: 3097–3117 [DOI] [PubMed] [Google Scholar]
- 39. Díaz-Orejas R, et al. 2010. Bacterial toxin-antitoxin systems targeting translation. J. Appl. Biomed. 8: 179–188 [Google Scholar]
- 40. Donegan NP, Cheung AL. 2009. Regulation of the mazEF toxin-antitoxin module in Staphylococcus aureus and its impact on sigB expression. J. Bacteriol. 191: 2795–2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dubnau D, Losick R. 2006. Bistability in bacteria. Mol. Microbiol. 61: 564–572 [DOI] [PubMed] [Google Scholar]
- 42. Durand P, Golinelli-Pimpaneau B, Mouilleron S, Badet B, Badet-Denisot MA. 2008. Highlights of glucosamine-6P synthase catalysis. Arch. Biochem. Biophys. 474: 302–317 [DOI] [PubMed] [Google Scholar]
- 43. Elowitz MB, Levine AJ, Siggia ED, Swain PS. 2002. Stochastic gene expression in a single cell. Science 297: 1183–1186 [DOI] [PubMed] [Google Scholar]
- 44. Engelberg-Kulka H, Amitai S, Kolodkin-Gal I, Hazan R. 2006. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet. 2: e135 doi:10.1371/journal.pgen.0020135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Engelberg-Kulka H, Glaser G. 1999. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 53: 43–70 [DOI] [PubMed] [Google Scholar]
- 46. Engelberg-Kulka H, Hazan R. 2003. Cannibals defy starvation and avoid sporulation. Science 301: 467–468 [DOI] [PubMed] [Google Scholar]
- 47. Epstein SS. 2009. Microbial awakenings. Nature 457: 1083 doi:10.1038/4571083a [DOI] [PubMed] [Google Scholar]
- 48. Evdokimov A, et al. 2009. New kinase regulation mechanism found in HipBA: a bacterial persistence switch. Acta Crystallogr. D Biol. Crystallogr. 65: 875–879 [DOI] [PubMed] [Google Scholar]
- 49. Falla TJ, Chopra I. 1998. Joint tolerance to beta-lactam and fluoroquinolone antibiotics in Escherichia coli results from overexpression of hipA. Antimicrob. Agents Chemother. 42: 3282–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fernandez-Lopez R, et al. 2005. Unsaturated fatty acids are inhibitors of bacterial conjugation. Microbiology 151: 3517–3526 [DOI] [PubMed] [Google Scholar]
- 51. Fineran PC, et al. 2009. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc. Natl. Acad. Sci. U. S. A. 106: 894–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fozo EM, Hemm MR, Storz G. 2008. Small toxic proteins and the antisense RNAs that repress them. Microbiol. Mol. Biol. Rev. 72: 579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Francuski D, Saenger W. 2009. Crystal structure of the antitoxin-toxin protein complex RelB-RelE from Methanococcus jannaschii. J. Mol. Biol. 393: 898–908 [DOI] [PubMed] [Google Scholar]
- 54. Gaál B, Pitchford JW, Wood AJ. 2010. Exact results for the evolution of stochastic switching in variable asymmetric environments. Genetics 184: 1113–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gamez G, Hammerschmidt S. 2012. Combat pneumococcal infections: adhesins as candidates for protein-based vaccine development. Curr. Drug Targets 13: 323–337 [DOI] [PubMed] [Google Scholar]
- 56. Garcia-Pino A, et al. 2008. Doc of prophage P1 is inhibited by its antitoxin partner Phd through fold complementation. J. Biol. Chem. 283: 30821–30827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Georgiades K, Raoult D. 2011. Genomes of the most dangerous epidemic bacteria have a virulence repertoire characterized by fewer genes but more toxin-antitoxin modules. PLoS One 6: e17962 doi:10.1371/journal.pone.0017962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gerdes K, Christensen KS, Lobner-Olensen A. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3: 371–382 [DOI] [PubMed] [Google Scholar]
- 59. Gerdes K, Gultyaev AP, Franch T, Pedersen K, Mikkelsen ND. 1997. Antisense RNA-regulated programmed cell death. Annu. Rev. Genet. 31: 1–31 [DOI] [PubMed] [Google Scholar]
- 60. Gerdes K, Rasmussen PB, Molin S. 1986. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. U. S. A. 83: 3116–3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gerdes K, Wagner EG. 2007. RNA antitoxins. Curr. Opin. Microbiol. 10: 117–124 [DOI] [PubMed] [Google Scholar]
