Abstract
The majority of multispanning inner mitochondrial membrane proteins utilize internal targeting signals, which direct them to the carrier translocase (TIM22 complex), for their import. MPV17 and its Saccharomyces cerevisiae orthologue Sym1 are multispanning inner membrane proteins of unknown function with an amino-terminal presequence that suggests they may be targeted to the mitochondria. Mutations affecting MPV17 are associated with mitochondrial DNA depletion syndrome (MDDS). Reconstitution of purified Sym1 into planar lipid bilayers and electrophysiological measurements have demonstrated that Sym1 forms a membrane pore. To address the biogenesis of Sym1, which oligomerizes in the inner mitochondrial membrane, we studied its import and assembly pathway. Sym1 forms a transport intermediate at the translocase of the outer membrane (TOM) complex. Surprisingly, Sym1 was not transported into mitochondria by an amino-terminal signal, and in contrast to what has been observed in carrier proteins, Sym1 transport and assembly into the inner membrane were independent of small translocase of mitochondrial inner membrane (TIM) and TIM22 complexes. Instead, Sym1 required the presequence of translocase for its biogenesis. Our analyses have revealed a novel transport mechanism for a polytopic membrane protein in which internal signals direct the precursor into the inner membrane via the TIM23 complex, indicating a presequence-independent function of this translocase.
INTRODUCTION
Transfer of metabolites or polypeptides across the inner mitochondrial membrane relies on dedicated and highly selective transporters. Mitochondria import the vast majority of proteins from the cytoplasm. While most of these proteins use amino-terminal-targeting signals (presequences) for transport, which direct the proteins across the outer and inner membranes of the mitochondria, many multispanning inner membrane proteins, such as the metabolite carriers and channel-forming subunits of the inner membrane protein translocases, utilize internal targeting signals. The transport of multispanning proteins into the inner membrane has been studied most for metabolite carriers, and five distinct transport stages have been identified (4, 11, 40, 46, 49). Initially, carrier proteins associate with chaperones in the cytosol that prevent aggregation of the hydrophobic molecules (stage I) (81, 83). Tom70 is the major receptor for internal targeting signals in the translocase of the outer membrane (TOM) complex (53, 61). In addition to the signal elements of the precursor, Tom70 also binds precursor-associated chaperones (stage II) (79). ATP hydrolysis by the substrate-carrying chaperones releases the carrier molecules for transfer across the TOM complex toward the intermembrane space. The carrier molecule traverses the Tom40 channel of the TOM complex in a hairpin-like conformation and associates with the small translocase of mitochondrial inner membrane (TIM) chaperone complex in the intermembrane space (stage III) (6, 12, 27, 34, 59, 67, 75). In in vitro import analyses, this stage can be stabilized when the membrane potential across the inner membrane is dissipated by ionophores. Inner membrane insertion of carriers is mediated by the carrier translocase (TIM22 complex). The small TIM complex delivers the carriers from the TOM to the TIM22 complex, which derives energy from the membrane potential to drive membrane insertion of the precursor (stage IV) (17, 31, 50, 58). Eventually, the carriers assemble into functional oligomers (stage V) (50). In contrast to metabolite carriers and channels of the inner mitochondrial membrane, a group of multispanning membrane proteins, such as the ABC transporters and the insertase OxaI, contain N-terminal presequences (2, 16, 20). The insertion of these proteins into the lipid phase requires the presequence translocase (TIM23 complex), which initially recognizes the precursor through an interaction between the presequence and the receptor Tim50 (35, 56). Recognition of the presequence is thought to activate the translocase and thus to initiate membrane translocation (66).
MPV17 (Sym1 in Saccharomyces cerevisiae) belongs to a family of proteins conserved from hydrogenosomes in the anaerobic ciliate Nyctotherus ovalis to the mitochondria of yeast and mammals (3). All family members are multispanning membrane proteins with four predicted transmembrane spans. MPV17 was first identified in a mouse strain generated by random retrovirus integration, which was affected by nephrotic syndrome (72). Defects in human MPV17 cause a dramatic reduction of mitochondrial DNA, described as the hepatocerebral form of the inherited autosomal mitochondrial DNA depletion syndrome (MDDS) (64). As a result of MPV17 loss of function, respiration and other mitochondrial activities are dramatically reduced in tissues (69). Patients with MDDS display liver failure, hypoglycemia, growth retardation, and neuronal disorders within the first year of their life. The molecular functions of MPV17 and its yeast homolog Sym1 have remained enigmatic.
In S. cerevisiae, two proteins show similarity to MPV17: Sym1, residing in the inner mitochondrial membrane (similar to MPV17), and the product of the uncharacterized open reading frame YOR292c, which has been localized to the vacuole (21). Complementation of the yeast SYM1 deletion by expression of the human MPV17 gene defines these two proteins as functional orthologs and has established Sym1 as a model for MPV17 function (63). The observed reduction of steady-state protein levels of mutated MPV17 in patient cells has raised questions regarding the stability of the protein or, alternatively, could indicate mistargeting within patient cells (63). Therefore, we studied the import and assembly of Sym1 in yeast mitochondria. Surprisingly, Sym1 does not carry a cleavable presequence, as N-terminal truncations do not affect translocation and integration into the inner membrane. We found that purified Sym1 forms an aqueous channel upon reconstitution in black lipid membranes. Based on this observation, we speculated that, similar to the Tim23 and Tim22 channels, Sym1 could follow the carrier pathway into the mitochondria. Although our analyses revealed transport intermediates, which strongly resembled those of carrier proteins, Sym1 assembled independently of the carrier pathway. Surprisingly, we found that Sym1 is transported and integrated into the inner membrane in a presequence-independent manner via the presequence translocase (TIM23). Thus, our analysis identified a novel role of the presequence translocase complex in the insertion of a protein with internal signals into the inner membrane.
MATERIALS AND METHODS
Yeast strains, human cell lines, and isolation of mitochondria.
Saccharomyces cerevisiae sym1Δ and SYM1ZZ strains used in this study were derived from YPH499 by chromosomal integration (22, 25). The Tim50HA and Tim50PBD-HA yeast strains, expressing TIM50 under the GAL1 promoter and plasmid-encoded C-terminally hemagglutinin (HA)-tagged full-length Tim50HA (amino acids [aa] 1 to 476) or truncated Tim50ΔPBD-HA (aa 1 to 365) under the control of the MET25 promoter, have been described previously (56). The tom22His, tim5-1, tim10-2, tim54-11, and tim12-4 strains and the strain expressing TIM23 under the control of the GAL1 promoter have been described previously (5, 17, 38, 56, 67, 71). For in vivo studies on the membrane integration of Sym1ZZ and Sym1ZZ lacking the N terminus (amino acids [aa] 1 to 16), the constitutive ADH promoter was integrated either directly before or 16 codons downstream of the SYM1 start codon in the SYM1ZZ yeast strain. Yeast cultures were grown at 24°C or 30°C, unless otherwise indicated, in rich medium (1% yeast extract, 2% peptone, and 2% dextrose or 3% glycerol). Yeast mitochondria were isolated as described previously (37). Human mitochondria were isolated from HEK293T cells (32) and cultured in Dulbecco's modified Eagle medium (Gibco; Invitrogen) supplemented with 10% fetal bovine serum (Biochrom) at 37°C in a 5% CO2 atmosphere.
In vitro import and assembly into isolated mitochondria.
For synthesis of 35S-labeled precursor proteins, the respective open reading frames were cloned downstream of the SP6 promoter in pGEM4Z (Promega) or pCR-Blunt2-TOPO (Invitrogen). Plasmids were used to synthesize 35S-labeled precursor proteins by coupled in vitro transcription-translation (TNT quick coupled transcription-translation system; Promega). Alternatively, the open reading frames were amplified from plasmid DNA by PCR, transcribed and purified (mMessage mMachine SP6 system and MEGAclear kit; Ambion), and subsequently used for in vitro translation (Flexi rabbit reticulocyte lysate system; Promega).
In vitro imports of radiolabeled precursor proteins into isolated yeast or human mitochondria were performed as described previously (32, 74). After import, samples were analyzed by SDS-PAGE or blue-native PAGE (BN-PAGE), and 35S-labeled proteins were detected by digital autoradiography and quantified using ImageQuant TL software (GE Healthcare).
Antibody shift and depletion assays.
An in-gel antibody shift assay of protein complexes was performed by the addition of 2% (vol/vol) antiserum after in vitro import. After incubation for 30 min at 4°C, mitochondria were sedimented by centrifugation, and samples were processed for BN-PAGE. For complex depletion analysis, [35S]Sym1 was imported in vitro into tom22His mitochondria. Subsequently, mitochondria were solubilized for 30 min in 1% digitonin, 20 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 50 mM NaCl, 10% (wt/vol) glycerol, and 1 mM phenylmethylsulfonyl fluoride (PMSF). We then added 5% (vol/vol) (bed volume) Ni-nitrilotriacetic acid (NTA) agarose (Qiagen), and after incubation for 30 min at 4°C, beads were sedimented and the supernatant was analyzed by BN-PAGE and digital autoradiography.
Triplex isolation of Sym1 from yeast mitochondria.
