Abstract
Metal ion homeostasis is a critical function of many integral and peripheral membrane proteins. The genome of the etiologic agent of syphilis, Treponema pallidum, is compact and devoid of many metabolic enzyme genes. Nevertheless, it harbors genes coding for homologs of several enzymes that typically require either iron or zinc. The product of the tp0971 gene of T. pallidum, designated Tp34, is a periplasmic lipoprotein that is thought to be tethered to the inner membrane of this organism. Previous work on a water-soluble (nonacylated) recombinant version of Tp34 established that this protein binds to Zn2+, which, like other transition metal ions, stabilizes the dimeric form of the protein. In this study, we employed analytical ultracentrifugation to establish that four transition metal ions (Ni2+, Co2+, Cu2+, and Zn2+) readily induce the dimerization of Tp34; Cu2+ (50% effective concentration [EC50] = 1.7 μM) and Zn2+ (EC50 = 6.2 μM) were the most efficacious of these ions. Mutations of the crystallographically identified metal-binding residues hindered the ability of Tp34 to dimerize. X-ray crystallography performed on crystals of Tp34 that had been incubated with metal ions indicated that the binding site could accommodate the metals examined. The findings presented herein, coupled with bioinformatic analyses of related proteins, point to Tp34's likely role in metal ion homeostasis in T. pallidum.
INTRODUCTION
Transition metal ions are essential structural components and cofactors for many proteins in virtually all biological systems (36, 47, 65). As such, bacteria have evolved numerous systems for obtaining metal ions from their environments and hosts and for transporting these ions against steep concentration gradients. These mechanisms include TonB-type and ABC transporters, P-type ATPases, proton/ion-driven symporters, and receptor/transporter systems for host metal-sequestering proteins such as transferrin and lactoferrin (31, 47, 60). However, because metal homeostasis involves maintaining a tight balance between physiological needs and undesirable metal toxicity, bacteria also harbor systems to expel excess metal ions, such as RND efflux pumps and export P-type ATPases (47, 60).
Treponema pallidum, the bacterial agent of syphilis, remains a significant threat to human health worldwide (11, 13, 30, 61). T. pallidum is among the most poorly understood of all sexually transmitted pathogens, due largely to the historic inability to cultivate the spirochete in vitro and the consequent inability to apply contemporary genetic approaches to understand many aspects of treponemal biology and virulence. To circumvent these severe experimental obstacles, determining the structures and functions of selected treponemal proteins has been one fruitful avenue of investigation to garner new insights into T. pallidum membrane biology (7, 17–20, 44, 46, 48, 64).
In comparison with other bacterial pathogens, one particularly enigmatic feature of T. pallidum appears to be its reduced need for metal ions (29). Understanding the molecular aspects of T. pallidum's apparently reduced metal ion requirement is an important aspect of better characterizing this human pathogen physiologically. The genome of T. pallidum contains only a few genes that encode iron-requiring proteins: tp0152, tp0612, tp0613, tp0615, tp0972, tp0991, and tp1038 (from UniProtKB annotations), as well as tp0053, tp0080, tp0735, tp0823, tp0842, tp0939, tp0991, and tp1038 (27, 29, 32, 33, 38, 54, 64). Also, the genes for several Zn2+-requiring enzymes are present in the T. pallidum genome, and at least two ABC-type transporters for Zn2+ have been characterized (21, 33, 44).
We previously presented evidence to support the hypothesis that the ca. 22-kDa Tp34 membrane lipoprotein of T. pallidum likely participates in metal ion homeostasis (17). Crystallographic studies of a water-soluble recombinant derivative of this protein (rTp34) demonstrated that it adopts an immunoglobulin-like fold and binds Zn2+ (17). X-ray diffraction data from rTp34 crystals that had been incubated in the presence of Zn2+ indicated that metal binding occurs at the interface of a Tp34 dimer. In solution, apo-rTp34 exists mostly as a monomer, but the addition of divalent cations induces dimerization of the protein (17). Further support that Tp34 may facilitate metal acquisition can be found in the fact that the tp34 (also known as tp0971) gene is located directly adjacent to a gene that encodes a putative membrane protein (Tp0972) whose sequence is similar to those of a family of high-affinity Fe2+/Pb2+ permeases (16). Furthermore, Tp34 was shown to bind specifically to the mucosal iron-carrying protein lactoferrin. Though there was some evidence that Tp34 can bind iron, these studies were complicated by the poor solubility of the protein in the presence of these ions.
In the current study, we employed a number of biophysical approaches to extend our knowledge of the functional characteristics of Tp34. Additional X-ray crystallographic studies revealed the manner in which the protein binds to several different transition metal ions, and site-directed mutagenesis experiments combined with analytical ultracentrifugation (AUC) unequivocally established that the crystallographically observed metal-binding site is responsible for metal-induced dimerization of Tp34. In addition, the results of the current study reveal the relative contributions of metal-binding residues to the dimerization activity. Finally, new analyses of the genes cotranscribed within the Tp34 operon offer additional insights into the most plausible functions of this enigmatic periplasmic membrane lipoprotein.
MATERIALS AND METHODS
Protein preparation.
Recombinant Tp34 and its variants were expressed in Escherichia coli and purified as described previously (17). The His6 tag was removed as described previously (8) when Tp34 was to be used in the presence of transition metal ions. Site-directed mutagenesis of Tp34 was performed by the QuikChange site-directed mutagenesis method (Stratagene) and with the following primer pairs: for the H70A mutation, 5′-GAAGAGGCGGACTGTGCCATAGAAGCGGATATC-3′ and 5′-GATATCCGCTTCTATGGCACAGTCCGCCTCTTC-3′; for the E72A mutation, 5′-GCGGACTGTCACATAGCAGCGGATATCCACGCA-3′ and 5′-TGCGTGGATATCCGCTGCTATGTGACAGTCCGC-3′; for the M117A mutation, 5′-GTGATGTTTGCGCCCGCGAACGCAGGGGACGGT-3′ and 5′-ACCGTCCCCTGCGTTCGCGGGCGCAAACATCAC-3′; for the H124A mutation, 5′-GCAGGGGACGGTCCGGCTTATGGGGCGAACGTG-3′ and 5′-CACGTTCGCCCCATAAGCCGGACCGTCCCCTGC-3′; and for the H155A mutation, 5′-GATGAGTACTCGCTAGCTATTGATGAGCAAACT-3′ and 5′-AGTTTGCTCATCAATAGCTAGCGAGTACTCATC-3′ (underlined nucleotides indicate mutations). All mutant genes produced were verified by DNA sequence analysis.
Operon mapping of the Tp34 gene cluster.
Treponemal RNA extraction and reverse transcription-PCR (RT-PCR) methods were described previously (18). Approximately 1 × 109 T. pallidum bacteria were freshly harvested from rabbit testicular tissue. Briefly, infected rabbit testes were minced, and 10 ml of saline was added per testis. The treponemes were extracted for 30 min at room temperature and centrifuged for 10 min (∼1,000 × g) to remove the rabbit-derived cellular debris. This step was repeated once to remove any residual particulates from the first spin. Finally, treponemes were pelleted by centrifugation (∼16,000 × g for 20 min at 4°C) and immediately resuspended in 1 ml of TRIzol (Invitrogen) for RNA extraction. RNA was treated with DNase I (GenHunter) to remove contaminating genomic DNA. DNase I-treated RNA was then purified using TRIzol reagent and checked for residual DNA contamination by qualitative amplification using the T. pallidum-specific primers listed in Table 1. SuperScript III reverse transcriptase (Invitrogen) was used to synthesize treponemal cDNA via random priming. Approximately 10 ng of cDNA was then used as the template for subsequent PCR amplifications, using the specific primer pairs listed in Table 1, for genes near tp0971. As a positive control, each DNA fragment was also amplified with the same primers, with treponemal DNA as the template. As a negative control, PCRs (without previous reverse transcription) were performed on the same RNA templates to confirm the absence of contaminating treponemal DNA. PCR products were electrophoresed through 1.5% (wt/vol) agarose gels containing ethidium bromide and visualized with UV light.
Table 1.
