Abstract
Protein lysine methylation occurs extensively in the Crenarchaeota, a major kingdom in the Archaea. However, the enzymes responsible for this type of posttranslational modification have not been found. Here we report the identification and characterization of the first crenarchaeal protein lysine methyltransferase, designated aKMT, from the hyperthermophilic crenarchaeon Sulfolobus islandicus. The enzyme was capable of transferring methyl groups to selected lysine residues in a substrate protein using S-adenosyl-l-methionine (SAM) as the methyl donor. aKMT, a non-SET domain protein, is highly conserved among crenarchaea, and distantly related homologs also exist in Bacteria and Eukarya. aKMT was active over a wide range of temperatures, from ∼25 to 90°C, with an optimal temperature at ∼60 to 70°C. Amino acid residues Y9 and T12 at the N terminus appear to be the key residues in the putative active site of aKMT, as indicated by sequence conservation and site-directed mutagenesis. Although aKMT was identified based on its methylating activity on Cren7, the crenarchaeal chromatin protein, it exhibited broad substrate specificity and was capable of methylating a number of recombinant Sulfolobus proteins overproduced in Escherichia coli. The finding of aKMT will help elucidate mechanisms underlining extensive protein lysine methylation and the functional significance of posttranslational protein methylation in crenarchaea.
INTRODUCTION
Protein lysine methylation, first reported in the flagellum protein of Salmonella enterica serovar Typhimurium about half a century ago (2), is now known to occur in all three domains of life (19, 32, 41). Like other forms of posttranslational modifications, e.g., phosphorylation, acetylation, and ubiquitilation, covalent methylation of lysine residues may serve to alter the physicochemical properties and regulate the biological functions of a protein (27, 49). In Eukarya, histones H3 and H4 are posttranslationally methylated at specific lysine residues of the N- and C-terminal extensions by site-specific methyltransferases, most of which contain the catalytic SET domain or dot1 domain. Depending on the sites of methylation, transcriptional activation or repression may occur (21, 32, 47, 50). Nonhistone proteins, such as cytochrome c, Rubisco, several ribosomal proteins, and transcriptional factors p53, TAF10, and AML1, are also targets for lysine methylation, but the roles of these modifications are less well known (10, 13, 28, 30, 31, 44, 54). In Bacteria, known lysine methylation is restricted to very few proteins, such as flagellar and ribosomal proteins (2, 11). The roles of these modifications are unclear, but the methylated proteins appear to be more resistant to proteolysis (42). Notably, unlike eukaryal histones, no bacterial chromatin proteins have been shown to be methylated.
An increasing number of archaeal proteins, especially those from hyperthermophilic crenarchaea, have been shown to undergo posttranslational lysine methylation. The early examples included ferredoxin from Sulfolobus acidocaldarius and glutamate dehydrogenase, aspartate aminotransferase, β-glycosidase, and ribosomal proteins from Sulfolobus solfataricus (20, 38, 40, 45, 46, 55). Methylation of lysine residues in the S. solfataricus glutamate dehydrogenase and β-glycosidase has been suggested to enhance thermal stability of the enzymes or reduce their susceptibility to denaturation and aggregation. The Euryarchaeota and the Crenarchaeota, two major archaeal kingdoms, employ histone homologs and Cren7/Sul7d, respectively, as the chromatin proteins (25, 52). Intriguingly, none of the archaeal histones is known to be methylated, consistent with their lack of N- and C-terminal tails, whereas both Sul7d and Cren7 are methylated to various extents (5, 25). Little difference was observed between the methylated and the unmethylated versions of Sul7d or Cren7 in interaction with DNA, but the level of lysine methylation of Sul7d proteins increased during heat shock, suggesting an undetermined functional role of the modification (5). More recently, Botting et al. showed that lysine methylation may occur more extensively in crenarchaea than previously thought (7). They identified 21 methyllysine residues from nine subunits of S. solfataricus RNA polymerase. In a limited survey of the Thermoproteus tenax proteome, they found 52 methyllysine residues in 30 different proteins. The widespread methylation in crenarchaea has been speculated to represent an adaptation of these organisms to growth in hyperthermal environments (7).
Although the list of archaeal proteins methylated at lysine residues has been growing rapidly, very little is currently known about the enzymes that catalyze the modification. The euryarchaeon Methanosarcina mazei synthesizes a SET domain protein capable of methylating a lysine residue in the chromatin protein MC1-α (37). However, homologs of this methyltransferase exist in only a few methanogens. Furthermore, no SET domain proteins have been identified in the sequenced genomes of crenarchaea, where lysine methylation is prevalent. In this report, we have identified and isolated the first crenarchaeal protein lysine methyltransferase, designated aKMT, from Sulfolobus islandicus by employing a purification scheme involving the use of SDS-PAGE separation of a partially purified protein sample, elution of individual proteins from the gel slices, and recovery of the active target protein by denaturation and renaturation. This methyltransferase is highly conserved among crenarchaea. Although it was identified for its ability to methylate Cren7, aKMT exhibits broad substrate specificity. The discovery of aKMT will allow a better understanding of mechanisms underlining extensive lysine methylation in crenarchaea and shed light on evolutionary relationships among methyltransferases from the three domains of life.
MATERIALS AND METHODS
Purification and identification of aKMT from S. islandicus.
S. islandicus Rey15A was grown at 75°C with shaking to an optical density at 600 nm of 1.0 in basal salt solution supplemented with 0.1% tryptone, 0.05% yeast extract, 0.2% Casamino Acids, and 0.2% sucrose (14). The cells were harvested by centrifugation, resuspended in 20 mM Tris-Cl (pH 6.8), 50 mM KCl, 1 mM EDTA, 1 mM dithiothreitol (DTT), and 10% (wt/vol) glycerol, and sonicated on ice. The cell extract was clarified by centrifugation at 20,000 × g for 30 min at 4°C. The supernatant was subjected to stepwise fractionation by ammonium sulfate precipitation at 25%, 50%, 75%, and 100% saturation. The precipitates were dialyzed against buffer A (20 mM Tris-Cl [pH 8.8], 50 mM KCl, 1 mM EDTA, 1 mM DTT, and 10% [wt/vol] glycerol) for 12 h at 8°C. The sample showing peak methyltransferase activity was loaded onto a HiTrap Q column (5 ml; GE Healthcare) preequilibrated with buffer A, and the column was washed with buffer A. The flowthrough fractions were pooled and subjected to a HiTrap S column (5 ml; GE) preequilibrated with buffer B (20 mM Tris-Cl [pH 6.8], 50 mM KCl, 1 mM EDTA, 1 mM DTT, and 10% [wt/vol] glycerol), and the column was washed with buffer B. The flowthrough fractions were pooled and loaded onto a Superdex G200 column (10/300; GE). Proteins were eluted with buffer A. Active fractions were combined, and an aliquot of the sample was subjected to SDS-PAGE. Following gel electrophoresis, the sample lane of the gel was cut into the indicated slices from top to bottom, covering the entire protein-containing range, and each gel slice was soaked at 37°C overnight in 0.5 M ammonium acetate, 10 mM magnesium acetate, 1 mM EDTA, and 0.1% SDS in a solution volume of 300 μl. Proteins in the eluates were processed for denaturation and renaturation as described previously (9) with modifications. The samples were precipitated with trichloroacetic acid (TCA), and the precipitates were dissolved in 6 M guanidine hydrochloride. After 2 h, the samples were dialyzed against renaturation buffer (20 mM Tris-Cl [pH 8.8], 50 mM KCl, 1 mM EDTA, 1 mM DTT, 10% [wt/vol] glycerol, 0.04% Tween 40). The dialyzed samples were assayed for methyltransferase activity. Each protein band in a gel slice, corresponding to that containing proteins active in the assay, was recovered from a Coomassie brilliant blue-stained gel run in parallel and identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS).