Sym1 was purified from Sym1ZZ mitochondria by double-affinity chromatography followed by tobacco etch virus (TEV) protease cleavage (30). In brief, mitochondria were resuspended to a protein concentration of 10 mg/ml in buffer A (1% SDS, 10% [wt/vol] glycerol, 100 mM NaCl, 10 mM imidazole, 1 mM PMSF, 50 mM NaH2PO4). Proteins were denatured by incubation for 15 min at 25°C. Subsequently, proteins were 20× diluted with buffer B (0.2% Triton X-100, 10% [wt/vol] glycerol, 100 mM NaCl, 10 mM imidazole, 1 mM PMSF, 50 mM NaH2PO4) and incubated for 30 min at 4°C. After a clarifying spin (15 min, 4°C, 20,000 × g), the supernatant was incubated with Ni-NTA agarose beads. Beads were washed with buffer C (0.1% Triton X-100, 10% [wt/vol] glycerol, 100 mM NaCl, 10 mM imidazole, 1 mM PMSF, 50 mM NaH2PO4), and bound proteins were eluted using the same buffer supplemented with 300 mM imidazole. Subsequently, imidazole was diluted using buffer C, and samples were incubated with IgG-Sepharose beads. Beads were washed extensively with buffer C, and bound protein was released from the Sepharose by TEV protease cleavage. His-tagged TEV protease was removed by incubation with Ni-NTA agarose beads.
Electrophysiological characterization of Sym1 in planar lipid bilayers.
Planar lipid bilayers were formed using the painting technique. In brief, a lipid solution of purified l-α-phosphatidylcholine (60 mg/ml, type IV-S) (Sigma) in decan (purity > 99%) (Sigma) was applied to a hole in the polytetrafluoroethylene (PTFE) membrane, which separates two chambers. After successful bilayer formation, which was monitored optically, Sym1 was inserted into the membrane by osmotic fusion of proteoliposomes with the bilayer under asymmetrical buffer conditions (250 mM KCl and 10 mM MOPS [morpholinepropanesulfonic acid]-Tris [pH 7.0] in the cis chamber and 20 mM KCl and 10 mM MOPS-Tris [pH 7.0] in the trans chamber). Ag/AgCl electrodes were connected to the chambers using 2 M KCl-agar bridges. The membrane potentials refer to the trans compartment, as the electrode of this compartment was directly connected to the headstage of a current amplifier (Axon Gene Clamp 500; Axon Instr.). A self-created analysis software in combination with Origin 6.0 (MicroCal Software Inc.) was used to analyze the current recordings.
Miscellaneous.
Standard techniques were used for SDS-PAGE and Western blotting of proteins to polyvinylidene difluoride (PVDF) membranes. Proteins were probed using primary antibodies raised in rabbits and secondary antibodies coupled to horseradish peroxidase (HRP). Signals were detected using an enhanced chemiluminescence (ECL) detection system (GE Healthcare) and X-ray films.
Blue-native PAGE was performed essentially as described previously (9). Mitochondria were solubilized in 1% digitonin, 20 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 50 mM NaCl, 10% (wt/vol) glycerol, and 1 mM PMSF for 30 min at 4°C. After a clarifying spin (15 min, 20,000 × g, 4°C) and the addition of 10× loading dye (5% Coomassie brilliant blue G-250, 500 mM ε-amino n-caproic acid, 100 mM Bis-Tris [pH 7.0]), the supernatant was separated on a 6 to 16% polyacrylamide gradient gel. For 2-dimensional BN-PAGE/SDS-PAGE, a lane was cut out after the BN-PAGE run and mounted on top of an SDS-PAGE gel.
Digital autoradiography of radiolabeled proteins was performed by exposition of dried SDS or BN gels to storage phosphor screens (GE Healthcare). Signals were detected using the Storm 820 scanner (Storm imaging system; GE Healthcare).
For submitochondrial protein localization, mitochondria were osmotically stabilized in SEM buffer (250 mM sucrose, 1 mM EDTA, 10 mM MOPS [pH 7.2]) or the outer mitochondrial membrane was disrupted by osmotic swelling in EM buffer (1 mM EDTA, 10 mM MOPS [pH 7.2]) prior to treatment with the indicated concentrations of proteinase K for 10 min on ice. Subsequently, proteinase K was inactivated, and samples were analyzed by SDS-PAGE and Western blotting. For carbonate extraction of mitochondrial membranes, isolated mitochondria were incubated in 100 mM Na2CO3 at pH 10.8 or pH 11.5 for 20 min on ice. Extracted membranes were separated by ultracentrifugation at 100,000 × g for 30 min. Proteins from total samples and supernatant and pellet fractions were precipitated in 13% trichloroacetic acid (TCA) and washed with ice-cold acetone before separation by SDS-PAGE and Western blotting.
RESULTS
Purification and biochemical characterization of Sym1.
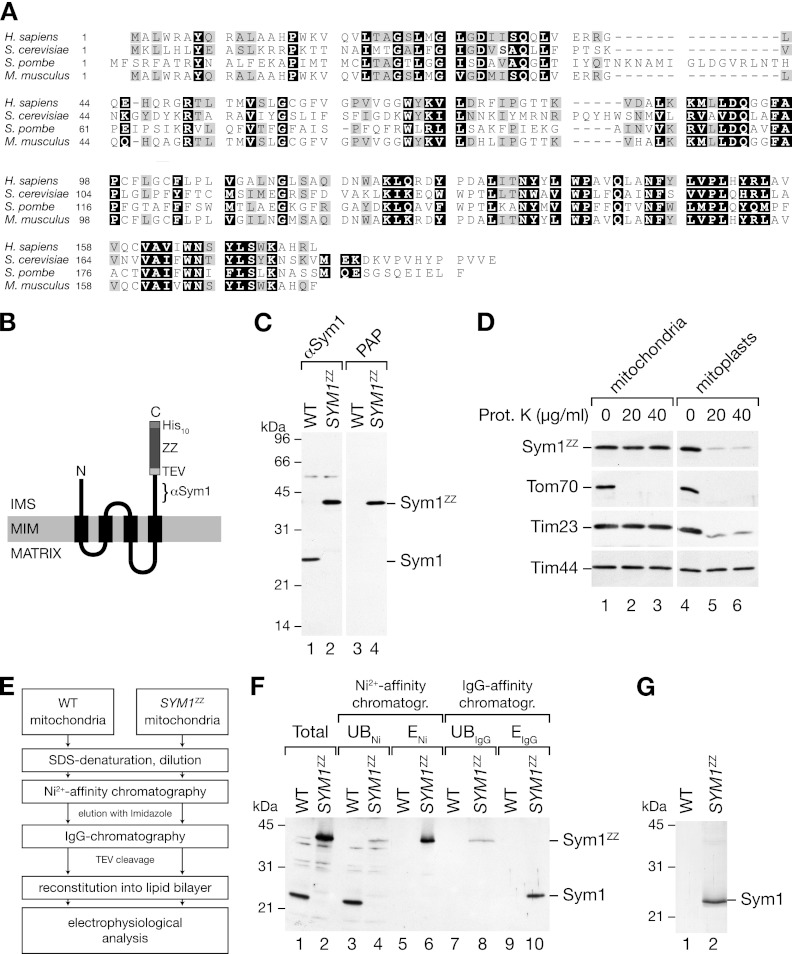
The yeast Sym1 protein shares high sequence similarities with its human and mouse orthologues (Fig. 1A). Both human MPV17 and yeast Sym1 have been localized to the inner mitochondrial membrane and display similar membrane topologies, with four predicted transmembrane spans (63, 65, 77). For biochemical characterization of Sym1, we generated a yeast strain (SYM1ZZ) expressing Sym1 from its own promoter, which was fused to two repeats of the IgG-binding domain of protein A (the Z domain) followed by a polyhistidine tag at its C terminus. A TEV cleavage site between Sym1 and the first Z domain allowed specific cleavage of the tag by TEV protease treatment (Fig. 1B). To assess the expression of SYM1ZZ, we isolated mitochondria and subjected them to Western blot analysis. Detection of the ZZ tag with peroxidase antiperoxidase (PAP) antibody (HRP–anti-HRP) revealed a protein of the predicted size, indicating stable expression of SYM1ZZ in yeast (Fig. 1C, lanes 3 and 4). An antiserum against a C-terminal peptide of Sym1 revealed one predominant signal in wild-type mitochondria which was shifted to a higher molecular mass in mitochondria from SYM1ZZ yeast (Fig. 1C, lanes 1 and 2). We confirmed the correct localization of Sym1ZZ within mitochondria by using protease protection assays to compare mitochondria and mitoplasts (generated by rupture of the outer membrane in a hypo-osmotic buffer). In mitochondria and mitoplasts, the outer membrane proteins (e.g., Tom70) were degraded by externally added proteases, whereas matrix proteins, such as Tim44, remained protected in both cases. The intermembrane space domain of Tim23 became accessible to protease treatment only in mitoplasts (Fig. 1D). We detected Sym1ZZ with the Sym1 antibody (data not shown) and the PAP antibody (Fig. 1D) and observed protease accessibility only after removal of the outer membrane. Neither of the antibodies detected protease-stable fragments of Sym1, indicating that the C terminus was exposed to the intermembrane space (IMS). We concluded that Sym1ZZ, like endogenous Sym1, was integrated in the inner membrane with an Nout/Cout orientation (65).
Fig 1.
Isolation of Sym1 from mitochondrial membranes. (A) ClustalW2 sequence alignment of Sym1 homologues in fungi and mammals. Black boxes indicate 100% similarity, and gray boxes indicate 75% similarity. (B) Schematic representation of the ZZ tag fused to Sym1 (ZZ, two repeats of the Z domain of protein A; TEV, cleavage site of the tobacco etch virus protease; His10, decahistidine tag). The epitope recognized by the Sym1 antibody is indicated (αSym1). IMS, intermembrane space; MIM, mitochondrial inner membrane. (C) Isolated mitochondria from the wild type (WT) and the SYM1ZZ strain were subjected to SDS-PAGE and Western blotting using antibodies against Sym1 and PAP (HRP–anti-HRP). (D) Mitochondria were left untreated or subjected to swelling in hypo-osmotic buffer, treated with the indicated amounts of protease, and subjected to SDS-PAGE and Western blotting. Sym1ZZ was detected using PAP. Prot. K, proteinase K. (E) Flow diagram of the isolation strategy for Sym1ZZ. (F) Total (5%), unbound (UB) (5%), and eluate (E) (ENi, 5%; EIgG, 100%) fractions of Ni-NTA affinity and IgG affinity chromatography were separated by SDS-PAGE and subjected to Western blot analyses using the Sym1 antibody. (G) Coomassie brilliant blue-stained SDS-PAGE gel of the IgG chromatography eluate.
