Abstract
Summary: Intuitively, it may seem that from the perspective of an individual bacterium the ocean is a vast, dilute, and largely homogeneous environment. Microbial oceanographers have typically considered the ocean from this point of view. In reality, marine bacteria inhabit a chemical seascape that is highly heterogeneous down to the microscale, owing to ubiquitous nutrient patches, plumes, and gradients. Exudation and excretion of dissolved matter by larger organisms, lysis events, particles, animal surfaces, and fluxes from the sediment-water interface all contribute to create strong and pervasive heterogeneity, where chemotaxis may provide a significant fitness advantage to bacteria. The dynamic nature of the ocean imposes strong selective pressures on bacterial foraging strategies, and many marine bacteria indeed display adaptations that characterize their chemotactic motility as “high performance” compared to that of enteric model organisms. Fast swimming speeds, strongly directional responses, and effective turning and steering strategies ensure that marine bacteria can successfully use chemotaxis to very rapidly respond to chemical gradients in the ocean. These fast responses are advantageous in a broad range of ecological processes, including attaching to particles, exploiting particle plumes, retaining position close to phytoplankton cells, colonizing host animals, and hovering at a preferred height above the sediment-water interface. At larger scales, these responses can impact ocean biogeochemistry by increasing the rates of chemical transformation, influencing the flux of sinking material, and potentially altering the balance of biomass incorporation versus respiration. This review highlights the physical and ecological processes underpinning bacterial motility and chemotaxis in the ocean, describes the current state of knowledge of chemotaxis in marine bacteria, and summarizes our understanding of how these microscale dynamics scale up to affect ecosystem-scale processes in the sea.
INTRODUCTION
Natural ecosystems are typically heterogeneous (105). Consequently, organisms must either tune their metabolism to prevailing local conditions (208) or migrate to favorable environments (83). Migratory responses to heterogeneous resources require the ability to acquire information on the resource landscape and direct locomotion accordingly. Bacteria often inhabit highly heterogeneous realms and employ a wide range of strategies to navigate their environment, with the ultimate objective of locating and exploiting favorable niches. Many bacteria are motile and propel themselves by rotating helical flagella driven by molecular motors (51, 113). Since the initial observations of bacterial motility, a rich set of “taxes” (i.e., directional movement responses) in response to external stimuli have been identified (70), including motion directed by light (phototaxis), temperature (thermotaxis), magnetic fields (magnetotaxis), electric fields (galvanotaxis), pH (pH-taxis), oxygen (aerotaxis), and chemical cues (chemotaxis) (29, 59, 63, 91). The most ubiquitous and best studied of these behaviors is chemotaxis (112).
Chemotaxis is the ability to sense and direct movement in response to chemical gradients. By using transmembrane chemoreceptors to measure chemical concentrations in their immediate vicinity and signal transduction systems to subsequently process this information, bacteria can sense chemical gradients and tune motility accordingly (202). This chemotactic behavior, which allows bacteria to swim toward favorable chemicals or away from noxious ones, was first described in the late 1800s (47, 149). Since the 1960s, chemotaxis has grown to be one of the best-described sensory systems in biology, chiefly in well-defined model organisms such as Escherichia coli (25, 112, 152, 202). It is now recognized that most motile bacteria are capable of chemotaxis (112). At a broad level, the biochemical control mechanisms of chemotaxis appear to be relatively conserved across bacteria (152, 187, 202), but growing evidence suggests that chemotactic capabilities can differ, often deviating substantially from those of well-described model bacteria (130, 152).
Owing to the wide range of bacterial habitats, the ecological drivers and implications of bacterial chemotaxis are extremely diverse. Many pathogenic bacteria use chemotaxis to find suitable colonization sites: chemotaxis can guide Helicobacter pylori to the mucus lining of the human stomach (150), Vibrio cholerae toward the intestinal mucosa (57), and Vibrio anguillarum from seawater to the skin of fish (139). In the wider natural environment, bacterial chemotaxis affects the transformation and cycling of chemicals (142, 175) and can favor mutualistic microbe-microbe (33), microbe-plant (146), and microbe-animal (43) relationships. Underpinning the diversity of chemotactic expression and function is the bacteria's ability to detect and locate more favorable, spatially localized microenvironments. Even within the vast and seemingly well-mixed ocean, bacteria can use chemotaxis to exploit localized dissolved substrate patches (28), attach to biotic and abiotic surfaces (39), find suspended and sinking biogenic particles (94), and colonize animal hosts (17).
The ocean is one of the largest reservoirs of bacteria. On average, every liter of seawater contains ∼1 billion bacteria, and there are a total of ∼1029 bacteria in the world's oceans (204), constituting the bulk of ocean biomass (151). Each liter of seawater may contain several thousand different bacterial “species” (185), and the staggering diversity of their genetic complement (197, 209) allows marine bacteria to perform an extremely broad suite of functions (48). For example, marine bacteria are the primary biotic drivers of ocean biogeochemistry, controlling most of the oceans' major chemical cycles (carbon, nitrogen, and sulfur) (13, 32, 48, 181). Marine bacteria also act as the main conduit for the transfer of energy and matter from the oceans' large pools of dissolved organic matter (DOM) to higher trophic levels through the “microbial loop” (11, 100, 101). Finally, marine bacteria exhibit important symbiotic, mutualistic, and pathogenic interactions with many species of marine animals and plants (60, 73, 154, 159). In all of these functions, microscale behavior—including chemotaxis—may play a pivotal role that has been largely underappreciated, because oceanographers have traditionally considered the ecology of marine bacteria over large spatiotemporal scales.
Here we review the occurrence, characteristics, and consequences of bacterial chemotaxis in the ocean. Although recent genomic (5, 131), metagenomic (45, 196), and metatranscriptomic (61, 119, 179, 200) studies have revealed ecologically relevant shifts in the abundance and nature of bacterial motility and chemotaxis genes in the ocean, these will not be covered in this review, where we instead focus on a detailed treatment of the dynamic physicochemical landscape experienced by marine bacteria and a description of how this influences bacterial motility and chemotaxis. We will also examine the role of bacterial chemotaxis in interdomain interactions and how the chemotaxis of marine bacteria can shape ecosystem-level processes in the ocean, including large-scale biogeochemical cycling.
MARINE MICROENVIRONMENTS: A BACTERIUM'S VIEW OF THE OCEAN
The strong emphasis of chemotaxis research on model organisms, such as E. coli, has resulted in a blueprint against which the chemotactic behavior of other species can be compared. Despite the presence of highly conserved design features at the molecular level (187, 202), the diversity of environments experienced by different species of bacteria has resulted in specific molecular, mechanical, and behavioral adaptations. In comparison to enteric bacteria such as E. coli, marine bacteria are faced with a very different set of environmental challenges. Seawater is characterized by generally low (pM to nM) bulk concentrations of growth substrates, which are often injected into the water column by a plethora of point source events or through exudation from biotic or abiotic surfaces. These nutrients, and the organisms that consume them, are exposed to steady or intermittent fluid motion driven by currents and turbulence. Although seawater may ostensibly seem to be a dilute homogeneous soup and has long been modeled as such by oceanographers (76), the world experienced by marine bacteria inhabiting both eutrophic and oligotrophic ocean habitats can be tremendously heterogeneous, with a broad suite of discrete niches where growth may be enhanced by favorable chemical conditions (Fig. 1). This microspatial architecture likely influences the behavior of individual bacteria and the competitive interactions between populations.
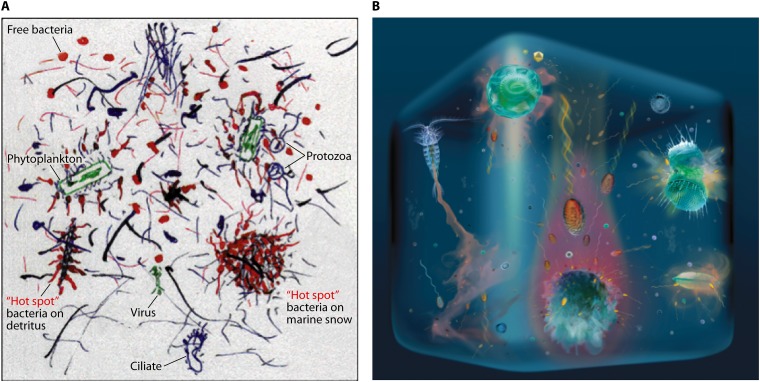
Fig 1.
Two views of the ocean at the microscale, emphasizing the heterogeneity of resources and related microbial interactions and distributions. (A) “Hot spots” of microbial activity occur in association with detritus, marine snow particles, and phytoplankton cells. Panel A further emphasizes the rich texture of particles, filaments, and polymers of various sizes and origins, which contribute to microscale heterogeneity. (Reprinted from reference 13 with permission from AAAS.) (B) Organic substrates diffuse from a range of sources, including zooplankton excretions (left), phytoplankton exudation (the “phycosphere”) (top; bottom right), phytoplankton lysis (top right), and settling marine snow particles (center bottom). The last, in particular, can produce intense, comet-like plumes of dissolved matter. (Adapted from reference 186a with permission.) Both panels emphasize the distinction between nonmotile cells and flagellated, motile cells; the latter often are able to cluster at hot spots by chemotaxis. We estimate an approximate scale for each image to be 1 cm.
The Phycosphere and Other Pulses of Dissolved Organic Substrates
In the water column, the chemical seascape experienced by planktonic bacteria can be highly heterogeneous, with ingestion, digestion, egestion, excretion, and exudation by other marine organisms producing a habitat characterized by the frequent and pervasive occurrence of microscale chemical gradients. For example, bacteria experience locally elevated concentrations of dissolved organic carbon (DOC) in the neighborhood of individual phytoplankton cells due to the exudation of photosynthetic products (22). Both healthy and stressed marine phytoplankton release significant amounts of simple sugars, amino acids, organic acids, complex polysaccharides, and lipids into the surrounding water (1, 54, 72, 84). From the perspective of a marine bacterium, the resultant microzone of exudates (or “phycosphere” [22]), which extends a few cell diameters from the surface of the phytoplankton cell, represents a nutrient-rich microenvironment (22) (Fig. 2). Exploitation of this microenvironment has been proposed to substantially enhance the growth rate of heterotrophic bacteria in the vicinity of phytoplankton cells (12, 31, 79, 124). Even the plume of chemical exudates left in the wake of sinking (80) or swimming (19) phytoplankton cells is believed to generate a nutrient-rich niche that bacteria may be able to exploit, provided that the size of the phytoplankton cell is at least a few micrometers (79).
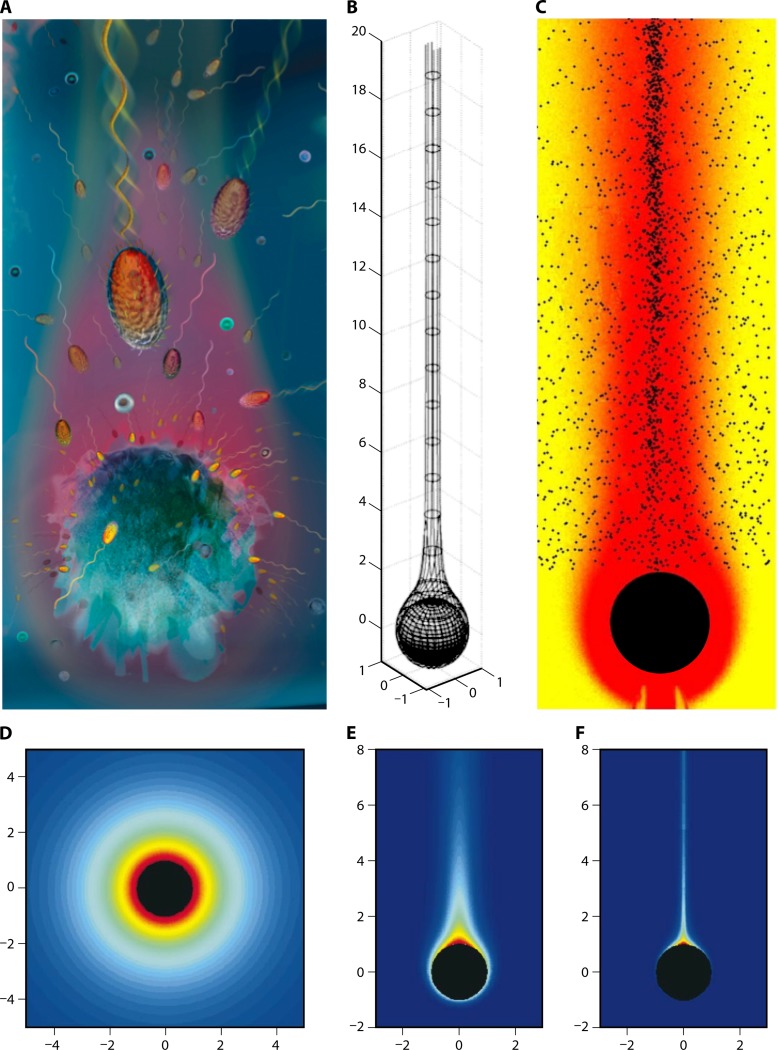
Fig 2.
The phycosphere. (A and B) An artist's view of the diffusion boundary layer around individual phytoplankton cells. Motile bacteria can use chemotaxis to respond to the nutrient gradients associated with the enhanced dissolved organic matter concentrations within the phycosphere. Magnified from Fig. 1B. (C) Experimental observation of the phycosphere. A coastal seawater sample was collected via net tow to concentrate planktonic particles. The particles and entrained seawater were incubated for 24 h at 22°C, which enriched the bacterial community and stimulated motility. Settled particles were gently resuspended just prior to video capture via dark-field microscopy. The swimming trajectories shown here illustrate dense accumulations of chemotactic cells near the surface of an individual Chaetoceros diatom (S. Smriga and R. Stocker, unpublished). (D) Trajectories of individual bacteria in the same observation shown in panel C can be obtained using digital image analysis, here with an in-house automated algorithm that removes those shorter than 5 μm. This revealed a mean swimming speed of 25 μm/s. Crosses denote starting points of individual trajectories (S. Smriga, K. Son, and R. Stocker, unpublished). (E) Decay of DOM concentration with distance r from the center of a DOM-exuding phytoplankton. The curves refer to two phytoplankton radii, 10 μm (lower, red curve) and 50 μm (upper, blue curve). The black dashed line provides a reference to a bulk background concentration of 10 nM (typical of many organic solutes in the ocean). The DOM concentration field was obtained by solving the steady-diffusion equation for a constant source, following what was done by Seymour et al. (175). The phytoplankton cell was assumed to have a 100 mM internal concentration and to exude 100% of its daily production of the solute. This upper limit of the exudation rate is most applicable to stressed or senescent cells. The diffusion coefficient for the solute was 7.2 × 10−10 m2/s, consistent with values for many organic substrates, such as DMSP.
Plumes of organic and inorganic substrates are also generated by the excretions of swimming zooplankton (Fig. 1B), which increase bulk concentrations of organic and inorganic substrates in the water column (99, 148). Individual excretion events have previously been modeled as diffusing pulses of ammonium, having an initial concentration of 0.2 to 5 μM and an initial width of ∼100 μm (78, 120). While the importance of these pulses for the growth of phytoplankton has been recognized and investigated (64, 78, 103, 104), zooplankton excretions may also represent a significant nutrient source for heterotrophic bacteria (27). Nutrient patches produced by sloppy feeding processes, whereby zooplankton and other animals release organic material into the surrounding water during feeding (99), also enhance bacterial growth by producing ephemeral diffusing patches of DOC (148). Short-lived microscale nutrient patches further arise following the lysis of other microbes that have been infected by viruses (28): viral infection and lysis result in the pulse release of host organic material, creating ephemeral, nutrient-rich micropatches for bacteria in the surrounding water (156) (Fig. 3).
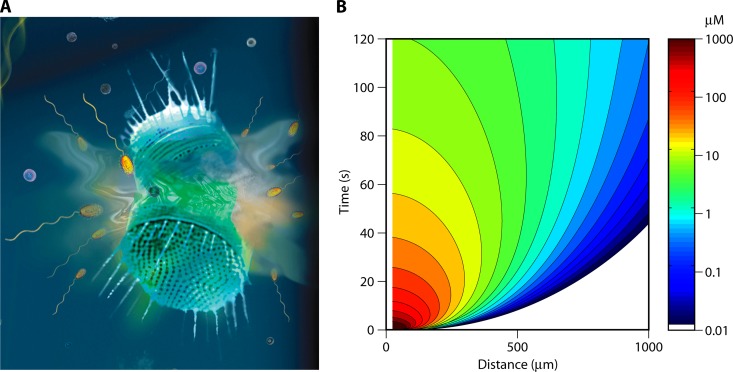
Fig 3.
