Abstract
Bile acids are highly abundant steroids with important functions in vertebrate digestion. Their catabolism by bacteria is an important component of the carbon cycle, contributes to gut ecology, and has potential commercial applications. We found that Rhodococcus jostii RHA1 grows well on cholate, as well as on its conjugates, taurocholate and glycocholate. The transcriptome of RHA1 growing on cholate revealed 39 genes upregulated on cholate, occurring in a single gene cluster. Reverse transcriptase quantitative PCR confirmed that selected genes in the cluster were upregulated 10-fold on cholate versus on cholesterol. One of these genes, kshA3, encoding a putative 3-ketosteroid-9α-hydroxylase, was deleted and found essential for growth on cholate. Two coenzyme A (CoA) synthetases encoded in the cluster, CasG and CasI, were heterologously expressed. CasG was shown to transform cholate to cholyl-CoA, thus initiating side chain degradation. CasI was shown to form CoA derivatives of steroids with isopropanoyl side chains, likely occurring as degradation intermediates. Orthologous gene clusters were identified in all available Rhodococcus genomes, as well as that of Thermomonospora curvata. Moreover, Rhodococcus equi 103S, Rhodococcus ruber Chol-4 and Rhodococcus erythropolis SQ1 each grew on cholate. In contrast, several mycolic acid bacteria lacking the gene cluster were unable to grow on cholate. Our results demonstrate that the above-mentioned gene cluster encodes cholate catabolism and is distinct from a more widely occurring gene cluster encoding cholesterol catabolism.
INTRODUCTION
Bile salts are surface-active steroids with an important role in the uptake and metabolism of lipophilic substrates in vertebrates. These steroids, which include cholate and chenodeoxycholate, are synthesized in the liver from cholesterol, and their eventual fate is excretion in feces or urine. Bile salts may be modified, either by microbiological activity in the duodenum or by host cell bioactivity, leading to their conjugation to glycine, taurine, or sulfate. As such, biodegradation of the various bile salts is a significant process in carbon cycling in soil and aquatic environments. The processes involved in microbial transformation of steroids are also relevant for biotechnological applications in the synthesis and/or selective modification of steroid-based drugs (29).
Despite the cytotoxicity of cholate toward various prokaryotic and eukaryotic cells, several bacterial species, especially members of the Proteobacteria (27, 28) and Actinomycetales (9, 32), are able to efficiently metabolize this substrate to sustain growth. Recent studies on microbial bile salts degradation have focused on the Proteobacteria. Genes encoding several steps in cholate degradation were identified, mainly in Comamonas testosteroni TA441 and Pseudomonas sp. strain Chol1. In the former strain, genes responsible for oxidation of the steroid nucleus were found (10–13), while in the latter, genes responsible for degradation of the cholate side chain were identified, including acad, encoding an acyl coenzyme A (acyl-CoA) dehydrogenase (2), and skt, encoding a thiolase (3).
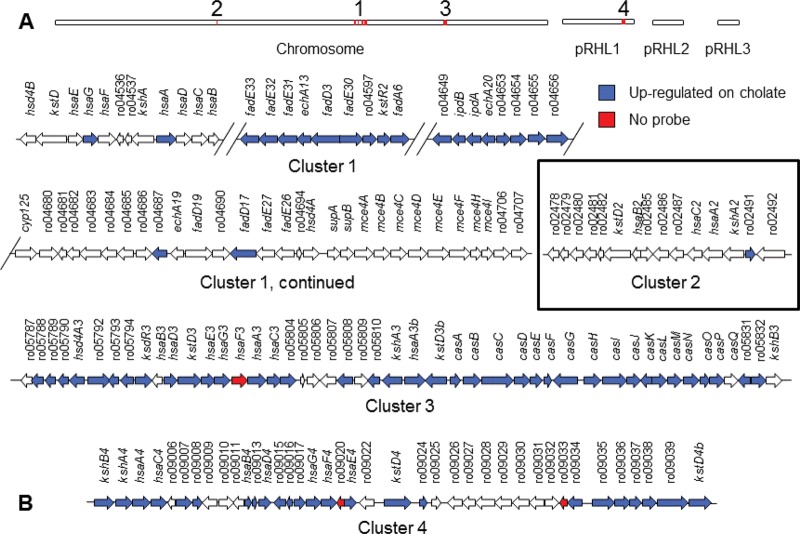
We recently identified a gene cluster encoding cholesterol catabolism in Rhodococcus jostii RHA1 (32). This process has been the focus of much investigation, ultimately leading to the discovery that host cholesterol is likely essential for survival of Mycobacterium tuberculosis in the host (14, 21, 34). The genome of strain RHA1 contains four clusters of genes putatively encoding steroid catabolism: ro02478 to ro02492 (cluster 2), ro04531 to ro04705 (cluster 1), ro05788 to ro05832 (cluster 3), and ro09002 to ro09040 (cluster 4) (Fig. 1). Each cluster includes genes encoding a 3-ketosteroid-Δ1-dehydrogenase (KstD) and a 3-ketosteroid-9α-hydroxylase (KshAB), both of which are essential for degradation of the A and B rings of steroid nuclei (4, 19). Cluster 1 was shown to encode cholesterol catabolism specifically, while the roles of the other putative steroid gene clusters remain unclear.
Fig 1.
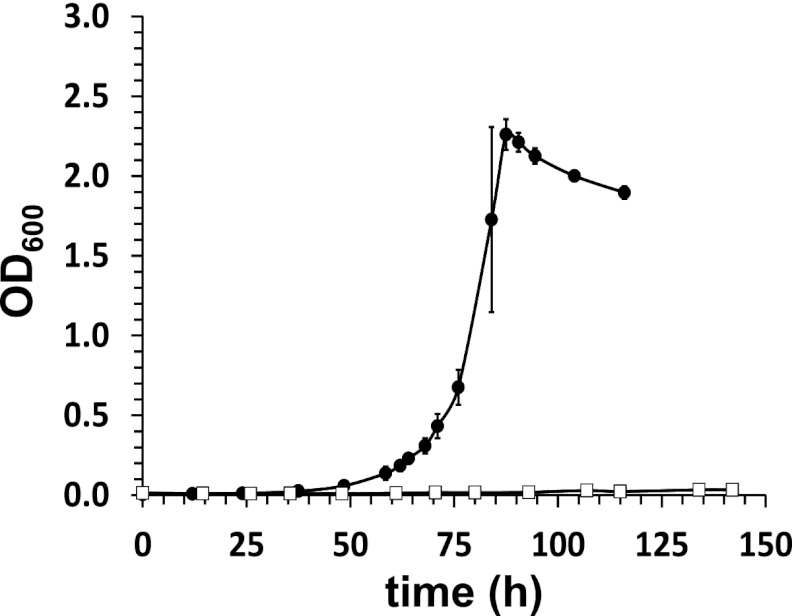
Growth of RHA1 and ΔkshA3 on cholate. RHA1 (solid symbols) and ΔkshA3 (open symbols) were grown in mineral medium supplemented with 2.0 mM cholate at 30°C. The error bars indicate the range in duplicate experiments.