Sym1-related Pxmp2 is a peroxisomal membrane protein in mice which was suggested to form a largely unregulated diffusion pore (52). To investigate if Sym1 forms a channel in the mitochondrial inner membrane, we established a protocol for purification and reconstitution of Sym1ZZ into lipid bilayers (Fig. 1E). We isolated mitochondria from Sym1ZZ yeast cells and solubilized mitochondrial membranes under denaturing conditions. The His tag on Sym1ZZ was used to isolate Sym1 by Ni affinity chromatography. Eluted proteins were diluted to allow refolding of the ZZ tag. Sym1ZZ was recovered in a second purification step using IgG affinity chromatography followed by TEV protease cleavage of the fusion protein (see above). The purification procedure was monitored by Western blot analysis (Fig. 1F), and the purity of isolated Sym1 was confirmed by SDS-PAGE and Coomassie brilliant blue staining (Fig. 1G). Sym1 was inserted into small unilamellar vesicles by detergent-mediated reconstitution (66). For the electrophysiological measurements, we fused the Sym1-containing proteoliposomes with planar lipid bilayers. A mock purification sample from wild-type mitochondria served as a control.
Sym1 forms a channel in lipid membranes.
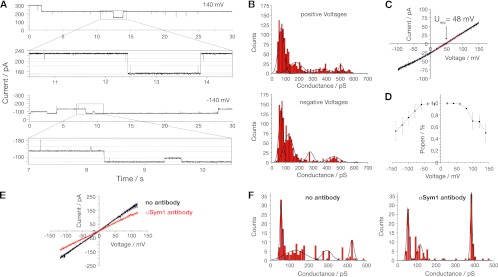
Analysis of single-channel recordings of bilayers containing Sym1 (Fig. 2A) in 250 mM KCl showed rectifying conductivity with main conductance states (G) of ∼510 pS when positive membrane potentials were applied and ∼450 pS with negative membrane potentials applied (Fig. 2B). No signs of ion channel activity were detected in the mock control. The channel exhibited a reversal potential (Urev) equaling ∼48 mV (cis, 250 mM KCl; trans, 20 mM KCl), from which, according to the Goldman-Hodgkin-Katz (GHK) approach, a preference for cations with a permeability ratio of PK+/PCl− equal to 14/1 can be calculated (Fig. 2C). Interestingly, a voltage-dependent closure of the channel was detected. At membrane potentials above ±150 mV, the open probability was reduced by 50% (Fig. 2D). To unambiguously show that the measured channel activity was attributable to Sym1, we performed experiments with antibodies raised against Sym1. The addition of antibodies led to a decrease of the channel conductivity (Fig. 2E). A significant change in the pattern of gating events was observed before and after the addition of antibody. This change was manifested in a shift of the amplitude and frequency of conductance states as calculated from single gating events (Fig. 2F). In summary, Sym1 constitutes a rectifying high-conductance cation-selective ion channel in the inner mitochondrial membrane which is voltage regulated. Our results suggest that this channel closes at the physiological membrane potential of the inner mitochondrial membrane. This voltage sensitivity would prevent ion leakage across the inner membrane.
Fig 2.
Sym1 forms a channel after reconstitution into planar lipid bilayers. (A) Sym1-containing proteoliposomes were fused with lipid bilayers, and single-channel activity was recorded in 250 mM KCl; (B) conductance state histograms deduced from gating events at positive and negative membrane potentials; (C) current-voltage relationship under asymmetrical buffer conditions (Urev, reversal potential); (D) voltage-dependent open probability (Popen); (E) current-voltage relationship under symmetrical buffer conditions in the presence or absence of a specific antibody against Sym1; (F) conductance states in the presence or absence of an anti-Sym1 antibody.
Internal targeting signals direct the import of Sym1 into mitochondria.
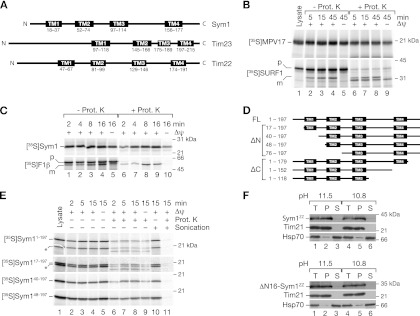
The architectures of the transmembrane helices of Sym1 and MPV17 are highly conserved and share topological similarities with two other channel proteins of the inner mitochondrial membrane, Tim23 and Tim22. Based on the characteristic topology of their four transmembrane spans in an Nout/Cout orientation, Tim22 and Tim23 and the chloroplast Oep16 have been described as founding members of the preprotein and amino acid transporter (PRAT) protein family (48). Sym1 not only shares the same topology as Tim23, Tim22, and Oep16 (Fig. 3A) but also displays similar channel activity (28, 36, 47). Tim22 and Tim23 utilize internal targeting signals for import into mitochondria and are transported along the carrier pathway. In contrast, previous studies have suggested that a presequence at the N terminus of MPV17 is responsible for its import into mitochondria (63), indicating a transport mechanism via the TIM23 complex. Indeed, with MitoProtII, a presequence was predicted for MPV17 and Sym1 (human MPV17, 86.0%; mouse MPV17, 85.4%; Sym1, 82.8%). As only a limited number of polytopic TIM23 substrates are known, we studied the import pathway of MPV17 and Sym1 directly.
Fig 3.
Sym1 is directed to mitochondria by internal targeting signals. (A) Schematic representation of the transmembrane (TM) domains of Sym1 and inner membrane channels of similar topology. (B) Radiolabeled MPV17 and SURF1 were imported into mitochondria isolated from HEK293T cells. In vitro import was performed for the indicated times in the presence or absence of a membrane potential (Δψ). Reisolated mitochondria were treated with proteinase K (Prot. K) where indicated and analyzed by SDS-PAGE and digital autoradiography. p, precursor; m, mature. (C) 35S-labeled proteins were imported into isolated yeast mitochondria and analyzed by SDS-PAGE and digital autoradiography. (D) Constructs for analysis of targeting signals. FL, full-length protein; ΔN, N-terminal truncation; ΔC, C-terminal truncation; TM, transmembrane span. (E) In vitro import of truncation constructs into isolated yeast mitochondria as described for panel C. Indicated samples were sonicated in the presence or absence of proteinase K. Truncated Sym1s originating from a second start codon in SYM1 are indicated with asterisks. (F) Carbonate extraction of mitochondrial membranes at the indicated pH. After centrifugation, total fractions of samples (T), pellets (P), and supernatants (S) were analyzed by Western blotting.
MPV17 was synthesized in an in vitro transcription-translation system in the presence of radiolabeled [35S]methionine and imported into energized human mitochondria. MPV17 was efficiently imported into mitochondria and protected against protease treatment (Fig. 3B). Interestingly, the MPV17 precursor was not processed to a faster-migrating form during import. In contrast, the presequence-containing precursor of SURF1 was processed to a mature form in the presence of a membrane potential (Δψ) (51, 78). In the absence of a Δψ, only a partial reduction of MPV17 import was observed, suggesting that a fraction of MPV17 was transported to a protease-protected location within mitochondria, independent of the membrane potential. This import behavior was reminiscent of carrier pathway substrates (45, 53, 82). This surprising finding led us to investigate the import of Sym1 into yeast mitochondria. We synthesized the Sym1 precursor in a coupled in vitro transcription-translation system. Sym1 was efficiently imported into isolated yeast mitochondria (Fig. 3C). Similar to MPV17, Sym1 was not processed during import, while in a control experiment, the precursor of F1-ATPase β subunit (F1β) was imported and processed in a Δψ-dependent manner. In contrast to F1β, for Sym1 in the absence of Δψ, only a fraction was accessible to protease, results similar to those for the MPV17 import. To exclude the possibility that Sym1 displayed intrinsic protease resistance, sonicated mitochondria were protease treated (Fig. 3E, lanes 10 and 11), resulting in efficient degradation of Sym1. Interestingly, when purified SYM1 mRNA was synthesized in an in vitro translation system, we observed an additional translation product, likely originating from a second start codon in SYM1, which was also imported into mitochondria despite the lack of its N terminus (Fig. 3E). Thus, both MPV17 and Sym1 showed no processing of the N terminus. To validate this finding, we purified Sym1ZZ from mitochondria and analyzed the N terminus by Edman sequencing and mass spectrometry. These analyses confirmed that the mature N terminus corresponded to the nonprocessed protein. We therefore asked if the predicted presequence at the N terminus of the protein was required for import, and we generated truncation constructs with increasing N-terminal deletions of Sym1 (Fig. 3D). Similar to the full-length protein (and the fragment generated from the second start codon), all Sym1ΔN constructs were imported into mitochondria. Like full-length Sym1, a fraction of the truncation constructs were transported to a protease-inaccessible location in a Δψ-independent manner but became accessible to protease by sonication (Fig. 3E). To ensure that the N terminus of Sym1 was not required for import and membrane integration in vivo, we expressed Sym1ZZ and Sym1ZZ lacking the N terminus (aa 1 to 16) in yeast cells. Both proteins were recovered in mitochondria and displayed resistance to carbonate extraction, indicating that they were integral membrane proteins (Fig. 3F). Taken together, our data show that internal targeting signals are responsible for the import of Sym1 and MPV17 into mitochondria. Moreover, in the absence of a Δψ, the observed import resembled the protease-protected stage III intermediate of carrier transport (45, 53, 80).
