Abstract
LapF is a large secreted protein involved in microcolony formation and biofilm maturation in Pseudomonas putida. Its C-terminal domain shows the characteristics of proteins secreted through a type I secretion system and includes a predicted calcium binding motif. We provide experimental evidence of specific binding of Ca2+ to the purified C-terminal domain of LapF (CLapF). Calcium promotes the formation of large aggregates, which disappear in the presence of the calcium chelator EGTA. Immunolocalization of LapF also shows the tendency of this protein to accumulate in vivo in certain extracellular regions. These findings, along with results showing that calcium influences biofilm formation, lead us to propose a model in which P. putida cells interact with each other via LapF in a calcium-dependent manner during the development of biofilms.
INTRODUCTION
Bacteria are able to form surface-attached multicellular communities, called biofilms, which represent a protected mode of growth that allows cells to survive in hostile environments and colonize new niches (8). Because biofilms can form on virtually any surface, they have a profound impact in medicine, industry, and agriculture. Intensive research over the last decade has led to the identification of key factors for biofilm formation. Among these, the presence of an extracellular matrix produced by the bacterial cells contributes to the architecture, organization, and maintenance of the three-dimensional structure of biofilms. The composition of the matrix depends on the bacterial species and the environmental conditions but it is generally made up of different exopolysaccharides (EPS) (3, 18, 19), extracellular DNA (27), and secreted proteins (13, 15, 22). The involvement of cell surface proteins in biofilm development has been described in detail for a variety of bacterial species. They include BapA in Salmonella enterica (13), Bap in Staphylococcus aureus (12), TasA in Bacillus subtilis (22), and LapA in Pseudomonas putida and Pseudomonas fluorescens (9, 29). In a recent publication (15), we described that the adhesin LapF mediates cell-cell interactions which lead to microcolony formation and biofilm maturation in Pseudomonas putida KT2440. However, the molecular mechanisms by which LapF exerts its function remain unknown.
A significant number of large proteins involved in cell surface and cell-cell interactions contain putative calcium binding domains (1, 11, 28). Calcium has been related to a variety of biological processes in bacteria (4, 7, 10), but its role in biofilm development is somewhat controversial. Previous research has shown that increasing amounts of calcium disable Bap-mediated biofilm formation and intercellular adhesion of Staphylococcus aureus (1). However, recent reports indicate that calcium promotes biofilm formation in other microorganisms, such as Xyllela fastidiosa or Vibrio vulnificus (4, 7). Therefore, calcium may modulate biofilm formation in opposite ways in different bacteria, but the molecular basis for its role remains unclear. Its potentially direct connection with surface proteins has been explored in Shewanella oneidensis, where the presence of calcium within a certain concentration range promotes biofilm formation mediated by the Bap-related surface protein BfpA (25). In contrast, recent data indicate a negative influence of calcium on the function of LapA, the main adhesin of Pseudomonas fluorescens (2). This effect would be indirect, through the activity of the protease LapG, which is modulated by calcium.
In this report we present evidence for the direct interaction of Ca2+ with the C-terminal domain of LapF (CLapF). This interaction results in the aggregation of the protein. Interestingly, calcium depletion has a negative effect on biofilm formation by P. putida. This leads us to suggest that this cation promotes the interaction between the C-terminal domains of LapF molecules from adjacent cells and thus promotes the cell-cell interactions needed for microcolony formation and the development of the three-dimensional structure of the biofilm.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Pseudomonas putida KT2440 is a plasmid-free derivative of the original soil isolate Pseudomonas putida mt-2 (21). Escherichia coli DH5α and BL21 were used as hosts in cloning and expression experiments, respectively. Unless otherwise specified, bacteria were grown in LB medium at 30°C (P. putida) or 37°C (E. coli). When appropriate, ampicillin (100 μg/ml) was added.
Construction of expression plasmid pMMGCa2.
A 2-kb DNA fragment corresponding to the C-terminal domain of LapF (amino acids 5622 to 6310) was PCR amplified from the chromosome of Pseudomonas putida KT2440 using primers CaFw2 forward (5′-GGGCAGCCATATGGATGCCAACGCGCCGGTCA-3′) and CaRev reverse (5′-AAAACTCGAGTCAAACCACATGGATACC-3′) with Expand high-fidelity polymerase (New England). The resulting PCR product was digested with NdeI and XhoI (underlined sites) and cloned in frame into the plasmid pET-15b (Novagen) cut with the same enzymes. The resulting plasmid, pMMGCa2, was transformed into E. coli DH5α, and the insert and flanking regions were verified by DNA sequencing.
Expression and purification of the recombinant C-terminal domain of LapF.
The plasmid pMMGCa2 was used for the production of a fusion protein with 6 histidine residues at the N terminus. The plasmid was transformed in Escherichia coli BL21 competent cells for protein expression. Cultures were grown in 250 ml LB with orbital shaking to an absorbance of 0.5 at 600 nm. The cells were induced with isopropyl-1-thio-β-galactopyranoside (IPTG) to a final concentration of 1 mM, and the cultures were incubated for additional 4 h at 37°C. Cells were harvested by centrifugation, resuspended in 20 ml of lysis buffer consisting of 1× CelLytic B cell lysis reagent (Sigma) diluted in 20 mM Tris, 500 mM NaCl, 20 mM imidazole, and 1 mM phenylmethylsulfonyl fluoride (PMSF) and supplemented with 100 μg/ml of freshly prepared lysozyme solution, and incubated for 20 to 30 min. Further disruption and reduction of viscosity were done by sonication.