Ephemeral nutrient pulses. (A) Artist's view of the lysis of a phytoplankton cell, resulting in a strong yet ephemeral pulse of dissolved organic matter, to which marine bacteria can respond by chemotaxis if sufficiently rapid. Magnified from Fig. 1B. (B) The DOM concentration field associated with a lysis event, computed by a mathematical model of diffusion from a pulse source, following what was done by Seymour et al. (175). The abscissa shows the distance from the center of the lysing cell, and the ordinate shows the time after the lysis event. Colors indicate the concentration in μM. This computation applies to a cell with a 25-μm radius, an internal concentration of 100 mM, and a diffusion coefficient of the solute of 7.2 × 10−10 m2/s. These parameters are appropriate, for example, for a medium-size, DMSP-producing phytoplankton.
Particles as Resource Islands
Marine bacteria can exploit microscale hot spots of particulate organic carbon (POC) associated with suspended and sinking particles. These organic particles range in size from micrometer-sized colloids (97) to millimeter-sized marine snow aggregates (9). Marine snow refers to a conspicuous collection of particles, often operationally defined to be larger than 500 μm, derived from a number of sources, including the coagulation of dead or senescent phytoplankton cells (9, 78) and zooplankton fecal pellets (88, 195). Marine snow and other organic particles can contain concentrations of organic and inorganic molecules that often exceed background concentrations by 2 to 4 orders of magnitude (153, 177). Consequently, these particles are heavily colonized by heterotrophic bacteria (6, 7), which use a suite of surface-bound ectohydrolases, including proteases, lipases, chitinases, and phosphatases, to hydrolyze particulate organic material into dissolved form (184).
As a consequence of this hydrolysis, the nutritional benefits provided by organic particles may extend beyond those experienced by particle-attached bacteria (14). Due to diffusion, a substantial fraction of the hydrolyzed material escapes uptake by the attached community (95). The excess solutes leak from the particles to form nutrient-rich gradients in the surrounding water. Using microelectrodes, Alldredge and Cohen (8) directly demonstrated the existence of these chemical gradients extending from the surfaces of individual particles. When the particle is heavier than water, it sinks through the water column, and the associated flow distorts gradients into comet-like plumes in the particle's wake (65, 95, 199) (Fig. 4). These plumes can be exploited by free-living microbes, potentially increasing bacterial production by up to an order of magnitude (14, 93, 186).
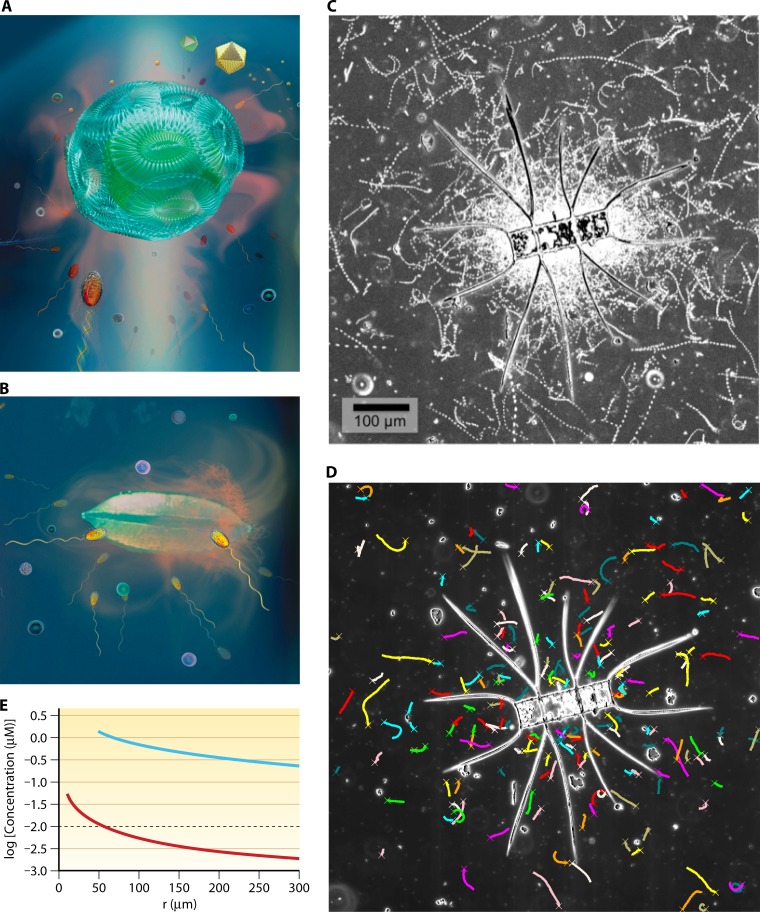
Fig 4.
Marine particles. (A) Artist's view of the plume of dissolved organic matter emanating from a sinking marine snow particle. Magnified from Fig. 1B. (B) Mathematical model illustrating the large vertical extent that these plumes can achieve. Shown is a 0.5-cm-radius particle sinking at 1 mm/s. The plume corresponds to a concentration of amino acids of 30 nM above background. Distances are in units of particle radii. (Reproduced from reference 95 with permission.) (C) Chemotactic accumulation of P. haloplanktis bacteria in the plume of a particle, obtained by video microscopy using a microfluidic experimental model of a marine particle and its plume. Colors indicate concentration of solutes, and black dots are individual bacteria. The particle size was 500 μm and its sinking speed 110 μm/s, corresponding to a Peclet number of 110 (see panels D to F). (Reproduced from reference 186 with permission.) (D to F) The shape of the plume for different Peclet numbers. The Peclet number is a dimensionless index, obtained as the product of the particle radius, its sinking speed, and the inverse of the molecular diffusivity of the solutes forming the plume. Shown are plumes for Peclet numbers of 0 (D), 100 (E), and 10,000 (F). (Reproduced from reference 95 with permission.)
In between the dissolved and particulate phases of organic matter lies a suite of colloids, transparent polymers, and organic gels that add further structure to microbial microenvironments and are also heavily colonized by bacteria (10, 110). These different phases constitute an organic matter continuum, with rich microarchitecture at multiple scales (13, 198) (Fig. 1A). Superimposed upon this physical structure is the mélange of DOC gradients derived from multiple microbial production and uptake processes (15, 71).
As a result, from a bacterium's perspective, seawater is a highly heterogeneous landscape, dotted by diverse microenvironments that stand out against an otherwise nutrient-depleted background. The existence of this heterogeneous seascape is supported by studies that have examined the microscale spatiotemporal distributions of bacteria in the ocean and demonstrated the existence of substantial heterogeneity in the abundance, activity, and composition of microbial communities across millimeter scales (110, 167–171).
Animals and Plants as Microbial Niches
Both sedentary and swimming animals can provide discrete islands for microbial colonization. Zooplankton act as hot spots for bacteria, with individual copepods supporting populations of more than 10,000 bacteria (188). The surfaces and gastrointestinal tracts of fish and marine mammals also represent nutrient-rich microhabitats that accordingly contain dense populations of bacteria (137, 160, 180). Steep chemical gradients occur on the external surfaces of benthic organisms, including corals, sponges, and marine plants, which often host microbial assemblages that are species specific and distinct from the free-living community (157, 158, 203). For instance, within the 10- to 1,000-μm-thick layer of mucus that covers the surface of many coral species, concentrations of organic and inorganic substrates are often up to 3 to 4 orders of magnitude higher than in the surrounding seawater (34, 205), and this microenvironment typically hosts highly active and diverse microbial assemblages (147, 157, 158). Chemical gradients also occur on the surfaces of benthic marine plants and macroalgae, which exude high concentrations of organic carbon into the surrounding water and sediments via fronds and roots (69, 132).
Gradients at the Sediment-Water Interface
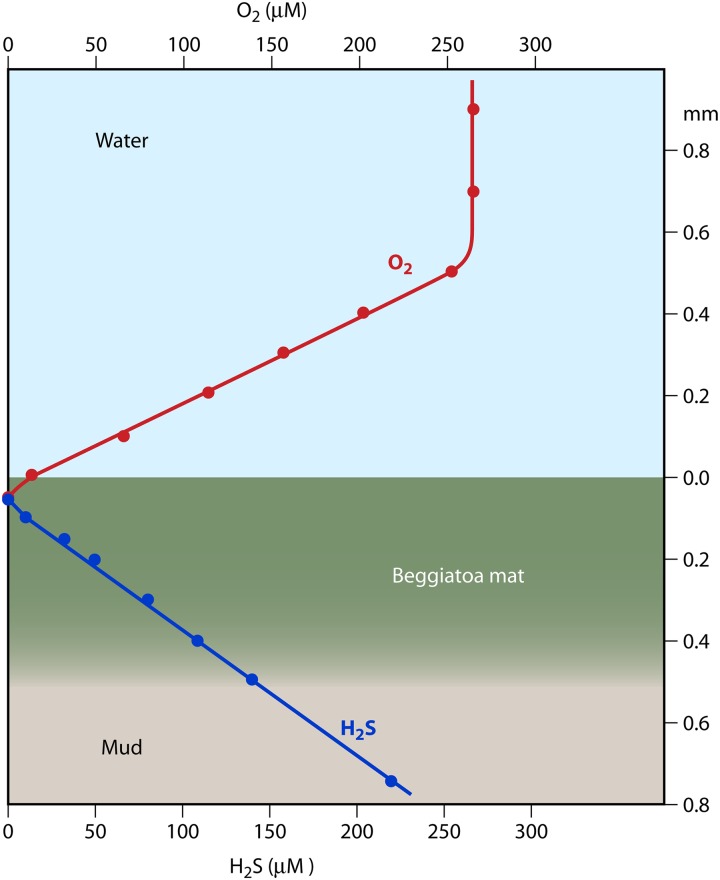
As a consequence of the high concentrations of organic material and the intense microbial activity in marine sediments, steep microgradients also occur at the sediment-water interface (165) (Fig. 5). These chemical gradients are often geometrically “simpler” than those found in the water column, because they extend primarily in the vertical direction, producing a largely one-dimensional chemical landscape. Flow and turbulence can disrupt the smoothness of this chemical environment but typically not the layering of chemistry and biology (87, 165). Microelectrode measurements at high vertical resolution (50-μm depth increments) have revealed linear countergradients of oxygen and hydrogen sulfide at the sediment-water interface (86) (Fig. 5).
Fig 5.
Countergradients of oxygen and sulfide at the sediment-water interface, measured using microelectrodes. Respiration in the sediments depletes oxygen, which in the absence of turbulence exhibits a linear concentration profile from the oxygenated, circulating water above to the anoxic conditions near the interface. The region of transition is the diffusion boundary layer, here ∼500 μm thick. Simultaneously, sulfide diffuses upwards and is respired near the interface, also leading to a linear concentration profile. Shown here is also the location of a mat of the bacteria Beggiatoa, which use gliding motility to adjust their position at the oxygen-sulfide interface. (Reproduced from reference 86 with permission.)
THE PHYSICS OF BACTERIAL MICROENVIRONMENTS
We have seen above that chemical heterogeneity in the ocean arises from a diverse range of microscale sources, from which solutes spread by molecular diffusion. Importantly, this heterogeneity is not erased by ocean turbulence, contrary to one's intuition that turbulence always mixes chemical gradients. Chemical gradients are affected by turbulence only above a minimum length scale, typically hundreds of micrometers to a few millimeters in the ocean (92, 190), and even then the effect of flow is considerably richer than simple mixing, as we highlight below. Before addressing the role of flow, we illustrate the different facets of diffusion and the diversity of gradients it creates.
A Diffusion-Dominated World: the Diffusion Boundary Layer
The sediment-water interface provides a convenient starting point for this discussion, because the physicochemical microenvironment has been characterized with great precision using microelectrodes and has been found to consist largely of one-dimensional gradients along the vertical direction (86, 165) (Fig. 5). Assuming a constant and uniform release rate (for example, of hydrogen sulfide) from the sediment surface, molecular diffusion establishes a linear concentration gradient reaching a fraction of a millimeter (0.1 to 1 mm) into the water above the sediments. This region is the “diffusion boundary layer” (DBL) (165, 192). The DBL harbors steep gradients not only of substances released from the sediments but also of those (e.g., oxygen) diffusing in from the water above and consumed at the sediments (165, 192). The DBL is contained within a region of water adjacent to the sediment-water interface (the “momentum boundary layer”) where turbulence is quenched, due to the proximity of the solid surface, and flow is thus laminar, preventing the rapid erosion of gradients. Therefore, within the DBL vertical transport occurs by molecular diffusion rather than by turbulent dispersion.
DBLs surround all solid surfaces in the ocean, including coral surfaces and animal skins, and in some cases include a mucus layer that further quenches flow (205). Flow over the surface has the effect of thinning the DBL, sharpening the gradients, and thus increasing solute transport to or from the surface. This happens, for example, at the sediment-water interface (165, 192) and over corals (98), where the DBL becomes thinner as the speed of the flow increases. DBLs are important because they control the exchange of solutes at those interfaces (192) and because they harbor largely steady gradients that can serve as robust chemical cues for chemotactic microorganisms.
DBLs also occur around microorganisms. The phycosphere (see “The Phycosphere and Other Pulses of Dissolved Organic Substrates” above) (Fig. 2), for example, is the DBL surrounding a phytoplankton cell, where the release of DOC or oxygen occurs at the cell surface (22, 28). Here, too, transport is dominated by molecular diffusion, but the concentration profile is not linear as in the case of the sediment-water interface. Rather, concentration decreases as the inverse of the distance from the phytoplankton cell surface, a consequence of the three-dimensional nature of the phycosphere (92) (Fig. 4D). Gradients in the phycosphere are steady as long as the release rate is constant.
Ephemeral Nutrient Pulses
Microscale gradients are not always steady; unsteady gradients are also very common. For instance, the lysis of a microorganism following viral infection results in a cell bursting and suddenly releasing its contents into the surrounding water (156) (Fig. 3A). A sloppy feeding event, where a predator only partially consumes a microbial prey (27), also produces highly unsteady gradients, as does the excretion of solutes by larger organisms such as zooplankton (103, 104). In all of these cases, the sudden pulse of nutrients creates strong solute gradients, which eventually dissipate by molecular diffusion (27). As intracellular concentrations (∼mM) of organic matter (for instance, in phytoplankton) can be 3 to 6 orders of magnitude higher than those under background conditions (∼nM), enhanced DOM concentrations can linger in the region surrounding the lysis event for considerable time, enabling chemotactic bacteria to respond (Fig. 3B). For example, concentrations of a solute with diffusivity D around a lysing cell of radius R will be reduced 1,000-fold relative to intracellular concentrations over a time on the order of (10R)2/D. This amounts to ∼20 min for R = 100 μM and D = 10−9 m2/s. The dependence of this time scale on the solute's diffusivity indicates that the multitude of compounds making up the intracellular space of a phytoplankton cell (72) do not diffuse at the same rate. Instead, higher-molecular-weight compounds diffuse more slowly, and their gradients will linger for longer. This difference can be substantial, because diffusivities can vary by 2 to 4 orders of magnitude.
A further source of unsteadiness arises when the nutrient source moves relative to the surrounding water. This is the case for sinking marine snow particles and fecal pellets (93) but potentially also for swimming phytoplankton cells (19, 108). The release of solutes, coupled with the movement relative to the water, distorts the otherwise nearly spherical diffusion boundary layer into a comet-like plume (Fig. 4A and B); steep gradients occur over a compressed region in front of the particle, and a long tail of shallower gradients characterizes the wake behind the particle (93). The plume becomes progressively more compressed in front of the particle and more slender in its wake as the particle's Peclet number increases. The Peclet number, Pe = UR/D, is a dimensionless parameter that captures the combined effect of particle size R, sinking speed U, and solute diffusivity D on the shape of the chemical plume (68, 93) (Fig. 4D to F).
A Turbulent World: the Batchelor Scale
Turbulence can also inject considerable heterogeneity into small-scale chemical landscapes. Although everyday intuition tells us that turbulent flow homogenizes solutes, underlying this mixing is the process of stirring, whereby turbulent whirls stretch and fold solutes such as DOM into ever-finer sheets and filaments (Fig. 6). This erratic weaving proceeds down to the Batchelor scale (190), the length scale below which molecular diffusion dissipates gradients, truly mixing solutes. The Batchelor scale, LB = (νD2/ε)1/4, increases with the kinematic viscosity of water, ν, and with the diffusivity of the solute, D, and decreases with the turbulent dissipation rate, ε, a measure of the strength of turbulence. For typical marine conditions, the Batchelor scale ranges from 30 to 300 μm (68). This should be considered a “universal” scale in microbial oceanography, because any source of solutes, whether microscopic or macroscopic, will ultimately be stirred into Batchelor-scale filaments (Fig. 6), provided that its initial size is larger than LB. For solute patches smaller than LB, turbulence amounts to a simple stretching of the patch. For solute patches larger than LB, such as the plumes of settling particles, turbulence acts to fragment the patch into smaller, disconnected patches, increasing the heterogeneity of the nutrient field (201). Beyond its effect in ultimately mixing solutes, turbulence can therefore contribute to produce a rich fabric of microscale gradients.