Overproduction and purification of recombinant wild-type and mutant aKMT proteins.
The wild-type aKMT gene (SiRe_1449) was amplified by high-fidelity PCR using the S. islandicus genomic DNA as the template (for primer sequences, see Table S1 in the supplemental material) and cloned into expression plasmid pET30a(+) (Novagen) between the NdeI and NotI sites. The resultant expression vector (pET30a-MT) was transformed into E. coli Rosetta(DE3). The sequence of the cloned insert was verified by DNA sequencing. The recombinant aKMT protein was overproduced after induction with 0.8 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG) when the culture was grown to an optical density at 600 nm of 0.7 at 37°C in LB medium supplemented with 50 μg/ml kanamycin and 34 μg/ml chloramphenicol. Following a 3-h induction, the cells were harvested by centrifugation, resuspended in lysis buffer (20 mM Tris-HCl [pH 8.8], 1 mM DTT, 0.1 mM EDTA, 50 mM KCl, 10% [wt/vol] glycerol) supplemented with protease inhibitors (50 μg/ml phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, 0.2 μg/ml benzamidine, 0.1 μg/ml leupeptin, 0.5 μg/ml pepstatin A) and sonicated on ice. The lysate was centrifuged at 20,000 × g for 30 min at 4°C, and the supernatant was heat treated at 70°C for 20 min. The sample was centrifuged at 20,000 × g for 30 min at 4°C. The supernatant was applied to a preequilibrated HiTrap Q column (5 ml; GE) and washed with buffer A. The flowthrough fractions were concentrated and loaded onto a Superdex G200 column (10/300; GE). Proteins were eluted with buffer A. Fractions were analyzed by 15% SDS-PAGE, and those containing pure methyltransferase were pooled.
Expression vectors for aKMT mutant proteins containing a single point mutation (L1A, S2A, Y3A, V4A, P5A, H6A, V7A, P8A, Y9A, V10A, P11A, T12A, P13A, E14A, K15A, V16A, V17A, R18A, R19A, D34K, or G36R) were constructed by introducing corresponding mutations into the wild-type aKMT gene on pET30-MT by using the Fast Mutagenesis system (TransGen Biotech, China) (for sequences of the mutagenic primers, see Table S1 in the supplemental material). N-terminal 6- and 12-amino-acid-residue deletion mutants (ND6 and ND12) were prepared by PCR using the S. islandicus genomic DNA as the template (for primer sequences, see Table S1). Overproduction and purification of the mutant proteins were carried out as described for wild-type aKMT.
Protein concentrations were determined by the Lowry method with bovine serum albumin (BSA) as the standard (36).
Preparation of Sulfolobus proteins.
Native Cren7 was purified from S. islandicus as described previously (25). Genes encoding 50S ribosomal proteins L11 from S. islandicus and C56 from S. solfataricus plasmid pSSVi (51) were cloned into expression plasmid pET30a(+) (Novagen) and overexpressed in E. coli Rosetta(DE3). The recombinant proteins were purified on a HiTrap S column (5 ml; GE) preequilibrated with buffer B, and the column was washed with buffer C (20 mM Tris-Cl [pH 6.8], 1 M KCl, 1 mM EDTA, 1 mM DTT, and 10% [wt/vol] glycerol). S. solfataricus chromatin and replication proteins RFC, Pri1/Pri2, Dpo4, PCNA, Topo III, PolB1, GINS, FEN1, Cren7, and Sso7d2 were overproduced in E. coli and purified as described previously (8, 12, 15, 17, 25, 26, 35, 39, 43, 53).
Methyltransferase activity assays.
The standard methyltransferase assay mixture (50 μl) contained 20 mM HEPES-NaOH (pH 8.0), 2 mM MgCl2, 250 nM methyltransferase, 8 μM Cren7, and 4 μM S-adenosylmethionine (SAM; Sigma) including 200 nM S-[methyl-3H]adenosyl-l-methionine (10 Ci/mmol; Perkin-Elmer). Reaction mixtures were incubated for 90 min at 50°C. Proteins were precipitated with 20% TCA, and the pellets were washed with acetone and left to air dry. The samples were resuspended in 20 mM Tris-HCl (pH 8.8) and counted in Ultima Gold cocktails (Perkin-Elmer) on a MicroBeta2 LumiJET liquid scintillation counter (2460-0010; Perkin-Elmer).
For fluorographic analysis, the proteins from the above reactions were resolved by 15% SDS-PAGE. The gel was immersed for 1 h in EN3HANCE (Perkin-Elmer), washed for 25 min in deionized water, and soaked for 10 min in 10% acetic acid and 2% glycerol. The gel was then exposed to X-ray film.
Mass spectrometry.
The extent of methylation of Cren7 from S. islandicus was determined by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS). Sites of methylation of proteins by aKMT in vitro were determined as described previously (25, 29). Briefly, methylation reaction samples were subjected to 15% SDS-PAGE. The gel was stained with ZnSO4-imidazole, and proteins were digested in-gel with trypsin (Promega). The tryptic fragments were subjected to LC-MS/MS on an AB SCIEX TripleTOF 5600 system.
RESULTS
Purification and identification of aKMT from S. islandicus.
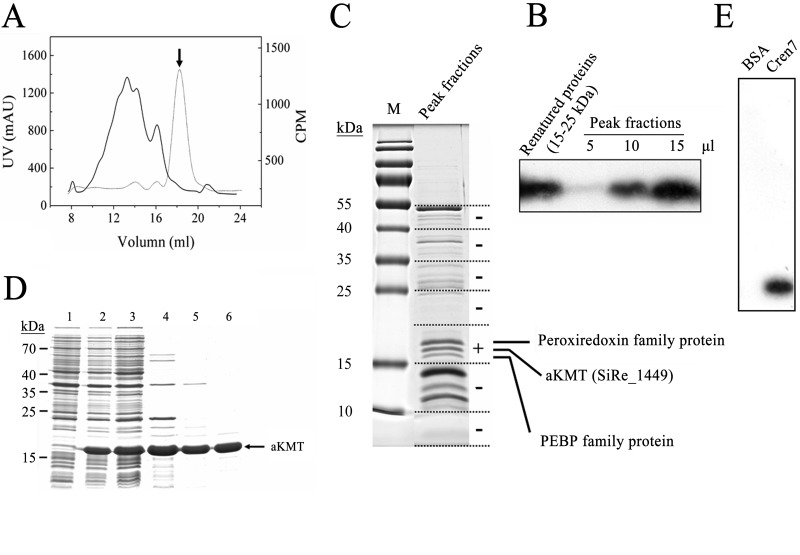
We showed previously that Cren7 from the hyperthermophilic archaeon Sulfolobus shibatae was methylated to various degrees at some of its 12 lysine residues (25). In a subsequent study, we found that a total of five lysine residues, i.e., K5, K11, K16, K31, and K42, were methylated in S. solfataricus Cren7 (see Table S2 in the supplemental material), which is identical to its S. shibatae homolog in amino acid sequence. At least one of these residues, i.e., K42, was also found to exist in an unmethylated form. To search for the lysine methyltransferase capable of methylating Cren7, we established a methyltransferase assay using unmethylated recombinant Cren7, overproduced in E. coli, as the substrate and radiolabeled SAM as the methyl donor. We found that the crude cell extract of S. islandicus was active in the methylation assay (see Fig. S1A in the supplemental material). The substrate Cren7 was methylated as shown by subjecting the reaction mixture to SDS-PAGE followed by fluorography (see Fig. S1B). The cell extract was subsequently fractionated by ammonium sulfate precipitation. The methyltransferase activity precipitated between 50 and 75% (NH4)2SO4 saturation. The precipitated proteins were then subjected sequentially to chromatography on HiTrap Q and HiTrap S columns. The activity bound to neither column under the experimental conditions. However, successive passage through the two columns drastically reduced the number of other proteins in the sample. The partially purified sample was further fractionated on a Superdex G200 gel filtration column (Fig. 1A and B). Proteins in the active fraction from the gel filtration column were separated by SDS-PAGE, and the gel-resolved proteins were assayed for methyltransferase activity following protein elution, denaturation, and refolding. A protein capable of methylating Cren7 was finally purified and identified as the product of SiRe_1449, a gene predicted to encode a homolog of ribosomal protein L11 methyltransferase in bacteria (24), by mass spectrometry (Fig. 1C).