Sym1 assembles into high-molecular-mass complexes in the inner membrane.
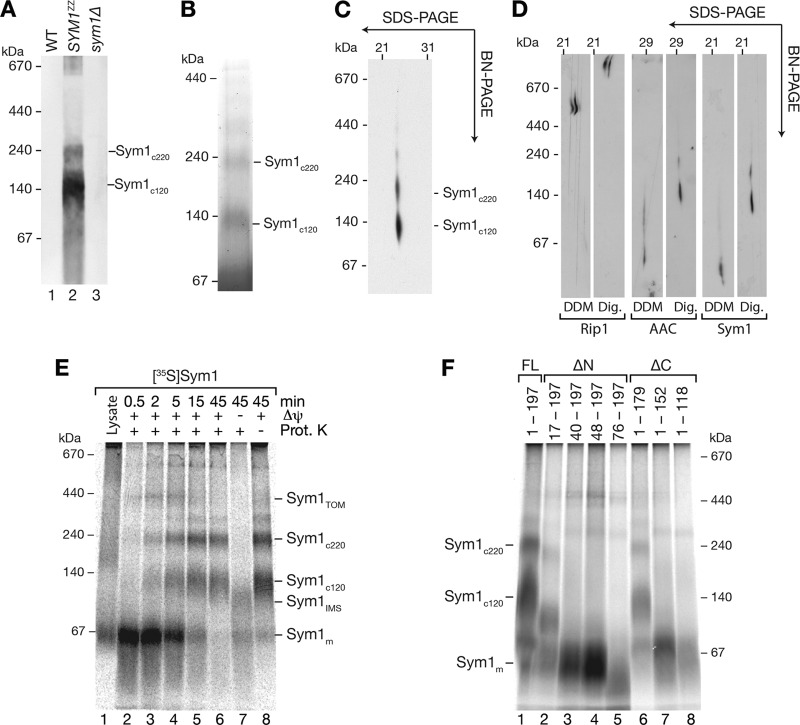
A common feature of carrier proteins in the inner membrane is their organization into high-molecular-mass complexes, which can be analyzed by BN-PAGE (42, 43). To assess complex formation, Sym1ZZ mitochondria were solubilized, and the complexes were analyzed by BN-PAGE and Western blotting. Wild-type and sym1Δ mitochondria were analyzed as controls. Sym1ZZ-specific signals were detected at molecular masses of 120 and 220 kDa (Fig. 4A). We enriched these complexes by native isolation of Sym1-containing complexes by IgG chromatography. Eluates were analyzed by BN-PAGE and stained with Coomassie brilliant blue. Two bands (120 and 220 kDa) were clearly visible and resembled the results of the Western blot analysis (Fig. 4B). To verify complex formation of endogenous (nontagged) Sym1, we used 2-dimensional polyacrylamide gel electrophoresis (2D-PAGE) (1st dimension, BN-PAGE; 2nd dimension, SDS-PAGE), as the Sym1 antibody did not recognize Sym1 in complexes on native gels. We detected 120-kDa and 220-kDa complexes of endogenous Sym1 (Fig. 4C). According to their molecular masses, the two complexes were termed Sym1c120 and Sym1c220. Inner membrane complex oligomers, such as respiratory chain supercomplexes and ADP-ATP carrier (AAC) oligomers, have been found to be stabilized by the inner membrane lipid environment. Such complexes are best maintained for biochemical analyses in the detergent digitonin, but they dissociate under harsher conditions (55, 76). Thus, we compared the solubilization behavior of Sym1 to AAC and respiratory chain supercomplexes in digitonin with dodecyl maltoside. Similar to AAC and respiratory chain supercomplexes, Sym1 complexes were shifted to a lower molecular-mass range (Fig. 4D) when dodecyl maltoside was used as the detergent.
Fig 4.
Assembly of Sym1 into mature complexes. (A) Yeast mitochondria were solubilized in digitonin buffer, separated by BN-PAGE, and analyzed by Western blotting using the PAP antibody; (B) Sym1ZZ isolated from mitochondria by IgG chromatography was analyzed by BN-PAGE and stained with Coomassie brilliant blue; (C) mitochondria were solubilized in digitonin-containing buffer, and complexes were separated by 1st-dimension BN-PAGE and 2nd-dimension SDS-PAGE and analyzed by Western blotting with the anti-Sym1 antibody; (D) mitochondria were solubilized in buffers containing digitonin (Dig.) or dodecyl maltoside (DDM), and the complexes were separated by 1st-dimension BN-PAGE and 2nd-dimension SDS gel and analyzed by Western blotting; (E) Sym1 import was monitored with BN-PAGE and subsequent digital autoradiography; (F) import and BN-PAGE analysis of truncation constructs described for Fig. 3D. Complexes were visualized by digital autoradiography.
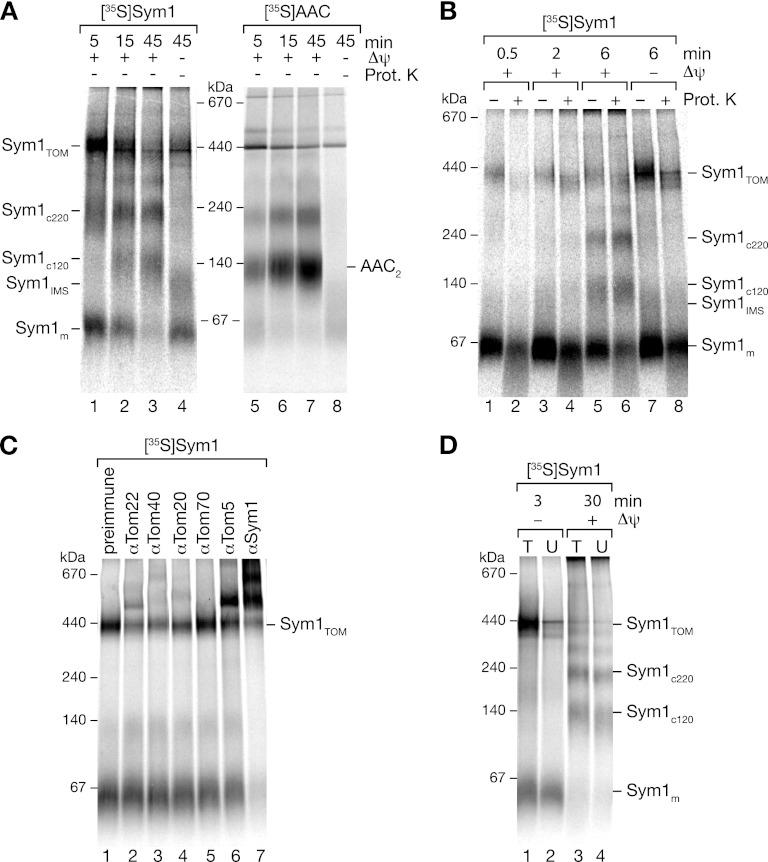
The visualization of complexes by BN-PAGE in combination with the import of Sym1 allowed us to study the kinetics of assembly. We therefore imported Sym1 into isolated mitochondria and analyzed the complexes after solubilization by BN-PAGE. Sym1 formed complexes of different molecular masses in a time- and Δψ-dependent manner (Fig. 4E). After short import times (0.5 to 2 min), a signal in the low-molecular-mass range transiently peaked and subsequently decreased (Fig. 4E, lanes 2 and 3). The small molecular size and comigration with in vitro-synthesized Sym1 (Fig. 4E, lane 1) and its transient appearance led us to conclude that it represented monomeric Sym1 (Sym1m). With a slight delay, a second complex of 450 kDa peaked after 2 min (Fig. 4E, lane 3). Based on its size, we speculated that this form could represent an assembly intermediate in the TOM complex (Sym1TOM). The final products of the import reactions were the two mature complexes, Sym1c120 and Sym1c220. These two complexes formed last and coincidentally with the decrease of the intermediate products. The assemblies of both complexes were dependent on the Δψ. Only in the absence of Δψ was a protease-inaccessible complex of approximately 100 kDa formed in addition to the monomeric Sym1. We termed this 100-kDa complex Sym1IMS because it was resistant to protease treatment and had thus passed through the outer membrane, while at the same time, due to the lack of Δψ, it was not inserted into the membrane. The formation of protease-resistant complexes in the absence of a Δψ resembled the well-characterized stage III intermediates in the import pathway of carrier proteins. These intermediates are released from the TOM complex on BN-PAGE and thus run in the low-molecular-mass range (10, 34, 45, 53). Based on the ability of Sym1 to form a complex under similar conditions, we hypothesized that the 100-kDa complex shown in Fig. 4E represents a Sym1 transport intermediate detached from the TOM complex during the solubilization of mitochondria.
To analyze which regions of Sym1 were necessary for import and assembly, we used truncation constructs with increasing N- and C-terminal deletions (Fig. 3D). Sym1 constructs were imported into mitochondria, and the assemblies were monitored by BN-PAGE analyses after treatment of the samples with proteinase K (Fig. 4F). Truncations at the N or C terminus, even those that eliminated one transmembrane segment from the N terminus or the C terminus, were still efficiently imported and protected against protease, indicating that neither the N nor the C terminus was necessary for the import of Sym1. However, constructs lacking one transmembrane segment did not assemble into mature complexes and remained in a monomeric form. N-terminal truncations exceeding one transmembrane domain displayed decreased amounts of monomers, indicative of reduced import efficiencies or increased turnover rates.
Sym1 forms an assembly intermediate in the TOM complex.