A 1.5-ml portion of nickel chelating resin (G-Bioscience) was washed and conditioned as indicated by the manufacturer prior to addition to the samples and incubation with gentle agitation for 1 h at room temperature. The mixture was centrifuged, and after decanting the supernatant, the lysate-resin mixture was washed with 5 volumes of binding buffer (20 mM Tris, 500 mM NaCl, 1 mM imidazole, 1 mM PMSF) and 3 volumes of washing buffer I (20 mM Tris, 500 mM NaCl, 30 mM imidazole, 1 mM PMSF). The proteins were eluted with elution buffer (20 mM Tris, 500 mM NaCl, 500 mM imidazole, and 1 mM PMSF). The purified protein was treated with 10 mM EGTA and cleaned by Sephadex G-25 gel filtration chromatography, followed by overnight dialysis against 10 mM Tris-HCl–50 mM NaCl, pH 7.5. The protein concentration was determined as previously described (26). The integrity and homogeneity of the recombinant protein were confirmed by SDS-PAGE (4% stacking gel and 12% separating gel) and Western blot analysis using anti-His tag antibodies (Santa Cruz Biotechnology, Inc.). The identity of the protein was confirmed by electrospray ionization-tandem mass spectrometry (ESI-MS/MS) analysis.
Molecular mass estimation by analytical FPLC.
The molecular masses of the protein before and after treatment with 10 mM EGTA were estimated using a Superdex-200 HR 10/30 column with an AKTA Explorer fast protein liquid chromatography (FPLC) system (Amersham Biosciences). The column was equilibrated with 20 mM Tris–50 mM NaCl buffer, pH 7.5. The molecular mass of the eluted protein was estimated by a calibration curve prepared using the elution times measured for standards (Bio-Rad, Hercules, CA) ranging from 1.35 to 670 kDa (thyroglobulin [670 kDa], α-globulin [158 kDa], albumin [67 kDa], ovalbumin [44 kDa], myoglobin [17 kDa], and vitamin B12 [1.35 kDa]).
Dynamic light scattering (DLS).
Experiments were performed at 25°C using the DynaPro NanoStar (Wyatt). Protein solutions were dialyzed overnight against a filtered buffer (10 mM Tris-HCl, 50 mM NaCl, pH 7.5) and filtered before each experiment using a sterile syringe filter with a 0.45-μm cellulose acetate membrane (VWR). The protein solution was adjusted to a final concentration of 5 mg/ml of protein, injected into a 50-μl cuvette, and placed into the sample cell. The total sample injection volume was 60 μl. To test the effect of calcium in the protein solution, a filtered CaCl2 solution in 10 mM Tris-HCl–50 mM NaCl (pH 7.5) was injected into the sample to a final concentration of 10 mM, using a Hamilton syringe (Hamilton Co). A filtered EGTA solution was subsequently added to the sample to a final concentration of 20 mM. Data were collected from three independently prepared samples. Each sample measurement consisted of 4 repetitions, each one of 20 acquisitions of 15 s. Raw data were analyzed with the DYNAMICS software package. The dispersity of the solution was assessed, and the average of the hydrodynamic radius (RH) was calculated.
Isothermal titration calorimetry (ITC).
Reaction enthalpy measurements were done on a VP-ITC microcalorimeter (MicroCal) at 30°C. The Ca2+ in the purified protein was removed with 10 mM EGTA as described above and subsequently dialyzed overnight against 10 mM Tris-HCl–50 mM NaCl (pH 7.5), adjusted to a final concentration of 8.5 μM, and placed into the sample cell. A 100 μM CaCl2 solution was prepared using the dialysis buffer and placed into the syringe injector. A typical experiment involved the automatic injection of 10 μl ligand into the protein solution over 25 injections with stirring at 300 rpm. Heat produced due to dilution was measured by injecting the ligand solution into the sample cell without protein. For each titration the heat of dilution was subtracted from the corresponding Ca2+ binding data for the protein. Data were fit to appropriate binding models and thermodynamic parameters determined from nonlinear least-squares fits, using the MicroCal version of the ORIGIN software.
Transmission electron microscopy and immunogold labeling.
Protein samples were treated with 10 mM CaCl2 and incubated for 2 h or 3 days at room temperature. After treatment with calcium, protein samples were diluted with distilled water and adsorbed onto a Formvar-carbon-coated grid (the grid surface was previously made hydrophilic with glow discharge in a vacuum evaporator) for approximately 10 min at room temperature. Excess sample was discarded on a filter paper, the grid was incubated with 5 μl of negative staining solution (2% aqueous uranyl acetate) for 2 min at room temperature, and the excess was discarded with filter paper. Samples were dried at room temperatures for 5 to 10 min and were visualized in a Tecnai G2 Spirit BioTWIN microscope at an accelerating voltage of 80 kV. Images were taken with an AMT 2k charge-coupled device (CCD) camera.
For immunolocalization studies of LapF, biofilms of P. putida KT2440 were grown on nickel grids for 8 h in LB medium in a 6-well microtiter plate. Grids were recovered from the culture and treated with blocking buffer consisting of 1% nonfat dry milk in phosphate-buffered saline with 0.1% Tween 20 (PBST) for 30 min, incubated for 2 h with anti-LapF primary antibody (15) diluted 1:150 in blocking buffer, rinsed in PBST, exposed to goat anti-rabbit 20-nm gold-conjugated secondary antibody diluted 1:50 (TedPella, Inc.) for 1 h, and rinsed. All grids were stained with uranyl acetate and lead citrate and viewed as described above.
For subcellular localization studies of LapF, cells were grown for 8 h in LB medium in a 6-well microtiter plate. Cells were harvested and fixed with an equal volume of 4% paraformaldehyde in 0.1 M sodium phosphate buffer (pH 7.4). Small pieces of cell pellet were infiltrated in 2.3 M sucrose in PBS with 0.15 M glycine. Samples were high-pressure frozen in a Leica EM Pact2 high-pressure freezer and freeze-substituted in 0.2% glutaraldehyde and 0.1% uranyl acetate in acetone at −90°C for 72 h. Samples were warmed at room temperature and embedded in LR White resin. Ultrathin sections were cut at −120°C with a cryo-diamond knife. Sections were transferred to a Formvar-carbon-coated grid for immunogold labeling. Grids were contrasted and visualized as described above.
Biofilm formation analysis.