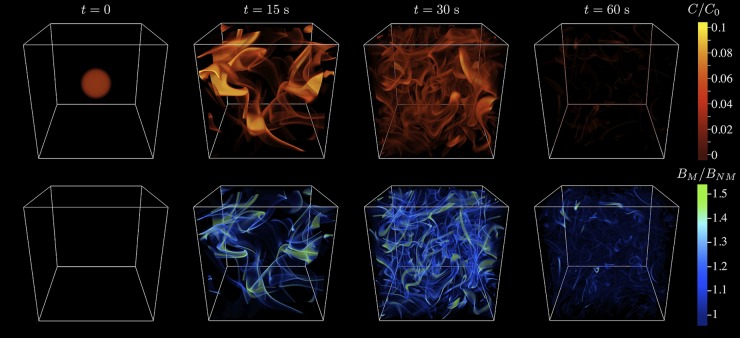
Fig 6.
Effect of turbulence on the chemotaxis of marine microbes. Top row, a microscale nutrient patch is stretched, folded, and stirred by turbulence to create a tangled web of sheets and filaments as small as the Batchelor scale (30 to 300 μm). C denotes the nutrient concentration and is shown relative to the initial concentration C0. Bottom row, motile bacteria with a chemotactic velocity of 20 μm/s, initially distributed uniformly, accumulate within the nutrient sheets and filaments by chemotaxis, achieving a significant nutrient gain compared to nonmotile species. The characteristic time scale of this process, for a 2.5-mm patch in moderately strong turbulence (turbulent dissipation rate, 10−6 W/kg), is ∼1 min. The domain size is 5.65 cm. BM denotes the concentration of motile bacteria and is shown relative to the initial uniform concentration, BNM. (Reproduced from reference 190 with permission.)
The simple consideration that only relative flow matters has widespread implications that are often overlooked. In particular, it applies not only to the spread of solutes but also to the motility of microorganisms. If a phytoplankton lyses 1 mm away from a bacterium within the Gulf Stream, the fact that the latter is traveling at several tens of centimeters per second has no bearing whatsoever on the ability of the bacterium to chemotax toward the nutrient pulse: the entire system is traveling at the same speed and only the motility of the bacterium relative to that of the water matters. Discarding bacterial motility as inconsequential on the basis that the bacterial swimming speed is negligible compared with typical speeds of macroscopic marine flows is therefore a mistake. Similarly, bacteria in turbulence travel largely in synchrony with the solutes immediately surrounding them; only local velocity gradients and motility can cause relative motion, favoring or hindering chemotaxis. The accumulation of bacteria within the nutrient filaments produced by turbulence, recently demonstrated in numerical simulations (201) (Fig. 6), is an example of this process. More broadly, despite the ubiquity of flow in the ocean and its many guises (from currents to shear to turbulence), the effect of flow on chemotaxis has received little attention to date and represents a fruitful research avenue with potentially important implications for the ecology of marine microbes.
Clearly, the ocean's water column can have a rich physical, chemical, and biological microarchitecture. Furthermore, marine microscale gradients are frequently ephemeral, either because they diffuse (e.g., cell lysis) or because relative motion limits the contact window between gradients and organisms (e.g., settling particles). This imposes a temporal constraint on chemotaxis, whereby the response must be sufficiently rapid to allow bacteria to move into the patch before it is dissipated. This contrasts with classical studies of chemotaxis, which have typically been performed for steady gradients or, as in the inherently unsteady capillary assay (2), without explicit regard to the temporal component of the bacterial response. More importantly, this temporal constraint has likely resulted in specific adaptations in marine bacteria, which we discuss below.
CHEMOTAXIS BY MARINE BACTERIA
Bacterial Motility and Chemotaxis
The shift in our view of the ocean's microscale from that of a homogeneous environment to that of a heterogeneous environment has important implications for our understanding of the behavior of marine bacteria and the way in which they interact with and modify ocean chemistry. If resources are heterogeneous (e.g., as shown in Fig. 1), motility can confer an important fitness advantage (186), whereas swimming in a homogeneous environment would entail only an energetic cost (125) and an increased risk of predation (96) and viral infection (135).
Bacterial motility can be understood as a combination of two processes: exploration and exploitation. In a homogeneous environment, motile cells explore space by propelling themselves in random directions using one or more helical flagella. Each flagellum is anchored to the cell wall and rotated by a molecular motor, driven by either proton or sodium gradients (25). Rotation of the flagellum, in the shape of a corkscrew motion, causes a propulsive force, as each segment of the helical flagellum “pushes off” against the water. This is an effective means of propulsion at these minute scales, where movement is devoid of inertia and where propulsion strategies that work at larger scales (such as the formation of jets or vortices) are physically impossible. A corollary of the absence of inertia is the lack of coasting: as soon as a bacterium stops rotating its flagella, it stops instantly (within a distance of less than one hydrogen atom!) (155). This counterintuitive result is a direct consequence of bacteria operating at low Reynolds numbers, where the Reynolds number is a dimensionless parameter that measures the relative importance of inertial forces and viscous forces.
The motility pattern of the classic model bacterium E. coli, which swims using multiple (∼4 to 8) helical flagella (Fig. 7A) (29), can be described as a sequence of relatively straight, 1- to 4-s long “runs,” executed at a speed of 10 to 30 μm/s, interrupted by nearly random reorientations or “tumbles” (23) (Fig. 7B). Run-and-tumble motility results in a random-walk exploration of the environment, which E. coli can bias to follow spatial gradients of a chemoeffector (a chemical that attracts or repels cells). Gradient sensing is typically achieved by temporal comparisons: in E. coli, the receptor occupancy in the recent past is compared to that in the more distant past (166). Spatial sensing, where a bacterium instantaneously compares concentrations over the length of its body, is believed to be more rare but not impossible (46, 193). The intracellular transfer of information involved in gradient sensing has been remarkably well characterized and is reviewed elsewhere (152, 187, 202).
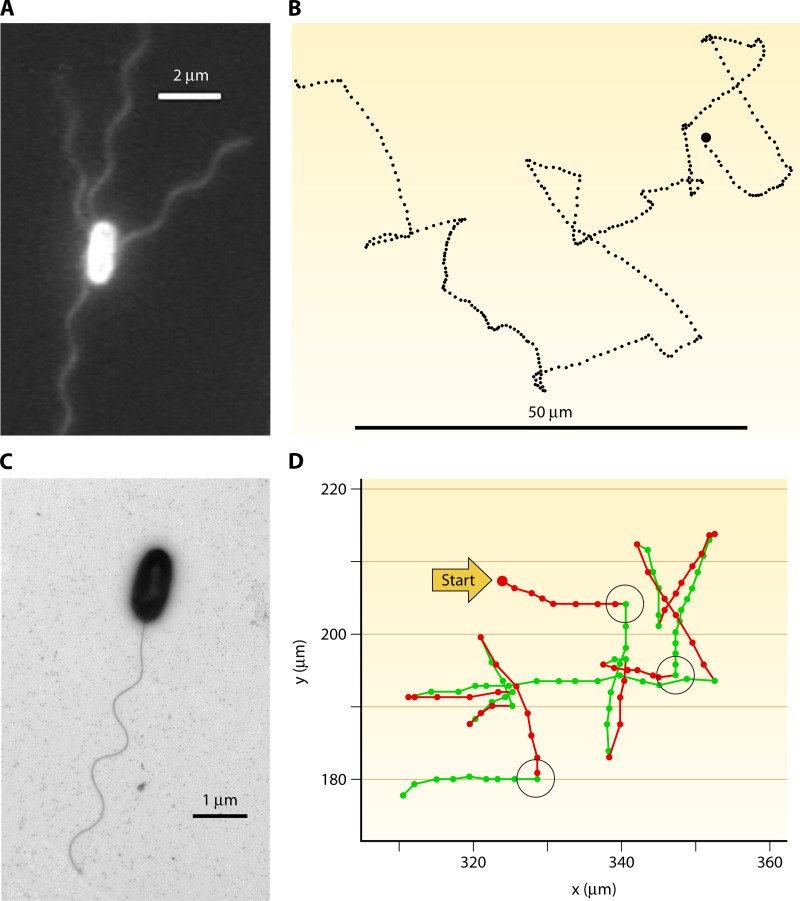
Fig 7.
Flagellation and motility patterns, illustrating fundamental differences between enteric model organisms and some marine bacteria. (A) Epifluorescent image of Escherichia coli, showing multiple flagella. Reproduced from reference 194 with permission. (B) E. coli swims in a run-and-tumble pattern, where each nearly straight swimming segment (run) is interrupted by a nearly random change in direction (tumble). The image shows a 30-s trajectory containing 26 runs and tumbles. The trajectory spans ∼100 μm from left to right. (Reproduced from reference 25a by permission from Macmillan Publishers Ltd. Copyright 1972.) (C) Transmission electron microscopy image of Vibrio alginolyticus, showing the single polar flagellum (K. Son, J. S. Guasto, and R. Stocker, unpublished). (D) The “hybrid” swimming pattern of Vibrio alginolyticus, alternating reversals (180-degree changes in directions) after each forward run (green segments) and large reorientations after each backward run (red segments). The reorientations, some of which are highlighted by black circles, are caused by a whip-like deformation of the flagellum (a “flick”). Dots are 1/30th of a second apart. (Reproduced from reference 207 with permission.)
The adjustment of flagellar rotation in response to chemoeffectors can result in two processes that are often lumped under the label “chemotaxis”: actual chemotaxis and chemokinesis (102). Chemotaxis proper is a directional response driven by the presence of a chemoeffector gradient, whereas chemokinesis is the modulation of the swimming speed in response to the concentration (not the gradient) of the chemoeffector. For example, cells can accumulate in regions of high chemoeffector concentration by reducing their swimming speed, even if they do not sense a gradient per se. Here we interpret chemotaxis in the more restrictive sense of a gradient response.
The “explore and exploit” strategy is common to most chemotactic bacteria, yet specific implementations vary greatly (130), likely in response to different environmental conditions. The ocean provides unique environmental constraints for chemotaxis, including low nutrient concentrations, ephemeral gradients, and pervasive flow. It is thus not surprising that marine bacteria (see, e.g., Fig. 7C), as we will see, exhibit strong phenotypic differences from enteric models like E. coli, including higher swimming speeds, drastically different motility patterns (Fig. 7D), and higher levels of chemotactic performance (186).
While some of the most abundant groups of marine bacteria, such as SAR11 (133) and Prochlorococcus (107), are nonmotile (62), there is evidence that bacterial motility is widespread in the ocean. Mitchell and coworkers (126, 127) found that the fraction of cells swimming is often low (∼10%) in coastal seawater samples, but that enrichment with organic substrates increases motility levels to 80% within a few (∼12) hours. They concluded that a large proportion of water column bacteria are capable of motility, although the possibility remains that enrichment resulted in a shift in community composition toward more rapidly growing motile species. No enrichment was applied in the experiments of Grossart and coworkers (66) and Fenchel (50), who showed that the fraction of motile bacteria in the water column ranges between <5 and 70%. Motility levels further appear to be subject to large natural variability, with the fraction of motile cells larger in the summer than in the winter, larger by day than by night, and increasing with elevated levels of particulate organic carbon, for example, that associated with phytoplankton blooms (66). The last point fits well with the view that bacterial motility is most beneficial in marine environments characterized by a landscape of small-scale chemical gradients that bacteria can navigate using chemotaxis (128).
Chemotaxis is widespread among diverse taxa in the ocean. Many of the marine bacteria that have been isolated from seawater, including representatives from diverse genera such as Vibrio, Silicibacter, Roseobacter, Pseudoalteromonas, Pseudomonas, Synechococcus, and Alcaligenes, exhibit chemotaxis toward a wide range of attractants, including amino acids, sugars, carboxylic acids, organic sulfur compounds, oxygen, nitrate, nitrite, ammonium, urea, and phosphate (18, 77, 117, 122, 136, 172, 175, 183, 186, 206, 213). As discussed below, the ecological drivers of this chemotactic behavior are wide and varied.
Chemotaxis toward Phytoplankton
Some of the earliest evidence of chemotaxis in marine bacteria revealed that this phenotype may play an important role in bacterium-phytoplankton interactions. The classic study by Bell and Mitchell (22) demonstrated that several unidentified marine bacteria exhibited strong chemotaxis toward the extracellular products of phytoplankton cultures. Chemotaxis was particularly strong toward the exudates of nutrient-stressed or dead phytoplankton. Bell and Mitchell proposed that this chemotactic response may allow for the spatial coupling between bacteria and phytoplankton in the ocean, with bacteria clustering within the DOC-rich “phycosphere” surrounding individual algal cells (Fig. 2).
Several theoretical studies have attempted to predict whether chemotaxis toward phytoplankton allows bacteria to cluster within and exploit the phycosphere. Mitchell and coworkers (124) predicted that chemotaxis toward the phycosphere would provide only moderate nutrient gains (10%) for bacteria under the most favorable conditions, where flow and phytoplankton motility are negligible. Jackson (79) calculated that chemotaxis allows bacteria to locate and exploit the phycosphere of stationary phytoplankton cells only if these are sufficiently large (radius of >2 μm) and have high leakage rates. Bowen and coworkers (31) included the effect of turbulence and predicted that, for realistic phytoplankton exudation rates and ocean turbulence levels, up to 20% of chemotactic bacteria in the water column can cluster around phytoplankton cells but that clustering decreases substantially when shear exceeds moderate ocean levels (0.2 s−1).
These modeling studies suggest that bacterial chemotaxis toward the phycosphere can occur in the ocean but is limited to favorable conditions. However, due to limited data on the swimming and chemotaxis abilities of marine bacteria at the time, these studies generally modeled chemotaxis using parameters largely derived from E. coli (with the exception of that by Bowen and coworkers [31], who also considered higher swimming speeds). It has since been established that the chemotactic performance of marine bacteria can be substantially higher than that of E. coli (126–128, 186) (see Chemotaxis Adaptations of Marine Bacteria below), indicating that these early modeling results might underestimate the potential of marine bacteria to employ chemotaxis to cluster within phycospheres.
By imaging natural communities of marine bacteria with video microscopy, Blackburn and coworkers (28) provided the first direct evidence for the chemotactic clustering of bacteria around particles and phytoplankton cells (Fig. 8A). They observed that bacteria from seawater enrichments under conditions of low oxygen saturation formed intense accumulations around individual Pavlova lutheri cells and speculated that the clustering occurred in response to oxygen gradients produced by the alga. Alternatively, it is also plausible that clustering occurred as a result of chemotaxis toward the phytoplankton's organic exudates, as marine bacteria are highly chemotactic to the chemical products of both eukaryotic and prokaryotic phytoplankton, including diatoms, dinoflagellates, chlorophytes, raphidophytes, and cyanobacteria (37, 122, 123, 172, 173, 176, 186) (Fig. 8B). Separate experiments have shown that several compounds commonly found in phytoplankton exudates elicit chemotaxis, including glycolate, acrylate, specific amino acids, and dimethylsulfoniopropionate (DMSP) (22, 94, 122, 175, 182).
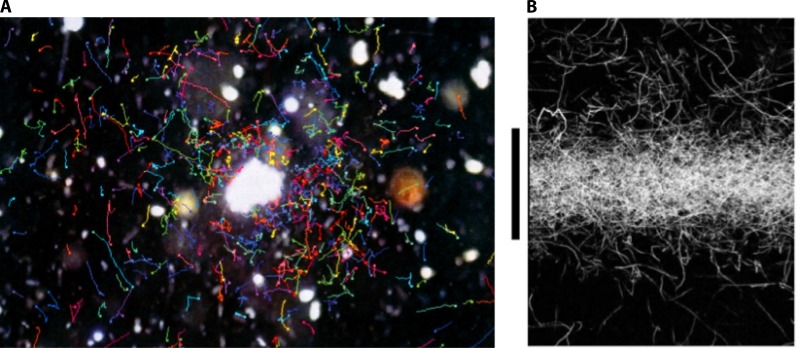
Fig 8.
Bacterial accumulation by chemotaxis. (A) Chemotaxis of a natural bacterial community toward a detritus particle. (Reproduced from reference 28a with permission.) (B) Chemotaxis of Pseudoalteromonas haloplanktis toward a pulse of phytoplankton exudates. The original footprint of the pulse corresponds to a band of 300-μm width, extending across the image from left to right, in correspondence to the black vertical bar. The image was acquired 2 min after release of the pulse and shows intense accumulation of trajectories (white paths) at the center of the pulse. (Reproduced from reference 186 with permission.)