The transient formation of the 450-kDa assembly intermediate suggested the formation of a complex of Sym1 with the TOM complex. Therefore, we omitted proteinase K treatment after import of Sym1 into mitochondria and monitored the assembly of the ADP/ATP carrier (AAC) as a control (Fig. 5A). During BN-PAGE, the 450-kDa intermediate clearly accumulated after short import times when protease treatment was omitted (Fig. 5A; compare to Fig. 4E). To investigate a potential interaction of Sym1 with the TOM complex, we compared the accessibility of the 450-kDa intermediate to externally added protease during optimized import times between 0.5 min and 6 min. As shown in Fig. 5B, at import times between 2 and 6 min, increasing amounts of precursor forming the 450-kDa intermediate were progressing into a state where they were protected from protease. After 6 min of import, the 450-kDa intermediate strongly accumulated upon dissipation of the membrane potential. Protease treatment revealed that both protease-accessible and -nonaccessible forms were increased. We used these conditions in the following experiments to analyze our hypothesis that the 450-kDa complex represents an intermediate of Sym1 associated with the TOM complex.
Fig 5.
TOM complex intermediates formed during Sym1 import. (A) Analysis of [35S]Sym1 and [35S]AAC import into mitochondria without proteinase K treatment; (B) comparison of short-time import of [35S]Sym1 proteinase K-treated and nontreated samples; (C) mitochondria were solubilized after import of [35S]Sym1 for 3 min and incubated with the indicated antibodies before separation by BN-PAGE and digital autoradiography; (D) radiolabeled Sym1 was imported into tom22His mitochondria. After solubilization, Tom22His-containing complexes were eliminated by incubation with Ni-NTA beads. Total (T) and unbound (U) fractions were separated by BN-PAGE and analyzed by digital autoradiography.
In order to show an interaction of the imported precursor with the TOM complex, we performed antibody shift assays. After the import of Sym1, mitochondria were reisolated, solubilized, and incubated with antisera directed against Sym1, the central TOM subunits Tom22, Tom20, Tom5, and Tom40, and the peripheral TOM component Tom70. Binding of antibodies against Sym1 to the assembled [35S]Sym1 dramatically affected the migration of complexes on BN-PAGE, whereas preimmune serum had no effect (Fig. 5C, lanes 1 and 7). Antibodies against the central TOM components strongly affected the migration of a substantial fraction of the 450-kDa intermediate, indicating that the 450-kDa Sym1-containing complex represented an import intermediate of Sym1 associated with the TOM complex (Fig. 5C). However, the peripheral TOM component Tom70 largely dissociated from the TOM complex upon solubilization with digitonin; therefore, no shift was observed with Tom70 antiserum. The low-molecular-mass complexes were shifted only by Sym1 antiserum, indicating that these intermediates were detached from the tested Tom components.
In a complementary approach, we performed a depletion assay using mitochondria from a yeast strain expressing the central TOM component Tom22 with a 10-histidine extension (tom22His) (38, 57). Sym1 was imported into Tom22His-containing mitochondria. Solubilized mitochondrial extracts were then incubated with Ni-NTA agarose to remove Tom22His and associated proteins from the extract. Binding of Tom22 to the resin efficiently depleted the 450-kDa intermediate (Fig. 5D). Neither the 100-kDa intermediate nor the mature complexes were affected by the treatment. We conclude that the 450-kDa intermediate represents an intermediate stage of Sym1 import, in which the precursor is bound to the TOM complex and is partially accessible to protease treatment from the cytosolic side of the outer membrane.
Sym1 is transported independently of the TIM22 translocase.
Precursor proteins are transported on different pathways from the TOM complex into the inner membrane. Considering that the N terminus of Sym1 was not required for import or assembly, Sym1 displayed channel activity, a stage III-like TOM intermediate was formed in the absence of a Δψ, and four predicted transmembrane domains were present, we speculate that Sym1, like Tim22 and Tim23, follows the carrier pathway for import (23, 24). After translocation across the TOM complex, carrier precursors typically bind to the small TIM complex (Tim9/Tim10), leading to stage III formation. Concomitantly, in the absence of a functional Tim9/Tim10 complex, carriers accumulate in the TOM complex in a protease-sensitive manner (6, 7, 26, 27, 44, 67). In order to investigate an involvement of small TIM proteins in the transport of Sym1, we performed import and assembly assays in mutant mitochondria.
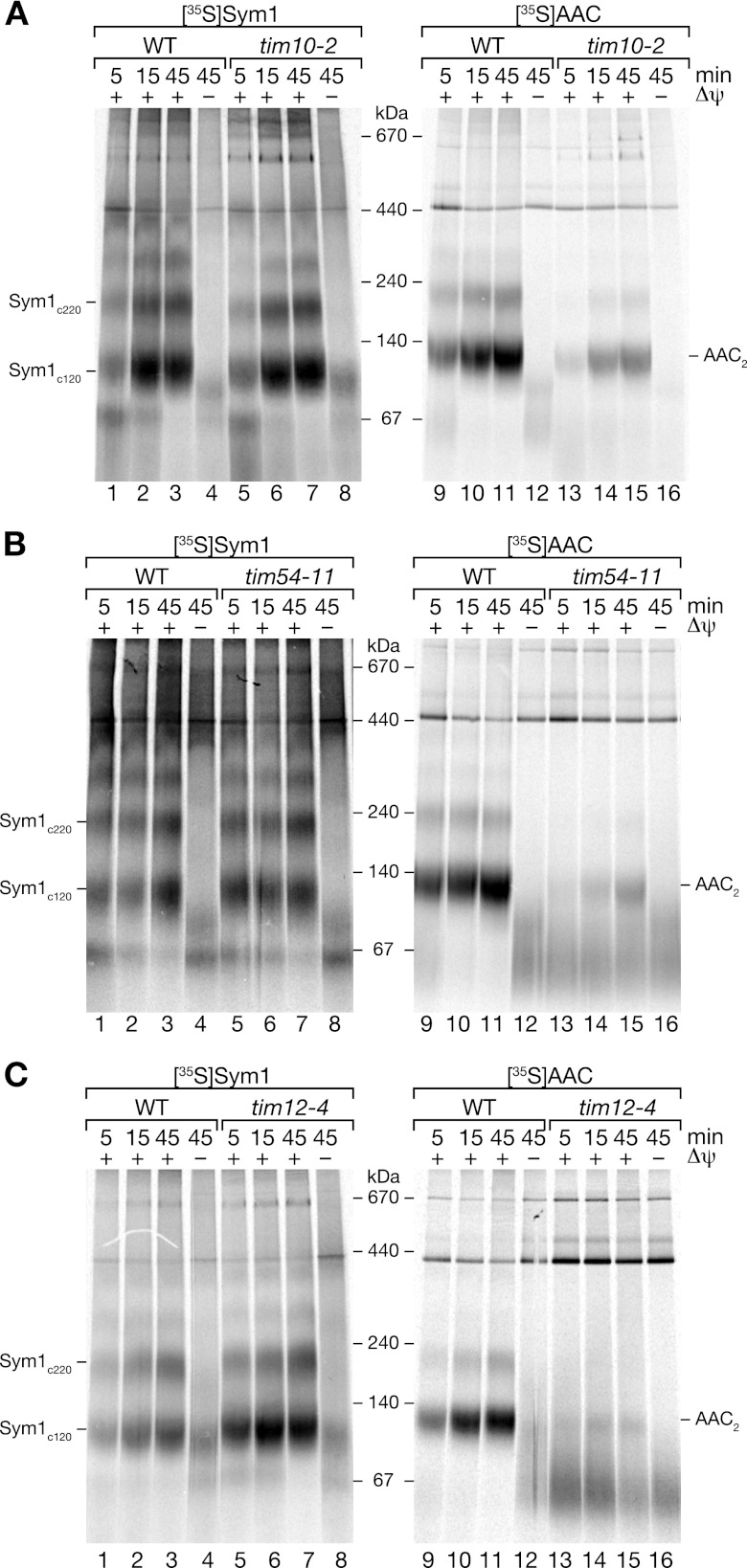
Temperature-conditional tim10-2 mitochondria were isolated from cells grown under permissive conditions and shifted to nonpermissive conditions prior to import (67). While the ADP/ATP carrier, a typical substrate of the carrier pathway, displayed a strong decrease in these mutant mitochondria (Fig. 6A), Sym1 assembly was not affected. This surprising result indicated that the Tim9/Tim10 complex was not involved in the transport of Sym1.
Fig 6.
Sym1 is imported independent of Tim9/Tim10 and TIM22 complexes. Radiolabeled proteins were imported into mitochondria isolated from the WT and the tim10-2 (A), tim54-11 (B) and tim12-4 (C) mutant strains. In vitro import was performed for the indicated times in the presence or absence of a membrane potential (Δψ). Reisolated mitochondria were solubilized in digitonin and analyzed by BN-PAGE and digital autoradiography.
Proteins transported via the carrier pathway are integrated into the inner membrane via the TIM22 complex. To investigate involvement of the carrier translocase in the insertion of Sym1 into the inner membrane, we used mitochondria isolated from temperature-conditional mutant strains. The temperature-sensitive allele tim54-11 impairs the function of the core constituent Tim54 (24, 28, 71). Tim12 is an essential constituent of the carrier translocase exposed to the intermembrane space (1, 17, 26, 59). The temperature-sensitive tim12-4 mutant displayed a strongly reduced efficiency of carrier import compared to the wild type (17). Mitochondria isolated from cells grown under permissive temperature conditions were shifted to the restrictive temperature prior to import. While AAC import and assembly were severely affected in both mitochondria mutants compared to in the wild-type control, Sym1 assemblies in the mutant mitochondria were similar to those in the wild-type mitochondria (Fig. 6B and C). In summary, these data demonstrate that neither the TIM22 complex nor the small TIM complex was required for Sym1 import and assembly into the inner membrane. We conclude that despite the lack of a cleavable N-terminal presequence, Sym1 is not imported via the carrier pathway.