Biofilm formation was examined during growth in liquid LB in polystyrene microtiter plates (Sterilin) under static conditions as described previously (20). Biomass attached to the surface was evaluated by staining with 0,5% crystal violet for 15 min (20). Biofilms were quantified by solubilization of crystal violet with 70% ethanol and measurement of absorbance at 580 nm on a spectrophotometer (Shimadzu UV-160A).
To visually follow biofilm formation, 40- by 20-mm glass coverslips were placed obliquely into wells of a 6-well plate with 5 ml of liquid LB diluted 1:10 and inoculated with the bacterial strain and the treatment to be tested (CaCl2 or EGTA). Cultures were allowed to grow at 30°C for 24 h and biofilm formation evaluated visually or by fluorescence microscopy using a Zeiss Axioscope microscope after briefly washing the glass coverslips with deionized water.
RESULTS
Calcium modulates biofilm formation by Pseudomonas putida.
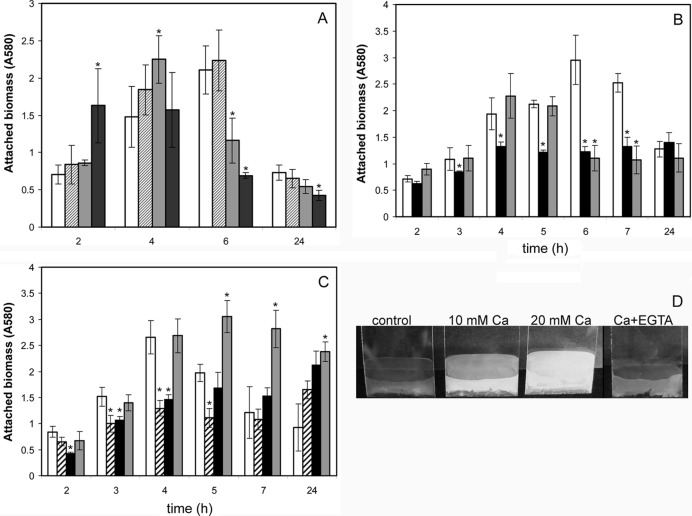
In order to analyze the role of calcium in biofilm formation by P. putida KT2440, this strain was first grown under static conditions in microtiter plates in LB with increasing concentrations of CaCl2. As shown in Fig. 1A, calcium caused a shift in the attachment-detachment kinetics of KT2440, with increasing concentrations promoting earlier attachment and early detachment of the cells. Addition of the calcium chelator EGTA, on the other hand, did not have a significant effect at early time points but limited biofilm development later on (Fig. 1B). EGTA also limited biofilm formation in minimal medium with glucose as a carbon source (not shown) and in 1:10 strength LB. This effect of the chelator was compensated for by addition of excess CaCl2 (Fig. 1B), even though cells began detaching earlier than in the control. To determine if the effect of EGTA could also be explained by its potential chelation of other ions, similar experiments were performed in LB in the presence of 10 mM MgCl2 with or without EGTA. The results are shown in Fig. 1C. Addition of magnesium relieved the negative effect of EGTA on biofilm formation. However, when MgCl2 was added alone, no increase in attachment was observed until 24 h after inoculation. This suggests that the effect of EGTA is indeed a consequence of calcium chelation, so that when Mg is added in excess it sequesters EGTA, and more free calcium is available. Surprisingly, the combination of MgCl2 and EGTA caused cells to remain attached, an effect that was not observed with magnesium alone.
Fig 1.
Influence of calcium, magnesium, and the chelator EGTA on biofilm formation by P. putida KT2440. (A to C) Attached biomass was determined by crystal violet staining, as described in Materials and Methods. Data are averages and standard deviations from four experiments. Asterisks indicate statistically significant differences with respect to the control. (A) Biofilm formation on microtiter plates during growth in LB with increasing concentrations of CaCl2 (white bars, control without addition; hatched bars, 10 mM; light gray bars, 20 mM; dark gray bars, 50 mM). (B) Effect of EGTA on biofilm formation, determined as described above (white bars, control; black bars, LB with 0.1 mM EGTA; gray bars, 0.1 mM EGTA and 10 mM CaCl2). (C) Biofilm formation in LB (white bars) and in the presence of 0.1 mM EGTA (hatched bars), 10 mM MgCl2 (black bars), or Mg plus EGTA (gray bars). (D) Biofilm formation on glass coverslips during growth in 1:10 strength LB. Coverslips were directly photographed at 24 h postinoculation.
The role of calcium was also tested in 6-well plates with glass coverslips partially immersed in the growth medium (1:10 LB with or without CaCl2). Biomass attached to the coverslip during growth was evaluated directly. As shown in Fig. 1D, addition of CaCl2 caused an increase in biofilm biomass at 24 h. Conversely, addition of the calcium chelator EGTA at a concentration that did not affect planktonic growth caused a reduction in the amount of biomass attached to the coverslips. In these experiments, no significant influence of either treatment was observed at earlier time points.
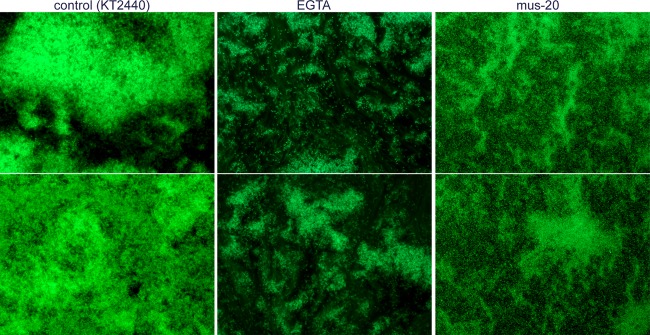
These data suggested that calcium has a time-dependent influence on attachment of P. putida. In this bacterium, two large adhesins showing putative calcium binding motifs, LapA and LapF, participate in biofilm formation. LapA is key for initial attachment, and LapF participates in later stages in a medium-dependent manner (15). We decided to focus our attention on LapF to define the potential role of calcium in its functionality. Initially, the biofilm phenotype previously observed for mus-20, a lapF mutant (6, 15), was compared to the effect caused on KT2440 by addition of EGTA. Biofilms of green fluorescent protein (GFP)-tagged strains grown on glass coverslips were examined by fluorescence microscopy. As shown in Fig. 2, KT2440 biofilms grown in the presence of EGTA showed an intermediate structure between the large, thick microcolonies formed by the wild type under regular conditions and the flat, less structured biofilm of the lapF mutant. EGTA caused a clear reduction in the size of microcolonies, with many cells attached individually, as in the lapF mutant. EGTA also appears to weaken the adhesive force of KT2440, since cells were easily swept away upon washing. It is worth mentioning that addition of CaCl2 did not increase attachment in the lapF mutant (data not shown).
