Abstract
Skeletal muscle exhibits a high plasticity and accordingly can quickly adapt to different physiological and pathological stimuli by changing its phenotype largely through diverse epigenetic mechanisms. The nuclear receptor corepressor 1 (NCoR1) has the ability to mediate gene repression; however, its role in regulating biological programs in skeletal muscle is still poorly understood. We therefore studied the mechanistic and functional aspects of NCoR1 function in this tissue. NCoR1 muscle-specific knockout mice exhibited a 7.2% higher peak oxygen consumption (VO2peak), a 11% reduction in maximal isometric force, and increased ex vivo fatigue resistance during maximal stimulation. Interestingly, global gene expression analysis revealed a high overlap between the effects of NCoR1 deletion and peroxisome proliferator-activated receptor gamma (PPARγ) coactivator 1α (PGC-1α) overexpression on oxidative metabolism in muscle. Importantly, PPARβ/δ and estrogen-related receptor α (ERRα) were identified as common targets of NCoR1 and PGC-1α with opposing effects on the transcriptional activity of these nuclear receptors. In fact, the repressive effect of NCoR1 on oxidative phosphorylation gene expression specifically antagonizes PGC-1α-mediated coactivation of ERRα. We therefore delineated the molecular mechanism by which a transcriptional network controlled by corepressor and coactivator proteins determines the metabolic properties of skeletal muscle, thus representing a potential therapeutic target for metabolic diseases.
INTRODUCTION
Improved muscle performance is directly linked to a lower prevalence of metabolic diseases (9, 50). In fact, while physical exercise and training can lower morbidity and mortality, physical inactivity has been recognized as one of the main risk factors for these pathologies (8). Lower whole-body aerobic capacity, muscle mitochondrial content, and oxidative activity, which all correlate with a sedentary lifestyle, contribute to the development of metabolic disorders (9, 25, 34, 38). Therefore, maintenance or improvement of skeletal muscle function, especially its oxidative metabolism, should be considered among the first interventions in the treatment and prevention of metabolic diseases.
Skeletal muscle is a highly plastic tissue that can quickly adapt to different physiological (e.g., exercise) and pathological (e.g., overnutrition) stimuli. In fact, muscle fibers can change their gene expression profile and phenotype to a great extent through diverse epigenetic mechanisms (3, 6, 31). Accordingly, muscle remodeling is highly regulated by different transcription factors and coregulator complexes, which are able to modify chromatin structure and thereby regulate gene transcription (27, 41). The nuclear receptor corepressor 1 (NCoR1) is a ubiquitously expressed corepressor, originally identified as the mediator of ligand-independent transcriptional repression of the thyroid hormone and the retinoic acid receptor (22). NCoR1 interacts with several transcription factors through its receptor interaction domains located in the C terminus (48). However, because NCoR1 lacks intrinsic histone deacetylase (HDAC) activity, it regulates gene transcription by forming a large protein complex in which G protein pathway suppressor 2 (GPS2), transducin β-like 1 (TBL1), TBL-related 1 (TBLR1), and HDAC3 represent the core subunits (52). In fact, the NCoR1-HDAC3 interaction plays an essential role in the control of gene transcription, since HDAC3 is directly activated by the deacetylase activation domain (DAD) of NCoR1 (23).
NCoR1 interacts with different proteins that play an important role in muscle physiology, such as peroxisome proliferator-activated receptors (PPAR) and p85α (15, 32), although its role in skeletal muscle remains largely enigmatic. Cell culture experiments implied that NCoR1 modulates myoblast differentiation through the regulation of the expression and transcriptional activity of several transcription factors, e.g., MyoD, TRα1, and Csl (5, 10, 26). The role of NCoR1 in vivo is not well understood because Ncor1−/− mice embryos die during gestation (24). Recently, conditional knockout models revealed that NCoR1 is an important player in skeletal muscle and adipose tissue energy metabolism (28, 51). In skeletal muscle, NCoR1 deletion enhances oxidative metabolism and slightly improves insulin tolerance under a high-fat diet (51). Specific genetic ablation of NCoR1 in white adipose tissue lowers inflammation and improves whole-body insulin sensitivity (28). However, the mechanism by which NCoR1 deletion results in these effects is not well understood.
NCoR1 is expressed in adult glycolytic and oxidative muscles at equal levels (42, 43). Chronic low-frequency stimulation of rat hind limb and acute endurance exercise in mice repress the expression of NCoR1 in this tissue (42, 51). Interestingly, by using knockout mice, it has been demonstrated that the formation of slow oxidative muscle fibers is negatively regulated by class IIa HDACs (39), which depend on interaction with the NCoR1-HDAC3 complex in order to induce protein deacetylation (14). Consistent with this, the global disruption of the NCoR1-HDAC3 complex in mice results in improved energy metabolism (e.g., higher insulin sensitivity and oxygen consumption) and altered circadian behavior (1). The aim of this study was to further investigate the role of NCoR1 in different aspects of skeletal muscle function (e.g., force generation) and, importantly, to elucidate the elusive mechanism by which NCoR1 regulates oxidative metabolism in this tissue.
MATERIALS AND METHODS
Animal housing and NCoR1 MKO mouse generation.
Mice were housed in a conventional facility with a 12-h night/12-h day cycle with free access to food and water. All experiments were performed on adult male mice with the approval of the Swiss authorities. NCoR1 muscle-specific knockout (MKO) animals were generated as previously described (51). Briefly, Ncor1loxP/loxP mice were crossed with HSA-Cre transgenic mice to generate NCoR1 MKO mice. Ncor1loxP/loxP animals without Cre expression were used as control (CON) mice. No overt phenotypic differences between CON and wild-type (WT) mice were observed. Genotyping was performed from tail biopsy specimens by PCR using specific primer pairs to detect the presence of the 5′ and 3′ loxP sites. The presence of the 5′ loxP site resulted in an amplicon of 450 bp (WT allele, 403 bp), while the presence of the 3′ loxP site resulted in an amplicon of 346 bp (WT allele, 207 bp) (see Fig. S1A in the supplemental material). Specific primer pairs to detect Cre recombinase resulted in an amplicon of 320 bp in NCoR1 MKO animals (see Fig. S1A). In addition, using muscle samples, recombination was confirmed by PCR using the forward and reverse primers used to detect the 5′ and 3′ loxP sites, respectively. Consequently, a 246-bp band was detected exclusively in NCoR1 MKO animals (see Fig. S1B). The recombination of the Ncor1 floxed allele decreased its mRNA specifically in skeletal and, to a lesser extent, cardiac muscle compared to that in CON mice (see Fig. S1C). Importantly, previous work has indicated that the slight decrease of NCoR1 mRNA in the heart of MKO mice does not affect cardiac morphology and function (51).
The generation and characterization of transgenic mice expressing peroxisome proliferator-activated receptor gamma (PPARγ) coactivator 1α (PGC-1α) under the control of the MCK promoter has been published (30). These PGC-1α muscle-specific transgenic (mTg) mice exhibit a 9.5-fold increase in skeletal muscle PGC-1α mRNA (see Fig. S3A in the supplemental material), higher oxidative metabolism, and improved exercise performance (11, 30).
Exercise performance assessment.