Deoxyoligonucleotide primers used in this study
| Primer name | Sequence (5′ to 3′) |
|---|---|
| TP0958F | CAAAGAGCGCACCTAAGAGC |
| TP0959R | CGCTGTTTGTGAGAGTGATG |
| TP0959R | CGCATAGACTGCACCATCAC |
| TP0960F | AGATCCGGTTGAGGTAGGAC |
| TP0960R | TACAACAGGTCCTCGAACTC |
| TP0961F | GAATGATGGGCAGATTGTGG |
| TP0961R | CCTCAACACCGAGGCCTAAC |
| TP0962F | GTCTGAGCACTGCGGGTATG |
| TP0962R | TCGCGTCCGATACGACTCAG |
| TP0963F | CGGGTCTTGACCCAATCCAG |
| TP0963R | GCCACGCGCTCTTTAAGATG |
| TP0964F | TCGTTGCAGACGAACCTACC |
| TP0964R | ACCCGAAGGACCAAGGATAG |
| TP0965F | GCAGAACCAAGCGTGAAAGC |
| TP0965R | CAAACTGGGCAGTGACGTAG |
| TP0966F | TACAGGCGCTCATCACGTTG |
| TP0966R | TGTTCGCGGCAATTTCGTAG |
| TP0967F | TTTGGCGAAGGCGAATGTAG |
| TP0967R | CGTACTGGCTCTGTGTTATC |
| TP0968F | TGTTCGCGGCAATTTCGTAG |
| TP0968R | TAGCTGCTGGTACGGATTGG |
| TP0969F | TATCGCTCGCTGGACTACAC |
| TP0969R | GGTGCTCGAGGAGTTGTTGG |
| TP0971F | TCGAAGGAAGAGGCGGACTG |
| TP0971R | TCACCGTGCTGATGCTCTCC |
| TP0972F | CAGTGCGACTTCCGTTGAGG |
| TP0972R | GTGCAACCCATACCATAGAC |
| TP0973F | CTGGTGCTCTTCGATGATTC |
Analytical ultracentrifugation.
All AUC experiments were carried out in a Beckman-Coulter (Brea, CA) model XL-I ultracentrifuge. An An-50Ti rotor (Beckman-Coulter) was used, and all such studies were performed at a rotor speed of 50,000 rpm (∼182,000 × g). Solutions (390 μl) were placed in dual-sectored charcoal-filled Epon centerpieces that were sandwiched between sapphire windows. For the metal ion titration experiments (see Results), the two sectors were both filled with sample (i.e., no reference was used) that comprised 2.6 μM Tp34 and various concentrations of transition metal ions. The data were collected in intensity mode at 280 nm. The resulting data, i.e., readouts of transmitted intensity counts versus radius, were treated according to the “pseudoabsorbance” procedure of Schuck and colleagues (39). Briefly, an arbitrary intensity was chosen as the pseudoreference value, and the data were transformed to their radially dependent pseudoabsorbance values [Ap(r)] by use of equation 1:
| (1) |
where I(r) is the observed intensity as a function of radius, Ip is the pseudoreference intensity, and r is the radius. In all cases in this report, Ip = 1,000. A function that performs this transformation is built into the public domain computer program SEDFIT (39, 57, 59). The transformation results in a signal that is proportional to the concentration of the protein, but with significant baseline offsets, scan-to-scan variations, and underlying slopes. In addition, all absorbance or pseudoabsorbance data may have significant time-invariant noise features resulting from, for example, imperfections of the cell windows. All of the aforementioned noise features were algebraically decomposed from the sedimentation signals by use of SEDFIT (58). This program was also used to calculate sedimentation-coefficient distributions [c(s)] and to quantify the amount of material sedimenting at a particular sedimentation coefficient. The c(s) plots presented in this paper were made using GUSSI (biophysics.swmed.edu/MBR/software.html). The titrations were analyzed by least-squares fitting of the response of the signal-average sedimentation coefficient (ssig) to metal ion concentrations by use of the following sigmoidal dose-response equation:
| (2) |
where smin and smax are the minimum and maximum expected s-values, respectively, and EC50 is the concentration at which a 50% response is observed. The values of smax and EC50 were fitted; smin was fixed at 1.96 S, the known sedimentation coefficient of the protein in the absence of metal ions. The same equation (equation 2) was used to fit the data for the mutants of Tp34 in the presence of Co2+, except that smax was fixed at 3.20 S (the fitted smax for the experiment with the wild-type protein and Co2+).
X-ray crystallography.
Crystals of Tp34 and its mutants were obtained as described previously (17). The crystals were transferred to a solution composed of 100 mM Tris, pH 7.5, 2.4 M ammonium sulfate, and 10 mM sulfate salt of a chosen metal ion (Zn2+ or Co2+). Following a 1-min incubation in this solution, the crystals were serially transferred to solutions having the same composition plus increasing concentrations of ethylene glycol; the final concentration of ethylene glycol was 15% (vol/vol). The total time that the crystals spent in the metal-containing buffers was between 5 and 10 min. Next, the crystals were supported on a nylon loop (Hampton Research) and plunged into liquid nitrogen. X-ray diffraction data were collected at beamline 19-BM at the Advanced Photon Source, Argonne, IL. The data were indexed, integrated, and scaled using HKL2000 (50). Negative intensities were corrected, and the data were put on an absolute scale by use of the TRUNCATE procedure in CCP4 (28). All of the crystals were isomorphous to the native crystal of Tp34 (17). The Tp34 model, denuded of heterogens, was “shaken” such that the atoms had new, random positions that differed from their original positions. The new positions compared to the old resulted in a root mean square deviation (RMSD) of 0.5 Å; this step was undertaken to remove statistical bias from the starting model and to allow an assignment of reflections to the Rfree set independently of the set used for refining the parent structure. The models thus treated were rigid-body refined using the newly acquired data sets. The new structures were further refined using the positional, simulated-annealing, individual ADP, and TLS refinement protocols available in Phenix (1). The model was adjusted manually between rounds of refinement by use of Coot (23). The identity and positions of metal ions were confirmed using anomalous difference Fourier maps calculated in CNS, version 1.1 (8), and simulated-annealing omit maps were calculated using CNS, version 1.2 (8, 35).
Protein structure accession numbers.
The coordinates and structure factors of the Co2+- and Zn2+-containing Tp34 structures have been deposited in the Protein Data Bank (PDB) (www.rcsb.org) under accession numbers 3PJL and 3PJN, respectively.
RESULTS
Metal ion-induced dimerization of Tp34.
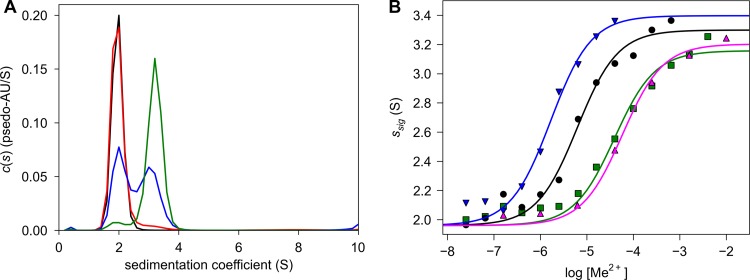
Previously, we observed that transition metal ions stabilized the dimeric form of a water-soluble construct of Tp34 (17). To ascertain which metals were the most efficacious inducers of dimerization, we performed AUC sedimentation velocity (SV) experiments on the protein under conditions that are likely to mimic the environment of T. pallidum, i.e., the human mucosa (20 mM HEPES, pH 6.8, 100 mM NaCl) (Fig. 1). In these experiments, the concentration of Tp34 (2.6 μM) was held constant and the concentration of the transition metal ion was varied. Four metal ions were chosen for study: Ni2+, Co2+, Zn2+, and Cu2+. Fe ions were not studied because they tended to precipitate or to cause the precipitation of Tp34 under these solution conditions. Experiments with Tp34 and ferric citrate were attempted, but very little dimerization was observed, and aggregation was evident at high concentrations of the latter (not shown). For these experiments, graphs of the signal-weighted sedimentation coefficient (ssig) versus the log10 of the metal concentration were evaluated (Fig. 1B). The ssig value is a measure of the bulk transport properties of all species in solution; if all Tp34 molecules were present in solution as monomers, ssig should be ∼2 S, the sedimentation coefficient of monomeric Tp34 (17). Conversely, if all of the protein had been converted completely to a dimer, the ssig value should assume the sedimentation coefficient of the dimer, i.e., ∼3.2 S. In solutions containing both species, the ssig value should be an intermediate value weighted by the amount of signal detected for each component. The data could be fit to an equation describing a Hill-type binding isotherm (see Materials and Methods), yielding an EC50 for each metal ion that is indicative of its ability to induce dimerization (Table 2). Figure 1A shows that Tp34 was a monomer under the experimental conditions and without added transition metal ions. Cu2+ most effectively induced the dimerization of Tp34, with an EC50 of 1.7 μM. Zn2+ was the second-most effective ion (EC50 = 6.2 μM), and Co2+ and Ni2+ had nearly identical titration profiles (EC50s of 58 and 39 μM, respectively). Notably, although our earlier study (17) suggested that alkali and alkaline earth metals might be able to induce Tp34 dimerization, our more recent protein preparations have not uniformly exhibited this activity (data not shown). We thus contend that our earlier results likely were the result of transition metal contamination of the protein preparation, the AUC apparatus, or the buffers used in those experiments.
Fig 1.