Fig 2.
Biofilm formation on glass coverslips by KT2440 in the absence (control) or presence of EGTA, compared to that by the lapF mutant strain mus-20. Biofilms were grown in 1:10 strength LB as described in Materials and Methods and observed after 24 h by fluorescence microscopy. Two different fields are shown in each case. Magnification, ×400.
CLapF selectively binds calcium.
LapF consists of three domains: an N-terminal domain of 174 amino acids, followed by a long, repetitive region spanning most of the protein, and finally a C-terminal domain typical of proteins secreted through a type I secretion system (15). Analysis of the sequence of LapF revealed the presence in the C-terminal region of the protein (CLapF) of putative calcium binding sites related to the hemolysin type and those of the NodO protein (5). The presence of these sites was further supported by in silico modeling of the three-dimensional structure of a region spanning the 300 C-terminal amino acids of LapF (Fig. 3). The analysis with HHPred (24) returned as best templates calcium binding proteins and resulted in a predicted structure containing antiparallel β-sheets that closely resemble those that are characteristically found in RTX toxins and known to bind calcium (14).
Fig 3.
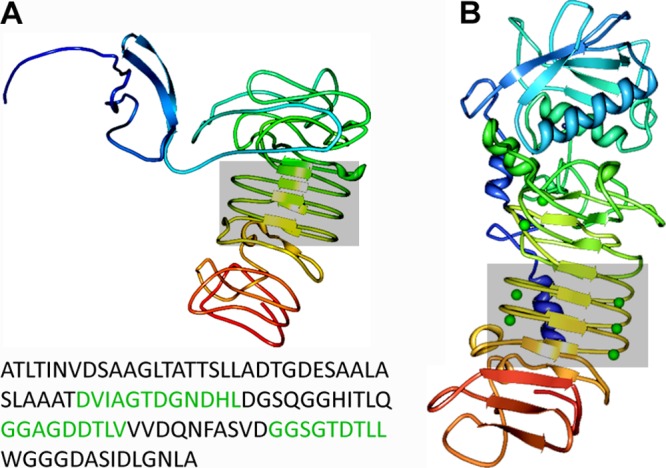
Bioinformatic analysis of the structure of the 300 C-terminal amino acids of LapF (A) compared to the Serratia marcescens metalloprotease (1SAT in the RCSB PDB database, which shows bound calcium as green balls) (B). The calcium binding motifs are shown in green in the sequence, and the corresponding region is boxed in the structural prediction. Model prediction was obtained using HHPred (24), and the Protein Workshop toolkit (16) was used for visualization.
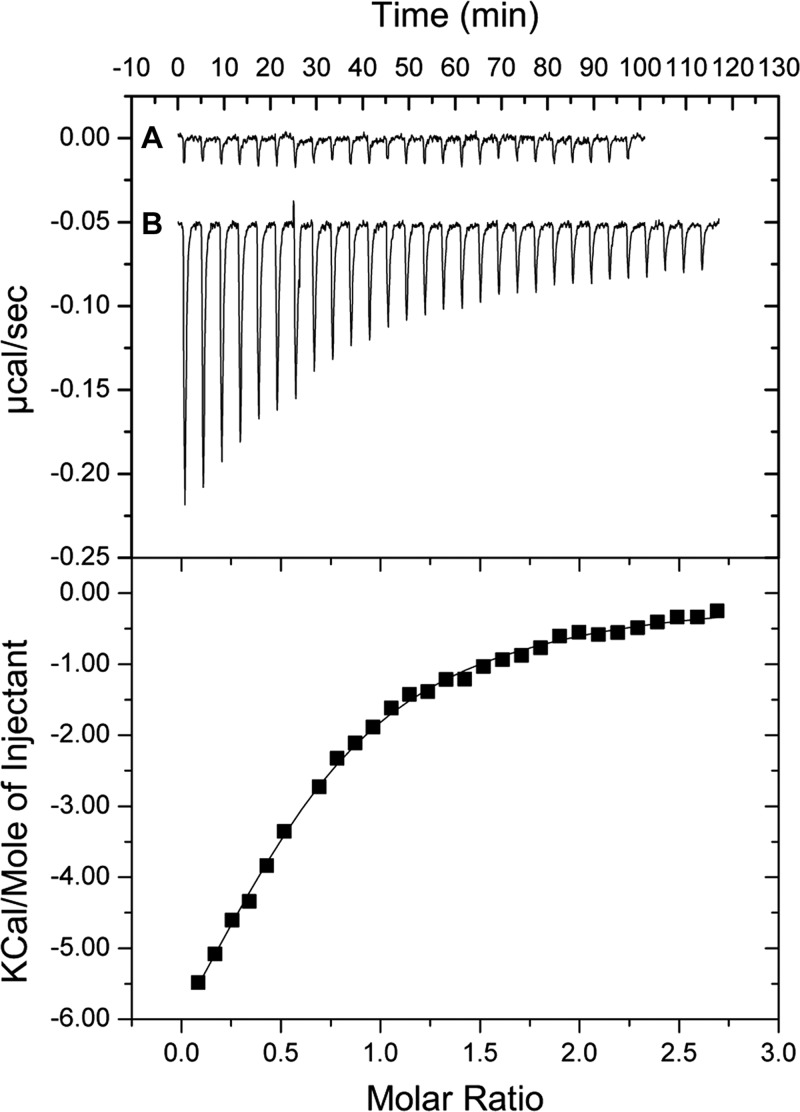
To begin defining the role of calcium in the functionality of the protein, a DNA fragment encoding CLapF was cloned in the expression vector pET15b. This construct was used to express and purify CLapF as a 690-amino-acid fusion protein with a 6-His tag (see Fig. S1 in the supplemental material). The purified polypeptide was treated with EGTA to remove any potential traces of calcium, and isothermal titration calorimetry (ITC) assays were performed in order to test the interaction of CLapF with this cation. As shown in Fig. 4 and Table 1, CLapF binds Ca2+ with moderate affinity (KD [equilibrium dissociation constant] = 3.54 ± 0.016 μM), calculated by fitting the experimental data to a single-event model (n = 1) for Ca2+. No binding could be observed with other divalent cations such as Mg2+ or Mn2+ (data not shown), suggesting that the interaction of CLapF with Ca2+ is specific for this cation.
