Abstract
Streptomyces davawensis JCM 4913 synthesizes the antibiotic roseoflavin, a structural riboflavin (vitamin B2) analog. Here, we report the 9,466,619-bp linear chromosome of S. davawensis JCM 4913 and a 89,331-bp linear plasmid. The sequence has an average G+C content of 70.58% and contains six rRNA operons (16S-23S-5S) and 69 tRNA genes. The 8,616 predicted protein-coding sequences include 32 clusters coding for secondary metabolites, several of which are unique to S. davawensis. The chromosome contains long terminal inverted repeats of 33,255 bp each and atypical telomeres. Sequence analysis with regard to riboflavin biosynthesis revealed three different patterns of gene organization in Streptomyces species. Heterologous expression of a set of genes present on a subgenomic fragment of S. davawensis resulted in the production of roseoflavin by the host Streptomyces coelicolor M1152. Phylogenetic analysis revealed that S. davawensis is a close relative of Streptomyces cinnabarinus, and much to our surprise, we found that the latter bacterium is a roseoflavin producer as well.
INTRODUCTION
The Gram-positive bacterium Streptomyces davawensis was first isolated from a Philippine soil sample within the framework of a screening program for novel antibiotics (45). The species name refers to the site of sampling, which was near Davao City in the Philippines (53). S. davawensis was found to produce a compound which exhibited antibiotic activity against Bacillus subtilis, Staphylococcus aureus, Bacillus cereus, Bacillus cereus var. mycoides, and Micrococcus luteus (Sarcina lutea) (45). Due to its red color and its structural similarity to riboflavin (vitamin B2), the novel antibiotic was named roseoflavin (45) (see Fig. S1 in the supplemental material). Roseoflavin is taken up by many bacteria via riboflavin transporters (18, 20, 35, 59). Subsequently, roseoflavin is activated to roseoflavin mononucleotide (RoFMN) and roseoflavin adenine dinucleotide (RoFAD) by promiscuous flavokinases (EC 2.7.1.26)/FAD synthetases (EC 2.7.7.2) (17, 48) (see Fig. S1). RoFMN and RoFAD have different physicochemical properties than the cofactors flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD) and very likely reduce the activity of some (if not all) flavoproteins present within a cell (52, 60). In addition, non-protein cellular targets for RoFMN were discovered. According to these works, RoFMN reduces expression of genes controlled by FMN riboswitches in B. subtilis, Streptomyces coelicolor, and the human pathogen Listeria monocytogenes (33, 35, 46, 47). FMN riboswitches are genetic elements involved in the regulation of riboflavin biosynthesis and transport (62). Blocking of FMN riboswitches leads to riboflavin auxotrophy of target cells (33). Notably, S. davawensis carries a highly specialized FMN riboswitch which is not negatively affected by RoFMN and thus confers roseoflavin resistance to S. davawensis (47).
The biosynthesis of roseoflavin remains to be elucidated. It was postulated that roseoflavin is synthesized from riboflavin through 8-amino-8-demethyl-riboflavin and 8-methylamino-8-demethyl-riboflavin (25, 38). Recently, a novel N,N-dimethyltransferase (encoded by rosA) was reported to catalyze the two sequential S-adenosylmethionine-dependent methylation reactions necessary to convert 8-amino-8-demethyl-riboflavin into roseoflavin (24). The remaining genes (enzymes) of the roseoflavin biosynthetic pathway, however, could not be localized. Riboflavin is synthesized in a series of enzymatic reactions starting from guanosine-5′-triphosphate (GTP) and two molecules of ribulose 5-phosphate (14). The enzymes involved are a bifunctional GTP cyclohydrolase II/3,4-dihydroxy-2-butanone-4-phosphate synthase (RibA; EC 3.5.4.25 and EC 4.1.99.12), a bifunctional riboflavin specific deaminase/reductase (RibG; EC 3.5.4.26 and EC 1.1.1.193), a lumazine synthase (RibH; EC 2.5.1.9), and a riboflavin synthase (RibB; EC 2.5.1.9) (see Fig. S2 in the supplemental material).
We now report the chromosomal sequence of S. davawensis and a large plasmid. The sequence was compared to the genomic sequences of other Streptomyces strains, and special attention was paid to the biosynthesis of riboflavin and roseoflavin.
MATERIALS AND METHODS
Strains and plasmids.
S. davawensis (JCM 4913) (53) was obtained from the Japan Collection of Microorganisms (JCM). Streptomyces cinnabarinus (DSM 40467) was obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ).
Culture conditions and DNA isolation.
S. davawensis and S. cinnabarinus were aerobically grown in a yeast-starch (YS) nutrient broth containing yeast extract (2 g/liter) and soluble potato starch (10 g/liter). S. davawensis was grown at 37°C. S. cinnabarinus was grown at 30°C. Preparation of S. davawensis genomic DNA was performed using a modified protocol of the Kirby mix procedure (28). Precipitation of genomic DNA was done by adding polyethylene glycol 6000 (39% [wt/vol] in water).
Genome sequencing and assembly.
Sequencing was performed using the ultrafast pyrosequencing technology with special chemistry for G+C-rich organisms as described earlier (51). For sequencing, two genomic libraries were constructed, a standard 454 WGS library and an 8-kb paired-end library, using standard kits (454 Life Sciences, MD). High-throughput pyrosequencing was carried out on a Genome Sequencer FLX system (454 Life Sciences). The subsequent assembly of the generated reads was performed using the Newbler assembly software, version 2.0.00.22 (454 Life Sciences). Details of the sequencing and assembly procedures are described elsewhere (51).
Prediction of open reading frames and functional annotation.
Potential protein-coding sequences (CDSs) were identified using the GenDB genome annotation tool. This program combines a series of programs in an automated annotation pipeline (42). For the identification of CDSs, the prokaryotic gene finders Prodigal (21) and GISMO (29) were employed. To optimize the results and to allow for easy manual correction, further methods were applied by means of the Reganor software (34, 39) utilizing the gene prediction tools Glimmer (11) and CRITICA (3). To link the identified open reading frames to potential functions, different software packages were used to analyze DNA and amino acid sequences. To identify conserved sequences, similarity-based searches against public and/or proprietary nucleotide and protein databases were performed by employing BLASTp (10) and RPS-BLAST (36, 37). From significant sequence similarities of the major section of a gene, similar functions in S. davawensis were determined. Enzymatic classification was carried out on the basis of enzyme commission (EC) numbers (4). Primarily, they were derived from the PRIAM database (9) using the PRIAM search tool (May 2011 version). As a second approach, when no PRIAM result was available, EC number annotations were derived from searches against the Kyoto Encyclopedia of Genes and Genomes databases (5, 26, 27). Further functional gene annotations were performed using the database (March 2003 version) of the cluster of orthologous groups of proteins (COG) classification system (56–58). Secondary metabolite gene clusters were identified using the antiSMASH software pipeline, which allows automated identification of gene clusters of all known secondary metabolite compound classes (40). To identify potential transmembrane proteins, the software TMHMM (30, 54) was used. Finally, the genome was examined manually using the BLASTp program (10) and Artemis (8).
Phylogenetic analysis.
The 16S rRNA gene-based phylogenetic analysis was performed on DNA sequences retrieved by public BLAST (1, 2) searches against the complete 16S rRNA gene of S. davawensis as predicted by the online tool RNAmmer (31). From the best hits, a multiple-sequence alignment was built and phylogeny was derived by applying the ClustalW2 program (16, 32). Genome-based phylogenetic analysis was performed with the comparative genomics tool EDGAR (7). Employing this software pipeline, a core genome of all selected Streptomyces strains was calculated and phylogenetic distances were derived from multiple-sequence alignments. The phylogenetic trees were generated from concatenated core gene alignments.
Comparative analysis of Streptomyces genomes.