Titration of Tp34 with metal ions in solution. (A) c(s) distributions of Tp34 sedimented in the absence or presence of different total concentrations of Ni2+. The [Ni2+]total values for the four experiments were 0 nM (black line), 25 nM (red line), 40 μM (blue line), and 1.6 mM (green line). (B) Binding isotherms for four metal ions. The circles represent individual data points, while the lines are fits to those data as described in the text. Data for Cu2+ (blue inverted triangles and line), Zn2+ (black circles and line), Co2+ (magenta triangles and line), and Ni2+ (green squares and line) are shown.
Table 2.
EC50s obtained from centrifugation in the presence of various metal ions
| Metal | EC50 (μM)a |
|---|---|
| Cu2+ | 1.7 ± 0.3 |
| Zn2+ | 6.2 ± 1.0 |
| Ni2+ | 39 ± 7 |
| Co2+ | 58 ± 9 |
Error values shown are the square roots of the respective variances for the fitted EC50s.
Two routes to dimerization are readily conceivable in this system. In the first, a metal ion binds to one molecule of monomeric Tp34, prompting a second to interact by overcoming presumably repulsive forces that are present without the metal ion bound. In the second route, Tp34 transiently dimerizes prior to metal ion binding; a metal ion or ions bind to this dimer, stabilizing it. As designed, the experiments in this study cannot readily distinguish between these possibilities; in the absence of other data, we assume that both routes are possible and that they form a thermodynamic cycle. Despite this ambiguity, the EC50 studies presented herein clearly establish the relative dimerization efficacies of the transition metals that we studied.
Mutagenesis of the metal-ion-binding site.
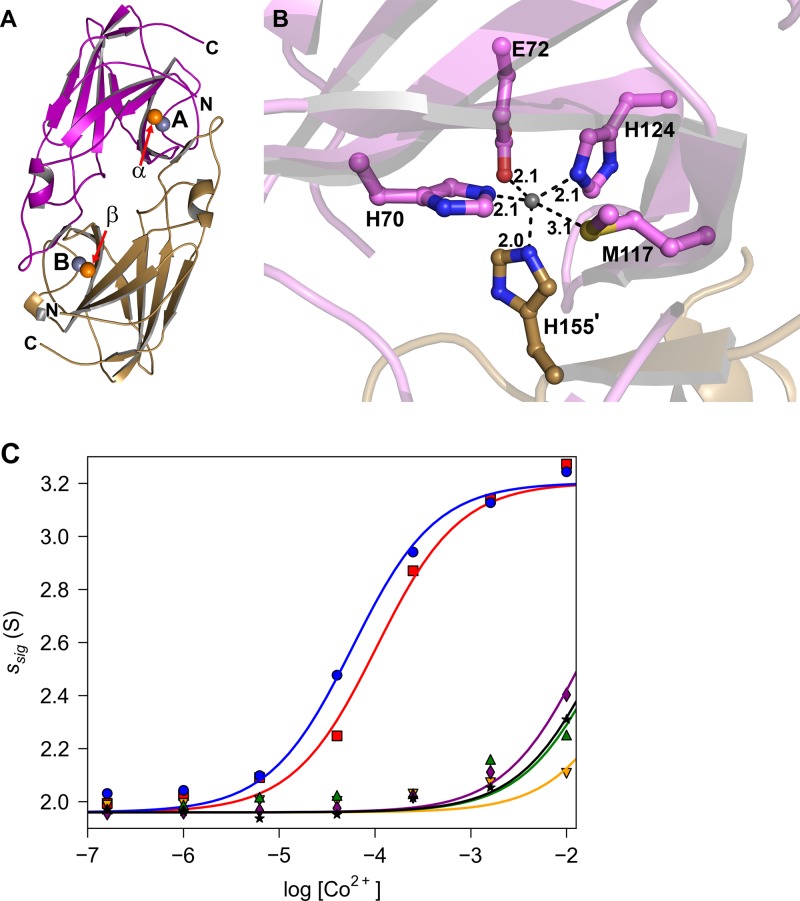
The crystallographically observed metal-ion-binding site of Tp34 is located at the dimer interface (Fig. 2A); it comprises the side chains of amino acid residues H70, E72, M117, H124, and H155′, where the prime symbol denotes that this residue originates from a neighboring monomer (Fig. 2B). Although it is highly likely that this metal-binding site is directly responsible for the solution phenomena described above, there has been no direct evidence for this hypothesis. To provide such evidence and to define the roles that these residues may play at the metal-binding site, each was individually mutated to alanine. The mutated proteins were subjected to SV as delineated above (Fig. 2C; Table 3); Co2+ was chosen to test the ability of these proteins to form metal-ion-induced dimers because it efficaciously induces dimerization and is easily observable crystallographically (see below), and unlike Cu2+ and Zn2+, it does not cause Tp34 to precipitate at high metal ion concentrations. To quantify the dimerization activities of these proteins, the same approach as that described above was taken (i.e., EC50 values were calculated). The mutation of histidine 70 to alanine (H70A) demonstrated the most severe effect, raising the apparent EC50 by about 1,000-fold (Table 3). The dimerization activities of the E72A, H124A, and H155A mutants were also severely reduced (raising the apparent EC50s by ∼500-, ∼300-, and ∼400-fold, respectively) compared to that of the wild-type protein. The apparent EC50s for these mutant proteins should be considered only estimates, because the full dimerization isotherm was not observed. Changing M117 to alanine had only a mild effect on dimerization in the presence of Co2+. These data clearly demonstrate that the crystallographically characterized metal-ion-binding site is directly responsible for the metal-induced dimerization of Tp34. The side chains of residues H70, E72, H124, and H155′ were especially critical for the formation of metal-induced dimers. The side chain of M117 appears to be nearly dispensable for the formation of Co2+-induced dimers of Tp34; similar results to those presented above were obtained for Ni2+ (data not shown).
Fig 2.
Overall structure and metal binding by Tp34. (A) Dimeric structure of Tp34 (the structure under PDB accession no. 2O6E is shown). One monomer of Tp34 is shown in tan, and the other is shown in purple. The spheres represent bound metal ions, with those bound at sites A and B (labeled; these are the main interfacial metal-binding sites) shown in gray and those bound at sites α and β (labeled; these are the most prominent “peripheral” binding sites) shown in orange. Residues 28 to 185 are resolved in the purple monomer (chain A), while residues 29 to 185 are resolved in the tan monomer (chain B). (B) The Zn2+-binding environment of Tp34 site A. The secondary structure is shown semitransparently for clarity. Dashed lines represent inner sphere contacts of the amino acid atoms with the metal ion. Distances are shown in angstroms. Carbon atoms from monomer A are shown in purple, and those from monomer B are shown in tan. All nitrogens are blue, oxygens are red, sulfur is yellow, and Zn2+ is gray. (C) Co2+-binding isotherms of mutant Tp34 proteins. The graph was constructed as described in the legend to Fig. 1B. The data and fitted lines are for the wild-type protein (blue circles and line) and the H70A (orange inverted triangles and line), E72A (green triangles and line), M117A (red squares and line), H124A (purple diamonds and line), and H155A (black stars and line) mutants.
Table 3.
EC50s obtained from centrifugation of Tp34 mutants in the presence of Co2+
| Protein | EC50 (μM)a |
|---|---|
| Wild type | 58 ± 7 |
| H70A mutant | 60,000 ± 20,000b |
| E72A mutant | 27,000 ± 8,000b |
| M117A mutant | 103 ± 15 |
| H124A mutant | 16,900 ± 1,500b |
| H155A mutant | 25,000 ± 2,000b |
Error values shown are the square roots of the respective variances for the fitted EC50s.
The entire binding isotherm for these titrations could not be observed. These values should be considered estimates.
Crystallographic examination of metal ion binding.
Previously, the crystal structure of Tp34 was reported with Zn2+ bound at the metal-binding site (17). This structure was obtained by incubating crystals of Tp34 in a solution containing 2.4 M ammonium sulfate, 100 mM Bicine, pH 9.0, and 10 mM zinc acetate. Other metal ions were refractory to this treatment owing to the facile formation of metal hydroxide crystals under these conditions (not shown). For the current study, it was observed that the Tp34 crystals were stable in a solution comprising 2.4 M ammonium sulfate, 100 mM Tris, pH 7.5, and 10 mM metal sulfate. In this solution, the crystals could be incubated without visible precipitation or crystallization of the hydroxide salt of the metal ion. This procedure resulted in Tp34 crystals that diffracted well and resulted in well-refined structures (Table 4) (12).
Table 4.