Fig 4.
Isothermal titration of the C-terminal domain of LapF in the absence and presence of Ca2+. An injection volume of 10 μl and a total number of 25 injections were used. (A) Injection of the ligand CaCl2 into buffer. (B) Injection of the ligand into a 8.5 μM protein solution. Ligand and protein were in 0.1 M Tris–0.5 M NaCl, pH 7.5. Experiments were carried out at 30°C. Upper panel, raw titration data. Lower panel, integrated and dilution-corrected peak areas of raw data. Data were fitted using the “one-binding-site model” of the MicroCal version of ORIGIN.
Table 1.
Binding parameters derived from isothermal titration calorimetry experimentsa
| Parameter (unit) | Value (mean ± SD) |
|---|---|
| KA (M−1) | 2.82 (± 0.16) × 105 |
| KD (μM) | 3.54 ± 0.016 |
| ΔH (kcal/mol) | −9.8 ± 0.4 |
| ΔS (cal/mol/°) | −7.35 |
| N (sites) | 0.603 ± 0.0206 |
All experiments were conducted at 30°C. Ligands were placed in the syringe and CLapF in the sample cell. The buffer was 10 mM Tris-HCl–50 mM NaCl2 adjusted to a pH 7.5 by the addition of concentrated HCl.
Multimerization of CLapF in the presence of calcium.
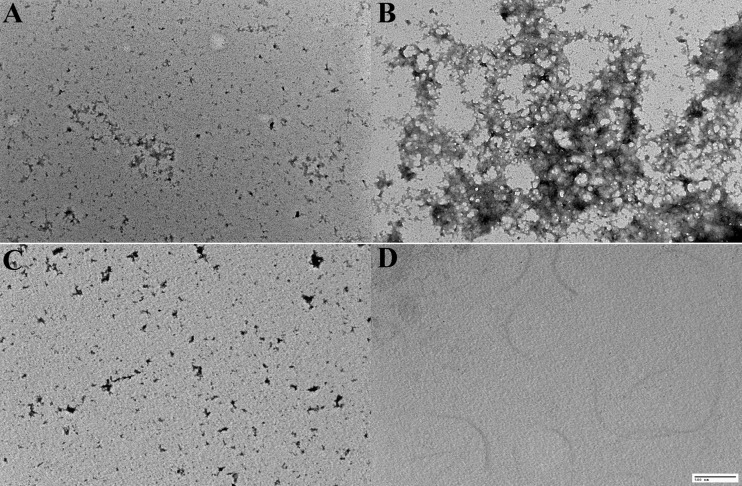
During these in vitro experiments with purified CLapF, we noticed that prolonged incubation of CLapF in the presence of high calcium concentrations resulted in faintly visible precipitates in the samples, suggesting that calcium might cause aggregation of the protein. After purification and treatment of CLapF with 10 mM EGTA, the protein was incubated for 3 days at room temperature with 10 mM CaCl2 and a control without calcium. Samples were analyzed by transmission electron microscopy. In samples treated with calcium, aggregates were clearly visible (Fig. 5). Controls without calcium or with calcium in the absence of protein were devoid of any aggregates. Addition of EGTA in the presence of calcium resulted in lack of aggregation (Fig. 5), whereas increasing concentrations of calcium caused faster and more compact clustering of the protein, so that with 100 mM CaCl2, large aggregates were visible after 3 h of incubation (data not shown).
Fig 5.
(A to C) Transmission electron microscopy of CLapF incubated for 3 days at room temperature without CaCl2 (A), with 10 mM CaCl2 (B), or with CaCl2 and EGTA (C). (D) A control with 10 mM CaCl2 without protein, to ensure that the aggregates in panel B are not artifacts due to calcium precipitation. Scale bar, 500 nm.
Dynamic light scattering (DLS) was then used to analyze the size distribution of the hydrodynamic radius and the polydispersity of CLapF in the presence or absence of Ca2+ and EGTA. The polydispersity (Pd) value gives information about the homogeneity or heterogeneity of a given population (peak). The level of homogeneity is considered high when the population has a Pd value of less than 15%. Purified CLapF shows three different protein subpopulations in solution (Fig. 6). The hydrodynamic radius (RH) of each subpopulation is described in Table 2. After addition of 10 mM CaCl2 to the same sample, a new subpopulation of large aggregates with an RH of 5,262 nm, nearly six times higher than that of the largest aggregate in the absence of calcium, is generated. The new subpopulation of aggregates corresponds to more than 50% of the total mass of the protein sample (Table 2). This effect was completely reversed when EGTA was added to a final concentration of 20 mM, with the subpopulation with higher RH disappearing after 10 min of incubation with the chelator. Data from buffer and buffer with CaCl2 and EGTA were acquired as control (data not shown).
Fig 6.