Comparisons on a genomic scale were performed using EDGAR (7). For all calculations, BLASTp was used with BLOSUM62 as the similarity matrix. To discriminate between nonspecific hits and orthologous genes, EDGAR calculates score ratio values (SRVs) for all BLAST hits of one genome versus another. The basis for all of the analyses in this work was a calculated SRV of 0.935. For the calculation of the core genome and genes unique to S. davawensis, the complete genome sequences of the following Streptomyces species were used: S. davawensis, Streptomyces avermitilis MA-4680 (NC 003155) (22), Streptomyces coelicolor A3 (NC 003888) (2, 6), Streptomyces sp. strain SirexAA-E (NC 015953), Streptomyces scabiei 8722 (NC 013929), Streptomyces flavogriseus ATCC 33331 (NC 016114), and Streptomyces griseus subsp. griseus NBRC 13350 (NC 010572) (44). For the comparison of the riboflavin gene clusters, additional genomes of the following Streptomyces species were employed: Streptomyces violaceusniger Tu 4113 (NC 015957), Streptomyces cattleya NRRL 8057 (NC 016111), and Streptomyces bingchenggensis BCW-1 (NC 016582) (61).
Computational identification of riboswitches in S. davawensis was done by performing profile searches. For this purpose, the covariance models (CMs) of known riboswitch families were downloaded from the Rfam database (version 10.0) (15), and cmsearch, which is part of infernal 1.0.2 (43), was used to search the genome of S. davawensis for sequences that match the CMs. Matches that exceeded the gathering threshold of the CMs were reported as family members.
Analysis of roseoflavin.
The high-performance liquid chromatography-mass spectrometry (HPLC-MS) analysis of roseoflavin and other flavins was carried out as described previously (24).
Construction of a genomic S. davawensis library.
A Streptomyces/Escherichia coli P1-derived artificial chromosome (PAC) library was constructed using pESAC13. This plasmid is a derivative of pPAC-S1 (55) that can be transferred from E. coli into actinomycetes by conjugation (M. Sosio et al., unpublished data). High-molecular-weight DNA was isolated from S. davawensis and was partially digested to yield fragments with a size of about 120 kb. The fragments were ligated to BamHI-treated PACs. Notably, pESAC13 contains an attP-int-tsr cassette which allows integration of DNA molecules into the chromosomes of Streptomyces species. E. coli DH10B was used as a host for the construction of the PAC library.
Library screening and heterologous expression of S. davawensis genes in Streptomyces coelicolor.
The S. davawensis PAC library obtained as described above was screened by PCR for clones containing the gene rosA, which is involved in roseoflavin biosynthesis (24). First, the PAC library clones were screened for the presence of a 500-bp internal fragment using the oligonucleotides 100rosmfw (5′-GGA GGA ATG GGA CGA GGT GAT GG-3′) and 100rosmrv (5′-GGT TGT CGT AGT AGC GGG CGA T-3′). In a second step, positively reacting clones from step 1 were screened for the presence of a 500-bp fragment about 50 kbp upstream of rosA using the oligonucleotides 100ros5′fw (5′-GCC AGG TGA CGA AGG AAG GGA A-3′) and 100ros5′rv (5′-GGA ACT GGA TGG GGT CGG TGT A-3′). In a third step, positively reacting clones from step 2 were screened for the presence of a 500-bp fragment about 50 kbp downstream of rosA using the oligonucleotides 100ros3′fw (5′-GGC TTC GAG GGT GTG CAG GTT G-3′) and 100ros3′rv (5′-TCA ATG AGG TGG GGG TAG GCG G-3′). Finally, two positive clones were identified and used for triparental conjugation (M. Bibb, unpublished data) to move the PAC DNA together with the helper plasmid pR9406 from E. coli DH10B to the methylation-deficient E. coli strain GM2163, which is needed for conjugation with Streptomyces species. The resulting E. coli strains were used for intergeneric conjugation with S. coelicolor M1152 (Δact Δred Δcpk Δcda rpoB [C1298T]). Exconjugants were grown in liquid YS for 7 days. Production of roseoflavin was monitored by HPLC-MS.
Nucleotide sequence accession numbers.
The complete annotated chromosomal sequence was deposited at the European Nucleotide Archive (EBI) under EBI HE971709. The annotated sequence of the plasmid pSDA is available under EBI HE971710.
RESULTS
High-throughput pyrosequencing of the S. davawensis genome.
The determination of the S. davawensis genomic sequence was accomplished by combining the data generated by paired-end and whole-genome shotgun pyrosequencing strategies (51). Utilizing the Newbler software, the combined assembly of both runs resulted in a draft genome sequence comprising 184 contigs (150 contigs, ≥500 bases) and 9,538,124 nucleotides at a 34.1-fold coverage assembled from 1,078,304 reads. Gap closure between the remaining contigs was carried out by genomic PCR technology (341 reads). Two small gaps in the chromosomal sequence could not be closed. The first gap of an estimated 1,247 bp is located between positions 1,520,805 and 1,522,053, and the second gap of an estimated 200 bp is located between positions 7,358,125 and 7,358,325.
General features of the S. davawensis genome.
S. davawensis contains a linear chromosome of 9,466,619 bp and a linear plasmid of 89,331 bp. General features of the chromosome and plasmid sequence are shown in Table 1 and Fig. 1. Both arm regions of the S. davawensis genome show a remarkably lower G+C content than the overall sequence (Fig. 1A and B). The S. davawensis chromosome contains 8,503 CDSs with a coding percentage of 90.1%. The hypothetical gene products of 6,144 CDSs were identified as conserved proteins with known or putative function, and 2,032 CDSs were identified as conserved gene products with unknown function. For the hypothetical gene products of 327 CDSs, no significant similarity was found with any of the applied methods for functional annotation. Classification of 4,080 CDSs with an annotated COG category is shown in Fig. S3 in the supplemental material.
Table 1.
General features of the Streptomyces davawensis genome
| Chromosome or plasmid parameter | Value |
|---|---|
| Linear chromosome | |
| Length (bp) | 9,466,619 |
| G+C content (%) | 70.58 |
| CDSs (n) | |
| Conserved with protein function assigned | 6,144 |
| Conserved with unknown protein function | 2,032 |
| Nonconserved | 327 |
| Total | 8,503 |
| Avg CDS size (bp) | 1,003 |
| Coding density (per kbp) | 0.90 |
| Coding percentage | 90.1 |
| RNA (n) | |
| rRNA (16S-23S-5S) | 6 |
| tRNA | 69 |
| tmRNA | 1 |
| 4.5S RNA | 1 |
| Linear plasmid pSDA | |
| Length (bp) | 89,331 |
| G+C content (%) | 69.89 |
| CDSs (n) | |
| Conserved with protein function assigned | 23 |
| Conserved with unknown protein function | 44 |
| Nonconserved | 46 |
| Total | 113 |
| Avg CDS size (bp) | 676 |
| Coding density (per kbp) | 1.26 |
| Coding percentage | 85.5 |
| RNA (n) | 0 |
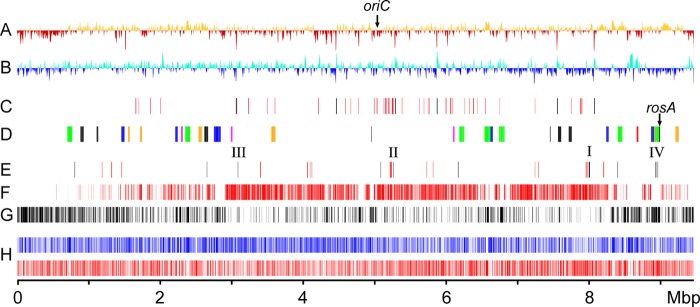
Fig 1.