In this study, we used transcriptomic analysis to identify RHA1 genes associated with cholate catabolism. Bioinformatic, molecular genetic, and enzymological approaches were then used to demonstrate the role of one of the four above-mentioned gene clusters in cholate catabolism. The occurrence of this gene cluster in bacterial genomes was evaluated and compared to the ability of organisms to grow on cholate. In a companion paper (26), we identify metabolites accumulating during growth on cholate and transporters functioning in the reassimilation of those metabolites.
MATERIALS AND METHODS
Bacterial strains, plasmids, and chemicals.
R. jostii RHA1 was grown at 30°C in W medium mineral salts (25) supplemented with a carbon source, as indicated. For transcriptome analysis, cultures of RHA1 were grown in W medium with 2.0 mM cholate or 20 mM pyruvate as sole organic substrates and harvested at midexponential phase (optical densities at 600 nm [OD600] of 1.5 for cholate and 1.0 for pyruvate). For reverse transcriptase quantitative PCR (RT-QPCR) assays, cultures of RHA1 were grown as described above or with 2.0 mM cholesterol and harvested at midexponential phase. Prior to these analyses, cells were stored at −80°C as previously described (7).
Escherichia coli DH5α and plasmid pBlueScript II KS were used for cloning purposes. E. coli BL21(DE3) and pET15b were used for heterologous expression studies. E. coli S17-1 and pK18mobsacB were used for targeted gene inactivation in Rhodococcus (31). All E. coli strains were grown in LB broth (Lennox [Difco]) at 37°C unless otherwise specified. If applicable, agar (Becton, Le Pont de Claix, France) was added at a final concentration of 1.5% (wt/vol).
Coenzyme A, cholic acid, chenodeoxycholic acid, deoxycholic acid, glycocholic acid, and taurocholic acid were from Sigma. Deoxycholenic acid, 5-cholenic acid-3β-ol, 23,24-bisnor-4-cholenic acid-3-one, and 23,24-bisnor-5-cholenic acid-3β-ol were obtained from Steraloids Inc. (Newport, RI). ATP was purchased from Duchefa (Haarlem, The Netherlands). 4-Androstene-3,17-dione was provided by MSD (Oss, The Netherlands). All chemicals were of the highest purity available.
RT-QPCR.
RNA was extracted as previously described (17), treated twice with DNase I (Ambion; Turbo), and purified with RNeasy columns (Qiagen). cDNA was synthesized with the ThermoScript RT-PCR System (Invitrogen) using random hexamers. The primer and probe (6-carboxyfluorescein [FAM]-labeled and ZEN-double-quenched) sequences (see Table S1 in the supplemental material) were designed either with PrimeTime Assay (Integrated DNA Technologies, San Diego, CA) or by custom design. The specificity of each set of primers and probe was checked to avoid cross-reactivity with transcripts from paralogous genes in RHA1. QPCRs were prepared and performed as previously described (17). The sigA gene, encoding sigma factor A, was used as the reference gene. A standard curve of threshold cycle (Ct) versus gene copy number for each gene was established by using different amounts of genomic DNA as the template. Since all the genes tested have a single copy on the chromosome, gene copy numbers were assumed to be the same as the genome copy number.
Construction and functional complementation of mutant RHA1ΔkshA3.
Mutant RHA1ΔkshA3, carrying a 1,010-bp in-frame deletion in kshA3 (ro05811), was constructed using a sacB counterselection system (31) with mutagenic plasmid pDelkshA3. Plasmid pDelkshA3 was constructed as follows. The up- and downstream regions of ro05811 were amplified by PCR using the primers listed in Table S1 in the supplemental material and cloned into the EcoRV site of pBluescript II KS, yielding pBS_ro05811up and pBS_ro05811down, respectively. A 1.1-kb fragment obtained by HindIII/NdeI digestion of pBS_ro05811down was ligated into HindIII/NdeI-digested pBS_ro05811up, yielding pBS_ro05811. Following digestion of pBS_ro05811 with EcoRI/HindIII, a 2.2-kb fragment containing the ro05811 deletion cassette was ligated into EcoRI/HindIII-digested pK18mobsacB, yielding plasmid pDelkshA3. Mutants obtained after the second recombination event were verified by PCR using primers ro05811cont-F and ro05811cont-R (Table S1) to confirm deletion of kshA3 at the correct genomic locus.
For functional complementation of RHA1ΔkshA3, plasmid pSET_kshA3 was constructed as follows. A 2.2-kb amplicon containing full-length kshA3 was obtained by PCR using primers 5811UP-F and 5811comp-R (Table S1) cloned into EcoRV-digested pSET152 (33), and its sequence was verified.
Cloning and expression of casG and casI.
The casG (ro05820) and casI (ro05822) genes were amplified from chromosomal DNA of strain RHA1 using the primers listed in Table S1 and cloned into EcoRV-digested pBlueScript II KS, yielding pBScasG and pBScasI, respectively. A 1.7-kb fragment containing full-length casG was obtained by NdeI/BamHI digestion of pBScasG and cloned into NdeI/BamHI-digested pET15b, yielding pETCasG. The pBScasI plasmid was digested with NdeI and BglII, and the 1.7-kb DNA fragment containing casI was ligated into NdeI/BamHI-digested pET15b, resulting in pETCasI.