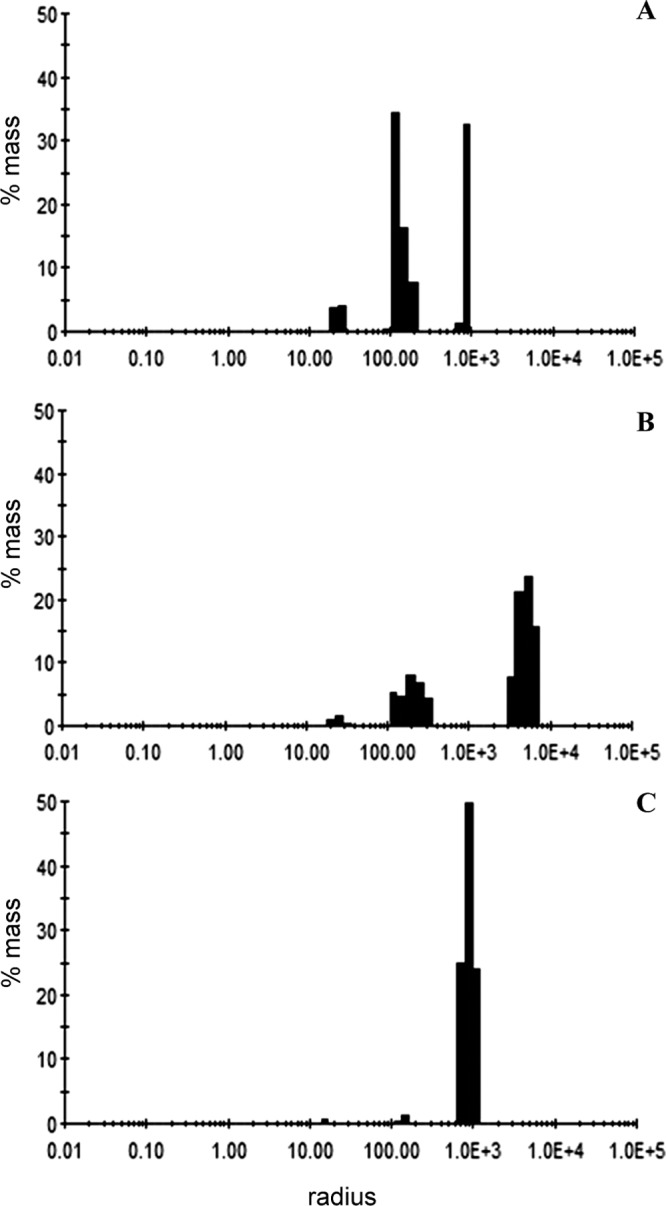
Size distribution histograms for purified CLapF treated with 10 mM EGTA and dialyzed overnight against 10 mM Tris-HCl–50 mM NaCl, pH 7.5 (A), protein from panel A with 10 mM CaCl2 (B), and protein from panel B treated with EGTA to a final concentration of 20 mM (C).
Table 2.
Dynamic light scattering results for CLapF
| Peak | Control |
With CaCl2 |
With CaCl2 + EGTA |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Hydrodynamic radius (nm) | Polydispersity (%) | Mass (%) | Hydrodynamic radius (nm) | Polydispersity (%) | Mass (%) | Hydrodynamic radius (nm) | Polydispersity (%) | Mass (%) | |
| 1 | 24.313 | 11.2 | 7.8 | 25.233 | 17 | 3 | 16.183 | 8.8 | 0.6 |
| 2 | 148.346 | 18.7 | 58.4 | 136.969 | 12.8 | 9.9 | 153.293 | 5.7 | 1.3 |
| 3 | 917.719 | 4.7 | 33.8 | 251.616 | 20.5 | 19 | 913.999 | 18.5 | 98.1 |
| 4 | 5262 | 23.7 | 68 | ||||||
Based on the DLS results, analytical gel filtration was used to determine the molecular masses of CLapF and the different aggregates. Purified CLapF after treatment with EGTA appeared in only two different forms that eluted in the void volume of an analytical gel filtration column with an exclusion size of 600 kDa (Fig. 7). The smallest peak detected (∼77 kDa) corresponds to the monomer of the protein. A peak of approximately 630 kDa was also found in the purified protein solution. This peak may correspond to aggregates of eight CLapF monomers. After incubation with 10 mM CaCl2, the 77-kDa peak practically disappears and two new peaks corresponding to sizes of around 180 and larger than 900 kDa appear (Fig. 7). These data further support multimerization of the protein in the presence of calcium.
Fig 7.
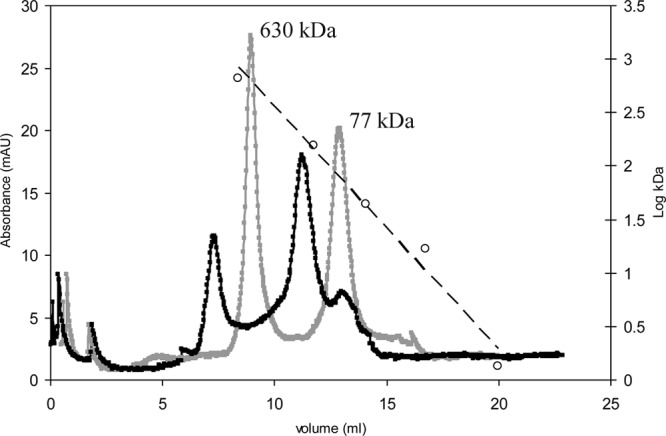
Study of the molecular mass of CLapF by analytical FPLC. Purified CLapF after treatment with EGTA (gray line) and CLapF incubated with 10 mM CaCl2 (black line) are shown, as well as the molecular masses (open circles) and linear trend (broken line) of the standards. The ∼77-kDa and ∼630-kDa peaks are indicated.
LapF clusters on the cell surface and between cells in biofilms.