Fig 1.
Identification and purification of aKMT from Sulfolobus islandicus. (A) Gel filtration of a partially purified S. islandicus protein sample active in Cren7 methylation assays on Superdex G200. The sample had been through (NH4)2SO4 fractionation and successive chromatography on HiTrap Q and HiTrap S columns. The UV absorbance at 280 nm (solid line) and the methyltransferase activity (dotted line) of the column fractions are shown. The peak fraction, indicated by an arrow, displayed the highest methyltransferase activity. (B) Fluorographic analysis of methylation of Cren7 by the peak fractions and refolded proteins from the 15–25 kDa gel slice. Aliquots of the peak fractions and refolded samples from the 15-to-25-kDa gel slice were incubated with recombinant Cren7 in the presence of [3H]SAM. Proteins were precipitated with TCA, and the pellets were subjected to SDS-PAGE. The gel was processed for fluorography. (C) Separation of proteins in the peak fractions by SDS-PAGE and identification of proteins in a gel slice containing proteins active in Cren7 methylation assays by mass spectrometry. A sample of the peak fractions was resolved by 15% SDS-PAGE. The protein-containing portion of a sample lane was cut into slices, as shown by the dashed lines. Proteins in each gel slice were eluted, denatured, and renatured. The renatured samples were assayed for methyltransferase activity. The results of the assays are shown on the right. The gel slice that contained proteins active in the assay is indicated by a plus sign. The three visible protein bands in the slice showing methyltransferase activity were individually recovered from a separate gel, and each protein was identified by LC-MS/MS. (D) Overproduction and purification of recombinant aKMT protein. Samples taken at various steps during the overproduction and purification of aKMT were subjected to SDS-PAGE. Lane 1, total cellular proteins from cells before induction; lane 2, total cellular proteins from cells after induction; lane 3, supernatant of the cell lysate; lane 4, supernatant of the heat-treated sample; lane 5, flowthrough fractions of the HiTrap Q column; lane 6, fractions of the Superdex G75 column. Molecular mass markers are indicated on the left. (E) Methylation of Cren7 by recombinant aKMT. Recombinant aKMT was incubated with recombinant Cren7 in the presence of [3H]SAM. The samples were processed for fluorography as described above.
To verify that SiRe_1449 encodes a protein lysine methyltransferase, the gene was cloned and overexpressed in E. coli. The purified recombinant protein was found to methylate recombinant Cren7, primarily at the K11, K16, and K31 residues under the assay conditions (Fig. 1D and E and 2A; see also Fig. S2A to C in the supplemental material). Furthermore, we coexpressed SiRe_1449 with the Cren7 gene in E. coli. The recombinant Cren7 protein was found to be methylated also at these three lysine residues (Fig. 2B; see also Fig. S2D to F). Since Cren7 was not methylated when overproduced alone in the same E. coli host, as shown previously (56), the observed methylation of Cren7 should have resulted from the action of the protein encoded by SiRe_1449. Therefore, we conclude that the protein encoded by SiRe_1449 is capable of methylating Cren7. We have designated this protein archaeal protein lysine methyltransferase (aKMT).
Fig 2.
Methylation of recombinant Cren7 proteins by aKMT. (A) MALDI-TOF MS spectrum of the recombinant S. solfataricus Cren7 protein methylated by aKMT in vitro. Recombinant Cren7 was incubated with recombinant aKMT in the presence of SAM and analyzed by MALDI-TOF MS. Methylated lysine residues are labeled. (B) MALDI-TOF MS spectrum of the recombinant S. islandicus Cren7 protein coproduced with aKMT in E. coli. Genes encoding Cren7 and aKMT from S. islandicus were coexpressed in E. coli. The recombinant Cren7 was purified and subjected to MALDI-TOF MS. Molecular masses (as the mass/charge ratio) for peaks corresponding to unmethylated Cren7 (Um) and Cren7 containing one, two, and three methylated lysine residues are indicated. (Insets) Methylated lysine residues in the partial amino acid sequences of Cren7 are labeled.
Three of the five lysine residues methylated in native Cren7 were methylated by aKMT in vitro and in a heterologous host. The discrepancy in the extent of Cren7 methylation presumably arose from the differences in methylation efficiencies under the different conditions. An alternative possibility is that there are additional methyltransferases capable of methylating Cren7 in Sulfolobus, although aKMT appeared to be the primary methylase serving this role in the cell (see below). The observation that Cren7 was methylated at the same lysine residues both in vitro and in E. coli agrees with the suggestion that lysine methylation by aKMT proceeds in a posttranslational manner in S. islandicus (7).
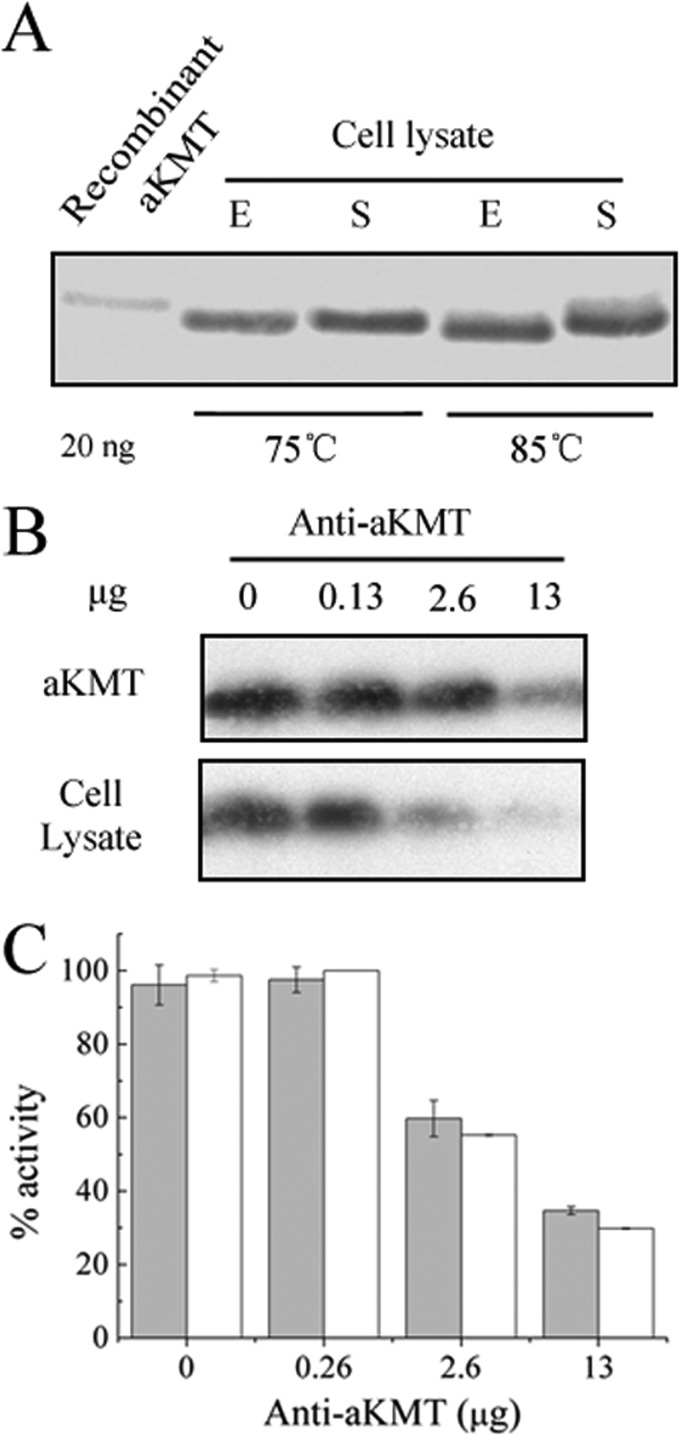
We then determined the cellular contents of aKMT in S. islandicus during different growth phases and at different temperatures by immunoblotting with rabbit antibodies against purified recombinant aKMT. As shown in Fig. 3A, whether grown at 75 or 85°C, or in the exponential or the stationary phase, the intracellular levels of the protein appeared to be similar.