Schematic representation of the Streptomyces davawensis genome. (A) G+C content variation for nonoverlapping 10-kb windows and a 200-bp step size. Yellow and red are above and below the mean, respectively. (B) GC skew for nonoverlapping 10-kb windows and a 200-bp step size. Light blue and blue are above and below the mean, respectively. (C) Distribution of rRNA gene clusters (black) and tRNA genes (red). (D) Distribution of putative secondary metabolite gene clusters (green, nonribosomal peptide synthetases [NRPS]; blue, polyketide synthases [PKS]; yellow, terpenes; red, lantibiotics; magenta, siderophores; black, others). The gene rosA (BN159_8032) encodes a roseoflavin-specific N,N-8-amino-8-demethyl-d-riboflavin dimethyltransferase (24). (E) Distribution of riboflavin/FMN/FAD biosynthesis genes (black). I, ribBMAH gene cluster (BN159_7144, BN159_7145, BN159_7146, and BN159_7147); II, ribA-ribG cluster (BN159_4727 and BN159_4729); III, ribC (BN159_2715); IV, additional bifunctional ribA (BN159_7984) and ribB (BN159_7986) and a monofunctional gene, ribA (BN159_8007), are located close to the rosA gene cluster. Putative GTP biosynthesis genes are shown (red). (F) Core Streptomyces genome compared to the genomes of S. davawensis, Streptomyces avermitilis, Streptomyces coelicolor, Streptomyces sp. strain SirexAA-E, Streptomyces scabiei, Streptomyces flavogriseus, and Streptomyces griseus subsp. griseus. (G) Singletons compared to the genomes listed in line F. (H) Distribution of CDSs according to the direction of transcription (blue, negative strand; red, positive strand).
The chromosome contains six rRNA operons with an average G+C content of 57.48% and 69 tRNA (51 different species) genes with an average G+C content of 62.23% (Fig. 1C). The chromosome has long terminal inverted repeats of 33,255 bp each, and the corresponding telomere ends contain several palindromes whose loop sequences are 5′-GGA-3′ (see Fig. S4A in the supplemental material), as in S. griseus IFO (44).
The S. davawensis origin of replication is highly similar to corresponding sequences of S. lividans, S. coelicolor, S. avermitilis, and S. scabiei. It is located between the genes dnaN and dnaA (positions 5,046,456 to 5,047,210) and contains 19 DnaA box-like sequences (23).
The linear S. davawensis plasmid pSDA contains 113 CDSs with a coding percentage of 85.5%. The hypothetical gene products of 23 CDSs were identified as conserved proteins with known or putative function, and 44 CDSs were identified as conserved proteins with unknown function. For 46 CDSs, no significant similarity to any other protein was found. The plasmid pSDA does not harbor genes for stable RNAs. pSDA contains imperfect terminal inverted repeats of 191 bp (with 9 differing nucleotides). The telomeres contain several palindromes that form stable 5′-GCA-3′ loops (see Fig. S4B in the supplemental material). A potential origin of replication was not identified in the sequence of pSDA.
Potential regulatory riboswitch sequences present in the genome of S. davawensis are shown in Table S1 in the supplemental material.
Comparative analysis of the S. davawensis genome.
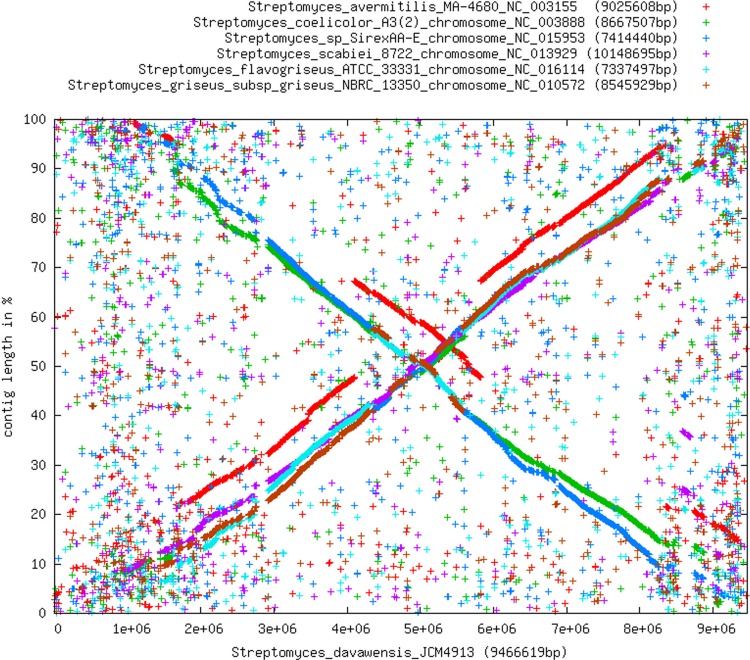
All CDSs of seven different Streptomyces genomes (S. davawensis, S. avermitilis, S. coelicolor, Streptomyces sp. strain SirexAA-E, S. scabiei, S. flavogriseus, and S. griseus subsp. griseus) were compared using the software platform EDGAR (7). To create synteny plots, every CDS of the S. davawensis genome contig was examined for a reciprocal best BLASTp hit in the genome contig for comparison. If such a hit was found, the stop positions of both CDSs were read from the database and used as coordinates for a dot. The synteny plots illustrate a highly conserved internal core region and several large inversions centered around oriC (Fig. 2).
Fig 2.
Synteny plots of all annotated proteins of seven Streptomyces chromosomes (see species listed in the legend to Fig. 1F). The software pipeline EDGAR was used to create the plots. Every CDS of the S. davawensis contig (genome) was examined for a reciprocal best BLASTp hit in the genome contig for comparison. If a hit was found, the stop positions of both CDSs were read from the database and used as coordinates for a dot.
The CDSs of S. davawensis were also used as a reference to calculate a core genome from the seven Streptomyces species. The latter analysis revealed a core genome consisting of 2,932 CDSs (Fig. 1F). The corresponding CDSs are more frequently located in the central region of the genome than in the peripheral arm regions and most likely represent highly conserved proteins among all Streptomyces species with essential cell functions. A total of 1,694 CDSs did not have a reciprocal BLASTp analysis match with an SRV higher or equal to the master cutoff compared to any of the other six Streptomyces genomes (Fig. 1G). These singleton genes are found more frequently in the arm regions of the genome and in some cases coincide with putative secondary metabolite gene clusters (Fig. 1D). The EDGAR tool was also used to calculate a pangenome from the seven Streptomyces species. The pangenome contains the core genome, genes present in two or more genomes, and the genes that are only present within one genome (41). The pangenome was calculated to contain 20,955 genes.
Distribution of putative riboflavin biosynthesis genes in S. davawensis.
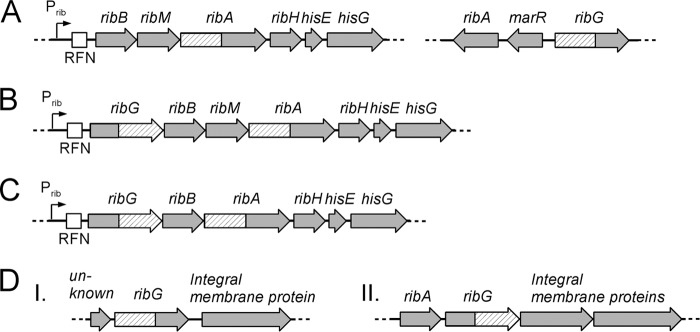
S. davawensis naturally produces and secretes the riboflavin analog roseoflavin. Riboflavin is the precursor of roseoflavin, and the purine nucleotide GTP in turn is the precursor of riboflavin. Therefore, the genomic data of S. davawensis in particular were analyzed with regard to riboflavin and purine metabolism (Fig. 1E). The riboflavin biosynthetic genes are distributed over the chromosome, forming two gene clusters. One cluster contains the genes ribBMAH (BN159_7144, BN159_7145, BN159_7146, and BN159_7147), coding for riboflavin synthase (RibB), a flavin transporter (RibM), bifunctional 3,4-dihydroxy-2-butanone-4-phosphate synthase/GTP cyclohydrolase II (RibA), and lumazine synthase (RibH) (Fig. 3A). On another subgenomic fragment, ribG (BN159_4729), coding for bifunctional riboflavin-specific deaminase/reductase, and ribA, coding for a monofunctional GTP cyclohydrolase II (BN159_4727), are present (Fig. 3A). The functions of the genes ribB-ribM, ribG, and monofunctional ribA have been verified experimentally in an earlier work (18). Moreover, it was recently shown that the ribBMAH genes are controlled at the level of translation by an FMN riboswitch (ribB FMN riboswitch, or RFN element) located directly upstream of ribBMAH (47).