CasG and CasI were heterologously produced as affinity-tagged proteins using E. coli BL21(DE3) containing pETCasG and pETCasI, respectively. Cells were grown in LB broth supplemented with sorbitol (500 mM), ampicillin (100 μg/ml), and isopropyl-d-1-thiogalactopyranoside (0.1 mM). A single colony was used to inoculate 50 ml of liquid medium, which was incubated at 25°C with shaking (220 rpm). After 20 h of incubation, cells were harvested by centrifugation (4,600 × g; 4°C; 10 min) and washed with 50 mM Tris-HCl, pH 8, containing 1 mM dl-dithiothreitol (DTT). Cell pellets were stored at −80°C until use.
Purification of CasG and CasI.
Cell pellets were resuspended in 50 mM Tris-HCl, pH 8, containing 1 mM DTT. The cells were lysed by ultrasonication in a Branson sonifier, and cell debris was removed by centrifugation at 40,000 × g for 20 min at 4°C. Cell extracts were applied to an Ni-nitrilotriacetic acid (NTA) column (Sigma). The resin was then washed, first with 50 mM Tris-HCl, pH 8, containing 0.5 M NaCl and 1 mM DTT and then with 50 mM Tris-HCl, pH 8, containing 0.5 M NaCl, 5 mM imidazole, and 1 mM DTT. CasG and CasI eluted using 50 mM Tris-HCl, pH 8, containing 0.5 M NaCl, 30 mM imidazole, and 1 mM DTT. SDS-PAGE analysis of the proteins obtained showed a single band at approximately 65 kDa after Coomassie brilliant blue staining for both CasG (expected size, 62.6 kDa) and CasI (expected size, 63.3 kDa). Purified proteins were stored on ice and remained active up to 48 h.
CoA synthetase assay.
Reaction mixtures containing 1.5 μM CasG or CasI, 0.75 mM carboxylate substrate, 1.0 mM free coenzyme A (CoASH), 5 mM MgCl2, and 2.5 mM ATP were incubated for 2 h at 22°C. Substrate depletion and product accumulation were determined using high-performance liquid chromatography (HPLC), essentially as described previously (4). Briefly, reaction substrates and products were separated using a Waters 2695 Separations HPLC module (Milford, MA) equipped with a Waters 2996 photodiode array detector and a Luna 3-μm PFP(2) 50- by 4.6-mm column (Phenomenex, Torrance, CA). Compounds were eluted using a linear gradient from 0 to 90% methanol in 0.1 M ammonium acetate, pH 4.5, over 20 min, and the eluate was monitored at 248 nm. Peaks corresponding to ATP, AMP, and CoASH were identified according to the retention times and spectra of pure standards. The CoA thioester obtained upon incubating cholate with CasG was isolated by HPLC, dried, and analyzed via electrospray ionization-mass spectrometry (ESI-MS).
Sequence analyses.
Sequences were aligned using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) and the default parameters. Phylogenetic trees were built using the maximum-likelihood model of the PHYLIP 3.69 package.
Microarray data accession number.
Details of the microarray design, transcriptomic experimental design, and transcriptomic data have been deposited in the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO series accession number GSE28048.
RESULTS
Growth on bile salts.
RHA1 grew on cholate as a sole organic substrate with a doubling time of 7.8 h (Fig. 1), consistent with previous results (20). RHA1 also grew on the conjugates, taurocholate, and glycocholate. However, RHA1 failed to grow on two other bile salts, chenodeoxycholate and deoxycholate.
Transcriptomic analysis.
To identify genes encoding cholate catabolism, we used a microarray to compare the transcriptome of RHA1 growing on cholate versus growing on pyruvate. In cells harvested at midlog phase, a total of 389 genes were differentially expressed (expression ratio ≥ 2-fold and P value ≤ 0.05). Of these genes, 284 were upregulated on cholate (see Table S2 in the supplemental material) and 145 were downregulated. Many of the upregulated genes occurred in three of four previously identified gene clusters containing steroid catabolism genes (32) (Fig. 2). Genes in cluster 3 were most highly upregulated, with maximal expression ratios (cholate/pyruvate) of 114, 42, and 22 for genes in clusters 3, 1, and 4, respectively. Cluster 1, previously shown to encode cholesterol catabolism (32), revealed an interesting pattern of expression. Only some groups of genes within that cluster were upregulated, and most of the genes with demonstrated functions in cholesterol catabolism were not upregulated on cholate.
Fig 2.
Gene clusters in RHA1 encoding steroid catabolism. (A) Red regions indicate locations of gene clusters within the 9.7-Mb genome, comprising one linear chromosome plus three linear plasmids. (B) Organization and expression of genes.
RT-QPCR.
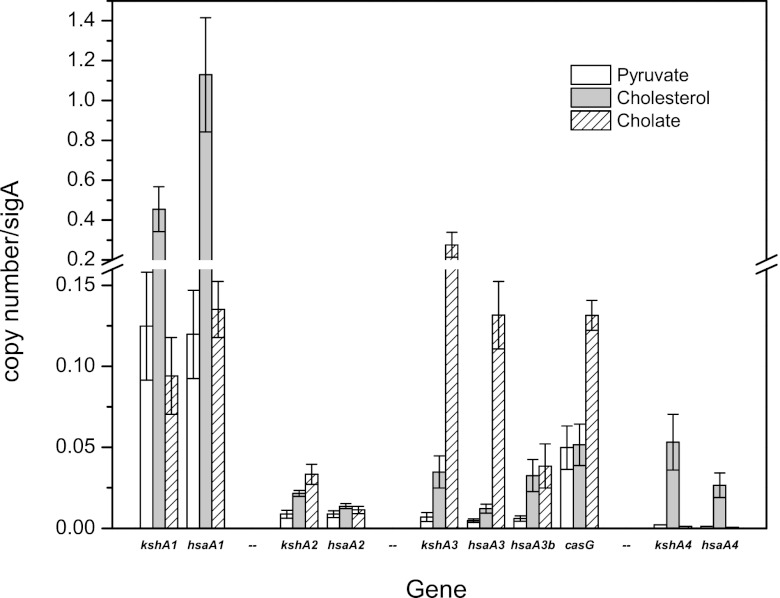
To verify the expression in RHA1 of genes in the various steroid catabolism gene clusters during growth on cholate, RT-QPCR was used to measure expression of 10 selected genes. The genes selected include two sets of paralogs represented in each of the four gene clusters predicted to encode degradation of the A and B rings of steroid nuclei. Thus, we selected four paralogs of kshA, encoding the oxygenase component of 3-ketosteroid 9α-hydroxylase, and five paralogs of hsaA, encoding the oxygenase component of 3-hydroxy-9,10-seconandrosta-1,3,5(10)-trien-9,17-dione 4-hydroxylase. We also selected casG, encoding a CoA synthetase that was further characterized in this study. Gene expression levels in RHA1 growing on pyruvate, cholesterol, or cholate were compared. Estimates of gene expression by a common method using relative expression ratios resembling the microarray analysis were confounded by extreme variation in the constitutive levels of gene expression on pyruvate. Therefore, we instead calculated copy numbers of gene transcripts, which were normalized on the basis of copy numbers of sigA transcripts (Fig. 3). This analysis avoided misleading cases where either (i) expression ratios were high but transcript levels were actually low or (ii) expression ratios were low but the magnitude of changes in expression levels were actually high.