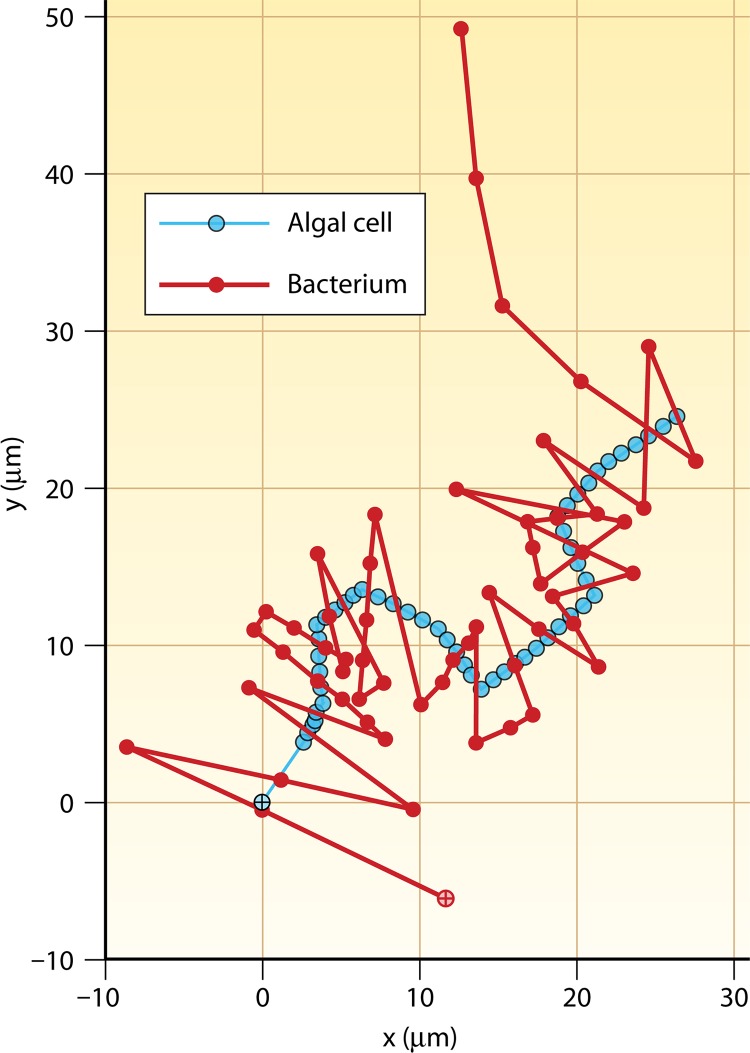
Beyond the observations of Blackburn and coworkers (28), one of the only other studies to experimentally investigate bacterial spatial interactions with live phytoplankton cells showed that marine bacteria may even use chemotaxis to chase swimming phytoplankton cells (19). The marine bacterium Pseudoalteromonas haloplanktis and a marine Shewanella putrefaciens strain were shown to use fast swimming speeds and rapid changes in direction to pursue a moving phytoplankton cell for 1 to 2 s, presumably in response to nutrient or oxygen gradients emanating from the phytoplankton and forming a plume behind it (Fig. 9). Interestingly, numerical models investigating the similar scenario of bacteria tracking a sinking phytoplankton cell indicate that only in exceptional circumstances (very large and leaky phytoplankton cells) would bacterial chemosensory movement relative to the moving phytoplankton cell enhance bacterial nutrition (80). Consequently, these experimental observations indicate that the chemosensory capabilities of marine bacteria may exceed what previous theory had predicted. However, while this impressive “chasing” ability was initially attributed to chemotaxis alone, one subsequent theoretical study has suggested that the origin of the phenomenon might be purely hydrodynamic, whereby the fluid velocity gradients in the wake of the swimming alga reorient bacteria so that they remain in the wake (108). While this remains an interesting possibility, the very sharp turns executed by the chasing bacteria still suggest an active behavior, leaving this topic open for further investigation.
Fig 9.
Bacteria chasing phytoplankton. A bacterium (Pseudoalteromonas haloplanktis) (red) tracking a swimming phytoplankton cell (Pavlova lutheri) (cyan) is shown. Starting locations are shown by crosses within circles. The reversals in swimming direction enabled the bacterium to pursue the algal cell for many correct consecutive turns. Symbols on each trajectory are 0.04 s apart, and the total time is 2.2 s. (Adapted from reference 19 with permission. [Copyright 2002 Federation of European Microbiological Societies.] We acknowledge Greg Barbara for providing the cell positions used to redraw the original image in color.)
Chemotaxis toward Nutrient Pulses
Whereas the chemical gradients associated with phycospheres are largely steady, as long as phytoplankton exudation rates do not vary too rapidly, several microscale features of the marine chemical landscape are highly unsteady. Typically, these are events that begin with a very concentrated nutrient pulse, which diffuses away over minutes or tens of minutes. A rapid response becomes of the essence to make chemotaxis feasible under these conditions, but the response can be worthwhile owing to the high nutrient concentrations in the pulse. For instance, microscale nutrient pulses resulting from zooplankton excretions and cell lysis events (27, 28, 103, 104) can represent ephemeral growth environments for chemotactic cells (Fig. 1B). Microscopic observations of intense aggregations of bacterial cells that persist for several minutes in seawater suspensions have been explained by chemotactic responses toward microbial lysis events and pulses of nutrients excreted by protozoa (28). This idea was tested experimentally by Stocker and coworkers (186), who used a microfluidic system to create diffusing microscale chemical pulses and video microscopy to observe bacterial responses to the pulses. These experiments showed that the marine gammaproteobacterium P. haloplanktis exhibits a very rapid chemotactic response, accumulating within 300-μm-wide pulses of phytoplankton (Dunaliella tertiolecta) exudates in under 30 s and resulting in a >3-fold increase in potential nutrient uptake (Fig. 8B). This chemotactic response was considerably faster than the typical lifetime of nutrient pulses in the ocean (several minutes) (27, 186), indicating that marine bacteria are well equipped to exploit the gamut of short-lived pulses that dot the microbial nutrient landscape.
Chemotaxis to Particles and Their Plumes
Motility and chemotaxis can be important in shaping the interactions of bacteria with particles in at least three respects: increasing rates of encounters with particles, utilizing the particulate organic matter within the particles, and exploiting the dissolved organic matter leaking out of the particles. Kiørboe and coworkers (95) quantified the colonization of particles using millimeter-size agar spheres as model particles. They found that motile bacteria colonize particles at rates that are much higher than those of nonmotile bacteria and that strains capable of tumbling (i.e., of actively changing swimming direction) colonized aggregates enriched with organic substrates faster than unenriched aggregates, whereas a nontumbling strain did not. Because tumbling is essential for chemotaxis, these observations highlight the importance of chemotaxis in the colonization of particles. They are also in line with results from a mathematical model predicting that chemotaxis enhances rates of encounters with particles by 2- to 5-fold for particles ranging from 200 μm to 1.5 cm in radius (93).
Bacteria on particles use ectoenzymes to solubilize particulate organic matter (184). Only part of the resulting DOM is taken up by the particle-attached bacteria, with the remainder diffusing out and forming DOM plumes in the wake of sinking particles (see “Particles as Resource Islands” above) (Fig. 4). Using a mathematical model, Kiørboe and Jackson (93) quantified the utilization of this plume by bacteria, finding that, under the assumption of optimal chemotactic behavior, chemotaxis increases the growth rate of free-living bacteria by 2-fold for large particles (1.5-cm radius) and by 20-fold for small particles (200-μm radius). When the quantification was scaled up to account for measured particle size spectra in the ocean, the increase in growth rate was a factor of ∼10. These predictions were tested by Stocker and coworkers (186), who created plumes in a microfluidic device and quantified the response of P. haloplanktis for particle sinking speeds ranging from 66 to 660 μm/s. Their observations revealed strong bacterial accumulations in the plumes (Fig. 4C), in particular for the lowest sinking speeds, resulting in a predicted nutrient gain of up to 4-fold compared to that by nonmotile cells. Taken together, results from these modeling and experimental studies indicate that motility and chemotaxis can greatly enhance the ability of marine bacteria to utilize particles and their plumes as resources.
Chemotaxis and Bacterium-Animal Associations
Chemotaxis is also an important phenotype in the establishment of marine bacterium-animal interactions and pathogenicity. One group of potentially pathogenic marine bacteria for which chemotaxis has been widely studied is the Vibrio genus. As a group, vibrios exhibit high levels of chemotaxis toward a variety of amino acids and sugars (56, 117, 139, 161, 172, 182, 211), as well as the chemical products from a wide range of larger organisms (16, 17, 30, 121). The human pathogen Vibrio cholerae, which typically inhabits estuarine or brackish environments (38, 89), is chemotactic toward amino acids (56), and chemotaxis is believed to play an important role in its ability to infect the host intestine (36). Vibrio fischeri exhibits chemotaxis toward squid-derived chitin oligosaccharides and N-acetylneuraminic acid, which is a component of the light-organ mucus in the bobtail squid (42, 43, 118). This behavior allows V. fischeri to colonize the squid's light organ and thus underpins a mutualistic relationship that provides the squid with bioluminescent properties (118). Vibrio furnissii exhibits chemotaxis toward chitinous material (20, 21, 211), which is suggestive of associations with marine crustaceans, cephalopods, or sinking particles and might consequently result in long-range dispersal by transport on zooplankton (67, 189).
A number of vibrios are pathogens of marine organisms and also exhibit high levels of chemotaxis (20, 21, 30, 161, 211). Both V. furnissii and V. parahaemolyticus are chemotactic toward fish skin, gill, and intestinal mucus and have been implicated in fish diseases (30). Other fish pathogens, including V. anguillarum, exhibit chemotaxis toward fish intestinal mucus (138, 139), where the expression of chemotaxis is essential for infection (139). Similarly, chemotaxis toward coral mucus by the coral pathogens Vibrio coralliilyticus and Vibrio shiloi plays an important role in the infection of some coral species (16, 17, 121). In the case of V. shiloi, chemotaxis to coral mucus depends on the presence of Symbiodinium organisms, the endosymbiotic dinoflagellates living within the coral (17), suggesting that the signal eliciting the chemotactic response is produced by these algae.
Chemotaxis at the Sediment-Water Interface
Many of the characteristics of bacterial motility and chemotaxis are markedly different at the sediment-water interface than in the water column. A principal reason for this difference is the nature of the chemical landscape, which is often one-dimensional (Fig. 5) (see “Gradients at the Sediment-Water Interface” above) and relatively stable over time, compared to the three-dimensional, often ephemeral microscale gradients occurring in the water column. A second reason relates to cell size. Bacteria from the sediment-water interface are often very large. For example, Thiovolum majus is up to 25 μm in diameter (49), and diameters of 10 to 20 μm are typical of several colorless sulfur bacteria living on the oxygen and sulfide gradients at the sediment-water interface (165). Because of their size, these bacteria are immune to Brownian rotational diffusion, whose effect scales with the inverse of the cell volume. Thus, these bacteria are considerably more effective at controlling their swimming direction than the small bacteria of the water column and are even able to actively “steer” (49). Unfortunately, experimental flexibility with bacteria from the sediment-water interface is limited by the lack of cultured species (165), yet observations of natural communities have revealed a host of interesting features.
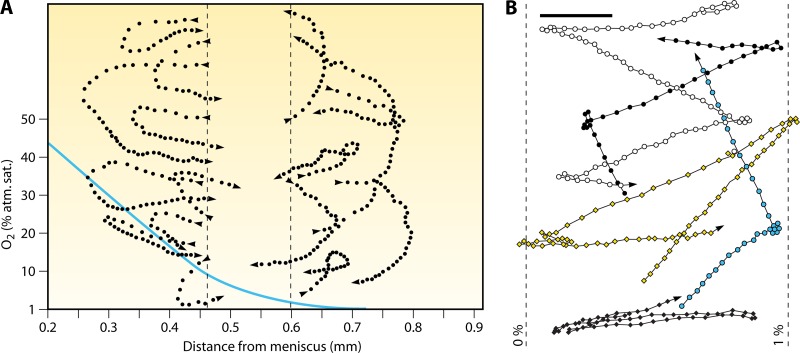
The most striking chemotactic motility is exhibited by T. majus, a microaerophilic spherical sulfur bacterium that swims at 600 μm/s (49). T. majus displays two strategies to retain position at its preferred oxygen concentration (4% of air saturation): it forms veils, and it exhibits phobic responses. In the first strategy, cells swarm to their preferred height above the sediments, which corresponds to the most favorable oxygen and hydrogen sulfide concentrations. There they stick together via 100-μm-long mucous stalks, forming ∼200-μm-thick, two-dimensional veils (86). The veil, which can develop an exquisite honeycomb structure (52), creates a sharp interface between the flowing, oxygenated water above and the stagnant, sulfide-rich water below. Concerted swimming allows the bacteria to adjust the vertical position of the veil and thereby control the fluxes of oxygen and sulfide (86). In the second strategy, T. majus accelerates and executes U-turns (Fig. 10A) when encountering an adverse environment, for example, when oxygen concentrations become too high (T. majus does not respond to sulfide [49]). Because it does not imply the ability to sense a gradient, this mechanism does not qualify as true chemotaxis but rather is a phobic response. This simple method for maintaining an optimal position is effective because of the stable, one-dimensional nature of the gradient and allows T. majus to return to zones of preferred oxygen concentrations within ∼1 s, or 200 μm (Fig. 10A). Phobic responses to high oxygen concentrations are also exhibited by other bacteria living at the sediment-water interface, including the fastest bacterium discovered to date, Ovobacter propellens, which swims at up to 1 mm/s and, unlike Thiovolum, exhibits sharp reversals rather than smooth turns upon encountering high (>1% air saturation) oxygen concentrations (53) (Fig. 10B).
Fig 10.
Swimming trajectories of bacteria living at the sediment-water interface. (A) Swimming trajectories of Thiovulum majus in a plane (dotted lines), with the profile of oxygen concentration (solid line; values provided on the y axis) as a function of the distance from the meniscus overlaid. Cells display a strong behavioral response to changes in oxygen concentration. Dots are cell positions at 40-ms intervals. When cells leave the band of optimal oxygen concentration (between the dashed lines), they rapidly (within 0.5 to 2 s) return to it by a U-turn. This controlled “steering” of the 10- to 25-μm large T. majus cell stands in contrast to the randomness inherent in the chemotaxis of smaller bacteria, which, unlike T. majus, are affected by Brownian rotational diffusion. Because it is not a response to a gradient but an avoidance of high oxygen concentrations, this behavior is not chemotaxis per se but rather is a phobic response. (Reproduced from reference 49 with permission of the Society for General Microbiology; permission conveyed through Copyright Clearance Center, Inc.) (B) Swimming trajectories of Ovobacter propellens in a plane, with the 0% and 1% oxygen isoclines overlaid. Cells display a strong phobic response for oxygen concentrations above 1% of air saturation. Dots are cell positions at 26-ms intervals. The scale bar is 50 μm. Note that, in contrast to Thiovulum, which executes a U-turn (panel A), Ovobacter reverses direction. (Reproduced from reference 53 with permission. Copyright 2004 Federation of European Microbiological Societies.)
While the gradients encountered by T. majus are often one-dimensional, an isolated yet intriguing observation suggests that their strategy for responding to gradients might be effective also for more complex chemical landscapes. Jørgensen and Revsbech (86) reported T. majus cells pursuing a phytoflagellate, a Euglena sp., presumably by tracking its oxygen plume. This hypothesis, which suggests an accurate aerotaxis strategy, was supported by the observation that T. majus lost its tracking ability upon light removal (because Euglena stops producing oxygen in the dark). This circumstantial evidence that the gradient response repertoire of T. majus extends beyond phobic responses and includes true chemotaxis has been subsequently confirmed by direct observations (191).
In contrast to the rapid chemotaxis displayed by T. majus, the chemotaxis by bacteria that partially penetrate or live attached to the seabed is often very slow. Two examples are represented by the filamentous sulfur bacteria Beggiatoa and Thioploca: the former responds with long delays (20 to 50 s) to changes in chemical conditions, while the latter exhibits very slow chemotactic speeds (<1.5 μm/s) (165). The difference between slow and fast strategies is likely a reflection of the sessile versus free-floating lifestyles (165): the stable chemical conditions within the diffusion boundary layer and the upper sediment layer make a slow response sufficient for organisms partially anchored to the seabed, whereas floating organisms can be transported away by flow to a potentially undesirable location and might thus need to shuttle rapidly among different positions in the gradient.
Beggiatoa lives at sulfide-oxygen interfaces (Fig. 5), where it forms mats that oxidize sulfide (165). Gliding motility allows Beggiatoa to adjust its position to fluctuations in the height of the sulfide-oxygen interface. By utilizing a phobic response, where cells reverse their gliding direction upon encountering oxygen concentrations above 5% air saturation, Beggiatoa can cluster tightly at the sulfide-oxygen interface.