- 62. Gomis-Ruth FX, et al. 1998. The structure of plasmid-encoded transcriptional repressor CopG unliganded and bound to its operator. EMBO J. 17: 7404–7415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. González-Pastor JE, Hobbs EC, Losick R. 2003. Cannibalism by sporulating bacteria. Science 301: 510–513 [DOI] [PubMed] [Google Scholar]
- 64. Gotfredsen M, Gerdes K. 1998. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol. Microbiol. 29: 1065–1076 [DOI] [PubMed] [Google Scholar]
- 65. Grady R, Hayes F. 2003. Axe-Txe, a broad-spectrum proteic toxin-antitoxin system by a multidrug-resistant, clinical isolate of Enterococcus faecium. Mol. Microbiol. 47: 1419–1432 [DOI] [PubMed] [Google Scholar]
- 66. Hanage WP, Fraser C, Tang J, Connor TR, Corander J. 2009. Hyper-recombination, diversity, and antibiotic resistance in pneumococcus. Science 324: 1454–1457 [DOI] [PubMed] [Google Scholar]
- 67. Harvey RM, et al. 2011. A variable region within the genome of Streptococcus pneumoniae contributes to strain-strain variation in virulence. PLoS One 6: e19650 doi:10.1371/journal.pone.0019650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hayes F. 2003. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301: 1496–1499 [DOI] [PubMed] [Google Scholar]
- 69. Hayes F, Van Melderen L. 2011. Toxins-antitoxins: diversity, evolution and function. Crit. Rev. Biochem. Mol. Biol. 46: 386–408 [DOI] [PubMed] [Google Scholar]
- 70. Heinrich J, Velleman M, Schuster H. 1995. The tripartite immunity system of phages P1 and P7. FEMS Microbiol. Rev. 17: 121–126 [DOI] [PubMed] [Google Scholar]
- 71. Hernandez-Rocamora VM, et al. 2009. Multivalent choline dendrimers as potent inhibitors of pneumococcal cell-wall hydrolysis. Angew. Chem. Int. Ed. Engl. 48: 948–951 [DOI] [PubMed] [Google Scholar]
- 72. Hoskins J, et al. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183: 5709–5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Huang F, He Z-G. 2010. Characterization of an interplay between a Mycobacterium tuberculosis MazF homolog, Rv1495 and its sole DNA topoisomerase I. Nucleic Acids Res. 38: 8219–8230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Iyer LM, Koonin EV, Aravind L. 2002. Extensive domain shuffling in transcription regulators of DNA viruses and implications for the origin of fungal APSES transcription factors. Genome Biol. 3: RESEARCH0012 doi:10.1186/gb-2002-3-3-research0012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Jayaraman R. 2008. Bacterial persistence: some new insights into an old phenomenon. J. Biosci. 33: 795–805 [DOI] [PubMed] [Google Scholar]
- 76. Jones RN. 2001. Resistance patterns among nosocomial pathogens: trends over the past few years. Chest 119(2 Suppl): 397S–404S doi:10.1038/msb.2011.98 [DOI] [PubMed] [Google Scholar]
- 77. Jørgensen MG, Pandey DP, Jaskolska M, Gerdes K. 2009. HicA of Escherichia coli defines a novel family of translation-independent mRNA interferases in bacteria and archaea. J. Bacteriol. 191: 1191–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kadioglu A, Weiser JN, Paton JC, Andrew PW. 2008. The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat. Rev. Microbiol. 6: 288–301 [DOI] [PubMed] [Google Scholar]
- 79. Kamada K, Hanaoka F. 2005. Conformational change in the catalytic site of the ribonuclease YoeB toxin by YefM antitoxin. Mol. Cell 19: 497–509 [DOI] [PubMed] [Google Scholar]
- 80. Kamada K, Hanaoka F, Burley SK. 2003. Crystal structure of the MazE/MazF complex: molecular bases of antidote-toxin recognition. Mol. Cell 11: 875–884 [DOI] [PubMed] [Google Scholar]
- 81. Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186: 8172–8180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Khoo SK, et al. 2007. Molecular and structural characterization of the PezAT chromosomal toxin-antitoxin system of the human pathogen Streptococcus pneumoniae. J. Biol. Chem. 282: 19606–19618 [DOI] [PubMed] [Google Scholar]