Fig 3.
RT-QPCR analysis of the expression of 10 genes during growth of RHA1 on three substrates. The error bars show standard deviations for triplicate cultures.
The RT-QPCR analysis indicated that the genes in cluster 1 were constitutively expressed at high levels on both pyruvate and cholate and, as anticipated, were upregulated on cholesterol (Fig. 3). In contrast, kshA3 and hsaA3 in cluster 3 were expressed at low levels on both pyruvate and cholesterol but were upregulated 9.7- and 10.0-fold, respectively, on cholate versus cholesterol. The other two genes in cluster 3 that were assayed exhibited distinct expression patterns: casG was constitutively expressed at relatively high levels but was nevertheless upregulated 4.5-fold on cholate versus cholesterol, whereas hsaA3b (a second homolog of hsaA) was not expressed at high levels under any of the conditions tested. The latter result suggests that hsaA3b may not play a significant role in cholate catabolism. None of the assayed genes in clusters 2 and 4 exhibited high expression levels under any of the tested growth conditions. Thus, the RT-QPCR analysis confirmed upregulation of cluster 3 on cholate but did not confirm upregulation of cluster 4, as initially suggested by the microarray analysis. In the case of cluster 4, expression ratios were high on cholate versus pyruvate, but absolute expression levels were actually low on both substrates. It is additionally possible that poor specificity of some of the microarray probes for cluster 4 genes, contributed to false-positive results in the microarray analysis. Thus, genes in cluster 3, but not cluster 4, appear to encode cholate catabolism in RHA1.
kshA3 deletion.
To test the hypothesis that cluster 3 genes are essential for cholate catabolism, we deleted kshA3 (ro05811), encoding a putative 3-ketosteroid-9α-hydroxylase. In contrast to the wild-type (WT) strain, the resulting ΔkshA3 mutant did not grow detectably on cholate, either in liquid (Fig. 1) or on solid medium (see Fig. S1 in the supplemental material). The mutant retained the ability to grow on 4-androstene-3,17-dione, and its ability to grow on cholate was restored by inserting the kshA3 gene with its native promoter into the att site of the RHA1 chromosome (see Fig. S1 in the supplemental material). Gas chromatography-coupled mass spectrometry (GC-MS) analyses failed to detect any cholate metabolites in solvent-extracted supernatants of cultures of the ΔkshA3 mutant (results not shown). This is in contrast to the metabolites detected in cultures of the WT strain, as described in the companion paper (26). Overall, these results demonstrate that at least one gene in cluster 3 is essential for growth on cholate and are consistent with the cluster specifically encoding cholate degradation. Notably, despite being constitutively expressed during growth on cholate, kshA in cluster 1 (ro04538) did not compensate for the deletion of kshA3.
Cluster 3 contains two steroid-CoA synthetases.
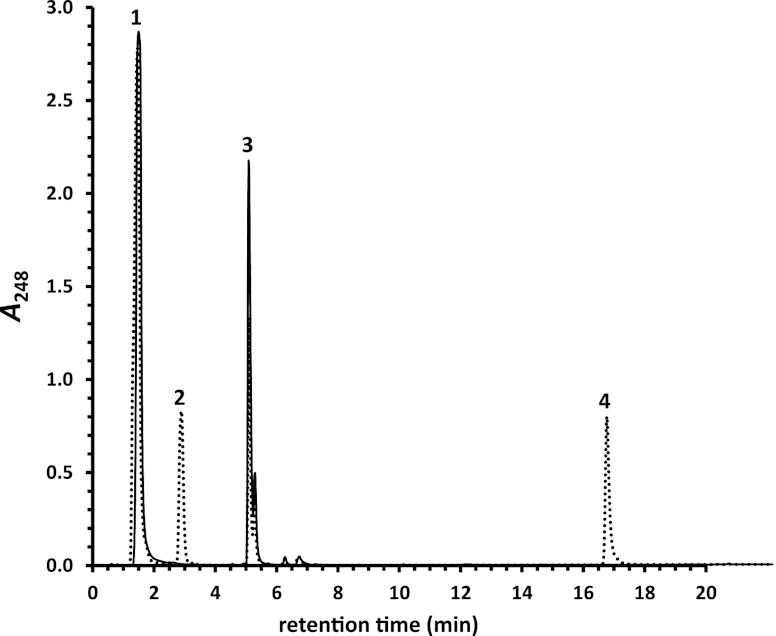
Two genes encoding putative AMP-forming acyl-CoA synthetases were identified in cluster 3. The products of casG (ro05820) and casI (ro05822) share 32% and 24% amino acid sequence identity with feruloyl-CoA synthetase (Fcs) of Amicolatopsis sp. strain HR167 (1) and FadD13 from M. tuberculosis H37Rv (16), respectively. Both genes were upregulated almost 9-fold during growth of RHA1 on cholate versus pyruvate. Heterologously produced purified CasG and CasI were tested for their respective abilities to catalyze the ATP-dependent formation of CoA thioesters of steroids bearing isopentanoyl and isopropanoyl side chains at C-17. For CasG, AMP formation was detected only in the presence of an isopentanoyl-bearing steroid (Table 1). Moreover, a CoA thioester was detected in reaction mixtures containing either cholate (Fig. 4), deoxycholate, or chenodeoxycholate. The CoA thioester formed in the presence of cholate was purified. ESI-MS analysis revealed that the compound had an m/z ratio of 1,158.5 (see Fig. S2 in the supplemental material), as expected for protonated cholyl-CoA, unambiguously identifying CasG as a cholyl-CoA synthetase. In contrast, AMP was detected only in CasI reactions in the presence of an isopropanoyl-bearing steroid. Moreover, a CoA thioester was detected in the reaction mixture containing 3-oxo-23,24-bisnor-4-cholenate.