Animals were acclimatized to treadmill running for 2 days. On the first day, mice ran in a closed treadmill (Columbus Instruments) for 5 min at 10 m/min with a 0° slope, followed by 5 min at 14 m/min with a 5° incline. On the second day, animals ran for 5 min at 10 m/min with a 5° incline, followed by 5 min at 14 m/min. To determine maximal exercise performance and peak oxygen consumption (VO2peak), indirect calorimetry was performed during a maximal exercise test. Thus, 2 days after the acclimatization, mice were placed in a closed treadmill for 6 min at 0 m/min with a 5° incline. Subsequently, the test started at 8 m/min for 3 min with a 5° incline and the speed was increased 2 m/min every 3 min until exhaustion.
To determine endurance performance, mice were placed in an open treadmill (Columbus Instruments) for 5 min at 0 m/min with a 5° incline, followed by 5 min at 8 m/min. Mice ran for 20 min at 60, 70, 80, and 90% of the maximal speed reached in the maximal exercise test (average of the group), and then the speed increased to 100% of the maximal speed until exhaustion. The endurance test was performed at least 3 days after the maximal test.
Blood lactate analysis.
Blood lactate was measured from the tail vein of mice fasted overnight (16 h) or fed animals before and after different time points after treadmill running (see the maximal exercise test) using a lactate meter (Nova Biomedical).
In vivo measurement of muscle contractility.
Grip strength of the fore and hind limbs was measured with a grip strength meter (Chatillon). To determine the maximal strength, three measurements were performed with at least 60 s of recovery between each repetition, and the maximum value obtained was used for the analysis. To assess isometric fatigue resistance, mice were placed on top of an elevated grid, and the maximum time that they could remain on the inverted grid was recorded.
Ex vivo determination of muscle contractility.
Maximal force and fatigue resistance of extensor digitorum longus (EDL) and soleus were measured with a muscle testing setup (Heidelberg Scientific Instruments). Contraction amplitude was digitalized at 4 kHz with an AD Instruments converter. After the determination of the optimal length, force generation of EDL and soleus during a single twitch was measured in response to a 15-V pulse for 0.5 ms. Tetanic contraction was assessed in response to a 15-V pulse at 150 Hz for 400 ms for EDL and 1,100 ms for soleus. Maximal force and kinetics of the single twitch and tetanus were recorded and analyzed.
After 10 min of recovery following the tetanic stimulation, fatigue resistance of EDL and soleus was assessed under two different protocols. First, a long-interval protocol was performed by stimulating the muscles with a 15-V pulse at 150 Hz for 350 ms at 3.6-s intervals during 6 min for EDL and 10 min for soleus, followed by 10 min of recovery. Subsequently, a short-interval protocol was performed by stimulating the muscles with a 15-V pulse at 150 Hz for 350 ms at 1-s intervals during 2 min for EDL and 3 min for soleus. Changes in force generation are expressed as a percentage of initial force (first tetanus) (18).
Cell culture experiments.
C2C12 myoblasts were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin (growth medium). To induce differentiation, growth medium of 90 to 95% confluent myoblasts was changed to DMEM supplemented with 2% horse serum and 1% penicillin-streptomycin (differentiation medium). Cells were maintained at 37°C, 95% O2, and 5% CO2.
Experiments using C2C12 cells were performed on fully differentiated myotubes 4 days after differentiation was induced. For knockdown experiments, different adenoviruses containing specific short hairpin RNAs (shRNAs) against NCoR1, PGC-1α, PGC-1β, and LacZ (control) were used. Cells were incubated with the corresponding adenovirus for 24 h, and then adenovirus-containing medium was exchanged for fresh differentiation medium for another 24 h. In addition, cells were treated for 48 h with 0.2% dimethylsulfoxide (DMSO) (as a control), 10 μM XCT790 (Sigma-Aldrich) to inhibit estrogen-related receptor α (ERRα), or 1 μM GSK0660 (Sigma-Aldrich) to inhibit PPARβ/δ, together with the corresponding adenovirus. Three independent experiments each were performed in triplicate.
Luciferase assays were performed on 12-well plates using COS-7 cells (African green monkey kidney fibroblast-like cells) grown in growth medium without antibiotics. COS-7 cells were used in our experiments as a heterologous experimental system because of their high transfection efficiency and suitability for expressing constructs containing simian virus 40 (SV40) promoters. Cells were transfected using Lipofectamine 2000 (Invitrogen) with 0.1 μg pRL-SV40 (E2231; Promega), 0.3 μg pPPRE X3-TK-luc (1015; Bruce Spiegelman; Addgene), 0.3 μg pERRE-luc (gift of Junichi Sadoshima) (36), 0.4 μg pBABE puro PPAR delta (8891; Bruce Spiegelman; Addgene), 0.4 μg pERRα (gift of Vincent Giguère), 0.4 μg pFlag-NCoR (gift of Christopher K. Glass), and 0.4 μg pAd-Track HA PGC-1α (14427; Pere Puigserver; Addgene). The total amount of plasmid DNA was kept constant at 1.6 μg per well by using the control plasmid pAdtrack-CMV (ATCC). Twenty-four hours after transfection, cells were lysed with 250 μl of 1× passive lysis buffer (Promega), and luciferase activity was measured in 75 μl of lysate in a 96-well plate using the Dual-Glo luciferase assay system (Promega). Renilla (pRL-SV40) luciferase activity was used for normalization. Five independent experiments each were performed in triplicate.
RNA isolation and real-time PCR.
Total RNA was isolated from C2C12 myotubes, liver, kidney, white adipose tissue, heart, gastrocnemius (GAS), soleus, plantaris, tibialis anterior, extensor digitorum longus, and quadriceps from NCoR1 MKO mice or gastrocnemius from PGC-1α mTg animals using lysing matrix tubes (MP Biomedicals) and TRI reagent (Sigma-Aldrich) according to the manufacturer's instructions. RNA concentration was measured with a NanoDrop 1000 spectrophotometer (Thermo Scientific). One microgram of RNA was treated with DNase I (Invitrogen) and then reversed transcribed using hexanucleotide mix (Roche) and SuperScript II reverse transcriptase (Invitrogen). The level of relative mRNA was quantified by real-time PCR on a StepOnePlus system (Applied Biosystems) using Power SYBR green PCR master mix (Applied Biosystems). The analysis of the mRNA was performed by the ΔΔCT method using TATA binding protein (TBP) as the endogenous control. Primers used for target genes and TBP had the same PCR efficiency. TBP transcript levels were not different between genotypes or different experimental conditions. Primer sequences can be found in Table S3 in the supplemental material.
Mitochondrial DNA measurement.
DNA was isolated from gastrocnemius using a NucleoSpin tissue kit (Macherey-Nagel). Real-time PCR analysis was performed to measure COX2 (mitochondrial DNA) and β-globin (nuclear DNA) levels.
Skeletal muscle staining.
Oxidative fibers were detected by NADH staining of 12-μm cross-sections from the tibialis anterior. Staining was performed by exposing the sections to 0.8 mg/ml NADH in the presence of 1 mg/ml nitroblue tetrazolium. Periodic acid-Schiff (PAS) staining was performed with a PAS kit by following the manufacturer's instructions (Sigma-Aldrich).
Protein isolation.