- 83. Kim Y, Wang X, Ma Q, Zhang XS, Wood TK. 2009. Toxin-antitoxin systems in Escherichia coli influence biofilm formation through YjgK (TabA) and fimbriae. J. Bacteriol. 191: 1258–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Knutsen E, Johnsborg O, Quentin Y, Claverys JP, Havarstein LS. 2006. BOX elements modulate gene expression in Streptococcus pneumoniae: impact on the fine-tuning of competence development. J. Bacteriol. 188: 8307–8312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kolodkin-Gal I, Hazan R, Gaathon A, Carmeli S, Engelberg-Kulka H. 2007. A linear pentapeptide is a quorum-sensing factor required for mazEF-mediated cell death in Escherichia coli. Science 318: 652–655 [DOI] [PubMed] [Google Scholar]
- 86. Korch SB, Hill TM. 2006. Ectopic overexpression of wild-type and mutant hipA genes in Escherichia coli: effects on macromolecular synthesis and persister formation. J. Bacteriol. 188: 3826–3836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kristoffersen P, Jensen GB, Gerdes K, Piškur J. 2000. Bacterial toxin-antitoxin gene system as containment control in yeast cells. Appl. Environ. Microbiol. 66: 5524–5526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kuchina A, et al. 2011. Temporal competition between differentiation programs determines cell fate choice. Mol. Syst. Biol. 7: 557 doi:10.1038/msb.2011.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kuwahara H, Soyer OS. 2012. Bistability in feedback circuits as a byproduct of evolution of evolvability. Mol. Syst. Biol. 8: 564 doi:10.1038/msb.2011.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Leplae R, et al. 2011. Diversity of bacterial type II toxin-antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Res. 39: 5513–5525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Leung V, Lévesque CM. 2012. A stress-inducible quorum-sensing peptide mediates the formation of persister cells with noninherited multidrug tolerance. J. Bacteriol. 194: 2265–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Lewis K. 2010. Persister cells. Annu. Rev. Microbiol. 64: 357–372 [DOI] [PubMed] [Google Scholar]
- 93. Li G-Y, Zhang Y, Inouye M, Ikura M. 2008. Structural mechanism of transcriptional autorepression of the Escherichia coli RelB/RelE antitoxin/toxin module. J. Mol. Biol. 380: 107–119 [DOI] [PubMed] [Google Scholar]
- 94. Lioy VS, et al. 2006. pSM19035-encoded zeta toxin induces stasis followed by death in a subpopulation of cells. Microbiology 152: 2365–2379 [DOI] [PubMed] [Google Scholar]
- 95. Lioy VS, Rey O, Balsa D, Pellicer T, Alonso JC. 2010. A toxin-antitoxin module as a target for antimicrobial development. Plasmid 63: 31–39 [DOI] [PubMed] [Google Scholar]
- 96. Liu M, Zhang Y, Inouye M, Woychik NA. 2008. Bacterial addiction module toxin Doc inhibits translation elongation through its association with the 30S ribosomal subunit. Proc. Natl. Acad. Sci. U. S. A. 105: 5885–5890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. López-Villarejo J, Diago-Navarro E, Hernández-Arriaga AM, Díaz-Orejas R. 2012. Kis antitoxin couples plasmid R1 replication and parD (kis,kid) maintenance modules. Plasmid 67: 118–127 [DOI] [PubMed] [Google Scholar]
- 98. Maisonneuve E, Shakespeare LJ, Jørgensen MG, Gerdes K. 2011. Bacterial persistence by RNA endonucleases. Proc. Natl. Acad. Sci. U. S. A. 108: 13206–13211 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 99. Makarova K, Wolf YI, Koonin EV. 2009. Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol. Direct 4: 19 doi:10.1186/1745-6150-4-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Mandell LA, et al. 2007. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin. Infect. Dis. 44: S27–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Marra A. 2004. Can virulence factors be viable antibacterial targets? Expert Rev. Anti Infect. Ther. 2: 61–72 [DOI] [PubMed] [Google Scholar]