Import and assembly of Sym1 require the presequence translocase.
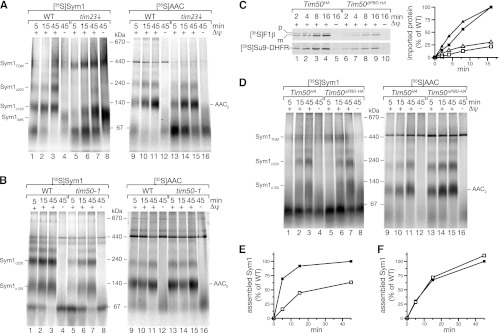
Presequence-containing proteins are imported by the presequence translocase (TIM23 complex). Since the carrier translocase (TIM22 complex) was not involved in the transport and integration of Sym1, we assessed if Sym1 was transported by the TIM23 complex despite the absence of a cleavable presequence. Therefore, we used a yeast strain in which the channel-forming component of the presequence translocase, Tim23, was expressed under the control of a galactose-inducible promoter. Growth of yeast cells in the presence of glucose decreases the expression of Tim23 and allows the isolation of mitochondria with significantly reduced Tim23 levels (56). When Sym1 was imported into mitochondria with reduced levels of Tim23, assembly was strongly affected (Fig. 7A). Compared to that of wild-type mitochondria, assembly of Sym1c120 was strongly delayed, and Sym1c220 was almost not detectable. Instead, Sym1IMS and monomeric Sym1 accumulated independently of the presence or absence of a Δψ. Moreover, the 450-kDa TOM complex intermediate was prominent in Tim23 shut-down mitochondria over the course of the assay (Fig. 7A). These data suggest that the presequence translocase plays a role in the transport of Sym1 and that, under conditions in which the amount of Tim23 becomes limiting, transport intermediates of Sym1 accumulate. In contrast, assembly of AAC was not significantly affected in the mutant mitochondria. However, due to a loss of membrane potential in the absence of a functional translocase, the stage III intermediate was more prominent in the mutant mitochondria at early time points.
Fig 7.
Sym1 is imported by the TIM23 translocase. (A and B) Import of radiolabeled proteins into isolated WT and mutant mitochondria (as indicated) and analysis by BN-PAGE and digital autoradiography. (C) [35S]F1β (squares) and [35S]Su9-DHFR (triangles) were imported into isolated mitochondria (Tim50HA, filled symbols; Tim50ΔPBD-HA, open symbols) and analyzed by SDS-PAGE followed by digital autoradiography (left) and quantification (right). (D) 35S-labeled proteins were imported into WT and mutant mitochondria and analyzed as described for panels A and B. (E and F) Quantification of the Sym1 120-kDa and 220-kDa complexes shown in panels B and D, respectively, comparing import into wild-type (WT) (filled symbols) and mutant (open symbols) mitochondria.
To exclude indirect effects, such as reduced Δψ, we imported Sym1 into mitochondria purified from a temperature-conditional strain carrying mutations in the import receptor Tim50. Tim50 is a central component of the presequence translocase that regulates Tim23 channel activity and is required for transport along the presequence pathway. In tim50-1 mutant cells, Tim50 function is impaired after a shift to nonpermissive temperature conditions, leading to selective defects in the transport of presequence-containing proteins through the TIM23 complex (5). Compared to wild-type mitochondria, tim50-1 mutant mitochondria displayed apparent defects in Sym1 assembly and an accumulation of Sym1 monomers (Fig. 7B, lanes 1 and 5). Quantification of the amounts of assembled Sym1 in the 120-kDa and 220-kDa complexes revealed drastically slower kinetics in the tim50-1 mutant (Fig. 7E), indicating that Tim50 is required for efficient assembly. In contrast to Sym1, import and assembly of AAC were comparable to those of the wild- type version (Fig. 7B), as reported previously (5, 56). The finding that the TIM23 complex was required for Sym1 biogenesis, as indicated by the defect in tim50-1 mitochondria, led us to investigate if the presequence receptor function of Tim50 was required for import or assembly. Therefore, we used mitochondria from a strain expressing TIM50 under the control of a galactose-inducible promoter from the chromosomal locus. This strain allows the selective reduction of Tim50 levels. We expressed a wild-type version of Tim50 (Tim50HA) or a version of Tim50 (Tim50ΔPBD-HA) lacking the essential presequence-binding domain in this background (56). The rates of import of the presequence-containing precursor F1β or the model matrix protein Su9-DHFR into the mutant mitochondria were reduced to 20% and 25%, respectively, after 16 min of import (Fig. 7C). In contrast, import and assembly of Sym1 and AAC were not affected (Fig. 7D and F). Altogether, these experiments showed that Sym1 is directed to the presequence translocase, where it is transported in a Tim50- and Tim23-dependent manner. However, in contrast to the drastic kinetic delay of Sym1 import in tim50-1 mutants, the assembly into the Tim50ΔPBD-HA mutant was comparable to that of the wild- type (Fig. 7E and F), indicating that the import reaction bypasses the requirement for presequence recognition at the translocase by the receptor Tim50. Thus, we conclude that the TIM23 complex is able to transport substrates, such as Sym1, that lack an N-terminal presequence segment, indicating a presequence-independent mode of action for this translocase.
DISCUSSION
Mutations in human MPV17 lead to the hepatocerebral form of mitochondrial DNA depletion syndrome (MDDS) (62). Seven of eight genes that have been linked to MDDS were found to play a direct role in mitochondrial DNA maintenance (13, 19, 41, 54, 62). The depletion of the mitochondrial DNA phenotype and the concomitant decrease of respiratory activity were recapitulated in MPV17 knockout mice. However, the function of MPV17 is still enigmatic. In this study we have demonstrated that Sym1, the orthologue of MPV17, forms a channel in the inner mitochondrial membrane. The conductance (G) of ∼450 pS corresponds to a pore size of about 1.6 nm (60), sufficient to allow the transport of large molecules such as metabolites across the inner mitochondrial membrane. However, the physiological function of the channel and the nature of the cargo remain unknown. Similar to the Tim23 and Tim22 channels of the inner membrane, Sym1 closure occurs toward the physiological membrane potential, indicating a means to prevent ion leakage. It is likely that specific factors exist in vivo that regulate channel gating.
As suggested for the channel-forming inner membrane proteins Tim22 and Tim23, four transmembrane domains do not suffice for pore formation (50, 66). In the case of Sym1, we were able to detect two complexes of 120 and 220 kDa. These findings agree with those of a previous study that indicated the presence of Sym1 in a high-molecular-mass complex (8). As we used denaturing conditions to purify Sym1 for our electrophysiological measurements, it remains to be analyzed if unknown constituents of the detected complexes might modulate the observed channel properties. It is also unclear if both complexes are functionally relevant, if they play different physiological roles within the membrane, and if further components are involved in complex formation.
The observed channel activity of Sym1 raised questions on its import and integration into the inner membrane. Two major import pathways direct nucleus-encoded proteins into the inner mitochondrial membrane. Presequence-containing proteins are transported by the presequence translocase (TIM23), which not only mediates full matrix translocation but also integrates proteins into the inner membrane (5, 15, 18, 29, 39, 68, 70). In contrast, metabolite carriers and the channel proteins Tim22 and Tim23 are directed into mitochondria by internal signals and use the TIM22 translocase for membrane integration. In silico analyses have suggested significant presequence probabilities for MPV17 and Sym1. However, we did not observe processing of the proteins after import into yeast or human mitochondria. Moreover, removal of N- or C-terminal segments of Sym1 did not block import of the precursor proteins, excluding an essential role of these regions for the transport process. We therefore conclude that internal targeting sequences are involved in Sym1 transport.
Analysis of Sym1 import into isolated mitochondria revealed intermediates in the transport process. A complex of 450 kDa represents an outer membrane transport intermediate in the TOM complex. A productive intermediate of a TIM23 substrate with the TOM complex has been shown for the multispanning protein OxaI, which utilizes an N-terminal presequence (16). This intermediate was identified in the absence of a membrane potential and remained accessible to external proteases, indicating that OxaI did not fully pass the TOM complex at this stage and required the membrane potential to be further translocated across the outer membrane (5, 16). Sym1TOM intermediates were identified in energized mitochondria and remained fully accessible to protease only at early stages of import. At later stages, an increasing fraction was protected from externally added protease. This indicates that the intermediate of 450 kDa represents more than one distinct import stage. This behavior was reminiscent of that of substrates of the carrier pathway that form protease-resistant stage III intermediates in the TOM complex in the absence of a membrane potential.
The small Tim proteins of the intermembrane space typically transport multispanning proteins with internal targeting sequences to the carrier translocase (6, 27, 59). Our analyses showed that neither the small Tim protein complex (Tim9/Tim10) nor the essential TIM22 complex constituents Tim12 and Tim54 are required for import and assembly of Sym1. Instead, we have demonstrated that the presequence translocase is required for inner membrane transport of Sym1. Import and assembly of Sym1 were strongly affected by depletion of the Tim23 channel and by the inactivation of Tim50. These data reveal an involvement of the presequence translocase in the transport of proteins with internal targeting signals. A small number of TIM23 substrates have been identified that lack an amino-terminal cleavable presequence. This holds true for Pam18, Mdj2, and Bcs1, which span the inner membrane with a single transmembrane segment (14, 39, 73). The current view is that presequence-like elements adjacent to the single membrane span direct these precursors to the TIM23 complex (40). In the case of the soluble helicase Hmi1, a cleavable C-terminal presequence is used for transport along the presequence pathway (33). In Sym1, neither terminus is essential for import, indicating the presence of internal targeting information. Moreover, in our in silico analyses, we did not detect internal segments adjacent to transmembrane spans with high presequence probability in Sym1. To experimentally assess if presequence-like elements of Sym1 are required for transport by the presequence translocase, we utilized a mutant of Tim50 which lacked the presequence-binding domain (56). While the import of classical presequence-containing precursors was blocked in these mitochondria, the import of Sym1 was not affected. Hence, our analyses revealed that the presequence translocase is able to transport a protein substrate with internal targeting signals in a presequence-independent manner (Fig. 8). Future work will have to clarify which segments in the Sym1 precursor are recognized by the TIM23 complex in order to promote membrane insertion.