Fig 3.
Inhibition of the Cren7-methylating activity of the S. islandicus cell extract by anti-aKMT antibodies. (A) Abundance of aKMT in S. islandicus. S. islandicus was grown to the exponential (E) or stationary (S) phase at 75° or 85°C. Samples were taken, centrifuged, and subjected to SDS-PAGE. aKMT was detected by immunoblotting using anti-aKMT antibodies. (B and C) Inhibition by anti-aKMT antibodies of the ability of recombinant aKMT and the S. islandicus cell extract to methylate recombinant Cren7. A sample of the S. islandicus cell extract or recombinant aKMT at a concentration similar to that in the extract was incubated with recombinant Cren7 and [3H]SAM in the presence of increasing amounts of anti-aKMT antibodies. Methyl transfer to Cren7 was determined by fluorography (B) and liquid scintillation counting (C). Reaction mixtures containing recombinant aKMT and the cell extract are shown in gray and white, respectively, in the fluorogram (C). All numbers are the averages of three independent measurements.
Methylation of recombinant Cren7 by aKMT was sensitive to inhibition by anti-aKMT antibodies (Fig. 3B and C). Based on this observation, we performed an antibody-based inhibition assay of Cren7 methylation by the S. islandicus cell extract. As with the purified aKMT protein, methylation of Cren7 by the cell extract was significantly reduced in the presence of the antibodies (Fig. 3B and C). These results suggested that aKMT plays a major role in Cren7 methylation in the cell.
aKMT is conserved among crenarchaea.
Pfam analysis shows that aKMT contains a single conserved methyltransferase domain belonging to the Methyltransf_31 family (3). Members of this protein family have been found in a large number of organisms from all three domains of life. The conserved DxGxGxG signature motif (34-DLGCGDG-40) responsible for SAM binding has also been identified in the protein (18).
To investigate the phylogenetic distribution of aKMT homologs, we conducted a sequence homology-based search of the protein database by using BLAST (1). As shown in Fig. 4, aKMT is highly conserved only among the crenarchaeal lineage in the Archaea. All of the genome-sequenced crenarchaea encode one or more homologs of this protein, most of which share 36 to 80% similarities to the S. islandicus protein at the amino acid sequence level. In other Archaea, aKMT homologs are only present in the genera Methanobacterium and Methanopyrus. In Bacteria, aKMT homologs with low sequence similarity to the S. islandicus protein (∼20 to 40%) are found in more than 27 genera, most of which are cyanobacteria and proteobacteria (see Table S4 in the supplemental material). Homologs of aKMT with 15 to 30% sequence similarity to the S. islandicus protein are also present in the Eukarya, mostly in animals, such as insects, birds, fish, and mammals, including humans (see Table S4). Species of the genus Pinus are the only plants, in addition to a few eukaryotic algae, that contain aKMT homologs. The wide distribution of aKMT homologs in the three domains of life suggests an ancient origin of this protein family. However, the properties and function of these proteins have yet to be understood.
Fig 4.
Sequence alignment of aKMT homologs from Crenarchaea (genera preceded by a C), Euryarchaea (A), Bacteria (B), and Eukarya (E). Sequences are from Sulfolobus islandicus M.14.25 (gi|227827841), Desulfurococcus kamchatkensis 1221n (gi|218884617), Thermosphaera aggregans DSM 11486 (gi|296243046), Pyrolobus fumarii 1A (gi|347524257), Thermofilum pendens Hrk 5 (gi|119719282), Thermoproteus neutrophilus V24Sta (gi|171185324), Salinibacter ruber M8 (gi|294507034), Nostoc sp. PCC 7120 (gi|17229012), Acidilobus saccharovorans 345-15 (gi|302349255), Methanobacterium sp. SWAN-1 (gi|333987018), Pinus radiate (gi|361066863), Volvox carteri f. nagariensis (gi|302837301), Gallus gallus (gi|363739652), Danio rerio (gi|165972475), Xenopus laevis (gi|148225045), Homo sapiens (gi|154240720), Drosophila mojavensis (gi|195113923), Fervidicoccus fontis Kam940 (gi|385805356), Methanopyrus kandleri AV19 (gi|20093891), Streptomyces roseosporus NRRL 15998 (gi|291445786), and Bacillus cereus 03BB108 (gi|196045290). The amino acids that are conserved or have similar properties are shown in red letters. Region I includes the putative active site residues. The invariant amino acid residues responsible for SAM binding (region II) are shown in white letters against a red background.
Physicochemical properties of aKMT.