Table 1.
Abilities of CasG (Ro05820) and CasI (Ro05822) to catalyze the ATP-dependent transformation of various steroid substrates bearing isopentanoyl and isopropanoyl side chainsa
| Substrate | CasG | CasI |
|---|---|---|
| Cholate | + | − |
| Lithocholate | + | − |
| Deoxycholate | + | − |
| Chenodeoxycholate | + | − |
| 3β-Hydroxy-5-cholenate | + | − |
| 3β-Hydroxy-23,24-bisnor-5-cholenate | − | + |
| 3-Oxo-23,24-bisnor-4-cholenate | − | + |
+, transformed; −, not transformed.
Fig 4.
Transformation of cholate to cholyl-CoA by CasG. Dotted chromatogram, CasG reaction after 2 h; solid chromatogram, no-enzyme control after 2 h. Substrate peaks: 1, ATP; 3, CoASH. Product peaks: 2, AMP; 4, cholyl-CoA. Cholate is not detected under these conditions.
Orthologous gene clusters and growth on cholate.
The genes in cluster 3 believed to be responsible for cholate catabolism were manually annotated on the basis of results from this study, as well as previous reports and database submissions (Table 2). We searched genomes for clusters of genes orthologous to cluster 3 of RHA1. Identification of orthologous genes was based on obtaining reciprocal best BLAST hits and further supported by gene synteny (see Fig. S3 in the supplemental material). A nearly identical gene cluster was found in Rhodococcus opacus B4. Other putatively orthologous gene clusters were found in Rhodococcus equi (103S and ATCC 33707), Rhodococcus erythropolis (PR4 and SK121), and Rhodococcus ruber Chol-4 (unpublished data), as well as Thermomonospora curvata DSM 43183. The clusters in the last six strains have some gene rearrangements and deletions compared to cluster 3 of RHA1. Notably, the cluster in the R. erythropolis strains lacks orthologs for hsaB3 and kshB3.
Table 2.
Annotation of the cholate catabolic gene cluster of R. jostii RHA1
| Gene name | RHA1 locus tag | Annotation of gene product | Best hita | Identity (%) |
|---|---|---|---|---|
| ro05790 | 2-Hydroxycyclohexanecarboxyl-CoA dehydrogenase | EHI45965 | 97 | |
| hsd4A3 | ro05791 | 17β-Hydroxysteroid dehydrogenase | ABG96481 | 54 |
| camM | ro05792 | 3,7(R),12(S)-Trihydroxy-9-oxo-9,10-seco-23,24-bisnorchola-1,3,5(10)-trien-22-oate uptake transporter; MFS superfamilyb | NA | |
| ro05793 | Conserved hypothetical protein | NA | ||
| ro05794 | Flavin F420-dependent oxidoreductase | NA | ||
| kstR3 | ro05795 | Transcriptional regulator; IclR family | BAA07185 | 37 |
| hsaB3 | ro05796 | 3-Hydroxy-9,10-seconandrosta-1,3,5(10)-trien-9,17-dione hydroxylase, reductase component | ABG96328 | 40 |
| hsaD3 | ro05797 | 4,5,9,10-Diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-dien-4-oic acid hydrolase | ABG96326 | 42 |
| kstD3 | ro05798 | 3-Ketosteroid-Δ1-dehydrogenase | ABW74859 | 45 |
| hsaE3 | ro05799 | 2-Hydroxypenta-2,4-dienoate hydratase | BAB97166 | 54 |
| hsaG3 | ro05800 | Acetaldehyde dehydrogenase | BAB97164 | 65 |
| hsaF3 | ro05801 | 4-Hydroxy-2-ketovalerate aldolase | BAB97165 | 50 |
| hsaA3 | ro05802 | 3-Hydroxy-9,10-seconandrosta-1,3,5(10)-trien-9,17-dione hydroxylase, oxygenase component | ABG96325 | 39 |
| hsaC3 | ro05803 | 3,4-Dihydroxy-9,10-seconandrosta-1,3,5(10)-trien-9,17-dione dioxygenase | ABG96327 | 45 |
| ro05810 | Short-chain alcohol dehydrogenase | NA | ||
| kshA3 | ro05811 | 3-Ketosteroid-9α-hydroxylase, oxygenase component | ACD11366 | 63 |
| hsaA3b | ro05812 | 3-Hydroxy-9,10-seconandrosta-1,3,5(10)-trien-9,17-dione hydroxylase, oxygenase component | ABG96325 | 48 |
| kstD3b | ro05813 | 3-Ketosteroid Δ1-dehydrogenase | AF096929_2 | 70 |
| casA | ro05814 | Transcriptional regulator; TetR family | AF096929_1 | 80 |
| casB | ro05815 | 3-Ketoacyl-CoA thiolase | CAB05060 | 64 |
| casC | ro05816 | Acyl-CoA dehydrogenase | NA | |
| casD | ro05817 | 3α,7α,12α-Trihydroxy-5β-chol-22-en-24-oyl-CoA hydratase | CAA55037 | 34c |
| casE | ro05818 | Acyl-CoA thioesterase | NA | |
| casF | ro05819 | Cholesterol oxidase | AAF64503 | 49d |
| casG | ro05820 | 3α,7α,12α-Trihydroxy-5β-cholanoyl-CoA synthetase (cholyl-CoA synthetase) | NA | |
| casH | ro05821 | NADH-dependent flavin oxidoreductase | NA | |
| casI | ro05822 | Steroid-22-oyl-CoA synthetase | NA | |
| casJ | ro05823 | Flavin-utilizing monooxygenase | NA | |
| casK | ro05824 | Conserved hypothetical protein | NA | |
| casL | ro05825 | 7α,12α-Dihydroxy-23,24-bisnorchola-1,4-dien-22-oyl-CoA dehydrogenase | ABQ10637 | 49 |
| casM | ro05826 | Conserved hypothetical protein | NA | |
| casN | ro05827 | Acyl-CoA dehydrogenase | NA | |
| casO | ro05828 | Conserved hypothetical protein | NA | |
| casP | ro05829 | 3-Ketoacyl-CoA thiolase 2 | AAD21068 | 29 |
| casQ | ro05830 | 3α,7α,12α-Trihydroxy-5β-chol-22-en-24-oyl-CoA hydratase | CAA55037 | 32c |
| ro05831 | Short-chain dehydrogenase/reductase | NA | ||
| ro05832 | Short-chain dehydrogenase/reductase | AAD21071 | 38 | |
| kshB3 | ro05833 | 3-Ketosteroid-9α-hydroxylase, reductase component | ACI62781 | 70 |
Accession number of functionally characterized best hit in NCBI database; NA, not available.