Gliding motility is also the basis of chemotaxis in the giant sediment-dwelling bacteria Thioploca spp., which can be up to 70 mm long and are composed of hundreds of cells. Thioploca spp. oxidize sulfide with nitrate and exhibit chemotaxis toward nitrate and low sulfide concentrations, whereas excessive oxygen and sulfide concentrations cause a phobic response resulting in a retreat into the sediments (75). Thioploca's motility is primarily confined to the top layer of the sediments, where cells shuttle up and down between nitrate-rich bottom water and sulfide-containing sediment layers a few centimeters beneath (55, 75, 165, 214). Within the sediments, they glide at up to 1.5 μm/s through sheaths, “highways” resulting from the slime excreted by the cells that facilitate their chemotactic migration (165). The ability to store both nitrate and sulfide, and to shuttle between the two, provides an effective strategy to use the two substrates, which do not typically coexist (164).
CHEMOTAXIS ADAPTATIONS OF MARINE BACTERIA
Considering the dynamic nature of many marine microenvironments, it is perhaps surprising that bacteria living in the ocean can so successfully use chemotaxis to exploit favorable chemical niches. This success stems from their often sophisticated chemotactic strategies, which form the focus of this section.
Fast Swimming
The ephemeral nature of many nutrient sources in the ocean implies that fast responses are beneficial to increase nutrient uptake. The primary parameter affecting the chemotactic response rate is the swimming speed: its importance in different ecological processes, including the colonization of particles and the uptake of dissolved nutrients, has been established by means of both numerical simulations (93) and experiments (186).
Marine bacteria are often very fast. Observations of natural communities and isolates have revealed mean speeds of 45 to 230 μm/s, with the highest speeds occurring in microscale aggregations of cells and with burst speeds in excess of 500 μm/s for individual cells (66, 126–128). Stocker and coworkers (186) and Seymour and coworkers (175) recorded mean swimming speeds of 68 to 80 μm/s for P. haloplanktis, which contributed to this species' fast response to chemical gradients. Hütz and coworkers (77) measured mean speeds of up to 62 μm/s for bacteria of the genus Thalassospira. Muramoto and coworkers (134), Magariyama and coworkers (116), and Xie and coworkers (207) clocked Vibrio alginolyticus swimming at 45 to 116 μm/s. (The true magnitude of these speeds can be appreciated when swimming velocities are converted into body lengths per second, where one body length is on the order of 1 μm.) In contrast, a survey of 84 marine isolates revealed mean swimming speeds largely within the range of 25 to 35 μm/s (82). The origin of this discrepancy remains unclear and might be related to the growth stage or to long times in nutrient-rich culture media, which may ultimately lead to speed decreases. The speed record, however, belongs to marine bacteria living at the sediment-water interface, where T. majus swims at 600 μm/s (49) and O. propellens at a striking 1,000 μm/s (53).
The high swimming speeds of marine bacteria compared to, for example, E. coli (15 to 30 μm/s) might be partially due to differences in the molecular motors powered by sodium gradients across the cytoplasmic membrane versus those powered by proton gradients (106, 115). Muramoto and coworkers (134) demonstrated that the swimming speed of V. alginolyticus monotonically increases with sodium concentration, and sodium concentrations in the ocean are considerably higher than proton concentrations in most environments. Consistent with this difference, the rotation rate of V. alginolyticus' sodium-driven motor (∼1,100 to 1,700 Hz) is a factor of ∼4 to 6 higher than the rotation rate of E. coli's proton-driven motor (∼300 Hz) (210). Since swimming speed is to a first approximation linearly proportional to the motor's rotation rate, the high concentration of sodium in the ocean might contribute to explain the high swimming speeds of many marine bacteria.
Determining why high swimming speeds are optimal in the ocean is a question of energetics (125, 129): why would Vibrio not swim at the same speed as E. coli? Early viewpoints, developed based on findings with E. coli, painted bacterial motility as inexpensive (155) and have been corroborated by studies showing that the synthesis and operation of flagella require ∼2% and ∼0.1% of E. coli's total energy expenditure, respectively (114). However, E. coli is a comparatively slow swimmer living in a nutrient-rich environment (animal intestines). Nutrient concentrations are often 2 to 4 orders of magnitude lower in the ocean, and the energetic expenditure of marine bacteria, which swim ∼3- to 5-fold faster than E. coli, is ∼10- to 25-fold larger (because the propulsive power increases with the square of the swimming speed). This suggests that energetic requirements place a stringent fitness constraint on the evolution of bacterial motility in the ocean, a view supported by the large number of nonmotile marine bacterial species, and indicates that an energetic framework is required to understand the trade-offs of motility and chemotaxis in the sea.
Such a framework has been recently proposed by Taylor and Stocker (190), who used competition simulations in a turbulent flow to determine the fitness advantage of chemotaxis. By comparing the nutrient gain resulting from chemotaxis with the energetic expenditure due to swimming, they determined an optimal swimming speed of ∼60 μm/s for realistic marine conditions (bacterial concentrations, nutrient availability, and turbulence levels), which is in broad agreement with observed values for many species (see above). There is considerable additional scope for research in this direction, as a comprehensive treatment of motility trade-offs should include the costs of flagellar and motor synthesis, the costs of producing and running the signal transduction machinery, and ecological costs such as the higher rates of encounters with predators and viruses.
On the other hand, motile bacteria could save cost by not swimming at a constant speed, as assumed by Taylor and Stocker (190) in the absence of more specific experimental information. The hypothesis that swimming speed is adaptive, allowing cells to modulate it according to local or prevailing conditions, is intriguing but remains largely untested. Partial support for this hypothesis comes from microfluidic observations showing that P. haloplanktis increases its swimming speed by 20% when inside a patch of algal exudates (174) and experiments where P. haloplanktis and Shewanella putrefaciens both increase swimming speeds by between 3- and 4-fold when tracking algae (19). Some evidence suggests that marine bacterial motility varies over timescales of tens of seconds (66). In contrast, Mitchell and coworkers (127) found that motility among natural marine communities increases only upon several (∼12) hours of nutrient enrichment. Modulation timescales of hours would be insufficient for the organisms to respond to individual nutrient pulses, which last minutes, and could indicate that motility rather is modulated in response to prevailing nutrient conditions, for example, an algal bloom or a large dust deposition event.
At a mechanistic level, a further motivation for high speeds might relate to the small size of many bacteria in the water column, which is in turn related to oligotrophic conditions. The smaller a bacterium's size, the larger the effect of rotational Brownian motion. Just as random collisions with water molecules jostle any micrometer-size object in three dimensions, resulting in (translational) Brownian motion, they also cause the object to continuously reorient, a process called rotational Brownian motion. This effect is well known to affect bacteria by tending to randomize their swimming direction, thus hindering directional swimming and chemotaxis (25, 125). For a spherical bacterium of radius R, the rotational diffusivity is DR = kT/(8πμR3) (μ is the dynamic viscosity of water, k is Boltzmann's constant, and T is the temperature in degrees Kelvin). A spherical cell of radius R = 0.6 μm has DR = 0.76 rad2/s and thus experiences rotations of, e.g., (4DRt)1/2 = 100 degrees over a time (t) of 1 s; such a reorientation throws a cell completely off course. Although cell elongation and the presence of a flagellum stabilize bacteria against rotational diffusion (125), Brownian effects provide strong selective pressure for fast motility in small bacteria, because higher speeds allow cells to cover longer distances before being reoriented. Along these same lines, theoretical predictions indicate that bacteria smaller than R = 0.3 μm should not be motile (46).
An additional advantage for a fast-swimming cell is that, even in a uniform nutrient field, it could encounter more nutrient molecules per unit time. This effect is related to the fact that the fluid flow associated with swimming thins the concentration boundary layer around the cell, enhancing the nutrient flux to the cell. However, physical arguments show that such an advantage would be limited to large and fast cells and is negligible for the small bacteria living in the water column. This advantage could, however, apply to bacteria living at the water-sediment interface, such as T. majus and O. propellens (165). The relative increase in nutrient uptake afforded by swimming, over that due to diffusion alone, is measured by a dimensionless parameter called the Sherwood number (68). For 1-μm bacteria, swimming at even 100 μm/s does not increase the Sherwood number appreciably above 1.0, indicating no effect of swimming on uptake. In contrast, a 10-μm cell swimming at 1,000 μm/s has a Sherwood number of 2.2, indicating a 120% increase in uptake (68). This large effect of swimming on uptake—which is additional to the ability to move toward higher-concentration regions by chemotaxis—could be another element that sets large and fast bacteria apart from small ones, yet it remains a hypothesis to be tested experimentally.
Turning
Fast swimming speeds are certainly one reason for the high chemotactic performance of many marine bacteria but most likely are not the sole reason. This can be seen by considering a metric that is independent of the swimming speed, the chemotactic efficiency, VC/VS, which is the ratio between the chemotactic speed VC and the swimming speed VS. This parameter is independent of the swimming speed because, for a given geometrical pattern of swimming, VC increases linearly with VS (hence, the ratio is a constant). The chemotactic speed is the component of the swimming speed directed along the gradient. A chemotactic efficiency of 100% indicates a perfectly directional response (no randomness) up the gradient (attraction), VC/VS = −100% indicates a perfectly directional response down the gradient (repulsion), and VC/VS = 0 indicates a purely random motion (no response to the gradient). For marine bacteria, values of VC/VS close to 50% have been recorded (e.g., 44% for P. haloplanktis exposed to DMSP gradients [175]), whereas literature values for E. coli mostly range from 5 to 15% (see reference 4 and references therein). Under selected conditions, E. coli can also respond with high directionality (VC/VS = 35% [4]), but neither the level of directionality nor the breadth of chemical conditions under which this occurs appears to match those of marine bacteria.
In microfluidic experiments, Stocker and coworkers (186) compared the chemotaxis of E. coli and that of the marine bacterium P. haloplanktis to a nutrient pulse lasting on the order of 10 min. P. haloplanktis accumulated more strongly into the high-concentration region (Fig. 8B), and its chemotactic response was almost an order of magnitude faster than E. coli's, resulting in a 64 to 87% increase in potential nutrient uptake of the entire population and in a 10-fold increase for the fastest 20% of bacteria. Modeling indicated that the superior chemotactic performance of the marine bacteria could not be attributed exclusively to their higher swimming speed, VS (68 μm/s for P. haloplanktis versus 31 μm/s for E. coli), but also resulted from their more directional swimming strategy (i.e., higher VC/VS). It is therefore important to consider the overall movement behavior of a bacterium and not simply its swimming speed. One important feature of movement behavior relates to how bacteria change direction. E. coli's run-and-tumble swimming pattern (Fig. 7B), where reorientations have a broad distribution of angles with a mean of 68 degrees, is only one of several possible swimming strategies (130).
Many motile marine bacteria, such as most vibrios, swim in liquid media using a single polar flagellum (Fig. 7C). When the flagellum rotates in one direction, the cell swims forward: the flagellum pushes the cell. When the direction of rotation of the motor is inverted, the cell swims backward: the flagellum pulls the cell. In the absence of other effects, this mechanism leads to “run-and-reverse” motility, which has been reported for several species of marine bacteria (82, 128). In run-and-reverse motility, reorientations are all close to 180 degrees, in marked contrast with the case for E. coli. What prevents bacteria from constantly swimming along the same line is Brownian rotational diffusion, which introduces randomness in the swimming direction during each run.
Johansen and coworkers (82) found that run-and-reverse motility is the most prevalent movement pattern among 84 marine bacterial isolates. Reversals have also been suggested to be superior to tumbles in navigating more complex environments, in particular for retaining position near a phytoplankton cell in the presence of flow (111). In contrast, a mathematical model of bacterial chemotaxis toward particles and their plumes revealed no difference between reversals and tumbles (93), suggesting a sensitive dependence of the optimal chemotactic strategy on microenvironmental conditions.
Recent high-resolution observations of V. alginolyticus have revealed an unexpected twist on this mechanism. By tracking individual cells and visualizing their flagellum by fluorescent staining, Xie and coworkers (207) found that 180-degree reversals occur only every other change in direction, when the cell switches from forward to backward swimming. In contrast, the backward-to-forward transition is characterized by a larger reorientation, normally distributed around a mean of 90 degrees. This reorientation is associated with a “flick,” a large whip-like deformation of the flagellum, whose mechanism remains unknown. This newly discovered “hybrid” swimming pattern therefore mixes reversals traditionally associated with run-and-reverse motility and large reorientations more akin to run-and-tumble motility (Fig. 7D). Without having to rely on Brownian effects, V. alginolyticus achieves a result comparable to that of E. coli—runs interrupted by short reorientations that randomize the trajectory—and it does so with only one flagellum, saving the synthesis costs of multiple flagella. V. alginolyticus also displays very fast chemotaxis, indicating that a hybrid locomotion strategy is superior to a run-and-tumble strategy in terms of chemotactic performance, yet the fundamental mechanism for this difference remains to be determined. Because flicks are extremely rapid (∼ 20 ms) and often result in cells swimming out of the plane of focus in microscopy setups, they may have been missed in previous reports of run-and-reverse motility; it thus remains unclear whether Brownian motion or flicks are responsible for allowing bacteria with a single flagellum—among them, many marine bacteria—to effect changes in direction.
The multitude of bacterial swimming patterns indicates that the optimal pattern is environment dependent. Bacteria at the sediment-water interface rely on run-and-reverse or U-turn swimming strategies, because their environment is largely one-dimensional. Interestingly, the 180-degree change in direction executed by T. majus is achieved not by a reversal, as in V. alginolyticus, but by continuous steering into a U-turn. This behavior is made possible by the larger size of the cell (10 to 25 μm), which renders it virtually immune to the random Brownian motion that constrains the motility of smaller bacteria. The swimming patterns of these large bacteria are, in fact, closer to those of larger eukaryotes and are referred to as helical klinotaxis; even small misalignments of the propulsive system relative to the cell axis lead to helical trajectories. This motility pattern is believed to improve sensing, because cells swimming in a chemical gradient experience a periodic change in concentration and can respond by changing their rotational velocity to migrate along the gradient (191).
Differences in chemotactic performance might also result from differences in signal transduction. Currently, even simple speculations in this regard are impaired by the lack of information on signal transduction in marine bacteria. Mitchell and coworkers (128) speculated that signal processing times of marine bacteria would have to be considerably faster than E. coli's, in order to allow sensing of chemical gradients at the typically high swimming speeds observed. Circumstantial evidence for faster signal processing comes from observations showing that the turning frequencies of marine bacteria are often higher than those of E. coli (19, 174). We propose that their striking chemotactic performance makes marine bacteria promising candidate model organisms to advance our understanding of the physiological and molecular limits of bacterial sensing and locomotion. More generally, the identification of optimal movement strategies among marine bacteria represents an exciting area of research, which calls for integrated experimental and theoretical investigations that link signal transduction, ecology, and propulsion biophysics.
BIOGEOCHEMICAL IMPLICATIONS OF BACTERIAL CHEMOTAXIS IN THE OCEAN
Besides conferring motile bacteria a competitive advantage associated with higher nutrient uptake, bacterial chemotaxis in the ocean can have substantial ecological and biogeochemical implications.
Carbon Cycling
Bacterial chemotaxis plays a potentially pivotal role in marine chemical cycling processes. For example, the flux of particulate organic carbon sinking from the sunlit upper ocean to the deep ocean depends on the rate at which particles are degraded in the upper ocean by colonizing bacteria (14). As discussed above (see “Chemotaxis to Particles and Their Plumes”), chemotactic bacteria colonize sinking particles at a substantially higher rate than nonchemotactic bacteria. If these findings are representative of natural conditions, it follows that chemotaxis contributes to shape the oceanic carbon cycle by determining the amount of carbon retained within the upper ocean, which in turn influences the extent to which the ocean acts as a source or sink of atmospheric CO2 (14).
Marine bacteria also use chemotaxis to exploit microscale patches of DOC in the water column. Results from direct observations and mathematical models (28, 51, 186) indicate that chemotaxis can potentially at least double rates of bacterial uptake of organic carbon in these point-source events. However, in the absence of chemotactic bacteria that swim toward the core of the patch, the DOC from the patch would still ultimately become available to nonchemotactic bacteria in the surrounding water by diffusion. Thus, it is not clear whether the accelerated uptake associated with nutrient consumption by chemotactic bacteria translates into an increase in the actual amount of carbon that is transferred to the microbial food web. A hitherto untested hypothesis is that the bacterial growth efficiency (BGE) is higher under transiently elevated DOC concentrations (15). In this case, a larger proportion of the DOC would be channeled into bacterial biomass and a smaller proportion would be respired, directly affecting the productivity of the food web and carbon cycling. While increases in BGE with resource concentration over large-scale environmental gradients and across different oceanic provinces (41) provide some plausibility for the “concentration-dependent BGE hypothesis,” it remains unknown whether this pattern extends to the microscale spatiotemporal context.