Tissue samples were powdered on dry ice and homogenized with a polytron device in 300 μl of ice-cold lysis buffer (50 mM Tris-HCl, pH 7.5, 250 mM sucrose, 1 mM EDTA, 1 mM EGTA, 0.25% NP-40 substitute, 50 mM NaF, 5 mM Na4P2O7, 0.1% dithiothreitol [DTT], and fresh protease and phosphatase inhibitor cocktail). Samples then were shaken at 1,300 rpm for 30 min at 4°C. Samples were subsequently centrifuged at 13,000 × g for 10 min at 4°C, and the protein concentration of the supernatant was determined by the Bradford assay (Bio-Rad). Equal aliquots of protein were boiled for 5 min in Laemmli sample buffer (250 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 0.01% bromophenol blue, and 5% β-mercaptoethanol).
Western blotting.
Samples were separated on SDS-polyacrylamide gels and then transferred to nitrocellulose membranes for 60 min. Membranes were blocked for 1 h in 3% milk, Tris-buffered saline, and 0.1% Tween 20 (TBST) before overnight incubation at 4°C with the appropriate primary antibody in TBST (1:1,000 dilution). Proteins were detected with a primary antibody to p-AMPKαT172 (2535; Cell Signaling) and AMPKα (2603; Cell Signaling). As a loading control, eEF2 (2332; Cell Signaling) was used. Following incubation, membranes were washed 3 times with TBST before incubation with an appropriate peroxidase-conjugated secondary antibody in TBST (1:10,000 dilution). Antibody binding was detected using the enhanced chemiluminescence horseradish peroxidase (HRP) substrate detection kit (32106; Pierce).
Microarray and bioinformatic analysis.
RNA from gastrocnemius was isolated with an miRNeasy minikit (Qiagen), and microarray was performed using the GeneChip Gene 1.0 ST Array System (Affymetrix). In addition, gene ontology (GO) analysis was performed with the online tool FatiGO (http://babelomics.bioinfo.cipf.es/index.html) (2). Finally, microarray data were analyzed using Motif Activity Response Analysis (MARA; http://www.mara.unibas.ch/cgi/mara) (47).
Statistical analysis.
Values are expressed as means ± standard errors of the means (SEM). Statistical significance was determined with unpaired two-tailed t tests or one-way analysis of variance (ANOVA) with Tukey's post hoc test. A P value <0.05 was considered significant.
Microarray data accession number.
Microarray data can be found at the Gene Expression Omnibus (GEO) under accession number GSE40439.
RESULTS
Muscle NCoR1 deletion enhances VO2 during maximal exercise and decreases muscle contractility.
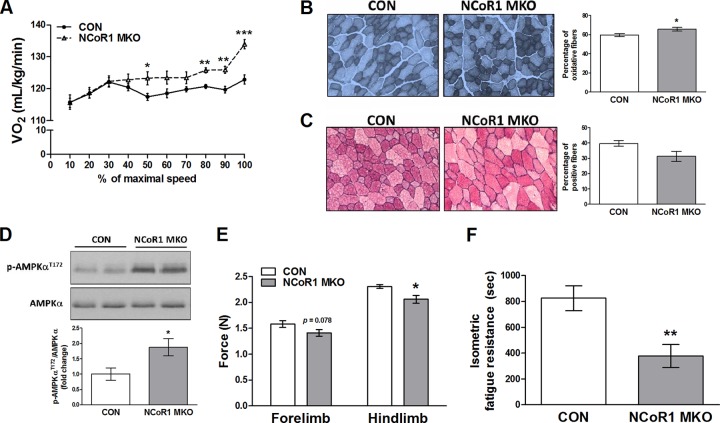
The role of NCoR1 in skeletal muscle was studied using NCoR1 MKO mice. A full description of this animal model has been recently published elsewhere (51). First, we assessed maximal oxidative capacity and treadmill running performance. Both genotypes exhibited the same exercise performance during maximal and endurance exercise tests, as reflected by equal speed, distance, time, work, and power (see Table S1 in the supplemental material). However, NCoR1 MKO mice reached a significantly higher (7.2%) VO2peak during the maximal exercise test (CON mice, 125 ± 1.2 ml/kg of body weight/min; MKO mice, 134 ± 1.5 ml/kg/min; P < 0.001). In fact, NCoR1 MKO animals showed a higher VO2 at 50, 80, 90, and 100% of maximal speed (Fig. 1A). Blood lactate measurement before and after maximal exercise, or during fasting, was not different in NCoR1 MKO mice (see Fig. S2A and B in the supplemental material). Consistent with this, respiratory exchange ratio (RER) measurement during the maximal exercise test did not show differences between CON and NCoR1 MKO mice (see Fig. S2C), indicating that energy substrate utilization was not altered by NCoR1 deletion. We then performed NADH and PAS staining of skeletal muscle to determine the proportion of oxidative fibers and glycogen content, respectively. NCoR1 MKO animals showed a 10% increase in oxidative fibers compared to CON mice (Fig. 1B), but glycogen content was not significantly affected (Fig. 1C). Furthermore, we have found higher levels of AMP-activated protein kinase (AMPK) phosphorylation in skeletal muscle of NCoR1 MKO mice (Fig. 1D). Altogether, these data show that NCoR1 deletion in striated muscle results in an enhanced oxidative metabolism, particularly during high-intensity exercise.
Fig 1.
Exercise performance and in vivo contractile properties of NCoR1 MKO mouse skeletal muscle. (A) VO2 during the maximal exercise test (n = 8 CON and n = 7 NCoR1 MKO mice). (B and C) Representative pictures and quantification of NADH (B) and PAS (C) staining of tibialis anterior (n = 3 to 4 CON and n = 3 to 4 NCoR1 MKO mice). (D) Representative blots and quantification of gastrocnemius AMPK phosphorylation levels (n = 7 CON and n = 4 NCoR1 MKO mice). (E and F) In vivo assessment of maximal grip strength and isometric muscle fatigue resistance (n = 13 CON and n = 12 NCoR1 MKO mice). Values represent means ± SEM. P < 0.05 (*), P < 0.01 (**), and P < 0.001 (**) for CON versus NCoR1 MKO mice.
An important aspect of muscle function besides endurance is the ability to generate force. To assess muscle contractility in vivo, we measured maximal grip strength of the fore and hind limbs. We observed a trend for lower peak isometric force generated by the fore limbs (P = 0.078) and a significant reduction by 11% in the hind limbs of NCoR1 MKO animals (Fig. 1E). We then quantified muscle fatigue resistance during predominantly isometric muscle contraction by measuring the maximal time that mice could remain on an inverted grid. Consistent with the lower maximal isometric force, isometric fatigue resistance was also decreased in NCoR1 MKO animals (Fig. 1F). To exclude systemic and neural factors that could affect force generation in vivo, we also determined muscle contractility in isolated muscles. Absolute (P < 0.05) and specific (P = 0.056) muscle contractility in response to a single twitch was decreased in the glycolytic muscle extensor digitorum longus (EDL) but not in the oxidative muscle soleus (Fig. 2A and B). Conversely, when EDL and soleus were subjected to maximal tetanic stimulation, no differences between genotypes were observed (Fig. 2C and D). Moreover, the contractile kinetics of EDL and soleus in response to a single twitch or a maximal tetanic stimulation were not significantly different between CON and NCoR1 MKO mice (see Table S1 in the supplemental material).