- 102. Martin B, et al. 1992. A highly conserved repeated DNA element located in the chromosome of Streptococcus pneumoniae. Nucleic Acids Res. 20: 3479–3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Masuda Y, Miyakawa K, Nishimura Y, Ohtsubo E. 1993. chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J. Bacteriol. 175: 6850–6856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Meinhart A, Alonso JC, Strater N, Saenger W. 2003. Crystal structure of the plasmid maintenance system epsilon/zeta: functional mechanism of toxin zeta and inactivation by epsilon2zeta2 complex formation. Proc. Natl. Acad. Sci. U. S. A. 100: 1661–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Moreno-Córdoba I, et al. 2012. The toxin-antitoxin proteins RelBE2Spn of Streptococcus pneumoniae: characterization and association to their DNA target. Proteins 80: 1834–1846 [DOI] [PubMed] [Google Scholar]
- 106. Moscoso M, Domenech M, García E. 2010. Vancomycin tolerance in clinical and laboratory Streptococcus pneumoniae isolates depends on reduced enzyme activity of the major LytA autolysin or cooperation between CiaH histidine kinase and capsular polysaccharide. Mol. Microbiol. 77: 1052–1064 [DOI] [PubMed] [Google Scholar]
- 107. Motiejunaite R, Armalyte J, Markuckas A, Suziedeliene E. 2007. Escherichia coli dinJ-yafQ genes act as a toxin-antitoxin module. FEMS Microbiol. Lett. 268: 112–119 [DOI] [PubMed] [Google Scholar]
- 108. Moyed HS, Bertrand KP. 1983. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 155: 768–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Mrazek J, Gaynon LH, Karlin S. 2002. Frequent oligonucleotide motifs in genomes of three streptococci. Nucleic Acids Res. 30: 4216–4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Muñoz-Gomez AJ, Santos-Sierra S, Berzal-Herranz A, Lemonnier M, Diaz-Orejas R. 2004. Insights into the specificity of RNA cleavage by the Escherichia coli MazF toxin. FEBS Lett. 567: 316–320 [DOI] [PubMed] [Google Scholar]
- 111. Murayama K, Orth P, de la Hoz AB, Alonso JC, Saenger W. 2001. Crystal structure of omega transcriptional repressor encoded by Streptococcus pyogenes plasmid pSM19035 at 1.5 A resolution. J. Mol. Biol. 314: 789–796 [DOI] [PubMed] [Google Scholar]
- 112. Musher DM. 2003. How contagious are common respiratory tract infections? N. Engl. J. Med. 348: 1256–1266 [DOI] [PubMed] [Google Scholar]
- 113. Mutschler H, Gebhardt M, Shoeman RL, Meinhart A. 2011. A novel mechanism of programmed cell death in bacteria by toxin-antitoxin systems corrupts peptidoglycan synthesis. PLoS Biol. 9: e1001033 doi:10.1371/journal.pbio.1001033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Mutschler H, Meinhart A. 2011. ϵ/ζ systems: their role in resistance, virulence, and their potential for antibiotic development. J. Mol. Med. 89: 1183–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Mutschler H, Reinstein J, Meinhart A. 2010. Assembly dynamics and stability of the pneumococcal Epsilon Zeta antitoxin toxin (PezAT) system from Streptococcus pneumoniae. J. Biol. Chem. 285: 21797–21806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Nieto C, et al. 2007. The yefM-yoeB toxin-antitoxin systems of Escherichia coli and Streptococcus pneumoniae: functional and structural correlation. J. Bacteriol. 189: 1266–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Nieto C, et al. 2006. The chromosomal relBE2 toxin-antitoxin locus of Streptococcus pneumoniae: characterization and use of a bioluminescence resonance energy transfer assay to detect toxin-antitoxin interaction. Mol. Microbiol. 59: 1280–1296 [DOI] [PubMed] [Google Scholar]
- 118. Nieto C, Sadowy E, de la Campa AG, Hryniewicz W, Espinosa M. 2010. The relBE2Spn toxin-antitoxin system of Streptococcus pneumoniae: role in antibiotic tolerance and functional conservation in clinical isolates. PLoS One 5: e11289 doi:10.1371/journal.pone.0011289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Overgaard M, Borch J, Gerdes K. 2009. RelB and RelE of Escherichia coli form a tight complex that represses transcription via the ribbon-helix-helix motif in RelB. J. Mol. Biol. 394: 183–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Overgaard M, Borch J, Jørgensen MG, Gerdes K. 2008. Messenger RNA interferase RelE controls relBE transcription by conditional cooperativity. Mol. Microbiol. 69: 841–857 [DOI] [PubMed] [Google Scholar]