Fig 8.
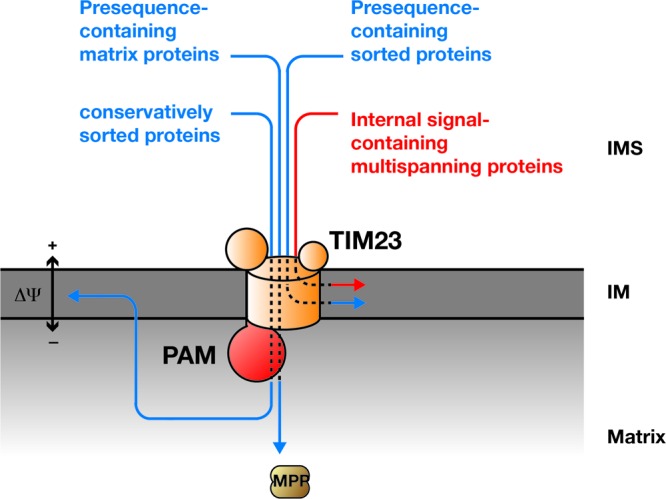
Schematic presentation of import substrates using the TIM23 complex. PAM, presequence translocase-associated protein import motor; MPP, mitochondrial processing peptidase; IM, inner mitochondrial membrane; IMS, intermembrane space.
ACKNOWLEDGMENTS
We thank M. Deckers and D. U. Mick for helpful discussions. We are indebted to Nicole Eiselt, Agnes Schulze-Specking, and Klaus Neifer for their expert technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 860 and SFB 746), the Max Planck Society (P.R.), and the Human Frontier Science Program (M.M.).
Footnotes
Published ahead of print 8 October 2012
REFERENCES
- 1. Adam A, et al. 1999. Tim9, a new component of the TIM22-54 translocase. EMBO J. 18:313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bohnert M, et al. 2010. Cooperation of stop-transfer and conservative sorting mechanisms in mitochondrial protein transport. Curr. Biol. 20:1227–1232 [DOI] [PubMed] [Google Scholar]
- 3. Boxma B, et al. 2005. An anaerobic mitochondrion that produces hydrogen. Nature 434:74–79 [DOI] [PubMed] [Google Scholar]
- 4. Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. 2009. Importing mitochondrial proteins: machineries and mechanisms. Cell 138:628–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chacinska A, et al. 2005. Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell 120:817–829 [DOI] [PubMed] [Google Scholar]
- 6. Curran S, Leuenberger D, Oppliger W, Koehler C. 2002. The Tim9p-Tim10p complex binds to the transmembrane domains of the ADP/ATP carrier. EMBO J. 21:942–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Curran S, Leuenberger D, Schmidt E, Koehler C. 2002. The role of the Tim8p-Tim13p complex in a conserved import pathway for mitochondrial polytopic inner membrane proteins. J. Cell Biol. 158:1017–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dallabona C, et al. 2010. Sym1, the yeast ortholog of the MPV17 human disease protein, is a stress-induced bioenergetic and morphogenetic mitochondrial modulator. Hum. Mol. Genet. 19:1098–1107 [DOI] [PubMed] [Google Scholar]
- 9. Dekker P, Müller H, Rassow J, Pfanner N. 1996. Characterization of the preprotein translocase of the outer mitochondrial membrane by blue native electrophoresis. Biol. Chem. 377:535–538 [PubMed] [Google Scholar]
- 10. Dietmeier K, et al. 1993. Targeting and translocation of the phosphate carrier/p32 to the inner membrane of yeast mitochondria. J. Biol. Chem. 268:25958–25964 [PubMed] [Google Scholar]
- 11. Dudek J, Rehling P, van der Laan M. Mitochondrial protein import: common principles and physiological networks. Biochim. Biophys. Acta, in press. doi:10.1016/j.bbamcr.2012.05.028 [DOI] [PubMed] [Google Scholar]
- 12. Endres M, Neupert W, Brunner B. 1999. Transport of the ADP/ATP carrier of mitochondria from the TOM complex to the TIM22-54 complex. EMBO J. 18:3214–3221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ferrari G, et al. 2005. Infantile hepatocerebral syndromes associated with mutations in the mitochondrial DNA polymerase-gammaA. Brain 128:723–731 [DOI] [PubMed] [Google Scholar]
- 14. Fölsch H, Guiard B, Neupert W, Stuart RA. 1996. Internal targeting signal of the BCS1 protein: a novel mechanism of import into mitochondria. EMBO J. 15:479–487 [PMC free article] [PubMed] [Google Scholar]
- 15. Frazier A, et al. 2004. Pam16 has an essential role in the mitochondrial protein import motor. Nat. Struct. Mol. Biol. 11:226–233 [DOI] [PubMed] [Google Scholar]
- 16. Frazier AE, et al. 2003. Mitochondria use different mechanisms for transport of multispanning membrane proteins through the intermembrane space. Mol. Cell. Biol. 23:7818–7828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gebert N, et al. 2008. Assembly of the three small Tim proteins precedes docking to the mitochondrial carrier translocase. EMBO Rep. 9:548–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Glick BS, et al. 1992. Cytochromes c1 and b2 are sorted to the intermembrane space of yeast mitochondria by a stop-transfer mechanism. Cell 69:809–822 [DOI] [PubMed] [Google Scholar]
- 19. Hakonen AH, et al. 2007. Recessive Twinkle mutations in early onset encephalopathy with mtDNA depletion. Brain 130:3032–3040 [DOI] [PubMed] [Google Scholar]
- 20. Herrmann J, Neupert W, Stuart R. 1997. Insertion into the mitochondrial inner membrane of a polytopic protein, the nuclear-encoded Oxa1p. EMBO J. 16:2217–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huh WK, et al. 2003. Global analysis of protein localization in budding yeast. Nature 425:686–691 [DOI] [PubMed] [Google Scholar]
- 22. Janke C, et al. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21:947–962 [DOI] [PubMed] [Google Scholar]
- 23. Kaldi K, Bauer MF, Sirrenberg C, Neupert W, Brunner M. 1998. Biogenesis of Tim23 and Tim17, integral components of the TIM machinery for matrix-targeted preproteins. EMBO J. 17:1569–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kerscher O, Holder J, Srinivasan M, Leung RS, Jensen RE. 1997. The Tim54p-Tim22p complex mediates insertion of proteins into the mitochondrial inner membrane. J. Cell Biol. 139:1663–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Knop M, et al. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15:963–972 [DOI] [PubMed] [Google Scholar]
- 26. Koehler C, et al. 1998. Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science 279:369–373 [DOI] [PubMed] [Google Scholar]
- 27. Koehler C, et al. 1998. Tim9p, an essential partner subunit of Tim10p for the import of mitochondrial carrier proteins. EMBO J. 17:6477–6486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kovermann P, et al. 2002. Tim22, the essential core of the mitochondrial protein insertion complex, forms a voltage-activated and signal-gated channel. Mol. Cell 9:363–373 [DOI] [PubMed] [Google Scholar]
- 29. Kozany C, Mokranjac D, Sichting M, Neupert W, Hell K. 2004. The J domain-related cochaperone Tim16 is a constituent of the mitochondrial TIM23 preprotein translocase. Nat. Struct. Mol. Biol. 11:234–241 [DOI] [PubMed] [Google Scholar]
- 30. Krueger V, et al. 2012. The mitochondrial oxidase-assembly-protein1 (OxaI) insertase forms a membrane pore in lipid bilayers. J. Biol. Chem. 287:33314–33326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kurz M, Martin H, Rassow J, Pfanner N, Ryan MT. 1999. Biogenesis of Tim proteins of the mitochondrial carrier import pathway: differential targeting mechanisms and crossing over with the main import pathway. Mol. Biol. Cell 10:2461–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lazarou M, Smith SM, Thorburn DR, Ryan MT, McKenzie M. 2009. Assembly of nuclear DNA-encoded subunits into mitochondrial complex IV, and their preferential integration into supercomplex forms in patient mitochondria. FEBS J. 276:6701–6713 [DOI] [PubMed] [Google Scholar]
- 33. Lee CM, Sedman J, Neupert W, Stuart RA. 1999. The DNA helicase, Hmilp, is transported into mitochondria by a C-terminal cleavable targeting signal. J. Biol. Chem. 274:20937–20942 [DOI] [PubMed] [Google Scholar]
- 34. Luciano P, et al. 2001. Functional reconstitution of the import of the yeast ADP/ATP carrier mediated by the TIM10 complex. EMBO J. 20:4099–4106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marom M, et al. 2011. Direct interaction of mitochondrial targeting presequences with purified components of the TIM23 protein complex. J. Biol. Chem. 286:43809–43815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meinecke M, et al. 2006. Tim50 maintains the permeability barrier of the mitochondrial inner membrane. Science 312:1523–1526 [DOI] [PubMed] [Google Scholar]
- 37. Meisinger C, Pfanner N, Truscott K. 2006. Isolation of yeast mitochondria. Methods Mol. Biol. 313:33–39 [DOI] [PubMed] [Google Scholar]
- 38. Meisinger C, et al. 2001. Protein import channel of the outer mitochondrial membrane: a highly stable Tom40-Tom22 core structure differentially interacts with preproteins, small Tom proteins, and import receptors. Mol. Cell. Biol. 21:2337–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mokranjac D, Sichting M, Neupert W, Hell K. 2003. Tim14, a novel key component of the import motor of the TIM23 protein translocase of mitochondria. EMBO J. 22:4945–4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Neupert W, Herrmann JM. 2007. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76:723–749 [DOI] [PubMed] [Google Scholar]
- 41. Nguyen KV, et al. 2005. POLG mutations in Alpers syndrome. Neurology 65:1493–1495 [DOI] [PubMed] [Google Scholar]