Crystallographic and refinement data for structures presented in this work
| Parameter | Value or descriptiona |
|
|---|---|---|
| Co2+ | Zn2+ | |
| Crystallographic data | ||
| PDB accession no. | 3PJL | 3PJN |
| Space group | P212121 | P212121 |
| Unit cell dimensions (Å) | ||
| a | 34.4 | 34.6 |
| b | 66.0 | 65.4 |
| c | 152.4 | 152.4 |
| α, β, γ | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 49.9–1.7 (1.76–1.70) | 32.9–1.7 (1.76–1.70) |
| Completeness (%) | 99.8 (99.6) | 99.4 (99.4) |
| Multiplicity | 4.9 (4.8) | 5.6 (4.9) |
| No. of unique reflections | 39,218 (3,822) | 38,964 (3,828) |
| Rsymb | 0.039 (0.587) | 0.053 (0.537) |
| I/σI | 35.6 (2.2) | 24.6 (2.9) |
| Wilson B value (Å2) | 23.1 | 21.3 |
| Refinement data | ||
| Resolution (Å) | 49.9–1.7 | 32.9–1.7 |
| No. of nonsolvent atoms | 2,541 | 2,537 |
| No. of solvent atoms | 218 | 200 |
| Cutoff Fo/σFo | 0 | 0 |
| Avg no. of B factors (Å2) | ||
| Nonsolvent | 28.8 | 27.2 |
| Solvent | 36.4 | 35.2 |
| Metal ions | 43.3 | 52.0 |
| R values | ||
| Rwork | 0.171 | 0.169 |
| Rfree | 0.201 | 0.195 |
| Ramachandran statisticsc | ||
| No. of outliers | 0 | 0 |
| Most favored region (%) | 97.2 | 97.7 |
| RMSD | ||
| Bonds (Å) | 0.012 | 0.011 |
| Angles (°) | 1.4 | 1.4 |
Values in parentheses are for the highest-resolution shell.
Rsym = ΣhΣi|Ih,i − 〈Ih〉|/ΣhΣiIh,i, where the outer sum (h) is over the unique reflections and the inner sum (i) is over the set of independent observations of each unique reflection.
From MolProbity (12).
The Tp34 dimer has two metal-binding sites at the dimer interface. Because of the 2-fold rotational noncrystallographic symmetry in the dimer, the two sites occur at symmetrically equivalent positions (Fig. 2A). In the crystallographic asymmetric unit, the two sites may be distinguished by considering whether most of the side chains are donated by chain A or chain B of the dimer. We term these two sites “A” and “B,” respectively. The metal-binding characteristics of the two sites differ (not shown): site B has poorer metal-binding characteristics than site A. This phenomenon is likely caused by interference at the positions of amino acids near site B by a symmetry-related dimer of Tp34 (not shown). We therefore focus on the metal-binding characteristics of site A in the following discussion.
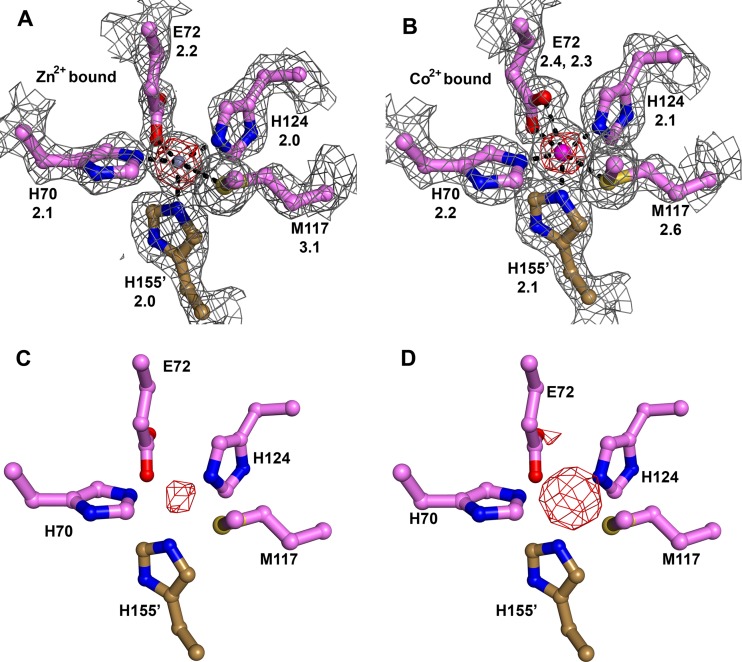
The configuration of metal-binding site A of Tp34 in the presence of Zn2+ and Tris, pH 7.5, buffer (see above) is shown in Fig. 3A. This structure was determined to ascertain whether the different incubation conditions of the crystal, i.e., the lower pH, would cause changes to the metal-binding site. No significant differences in the ligand-binding environment or occupancy of Zn2+ were observed in this structure compared to that obtained from crystals stabilized in the presence of the Bicine-buffered solution (17). Inner sphere ligands for Zn2+ are detailed above and shown in Fig. 3A; they form a distorted square pyramid around the ion. Significant difference electron density (not shown) around the refined position of the Zn2+ necessitated the refinement of anisotropic atomic displacement factors for the zinc ion. An important aspect of metal ion binding at site A is that the Zn2+ ion has no solvent-accessible surface, i.e., it is completely buried at the dimer interface.
Fig 3.
Metal binding in crystalline Tp34. For this figure, all coloration and annotation are as described in the legend to Fig. 2B. In panels A and B, for residues that contact a bound metal ion, the distance in angstroms between the metal ion and the ligand atom is given, and a σA-weighted simulated-annealing omit map (mFo − DFc) contoured at the 3-σ level (gray mesh) is superposed on the final refined models. All atoms shown were omitted for this calculation in these two panels. The positions of the side chains in panels C and D are the same as those in panel A; they are not refined and are shown only for reference. In addition, all parts show an anomalous difference Fourier map (red mesh). The contour level of this map is noted in the legend for each part, with the phrase “anomalous map contour = x.” (A) Zn2+-binding environment. The Zn2+ ion is shown in gray. Secondary structure was omitted for clarity. Anomalous map contour = 15σ. (B) Co2+-binding environment. The Co2+ ion is shown in magenta. Anomalous map contour = 20σ. (C) Anomalous difference Fourier map for Ni2+ bound to Tp34. Map contour level = 5σ. (D) Anomalous difference Fourier map for Cu2+ bound to Tp34. Map contour level = 6σ. The peak at the rear, partially occluded by the carboxylate group of E72, is evidence for Cu2+ binding at site α.
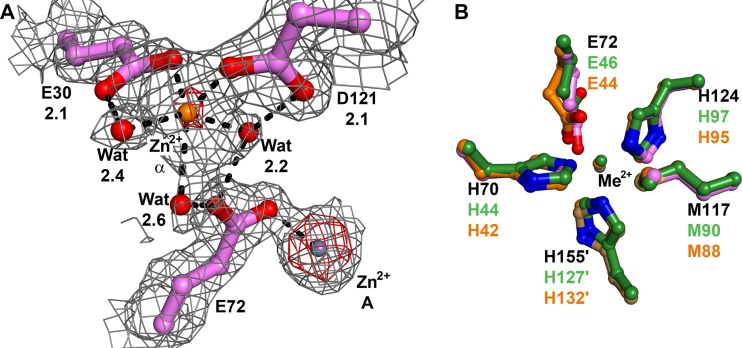
The Tp34 data set for Zn2+ was collected using a wavelength (Table 4) of X-radiation at which Zn atoms diffract anomalously. Therefore, an anomalous difference Fourier map could be calculated using these data; any strong peaks in this map would betray the position of Zn2+ ions. In this way, we established that 13 additional zinc(II) ions are bound to Tp34 under these conditions, and they are included in the model (not shown). All of them are located on the periphery of the Tp34 dimer. To distinguish these sites from metal sites A and B, we labeled the peripheral sites with the Greek alphabet, beginning with α. Sites α and β are the peripheral metal-binding sites that are most proximal to metal sites A and B, respectively. Sites α and β are related by the dyad axis of the Tp34 dimer. Metal site α is shown in Fig. 4A. Inner sphere contacts with the metal ion are made by E30 and D121. An outer sphere contact is made by E72, which is a metal-ligating residue for site A (Fig. 4A). Metal sites α and β are solvent exposed, and the Zn2+ that occupies site α has three water molecules making inner sphere contacts to it (Fig. 4A). Although the anomalous difference peak for the Zn2+ ion at site α was clearly weaker than that for the Zn2+ ion at site A, the site α metal was refined well at full occupancy, i.e., its B factor (37.5 Å2) matched well with those of the two contacting protein atoms (30.7 Å2 and 34.3 Å2). Thus, we concluded that the weaker density for this atom than for the site A metal was due to a higher degree of static or dynamic disorder for the site α metal.
Fig 4.
Metal binding in Tp34 at site α and a comparison of sites A from three proteins. (A) Metal binding at site α in Tp34. The color, map, and notation conventions are as described in the legend to Fig. 3. The site α Zn2+ is shown as an orange sphere. Anomalous map contour = 15σ. (B) Sites A from three proteins. Shown are residues from Tp34 with Zn2+ bound (colored as in Fig. 2), CJp19 (orange carbon atoms), and FetP (green carbon atoms). The metal ions from CJp19 and FetP are coppers; they are colored the same as the carbon atoms from their respective structures. The superpositions were performed using all Cα atoms from the respective proteins. The residue numbers are noted; those belonging to Tp34 are black, while those belonging to the other two structures are colored according to their respective carbon atom colors.