Fig 2.
Ex vivo contractile properties of NCoR1 MKO mouse skeletal muscle. (A to D) Ex vivo assessment of extensor digitorum longus (EDL) and soleus (SOL) contractility in response to a single twitch or a maximal tetanic stimulation (n = 10 CON and n = 14 NCoR1 MKO muscles per group). (E and F) Ex vivo assessment of EDL and SOL fatigue resistance in response to repeated tetanic stimulation through a long- and short-interval protocol. The inset in panel F shows the section with significant differences (n = 10 CON and n = 14 MKO muscles per group). Values represent means ± SEM. P < 0.05 (*) and P < 0.01 (**) for CON versus MKO mice.
Finally, muscle fatigue resistance was determined ex vivo by a long- and short-interval protocol. As expected, soleus exhibited a lower decrease in force than EDL in the long-interval protocol, but no differences between genotypes were found (Fig. 2E). Similarly, soleus had higher fatigue resistance than EDL during the short-interval protocol. However, in this protocol, EDL from NCoR1 MKO animals exhibited a lower decrease in force generation, while soleus of NCoR1 MKO and CON mice were identical (Fig. 2F). In fact, in the short-interval protocol, EDL from NCoR1 MKO mice generated ∼43% more force from tetanic stimulation no. 35 to 100 (Fig. 2F, inset), indicating a higher muscle fatigue resistance. Therefore, it seems that glycolytic muscles are more susceptible to the effect of NCoR1 deletion on muscle contractility, resulting in a decreased maximal isometric force and increased fatigue resistance during maximal stimulation.
NCoR1 and PGC-1α target a common subset of genes involved in oxidative metabolism.
The phenotype exhibited by NCoR1 MKO animals implies a direct link between NCoR1 and oxidative metabolism. At the transcriptional level, mitochondrial function is mainly regulated by the PPARγ coactivator 1 (PGC-1) family of coactivators, which are able to activate different transcription factors, such as ERRα and NRF-1 (29). Interestingly, NCoR1 MKO mice mirror some aspects of the phenotype exhibited by PGC-1α mTg mice, such as the enhanced oxidative metabolism and decreased maximal force (11, 30, 45). To explore the idea of an NCoR1-PGC-1α cross talk and to further characterize the oxidative phenotype of NCoR1 MKO mice, microarray analyses of gene expression patterns in NCoR1 MKO and PGC-1α mTg skeletal muscles were compared. GO enrichment analysis revealed overrepresentation of transcripts related to metabolic pathways, such as oxidative phosphorylation and the citrate cycle (tricarboxylic acid [TCA] cycle), in both NCoR1 MKO and PGC-1α mTg animals (Fig. 3A and B; also see Fig. S4A to F in the supplemental material). Interestingly, when both microarray data sets were compared, we observed that 188 of the genes affected by NCoR1 deletion were also found in the PGC-1α mTg data set, suggesting that ∼50% of the genes regulated by NCoR1 are also targets of PGC-1α (Fig. 3C). When only the genes found with the GO terms oxidative phosphorylation and citrate cycle (TCA cycle) were compared, we found that all of the genes present in the NCoR1 MKO data set were also targets of PGC-1α (Fig. 3C). Interestingly, we observed that in both microarrays all of these common genes were actually upregulated (Fig. 3D and E). Several of these transcripts were validated by real-time PCR analysis of GAS, EDL, and soleus (see Fig. S3B and C in the supplemental material), confirming the results obtained with the microarrays. However, no fiber type-specific (oxidative versus glycolytic) differences in terms of regulation of gene expression were observed in NCoR1 MKO mice (see Fig. S3C). Furthermore, skeletal muscle from NCoR1 MKO and PGC-1α mTg mice had higher transcript levels of mitochondrial genes involved in oxidative metabolism, such as cytochrome c oxidase subunit I (COX1), ATP synthase F0 subunit 6 (ATP6), and NADH dehydrogenase subunit 1 (ND1) (Fig. 3F; also see Fig. S3B). However, no differences in transcription factor A, mitochondrial (TFAM) mRNA levels, and mitochondrial DNA content were found as a consequence of NCoR1 deletion (Fig. 3G; also see Fig. 6A). These results further demonstrate the role of NCoR1 in the transcriptional control of different mitochondrial pathways and strongly suggest a role of PGC-1α in the control of the oxidative phenotype of the NCoR1 MKO animals.
Fig 3.
Similarities between NCoR1 MKO and PGC-1α mTg mice on oxidative metabolism. (A and B) Top 5 KEGG pathways from GO analysis of the up- and downregulated genes from the NCoR1 MKO and PGC-1α mTg microarray data sets (n = 5 per group). (C) Venn diagrams showing the overlap between NCoR1 MKO and PGC-1α mTg microarray data sets. (D and E) Heat maps generated using probe set intensities of the overlapping transcripts between NCoR1 MKO and PGC-1α mTg related to the GO terms oxidative phosphorylation (OXHPOS) and citrate cycle (TCA cycle). (F) Real-Time PCR analysis of mRNA levels of mitochondrial genes of gastrocnemius from CON (n = 7) and NCoR1 MKO (n = 5) animals. (G) Measurement of mitochondrial DNA (mtDNA) content of gastrocnemius from CON (n = 7) and NCoR1 MKO (n = 5) animals. Values represent means ± SEM. P < 0.05 (*) and P < 0.01 (**) for CON versus NCoR1 MKO mice.
Fig 6.
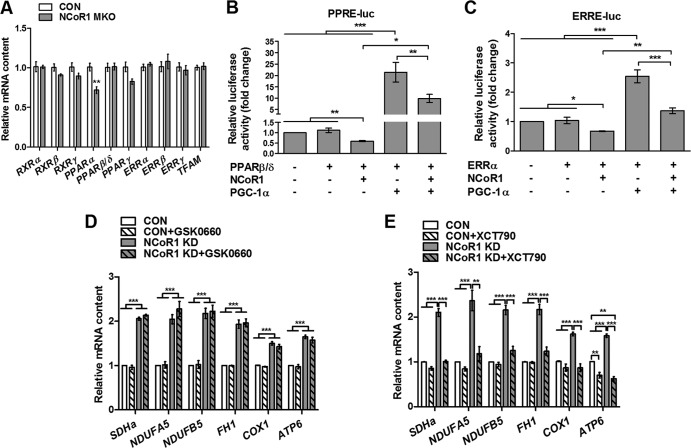
Role of PPARβ/δ and ERRα in NCoR1 regulation of oxidative metabolism. (A) Gastrocnemius mRNA levels of different transcription factors (n = 7 CON and n = 5 NCoR1 MKO mice). RXR, retinoid X receptor; PPAR, peroxisome proliferator activated receptor; ERR, estrogen-related receptor; TFAM, transcription factor A, mitochondrial. (B and C) Luciferase activity of PPRE-luc and ERRE-luc reporter plasmids in COS-7 cells cotransfected with PPARβ/δ, ERRα, NCoR1, and PGC-1α as indicated in each figure (n = 5 independent experiments each performed in triplicate). (D and E) Adenoviral knockdown (KD) of LacZ (CON) or NCoR1 alone or in combination with 10 μM XCT790 or 1 μM GSK0660 for 48 h in C2C12 myotubes (n = 3 independent experiments each performed in triplicate). Values represent means ± SEM. P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***) for CON versus NCoR1 MKO mice or as indicated.