Nitrogen Cycling
Nitrogen cycling processes in the ocean frequently occur in the context of microscale chemical gradients or require specific spatial relationships among different microbial populations (143–145). Both are situations where chemotaxis can be important. For instance, nitrogen fixation often relies on symbiotic relations maintained by chemotaxis (35). Nitrogen-fixing cyanobacteria, including Trichodesmium and Anabaena, live in close spatial association with heterotrophic bacteria (140, 141, 178). Chemotaxis by heterotrophic bacteria is directly responsible for maintaining their spatial association with Anabaena (58, 142) and has been demonstrated to ultimately enhance nitrogen fixation (142), which in turn supports the productivity of aquatic environments (212). Chemotaxis is also involved in other important parts of the marine nitrogen cycle, including nitrification and denitrification, where bacteria respond to microscale gradients of ammonium, nitrite, nitrate, and oxygen (90), allowing specific bacterial groups to migrate to, or maintain position within, the precise microzonal conditions (e.g., oxic/anoxic interfaces) required for these processes (144).
Sulfur Cycling
Key sulfur cycling processes, including the chemolithotrophic use of reduced sulfur and the heterotrophic assimilation of organic sulfur compounds, involve bacterial chemotaxis. Chemotaxis is used by both sulfur-reducing and sulfur-oxidizing bacteria to locate optimal chemical conditions. Sulfate-reducing bacteria use aerotaxis to remain within anoxic zones and are capable of chemotaxis toward sulfate, thiosulfate, nitrate, and lactate (162). Sulfur-oxidizing bacteria employ chemotaxis to position themselves within narrow bands corresponding to optimal sulfide and oxygen concentrations (49, 86, 191). Because the balance of sulfate reduction and sulfur oxidation drives the cycling of elemental sulfur in marine sediments and, furthermore, affects the cycling of carbon, nitrogen, and phosphorus at the seafloor (55, 85, 163), bacterial chemotaxis can significantly impact biogeochemistry at the sediment-water interface.
At the opposite end of the water column, in the sunlit upper ocean, several species of phytoplankton produce dimethylsulfoniopropionate (DMSP), which represents a significant source of reduced sulfur and carbon for heterotrophic bacteria (181) and can be degraded to volatile dimethylsulfide (DMS). DMS is the primary contributor to the ocean-atmosphere sulfur flux and a precursor to aerosol particles that are involved in cloud condensation. Several studies have shown that DMSP is a potent chemoattractant for different species of marine bacteria (94, 122, 175, 213). Because DMSP is released into the water column in ephemeral point source events and because there are two competing DMSP transformation pathways, only one of which results in DMS production, chemotaxis can have a major impact on the fate of DMSP by determining which bacteria gain access to newly released DMSP first. Some marine bacteria cleave DMSP to yield acrylate and release DMS (40), whereas others convert DMSP to methanethiol without the production of DMS (74). Both groups of bacteria, irrespective of the DMSP degradation pathway, exhibit chemotaxis toward DMSP (122, 175, 213). The consequences of who “wins the race” go beyond the competitive advantage of specific species and affect the transformation pathways of sulfur, the release of sulfur into the atmosphere as DMS, and ultimately the local climate through cloud formation.
The processes described above provide several examples of how chemotaxis may ultimately have ocean- or global-scale biogeochemical effects. As new information on mechanisms, interactions, and rates becomes available from microscale observations, the next challenge will be to incorporate this evidence into large-scale models of chemical flux. The complexities of this undertaking notwithstanding, the reward of this upscaling effort will be a new understanding of how microbial ecology influences the biogeochemistry of our planet.
CONCLUSIONS AND FUTURE DIRECTIONS
The importance of microscale behaviors, such as bacterial chemotaxis, are not immediately apparent within the vast and turbulent ocean and have consequently been largely overlooked by oceanographers and marine microbiologists. However, bacterial chemotaxis not only is widespread in the marine environment but often plays critical roles in ocean ecology and biogeochemistry. Chemotaxis by marine bacteria provides motile species with competitive advantages under appropriate nutrient conditions; it establishes and maintains symbiotic and pathogenic associations between bacteria and many other micro- and macro-organisms in the ocean; it affects the rates and directions of several chemical cycling processes, thus influencing ocean productivity; and it impacts the biogeochemical links between the ocean and the atmosphere, ultimately having implications for the global climate.
Marine environments harbor a diverse range of microscale chemical niches, from ephemeral solute pulses to animal surfaces, particles, and one-dimensional gradients, which present specific challenges to motile, chemotactic bacteria. Marine bacteria have evolved specific adaptations, including high-speed motility and run-and-reverse or run-reverse-and-flick movement patterns, to successfully employ chemotaxis in the ocean. We still know very little about the signaling transduction and the biophysical features of marine bacterial chemotaxis, especially in comparison to well-defined enteric model systems such as E. coli. We do know that marine bacteria often outperform E. coli in chemotaxis, but we only have relatively rudimentary insights into how and why. A concerted effort to understand chemotaxis of marine bacteria will be beneficial in two respects. First, the speed and precision of chemotaxis among marine bacteria promise to shed light on the physiological limits of gradient sensing among bacteria. In this respect, species such as V. alginolyticus, for which considerable information on swimming kinematics and physiology is already at hand, are excellent candidates for becoming model organisms for the study of chemotaxis. Second, a better understanding of the dependence of motility and chemotaxis in marine bacteria on environmental conditions will enable these phenotypes to be more accurately incorporated into ecological and biogeochemical models, without the need to resort to motility parameters obtained from enteric bacteria.
Biographies

Roman Stocker is a civil engineer by training, specializing in small-scale biological fluid mechanics, and has pioneered the use of microfluidics in microbial oceanography. His research interests focus on motility and chemotaxis, on the behavioral responses of microbes to gradients, and on the effect of physical forces and biochemical cues on the ecology of microorganisms. To address these questions, he integrates microfluidic technology, high-resolution video microscopy, image analysis, cell tracking, and mathematical modeling. Among his primary contributions are (i) the first experimental demonstration that marine bacteria can exploit the nutrient plumes emanating from sinking marine particles (186), (ii) a novel hydrodynamic mechanism for the formation of thin layers of phytoplankton in the ocean (45a), and (iii) the demonstration that organic sulfur compounds, and primarily DMSP, act as widespread infochemicals among marine microorganisms (175). As a hobby, and departing from his usual microbial focus, he has studied the fluid mechanics of how cats (in particular, his own) lap (155a).

Justin R. Seymour is a marine microbial ecologist whose research considers the ocean from a microbe's perspective, by combining oceanographic and microbiological approaches to examine microbial community dynamics and behavior at the ocean's microscale. His research has demonstrated that bacteria, viruses, and phytoplankton exhibit intense microscale heterogeneity in numbers, composition, and activities in the ocean (see, e.g., references 167 to 172) and that patchiness at the microbial scale often far exceeds that observed over the larger scales typically considered in oceanographic research. To examine the mechanisms underlying these patterns, he helped to pioneer the use of microfluidic tools to decipher the foraging ecology of aquatic microbes and demonstrated that marine bacteria use high-performance chemotaxis to survive in a patchy ocean (see, e.g., references 174 and 186). Using these approaches, Seymour and colleagues also demonstrated widespread chemotactic responses by marine microbes to the climatically important compound DMSP, revealing direct links between marine microbial behavior and global-scale chemical cycling processes (175).
REFERENCES
- 1. Aaronson S. 1978. Excretion of organic matter by phytoplankton in vitro. Limnol. Oceanogr. 23: 838 [Google Scholar]
- 2. Adler J. 1973. A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli. J. Gen. Microbiol. 74: 77–91 [DOI] [PubMed] [Google Scholar]
- 3. Reference deleted.
- 4. Ahmed T, Stocker R. 2008. Experimental verification of the behavioral foundation of bacterial transport parameters using microfluidics. Biophys. J. 95: 4481–4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alexandre G, Greer-Phillips S, Zhulin IB. 2004. Ecological role of energy taxis in microorganisms. FEMS Microbiol. Rev. 28: 113–126 [DOI] [PubMed] [Google Scholar]
- 6. Alldredge AL, Youngbluth MJ. 1985. The significance of macroscopic aggregates (marine snow) as sites of heterotrophic bacterial production in the mesopelagic zone of the subtropical Atlantic. Deep Sea Res. 32: 1445–1456 [Google Scholar]
- 7. Alldredge AL, Cole JJ, Caron DA. 1986. Production of heterotrophic bacteria inhabiting macroscopic aggregates (marine snow) from surface waters. Limnol. Oceanogr. 31: 68–78 [Google Scholar]
- 8. Alldredge AL, Cohen Y. 1987. Microscale chemical patches persist in the sea? Microelectrode study of marine snow, fecal pellets. Science 235: 689–691 [DOI] [PubMed] [Google Scholar]
- 9. Alldredge AL, Silver MW. 1988. Characteristics, dynamics and significance of marine snow. Progr. Oceanogr. 20: 41–82 [Google Scholar]
- 10. Alldredge AL, Passow U, Logan B. 1993. The abundance and significance of a class of large, transparent organic particles in the ocean. Deep Sea Res. 40: 1131–1140 [Google Scholar]
- 11. Azam F, et al. 1983. The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10: 257–263 [Google Scholar]
- 12. Azam F, Ammerman JW. 1984. Cycling of organic matter by bacterioplankton in pelagic marine ecosystems: microenvironmental considerations, p 345–360. In Fasham MJR. (ed), Flows of energy and materials in marine ecosystems. Plenum, New York, NY [Google Scholar]
- 13. Azam F. 1998. Microbial control of oceanic carbon flux: the plot thickens. Science 280: 694–696 [Google Scholar]
- 14. Azam F, Long RA. 2001. Sea snow microcosms. Nature 414: 495–498 [DOI] [PubMed] [Google Scholar]
- 15. Azam F, Malfatti F. 2007. Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 5: 782–791 [DOI] [PubMed] [Google Scholar]
- 16. Banin E, et al. 2000. Penetration of the coral-bleaching bacterium Vibrio shiloi into Oculina patagonica. Appl. Environ. Microbiol. 66: 3031–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Banin E, Israely T, Fine M, Loya Y, Rosenberg E. 2001. Role of endosymbiotic zooxanthellae and coral mucus in the adhesion of the coral-bleaching pathogen Vibrio shiloi to its host. FEMS Microbiol. Lett. 199: 33–37 [DOI] [PubMed] [Google Scholar]
- 18. Barbara GM, Mitchell JG. 2003. Marine bacterial organisation around point-like sources of amino acids. FEMS Microbiol. Ecol. 43: 99–109 [DOI] [PubMed] [Google Scholar]
- 19. Barbara GM, Mitchell JG. 2003. Bacterial tracking of motile algae. FEMS Microbiol. Ecol. 44: 79–87 [DOI] [PubMed] [Google Scholar]
- 20. Bassler BL, Gibbons PJ, Yu C, Roseman S. 1991. Chitin utilization by marine bacteria. Chemotaxis to chitin oligosaccharides by Vibrio furnissii, J. Biol. Chem. 266: 24268–24275 [PubMed] [Google Scholar]
- 21. Bassler BL, Gibbons PJ, Roseman S. 1989. Chemotaxis to chitin oligosaccharides by Vibrio furnissii, a chitinivorous marine bacterium. Biochem. Biophys. Res. Com. 161: 1172–1176 [DOI] [PubMed] [Google Scholar]
- 22. Bell W, Mitchell R. 1972. Chemotactic and growth responses of marine bacteria to algal extracellular products. Biol. Bull. 143: 265–277 [Google Scholar]
- 23. Berg HC. 1993. Random walks in biology. Princeton University Press, Princeton, NJ [Google Scholar]
- 24. Reference deleted.
- 25. Berg HC. 2004. E. coli in motion. Springer, Berlin, Germany [Google Scholar]
- 25a. Berg HC, Brown DA. 1972. Chemotaxis in Escherichia coli analysed by three-dimensional tracking. Nature 239: 500–504 [DOI] [PubMed] [Google Scholar]
- 26. Reference deleted.
- 27. Blackburn N, Azam F, Hagstrom Å 1997. Spatially explicit simulations of a microbial food web. Limnol. Oceanogr. 42: 613–622 [Google Scholar]
- 28. Blackburn N, Fenchel T, Mitchell JG. 1998. Microscale nutrient patches in plankton habitats shown by chemotactic bacteria. Science 282: 2254–2256. [DOI] [PubMed] [Google Scholar]
- 28a. Blackburn N. 2002. Cell structure/function, chapter 4, p 55–101. In Madigan MT, Martinko JM, Parker J. (ed), Brock biology of microorganisms, 10th ed. Pearson, Upper Saddle River, NJ [Google Scholar]
- 29. Blakemore RP. 1982. Magnetotactic bacteria. Annu. Rev. Microbiol. 36: 217–238 [DOI] [PubMed] [Google Scholar]
- 30. Bordas MA, Balebona C, Rodriguez-Maroto JM, Borrego JJ, Moriñigo MA. 1988. Chemotaxis of Vibrio strains towards mucus surfaces of guilt-head sea bream (Sparus aurata L.) Appl. Environ. Microbiol. 64: 1573–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bowen JD, Stolzenbach KD, Chisholm SW. 1993. Simulating bacterial clustering around phytoplankton cells in a turbulent ocean. Limnol. Oceanogr. 38: 36–51 [Google Scholar]
- 32. Brandes JA, Devol AH, Deutsch C. 2007. New developments in the marine nitrogen cycle. Chem. Rev. 107: 577–589 [DOI] [PubMed] [Google Scholar]
- 33. Brehm U, Krumbein WE, Palinska KA. 2003. Microbial spheres: a novel cyanobacterial-diatom symbiosis. Naturwissenschaften 90: 136–140 [DOI] [PubMed] [Google Scholar]
- 34. Broadbent AD, Jones GB. 2004. DMS and DMSP in mucus ropes, coral mucus, surface films and sediment pore waters from coral reefs in the Great Barrier Reef. J. Mar. Fresh. Res. 55: 849–855 [Google Scholar]
- 35. Broek Vande A, Lambrecht M, Vanderleyden J. 1998. Bacterial chemotactic motility is important for the initiation of wheat root colonization by Azospirillum brasilense. Microbiology 144: 2599–2606 [DOI] [PubMed] [Google Scholar]
- 36. Butler SM, Camilli A. 2005. Going against the grain: chemotaxis and infection in Vibrio cholerae. Nat. Rev. Microbiol. 3: 611–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chet I, Fogel S, Mitchell R. 1971. Chemical detection of microbial prey by bacterial predators. J. Bacteriol. 106: 863–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Colwell RR, et al. 1981. Occurrence of Vibrio cholerae serotype O1 in Maryland and Louisiana estuaries. Appl. Environ. Microbiol. 41: 555–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cooksey KE, Wigglesworth-Cooksey B. 1995. Adhesion of bacteria and diatoms to surfaces in the sea: a review. Aquat. Microb. Ecol. 9: 87–96 [Google Scholar]
- 40. Curson ARJ, Todd JD, Sullivan MJ, Johnston AWB. 2011. Catabolism of dimethylsulfoniopropionate: microorganisms, enzymes and genes. Nat. Rev. Microbiol. 9: 849–859 [DOI] [PubMed] [Google Scholar]
- 41. del Giorgio PA, Cole JJ. 1998. Bacterial growth efficiency in natural aquatic systems. Annu. Rev. Ecol. Syst. 29: 503–541 [Google Scholar]
- 42. DeLoney-Marino CR, Visick KL. 2012. Role for cheR of Vibrio fischeri in the Vibrio-squid symbiosis. Can. J. Microbiol. 58: 29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. DeLoney-Marino CR, Wolfe AJ, Visick KL. 2003. Chemoattraction of Vibrio fischeri to serine, nucleosides, and N-acetylneuraminic acid, a component of squid light-organ mucus. Appl. Environ. Microbiol. 69: 7527–7530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Reference deleted.