Also related by Tp34's dyad axis are sites ι and κ. These Zn2+-binding sites feature residues from both monomers of the dimer: H55 and H149′. These residues were not mutated for the hydrodynamic experiments presented above, and it is therefore not clear whether they affect the Zn2+-induced dimerization of Tp34. Metal sites ι and κ were not observed in the presence of the other metal ions in this study (not shown), yet those metals were able to induce dimerization. Thus, these sites do not appear to be essential to metal-induced dimerization.
The crystal structures of two Tp34 homologs, CJp19 (10) and FetP (41), are very similar to that of Tp34: the RMSD of 143 comparable Cα atoms of Tp34 and CJp19 (∼35% sequence identity) is 1.2 Å, and that of 142 comparable Cα atoms of Tp34 and FetP (∼40% identity) is also 1.2 Å. The structures of both CJp19 and FetP were determined with copper ions bound at the analogs of site A. A comparison of the sites A of these three proteins is shown in Fig. 4B. The side chains that contact the metal ions are identical, and their positions are very similar. The only side chain that shows a significant difference is that of E44 of CJp19 (the equivalent of E72 in Tp34). It adopts a conformation that allows a bidentate interaction between its carboxylate oxygen atoms and the bound metal ion. Indeed, this conformation is similar to that observed for E72 of Tp34 when Co2+ is bound (Fig. 3B). These results indicate that the disparate crystallization conditions and bound-metal identities among these three proteins do not significantly alter the disposition of the side chains in the active site.
The binding of Co2+ to metal-binding site A is very similar to that of Zn2+ (Fig. 3B). As in the Zn2+-bound structure, the ion is completely buried at the dimer interface. There is one major difference, however. The carboxylate moiety of E72 takes up a different position, allowing both of its oxygen atoms to ligate the Co2+ ion. Such a bidentate ligand interaction is common with acidic amino acid residues (e.g., see references 6, 26, and 55). The geometry of the site is therefore changed from a distorted square pyramidal (pentacoordinate) to a distorted octahedral (hexacoordinate) geometry. Although the inner sphere contact of S-δ of M117 to zinc is, to our knowledge, unprecedented (17), there are at least two structures in the current release of the Protein Data Bank (www.rcsb.org) wherein contacts between Co2+ and a methionine are observed. The crystal structures of the iron-dependent regulator proteins of Mycobacterium tuberculosis and Corynebacterium diphtheriae each feature a cobalt(II) ion that has the respective sulfur atoms of a cysteine and a methionine as inner sphere ligands (25, 52). These proteins bind to and are activated by several transition metal ions, including Fe2+, Zn2+, Co2+, and Ni2+ (56, 63). The chemical nature of the metal-binding sites in these proteins, like that of sites A and B of Tp34, is mixed, containing sulfur, oxygen, and nitrogen ligands. Hexacoordinate cobalt(II) is commonly encountered in proteins and nucleic acids crystallized in the presence of this cation; indeed, cobalt hexamine is a standard heavy-atom compound used for obtaining phases in nucleic acid crystal structures (40). However, a Co2+ ion coordinated by six protein ligands is rare. In a database current to 2003 (9), we found only one example: a second cobalt-binding site in the iron-dependent regulator protein (DtxR) from C. diphtheriae (52). In this case, a carboxylate moiety also serves as a bidentate ligand for the Co2+ ion.
In separate experiments, crystalline Tp34 was also incubated with Cu2+ and Ni2+. Anomalous difference Fourier maps clearly indicate that these metal ions bind at sites A (Fig. 3C and D). However, attempts to refine these structures indicated that the metal ions are not present at full occupancy. Thus, despite efficaciously inducing dimerization in our solution experiments (Fig. 1B), Cu2+ and Ni2+ are hindered from fully occupying sites A by some factor in the treatment of the crystals (e.g., high salt concentration, higher pH, or shorter incubation). Because of the low occupancies, we cannot draw definitive conclusions regarding the ligand-binding environment of these metals in Tp34. For Cu(II), the anomalous difference Fourier maps also indicated that this metal bound at sites B, α, β, ι, and κ. For Ni(II), these maps indicated significant occupancy of the metal ion at sites β and ι, in addition to site A.
Genes cotranscribed with the Tp34 gene (tp0971).
Genomic analysis of T. pallidum suggested that the tp0959 to tp0972 genes (tp34 gene cluster) are oriented in the same direction and thus may be cotranscribed (4). Therefore, we used RT-PCR to assess the transcriptional linkage of the putative tp34 gene cluster. The transcripts were mapped from RNA isolated from rabbit tissue-extracted T. pallidum. As shown in Fig. 5, the tp34 gene cluster was demonstrated by RT-PCR to be linked transcriptionally. These results confirm the transcriptional organization of the tp34 gene cluster in an operon (tp34 operon). As expected, the nonoperonic gene pair of tp0958 and tp0959 yielded no RT-PCR product. However, the RT-PCR product observed for the tp0972 and tp0973 pair may have been due to the presence of noncoding mRNA that is transcribed in the opposite direction of the tp34 operon.
Fig 5.
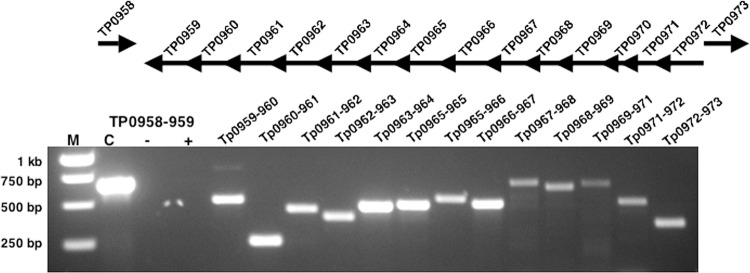
Schematic representation of the putative tp34 operon in the T. pallidum genome and results of operon mapping by RT-PCR. RT-PCR was performed on T. pallidum RNA by use of the primer pairs listed in Table 1, specific for either the intergenic regions of listed gene pairs or the entire tp0970 gene (pseudogene) between tp0969 and tp0971 (tp34). Lane M, DNA molecular size markers; lane C, PCR with the indicated primer pair, using T. pallidum genomic DNA as the template in place of RNA, served as a positive control; lane −, PCR with the indicated primer pair, using RNA as the template (without RT), served as the negative control for DNA contamination; lane +, RT-PCR product with the indicated primer pair (nonoperonic). The rest of the lanes contained RT-PCR products amplified with the indicated primer pairs.
Because bacteria often cluster genes of similar function into operons, a detailed analysis of the products of the genes cotranscribed with Tp34 might shed light on the latter protein's function. Accordingly, bioinformatic analyses of the genes cotranscribed with Tp34 were performed, as summarized in Table 5. We note that this particular arrangement of genes is unique to T. pallidum; even closely related treponemal species, such as Treponema denticola, do not display the same operon organization. The proteins encoded by genes tp0959 through tp0961 are apparently flagellar components, as BLAST searches (2) consistently indicate a close correspondence between these sequences and those of known flagellar proteins. Database annotations concur with this assignment.
Table 5.
Genes cotranscribed with tp0971 and putative functions of their gene products
| Gene | Putative function of gene product |
|---|---|
| tp0959 | Flagellar protein |
| tp0960 | Flagellar protein |
| tp0961 | Flagellar protein |
| tp0962 | FtsX family ABC permease |
| tp0963 | FtsX family ABC permease |
| tp0964 | Nucleotide-binding protein |
| tp0965 | Membrane fusion protein |
| tp0966 | TolC-like protein |
| tp0967 | TolC-like protein |
| tp0968 | TolC-like protein |
| tp0969 | TolC-like protein |
| tp0970 | Small, likely untranslated gene |
| tp0971 | Tp34; metal import |
| tp0972 | Ftr1-like iron-lead permease |
The proteins encoded by tp0962 and tp0963 are sequentially similar (36% identity). Position-specific iterated BLAST queries (3) suggest a close relationship between them and macrolide-exporting ABC-type permeases. Using algorithms that predict the presence of transmembrane α-helices (34), we confirmed that these proteins are predicted to have a similar topology to that of known macrolide exporter permeases. That is, they are predicted to have four transmembrane α-helices, with a large, water-soluble domain between helices 1 and 2. Furthermore, a conserved domain search (49) identified a subset of residues in each protein (residues 13 to 250 in Tp0962 and residues 9 to 245 in Tp0963) as homologous to the “periplasmic core domain” of macrolide transport permeases. The same search also revealed that the C termini of these proteins bear homology to the FtsX family of export permeases, whose members include macrolide exporters. All of these facts constitute a body of evidence that identifies Tp0962 and Tp0963 as export permeases. Such permeases require the function of an associated ATPase; Tp0964 is annotated as such and seems likely to fulfill this role.