Interestingly, the mRNA level of PGC-1α was not different between NCoR1 MKO and CON mice (Fig. 4A; also see Fig. S5A in the supplemental material). In contrast, we found an increase in PGC-1β and a slight decrease in PGC-1-related coactivator (PRC) transcript levels in gastrocnemius and soleus from NCoR1 MKO mice (Fig. 4A; also see Fig. S5A). To dissect the contribution of the PGC-1 family members to the effects induced by NCoR1 knockdown, we studied C2C12 myotubes in culture that were virally transfected with two different shRNAs specifically targeting NCoR1 (Fig. 4B; also see Fig. S5B and C and S6A and B in the supplemental material). We then selectively induced the knockdown of PGC-1α or PGC-1β with specific shRNA constructs (Fig. 4B; also see Fig. S5B). Similar to the NCoR1 MKO animals, NCoR1 knockdown in C2C12 myotubes also induced an upregulation of succinate dehydrogenase complex, subunit A, flavoprotein (SDHa), NADH dehydrogenase 1α (ubiquinone) subcomplex 5 (NDUFA5), NADH dehydrogenase 1β (ubiquinone) subcomplex 5 (NDUFB5), and fumarate hydratase 1 (FH1) (Fig. 4C), in addition to the mitochondrial genes COX1 and ATP6 (Fig. 4C). Surprisingly, NCoR1 knockdown in C2C12 myotubes induced a big increase in PGC-1α mRNA (Fig. 4B). Importantly, however, although the simultaneous knockdown of PGC-1α partially prevented its upregulation, shRNA-mediated reduction of PGC-1α was sufficient to strongly inhibit the upregulation of SDHa, NDUFA5, NDUFB5, FH1, COX1, and ATP6 mRNA induced by NCoR1 knockdown (Fig. 4C). In the cultured muscle cells, NCoR1 knockdown did not alter PGC-1β gene expression (see Fig. S5B), while the opposite was observed in the NCoR1 MKO animals (Fig. 4A; also see Fig. S5A). In stark contrast to the knockdown of PGC-1α, the upregulation of the nuclear and mitochondrial genes induced by NCoR1 knockdown was not significantly affected by the simultaneous knockdown of PGC-1β in the cultured myotubes (Fig. 4D). Consistent with the microarray analysis, these results indicate that NCoR1 deletion enhances PGC-1α action on oxidative metabolism, probably due to a reduced competition between NCoR1 and PGC-1α for the regulation of common target genes.
Fig 4.
Role of PGC-1α/β in NCoR1 regulation of oxidative metabolism. (A) Gastrocnemius (GAS) mRNA levels of the PGC-1 family of coactivators (n = 7 CON and n = 5 NCoR1 MKO mice). (B to D) Adenoviral knockdown (KD) of LacZ (CON) or NCoR1 alone or in combination with PGC-1α or PGC-1β KD for 48 h in C2C12 myotubes (n = 3 independent experiments each performed in triplicate). Values represent means ± SEM. P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***) for CON versus MKO mice or as indicated.
NCoR1 and PGC-1α regulate oxidative metabolism through opposite modulation of ERRα.
Our data suggest that NCoR1 and PGC-1α target the same subset of transcription factors involved in the regulation of oxidative metabolism with opposite effects on transcriptional activation. In order to elucidate potential common transcription factors of NCoR1 and PGC-1α in skeletal muscle, we performed motif activity response analysis (MARA) (47) of our microarray data to predict the core set of transcription factors which significantly change their activity in response to NCoR1 deletion or PGC-1α overexpression. Consistent with the oxidative phenotype of these two mouse models, retinoid X receptor (RXR) and ERRα (also known as Esrra) were among the top 4 most significant motifs (z value, ≥1.5) (Fig. 5A and B; also see Table S2 in the supplemental material) and exhibited some of the highest levels of activity in response to NCoR1 deletion and PGC-1α overexpression (Fig. 5C to F). Given that RXRs are the main partners of PPARs (13), this suggests possible PPARβ/δ activation in MKO animals. In fact, PPARβ/δ or ERRα in complex with PGC-1α and PGC-1β are known to be key players in the regulation of muscle metabolism (13, 16, 29). In contrast, the role of the transcription factors with significantly decreased activity (z value, ≤−1.5) in NCoR1 MKO and PGC-1α mTg mice (Fig. 5A and B; also see Table S2) is not well known. Similar to the microarray analysis, the comparison of both MARA performances showed that 44% of the transcription factors found in the NCoR1 MKO data set were also predicted in PGC-1α mTg MARA; there is actually a larger overlap among motifs with increased activity (5 out of 9) than among ones with decreased activity (2 out of 7) (Fig. 5G). In summary, MARA-based biocomputational prediction strongly suggests ERRα and possibly the RXR heterodimerization partner PPARβ/δ as common targets of NCoR1 and PGC-1α.
Fig 5.
Identification of common transcription factor binding partners of NCoR1 and PGC-1α. (A and B) The top 4 transcription factor motifs exhibiting increased and decreased activity in MARA of the microarray data performed on gastrocnemius from NCoR1 MKO and PGC-1α mTg mice (n = 5 per group). (C to F) Changes in the activity of RXRs and ERRα (ESRRA) in NCoR1 MKO and PGC-1α mTg skeletal muscle predicted by MARA analysis of microarray data (n = 5 per group). (G) Venn diagrams showing the overlap between NCoR1 MKO and PGC-1α mTg MARA analysis.
The mRNA levels of PPARβ/δ, ERRα, and several transcription factors revealed by MARA were not increased in NCoR1 MKO mice (Fig. 6A), indicating that increased activity is the consequence of lower repression rather than of higher expression. In order to explore the potential repressive effect of NCoR1 on ERRα and PPARβ/δ transcriptional activity, we transfected COS-7 cells with a reporter plasmid containing PPAR response elements (PPRE-luc) or ERR response elements (ERRE-luc) together with expression plasmids for PPARβ/δ and ERRα, respectively. We next measured relative luciferase activity in the absence and presence of NCoR1 and PGC-1α. As predicted, NCoR1 decreased PPRE-luc and ERRE-luc luciferase activity by 43 and 36%, respectively (Fig. 6B and C). Inversely, PGC-1α induced a significant increase in PPRE-luc and ERRE-luc luciferase activity, 2,037 and 161%, respectively (Fig. 6B and C). Importantly, we have found that the activation of both PPRE-luc and ERRE-luc by PGC-1α was significantly decreased by NCoR1 (Fig. 6B and C). These data demonstrate that NCoR1 represses the transcriptional activity of both PPARβ/δ and ERRα, while PGC-1α competes with NCoR1 to exert a positive effect.