- 45. DeLong EF. 2006. Community genomics among stratified microbial assemblages in the ocean's interior. Science 31: 496–503 [DOI] [PubMed] [Google Scholar]
- 45a. Durham WM, Kessler JO, Stocker R. 2009. Disruption of motility by shear triggers formation of thin phytoplankton layers. Science 323: 1067–1070 [DOI] [PubMed] [Google Scholar]
- 46. Dusenbery DB. 2009. Living at micro scale. Harvard University Press, Cambridge, MA [Google Scholar]
- 47. Engelmann TW. 1881. Bacterium photometricum: an article on the comparative physiology of the sense for light and colour. Arch. Ges. Physiol. Bonn 30: 95–124 [Google Scholar]
- 48. Falkowski P, Fenchel T, DeLong EF. 2008. The microbial engines that drive Earth's biogeochemical cycles. Science 320: 1034–1039 [DOI] [PubMed] [Google Scholar]
- 49. Fenchel T. 1994. Motility and chemosensory behavior of the sulfur bacterium Thiovolum majus. Microbiology 140: 3109–3116 [Google Scholar]
- 50. Fenchel T. 2001. Eppur si muove: many water column bacteria are motile. Aquat. Microb. Ecol. 24: 197–201 [Google Scholar]
- 51. Fenchel T. 2002. Microbial behavior in a heterogeneous world. Science 296: 1068–1071 [DOI] [PubMed] [Google Scholar]
- 52. Fenchel T, Glud RN. 1998. Veil architecture in a sulfide-oxidizing bacterium enhances countercurrent flux. Nature 394: 367–369 [Google Scholar]
- 53. Fenchel T, Thar R. 2004. “Candidatus Ovobacter propellens”: A large conspicuous prokaryote with an unusual motility behavior. FEMS Microbiol. Ecol. 48: 231–238 [DOI] [PubMed] [Google Scholar]
- 54. Fogg GE. 1977. Excretion of organic matter by phytoplankton. Limnol. Oceanogr. 22: 576–577 [Google Scholar]
- 55. Fossing H, et al. 1995. Concentration and transport of nitrate by the mat-forming sulfur bacterium Thioploca. Nature 374: 713–715 [Google Scholar]
- 56. Freter R, O'Brien PCM, Macsai MS. 1979. Effect of chemotaxis in the interaction of cholera vibrios with intestinal mucosa. Am. J. Clin. Nutr. 32: 128–132 [DOI] [PubMed] [Google Scholar]
- 57. Freter R, O'Brien PCM. 1981. Role of chemotaxis in the association of motile bacteria with intestinal mucosa: chemotactic responses of Vibrio cholerae and description of motile nonchemotactic mutants. Infect. Immun. 34: 215–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gallucci KK, Paerl HW. 1983. Pseudomonas aeruginosa chemotaxis associated with blooms of N2-fixing blue-green algae (Cyanobacteria). Appl. Environ. Microbiol. 45: 557–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gest H. 1995. Phototaxis and other sensory phenomena in purple photosynthetic bacteria. FEMS Microbiol. Rev. 16: 287–294 [Google Scholar]
- 60. Getchell RJ. 1989. Bacterial shell disease in crustaceans: a review. J. Shellf. Res. 8: 1–6 [Google Scholar]
- 61. Gilbert JA, et al. 2010. The taxonomic and functional diversity of microbes at a temperate coastal site: a ‘multi-omic’ study of seasonal and diel temporal variation. PLoS One 5: e15545 doi:10.1371/journal.pone.0015545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Giovannoni SJ, et al. 2005. Genome streamlining in a cosmopolitan oceanic bacterium. Science 309: 1242–1245 [DOI] [PubMed] [Google Scholar]
- 63. Gluch MF, Typke D, Baumeister W. 1995. Motility and thermotactic responses of Thermotoga maritima. J. Bacteriol. 177: 5473–5479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Goldman JC, McCarthy JJ, Peavey DG. 1979. Growth rate influence on the chemical composition of phytoplankton in oceanic waters. Nature 279: 210–215 [Google Scholar]
- 65. Grossart H-P, Simon M. 1998. Bacterial colonization and microbial decomposition of limnetic organic aggregates (lake snow). Aquat. Microb. Ecol. 15: 127–140 [Google Scholar]
- 66. Grossart H-P, Rieman L, Azam F. 2001. Bacterial motility in the sea and its ecological implications. Aquat. Microb. Ecol. 25: 247–258 [Google Scholar]
- 67. Grossart H-P, Dziallasa C, Leunerta F, Tang KW. 2010. Bacteria dispersal by hitchhiking on zooplankton. Proc. Natl. Acad. Sci. U. S. A. 107: 11959–11964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Guasto JS, Rusconi R, Stocker R. 2012. Fluid mechanics of planktonic microorganisms. Annu. Rev. Fluid Mech. 44: 373–400 [Google Scholar]
- 69. Haas AF, Wild C. 2010. Composition analysis of organic matter released by cosmopolitan coral reef-associated green algae. Aquat. Biol. 10: 131–138 [Google Scholar]
- 70. Hazelbauer GL, Berg HC, Matsumura PM. 1993. Bacterial motility and signal transduction. Cell 73: 15–22 [DOI] [PubMed] [Google Scholar]
- 71. Hedges JI, et al. 2001. Evidence for non-selective preservation of organic matter in sinking marine particles. Nature 409: 801–804 [DOI] [PubMed] [Google Scholar]
- 72. Hellebrust JA. 1965. Excretion of some organic compounds by marine phytoplankton. Limnol. Oceanogr. 10: 192–206 [Google Scholar]
- 73. Higgins R. 2000. Bacteria and fungi of marine mammals: a review. Can. Vet. J. 41: 105–116 [PMC free article] [PubMed] [Google Scholar]
- 74. Howard E, et al. 2006. Bacterial taxa that limit sulfur flux from the ocean. Science 314: 649–652 [DOI] [PubMed] [Google Scholar]
- 75. Huettel M, Forster S, Kloser S, Fossing H. 1996. Vertical migration in the sediment-dwelling sulfur bacteria Thioploca spp. in overcoming diffusion limitations. Appl. Environ. Microbiol. 62: 1863–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hutchinson GE. 1961. The paradox of the plankton. Am. Nat. 95: 137–145 [Google Scholar]
- 77. Hütz A, Schubert K, Overmann J. 2011. Thalassospira sp. isolated from the oligotrophic eastern Mediterranean sea exhibits chemotaxis toward inorganic phosphate during starvation. Appl. Environ. Microbiol. 77: 4412–4421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Jackson GA. 1980. Phytoplankton growth and zooplankton grazing in oligotrophic oceans. Nature 284: 439–441 [Google Scholar]
- 79. Jackson GA. 1987. Simulating chemosensory responses of marine microorganisms. Limnol. Oceanogr. 32: 1253–1266 [Google Scholar]
- 80. Jackson GA. 1989. Simulation of bacterial attraction and adhesion to falling particles in an aquatic environment. Limnol. Oceanogr. 34: 514–530 [Google Scholar]
- 81. Reference deleted.
- 82. Johansen JE, Pinhassi J, Blackburn N, Zweifel UL, Hagstrom Å 2002. Variability in motility characteristics among marine bacteria. Aquat. Microb. Ecol. 28: 229–237 [Google Scholar]
- 83. Johnson AR, Wiens JA, Milne BT, Crist TO. 1992. Animal movements and population dynamics in heterogeneous landscapes. Land Ecol. 7: 63–75 [Google Scholar]
- 84. Jones AK, Cannon RC. 1986. The release of micro-algal photosynthate and associated bacterial uptake and heterotrophic growth. Br. Phycol. J. 21: 341–358 [Google Scholar]
- 85. Jørgensen BB. 1982. Mineralization of organic matter in the sea bed–the role of sulfate reduction. Nature 296: 643–645 [Google Scholar]
- 86. Jørgensen BB, Revsbech NP. 1983. Colorless sulfur bacteria, Beggiatoa spp. and Thiovulum spp., in O2 and H2S microgradients. Appl. Environ. Microbiol. 45: 1261–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Jørgensen BB, Revsbech NP. 1985. Diffusive boundary layers and the oxygen uptake of sediments and detritus. Limnol. Oceanogr. 30: 111–122 [Google Scholar]
- 88. Jumars PA, Penry DL, Baross JA, Perry MJ, Frost BW. 1989. Closing the microbial loop: dissolved carbon pathway to heterotrophic bacteria from incomplete ingestion, digestion and absorption in animals. Deep Sea Res. 36: 483–495 [Google Scholar]
- 89. Kaper J, Lockman H, Colwell RR, Joseph SW. 1979. Ecology, serology, and enterotoxin production of Vibrio cholerae in Chesapeake Bay. Appl. Environ. Microbiol. 37: 91–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kennedy MJ, Lawless JG. 1985. Role of chemotaxis in the ecology of denitrifiers. Appl. Environ. Microbiol. 49: 109–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Kihara M, Macnab RM. 1981. Cytoplasmic pH mediates pH taxis and weak-acid repellent taxis of bacteria. J. Bacteriol. 145: 1209–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kiørboe T. 2008. A mechanistic approach to plankton ecology. Princeton University Press, Princeton, NJ [Google Scholar]
- 93. Kiørboe T, Jackson GA. 2001. Marine snow, organic solute plumes, and optimal chemosensory behavior of bacteria. Limnol. Oceanogr. 46: 1309–1318 [Google Scholar]
- 94. Kiørboe T, Grossart H-P, Ploug H, Tang K. 2002. Mechanisms and rates of bacterial colonization of sinking aggregates. Appl. Environ. Microbiol. 68: 3996–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kiørboe T, Ploug H, Thygesen UH. 2001. Fluid motion and solute distribution around sinking aggregates. I. Small-scale fluxes and heterogeneity of nutrients in the pelagic environment. Mar. Ecol. Prog. Ser. 211: 1–13 [Google Scholar]
- 96. Kirchman DL. 2008. Microbial ecology of the oceans. Wiley & Sons, New York, NY [Google Scholar]
- 97. Koike I, Hara S, Terauch K, Kogure K. 1990. Role of submicrometre particles in the ocean. Nature 345: 242–244 [Google Scholar]
- 98. Kühl M, Cohen Y, Dalsgaard T, Jørgensen BB, Revsbech NP. 1995. Microenvironment and photosynthesis of zooxanthellae in scleractinian corals studied with microsensors for O2, pH and light. Mar. Ecol. Progr. Ser. 117: 159–172 [Google Scholar]
- 99. Lampert W. 1978. Release of dissolved organic carbon by grazing zooplankton. Limnol. Oceanogr. 23: 831–834 [Google Scholar]
- 100. Larsson U, Hagstrom Å 1979. Phytoplankton exudate release as an energy source for the growth of pelagic bacteria. Mar. Biol. 53: 199–206 [Google Scholar]
- 101. Larsson U, Hagstrom Å 1982. Fractionated phytoplankton primary production, exudate release, and bacterial production in a Baltic eutrophication gradient. Mar. Biol. 67: 57–70 [Google Scholar]
- 102. Lauffenburger D, Aris R, Keller K. 1982. Effects of cell motility and chemotaxis on microbial population growth. 40: 219–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Lehman JT, Scavia D. 1982a. Microscale nutrient patches produced by zooplankton. Proc. Natl. Acad. Sci. U. S. A. 79: 5001–5005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Lehman JT, Scavia D. 1982b. Microscale patchiness of nutrients in plankton communities. Science 216: 729–730 [DOI] [PubMed] [Google Scholar]
- 105. Levin SA. 1994. Patchiness in marine and terrestrial systems: from individuals to populations. Philos. Trans. R. Soc. B Biol. Sci. 343: 99–103 [Google Scholar]
- 106. Li N, Kojima D, Homma M. 2011. Sodium-driven motor of the polar flagellum in marine bacteria Vibrio. Genes Cells. 16: 985–999 [DOI] [PubMed] [Google Scholar]
- 107. Liu H, Nolla HA, Campbell L. 1997. Prochlorococcus growth rate and contribution to primary production in the equatorial and subtropical North Pacific Ocean. Aquat. Microb. Ecol. 12: 39–47 [Google Scholar]
- 108. Locsei JT, Pedley TJ. 2009. Bacterial tracking of motile algae assisted by algal cell's vorticity field. Microb. Ecol. 58: 63–74 [DOI] [PubMed] [Google Scholar]
- 109. Reference deleted.
- 110. Long RA, Azam F. 2001. Microscale patchiness of bacterioplankton assemblage richness in seawater. Aquat. Microb. Ecol. 26: 103–113 [Google Scholar]
- 111. Luchsinger RH, Bergersen B, Mitchell JG. 1999. Bacterial swimming strategies and turbulence. Biophys. J. 77: 2377–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Lux R, Shi W. 2004. Chemotaxis-guided movements in bacteria. Crit. Rev. Oral Biol. Med. 15: 207–220 [DOI] [PubMed] [Google Scholar]
- 113. Macnab RM, Aizawa SI. 1984. Bacterial motility and the bacterial flagellar motor. Annu. Rev. Biophys. Bioeng. 13: 51–83 [DOI] [PubMed] [Google Scholar]
- 114. Macnab RM. 1996. Flagella and motility, p 123–143.In Neidhardt FC. (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed ASM Press, Washington, DC [Google Scholar]
- 115. Magariyama Y, et al. 1994. Very fast flagellar rotation. Nature 371: 752. [DOI] [PubMed] [Google Scholar]
- 116. Magariyama Y, Sugiyama S, Kudo S. 2001. Bacterial swimming speed and rotation rate of bundled flagella. FEMS Microbiol. Lett. 199: 125–129 [DOI] [PubMed] [Google Scholar]
- 117. Malmcrona-Friberg K, Goodman A, Kjelleberg S. 1990. Chemotactic responses of marine Vibrio sp. strain S14 (CCUG 15956) to low-molecular weight substances under starvation and recovery conditions. Appl. Environ. Microbiol. 56: 3699–3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Mandel MJ, et al. 2012. Squid-derived chitin oligosaccharides are a chemotactic signal during colonization by Vibrio fischeri. Appl. Environ. Microbiol. 78: 4620–4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. McCarren J, et al. 2010. Microbial community transcriptomes reveal microbe and metabolic pathways associated with dissolved organic matter turnover in the sea. Proc. Natl. Acad. Sci. U. S. A. 107: 16420–16427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. McCarthy JJ, Goldman JC. 1979. Nitrogenous nutrition of marine phytoplankton in nutrient depleted waters. Science 203: 670–672 [DOI] [PubMed] [Google Scholar]
- 121. Meron D, et al. 2009. Role of flagella in virulence of the coral pathogen Vibrio coralliilyticus. Appl. Environ. Microbiol. 75: 5704–5707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Miller TR, Hnilicka K, Dziedzic A, Desplats P, Belas R. 2004. Chemotaxis of Silicibacter sp. strain TM1040 toward dinoflagellate products. Appl. Environ. Microbiol. 70: 4692–4701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Miller TR, Belas R. 2006. Motility is involved in Silicibacter sp. TM1040 interaction with dinoflagellates. Environ. Microbiol. 8: 1648–1659 [DOI] [PubMed] [Google Scholar]
- 124. Mitchell JG, Okubo A, Fuhrman JA. 1985. Microzones surrounding phytoplankton form the basis for a stratified marine microbial ecosystem. Nature 316: 58–59 [Google Scholar]
- 125. Mitchell JG. 1991. The influence of cell size on marine bacterial motility and energetics. Microb. Ecol. 22: 227–238 [DOI] [PubMed] [Google Scholar]
- 126. Mitchell JG, et al. 1995. Long lag times and high velocities in the motility of natural assemblages of marine bacteria. Appl. Environ. Microbiol. 61: 877–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Mitchell JG, Pearson L, Dillon S, Kantalis K. 1995. Natural assemblages of marine bacteria exhibiting high speed motility and large accelerations. Appl. Environ. Microbiol. 61: 4436–4440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Mitchell JG, Pearson L, Dillon S. 1996. Clustering of marine bacteria in seawater enrichments. Appl. Environ. Microbiol. 62: 3716–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Mitchell JG. 2002. The energetics and scaling of search strategies in bacteria. Am. Nat. 160: 727–740 [DOI] [PubMed] [Google Scholar]
- 130. Mitchell JG, Kogure K. 2006. Bacterial motility: links to the environment and a driving force for microbial physics. FEMS Microbiol. Ecol. 55: 3–16 [DOI] [PubMed] [Google Scholar]
- 131. Moran MA, et al. 2004. Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature 432: 910–913 [DOI] [PubMed] [Google Scholar]
- 132. Moriarty DJW. 1986. Exudation of organic carbon by the seagrass Halodule wrightii Aschers and its effect on bacterial growth in the sediment. J. Exp. Mar. Biol. Ecol. 96: 115–126 [Google Scholar]
- 133. Morris RM, et al. 2002. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420: 806–810 [DOI] [PubMed] [Google Scholar]