Macrolide export systems generally comprise an inner membrane permease (possibly with associated ATPase proteins), a periplasmic “membrane fusion protein,” and a TolC-like outer membrane protein. In such systems, these proteins are thought to form a periplasm-spanning export complex (66). We found that genes for all of these components are in place in the Tp34 operon. Tp0965 is annotated as a membrane fusion protein, and further database searches confirmed this notion. The products of tp0966 through tp0969 are homologous to one another (see Table 6 for comparisons). They are all homologous to TolC-like proteins. This fact was ascertained using position-specific iterative searches (3) of amino acid sequence databases, which returned many other proteins annotated as TolC-like proteins. Furthermore, hidden Markov queries (62) of the Protein Data Bank using the amino acid sequences of Tp0966 to Tp0969 demonstrated that these four proteins are very likely (probability = 100%) to have structures similar to those of TolC (42), VceC (24), OprM (51), and CusC (43). All of the latter proteins are known TolC-like proteins. This result is surprising given the perceived paucity of outer membrane proteins in T. pallidum (53).
Table 6.
Pairwise amino acid identity comparisons of Tp0966 through Tp0969
| Protein | % Amino acid identity with: |
|||
|---|---|---|---|---|
| Tp0966 | Tp0967 | Tp0968 | Tp0969 | |
| Tp0966 | 26 | 22 | 21 | |
| Tp0967 | 19 | 21 | ||
| Tp0968 | 20 | |||
| Tp0969 | ||||
The combined evidence garnered by us and others thus suggests that a periplasm-spanning export system is encoded by the Tp34 operon, comprising permeases (Tp0962 and Tp0963), an ATPase (Tp0964), a membrane fusion protein (Tp0965), and TolC-like channels (Tp0966 to Tp0969). The presence of multiple permeases and channels suggests that T. pallidum may employ a “mix-and-match” strategy with these proteins to provide exporters with various specificities. The substrates for these systems, however, remain unknown. Although Tp0962 and Tp0963 appear to be similar to macrolide export permeases, macrolides would probably not be encountered commonly in any sustained fashion in the treponemal (human) environment. It is possible that other small molecules are exported, as members of this family also export lipids and peptides.
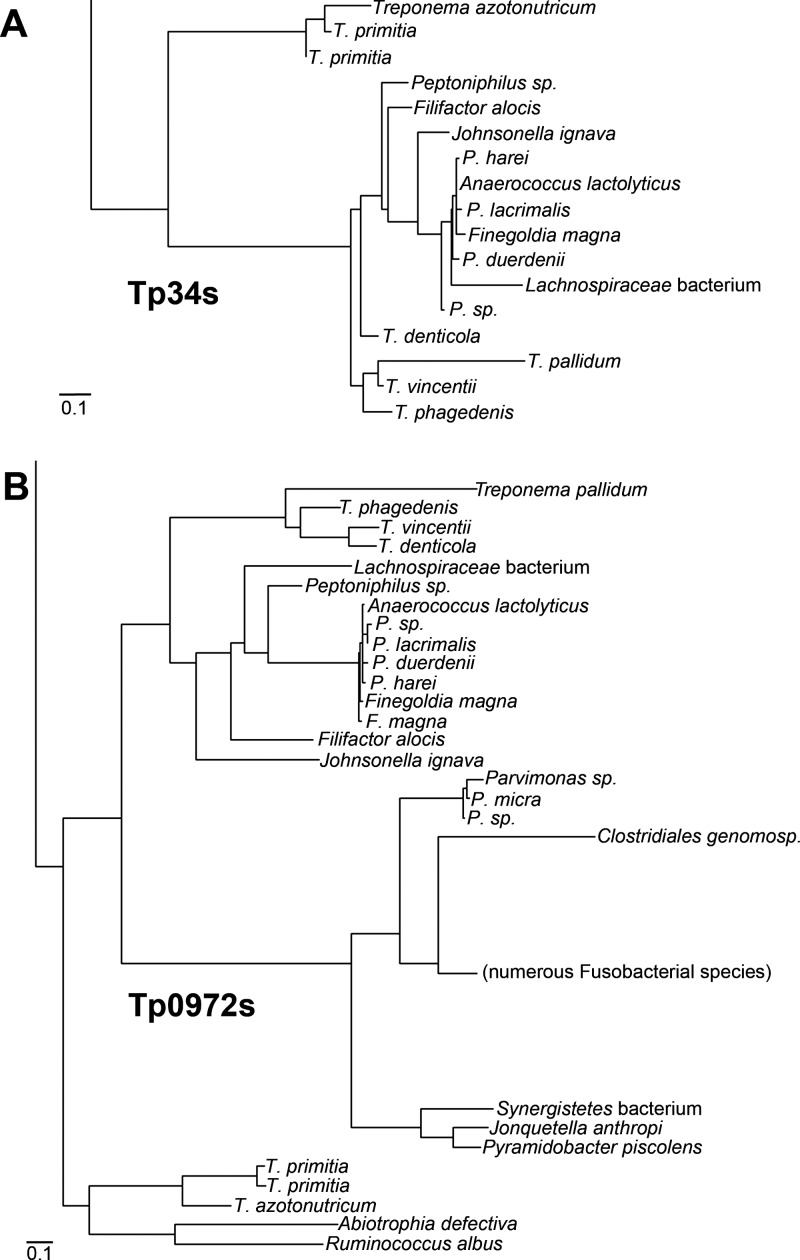
Tp0971 (Tp34) has some similarity to proteins that apparently aid in iron transport in other bacteria (10, 22, 41). It is unknown whether this sequence homology signifies functional similarity. To explore this question further, a molecular phylogeny of Tp34-like proteins was constructed (see Fig. S1 in the supplemental material). In this phylogeny, the relationships between 203 nonredundant Tp34-like proteins were examined. A few other Tp34-like proteins have been characterized functionally and structurally. They are the ChpA protein from a magnetotactic marine vibrio bacterium called MV-1 (22), a protein from Campylobacter jejuni (called CJp19 herein) (10), and the FetP protein from a uropathogenic strain (F11) of E. coli (41). All of these proteins were included in this analysis. These three proteins and Tp34 occupied distinct clades in the phylogram. Tp34 was placed in a clade containing Tp34 homologs from other treponemes (Fig. 6A). Unexpectedly, this clade also contained proteins from the anaerobic, nonspirochetal bacterial genera Johnsonella, Finegoldia, Peptoniphilus, and Anaerococcus (and an unnamed bacterium of the Lachnospiraceae). All of these nontreponemal bacteria are obligate anaerobes. Notably, T. pallidum is known to be sensitive to high-oxygen environments (15, 45).
Fig 6.
Molecular phylogenies of Tp34 and Tp0972. (A) Tp34-like proteins. (B) Tp0972-like proteins. Bar, number of substitutions per site.
In the genomes of nearly all of the species examined, an ftr1 homolog was found close to the tp34-like gene. This fact prompted a similar phylogenetic examination of Ftr1 (Tp0972) homologs. A subset (136) of the species examined for the Tp34 phylogeny was subjected to this analysis of Tp0972 homologs. The sequence of the Ftr1 homolog from MV-1 was not included because its gene is interrupted by several stop codons. The Tp0972-like proteins fall into two subgroups: those having about 400 to 450 amino acids (group I; Tp0972 belongs to this group) and those having about 650 amino acids (group II). The proteins in these subgroups are predicted to have 8 or 9 transmembrane helices, and their main differentiating features are the sizes of soluble domains that occur between two transmembrane helices. In the group I proteins, the domain is about 150 amino acids long, whereas in the group II proteins, it can comprise up to 400 amino acids. To avoid bias in the comparisons, only the amino acids in the C-terminal parts of the soluble domains and after were used to construct the phylogeny.
The results (see Fig. S2 in the supplemental material) indicate that the group I proteins occupy a distinct clade. There are outliers: the Tp0972 homologs from Magnetospirillum magneticum and Flexistipes sinusarabici are apparently slightly more closely related to the group II proteins than to the group I proteins, despite their short lengths (391 and 400 amino acids, respectively). Also, the 459-amino-acid Ftr homolog from Aggregatibacter actinomycetemcomitans serotype e strain SCC393 and the 498-amino-acid homolog from Campylobacter coli LMG 23344 fall into group II clades, despite their short sequences. Analyses of the latter proteins indicated that they are essentially N-terminal truncations of closely related group II proteins, and thus the phylogenetic analysis has placed them correctly.
Despite some differences in the overall organizational details of the phylograms (cf. Fig. S1 and S2 in the supplemental material), many of the details revealed in the Tp34 analysis are recapitulated in the Tp0972 phylogeny. For example, the same clustering of proteins from specific anaerobic genera with the treponemes is observed in both phylogenies (Fig. 6), and very similar phylogenetic organizations are found for the Tp34 and Tp0972 homologs from E. coli and C. jejuni. These facts suggest that the evolution of Tp34 homologs and their respective Tp0972 homologs is tightly correlated, and we infer from these data that the two proteins are functionally coupled.