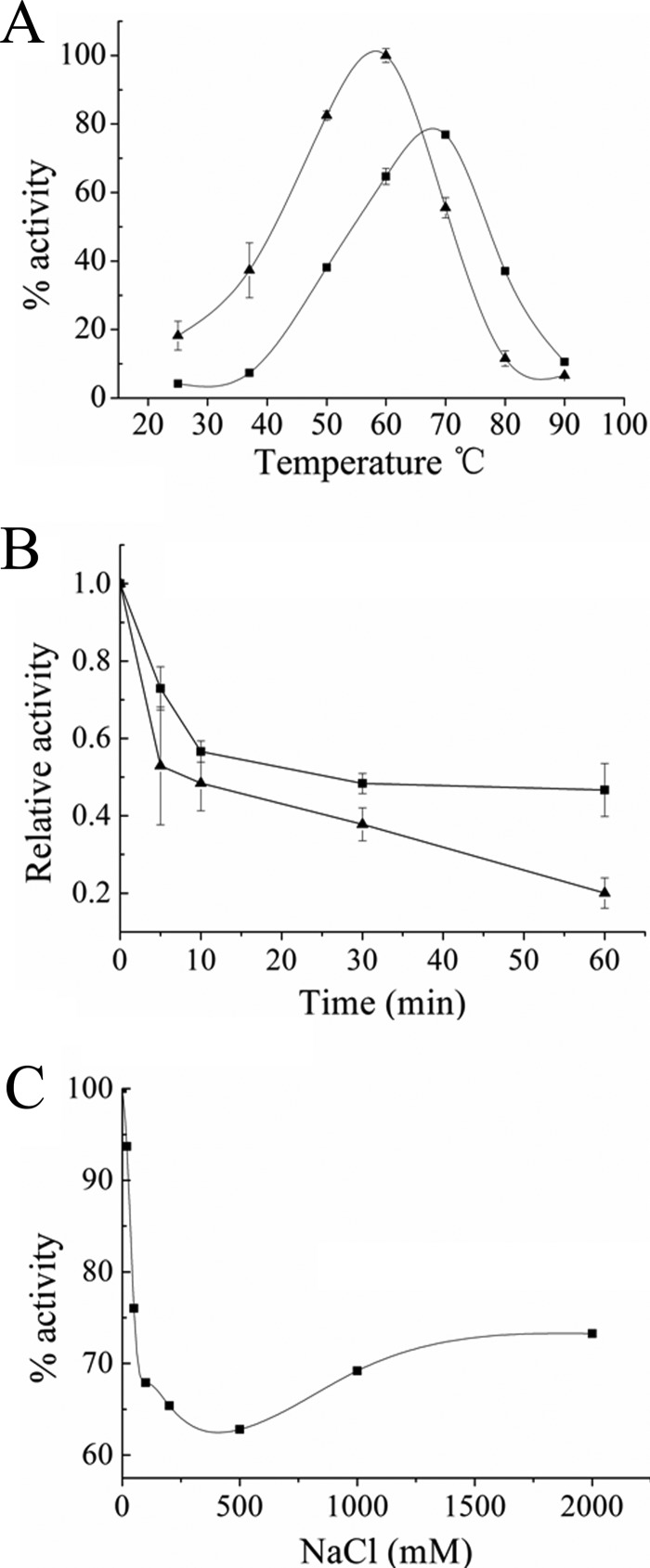
aKMT is a monomer in solution, as demonstrated by gel filtration (see Fig. S3 in the supplemental material). The enzyme was active over a wide temperature range, from 25 to 90°C, and showed the highest activity at ∼60°C when the reaction was allowed to proceed for 90 min (Fig. 5A). This is consistent with the finding that Cren7 was methylated by aKMT when both proteins were synthesized in E. coli growing at 37°C (Fig. 2B). The temperature optimum of aKMT was up-shifted to ∼70°C in a 10-min reaction (Fig. 5A), suggesting that the enzyme, while being capable of the most efficient methyl transfer near the physiological growth temperature of the organism, is not as thermostable as might be expected under the experimental conditions employed. Indeed, we found that the enzyme had half-lives of ∼30 and ∼10 min at 80 and 90°C, respectively (Fig. 5B). The aKMT protein required no divalent cations for its activity. The methylation activity of the enzyme was sensitive to inhibition by high levels of NaCl (Fig. 5C). This appears to be consistent with the low salt content in Sulfolobus cells (23).
Fig 5.
Physicochemical properties of recombinant aKMT protein. (A) Effect of temperature on the methyltransferase activity of aKMT. aKMT was incubated with recombinant Cren7 for 10 min (■) or 90 min (▲) at the indicated temperatures in the presence of [3H]SAM. The samples were precipitated with TCA, and radioactivity in the pellets was counted in a liquid scintillation counter. The highest activity obtained in the 90-min assay was defined as 100%. (B) Thermostability of aKMT. aKMT was incubated for the indicated lengths of time at 80°C (■) or 90°C (▲). The methyltransferase activity of the enzyme was then measured with recombinant Cren7 as the substrate at 50°C. (C) Effect of NaCl concentration on the methyltransferase activity of aKMT. aKMT was incubated with recombinant Cren7 and [3H]SAM for 90 min at 50°C in the presence of NaCl at the indicated concentrations. The samples were treated as described above for liquid scintillation counting. All numbers are the averages of three independent measurements.
Identification of key residues for the activity of aKMT.
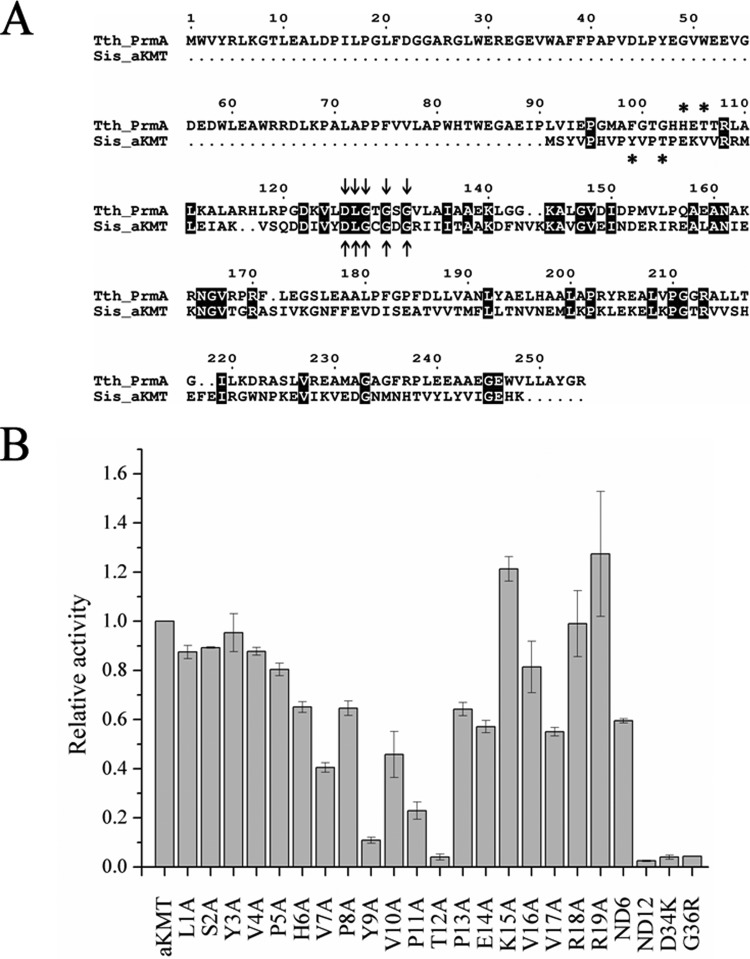
The aKMT protein from S. islandicus was originally annotated as a putative ribosomal protein L11 methyltransferase (24). Comparison between aKMT and the L11 methyltransferase PrmA from the hyperthermophilic bacterium Thermus thermophilus showed that the two proteins share low similarity at the amino acid sequence level (Fig. 6A). The N-terminal domain of PrmA responsible for L11 recognition is absent from aKMT, suggesting that the archaeal enzyme may have a less stringent substrate specificity than its bacterial counterpart. The amino acid sequence of aKMT is well aligned with that of the C-terminal catalytic domain of PrmA. Notably, both proteins contain the conserved DxGxGxG motif for SAM binding. To verify the importance of the motif for the methyltransferase activity of aKMT, we constructed two single mutants in this signature sequence (D34K and G36R). As expected, the two mutants were almost completely inactive in the methyl transfer reaction (Fig. 6B).
Fig 6.