See the companion paper (26).
Identity with central-domain amino acids 324 to 596.
Identity with C-terminal part, amino acids 79 to 270.
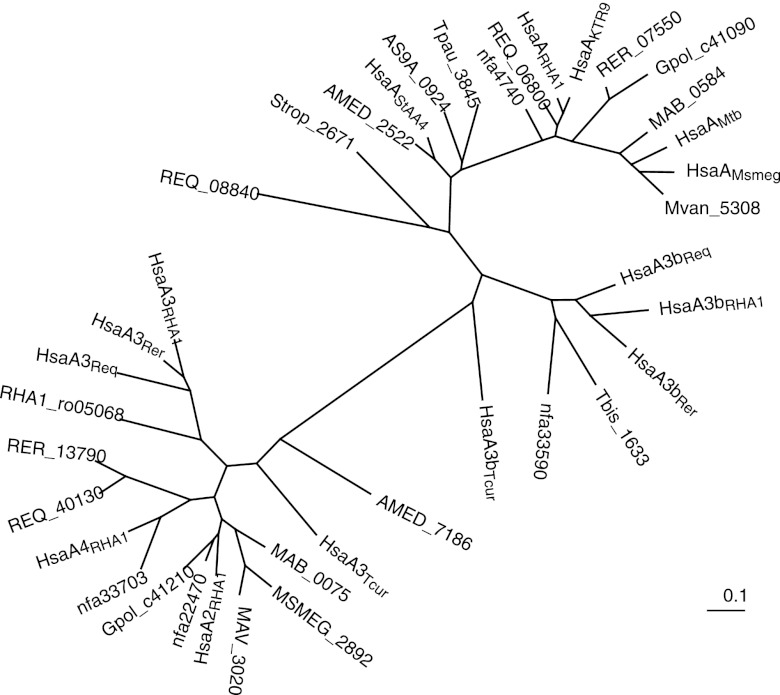
The phylogenies of individual steroid catabolism enzymes support the existence of distinct cholate and cholesterol catabolism gene clusters in the mycolic acid bacteria. HsaA3RHA1 and its orthologs among the rhodococci and T. curvata comprise a clade (Fig. 5), most of whose members are encoded by genes in the above-mentioned cholate catabolism gene clusters. Interestingly, another RHA1 protein, Ro05068, also falls within this clade. However, the ro05068 gene is not proximal to steroid catabolism genes, and it is not upregulated during growth on cholate. HsaA3bRHA1 and its orthologs comprise a distinct clade whose members are all encoded by genes not consistently linked to the cholate catabolism gene clusters. Finally, HsaARHA1 and its more numerous orthologs among diverse mycolic acid bacteria comprise a distinct clade whose members are all encoded by genes in putative cholesterol catabolism gene clusters. The above pattern is also found in the phylogeny of KshA (22).
Fig 5.
Dendrogram showing the phylogeny of HsaA homologs. AMED, Amycolatopsis mediterranei U32; AS9A, Amycolicicoccus subflavus DQS3-9A1; Gpol, Gordonia polyisoprenivorans VH2; KTR9, Gordonia sp. KTR9; MAB, Mycobacterium abscessus ATCC 19977; MAV, Mycobacterium avium 104; MSMEG, M. smegmatis strain MC2 155; Mtb, M. tuberculosis H37Rv; Mvan, Mycobacterium vanbaalenii PYR-1; nfa, Nocardia farcinica IFM 10152; RHA1, R. jostii RHA1; REQ, R. equi 103S; RER, R. erythropolis PR4; Strop, Salinispora tropica CNB-440; Tbis, Thermobispora bispora DSM 43833; Tcur, T. curvata DSM 43183; Tpau, Tsukamurella paurometabola DSM 20162. The following sequences from R. opacus B4 were omitted from the analysis due to their high amino acid identities with the RHA1 orthologs: ROP_44540 (99% identity with HsaARHA1), ROP_22160 (98% identity with HsaA2RHA1), ROP_58630 (97% identity with HsaA3RHA1), ROP_58740 (96% identity with HsaA3bRHA1), and ROP_51300 (96% identity with RHA1_ro05068). The RER_38340 sequence from R. erythropolis PR4 was omitted from the analysis due to its very high amino acid identity (98%) with HsaA3Rer.
We tested a number of strains, some containing the cholate catabolism gene cluster, for the ability to grow on cholate. In addition to RHA1, R. equi 103S and R. ruber Chol-4, both with the cholate cluster, grew on cholate. We were unable to obtain R. erythropolis PR4 for testing, but R. erythropolis SQ1, with very high genetic similarity to PR4 and predicted to have the cholate cluster, grew on cholate. On the other hand, Gordonia sp. strain KTR9, Mycobacterium smegmatis mc2155, and Mycobacterium bovis BCG, all lacking the cholate cluster, failed to grow on cholate. Thus, there was perfect correspondence between the cholate catabolism gene cluster and the ability to grow on cholate.
DISCUSSION
Cholate catabolism gene cluster.