Fig 3.
Riboflavin biosynthetic gene clusters among 10 Streptomyces species. Prib is a promoter and RFN denotes a regulatory element (FMN riboswitch). The genes ribA and ribG are bifunctional. (A) The ribBMAH cluster present in Streptomyces davawensis, Streptomyces coelicolor, Streptomyces avermitilis, and Streptomyces scabiei and the second cluster with ribA, marR, and ribG present in S. davawensis, S. avermitilis, and S. scabiei. (B) The ribGBMAH cluster present in Streptomyces griseus subsp. griseus, Streptomyces flavogriseus, Streptomyces sp. strain SirexAA-E, and Streptomyces cattleya. (C) The ribGBAH gene cluster present in Streptomyces violaceusniger and Streptomyces bingchenggensis (lacking ribM). (D) The two ribG gene loci of S. coelicolor. In panel I, RibG is similar to S. davawensis RibG (see panel A); in panel II, RibG is similar to RibG present in the ribGBMAH cluster of species listed in panel B. Interestingly, the two different bifunctional RibG enzymes differ with regard to the arrangement of their deaminase/reductase functions. In panel I the RibG reductase function is present in the N-terminal part, whereas the deaminase function is present in the C-terminal part. In panel II the situation is reversed. Bifunctional RibA always carries its 3,4-dihydroxy-2-butanone-4-phosphate synthase function at the N terminus and its GTP-cyclohydrolase function at the C terminus.
Interestingly, S. davawensis (like other Streptomyces species) was found to contain additional putative riboflavin biosynthesis genes (Table 2). The gene products of BN159_0742, BN159_2369, BN159_4557, BN159_5537, and BN159_7503 contain a RibG C-terminal domain that possibly is responsible for the reduction of the ribosyl side chain of 2,5-diamino-6-ribosylamino-4(3H)-pyrimidinone 5′-phosphate (riboflavin-specific reductase reaction) (50). Moreover, a second monofunctional GTP cyclohydrolase II gene (BN159_7984; ribA) is present, as is a unique putative monofunctional 3,4-dihydroxy-2-butanone-4-phosphate synthase gene (BN159_8007, ribA) which could complement the monofunctional GTP cyclohydrolase II genes (BN159_4727 and BN159_7984). The monofunctional BN159_8007 is located in a region not found in other Streptomyces species and is present within a 50-kb window 25 kb upstream of the roseoflavin biosynthesis gene rosA (BN159_8032) (24) (Fig. 4). Intriguingly, a second FMN riboswitch (ribA FMN riboswitch) was found directly upstream of BN159_8007. This functional riboswitch was found to control BN159_8007 at the level of translation (47). A second riboflavin synthase gene (BN159_7986; ribB) was found in a gene cluster 25 kb upstream of monofunctional BN159_8007 (Fig. 4). The putative gene product is very similar to archaeal riboflavin synthases, which do not show similarity to the corresponding eubacterial enzymes (14). The presence of BN159_7986 may be a result of horizontal gene transfer. Genes similar to BN159_7986 and BN159_8007 are not present in any of the other Streptomyces genomes (see Table S2 in the supplemental material).
Table 2.
Riboflavin biosynthesis genes in Streptomyces davawensis
| CDS | Protein and/or function | BLASTP best hit |
|---|---|---|
| BN159_2715 | RibC, bifunctional flavokinase (EC 2.7.1.26) (C-terminal amino acids)/FAD synthetase (EC 2.7.7.2) (N-terminal amino acids) (see reference 17 for experimental proof) | ref|ZP_09958537.1|; bifunctional riboflavin kinase/FMN adenylyltransferase; Streptomyces chartreusis NRRL 12338; identity, 87% |
| BN159_4727 | RibA, GTP cyclohydrolase II (EC:3.5.4.25), monofunctional RibA (see reference 18 for experimental proof) | ref|YP_006246117.1|; YP_006246117.1; unnamed protein product; Streptomyces hygroscopicus subsp. jinggangensis 5008; identity, 90% |
| BN159_4729 | RibG, bifunctional riboflavin specific reductase (EC 1.1.1.193) (N-terminal amino acids)/deaminase (EC 3.5.4.26) (C-terminal amino acids) (see reference 18 for experimental proof) | ref|ZP_06918110.1|; bifunctional enzyme deaminase/reductase; Streptomyces sviceus ATCC 29083; identity, 84% |
| BN159_7144 | RibB, riboflavin synthase (EC 2.5.1.9) (see reference 18 for experimental proof) | ref|NP_625724.1|; NP_625724.1 riboflavin synthase subunit alpha; Streptomyces coelicolor A3(2); identity, 91% |
| BN159_7145 | RibM, riboflavin transporter (see reference 20 for experimental proof) | ref|YP_006244025.1|; YP_006244025.1 unnamed protein product; Streptomyces hygroscopicus subsp. jinggangensis 5008; identity, 85% |
| BN159_7146 | RibA, bifunctional 3,4-dihydroxy-2-butanone 4-phosphate synthase (EC 4.1.99.12) (N-terminal amino acids)/GTP cyclohydrolase II (C-terminal amino acids) (EC 3.5.4.25), not functional as GTP cyclohydrolase II (see reference 18 for experimental proof) | ref|ZP_06706864.1|; 3,4-dihydroxy-2-butanone-4-phosphate synthase; Streptomyces sp. strain e14; identity, 88% |
| BN159_7147 | RibH, 6,7-dimethyl-8-ribityllumazine synthase (EC 2.5.1.78) | ref|ZP_05001584.1|; 6,7-dimethyl-8-ribityllumazine synthase; Streptomyces sp. strain Mg1; identity, 88% |
| BN159_7984 | RibA, monofunctional GTP cyclohydrolase II (EC 3.5.4.25) | gb|EIM43989.1|; bifunctional 3,4-dihydroxy-2-butanone 4-phosphate synthase/GTP cyclohydrolase II protein; Rhodococcus opacus M213; Length, 381 aa; identity, 51% |
| BN159_7986 | RibB, archaeal riboflavin synthase (EC 2.5.1.9) | ref|YP_003483117.1|; riboflavin synthase; Aciduliprofundum boonei T469; identity, 58% |
| BN159_8007 | RibA, monofunctional 3,4-dihydroxy-2-butanone 4-phosphate synthase (EC 4.1.99.12) | ref|YP_004404734.1|; unnamed protein product; Verrucosispora maris AB-18-032; identity, 74% |
| BN159_0742 | Riboflavin-specific reductase function (EC 1.1.1.193) | Identity of >50% to deaminase-reductase domain-containing proteins |
| BN159_2369 | ||
| BN159_4557 | ||
| BN159_5537 | ||
| BN159_7503 |
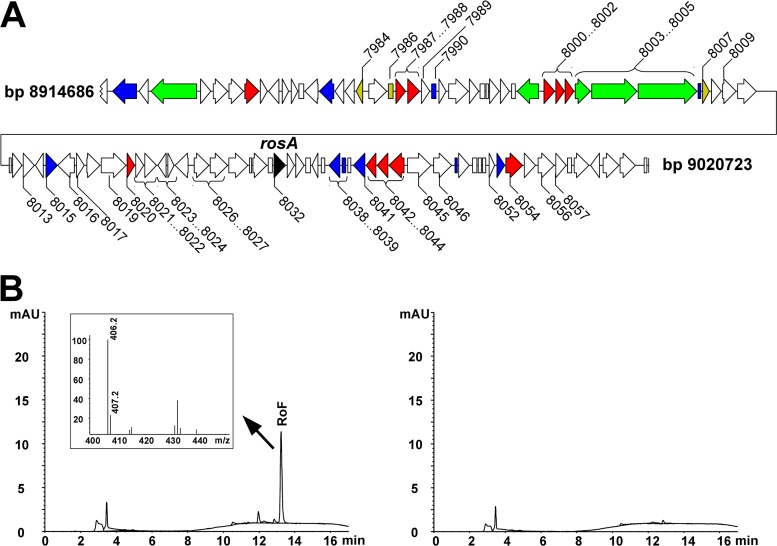
Fig 4.