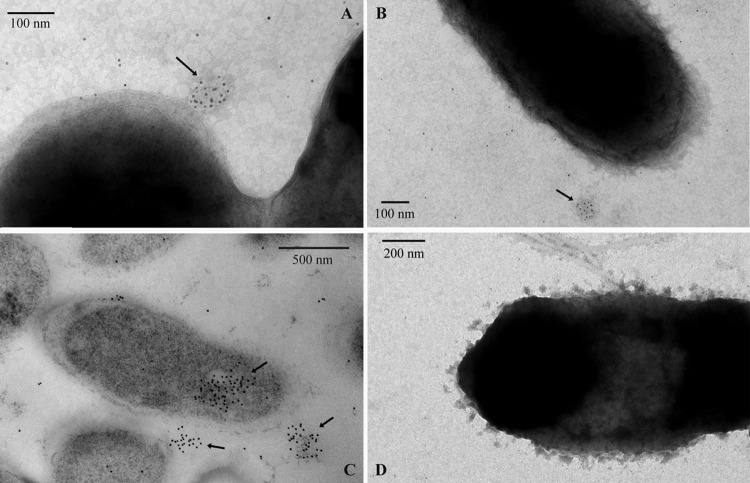
LapF had been previously localized in the cell surface of P. putida KT2440 and between cells in the biofilm using immunofluorescence and Western blot analysis (15). To get a more precise localization of LapF in cells within a biofilm, P. putida KT2440 was grown in 6-well microtiter plates in LB with a nickel grid in the middle of each well. Cells were incubated at 30°C, and after 8 h of incubation, grids were recovered and treated as described in Materials and Methods for electron microscopy observation with anti-LapF antibodies. As shown in Fig. 8, clustering of LapF on the cell surface can be observed. In ultrathin sections of the cells recovered from the well, LapF is detected in the cytoplasm but is localized mainly in large aggregates between cells, supporting its role in cell-cell interactions and as part of the extracellular matrix of the biofilm.
Fig 8.
Transmission electron microscopy and immunodetection of LapF. (A and B) Wild-type cells after 8 h growth and immunogold labeling with anti-LapF antibody; (C) thin sections of resin-embedded wild-type cells after 8 h growth in 6-well microtiter plates; (D) mutant strain mus-20 (lapF). Arrows point to the accumulation of gold particles in the cell surface and in the cytoplasm.
DISCUSSION
Calcium is essential for many biological processes and has been previously shown to influence the multicellular behavior of different microorganisms. In this work we have provided evidence of calcium causing accelerated biofilm formation by P. putida, followed by early detachment under static conditions. This effect might be in part due to calcium interaction with LapF, which causes the C-terminal portion of this adhesin to alter its conformation and form multimers. Among the variety of methods available to analyze the state of a macromolecular sample, DLS is a very sensitive technique that is frequently used to explore the size and aggregation properties of macromolecules in solution (17). In our results, the presence of calcium caused an increase in the polydispersity of the subpopulations of CLapF (Table 2). The effect of calcium on the sample heterogeneity is more evident in the new population of aggregates, whereas the homogeneity of the sample increases when EGTA is added. A lower proportion of multimers is present in the population of purified CLapF in the absence of calcium, and clustering can also be observed when the native protein is analyzed in vivo in biofilms through immunolocalization.
These results suggest a natural tendency of LapF to aggregate, an effect that is enhanced by increasing calcium concentrations. Thus, it seems possible that LapF contributes to the scaffolding of the extracellular matrix through its multimerization. This structural role based on polymerization would be somewhat similar to that reported for the formation of amyloid fibers by certain extracellular proteins. This is the case for TasA, which has been shown to polymerize to form amyloids and thus confer stability to Bacillus subtilis biofilms (22). Such an interpretation would be in contrast with the proposed influence of calcium on the functionality of other large surface proteins. In Staphylococcus aureus, for example, Ca2+ causes inhibition of biofilm formation in strains where the large adhesin Bap is present (1). This protein contains four EF-hand-like calcium binding motifs; when these are mutated, the inhibitory effect of Ca2+ is lost. It is worth noting that the Ca2+ binding sites in the C-terminal region of LapF do not correspond to EF-hand motifs but rather resemble those described in NodO (5), a Ca2+ binding secreted protein which has been proposed to mediate the interaction between Rhizobium leguminosarum cells and plant roots in a calcium-dependent way.
The influence of calcium in P. putida also contrasts with that observed in P. fluorescens, where this cation seems to have a negative effect on attachment mediated by the large adhesin LapA (2). It is worth noting that whereas LapA homologs are present in P. putida and P. fluorescens, LapF is found exclusively in the former. It could be that the role of calcium is different in the two species or that in P. putida calcium is somehow part of a “clock” for the attachment process. Our current model of a sequential requirement for LapA and LapF in biofilm formation and the fact that the calcium concentration alters the timing for attachment and detachment would favor this idea.
Calcium is a relatively abundant element in soils, with average concentrations ranging from 7 to 24 mg/g of soil, according to different surveys (23). It is generally present as a component of soil minerals or in its cationic form adsorbed to soil particles, from which it is available to plant roots to be incorporated by mass flow. It is therefore likely that the plant root-colonizing bacterium P. putida KT2440 encounters a gradient of this element in its natural niche and has partly adjusted its multicellular behavior to the concentrations found on the root surface.
ACKNOWLEDGMENTS
We thank Tino Krell for help with the interpretation of ITC data, G. O'Toole for helpful discussions and sharing of results prior to publication, M. Ericsson, and E. Benecchi for guidance with electron microscopy, D. Pheasant for assistance with ITC and DLS analysis in the Biophysical Instrumentation Facility at MIT, and J. Parker for help with the use of FPLC.
This work was supported by grant BFU2010-17946 from the Plan Nacional de I+D+I and FEDER funds, grant GM58213 from the NIH to R.K., a MEC/Fulbright postdoctoral fellowship to D.R., and an EMBO Short Term Fellowship to M.M.-G., who is also the recipient of an FPI Fellowship.