- 121. Pabo CO, Sauer RT. 1992. Transcription factors: structural families and principles of DNA recognition. Annu. Rev. Biochem. 61: 1053–1095 [DOI] [PubMed] [Google Scholar]
- 122. Pandey DP, Gerdes K. 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33: 966–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Paul D, Pandey G, Jain RK. 2005. Suicidal genetically engineered microorganisms for bioremediation: need and perspectives. Bioessays 27: 563–573 [DOI] [PubMed] [Google Scholar]
- 124. Pedersen K, Christensen KS, Gerdes K. 2002. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol. Microbiol. 45: 501–510 [DOI] [PubMed] [Google Scholar]
- 125. Pedersen K, et al. 2003. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112: 131–140 [DOI] [PubMed] [Google Scholar]
- 126. Ramage HR, Connolly LE, Cox JS. 2009. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet. 5: e1000767 doi:10.1371/journal.pgen.1000767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Ramos JL, et al. 1994. The behavior of bacteria designed for biodegradation. Biotechnology 12: 1349–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Rao CV, Wolf DM, Arkin AP. 2002. Control, exploitation and tolerance of intracellular noise. Nature 420: 231–237 [DOI] [PubMed] [Google Scholar]
- 129. Rimini R, et al. 2000. Global analysis of transcription kinetics during competence development in Streptococcus pneumoniae using high density DNA arrays. Mol. Microbiol. 36: 1279–1292 [DOI] [PubMed] [Google Scholar]
- 130. Rotem E, et al. 2010. Regulation of phenotypic variability by a threshold-based mechanism underlies bacterial persistence. Proc. Natl. Acad. Sci. U. S. A. 107: 12541–12546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Saavedra De Bast M, Mine N, Van Melderen L. 2008. Chromosomal toxin-antitoxin systems may act as antiaddiction modules. J. Bacteriol. 190: 4603–4609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Saluja SK, Weiser JN. 1995. The genetic basis of colony opacity in Streptococcus pneumoniae: evidence for the effect of box elements on the frequency of phenotypic variation. Mol. Microbiol. 16: 215–227 [DOI] [PubMed] [Google Scholar]
- 133. Schmidt O, et al. 2007. prlF and yhaV encode a new toxin-antitoxin system in Escherichia coli. J. Mol. Biol. 372: 894–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Schumacher MA, et al. 2009. Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science 323: 396–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Shah D, et al. 2006. Persisters: a distinct physiological state of Escherichia coli. BMC Microbiol. 6: 53 doi:10.1186/1471-2180-6-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Shao Y, et al. 2011. TADB: a Web-based resource for type 2 toxin-antitoxin loci in bacteria and archaea. Nucleic Acids Res. 39: D606–D611 doi:10.1093/nar/gkq908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Stocker W, Bode W. 1995. Structural features of a superfamily of zinc-endopeptidases: the metzincins. Curr. Opin. Struct. Biol. 5: 383–390 [DOI] [PubMed] [Google Scholar]
- 138. Takagi H, et al. 2005. Crystal structure of archaeal toxin-antitoxin RelE-RelB complex with implications for toxin activity and antitoxin effects. Nat. Struct. Mol. Biol. 12: 327–331 [DOI] [PubMed] [Google Scholar]
- 139. Tan Q, Awano N, Inouye M. 2011. YeeV is an Escherichia coli toxin that inhibits cell division by targeting the cytoskeleton proteins, FtsZ and MreB. Mol. Microbiol. 79: 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Tashiro Y, et al. 2012. RelE-mediated dormancy is enhanced at high cell density in Escherichia coli. J. Bacteriol. 194: 1169–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Tettelin H, et al. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293: 498–506 [DOI] [PubMed] [Google Scholar]
- 142. Thattai M, van Oudenaarden A. 2004. Stochastic gene expression in fluctuating environments. Genetics 167: 523–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Thomas CM. 2000. The horizontal gene pool. Harwood Academic Publishers, Amsterdam, Netherlands [Google Scholar]