Finally, to study the relative contribution of PPARβ/δ and ERRα to the regulation of the oxidative phenotype exhibited in response to NCoR1 deletion in muscle, we used the PPARβ/δ selective antagonist GSK0660 (44) and the ERRα inverse agonist XCT790 (33, 49). As expected, GSK0660 induced a strong decrease in the mRNA level of the PPARβ/δ target gene uncoupling protein 3 (UCP3) (see Fig. S6A in the supplemental material), demonstrating the efficiency of the antagonist and the presence of functional PPARβ/δ in C2C12 myotubes. Neither NCoR1 knockdown nor GSK0660 treatment changed PPARβ/δ mRNA levels (see Fig. S6A). Importantly, however, the induction of SDHa, NDUFA5, NDUFB5, FH1, COX1, and ATP6 mRNA were not inhibited by GSK0660 in cells with a knockdown of NCoR1 (Fig. 6D). Consistent with this, the mRNA levels of different PPARβ/δ target genes, like the carnitine palmitoyltransferase 1b gene (CPT1b), the lipoprotein lipase gene (LPL), and UCP3, were not significantly increased in NCoR1 MKO mice (see Fig. S3C in the supplemental material). In contrast, ERRα inhibition with XCT790 completely blocked the effects of NCoR1 knockdown on nuclear and mitochondrial genes (Fig. 6E). However, unlike the NCoR1 MKO animals, NCoR1 knockdown induced an upregulation of ERRα in C2C12 myotubes (see Fig. S6B). This upregulation of ERRα mRNA was only partially prevented by XCT790 (see Fig. S6B), indicating that ERRα activation, rather than its upregulation, is responsible for the effects of NCoR1 knockdown of oxidative metabolism in C2C12 myotubes. Interestingly, similar to NCoR1 and PGC-1α microarray analysis, GO enrichment analysis of previously published ERRα chromatin immunoprecipitation (ChIP)-on-chip data from mouse liver (12) also revealed overrepresentation of transcripts related to oxidative phosphorylation (see Fig. S4G in the supplemental material). Furthermore, we found a high overlap between the NCoR1 MKO microarray and the ERRα ChIP-on-chip data sets when comparing the genes found with the GO term oxidative phosphorylation (see Fig. S4H). Therefore, these data suggest that ERRα and PGC-1α are essential for the effects of NCoR1 deletion on oxidative metabolism in skeletal muscle, while PPARβ/δ and PGC-1β seem to play minor roles in this experimental context.
DISCUSSION
In stark contrast to obese and type 2 diabetic subjects, endurance athletes exhibit an increased metabolic fitness and consequently lower risk for metabolic disorders (9, 35). Importantly, most of the adaptations to muscle use (e.g., endurance training) and disuse (e.g., physical inactivity) are under the coordinated control of different transcription factors and coregulators (7, 19). NCoR1 and its homolog NCoR2 have been recently suggested as important modulators of energy metabolism in several tissues, including skeletal muscle (28, 40, 46, 51). Consistent with this, we observed that NCoR1 MKO animals exhibit an increased VO2 during high-intensity exercise. Surprisingly, while Yamamoto et al. (51) have reported improved exercise performance in NCoR1 MKO mice, we did not observe significant differences in distance, time, or work in the exercise trial in our experimental context. Considering the mild effect of NCoR1 deletion on muscle oxidative metabolism, it is possible that small differences in the exercise test protocol significantly affect exercise performance. In addition, as previously shown (51), we observed a higher proportion of oxidative fibers in NCoR1 MKO skeletal muscle. Interestingly, our data also indicate a higher activation of AMPK in NCoR1 MKO mice, which has been associated with enhanced oxidative metabolism (20, 21). Importantly, we have now demonstrated that NCoR1 controls skeletal muscle oxidative metabolism primarily through the regulation of ERRα and PGC-1α.
NCoR1 deletion in striated muscle led to an increase in the mRNA content of a broad range of mitochondrial enzymes, thus supporting its potential role as a negative regulator of oxidative metabolism. In agreement with this idea, NCoR1 MKO mice recapitulate many aspects of the phenotype exhibited by PGC-1α mTg mice, such as the increased expression of mitochondrial enzymes, higher levels of oxidative fibers, and enhanced VO2peak during maximal exercise (11, 30, 51). Moreover, as found in NCoR1 MKO animals, PGC-1α overexpression in skeletal muscle also results in a reduced maximal force and increased muscle fatigue resistance (30, 45). Curiously, only EDL exhibited a higher muscle fatigue resistance during the short-interval protocol in NCoR1 MKO mice. Since EDL is a highly glycolytic muscle, it might be more susceptible to benefitting from the mild improvement in oxidative metabolism as a consequence of NCoR1 deletion compared to the predominantly oxidative soleus. Analysis of NCoR1 MKO- and PGC-1α mTg-based microarrays also showed a high level of similarity between these mouse models, since all of the upregulated genes related to oxidative metabolism found in NCoR1 MKO mice were also increased by PGC-1α overexpression. Hence, our findings indicate that NCoR1 deletion facilitates PGC-1α action on its target genes. In fact, PGC-1α mRNA was not increased in NCoR1 MKO mice, suggesting that the higher transcription of its target genes is not associated with its upregulation. In contrast, NCoR1 knockdown in cultured cells induced a 133-fold increase in PGC-1α mRNA. Hence, it is possible that the bigger effects of NCoR1 deletion in C2C12 myotubes compared to those on the mice are a consequence of both upregulation of PGC-1α and lower competition between these coregulators. Importantly, however, we found that knockdown of PGC-1α, but not PGC-1β, in C2C12 myotubes completely blocked the effects of NCoR1 knockdown on oxidative metabolism, underlining the essential role of PGC-1α in this process. Similarly, prediction of the relative contribution of transcription factors to the gene expression pattern exhibited by NCoR1 MKO and PGC-1α mTg animals revealed a significant overlap between these mouse models. Interestingly, RXRs (heterodimerization partners for the PPARs) and ERRα stood out as top candidates, showing the strongest link to the control of energy metabolism (13, 16). In fact, Yamamoto et al. (51) have recently showed that in C2C12 myotubes, NCoR1 is recruited to the PPRE and nuclear receptor half-sites (ERR binding site) of UCP3 and PDK4 promoter, respectively, though whether the effects of NCoR1 deletion on gene expression actually depend on PPARβ/δ or ERRα activation was not studied. Here, through reporter gene assays, we have demonstrated that NCoR1 is a direct corepressor of both PPARβ/δ and ERRα. Importantly, given that NCoR1 and PGC-1α compete for the transcriptional regulation of PPARβ/δ and ERRα, our data indicate that these transcription factors are common targets of both coregulators. Interestingly, we observed a striking effect of the ERRα inverse agonist XCT790, since it completely inhibited the increase in the mRNA level of several mitochondrial enzymes triggered by NCoR1 knockdown in cultured myotubes. These data are consistent with MARA results and suggest that ERRα is the main target of NCoR1 in the control of muscle oxidative metabolism (Fig. 7).