Footnotes
Published ahead of print 5 October 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Arrizubieta MJ, Toledo-Arana A, Amorena B, Penadés JR, Lasa I. 2004. Calcium inhibits Bap-dependent multicellular behavior in Staphylococcus aureus. J. Bacteriol. 186:7490–7498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boyd CD, Chatterjee D, Sondermann H, O'Toole GA. 2012. LapG, required for modulating biofilm formation by Pseudomonas fluorescens Pf0-1, is a calcium-dependent protease. J. Bacteriol. 194:4406–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chang WS, et al. 2007. Alginate production by Pseudomonas putida creates a hydrated microenvironment and contributes to biofilm architecture and stress tolerance under water-limiting conditions. J. Bacteriol. 189:8290–8299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cruz LF, Cobine PA, De La Fuente L. 2012. Calcium increases surface attachment, biofilm formation, and twitching motility in Xylella fastidiosa. Appl. Environ. Microbiol. 78:1321–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Economou A, Hamilton WD, Johnston AW, Downie JA. 1990. The Rhizobium nodulation gene nodO encodes a Ca2(+)-binding protein that is exported without N-terminal cleavage and is homologous to haemolysin and related proteins. EMBO J. 9:349–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Espinosa-Urgel M, Salido A, Ramos JL. 2000. Genetic analysis of functions involved in adhesion of Pseudomonas putida to seeds. J. Bacteriol. 182:2363–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Garrison-Schilling KL, et al. 2011. Calcium promotes exopolysaccharide phase variation and biofilm formation of the resulting phase variants in the human pathogen Vibrio vulnificus. Environ. Microbiol. 13:643–654 [DOI] [PubMed] [Google Scholar]
- 8. Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95–108 [DOI] [PubMed] [Google Scholar]
- 9. Hinsa SM, Espinosa-Urgel M, Ramos JL, O'Toole GA. 2003. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol. 49:905–918 [DOI] [PubMed] [Google Scholar]
- 10. Johnson MD, et al. 2011. Pseudomonas aeruginosa PilY1 binds integrin in an RGD- and calcium-dependent manner. PLoS One 6:e29629 doi:10.1371/journal.pone.0029629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Josefsson E, O′Connell D, Foster TJ, Durussel I, Cox JA. 1998. The binding of calcium to the B-repeat segment of SdrD, a cell surface protein of Staphylococcus aureus. J. Biol. Chem. 273:31145–31152 [DOI] [PubMed] [Google Scholar]
- 12. Lasa I, Penadés JR. 2006. Bap: A family of surface proteins involved in biofilm formation. Res. Microbiol. 157:99–107 [DOI] [PubMed] [Google Scholar]
- 13. Latasa C, et al. 2005. BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar Enteritidis. Mol. Microbiol. 58:1322–1339 [DOI] [PubMed] [Google Scholar]
- 14. Linhartová I, et al. 2010. RTX proteins: a highly diverse family secreted by a common mechanism. FEMS Microbiol. Rev. 34:1076–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martínez-Gil M, Yousef-Coronado F, Espinosa-Urgel M. 2010. LapF, the second largest Pseudomonas putida protein, contributes to plant root colonization and determines biofilm architecture. Mol. Microbiol. 77:549–561 [DOI] [PubMed] [Google Scholar]
- 16. Moreland JL, Gramada A, Buzko OV, Zhang Q, Bourne PE. 2005. The molecular biology toolkit (MBT): a modular platform for developing molecular visualization applications. BMC Bioinformatics 6:21 doi:10.1186/1471-2105-6-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Murphy RM. 1997. Static and dynamic light scattering of biological macromolecules: what can we learn? Curr. Opin. Biotechnol. 8:25–30 [DOI] [PubMed] [Google Scholar]
- 18. Nielsen L, Li X, Halverson LJ. 2011. Cell-cell and cell-surface interactions mediated by cellulose and a novel exopolysaccharide contribute to Pseudomonas putida biofilm formation and fitness under water-limiting conditions. Environ. Microbiol. 13:1342–1356 [DOI] [PubMed] [Google Scholar]
- 19. Nilsson M, et al. 2011. Influence of putative exopolysaccharide genes on Pseudomonas putida KT2440 biofilm stability. Environ. Microbiol. 13:1357–1369 [DOI] [PubMed] [Google Scholar]
- 20. O'Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449–461 [DOI] [PubMed] [Google Scholar]
- 21. Regenhardt D, et al. 2002. Pedigree and taxonomic credentials of Pseudomonas putida strain KT2440. Environ. Microbiol. 4:912–915 [DOI] [PubMed] [Google Scholar]
- 22. Romero D, Aguilar C, Losick R, Kolter R. 2010. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. U. S. A. 107:2230–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shacklette HT, Boerngen JG. 1984. Element concentrations in soils and other surficial materials of the conterminous United States. U.S. Geological Survey professional paper 1270. U.S. Geological Survey, Reston, VA [Google Scholar]
- 24. Söding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Theunissen S, et al. 2010. The 285 kDa Bap/RTX hybrid cell surface protein (SO4317) of Shewanella oneidensis MR-1 is a key mediator of biofilm formation. Res. Microbiol. 161:144–152 [DOI] [PubMed] [Google Scholar]
- 26. Whitaker JR, Granum PE. 1980. An absolute method for protein determination based on difference in absorbance at 235 and 280 nm. Anal. Biochem. 109:156–159 [DOI] [PubMed] [Google Scholar]
- 27. Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. [DOI] [PubMed] [Google Scholar]
- 28. Yousef-Coronado F, Espinosa-Urgel M. 2007. In silico analysis of large microbial surface proteins. Res. Microbiol. 158:545–550 [DOI] [PubMed] [Google Scholar]
- 29. Yousef-Coronado F, Travieso ML, Espinosa-Urgel M. 2008. Different, overlapping mechanisms for colonization of abiotic and plant surfaces by Pseudomonas putida. FEMS Microbiol. Lett. 288:118–124 [DOI] [PubMed] [Google Scholar]