Identification of key residues for the activity of aKMT. (A) Sequence alignment of aKMT and L11 methyltransferase PrmA from Thermus thermophilus. The conserved DxGxGxG signature motif for the SAM binding site is labeled with arrows, and key residues for the activity are indicated by asterisks. (B) Methyltransferase activity assays of wild-type and mutant aKMT proteins. Wild-type or mutant aKMT was incubated with recombinant Cren7 and [3H]SAM. The samples were treated as described above for liquid scintillation counting. All numbers are the averages of three independent measurements.
Intriguingly, two key residues, i.e., H104 and T106, in the active site conserved in PrmA homologs are not present in aKMT (Fig. 6A). H6, a lone histidine residue in the N terminus of aKMT from S. islandicus, appears to be located too distal from the T12 residue and is not conserved among the aKMT homologs. Sequence alignment with other methyltransferases in the Methyltransf_31 family also revealed no known active site in aKMT (data not shown), raising the possibility that the aKMT homologs possess a unique active site. To gain insight into the active site of aKMT, we constructed two aKMT deletion mutants lacking the N-terminal 6 and 12 residues, termed ND6 and ND12, respectively. ND6 was about 70% as active as the wild-type enzyme, while ND12 was completely inactive in methyl transfer, indicating that residues from H6 to T12 may constitute the core active site (Fig. 6B). This is consistent with the suggestion that the first five residues at the N terminus of the S. islandicus aKMT are not required for the activity of the protein based on a sequence comparison of the N-terminal portions of aKMT proteins from S. islandicus and Pyrolobus fumarii 1A (Fig. 4). Indeed, as shown by alanine scanning of the 19 amino acid residues at the N terminus of S. islandicus aKMT, mutation of each of the first 5 residues did not affect the methyltransferase activity of the enzyme (Fig. 6B). However, single alanine substitution for each of the nine residues from H6 to E14 resulted in a significant decrease in methyl transfer by the mutant protein. Among the nine residues, Y9, P11, and T12 appeared to be the most critical, since their mutation reduced the methyltransferase activity of the protein by 89, 77, and 96%, respectively. By comparison, mutation at each of the other positions only decreased the activity of the protein by 30 to 55%. Single alanine substitution for the adjacent four residues, K15, V16, R18, and R19, had little effect on the activity of aKMT. However, mutation of V17 reduced the activity by ∼30%. V17 is highly conserved in the aKMT proteins and corresponds to T107 in PrmA, as shown by sequence alignment. It may be involved in SAM binding, as has been reported for T107 in PrmA (16). Taken together, these data suggest an important role for the 6-HVPYVPTPE-14 motif in methyl transfer by aKMT. In support of this notion, sequence analysis revealed that the motif 8-VPYVPTP-13 in the S. islandicus aKMT protein is highly conserved among aKMT homologs from the three domains of life. Consistent with their being essential for the activity of the aKMT, Y9 and T12 appear to be the most important of the seven conserved residues. Conservation of a threonine or serine residue at a position corresponding to T12 in aKMT proteins, or at an adjacent position in eukaryotic aKMT homologs, suggests that the hydroxyl group of the side chain of the amino acid residue is involved in methyl transfer. On the other hand, the presence of a tyrosine, phenylalanine, or tryptophan residue at a position corresponding to Y9 in aKMT points to a role for an aromatic ring in the reaction.
aKMT shows broad substrate specificity.
Since sequence analysis suggests that aKMT may have broad substrate specificity, we sought to verify this by performing the methyltransferase assay with aKMT on a range of recombinant S. solfataricus proteins overproduced in E. coli. As shown in Fig. 7, among the tested proteins, Sso7d2, ribosomal protein L11, both RFC subunits, Topo III, GINS15, both primase subunits (Pri1/Pri2), PolB1, PCNA1 and/or PCNA2, Dpo4, and FEN1 were all methylated by the enzyme to various extents in vitro. This was consistent with the observation that native L11, Sso7d2, and the RFC complex from S. solfataricus are methylated (5, 45, 46) (data not shown). By comparison, BSA and c56 from pSSVi did not appear to be measurably methylated. Our data suggest that aKMT is potentially capable of methylating a large number of proteins in crenarchaea.
Fig 7.
In vitro methylation by aKMT of the recombinant proteins encoded by S. solfotaricus. (A) aKMT was incubated with each of the tested proteins, as shown on a Coomassie brilliant blue-stained gel in the presence of [3H]SAM. (B) The samples were processed for fluorography as described above. Each panel comprises portions of separately run gels, as indicated by the dashed lines.
Taking advantage of the availability of the crystal structures of Sulfolobus Cren7 and Sso7d in complex with DNAs (22, 56), we looked into the structural context and the potential roles of lysine residues methylated in the native proteins (48). The modified lysine residues are located at the surface of the proteins, and the side chains of these residues are not involved in protein-DNA interactions. In other words, the amino group of each target lysine residue is solvent exposed and readily accessible to aKMT, even when the protein is bound to DNA. This agrees with the finding that Cren7 and Sul7d bind DNA equally well in both the methylated and unmethylated forms (4, 25). To learn if lysine methylation by aKMT shows sequence specificity, we analyzed 19 sites of methylation in recombinant Sulfolobus proteins (including Cren7, Sso7d2, FEN1, and Topo III) methylated by the enzyme in vitro by trypsin digestion and mass spectrometry. No apparent bias was detected in adjacent amino acid residues at these sites (see Fig. S4 in the supplemental material). Therefore, aKMT methylates lysine residues in a rather sequence-independent manner.
DISCUSSION
The finding of multiple methylated forms of Cren7 as well as the report of extensive protein methylation in crenarchaea prompted us to search for protein methyltransferases in this major kingdom of the Archaea. Sequence homology searches yielded no valuable clues, since no SET domain proteins were found in the crenarchaeal genomes. However, we found that the S. islandicus cell extract was able to transfer methyl groups from SAM to the lysine residues of Cren7. Inspired by a subsequent observation that the Cren7-methylating activity in the crude cell extract survived a protein denaturation/renaturation treatment, we designed a purification strategy involving separation of proteins by SDS-PAGE, elution of target proteins from the gel, and refolding of the denatured proteins. Use of this protocol led to the identification of aKMT, the first protein lysine methyltransferase to be discovered in crenarchaea.
Sequence analysis revealed that aKMT is evolutionarily highly conserved. Homologs of the protein are not only omnipresent in crenarchaea, but are also present in some lineages in Euryarchaea, Bacteria, and Eukarya. Examination of the DNA regions flanking the aKMT genes of various origins provided no evidence for cross-domain horizontal gene transfer. The wide distribution of these proteins among the three domains of life suggests that their ancestral form may have been present in the last universal common ancestor (LUCA). Presumably, gene loss has occurred in some lineages over the course of evolution. In the domain Archaea, the distribution of aKMT is distributed mainly in crenarchaea, with the genera Methanobacterium and Methanpyrus being the sole aKMT-containing noncrenarchaea. It would be of interest to learn if the lack of methylation of histones is due to the lack of aKMT in euryarchaea. All crenarchaeal genomes sequenced so far encode at least one copy of aKMT, suggesting an important role for the protein in cell physiology. Moreover, some crenarchaea encode distantly related aKMT paralogs (for example, five paralogs from Pyrolobus fumarii 1A share ∼40% sequence similarity), suggesting possible divergence of the proteins in function.