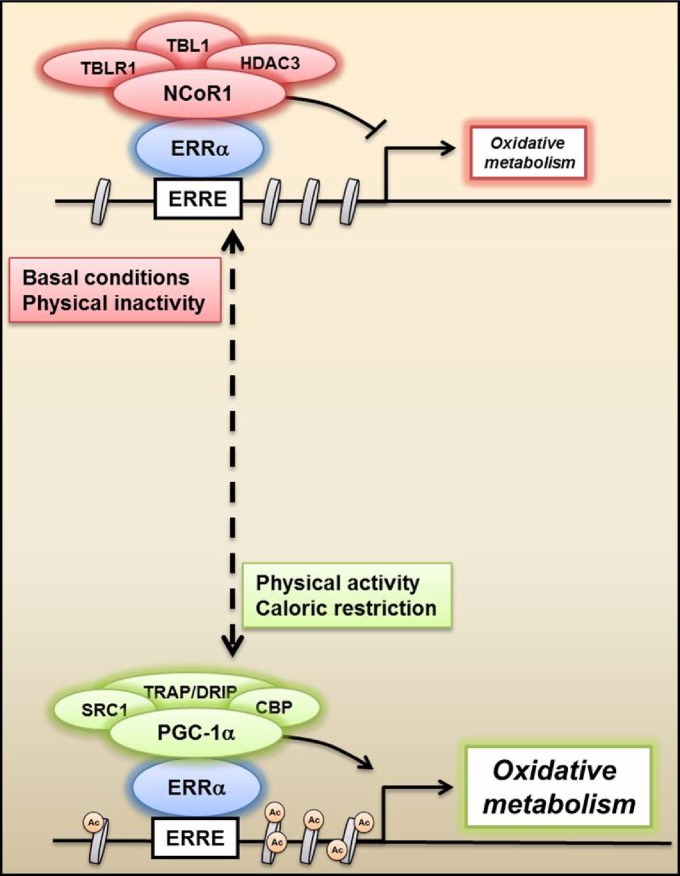
Fig 7.
Antagonistic regulation of oxidative metabolism by NCoR1 and PGC-1α. Proposed mechanism by which NCoR1 and PGC-1α complexes compete for the transcriptional regulation of ERRα and, as a consequence, oxidative metabolism. Under basal and possibly pathological conditions, NCoR1 represses ERRα and increases histone deacetylation, thereby decreasing metabolic gene transcription. In contrast, PGC-1α is activated by exercise and caloric restriction in skeletal muscle, coactivates ERRα, and consequently enhances mitochondrial function. ERRE, ERR response elements; Ac, histone acetylation.
Unexpectedly, we found that the PPARβ/δ selective antagonist GSK0660 did not affect the effects of NCoR1 deletion on oxidative phosphorylation and gene regulation. PPARβ/δ plays an important role in the transcriptional control of lipid metabolism (13). Intriguingly, our microarray analysis of NCoR1 MKO skeletal muscle shows a preferential enhancement of oxidative phosphorylation and the TCA cycle compared to the modulation of fatty acid β-oxidation. Thus, it is conceivable that in our experimental context of animals fed with normal chow, PPARβ/δ played less of a role than it did in studies using high-fat diets (51) or under conditions like obesity and physical inactivity. NCoR1 might control fatty acid metabolism through the transcriptional regulation of PPARβ/δ in an environment of elevated intramyocellular lipids that could act as PPARβ/δ ligands, but this hypothesis needs to be substantiated in future studies. Overall, it seems that a fully functional ERRα-PGC-1α complex is a prerequisite for the improved oxidative metabolism induced by muscle-specific NCoR1 deletion, at least in chow-fed mice.
Interestingly, NCoR1 MKO mice have a milder oxidative phenotype than PGC-1α transgenic mice. The weaker phenotype is also reflected in the quantitative differences in gene expression. Thus, while the muscle knockout of NCoR1 resulted in the upregulation of 21 genes involved in oxidative phosphorylation, 83 genes were increased in response to muscle PGC-1α overexpression. Consistent with this, PGC-1α mTg mice exhibit a higher increase in VO2peak (24%) (11) than NCoR1 MKO animals (7.2%). However, it is important to note that NCoR1 is a basal corepressor, thus it represses transcription factors under basal conditions, and it is exchanged by coactivators upon a positive stimulus (17, 37). Therefore, our data suggest that under basal conditions, NCoR1 deletion would not result in a full activation of PPARβ/δ and ERRα, which subsequently could be amplified by a positive stimulus, such as exercise, that is well known to increase PGC-1α expression (4) and thereby unleashes the full activity of these transcription factors.
In conclusion, our data indicate competition between NCoR1 and PGC-1α in the regulation of PPARβ/δ and ERRα transcriptional activity in skeletal muscle, of which the regulation of ERRα represents the predominant regulatory mechanism of oxidative metabolism during basal conditions (Fig. 7). The elucidation of different pharmacological or nonpharmacological (e.g., exercise training) strategies to modulate NCoR1 activity and thus facilitate PGC-1α action represents an attractive strategy for the treatment of metabolic diseases.
Supplementary Material
ACKNOWLEDGMENTS
We thank Christopher K. Glass (University of California, San Diego), Junichi Sadoshima (University of Medicine and Dentistry of New Jersey), and Vincent Giguère (McGill University, Montreal, Canada) for their generous contribution of pFlag-NCoR, pERRE-luc, and pERRα plasmids, respectively. We also thank Philippe Demougin (Biozentrum, University of Basel) for his help with microarray analysis and Hiroyasu Yamamoto for providing the LacZ and NCoR1 adenoviruses (Ecole Polytechnique Fédérale de Lausanne).
This project was funded by the Swiss National Science Foundation (SNF 310030_132900), the Muscular Dystrophy Association USA, the SwissLife Jubiläumsstiftung für Volksgesundheit und medizinische Forschung, the Swiss Society for Research on Muscle Diseases (SSEM), the Swiss Diabetes Association, the Roche Research Foundation, the United Mitochondrial Disease Foundation, the Association Française contre les Myopathies, the Ecole Polytechnique Féderale de Lausanne, the European Research Council, and the University of Basel.