Roseoflavin biosynthesis in Streptomyces davawensis. (A) Schematic representation of a subgenomic fragment that contains all genes/enzymes necessary for the heterologous production of roseoflavin in Streptomyces coelicolor M1152. The fragment spans the region from nucleotide 8914686 to nucleotide 9020723. Putative regulatory genes are presented in blue, nonribosomal peptide synthetase (NRPS) genes are presented in green, transporter genes are presented in red, riboflavin biosynthesis genes are presented in yellow, and rosA, coding for the roseoflavin-specific N,N-8-amino-8-demethyl-d-riboflavin dimethyltransferase (24), is shown in black. BN159_7984, bifunctional ribA. BN159_7986, archaeal riboflavin synthase gene ribB. BN159_7987…BN159_7988, genes for a putative ABC transport system. BN159_7989, gene for NADPH-dependent FMN reductase. BN159_7990, gene for a CheY-like response regulator. BN159_8000…BN159_8002, genes for a putative ABC transport system. BN159_8003…BN159_8005, cluster of NRPS genes. BN159_8007, ribA FMN riboswitch and monofunctional ribA gene. BN159_8009, gene for a putative cytochrome P450 protein. BN159_8013, gene for a putative oxidoreductase. BN159_8015, gene for a helix-turn-helix (HTH)-type transcriptional regulator. BN159_8016, gene for a putative dehydrogenase/reductase. BN159_8017, gene for alkaline phosphatase (ALPL). BN159_8019, gene for a predicted pyridoxal phosphate-dependent enzyme. BN159_8020, gene for ABC-type transport system followed by two putative dehydrogenase enzymes (BN159_8021 and BN159_8022). BN159_8023, gene for putative aminotransferase. BN159_8024, gene for putative adenylate kinase. BN159_8026…BN159_8027, genes for two Fe-S oxidoreductase enzymes. BN159_8038, BN159_8039, and BN159_8041, genes for HTH-like transcriptional regulators (purR). BN159_8042…BN159_8044, genes for the ABC-type transport system. BN159_8045 and BN159_8046, genes for conserved galactosidase-like enzymes. BN159_8052, gene for an uncharacterized bacitracin resistance protein (bacA). BN159_8054, gene for l-asparagine permease. BN159_8056, gene for aspartate ammonia lyase. BN159_8057, gene for glutaminase. (B) Heterologous production of roseoflavin by S. coelicolor M1152. Left panel, HPLC-MS analysis of a liquid culture supernatant of S. coelicolor M1152 transformed with a PAC containing the subgenomic fragment shown in panel A. Compared to the control, a novel signal was detected with a retention time identical to that of a roseoflavin standard. The most abundant mass-to-charge (m/z) ratio of 406.2 corresponds to roseoflavin (see the inset). Right panel, HPLC analysis of the liquid culture supernatant of the control strain S. coelicolor M1152 not containing the subgenomic fragment shown in panel A.
Only one gene (BN159_2715) coding for the essential bifunctional flavokinase/FAD synthetase function is present within the genome of S. davawensis, and this enzyme was characterized by us in an earlier work (17). The genes ribBMAH, ribG, and ribA, which were described above to be responsible for riboflavin biosynthesis, are located within the core genome region of S. davawensis.
Comparison of the organization of riboflavin biosynthetic gene clusters.
The comparison of riboflavin biosynthesis gene clusters among 10 different Streptomyces species revealed different patterns of organization (Fig. 3; also see Table S2 in the supplemental material). S. davawensis, S. coelicolor, S. avermitilis, and S. scabiei contain the cluster ribBMAH (Fig. 3A). A second cluster harboring a monofunctional ribA, marR (annotated as a MarR-family transcriptional regulator), and the missing ribG is located elsewhere in S. davawensis, S. avermitilis, and S. scabiei (Fig. 3). S. coelicolor has two different ribG clusters (Fig. 3D; also see below). One of the ribG clusters also harbors a monofunctional GTP cyclohydrolase II gene (ribA).
S. griseus, S. flavogriseus, Streptomyces sp. strain SirexAA-E, and S. cattleya contain a ribGBMAH cluster (Fig. 3B) in which all genes necessary for riboflavin synthesis (and transport) are present. All of the gene products encoded by the S. davawensis ribBMAH cluster are highly conserved at the amino acid level (>80%) among all analyzed Streptomyces species.
Only in S. violaceusniger and S. bingchenggensis is the flavin transporter ribM not present in the major ribGBAH gene cluster (Fig. 3C). Notably, the latter bacteria do not contain a gene similar to ribM at another location in their genome.
Two types of ribG genes seem to be present in streptomycetes. The enzyme encoded by ribG present in the clusters ribGBMAH and ribGBAH of S. griseus, S. flavogriseus, Streptomyces sp. strain SirexAA-E, S. cattleya, S. violaceusniger, and S. bingchenggensis (Fig. 3B and C) is different (similarity at the amino acid level, <50%) from that of RibG present in the ribA-marR-ribG clusters of S. davawensis, S. scabiei, and S. avermitilis (Fig. 3A). S. coelicolor is the only species to contain two ribG genes at two separate locations (Fig. 3D). RibG, encoded by the cluster shown in Fig. 3DI, is highly similar to RibG of S. davawensis, S. scabiei, and S. avermitilis, whereas RibG, shown in Fig. 3DII, is highly similar to the enzymes of S. griseus, S. flavogriseus, Streptomyces sp. strain SirexAA-E, S. cattleya, S. violaceusniger, and S. bingchenggensis (Fig. 3B and C). Notably, the two bifunctional RibG enzymes differ with regard to the arrangement of the reductase/deaminase functions (see the legend to Fig. 3).
Secondary metabolites in S. davawensis.
The genome of S. davawensis was analyzed with regard to secondary metabolism using the software tool antiSMASH (40). This analysis revealed the presence of 32 gene clusters carrying genes which may be involved in the synthesis of secondary metabolites in S. davawensis (Fig. 1D). Information with regard to these genes and the compounds which most likely are produced by the gene products is summarized in Table 3. Since the annotation was done automatically by antiSMASH, a few gene clusters seemingly overlap. Experimental evidence for the presence of any of these secondary metabolites currently is not available. Interestingly, many of the secondary metabolite gene clusters appear to be unique to S. davawensis and are not present in S. coelicolor, S. avermitilis, S. scabiei, S. griseus, S. flavogriseus, Streptomyces sp. strain SirexAA-E, or S. cattleya.
Table 3.