The diverse lines of evidence from this study consistently support the conclusion that cluster 3 of RHA1 harbors inducible genes that encode cholate catabolism. Orthologous clusters exist in all available Rhodococcus genomes, suggesting that the gene cluster and the ability to use cholate are widespread, if not ubiquitous, among members of the genus. Notably, the cluster was found in R. equi, which has a reduced genome. Thus, use of cholate appears to be a core metabolic feature of Rhodococcus spp. This suggests that cholate excreted by vertebrates is an important resource for Rhodococcus spp. in diverse environments. Conservation of cluster 3 in R. equi is consistent with the proposal that feces are an important reservoir for this animal pathogen (8). In at least the case of T. curvata, this gene cluster occurs beyond Rhodococcus spp. While Thermomonospora and Rhodococcus are both genera of the Actinomycetales, they are not closely related, and the former is not among the mycolic acid bacteria. Nevertheless, the cholate catabolism gene cluster appears to be less widespread among Actinobacteria than is the one encoding cholesterol metabolism (cluster 1 in RHA1).
It is unclear why in RHA1 cluster 3 is more tightly downregulated in the absence of cholate than cluster 1 in the absence of cholesterol. It is interesting that cluster 3 was also upregulated during growth on 7-keto-cholesterol (7KC) (18). However, only genes in cluster 1 were shown to be essential for 7KC catabolism, and it was concluded that induction of cluster 3 by 7KC is likely gratuitous. The current study provides a rationale for that observation, as 7KC and cholate both have substituents at C-7 that may be critical for differential regulation of clusters 1 and 3. Further, the high constitutive expression of cluster 1 observed in this study may be important for efficient degradation of 7KC, as the previous study found relatively little upregulation of cluster 1 during growth on 7KC.
We note two differences in the functions encoded by clusters 1 and 3, which relate to their respective catabolic substrates. Only cluster 1 encodes an Mce4 transporter. This transporter functions in the uptake of cholesterol and some other steroids, but not that of cholate (20). Cluster 3 appears to encode a single transporter, an MFS transporter, CamM (Fig. 2). We have determined that CamM is not essential for cholate uptake and, instead, functions in the uptake of a cholate metabolite (26). No uptake system for cholate has been identified in RHA1, and one possibility is that cholate enters the cell via passive diffusion. Another difference is that only cluster 1 encodes a P450 monooxygenase. CYP125, encoded in cluster 1, has been shown to initiate degradation of cholesterol by terminally oxidizing the aliphatic side chain (5, 24). This activity is not required for degradation of cholate, which has a terminally carboxylated side chain.
Ring A and B degradation.
Cluster 3 of RHA1 encodes degradation of rings A and B of the steroid nucleus, as well as the side chain of cholate. Many of the genes in cluster 3 are paralogous to genes in cluster 1 (Fig. 2 and 5) with demonstrated functions in degradation of rings A and B of cholesterol (4, 6, 34). These include the kst, ksh, and hsa genes of cluster 3. Thus, distinct, analogous pathways appear to exist for the two substrates. It is unclear at what point the additional hydroxyl substitutents at C-7 and C-12 of cholate are removed, but metabolites retaining these substituents suggest that they persist throughout much of the nucleus degradation process (26).
Our finding that the kshA3 gene in cluster 3 is essential for growth on cholate is consistent with investigation of the orthologous gene, kshA1, in Rhodococcus rhodochrous DSM43269 (23). Five kshA paralogs from R. rhodochrous were individually tested by a complementation assay, and only kshA1DSM43269 was able to support cholate catabolism. Accordingly, of the five paralogs, only kshA1DSM43269 exhibited high activity with the putative cholate metabolite, 7α,12α-dihydroxy-23,24-bisnorchola-1,4-dien-22-oate, as the substrate. However, we have not excluded the possibility that other genes in cluster 3 are nonessential, and paralogs in the other clusters may contribute to cholate catabolism by RHA1. In particular, the apparently high constitutive expression of cluster 1 in RHA1 creates the possibility that some of its genes might contribute to cholate catabolism. Detailed investigation of the expression of individual genes and the specificity of corresponding enzymes would be required to address this question.
Side chain degradation.
The available evidence indicates that genes from ro05814 to ro05830 encode degradation of the cholate side chain, so we have designated these genes cas, for cholate side chain. Orthologous genes with high synteny also occur in R. opacus, R. equi, R. erythropolis, and T. curvata (see Fig. S3 in the supplemental material). Our results demonstrate that CasG is a cholyl-CoA synthetase, which can initiate degradation of the side chain. Our results strongly suggest that CasI is a steroid-22-oyl-CoA synthetase. Presumably, this enzyme would not be required for uninterrupted beta-oxidation of the side chain, as a thiolase reaction removing the terminal C-2 unit from the side chain would normally transfer the CoA moiety to the remaining isopropanoyl chain. However, we have observed transient accumulation of metabolites with an isopropanoyl side chain in RHA1 cultures growing on cholate (26). The eventual degradation of these metabolites presumably requires the activity of CasI to activate the side chain for the final cycle of beta-oxidation. CasE is a thioesterase that may be involved in formation of the above-mentioned metabolites with isopropanoyl side chains, perhaps allowing the cells to excrete metabolites that accumulate and would otherwise have toxic effects.
The casG and casI genes are flanked by two putative operons, casABCDE and casLMNOPQ, that contain genes encoding beta-oxidation enzymes. Overall, the cas gene cluster appears to encode the two cycles of beta-oxidation required for cholate side chain degradation. Consistent with this function, two genes in the cluster have similarity to genes with demonstrated roles in cholate side chain degradation by Pseudomonas sp. Chol1. The first, casP, is similar to skt in Chol1, which encodes a steroid β-ketothiolase that completes the first round of beta-oxidation, cleaving the terminal C-2 unit from the side chain (3). The second, casL, is similar to acad in Chol1, which encodes an acyl-CoA dehydrogenase that initiates the second round, oxidizing the C-17–C-20 bond in preparation for cleavage of the remaining C-3 unit of the side chain (2). The authors identified similar genes in the genomes of two other Gram-negative bacteria capable of growing on cholate, Pseudoalteromonas haloplanktis TAC125 and C. testosteroni TA441. Despite the similarity of CasP to Skt and of CasL to ACAD, these are not orthologous pairs. Thus, the pathways for cholate catabolism identified in Gram-positive versus Gram-negative bacteria are distinct and probably not orthologous.
Ring C and D degradation.