The aKMT proteins differ significantly from other known SAM-dependent methyltransferases at the amino acid sequence level, except for the SAM binding domain. As shown by alanine scanning and deletion analysis, aKMT from S. islandicus appears to possess a novel active site that is well conserved among the homologs of the protein. The N-terminal motif, 6-HVPYVPTPE-14, plays an important role in methyl transfer by the protein. Among these residues, T12 is especially critical, since its mutation completely inactivated the enzyme. As revealed by sequence alignment, threonine and serine are the only residues found at this position in aKMT homologs. The T/S conservation raises the possibility that the side chain hydroxyl group of the amino acid is directly involved in the catalysis of the methyltransfer reaction. In the T. thermophilus PrmA protein, whose C-terminal catalytic domain shows similarity to aKMT in amino acid sequence, the active site contains two key residues, H104 and T106 (16). The role of the T/S residue in aKMT may resemble that of T106. Y9 is another key residue in aKMT. Since tyrosine is replaced by either phenylalanine or tryptophan at this position in some of the aKMT homologs, an aromatic ring may be required for the activity of the enzyme. Mutation of H6 also resulted in reduction of the methylase activity of aKMT. Like H104 in TthPrmA, this residue may function as a potential general base in the deprotonation of the target lysine side chain. However, the role of H6 may not be essential, since this residue is not conserved among aKMT homologs.
A surprising finding from the sequence analysis of aKMT is that the protein appears to lack a typical substrate recognition and binding domain, an integral part of other known protein methyltransferases. This prompted the speculation that aKMT may exhibit broad substrate specificity. Indeed, we found that aKMT was capable of methylating not only multiple lysine residues in Cren7 but also several other recombinant S. solfataricus proteins. In contrast, the SET domain protein from M. mazei, the only other protein lysine methyltransferase found in the Archaea, showed strict substrate specificity (37). Given its broad substrate specificity, aKMT is presumably involved in extensive lysine methylation in crenarchaea in general and in Sulfolobus species in particular.
Protein lysine methylation has been shown to modulate protein-protein interactions, chromatin structure, gene expression, and protein stability. Eukaryotic histone methylation has been studied most extensively. Highly regulated methylation of selected lysine residues on the N- and C-terminal tails of histones by site-specific protein lysine methyltransferases affects chromatin structure and, thereby, DNA transactions, such as replication, transcription, and DNA repair (6, 33, 34). Protein methylation has been shown to occur widely in the crenarchaeal lineage of the Archaea (19). Among the methylated proteins are those involved in chromosomal organization (e.g., Cren7 and Sul7d), DNA replication (e.g., Pri1/Pri2 and RFC), and transcription (RNA polymerase) (7). The roles of the modification are largely unknown, although it has been shown that methylated proteins are more stable to thermal denaturation (20, 38). Since protein lysine methyltransferases such as aKMT are likely responsible for the extensive protein methylation in crenarchaea, a better understanding of these enzymes will shed considerable light on the physiological function of the posttranslational protein modification.
ACKNOWLEDGMENTS
We are grateful to Wei Sun for his assistance in the MS analysis. We also thank Zhengyan Zhan, Yi Ren, Xuanxuan Xing, Bing Liu, Shiwei Lang, Junhua Zhang, and Fei Sun for the gifts of recombinant S. solfotaricus proteins.
This work was supported by the National Natural Science Foundation of China (30730003, 30921065, and 31000022).
Footnotes
Published ahead of print 19 October 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Ambler RP, Rees MW. 1959. Epsilon-N-methyl-lysine in bacterial flagellar protein. Nature 184:56–57 [DOI] [PubMed] [Google Scholar]
- 3. Bateman A, et al. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138–D141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baumann H, Knapp S, Karshikoff A, Ladenstein R, Hard T. 1995. DNA-binding surface of the Sso7d protein from Sulfolobus solfataricus. J. Mol. Biol. 247:840–846 [DOI] [PubMed] [Google Scholar]
- 5. Baumann H, Knapp S, Lundback T, Ladenstein R, Hard T. 1994. Solution structure and DNA-binding properties of a thermostable protein from the archaeon Sulfolobus solfataricus. Nat. Struct. Biol. 1:808–819 [DOI] [PubMed] [Google Scholar]
- 6. Berger SL. 2007. The complex language of chromatin regulation during transcription. Nature 447:407–412 [DOI] [PubMed] [Google Scholar]
- 7. Botting CH, Talbot P, Paytubi S, White MF. 2010. Extensive lysine methylation in hyperthermophilic crenarchaea: potential implications for protein stability and recombinant enzymes. Archaea 2010:106341 doi:10.1155/2010/106341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boudsocq F, Iwai S, Hanaoka F, Woodgate R. 2001. Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4): an archaeal DinB-like DNA polymerase with lesion-bypass properties akin to eukaryotic poleta. Nucleic Acids Res. 29:4607–4616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burridge K, Nelson A. 1995. An in-gel assay for protein tyrosine phosphatase activity: detection of widespread distribution in cells and tissues. Anal. Biochem. 232:56–64 [DOI] [PubMed] [Google Scholar]
- 10. Chakraborty S, Sinha KK, Senyuk V, Nucifora G. 2003. SUV39H1 interacts with AML1 and abrogates AML1 transactivity. AML1 is methylated in vivo. Oncogene 22:5229–5237 [DOI] [PubMed] [Google Scholar]
- 11. Chang FN, Chang CN, Paik WK. 1974. Methylation of ribosomal proteins in Escherichia coli. J. Bacteriol. 120:651–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Choli T, Henning P, Wittmann-Liebold B, Reinhardt R. 1988. Isolation, characterization and microsequence analysis of a small basic methylated DNA-binding protein from the Archaebacterium, Sulfolobus solfataricus. Biochim. Biophys. Acta 950:193–203 [DOI] [PubMed] [Google Scholar]
- 13. Chuikov S, et al. 2004. Regulation of p53 activity through lysine methylation. Nature 432:353–360 [DOI] [PubMed] [Google Scholar]
- 14. Contursi P, et al. 2006. Characterization of the Sulfolobus host-SSV2 virus interaction. Extremophiles 10:615–627 [DOI] [PubMed] [Google Scholar]
- 15. Dai P, Wang Y, Ye R, Chen L, Huang L. 2003. DNA topoisomerase III from the hyperthermophilic archaeon Sulfolobus solfataricus with specific DNA cleavage activity. J. Bacteriol. 185:5500–5507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Demirci H, Gregory ST, Dahlberg AE, Jogl G. 2007. Recognition of ribosomal protein L11 by the protein trimethyltransferase PrmA. EMBO J. 26:567–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dionne I, Nookala RK, Jackson SP, Doherty AJ, Bell SD. 2003. A heterotrimeric PCNA in the hyperthermophilic archaeon Sulfolobus solfataricus. Mol. Cell 11:275–282 [DOI] [PubMed] [Google Scholar]
- 18. Eddy SR. 1996. Hidden Markov models. Curr. Opin. Struct. Biol. 6:361–365 [DOI] [PubMed] [Google Scholar]