Footnotes
Published ahead of print 1 October 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Alenghat T, et al. 2008. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature 456:997–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Al-Shahrour F, Diaz-Uriarte R, Dopazo J. 2004. FatiGO: a web tool for finding significant associations of gene ontology terms with groups of genes. Bioinformatics 20:578–580 [DOI] [PubMed] [Google Scholar]
- 3. Baar K. 2010. Epigenetic control of skeletal muscle fibre type. Acta Physiol. 199:477–487 [DOI] [PubMed] [Google Scholar]
- 4. Baar K, et al. 2002. Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1. FASEB J. 16:1879–1886 [DOI] [PubMed] [Google Scholar]
- 5. Bailey P, et al. 1999. The nuclear receptor corepressor N-CoR regulates differentiation: N-CoR directly interacts with MyoD. Mol. Endocrinol. 13:1155–1168 [DOI] [PubMed] [Google Scholar]
- 6. Barres R, et al. 2012. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab. 15:405–411 [DOI] [PubMed] [Google Scholar]
- 7. Bassel-Duby R, Olson EN. 2006. Signaling pathways in skeletal muscle remodeling. Annu. Rev. Biochem. 75:19–37 [DOI] [PubMed] [Google Scholar]
- 8. Booth FW, Lees SJ. 2007. Fundamental questions about genes, inactivity, and chronic diseases. Physiol. Genomics 28:146–157 [DOI] [PubMed] [Google Scholar]
- 9. Booth FW, Roberts CK. 2008. Linking performance and chronic disease risk: indices of physical performance are surrogates for health. Br. J. Sports Med. 42:950–952 [DOI] [PubMed] [Google Scholar]
- 10. Busson M, et al. 2005. Coactivation of nuclear receptors and myogenic factors induces the major BTG1 influence on muscle differentiation. Oncogene 24:1698–1710 [DOI] [PubMed] [Google Scholar]
- 11. Calvo JA, et al. 2008. Muscle-specific expression of PPARgamma coactivator-1alpha improves exercise performance and increases peak oxygen uptake. J. Appl. Physiol. 104:1304–1312 [DOI] [PubMed] [Google Scholar]
- 12. Charest-Marcotte A, et al. 2010. The homeobox protein Prox1 is a negative modulator of ERRα/PGC-1α bioenergetic functions. Genes Dev. 24:537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ehrenborg E, Krook A. 2009. Regulation of skeletal muscle physiology and metabolism by peroxisome proliferator-activated receptor delta. Pharmacol. Rev. 61:373–393 [DOI] [PubMed] [Google Scholar]
- 14. Fischle W, et al. 2002. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell 9:45–57 [DOI] [PubMed] [Google Scholar]
- 15. Furuya F, et al. 2007. Nuclear receptor corepressor is a novel regulator of phosphatidylinositol 3-kinase signaling. Mol. Cell. Biol. 27:6116–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Giguere V. 2008. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr. Rev. 29:677–696 [DOI] [PubMed] [Google Scholar]
- 17. Glass CK, Rosenfeld MG. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121–141 [PubMed] [Google Scholar]
- 18. Gonzalez E, Delbono O. 2001. Age-dependent fatigue in single intact fast and slow fibers from mouse EDL and soleus skeletal muscles. Mech. Ageing Dev. 122:1019–1032 [DOI] [PubMed] [Google Scholar]
- 19. Handschin C, Spiegelman BM. 2008. The role of exercise and PGC1alpha in inflammation and chronic disease. Nature 454:463–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hardie DG, Ross FA, Hawley SA. 2012. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 13:251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hawley SA, et al. 2012. The ancient drug salicylate directly activates AMP-activated protein kinase. Science 336:918–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Horlein AJ, et al. 1995. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 377:397–404 [DOI] [PubMed] [Google Scholar]
- 23. Ishizuka T, Lazar MA. 2005. The nuclear receptor corepressor deacetylase activating domain is essential for repression by thyroid hormone receptor. Mol. Endocrinol. 19:1443–1451 [DOI] [PubMed] [Google Scholar]
- 24. Jepsen K, et al. 2000. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell 102:753–763 [DOI] [PubMed] [Google Scholar]
- 25. Kelley DE, He J, Menshikova EV, Ritov VB. 2002. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes 51:2944–2950 [DOI] [PubMed] [Google Scholar]
- 26. Kitamura T, et al. 2007. A Foxo/Notch pathway controls myogenic differentiation and fiber type specification. J. Clin. Investig. 117:2477–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kouzarides T. 2007. Chromatin modifications and their function. Cell 128:693–705 [DOI] [PubMed] [Google Scholar]
- 28. Li P, et al. 2011. Adipocyte NCoR knockout decreases PPARgamma phosphorylation and enhances PPARgamma activity and insulin sensitivity. Cell 147:815–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lin J, Handschin C, Spiegelman BM. 2005. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 1:361–370 [DOI] [PubMed] [Google Scholar]
- 30. Lin J, et al. 2002. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature 418:797–801 [DOI] [PubMed] [Google Scholar]
- 31. McGee SL, Hargreaves M. 2011. Histone modifications and exercise adaptations. J. Appl. Physiol. 110:258–263 [DOI] [PubMed] [Google Scholar]
- 32. McKenna NJ, O'Malley BW. 2010. SnapShot: NR coregulators. Cell 143:172–172.e1 doi:10.1016/j.cell.2010.09.032 [DOI] [PubMed] [Google Scholar]
- 33. Mootha VK, et al. 2004. Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc. Natl. Acad. Sci. U. S. A. 101:6570–6575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mootha VK, et al. 2003. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34:267–273 [DOI] [PubMed] [Google Scholar]
- 35. Nuutila P, et al. 1994. Different alterations in the insulin-stimulated glucose uptake in the athlete's heart and skeletal muscle. J. Clin. Investig. 93:2267–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oka S, et al. 2011. PPARalpha-Sirt1 complex mediates cardiac hypertrophy and failure through suppression of the ERR transcriptional pathway. Cell Metab. 14:598–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. 2004. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell 116:511–526 [DOI] [PubMed] [Google Scholar]
- 38. Petersen KF, Dufour S, Befroy D, Garcia R, Shulman GI. 2004. Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N. Engl. J. Med. 350:664–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Potthoff MJ, et al. 2007. Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers. J. Clin. Investig. 117:2459–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Reilly SM, et al. 2010. Nuclear receptor corepressor SMRT regulates mitochondrial oxidative metabolism and mediates aging-related metabolic deterioration. Cell Metab. 12:643–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Santos GM, Fairall L, Schwabe JW. 2011. Negative regulation by nuclear receptors: a plethora of mechanisms. Trends Endocrinol. Metab. 22:87–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schuler MJ, Buhler S, Pette D. 1999. Effects of contractile activity and hypothyroidism on nuclear hormone receptor mRNA isoforms in rat skeletal muscle. Eur. J. Biochem. 264:982–988 [DOI] [PubMed] [Google Scholar]
- 43. Schuler MJ, Pette D. 1998. Quantification of thyroid hormone receptor isoforms, 9-cis retinoic acid receptor gamma, and nuclear receptor co-repressor by reverse-transcriptase PCR in maturing and adult skeletal muscles of rat. Eur. J. Biochem. 257:607–614 [DOI] [PubMed] [Google Scholar]
- 44. Shearer BG, et al. 2008. Identification and characterization of a selective peroxisome proliferator-activated receptor beta/delta (NR1C2) antagonist. Mol. Endocrinol. 22:523–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Summermatter S, et al. 2012. Remodeling of calcium handling in skeletal muscle through PGC-1alpha: impact on force, fatigability, and fiber type. Am. J. Physiol. Cell Physiol. 302:C88–C99 [DOI] [PubMed] [Google Scholar]
- 46. Sutanto MM, et al. 2010. The silencing mediator of retinoid and thyroid hormone receptors (SMRT) regulates adipose tissue accumulation and adipocyte insulin sensitivity in vivo. J. Biol. Chem. 285:18485–18495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Suzuki H, et al. 2009. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat. Genet. 41:553–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Webb P, et al. 2000. The nuclear receptor corepressor (N-CoR) contains three isoleucine motifs (I/LXXII) that serve as receptor interaction domains (IDs). Mol. Endocrinol. 14:1976–1985 [DOI] [PubMed] [Google Scholar]
- 49. Willy PJ, et al. 2004. Regulation of PPARgamma coactivator 1alpha (PGC-1alpha) signaling by an estrogen-related receptor alpha (ERRalpha) ligand. Proc. Natl. Acad. Sci. U. S. A. 101:8912–8917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wisloff U, et al. 2005. Cardiovascular risk factors emerge after artificial selection for low aerobic capacity. Science 307:418–420 [DOI] [PubMed] [Google Scholar]
- 51. Yamamoto H, et al. 2011. NCoR1 is a conserved physiological modulator of muscle mass and oxidative function. Cell 147:827–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yoon HG, et al. 2003. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J. 22:1336–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.