DISCUSSION
The data presented herein and in our previous research (17) point to a role for Tp34 in transition metal import into the cytoplasm of T. pallidum. This inference is supported by the ability of the protein to bind to these metal ions (Fig. 3) and the fact that these cations efficiently induce the dimerization of Tp34 (Fig. 1). Further evidence of this role derives from the fact that the appearance of a tp34 homolog in a genome is tightly coupled with the appearance of an ftr1 homolog. The products of the latter genes have, in some cases, been shown to import the transition metal ions Fe3+ (4), Fe2+ (41), and Pb2+ (5) across the cytoplasmic membrane.
The conclusion regarding the putative function of Tp34 raises two important and related questions: (i) what is Tp34's role in metal import, and (ii) what metal ion is ordinarily imported? The answers to both questions currently remain obscure, but the results presented herein allow us to present cogent hypotheses. With respect to the first question, the behavior and functions of CJp19 and FetP deserve scrutiny, as they are structurally very similar to Tp34 (see above and Fig. 4B). Both CJp19 and FetP are dimeric; whereas the oligomerization status of CJp19 is at least somewhat responsive to metal ion concentrations (10), that of FetP is not (41). Also, both proteins are functionally associated with known iron import proteins (group II Ftr1 homologs). CJp19 is hypothesized to be a ferroxidase, and FetP a ferrireductase, although these putative activities have not been demonstrated in vitro. A third Tp34 homolog has been characterized functionally (but not crystallographically): ChpA from the MV-1 organism described above (22). ChpA isolated from its native source is dimeric and contains copper ions. This protein is known to assist in iron import into MV-1, but no enzymatic activity for ChpA was hypothesized. Rather, the authors contended that ChpA is a “copper-handling” protein that somehow aids in iron transport.
Despite the strong structural similarities to CJp19 and FetP, the distinctive solution behavior of Tp34 (see above) (17) seems most compatible with a role in transition metal homeostasis. Under nearly physiological, metal-free conditions, the protein is monomeric, but it assumes a slowly dissociating dimeric form in the presence of transition metal ions (Fig. 1). The metal ions bound at the dimer interface are completely solvent inaccessible (see above). The protein thus seems ideally suited to respond to local metal ion concentrations and to sequester these cations from the periplasm. How this activity complements the likely metal import role of Tp0971 (the Ftr1 homolog), however, remains unknown.
The second question, which regards the identity of the physiologically bound metal ion, is also currently unresolved. Notably, if the EC50s presented in this study (Table 2) are related to the metal affinities of Tp34, then the relative affinities (Cu2+ > Zn2+ > Ni2+ > Co2+) conform to those of general metal ion ligands (i.e., the Irving-Williams series [37]). Thus, the question of the in vivo identity of the metal ion at the dimerization interface likely depends largely on the relative concentrations of transition metal ions occurring naturally in the periplasm of T. pallidum. In turn, these availabilities will be driven by the microenvironment (i.e., the human host) and the actions of metal import and export systems in T. pallidum. It is perhaps noteworthy that zinc is the second-most abundant metal in the human body (14) and may be scavenged more easily than iron or copper, which tend to be bound very tightly to sequestering proteins. Thus, Zn2+ is a very plausible candidate to be bound to Tp34 in vivo.
Ultimately, a clarification of the remaining uncertainties surrounding this likely metal uptake system (Tp34/Tp0971) will probably require studies performed with other related but heterologous organisms, such as T. denticola or the other anaerobes mentioned earlier (Fig. 6). This necessity derives from the continued infeasibility of culturing and genetically manipulating T. pallidum in vitro.
ACKNOWLEDGMENTS
This study was supported by National Institutes of Health grant R01-AI056305 (to M.V.N.). Some results shown in this report derive from work performed at the Argonne National Laboratory, Structural Biology Center, at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research, under contract DE-AC02-06CH11357.
We acknowledge the technical assistance of Sandra J. Hill, Brent Biddy, and Martin Goldberg.
Footnotes
Published ahead of print 5 October 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Adams PD, et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure determination. Acta Crystallogr. D66:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403. [DOI] [PubMed] [Google Scholar]
- 3. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Askwith C, et al. 1994. The FET3 gene of S. cerevisiae encodes a multicopper oxidase required for ferrous iron uptake. Cell 76:403–410 [DOI] [PubMed] [Google Scholar]
- 5. Borremans B, Hobman JL, Provoost A, Brown NL, van der Lelie D. 2001. Cloning and functional analysis of the pbr lead resistance determinant of Ralstonia metallidurans CH34. J. Bacteriol. 183:5651–5658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brautigam CA, Aschheim K, Steitz TA. 1999. Structural elucidation of the binding and inhibitory properties of lanthanide (III) ions at the 3′-5′ exonuclease active site of the Klenow fragment. Chem. Biol. 6:901–908 [DOI] [PubMed] [Google Scholar]
- 7. Brautigam CA, Deka RK, Schuck P, Tomchick DR, Norgard MV. 2012. Structural and thermodynamic characterization of the interaction between two periplasmic Treponema pallidum lipoproteins that are components of a TPR-protein-associated TRAP transporter (TPAT). J. Mol. Biol. 420:70–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brunger AT, et al. 1998. Crystallography & NMR System: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905–921 [DOI] [PubMed] [Google Scholar]
- 9. Castagnetto JM, et al. 2002. MDB: the metalloprotein database and browser at The Scripps Research Institute. Nucleic Acids Res. 30:379–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chan ACK, et al. 2010. Structure and function of P19, a high affinity iron transporter of the human pathogen Campylobacter jejuni. J. Mol. Biol. 401:590–604 [DOI] [PubMed] [Google Scholar]
- 11. Chao JR, Khurana RN, Fawzi AA, Reddy HS, Rao NA. 2006. Syphilis: reemergence of an old adversary. Ophthalmology 113:2074–2079 [DOI] [PubMed] [Google Scholar]
- 12. Chen VB, et al. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66:12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cohen MS, Hawkes S, Mabey D. 2006. Syphilis returns to China…with a vengeance. Sex. Transm. Dis. 33:724–725 [DOI] [PubMed] [Google Scholar]
- 14. Coleman JE. 1992. Zinc proteins: enzymes, storage proteins, transcription factors, and replication proteins. Annu. Rev. Biochem. 61:897–946 [DOI] [PubMed] [Google Scholar]
- 15. Cover WH, Norris SJ, Miller JN. 1982. The microaerophilic nature of Treponema pallidum: enhanced survival and incorporation of tritiated adenine under microaerobic conditions in the presence or absence of reducing compounds. Sex. Transm. Dis. 9:1–8 [PubMed] [Google Scholar]
- 16. Debut AJ, Dumay QC, Barabote RD, Saier MH., Jr 2006. The iron/lead transporter superfamily of Fe3+/Pb2+ uptake systems. J. Mol. Biol. Biotechnol. 11:1–9 [DOI] [PubMed] [Google Scholar]
- 17. Deka RK, et al. 2007. Crystal structure of the Tp34 (TP0971) lipoprotein of Treponema pallidum: implications of its metal-bound state and affinity for human lactoferrin. J. Biol. Chem. 282:5944–5958 [DOI] [PubMed] [Google Scholar]
- 18. Deka RK, et al. 2006. The PnrA (Tp0319; TmpC) lipoprotein represents a new family of bacterial purine nucleoside receptor encoded within an ATP-binding cassette (ABC)-like operon in Treponema pallidum. J. Biol. Chem. 281:8072–8081 [DOI] [PubMed] [Google Scholar]
- 19. Deka RK, Machius M, Norgard MV, Tomchick DR. 2002. Crystal structure of the 47-kDa lipoprotein of Treponema pallidum reveals a novel penicillin-binding protein. J. Biol. Chem. 277:41857–41864 [DOI] [PubMed] [Google Scholar]
- 20. Deka RK, et al. 2004. Structural evidence that the 32-kilodalton lipoprotein (Tp32) of Treponema pallidum is an l-methionine-binding protein. J. Biol. Chem. 279:55644–55650 [DOI] [PubMed] [Google Scholar]