Synthesis of secondary metabolites in Streptomyces davawensis
| Type | Gene cluster | Compound which most likely is produced |
|---|---|---|
| Bacteriocin | BN159_1027...BN159_1041 | |
| Butyrolactone | BN159_4437...BN159_4443 | |
| Ectoine | BN159_6645...BN159_6653 | |
| Lantibiotic | BN159_7730...BN159_7752 | |
| NRPSa | BN159_5552...BN159_5613 | Melanin |
| NRPS | BN159_6018...BN159_6082 | Moenomycin |
| NRPS | BN159_7504...BN159_7544 | |
| NRPS | BN159_7971...BN159_8026 | Meilingmycin |
| Oligosaccharide | BN159_2334...BN159_2384 | Aclacinomycin |
| Other | BN159_0820...BN159_0862 | |
| Other | BN159_6747...BN159_6783 | |
| Other | BN159_6878...BN159_6920 | |
| PKSb (type 1) | BN159_1344...BN159_1381 | RK-682 (19) |
| PKS (type 3) | BN159_2002...BN159_2033 | |
| PKS (type 2) | BN159_2454...BN159_2482 | Oxytetracycline |
| PKS (type 1) | BN159_2472...BN159_2508 | Pyrrolomycin |
| PKS (type 2) | BN159_2498...BN159_2530 | Lankamycin/lankacidin |
| PKS (type 3) | BN159_5906...BN159_5930 | |
| PKS (type 3) | BN159_7364...BN159_7396 | Erythromycin |
| PKS (type 3) | BN159_7926...BN159_7957 | Furaquinocin A |
| PKS-NRPS MIX | BN159_2119...BN159_2158 | Tirandamycin |
| PKS-NRPS MIX | BN159_0658...BN159_0706 | Sanglifehrin A |
| PKS-NRPS MIX | BN159_5857...BN159_5906 | |
| Siderophore (?)c | BN159_2064...BN159_2088 | Geosmin |
| Siderophore | BN159_2640...BN159_2654 | 2-Methylisoborneol |
| Siderophore | BN159_5473...BN159_5494 | Desferrioxamine |
| Terpene | BN159_1431...BN159_1450 | |
| Terpene | BN159_1587...BN159_1611 | |
| Terpene/butyrolactone | BN159_2258...BN159_2301 | |
| Terpene/NRPS | BN159_3136...BN159_3188 | Albaflavenone |
| Terpene | BN159_79530...BN159_7981 | Furaquinocin A |
| Terpene/melanin | BN159_8242...BN159_8286 | |
| Vitamin analog | BN159_8007...BN159_8044 (?) | Roseoflavin |
NRPS, nonribosomal peptide synthetases.
PKS, polyketide synthases.
Question marks indicate hypothetical data.
The plasmid pSDA was also analyzed with regard to secondary metabolism; however, gene clusters of the known compound classes were not found.
Heterologous production of roseoflavin in S. coelicolor.
An S. davawensis expression library was constructed using a modified version of pPAC-S1 (56). A total of 1,920 different E. coli strains were isolated carrying PACs, and they had an average insert size of 120 kb. Using three different oligonucleotide pairs in separate PCRs, a specific strain was identified which contained a 100-kbp subgenomic S. davawensis fragment (nucleotides 8,914,686 to 9,020,723) in which the rosA gene (BN159_8032) was centrally located (Fig. 4A). By conjugation, the corresponding PAC was transferred to the heterologous expression host S. coelicolor M1152. One of the exconjugants was found to produce 1 μM roseoflavin, which was present in the supernatant of the culture after 7 days of growth (Fig. 4B). The control strain S. coelicolor M1152 did not produce roseoflavin.
Streptomyces cinnabarinus is a roseoflavin producer.
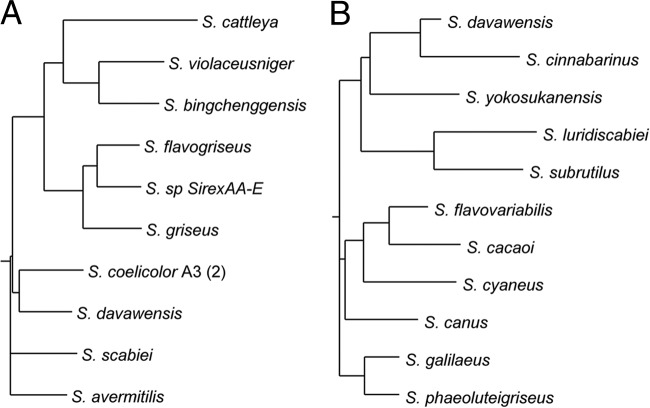
Whole-genome-based phylogenetic analysis of the completely sequenced Streptomyces species S. davawensis, S. coelicolor (6), S. avermitilis (22), S. scabiei, S. griseus (44), S. flavogriseus, Streptomyces sp. strain SirexAA-E, S. cattleya, S. violaceusniger, and S. bingchenggensis (61) was performed using EDGAR. According to these data, S. coelicolor is the closest relative of S. davawensis (Fig. 5A). Phylogenetic analysis based on 16S rRNA sequences, however, revealed that Streptomyces cinnabarinus (DSM 40467) was the closest relative of S. davawensis (Fig. 5B). When described in 1969 for the first time, S. cinnabarinus was reported to secrete a water-soluble red compound (12). Besides this description, no further data with regard to the secreted compound have been published.
Fig 5.
Phylogeny of Streptomyces davawensis. (A) Phylogeny based on a complete genome comparison (EDGAR). (B) Phylogeny based on the alignment of the complete 16S rRNA gene. All species shown are Streptomyces species.
S. cinnabarinus was grown in a liquid culture, and the supernatant was analyzed by liquid chromatography (LC)-MS. A major compound was detected with a mass-to-charge (m/z) ratio of 406.1, identical to the m/z ratio of a roseoflavin standard. Moreover, the UV-visible (UV-vis) spectrum of the major compound present in the supernatant was identical to the spectrum of roseoflavin (Fig. 6). Figure S5 in the supplemental material shows S. cinnabarinus and S. davawensis colonies on a solid growth medium.
Fig 6.
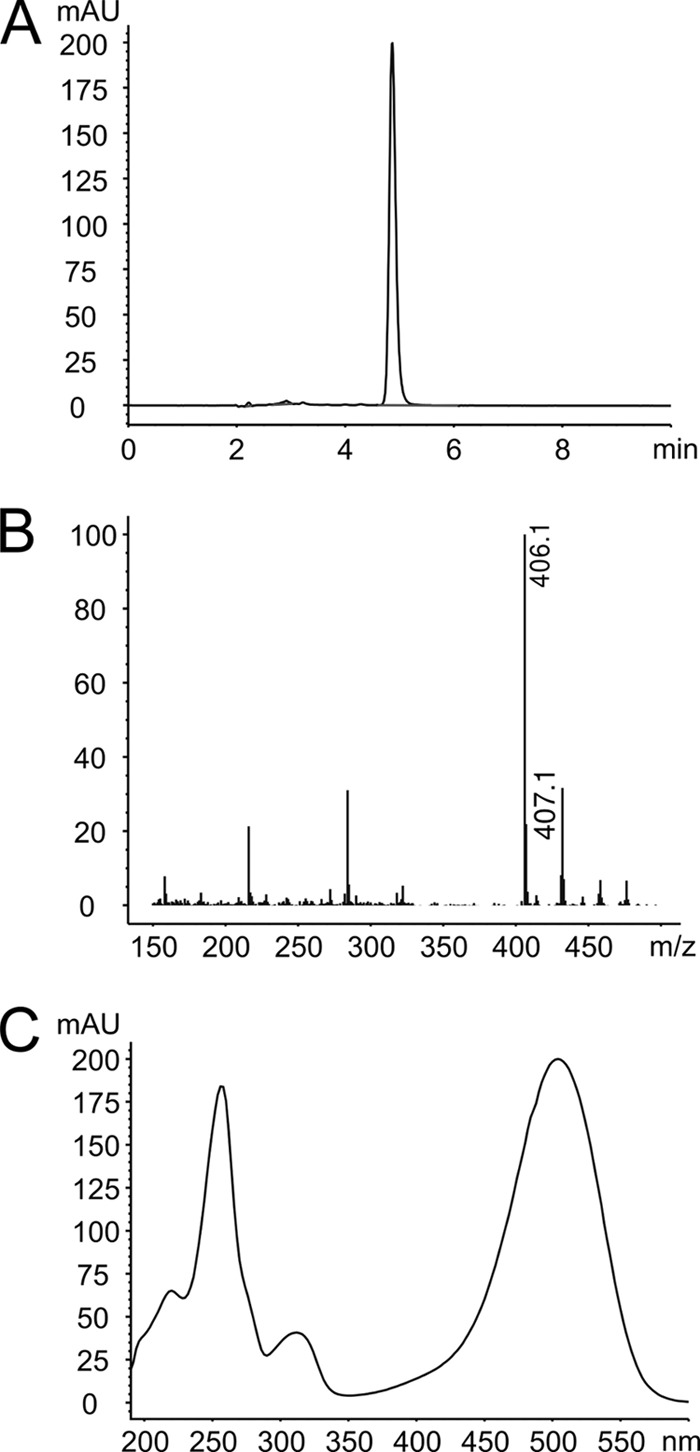
Identification of roseoflavin in Streptomyces cinnabarinus. (A) Retention time (HPLC) of the major compound present in the supernatant of actively growing cells of S. cinnabarinus. The retention time of the major compound is identical to that of a roseoflavin standard. (B) The mass (406.1 [M+H]+) of the major compound shown is identical to the mass of the roseoflavin standard. (C) UV-vis spectrum of the major S. cinnabarinus compound. The spectrum is identical to the spectrum of roseoflavin.