- 19. Eichler J, Adams MW. 2005. Posttranslational protein modification in Archaea. Microbiol. Mol. Biol. Rev. 69:393–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Febbraio F, et al. 2004. Thermal stability and aggregation of sulfolobus solfataricus beta-glycosidase are dependent upon the N-epsilon-methylation of specific lysyl residues: critical role of in vivo post-translational modifications. J. Biol. Chem. 279:10185–10194 [DOI] [PubMed] [Google Scholar]
- 21. Feng Q, et al. 2002. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 12:1052–1058 [DOI] [PubMed] [Google Scholar]
- 22. Gao YG, et al. 1998. The crystal structure of the hyperthermophile chromosomal protein Sso7d bound to DNA. Nat. Struct. Biol. 5:782–786 [DOI] [PubMed] [Google Scholar]
- 23. Green GR, Searcy DG, DeLange RJ. 1983. Histone-like protein in the Archaebacterium Sulfolobus acidocaldarius. Biochim. Biophys. Acta 741:251–257 [DOI] [PubMed] [Google Scholar]
- 24. Guo L, et al. 2011. Genome analyses of Icelandic strains of Sulfolobus islandicus, model organisms for genetic and virus-host interaction studies. J. Bacteriol. 193:1672–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guo L, et al. 2008. Biochemical and structural characterization of Cren7, a novel chromatin protein conserved among Crenarchaea. Nucleic Acids Res. 36:1129–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Horie M, et al. 2007. The N-terminal region is important for the nuclease activity and thermostability of the flap endonuclease-1 from Sulfolobus tokodaii. Biosci. Biotechnol. Biochem. 71:855–865 [DOI] [PubMed] [Google Scholar]
- 27. Jenuwein T, Allis CD. 2001. Translating the histone code. Science 293:1074–1080 [DOI] [PubMed] [Google Scholar]
- 28. Kluck RM, et al. 2000. Determinants of cytochrome c pro-apoptotic activity. The role of lysine 72 trimethylation. J. Biol. Chem. 275:16127–16133 [DOI] [PubMed] [Google Scholar]
- 29. Kolarich D, Jensen PH, Altmann F, Packer NH. 2012. Determination of site-specific glycan heterogeneity on glycoproteins. Nat. Protoc. 7:1285–1298 [DOI] [PubMed] [Google Scholar]
- 30. Kouskouti A, Scheer E, Staub A, Tora L, Talianidis I. 2004. Gene-specific modulation of TAF10 function by SET9-mediated methylation. Mol. Cell 14:175–182 [DOI] [PubMed] [Google Scholar]
- 31. Kruiswijk T, Kunst A, Planta RJ, Mager WH. 1978. Modification of yeast ribosomal proteins: methylation. Biochem. J. 175:221–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lachner M, Jenuwein T. 2002. The many faces of histone lysine methylation. Curr. Opin. Cell Biol. 14:286–298 [DOI] [PubMed] [Google Scholar]
- 33. Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116–120 [DOI] [PubMed] [Google Scholar]
- 34. Lake AN, Bedford MT. 2007. Protein methylation and DNA repair. Mutat. Res. 618:91–101 [DOI] [PubMed] [Google Scholar]
- 35. Lou H, Duan Z, Sun T, Huang L. 2004. Cleavage of double-stranded DNA by the intrinsic 3′–5′ exonuclease activity of DNA polymerase B1 from the hyperthermophilic archaeon Sulfolobus solfataricus at high temperature. FEMS Microbiol. Lett. 231:111–117 [DOI] [PubMed] [Google Scholar]
- 36. Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265–275 [PubMed] [Google Scholar]
- 37. Manzur KL, Zhou MM. 2005. An archaeal SET domain protein exhibits distinct lysine methyltransferase activity towards DNA-associated protein MC1-alpha. FEBS Lett. 579:3859–3865 [DOI] [PubMed] [Google Scholar]
- 38. Maras B, et al. 1992. The protein sequence of glutamate dehydrogenase from Sulfolobus solfataricus, a thermoacidophilic archaebacterium. Is the presence of N-epsilon-methyllysine related to thermostability? Eur. J. Biochem. 203:81–87 [DOI] [PubMed] [Google Scholar]
- 39. Marinsek N, et al. 2006. GINS, a central nexus in the archaeal DNA replication fork. EMBO Rep. 7:539–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Minami Y, et al. 1985. Amino acid sequence of a ferredoxin from thermoacidophilic archaebacterium, Sulfolobus acidocaldarius. Presence of an N6-monomethyllysine and phyletic consideration of archaebacteria. J. Biochem. 97:745–753 [DOI] [PubMed] [Google Scholar]
- 41. Paik WK, Paik DC, Kim S. 2007. Historical review: the field of protein methylation. Trends Biochem. Sci. 32:146–152 [DOI] [PubMed] [Google Scholar]
- 42. Pethe K, et al. 2002. Mycobacterial heparin-binding hemagglutinin and laminin-binding protein share antigenic methyllysines that confer resistance to proteolysis. Proc. Natl. Acad. Sci. U. S. A. 99:10759–10764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pisani FM, De Felice M, Carpentieri F, Rossi M. 2000. Biochemical characterization of a clamp-loader complex homologous to eukaryotic replication factor C from the hyperthermophilic archaeon Sulfolobus solfataricus. J. Mol. Biol. 301:61–73 [DOI] [PubMed] [Google Scholar]
- 44. Polevoda B, Martzen MR, Das B, Phizicky EM, Sherman F. 2000. Cytochrome c methyltransferase, Ctm1p, of yeast. J. Biol. Chem. 275:20508–20513 [DOI] [PubMed] [Google Scholar]
- 45. Ramirez C, Shimmin LC, Dennis PP, Matheson AT. 1991. Comparison of the structure of archaebacterial ribosomal proteins equivalent to proteins L11 and L1 from Escherichia coli ribosomes. Protein Seq. Data Anal. 4:75–79 [PubMed] [Google Scholar]
- 46. Ramirez C, Shimmin LC, Newton CH, Matheson AT, Dennis PP. 1989. Structure and evolution of the L11, L1, L10, and L12 equivalent ribosomal proteins in eubacteria, archaebacteria, and eucaryotes. Can. J. Microbiol. 35:234–244 [DOI] [PubMed] [Google Scholar]
- 47. Rea S, et al. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593–599 [DOI] [PubMed] [Google Scholar]
- 48. Snijders AP, Hung ML, Wilson SA, Dickman MJ. 2010. Analysis of arginine and lysine methylation utilizing peptide separations at neutral pH and electron transfer dissociation mass spectrometry. J. Am. Soc. Mass Spectrom. 21:88–96 [DOI] [PubMed] [Google Scholar]
- 49. Strahl BD, Allis CD. 2000. The language of covalent histone modifications. Nature 403:41–45 [DOI] [PubMed] [Google Scholar]
- 50. Tschiersch B, et al. 1994. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 13:3822–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang Y, et al. 2007. A novel Sulfolobus non-conjugative extrachromosomal genetic element capable of integration into the host genome and spreading in the presence of a fusellovirus. Virology 363:124–133 [DOI] [PubMed] [Google Scholar]
- 52. White MF, Bell SD. 2002. Holding it together: chromatin in the Archaea. Trends Genet. 18:621–626 [DOI] [PubMed] [Google Scholar]
- 53. Wu K, et al. 2007. Interplay between primase and replication factor C in the hyperthermophilic archaeon Sulfolobus solfataricus. Mol. Microbiol. 63:826–837 [DOI] [PubMed] [Google Scholar]
- 54. Ying Z, Mulligan RM, Janney N, Houtz RL. 1999. Rubisco small and large subunit N-methyltransferases: bi- and mono-functional methyltransferases that methylate the small and large subunits of Rubisco. J. Biol. Chem. 274:36750–36756 [DOI] [PubMed] [Google Scholar]
- 55. Zappacosta F, Sannia G, Savoy LA, Marino G, Pucci P. 1994. Post-translational modifications in aspartate aminotransferase from Sulfolobus solfataricus. Eur. J. Biochem. 222:761–767 [DOI] [PubMed] [Google Scholar]
- 56. Zhang Z, Gong Y, Guo L, Jiang T, Huang L. 2010. Structural insights into the interaction of the crenarchaeal chromatin protein Cren7 with DNA. Mol. Microbiol. 76:749–759 [DOI] [PubMed] [Google Scholar]