- 21. Desrosiers DC, et al. 2007. The general transition metal (Tro) and Zn2+ (Znu) transporters in Treponema pallidum: analysis of metal specificities and expression profiles. Mol. Microbiol. 65:137–152 [DOI] [PubMed] [Google Scholar]
- 22. Dubbels BL, et al. 2004. Evidence for a copper-dependent iron transport system in the marine, magnetotactic bacterium strain MV-1. Microbiology 150:2931–2945 [DOI] [PubMed] [Google Scholar]
- 23. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 24. Federici L, et al. 2005. The crystal structure of the outer membrane protein VceC from the bacterial pathogen Vibrio cholerae at 1.8 Å resolution. J. Biol. Chem. 280:15307–15314 [DOI] [PubMed] [Google Scholar]
- 25. Feese MD, Ingason BP, Goranson-Siekierke J, Holmes RK, Hol WGJ. 2001. Crystal structure of the iron-dependent regulator from Mycobacterium tuberculosis at 2.0-Å resolution reveals the Src homology domain 3-like fold and metal binding function of the third domain. J. Biol. Chem. 276:5959–5966 [DOI] [PubMed] [Google Scholar]
- 26. Forsén S, Kördel J. 1994. Calcium in biological systems, p 107–166 In Bertini I, Gray HB, Lippard SJ, Valentine JS. (ed), Bioinorganic chemistry. University Science Books, Sausalito, CA [Google Scholar]
- 27. Fraser CM, et al. 1998. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281:375–388 [DOI] [PubMed] [Google Scholar]
- 28. French S, Wilson K. 1978. On the treatment of negative intensity observations. Acta Crystallogr. A 34:517–525 [Google Scholar]
- 29. Gherardini FC, Boylan JA, Brett PJ. 2006. Metal utilization and oxidative stress, p 101–126 In Radolf JD, Lukehart SA. (ed), Pathogenic Treponema: molecular and cellular biology. Caister Academic Press, Norfolk, England [Google Scholar]
- 30. Golden MR, Marra CM, Holmes KK. 2003. Update on syphilis: resurgence of an old problem. JAMA 290:1510–1514 [DOI] [PubMed] [Google Scholar]
- 31. Gray-Owen SD, Schryvers AB. 1996. Bacterial transferrin and lactoferrin receptors. Trends Microbiol. 4:185–191 [DOI] [PubMed] [Google Scholar]
- 32. Hazlett KRO, et al. 2002. Contribution of neelaredoxin to oxygen tolerance by Treponema pallidum. Methods Enzymol. 353:150–156 [DOI] [PubMed] [Google Scholar]
- 33. Hazlett KRO, et al. 2003. The Treponema pallidum tro operon encodes a multiple metal transporter, a zinc-dependent transcriptional repressor, and a semi-autonomously expressed phosphoglyceride mutase. J. Biol. Chem. 278:20687–20694 [DOI] [PubMed] [Google Scholar]
- 34. Hirokawa T, Boon-Chieng S, Mitaku S. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378–379 [DOI] [PubMed] [Google Scholar]
- 35. Hodel A, Kim S-H, Brunger AT. 1992. Model bias in molecular crystal structures. Acta Crystallogr. A 48:851–859 [Google Scholar]
- 36. Hood MI, Skaar EP. 2012. Nutritional immunity: transition metals at the pathogen-host interface. Nat. Rev. Microbiol. 10:525–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Irving H, Williams RJP. 1948. Order of stability of metal complexes. Nature 162:746–747 [Google Scholar]
- 38. Jovanović T, et al. 2000. Neelaredoxin, an iron-binding protein from the syphilis spirochete, Treponema pallidum, is a superoxide reductase. J. Biol. Chem. 275:28439–28448 [DOI] [PubMed] [Google Scholar]
- 39. Kar SR, Kingsbury JS, Lewis MS, Laue TM, Schuck P. 2000. Analysis of transport experiments using pseudo-absorbance data. Anal. Biochem. 285:135–142 [DOI] [PubMed] [Google Scholar]
- 40. Keel AY, Rambo RP, Batey RT, Kieft JS. 2007. A general strategy to solve the phase problem in RNA crystallography. Structure 15:761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Koch D, et al. 2011. Characterization of a dipartite iron uptake system from uropathogenic Escherichia coli strain F11. J. Biol. Chem. 286:25317–25330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Koronakis V, Eswaran J, Hughes C. 2004. Structure and function of TolC: the bacterial exit duct for proteins and drugs. Annu. Rev. Biochem. 73:467–489 [DOI] [PubMed] [Google Scholar]
- 43. Kulathila R, Kulathila R, Indic M, van den Berg B. 2011. Crystal structure of Escherichia coli CusC, the outer membrane component of a heavy metal efflux pump. PLoS One 6:e15610 doi:10.1371/journal.pone.0015610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lee Y-H, Deka RK, Norgard MV, Radolf JD, Hasemann CA. 1999. Treponema pallidum TroA is a periplasmic zinc-binding protein with a helical backbone. Nat. Struct. Biol. 6:628–633 [DOI] [PubMed] [Google Scholar]
- 45. Lombard M, Touati D, Fontecave M, Nivière V. 2000. Superoxide reductase as a unique defense system against superoxide stress in the microaerophile Treponema pallidum. J. Biol. Chem. 275:27021–27026 [DOI] [PubMed] [Google Scholar]
- 46. Luthra A, et al. 2011. The transition from closed to open conformation of Treponema pallidum outer membrane-associated lipoprotein TP0453 involves membrane sensing and integration by two amphipathic helices. J. Biol. Chem. 286:41656–41668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ma Z, Jacobsen FE, Giedroc DP. 2009. Coordination chemistry of bacterial metal transport. Chem. Rev. 109:4644–4681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Machius M, et al. 2007. Structural and biochemical basis for polyamine binding to the Tp0655 lipoprotein of Treponema pallidum: putative role for Tp0655 (TpPotD) as a polyamine receptor. J. Mol. Biol. 373:681–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Marchler-Bauer A, et al. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37:D205–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326 [DOI] [PubMed] [Google Scholar]
- 51. Phan G, et al. 2010. Structural and dynamical insights into the opening mechanism of P. aeruginosa OprM channel. Structure 18:507–517 [DOI] [PubMed] [Google Scholar]
- 52. Pohl E, Holmes RK, Hol WGJ. 1999. Crystal structure of a cobalt-activated diphtheria toxin repressor-DNA complex reveals a metal-binding SH3-like domain. J. Mol. Biol. 292:653–667 [DOI] [PubMed] [Google Scholar]
- 53. Radolf JD. 1995. Treponema pallidum and the quest for outer membrane proteins. Mol. Microbiol. 16:1067–1073 [DOI] [PubMed] [Google Scholar]
- 54. Santos-Silva T, et al. 2006. The first crystal structure of class III superoxide reductase from Treponema pallidum. J. Biol. Inorg. Chem. 11:548–558 [DOI] [PubMed] [Google Scholar]
- 55. Sazinsky MH, Merkx M, Cadieux E, Tang S, Lippard SJ. 2004. Preparation and X-ray structures of metal-free, dicobalt and dimanganese forms of soluble methane monooxygenase from Methylococcus capsulatus (Bath). Biochemistry 43:16263–16276 [DOI] [PubMed] [Google Scholar]
- 56. Schmitt MP, Holmes RK. 1993. Analysis of diphtheria toxin repressor interactions and characterization of a mutant repressor with decreased activity for divalent metals. Mol. Microbiol. 9:173–181 [DOI] [PubMed] [Google Scholar]
- 57. Schuck P. 2000. Size distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 78:1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schuck P, Demeler B. 1999. Direct sedimentation analysis of interference optical data in analytical ultracentrifugation. Biophys. J. 76:2288–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schuck P, Perugini MA, Gonzales NR, Howlett GJ, Schubert D. 2002. Size-distribution analysis of proteins by analytical ultracentrifugation: strategies and application to model systems. Biophys. J. 82:1096–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Silver S, Phung LT. 2005. A bacterial view of the periodic table: genes and proteins for toxic inorganic ions. J. Ind. Microbiol. Biotechnol. 32:587–605 [DOI] [PubMed] [Google Scholar]
- 61. Simms I, et al. 2005. The re-emergence of syphilis in the United Kingdom: the new epidemic phases. Sex. Transm. Dis. 32:220–226 [DOI] [PubMed] [Google Scholar]
- 62. Söding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tao X, Murphy JR. 1992. Binding of the metalloregulatory protein DtxR to the diphtheria tox operator requires a divalent heavy metal ion and protects the palindromic sequence from DNase I digestion. J. Biol. Chem. 267:21761–21764 [PubMed] [Google Scholar]
- 64. Thumiger A, et al. 2006. Crystal structure of antigen TpF1 from Treponema pallidum. Proteins 62:827–830 [DOI] [PubMed] [Google Scholar]
- 65. Waldron KJ, Tutherford JC, Ford D, Robinson NJ. 2009. Metalloproteins and metal sensing. Nature 460:823–830 [DOI] [PubMed] [Google Scholar]
- 66. Xu Y, et al. 2011. Functional implications of an intermeshing cogwheel-like interaction between TolC and MacA in the action of macrolide-specific efflux pump MacAB-TolC. J. Biol. Chem. 286:13541–13549 [DOI] [PMC free article] [PubMed] [Google Scholar]