- 144. Trovatti E, Cotrim CA, Garrido SS, Barros RS, Marchetto R. 2008. Peptides based on CcdB protein as novel inhibitors of bacterial topoisomerases. Bioorg. Med. Chem. Lett. 18: 6161–6164 [DOI] [PubMed] [Google Scholar]
- 145. Van Melderen L. 2002. Molecular interactions of the CcdB poison with its bacterial target, the DNA gyrase. Int. J. Med. Microbiol. 291: 537–544 [DOI] [PubMed] [Google Scholar]
- 146. van Melderen L. 2010. Toxin-antitoxin systems: why so many, what for? Curr. Opin. Microbiol. 13: 781–785 [DOI] [PubMed] [Google Scholar]
- 147. Vazquez-Laslop N, Lee H, Neyfakh AA. 2006. Increased persistence in Escherichia coli caused by controlled expression of toxins or other unrelated proteins. J. Bacteriol. 188: 3494–3497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Walker JA, Allen RL, Falmagne P, Johnson MK, Boulnois GJ. 1987. Molecular cloning, characterization, and complete nucleotide sequence of the gene for pneumolysin, the sulfhydryl-activated toxin of Streptococcus pneumoniae. Infect. Immun. 55: 1184–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Weihofen WA, Cicek A, Pratto F, Alonso JC, Saenger W. 2006. Structures of Omega repressors bound to direct and inverted DNA repeats explain modulation of transcription. Nucleic Acids Res. 34: 1450–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Williams JJ, Hergenrother PJ. 2012. Artificial activation of toxin-antitoxin systems as an antibacterial strategy. Trends Microbiol. 20: 291–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Winther KS, Gerdes K. 2011. Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc. Natl. Acad. Sci. U. S. A. 108: 7403–7407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Wozniak RAF, Waldor MK. 2009. A toxin-antitoxin system promotes the maintenance of an integrative conjugative element. PLoS Genet. 5: e1000439 doi:10.1371/journal.pgen.1000439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Xu Y, Kanauchi A, von Arnim AG, Piston DW, Johnson CJ. 2003. Bioluminescence resonance energy transfer (BRET): a new technique for monitoring protein-protein interactions in living cells. Methods Enzymol. 360: 289–301 [DOI] [PubMed] [Google Scholar]
- 154. Yamaguchi Y, Inouye M. 2009. mRNA interferases, sequence-specific endoribonucleases from the toxin-antitoxin systems. Prog. Mol. Biol. Transl. Sci. 85: 467–500 [DOI] [PubMed] [Google Scholar]
- 155. Yamaguchi Y, Inouye M. 2011. Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nat. Rev. Microbiol. 9: 779–790 [DOI] [PubMed] [Google Scholar]
- 156. Yang M, Gao C, Wang Y, Zhang H, He Z-G. 2010. Characterization of the interaction and cross-regulation of three Mycobacterium tuberculosis RelBE modules. PLoS One 5: e10672 doi:10.1371/journal.pone.0010672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Zhang Y, Inouye M. 2009. The inhibitory mechanism of protein synthesis by YoeB, an Escherichia coli toxin. J. Biol. Chem. 284: 6627–6638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Zhang Y, Inouye M. 2011. RatA (YfjG), an Escherichia coli toxin, inhibits 70S ribosome association to block translation initiation. Mol. Microbiol. 79: 1418–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Zhang Y, Zhang J, Hoeflich KP, Ikura M, Inouye M. 2003. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 12: 913–923 [DOI] [PubMed] [Google Scholar]
- 160. Zhu L, Sharp JD, Kobayashi H, Woychik NA, Inouye M. 2010. Noncognate Mycobacterium tuberculosis toxin-antitoxins can physically and functionally interact. J. Biol. Chem. 285: 39732–39738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Zielenkiewicz U, Ceglowski P. 2005. The toxin-antitoxin system of the streptococcal plasmid pSM19035. J. Bacteriol. 187: 6094–6105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Zielenkiewicz U, Kowalewska M, Kaczor C, Ceglowski P. 2009. In vivo interactions between toxin-antitoxin proteins Epsilon and Zeta of streptococcal plasmid pSM19035 in Saccharomyces cerevisiae. J. Bacteriol. 191: 3677–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]