- 42. Palmieri L, et al. 1999. The mitochondrial dicarboxylate carrier is essential for the growth of Saccharomyces cerevisiae on ethanol or acetate as the sole carbon source. Mol. Microbiol. 31:569–577 [DOI] [PubMed] [Google Scholar]
- 43. Palmisano A, et al. 1998. Targeting and assembly of the oxoglutarate carrier: general principles for biogenesis of carrier proteins of the mitochondrial inner membrane. Biochem. J. 333:151–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Paschen S, et al. 2000. The role of the TIM8-13 complex in the import of Tim23 into mitochondria. EMBO J. 19:6392–6400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pfanner N, Neupert W. 1987. Distinct steps in the import of ADP/ATP carrier into mitochondria. J. Biol. Chem. 262:7528–7536 [PubMed] [Google Scholar]
- 46. Pfanner N, Tropschug M, Neupert W. 1987. Mitochondrial protein import: nucleoside triphosphates are involved in conferring import-competence to precursors. Cell 49:815–823 [DOI] [PubMed] [Google Scholar]
- 47. Pohlmeyer K, Soll J, Steinkamp T, Hinnah S, Wagner R. 1997. Isolation and characterization of an amino acid-selective channel protein present in the chloroplastic outer envelope membrane. Proc. Natl. Acad. Sci. U. S. A. 94:9504–9509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rassow J, Dekker PJ, van Wilpe S, Meijer M, Soll J. 1999. The preprotein translocase of the mitochondrial inner membrane: function and evolution. J. Mol. Biol. 286:105–120 [DOI] [PubMed] [Google Scholar]
- 49. Rehling P, Brandner K, Pfanner N. 2004. Mitochondrial import and the twin-pore translocase. Nat. Rev. Mol. Cell Biol. 5:519–530 [DOI] [PubMed] [Google Scholar]
- 50. Rehling P, et al. 2003. Protein insertion into the mitochondria inner membrane by a twin-pore translocase. Science 299:1747–1751 [DOI] [PubMed] [Google Scholar]
- 51. Reinhold R, et al. 2011. Mimicking a SURF1 allele reveals uncoupling of cytochrome c oxidase assembly from translational regulation in yeast. Hum. Mol. Genet. 20:2379–2393 [DOI] [PubMed] [Google Scholar]
- 52. Rokka A, et al. 2009. Pxmp2 is a channel-forming protein in mammalian peroxisomal membrane. PLoS One 4:e5090 doi:10.1371/journal.pone.0005090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ryan M, Müller H, Pfanner N. 1999. Functional staging of ADP/ATP carrier translocation across the outer mitochondrial membrane. J. Biol. Chem. 274:20619–20627 [DOI] [PubMed] [Google Scholar]
- 54. Saada A, et al. 2001. Mutant mitochondrial thymidine kinase in mitochondrial DNA depletion myopathy. Nat. Genet. 29:342–344 [DOI] [PubMed] [Google Scholar]
- 55. Schägger H. 2002. Respiratory chain supercomplexes of mitochondria and bacteria. Biochim. Biophys. Acta 1555:154–159 [DOI] [PubMed] [Google Scholar]
- 56. Schulz C, et al. 2011. Tim50's presequence receptor domain is essential for signal driven transport across the TIM23 complex. J. Cell Biol. 195:643–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sickmann A, et al. 2003. The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. U. S. A. 100:13207–13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sirrenberg C, Bauer MF, Guiard B, Neupert W, Brunner M. 1996. Import of carrier proteins into the mitochondrial inner membrane mediated by Tim22. Nature 384:582–585 [DOI] [PubMed] [Google Scholar]
- 59. Sirrenberg C, et al. 1998. Carrier protein import into mitochondria mediated by the intermembrane space proteins Tim10/Mrs11 and Tim 12/Mrs5. Nature 391:912–915 [DOI] [PubMed] [Google Scholar]
- 60. Smart OS, Breed J, Smith GR, Sansom MS. 1997. A novel method for structure-based prediction of ion channel conductance properties. Biophys. J. 72:1109–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Söllner T, Pfaller R, Griffiths G, Pfanner N, Neupert W. 1990. A mitochondrial import receptor for the ADP/ATP carrier. Cell 62:107–115 [DOI] [PubMed] [Google Scholar]
- 62. Spinazzola A, et al. 2008. Hepatocerebral form of mitochondrial DNA depletion syndrome: novel MPV17 mutations. Arch. Neurol. 65:1108–1113 [DOI] [PubMed] [Google Scholar]
- 63. Spinazzola A, et al. 2006. MPV17 encodes an inner mitochondrial membrane protein and is mutated in infantile hepatic mitochondrial DNA depletion. Nat. Genet. 38:570–575 [DOI] [PubMed] [Google Scholar]
- 64. Spinazzola A, Zeviani M. 2005. Disorders of nuclear-mitochondrial intergenomic signaling. Gene 354:162–168 [DOI] [PubMed] [Google Scholar]
- 65. Trott A, Morano K. 2004. SYM1 is the stress-induced Saccharomyces cerevisiae ortholog of the mammalian kidney disease gene Mpv17 and is required for ethanol metabolism and tolerance during heat shock. Eukaryot. Cell 3:620–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Truscott K, et al. 2001. A presequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by Tim23. Nat. Struct. Biol. 8:1074–1082 [DOI] [PubMed] [Google Scholar]
- 67. Truscott K, et al. 2002. Mitochondrial import of the ADP/ATP carrier: the essential TIM complex of the intermembrane space is required for precursor release from the TOM complex. Mol. Cell. Biol. 22:7780–7789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Truscott KN, et al. 2003. A J-protein is an essential subunit of the presequence translocase-associated protein import motor of mitochondria. J. Cell Biol. 163:707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Viscomi C, et al. 2009. Early-onset liver mtDNA depletion and late-onset proteinuric nephropathy in Mpv17 knockout mice. Hum. Mol. Genet. 18:12–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Voos W, Gambill B, Guiard B, Pfanner N, Craig E. 1993. Presequence and mature part of preproteins strongly influence the dependence of mitochondrial protein import on heat shock protein 70 in the matrix. J. Cell Biol. 123:119–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wagner K, et al. 2008. The assembly pathway of the mitochondrial carrier translocase involves four preprotein translocases. Mol. Cell. Biol. 28:4251–4260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Weiher H, Noda T, Gray DA, Sharpe AH, Jaenisch R. 1990. Transgenic mouse model of kidney disease: insertional inactivation of ubiquitously expressed gene leads to nephrotic syndrome. Cell 62:425–434 [DOI] [PubMed] [Google Scholar]
- 73. Westermann B, Neupert W. 1997. Mdj2p, a novel DnaJ homolog in the mitochondrial inner membrane of the yeast Saccharomyces cerevisiae. J. Mol. Biol. 272:477–483 [DOI] [PubMed] [Google Scholar]
- 74. Wiedemann N, Pfanner N, Rehling P. 2006. Import of precursor proteins into isolated yeast mitochondria. Methods Mol. Biol. 313:373–383 [DOI] [PubMed] [Google Scholar]
- 75. Wiedemann N, Pfanner N, Ryan MT. 2001. The three modules of ADP/ATP carrier cooperate in receptor recruitment and translocation into mitochondria. EMBO J. 20:951–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wittig I, Braun HP, Schägger H. 2006. Blue native PAGE. Nat. Protoc. 1:418–428 [DOI] [PubMed] [Google Scholar]
- 77. Wong LJC, et al. 2007. Mutations in the MPV17 gene are responsible for rapidly progressive liver failure in infancy. Hepatology 46:1218–1227 [DOI] [PubMed] [Google Scholar]
- 78. Yao J, Shoubridge EA. 1999. Expression and functional analysis of SURF1 in Leigh syndrome patients with cytochrome c oxidase deficiency. Hum. Mol. Genet. 8:2541–2549 [DOI] [PubMed] [Google Scholar]
- 79. Young J, Hoogenraad N, Hartl F. 2003. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 112:41–50 [DOI] [PubMed] [Google Scholar]
- 80. Zara V, Ferramosca A, Palmisano I, Palmieri F, Rassow J. 2003. Biogenesis of rat mitochondrial citrate carrier (CIC): the N-terminal presequence facilitates the solubility of the preprotein but does not act as a targeting signal. J. Mol. Biol. 325:399–408 [DOI] [PubMed] [Google Scholar]
- 81. Zara V, Ferramosca A, Robitaille-Foucher P, Palmieri F, Young JC. 2009. Mitochondrial carrier protein biogenesis: role of the chaperones Hsc70 and Hsp90. Biochem. J. 419:369–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zara V, Palmieri F, Mahlke K, Pfanner N. 1992. The cleavable presequence is not essential for import and assembly of the phosphate carrier of mammalian mitochondria but enhances the specificity and efficiency of import. J. Biol. Chem. 267:12077–12081 [PubMed] [Google Scholar]
- 83. Zimmermann R, Neupert W. 1980. Transport of proteins into mitochondria. Posttranslational transfer of ADP/ATP carrier into mitochondria in vitro. Eur. J. Biochem. 109:217–229 [DOI] [PubMed] [Google Scholar]