DISCUSSION
To the best of our knowledge, only two other examples for naturally occurring vitamin analogs with antibiotic/toxin function are known, bacimethrin and ginkgotoxin. The antibiotic bacimethrin is a structural thiamine (vitamin B1) analog naturally produced by Bacillus megaterium and Streptomyces albus (49). Ginkgotoxin is a neurotoxin occurring in Ginkgo biloba and is structurally related to vitamin B6 (pyridoxine) (13). The biosynthetic pathways for bacimethrin and ginkgotoxin are unknown.
The enzymatic conversion of the vitamin riboflavin into the antibiotic roseoflavin involves the mechanistically challenging replacement of a methyl group at C-8 of the isoalloxazine ring system of riboflavin by an amino group (25). As a first step toward understanding roseoflavin biosynthesis, we report here the genome of S. davawensis. The obtained sequence data were crucial for the previous identification of the novel N,N-8-amino-8-demethyl-riboflavin dimethyltransferase RosA that is responsible for the two terminal steps in roseoflavin biosynthesis (24). The introduction of a 100-kbp S. davawensis subgenomic fragment (containing 93 CDSs) into S. coelicolor M1152 led to the production of roseoflavin by a corresponding recombinant strain and constitutes another important step with regard to elucidation of roseoflavin biosynthesis. This subgenomic fragment contains the gene rosA, which in turn is located at the end of the rosA operon comprising a total of 10 genes (24) (Fig. 4). Notably, expression of the latter gene cluster did not support roseoflavin production (24). The remaining 83 CDSs on the 100-kb subgenomic fragment currently are being investigated with regard to their role in roseoflavin biosynthesis by gene inactivation experiments. The intermediates of the roseoflavin biosynthetic pathway (except for 8-amino-8-demethyl-riboflavin) are unknown. It is therefore difficult to speculate on which genes present on the 100-kbp subgenomic fragment are involved in roseoflavin biosynthesis. The working hypothesis now is that the region between the FMN riboswitch directly upstream of BN159_8007 and BN159_8044 contains the relevant genes.
Interestingly, putative riboflavin biosynthetic genes (BN159_7984, BN159_7986, and BN159_8007) (Table 2) were found to be present on the 100-kbp PAC responsible for roseoflavin biosynthesis. It is tempting to speculate that these genes support riboflavin biosynthesis when S. davawensis enters the roseoflavin production phase.
Traditionally, the names for riboflavin/FMN/FAD biosynthetic genes in Gram-positive and Gram-negative bacteria are different. Moreover, bifunctional riboflavin biosynthetic enzymes are employed by some species, which complicates genetic nomenclature even more. In order to avoid future confusion with regard to gene names, we suggest the following genetic nomenclature (also see Fig. S2 in the supplemental material): RibA, GTP cyclohydrolase II function (EC 3.5.4.25); RibB, 3,4-dihydroxy-2-butanone-4-phosphate synthase function (EC 4.1.99.12); RibD, riboflavin-specific deaminase function (EC 3.5.4.26); RibG, riboflavin-specific reductase function (EC 1.1.1.193); RibH, lumazine synthase function (ribH; EC 2.5.1.78); and RibE, riboflavin synthase function (EC 2.5.1.9). Moreover, flavokinase should be renamed to RibF (EC 2.7.1.26) and FAD synthetase to RibC (EC 2.7.7.2). According to this nomenclature, genes coding for bifunctional enzymes would be denoted as, e.g., ribCF if domains were present in the corresponding gene products that show sequence similarity to both FAD synthetase (RibC; N-terminal amino acid residues) and flavokinase (RibF; C-terminal amino acid residues).
S. davawensis is roseoflavin resistant. We hypothesized earlier that the mechanism of self resistance involved a flavokinase/FAD synthetase with a high substrate specificity for riboflavin. Such an enzyme would not accept roseoflavin as a substrate and thus would not produce toxic RoFMN and RoFAD. Our data on the S. davawensis flavokinase/FAD synthetase RibC (RibCF according to the new nomenclature), however, clearly showed that this enzyme does not discriminate between roseoflavin and riboflavin (17). A second flavokinase/FAD synthetase was not identified in the S. davawensis genome. Accordingly, roseoflavin resistance of S. davawensis seems to be due mainly to a specialized FMN riboswitch that is insensitive to RoFMN (47). However, since roseoflavin is found in the culture supernatant (and not within the cytoplasm), we postulate that an as-yet unknown roseoflavin exporter protein is present in S. davawensis which may support roseoflavin resistance. The corresponding gene may very well be located close to the roseoflavin biosynthesis genes (Fig. 4A).
The synthesis of roseoflavin may involve unusual enzymes. The present work represents major progress toward understanding roseoflavin biosynthesis and, moreover, is the prerequisite for the analysis of the regulation of flavin synthesis in S. davawensis.
The fact that a second roseoflavin-producing organism, S. cinnabarinus, was identified supports the idea that vitamin analogs with antibiotic function are more widespread in nature (47).
ACKNOWLEDGMENTS
We thank David Figurski (Columbia University, New York, NY) for sharing pR9406 and Juan Pablo Gomez-Escribano (John Innes Centre, Norwich, United Kingdom) for sharing S. coelicolor M1152.
This work was supported by the Federal Ministry for Education and Research (BMBF) (GenoMik) and B.R.A.I.N. AG (Zwingenberg). C.R. and J.K. acknowledge grants from the Ministry of Innovation and Research of the Federal State Northrhine-Westfalia (BIO.NRW) initiative and from the BMBF (GenoMik).