- 134. Muramoto K, et al. 1995. High-speed rotation and speed stability of the sodium-driven flagellar motor in Vibrio alginolyticus. J. Mol. Biol. 251: 50–58 [DOI] [PubMed] [Google Scholar]
- 135. Murray AG, Jackson GA. 1992. Viral dynamics: a model of the effects of size, shape, motion and abundance of single-celled planktonic organisms and other particles. Mar. Ecol. Progr. Ser. 89: 103–116 [Google Scholar]
- 136. Nealson KH, Moser DP, Saffarini DA. 1995. Anaerobic electron acceptor chemotaxis in Shewanella putrefaciens. Appl. Environ. Microbiol. 61: 1551–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Olsen MA, Aagnes TH, Mathiesen SV. 1994. Digestion of herring by indigenous bacteria in the minke whale forestomach. Appl. Environ. Microbiol. 60: 4445–4455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. O'Toole R, Milton DL, Wolf-Watz H. 1996. Chemotactic motility is required for invasion of the host by the fish pathogen Vibrio anguillarum. Mol. Microbiol. 19: 625–637 [DOI] [PubMed] [Google Scholar]
- 139. O'Toole R, et al. 1999. The chemotactic response of Vibrio anguillarum to fish intestinal mucus is mediated by a combination of multiple mucus components. J. Bacteriol. 181: 4308–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Paerl HW. 1978. Role of heterotrophic bacteria in promoting N2 fixation by Anabaena in aquatic habitats. Microb. Ecol. 4: 215–231 [DOI] [PubMed] [Google Scholar]
- 141. Paerl HW. 1982. Interactions with bacteria, p 441–461.In Carr NG, Whitton BA. (ed), The biology of cyanobacteria. Blackwell Scientific Publications, Oxford, United Kingdom [Google Scholar]
- 142. Paerl HW, Gallucci KK. 1985. Role of chemotaxis in establishing a specific nitrogen-fixing cyanobacterial-bacterial association. Science 227: 647–649 [DOI] [PubMed] [Google Scholar]
- 143. Paerl HW. 1996. Microscale physiological and ecological studies of aquatic cyanobacteria: macroscale implications. Microsc. Res. Technol. 33: 47–72 [DOI] [PubMed] [Google Scholar]
- 144. Paerl HW, Pinckney JL. 1996. A mini-review of microbial consortia: their roles in aquatic production and biogeochemical cycling. Microb. Ecol. 31: 225–247 [DOI] [PubMed] [Google Scholar]
- 145. Paerl HW, Steppe TF. 2003. Scaling up: the next challenge in environmental microbiology. Env. Microbiol. 5: 1025–1038 [DOI] [PubMed] [Google Scholar]
- 146. Pandya S, Poormina I, Gaitonde V, Parekh T, Desai A. 1999. Chemotaxis of Rhizobium sp. S2 towards Cajanus cajan root exudate and its major components. Curr. Microbiol. 38: 205–209 [DOI] [PubMed] [Google Scholar]
- 147. Paul JH, Deflaun MF, Jeffrey W. 1986. Elevated levels of microbial activity in the coral surface microlayer. Mar. Ecol. Prog. Ser. 33: 29–40 [Google Scholar]
- 148. Peduzzi P, Herndl GJ. 1992. Zooplankton activity fuelling the microbial loop: differential growth response of bacteria from oligotrophic and eutrophic waters. Limnol. Oceanogr. 37: 1087–1092 [Google Scholar]
- 149. Pfeffer W. 1884. Locomotorische Richtungsbewegungen durch chemische Reize. Untersuchung. Botanisch. Insti. Tubingen 1: 363–482 [Google Scholar]
- 150. Pittman MS, Goodwin M, Kelly DJ. 2001. Chemotaxis in the human gastric pathogen Helicobacter pylori: different roles for CheW and the three CheV paralogues, and evidence for CheV2 phosphorylation. Microbiology 147: 2493–2504 [DOI] [PubMed] [Google Scholar]
- 151. Pomeroy LR, Williams PJ, Azam F, Hobbie JE. 2007. The microbial loop. Oceanography 20: 28–33 [Google Scholar]
- 152. Porter SL, Wadhams GH, Armitage JP. 2011. Signal processing in complex chemotaxis pathways. Nat. Rev. Microbiol. 9: 153–165 [DOI] [PubMed] [Google Scholar]
- 153. Prezelin BB, Alldredge AL. 1983. Primary production of marine snow during and after an upwelling event. Limnol. Oceanogr. 28: 1156–1167 [Google Scholar]
- 154. Prieur D, Mevel G, Nicolas JL, Plusquellec A, Vigneulle M. 1990. Interactions between bivalve molluscs and bacteria in the marine environment. Oceanogr. Mar. Biol. Annu. Rev. 28: 277–352 [Google Scholar]
- 155. Purcell EM. 1977. Life at low Reynolds number, Am. J. Phys. 45: 3–11 [Google Scholar]
- 155a. Reis PM, Jung S, Aristoff JM, Stocker R. 2010. How cats lap: water uptake by Felis catus. Science 330: 1231–1234 [DOI] [PubMed] [Google Scholar]
- 156. Riemann L, Middelboe M. 2002. Viral lysis of marine bacterioplankton: implications for organic matter cycling and bacterial clonal composition. Mar. Biol. Res. 56: 57–68 [Google Scholar]
- 157. Rohwer F, Breitbart M, Jara J, Azam F, Knowlton N. 2001. Diversity of bacteria associated with the Caribbean coral Montastraea franksi. Coral Reefs 20: 85–91 [Google Scholar]
- 158. Rohwer F, Seguritan V, Azam F, Knowlton N. 2002. Diversity and distribution of coral-associated bacteria. Mar. Ecol. Progr. Ser. 243: 1–10 [Google Scholar]
- 159. Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I. 2007. The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5: 355–362 [DOI] [PubMed] [Google Scholar]
- 160. Sar N, Rosenberg E. 1987. Fish skin bacteria: colonial and cellular hydrophobicity. Microb. Ecol. 13: 193–202 [DOI] [PubMed] [Google Scholar]
- 161. Sar N, McCarter L, Simon M, Silverman M. 1990. Chemotactic control of the two flagellar systems of Vibrio parahaemolyticus. J. Bacteriol. 172: 334–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Sass AM, et al. 2002. Growth and chemosensory behavior of sulfate-reducing bacteria in oxygen-sulfide gradients. FEMS Microbiol. Ecol. 40: 47–54 [DOI] [PubMed] [Google Scholar]
- 163. Schulz HN, Schulz HD. 2005. Large sulfur bacteria and the formation of phosphorite. Science 307: 416–418 [DOI] [PubMed] [Google Scholar]
- 164. Schulz HN, et al. 1999. Dense populations of a giant sulfur bacterium in Namibian shelf sediments. Science 284: 493–495 [DOI] [PubMed] [Google Scholar]
- 165. Schulz HN, Jørgensen BB. 2001. Big bacteria. Annu. Rev. Microbiol. 55: 105–137 [DOI] [PubMed] [Google Scholar]
- 166. Segall JE, Manson MD, Berg HC. 1986. Temporal comparisons in bacterial chemotaxis. Proc. Natl. Acad. Sci. U. S. A. 83: 8987–8991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Seymour JR, Mitchell JG, Pearson L, Waters RL. 2000. Heterogeneity in bacterioplankton abundance from 4.5 millimetre resolution sampling. Aquat. Microb. Ecol. 22: 143–153 [Google Scholar]
- 168. Seymour JR, Seuront L, Mitchell JG. 2004. Microscale heterogeneity in the activity of coastal bacterioplankton communities. Aquat. Microb. Ecol. 35: 1–16 [Google Scholar]
- 169. Seymour JR, Patten N, Bourne DG, Mitchell JG. 2005. Spatial dynamics of virus-like particles and heterotrophic bacteria in a coral reef system. Mar. Ecol. Progr. Ser. 288: 1–8 [Google Scholar]
- 170. Seymour JR, Seuront L, Mitchell JG. 2005. Microscale and small-scale temporal dynamics of a coastal planktonic microbial community. Mar. Ecol. Progr. Ser. 300: 21–37 [Google Scholar]
- 171. Seymour JR, Seuront L, Doubell M, Waters RL, Mitchell JG. 2006. Microscale patchiness of virioplankton. J. Mar. Biol. Assoc. 86: 551–561 [Google Scholar]
- 172. Seymour JR, Ahmed T, Marcos Stocker R. 2008. A microfluidic chemotactic assay for assessing the behavior of microbes within diffusing nutrient patches. Limnol. Oceanogr. Methods 6: 477–488 [Google Scholar]
- 173. Seymour JR, Ahmed T, Stocker R. 2009. Bacterial chemotaxis towards the extracellular products of the toxic phytoplankton Heterosigma akashiwo. J. Plankton. Res. 31: 1557–1561 [Google Scholar]
- 174. Seymour JR, Marcos Stocker R. 2009. Resource patch formation and exploitation throughout the marine microbial food web. Am. Nat. 173: E15–E29 [DOI] [PubMed] [Google Scholar]
- 175. Seymour JR, Simó R, Ahmed T, Stocker R. 2010. Chemoattraction to dimethylsulfoniopropionate throughout the marine microbial food web. Science 329: 342–345 [DOI] [PubMed] [Google Scholar]
- 176. Seymour JR, Ahmed T, Stocker R. 2010. Chemotactic response of marine heterotrophic bacteria to the extracellular products of Synechococcus and Prochlorococcus. Aquat. Microb. Ecol. 59: 161–168 [Google Scholar]
- 177. Shanks AL, Trent JD. 1979. Marine snow: microscale nutrient patches. Limnol. Oceanogr. 24: 850–854 [Google Scholar]
- 178. Sheridan CC, Steinberg DK, Kling GW. 2002. The microbial and metazoan community associated with colonies of Trichodesmium spp.: a quantitative survey. J. Plankton. Res. 24: 913–922 [Google Scholar]
- 179. Shi Y, McCarren J, DeLong EF. 2012. Transcriptional responses of surface water marine microbial assemblages to deep-sea water amendment. Environ. Microbiol. 14: 191–206 [DOI] [PubMed] [Google Scholar]
- 180. Shotts EB, Albert TF, Wooley RE, Brown J. 1990. Microflora associated with the skin of the bowhead whale (Balaena mysticetus). J. Wildl. Dis. 26: 351–359 [DOI] [PubMed] [Google Scholar]
- 181. Simó R. 2001. Production of atmospheric sulfur by oceanic plankton: biogeochemical, ecological and evolutionary links. Trends Ecol. Evol. 16: 287–294 [DOI] [PubMed] [Google Scholar]
- 182. Sjoblad RD, Mitchell RJ. 1979. Chemotactic responses of Vibrio alginolyticus to algal extracellular products. Can. J. Microbiol. 25: 964–967 [DOI] [PubMed] [Google Scholar]
- 183. Slightom RN, Buchan A. 2009. Surface colonization by marine roseobacters: integrating genotype and phenotype. App. Environ. Microbiol. 75: 6027–6037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Smith DC, Simon M, Alldredge AL, Azam F. 1992. Intense hydrolytic enzyme activity on marine aggregates and implications for rapid particle dissolution. Nature 359: 139–142 [Google Scholar]
- 185. Sogin ML, et al. 2006. Microbial diversity in the deep sea and the underexplored ‘rare biosphere’. Proc. Natl. Acad. Sci. U. S. A. 103: 12115–12120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Stocker R, Seymour JR, Samadani A, Hunt DH, Polz MF. 2008. Rapid chemotactic response enables marine bacteria to exploit ephemeral microscale nutrient patches. Proc. Natl. Acad. Sci. U. S. A. 105: 4209–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186a. Stocker R, Seymour JR, Gorick G. 5 February 2010. Cover image. Science 327(5966). [Google Scholar]
- 187. Szurmant H, Ordal GW. 2004. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microb. Mol. Biol. Rev. 68: 301–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Tang KW. 2005. Copepods as microbial hotspots in the ocean: effects of host feeding activities on attached bacteria. Aquat. Microb. Ecol. 38: 31–40 [Google Scholar]
- 189. Tang WT, Turk V, Grossart H-P. 2010. Linkage between crustacean zooplankton and aquatic bacteria. Aquat. Microb. Ecol. 61: 261–277 [Google Scholar]
- 190. Taylor JR, Stocker R. 2012. Trade-offs of chemotactic foraging in turbulent water. Science 338: 675–679 [DOI] [PubMed] [Google Scholar]
- 191. Thar R, Fenchel T. 2005. Survey of motile microaerophilic bacterial morphotypes in the oxygen gradient above a marine sulfidic sediment. Appl. Environ. Microbiol. 71: 3682–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Thar R, Kühl M. 2002. Conspicuous veils formed by vibrioid bacteria on sulfidic marine sediment. Appl. Environ. Microbiol. 68: 6310–6320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Thar R, Kühl M. 2003. Bacteria are not too small for spatial sensing of chemical gradients: an experimental evidence. Proc. Natl. Acad. Sci. U. S. A. 100: 5748–5753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Turner L, Ryu WS, Berg HC. 2000. Real-time imaging of fluorescent flagellar filaments. J. Bacteriol. 182: 2793–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Urban-Rich J. 1999. Release of dissolved organic carbon from copepod fecal pellets in the Greenland Sea. J. Exp. Mar. Biol. Ecol. 232: 107–124 [Google Scholar]
- 196. Vega-Thurber R, et al. 2009. Metagenomic analysis of stressed coral holobionts. Env. Microbiol. 11: 2148–2163 [DOI] [PubMed] [Google Scholar]
- 197. Venter JC, et al. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304: 66–74 [DOI] [PubMed] [Google Scholar]
- 198. Verdugo P, et al. 2004. The oceanic gel phase: a bridge in the DOM-POM continuum. Mar. Chem. 92: 67–85 [Google Scholar]
- 199. Vetter YA, Deming JW, Jumars PA, Krieger-Brockett BB. 1998. A predictive model of bacterial foraging by means of freely released extracellular enzymes. Microb. Ecol. 36: 75–92 [DOI] [PubMed] [Google Scholar]
- 200. Vila-Costa M, et al. 2010. Transcriptomic analysis of a marine bacterial community enriched with dimethylsulfoniopropionate. ISME J. 4: 1410–1420 [DOI] [PubMed] [Google Scholar]
- 201. Visser AW, Jackson GA. 2004. Characteristics of the chemical plume behind a sinking particle in a turbulent water column. Mar. Ecol. Progr. Ser. 283: 55–71 [Google Scholar]
- 202. Wadhams GH, Armitage JP. 2004. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 5: 1024–1103 [DOI] [PubMed] [Google Scholar]
- 203. Webster NS, Taylor MW. 2012. Marine sponges and their microbial symbionts: love and other relationships. Environ. Microbiol. 14: 335–346 [DOI] [PubMed] [Google Scholar]
- 204. Whitman WB, Coleman DC, Wiebe WJ. 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. U. S. A. 95: 6578–6583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Wild C, et al. 2004. Coral mucus functions as an energy carrier and particle trap in the reef ecosystem. Nature 428: 66–70 [DOI] [PubMed] [Google Scholar]
- 206. Willey JM, Waterbury JB. 1989. Chemotaxis toward nitrogenous compounds by swimming strains of marine Synechococcus spp. Appl. Environ. Microbiol. 55: 1888–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Xie L, Altindal T, Chattopadhyay S, Wu X- L. 2011. Bacterial flagellum as a propeller and as a rudder for efficient chemotaxis. Proc. Natl. Acad. Sci. U. S. A. 108: 2246–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Yang Z, Midmore DJ. 2005. Modeling plant resource allocation and growth partitioning in response to environmental heterogeneity. Ecol. Model. 181: 59–77 [Google Scholar]
- 209. Yooseph S, et al. 2007. The Sorcerer II global ocean sampling expedition: expanding the universe of protein families. PLoS Biol. 5: e16 doi:10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Yorimitsu T, Homma M. 2001. Na+-driven flagellar motor of Vibrio. Biochim. Biophys. Acta 1505: 82–93 [DOI] [PubMed] [Google Scholar]
- 211. Yu C, Bassler BL, Roseman S. 1993. Chemotaxis of the marine bacterium Vibrio furnissii to sugars. A potential mechanism for initiating the chitin catabolic cascade. J. Biol. Chem. 268: 9405–9409 [PubMed] [Google Scholar]
- 212. Zehr JP, Paerl HW. 2008. Molecular ecological aspects of nitrogen fixation in the marine environment, p 481–525.In Kirchman DL. (ed), Microbial ecology of the oceans, 2nd ed. John Wiley & Sons, New York, NY [Google Scholar]
- 213. Zimmer-Faust RK, de Souza MP, Yoch DC. 1996. Bacterial chemotaxis and its potential role in marine dimethylsulfide production and biogeochemical sulfur cycling. Limnol. Oceanogr. 41: 1330–1334 [Google Scholar]
- 214. Zopfi J, Kjær T, Nielsen LP, Jørgensen BB. 2001. Ecology of Thioploca spp.: nitrate and sulfur storage in relation to chemical microgradients and influence of Thioploca spp. on the sedimentary nitrogen cycle. Appl. Environ. Microbiol. 67: 5530–5537 [DOI] [PMC free article] [PubMed] [Google Scholar]