Footnotes
Published ahead of print 5 October 2012
This work is dedicated to Karl Heimbs.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Badger JH, Olsen GJ. 1999. CRITICA: coding region identification tool invoking comparative analysis. Mol. Biol. Evol. 16:512–524 [DOI] [PubMed] [Google Scholar]
- 4. Bairoch A. 2000. The ENZYME database in 2000. Nucleic Acids Res. 28:304–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bairoch A, et al. 2005. The Universal Protein Resource (UniProt). Nucleic Acids Res. 33:D154–D159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bentley SD, et al. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147 [DOI] [PubMed] [Google Scholar]
- 7. Blom J, et al. 2009. EDGAR: a software framework for the comparative analysis of prokaryotic genomes. BMC Bioinformatics 10:154 doi:10.1186/1471-2105-10-154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. 2012. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28:464–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Claudel-Renard C, Chevalet C, Faraut T, Kahn D. 2003. Enzyme-specific profiles for genome annotation: PRIAM. Nucleic Acids Res. 31:6633–6639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coulson A. 1994. High-performance searching of biosequence databases. Trends Biotechnol. 12:76–80 [DOI] [PubMed] [Google Scholar]
- 11. Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Feng H, et al. 2010. Molecular characterization and expression of a heat shock protein gene (HSP90) from the carmine spider mite, Tetranychus cinnabarinus (boisduval). J. Insect Sci. 10:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fiehe K, et al. 2000. Biosynthesis of 4′-O-methylpyridoxine (ginkgotoxin) from primary precursors. J. Nat. Prod. 63:185–189 [DOI] [PubMed] [Google Scholar]
- 14. Fischer M, Bacher A. 2005. Biosynthesis of flavocoenzymes. Nat. Prod. Rep. 22:324–350 [DOI] [PubMed] [Google Scholar]
- 15. Gardner PP, et al. 2009. Rfam: updates to the RNA families database. Nucleic Acids Res. 37:D136–D140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goujon M, et al. 2010. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38:W695–W699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grill S, Busenbender S, Pfeiffer M, Kohler U, Mack M. 2008. The bifunctional flavokinase/flavin adenine dinucleotide synthetase from Streptomyces davawensis produces inactive flavin cofactors and is not involved in resistance to the antibiotic roseoflavin. J. Bacteriol. 190:1546–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grill S, et al. 2007. Identification and characterization of two Streptomyces davawensis riboflavin biosynthesis gene clusters. Arch. Microbiol. 188:377–387 [DOI] [PubMed] [Google Scholar]
- 19. Hamaguchi T, Sudo T, Osada H. 1995. RK-682, a potent inhibitor of tyrosine phosphatase, arrested the mammalian cell cycle progression at G1 phase. FEBS Lett. 372:54–58 [DOI] [PubMed] [Google Scholar]
- 20. Hemberger S, et al. 2011. RibM from Streptomyces davawensis is a riboflavin/roseoflavin transporter and may be useful for the optimization of riboflavin production strains. BMC Biotechnol. 11:119 doi:10.1186/1472-6750-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hyatt D, et al. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119 doi:10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ikeda H, et al. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21:526–531 [DOI] [PubMed] [Google Scholar]
- 23. Jakimowicz D, et al. 1998. Structural elements of the Streptomyces oriC region and their interactions with the DnaA protein. Microbiology 144(Pt 5):1281–1290 [DOI] [PubMed] [Google Scholar]
- 24. Jankowitsch F, et al. 2011. A novel N,N-8-amino-8-demethyl-d-riboflavin dimethyltransferase (RosA) catalyzing the two terminal steps of roseoflavin biosynthesis in Streptomyces davawensis. J. Biol. Chem. 286:38275–38285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Juri N, et al. 1987. Formation of roseoflavin from 8-amino- and 8-methylamino-8-demethyl-d-riboflavin. J. Biochem. 101:705–711 [DOI] [PubMed] [Google Scholar]
- 26. Kanehisa M, Goto S. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kanehisa M, et al. 2006. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 34:D354–D357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical streptomyces genetics, p 168–169 The John Innes Foundation, Norwich, United Kingdom [Google Scholar]
- 29. Krause L, et al. 2007. GISMO–gene identification using a support vector machine for ORF classification. Nucleic Acids Res. 35:540–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 31. Lagesen K, et al. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35:3100–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 33. Lee ER, Blount KF, Breaker RR. 2009. Roseoflavin is a natural antibacterial compound that binds to FMN riboswitches and regulates gene expression. RNA Biol. 6:187–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Linke B, McHardy AC, Neuweger H, Krause L, Meyer F. 2006. REGANOR: a gene prediction server for prokaryotic genomes and a database of high quality gene predictions for prokaryotes. Appl. Bioinformatics 5:193–198 [DOI] [PubMed] [Google Scholar]
- 35. Mansjo M, Johansson J. 2011. The riboflavin analog roseoflavin targets an FMN-riboswitch and blocks Listeria monocytogenes growth, but also stimulates virulence gene-expression and infection. RNA Biol. 8:674–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marchler-Bauer A, et al. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37:D205–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marchler-Bauer A, et al. 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39:D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Matsui K, Juri N, Kubo Y, Kasai S. 1979. Formation of roseoflavin from guanine through riboflavin. J. Biochem. 86:167–175 [PubMed] [Google Scholar]
- 39. McHardy AC, Goesmann A, Puhler A, Meyer F. 2004. Development of joint application strategies for two microbial gene finders. Bioinformatics 20:1622–1631 [DOI] [PubMed] [Google Scholar]
- 40. Medema MH, et al. 2011. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 39:W339–W346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. 2005. The microbial pan-genome. Curr. Opin. Genet. Dev. 15:589–594 [DOI] [PubMed] [Google Scholar]
- 42. Meyer F, et al. 2003. GenDB–an open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 31:2187–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nawrocki EP, Kolbe DL, Eddy SR. 2009. Infernal 1.0: inference of RNA alignments. Bioinformatics 25:1335–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ohnishi Y, et al. 2008. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 190:4050–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Otani S, Takatsu M, Nakano M, Kasai S, Miura R. 1974. Letter: roseoflavin, a new antimicrobial pigment from Streptomyces. J. Antibiot. (Tokyo) 27:88–89 [PubMed] [Google Scholar]
- 46. Ott E, Stolz J, Lehmann M, Mack M. 2009. The RFN riboswitch of Bacillus subtilis is a target for the antibiotic roseoflavin produced by Streptomyces davawensis. RNA Biol. 6:276–280 [DOI] [PubMed] [Google Scholar]
- 47. Pedrolli DB, et al. 2012. A highly specialized flavin mononucleotide riboswitch responds differently to similar ligands and confers roseoflavin resistance to Streptomyces davawensis. Nucleic Acids Res. 40:8662–8673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pedrolli DB, et al. 2011. The antibiotics roseoflavin and 8-demethyl-8-amino-riboflavin from Streptomyces davawensis are metabolized by human flavokinase and human FAD synthetase. Biochem. Pharmacol. 82:1853–1859 [DOI] [PubMed] [Google Scholar]
- 49. Reddick JJ, et al. 2001. The mechanism of action of bacimethrin, a naturally occurring thiamin antimetabolite. Bioorg. Med. Chem. Lett. 11:2245–2248 [DOI] [PubMed] [Google Scholar]
- 50. Richter G, et al. 1997. Biosynthesis of riboflavin: characterization of the bifunctional deaminase-reductase of Escherichia coli and Bacillus subtilis. J. Bacteriol. 179:2022–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schwientek P, Szczepanowski R, Ruckert C, Stoye J, Puhler A. 2011. Sequencing of high G+C microbial genomes using the ultrafast pyrosequencing technology. J. Biotechnol. 155:68–77 [DOI] [PubMed] [Google Scholar]
- 52. Shinkai S, et al. 1986. Spectral and reactivity studies of roseoflavin analogs: correlation between reactivity and spectral parameters. Bioorg. Chem. 14:119–133 [Google Scholar]
- 53. Shinobu R. 1974. Streptomyces davawensis nov. sp. Memoirs Osaka Kyoiku Univ. 23:1–8 [Google Scholar]
- 54. Sonnhammer EL, von Heijne G, Krogh A. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6:175–182 [PubMed] [Google Scholar]
- 55. Sosio M, et al. 2000. Artificial chromosomes for antibiotic-producing actinomycetes. Nat. Biotechnol. 18:343–345 [DOI] [PubMed] [Google Scholar]
- 56. Tatusov RL, et al. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41 doi:10.1186/1471-2105-4-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tatusov RL, Koonin EV, Lipman DJ. 1997. A genomic perspective on protein families. Science 278:631–637 [DOI] [PubMed] [Google Scholar]
- 58. Tatusov RL, et al. 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29:22–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vogl C, et al. 2007. Characterization of riboflavin (vitamin B2) transport proteins from Bacillus subtilis and Corynebacterium glutamicum. J. Bacteriol. 189:7367–7375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Walsh C, et al. 1978. Chemical and enzymatic properties of riboflavin analogues. Biochemistry 17:1942–1951 [DOI] [PubMed] [Google Scholar]
- 61. Wang XJ, et al. 2010. Genome sequence of the milbemycin-producing bacterium Streptomyces bingchenggensis. J. Bacteriol. 192:4526–4527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Winkler WC, Breaker RR. 2005. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 59:487–517 [DOI] [PubMed] [Google Scholar]