The results of this study suggest that the cholesterol and cholate catabolic pathways in Rhodococcus spp. converge on a common route for degradation of steroid rings C and D. Previously, a cluster of genes in R. equi was implicated in degradation of 3aα-H-4α(3′-propanoate)-7aβ-methylhexahydro-1,5-indanedione (HIP), a cholesterol degradation intermediate containing rings C and D (30). Within this cluster, ipdA and ipdB encode a heterodimeric CoA transferase proposed to initiate beta-oxidation of the propanoyl moiety of HIP. The proximal fadE30 gene encodes an acyl-CoA dehydrogenase proposed to catalyze the subsequent step in beta-oxidation of this moiety. The orthologous genes in RHA1 occur in the region of cluster 1, from ro04591 to ro04599 and from ro04649 to ro04656. Unlike the other genes in cluster 1 involved in degradation of the side chain and rings A and B of cholesterol, there are no obvious paralogs in cluster 3 or elsewhere in the RHA1 genome. Further, expression of these two groups of genes is regulated differently than that of the other genes in cluster 1. One group, ro04649 to ro04656, was upregulated during growth both on cholesterol (32) and on cholate (see Table S2 in the supplemental material). The other, ro04591 to ro04599, was upregulated only during growth on cholate but was likely expressed at a high constitutive level on cholesterol. Together, these two gene groups appear to correspond to the KstR2 regulon described in M. smegmatis (15). Thus, cluster 3 does not appear to encode the complete degradation of cholate, and a subset of genes in cluster 1 encoding degradation of rings C and D is upregulated during growth on cholate.
ACKNOWLEDGMENTS
This research was supported by operating grants from the Natural Sciences and Engineering Research Council (NSERC) to W.W.M. and the Canadian Institutes for Health Research (CIHR) to L.D.E. I.C. is the recipient of a fellowship from the Fonds de Recherche en Santé du Québec (FRSQ) and the Michael Smith Foundation for Health Research (MSFHR).
Footnotes
Published ahead of print 28 September 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Achterholt S, Priefert H, Steinbuchel A. 2000. Identification of Amycolatopsis sp. strain HR167 genes, involved in the bioconversion of ferulic acid to vanillin. Appl. Microbiol. Biotechnol. 54:799–807 [DOI] [PubMed] [Google Scholar]
- 2. Birkenmaier A, et al. 2007. Biochemical and genetic investigation of initial reactions in aerobic degradation of the bile acid cholate in Pseudomonas sp. strain Chol1. J. Bacteriol. 189:7165–7173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Birkenmaier A, Möller HM, Philipp B. 2011. Identification of a thiolase gene essential for β-oxidation of the acyl side chain of the steroid compound cholate in Pseudomonas sp. strain Chol1. FEMS Microbiol. Lett. 318:123–130 [DOI] [PubMed] [Google Scholar]
- 4. Capyk JK, Casabon I, Gruninger R, Strynadka NC, Eltis LD. 2011. Activity of 3-ketosteroid 9 alpha-hydroxylase (KshAB) indicates cholesterol side chain and ring degradation occur simultaneously in Mycobacterium tuberculosis. J. Biol. Chem. 286:40717–40724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Capyk JK, et al. 2009. Mycobacterial cytochrome P450 125 (Cyp125) catalyzes the terminal hydroxylation of C27 steroids. J. Biol. Chem. 284:35534–35542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dresen C, et al. 2010. A flavin-dependent monooxygenase from Mycobacterium tuberculosis involved in cholesterol catabolism. J. Biol. Chem. 285:22264–22275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gonçalves ER, et al. 2006. Transcriptomic assessment of isozymes in the biphenyl pathway of Rhodococcus sp. RHA1. Appl. Environ. Microbiol. 72:6183–6193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grimm MB, et al. 2007. Evaluation of fecal samples from mares as a source of Rhodococcus equi for their foals by use of quantitative bacteriologic culture and colony immunoblot analyses. Am. J. Vet. Res. 68:63–71 [DOI] [PubMed] [Google Scholar]
- 9. Hayakawa S. 1973. Microbiological transformation of bile acids. Adv. Lipid Res. 11:143–192 [DOI] [PubMed] [Google Scholar]
- 10. Horinouchi M, et al. 2008. Identification of genes involved in inversion of stereochemistry of a C-12 hydroxyl group in the catabolism of cholic acid by Comamonas testosteroni TA441. J. Bacteriol. 190:5545–5554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Horinouchi M, Hayashi T, Yamamoto T, Kudo T. 2003. A new bacterial steroid degradation gene cluster in Comamonas testosteroni TA441 which consists of aromatic-compound degradation genes for seco-steroids and 3-ketosteroid dehydrogenase genes. Appl. Environ. Microbiol. 69:4421–4430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Horinouchi M, Kurita T, Hayashi T, Kudo T. 2010. Steroid degradation genes in Comamonas testosteroni TA441: isolation of genes encoding a Δ4(5)-isomerase and 3α- and 3β-dehydrogenases and evidence for a 100 kb steroid degradation gene hot spot. J. Steroid Biochem. Mol. Biol. 122:253–263 [DOI] [PubMed] [Google Scholar]
- 13. Horinouchi M, et al. 2004. Steroid degradation gene cluster of Comamonas testosteroni consisting of 18 putative genes from meta-cleavage enzyme gene tesB to regulator gene tesR. Biochem. Biophys. Res. Commun. 324:597–604 [DOI] [PubMed] [Google Scholar]
- 14. Hu Y, et al. 2010. 3-Ketosteroid 9α-hydroxylase is an essential factor in the pathogenesis of Mycobacterium tuberculosis. Mol. Microbiol. 75:107–121 [DOI] [PubMed] [Google Scholar]
- 15. Kendall SL, et al. 2010. Cholesterol utilization in mycobacteria is controlled by two TetR-type transcriptional regulators: kstR and kstR2. Microbiology 156:1362–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Khare G, et al. 2009. Dissecting the role of critical residues and substrate preference of a fatty acyl-CoA synthetase (FadD13) of Mycobacterium tuberculosis. PLoS One 4:e8387 doi:10.1371/journal.pone.0008387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. LeBlanc JC, Gonçalves ER, Mohn WW. 2008. Global response to desiccation stress in the soil actinomycete Rhodococcus jostii RHA1. Appl. Environ. Microbiol. 74:2627–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mathieu J, et al. 2010. 7-Ketocholesterol catabolism by Rhodococcus jostii RHA1. Appl. Environ. Microbiol. 76:352–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McLeod MM, et al. 2006. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. U. S. A. 103:15582–15587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mohn WW, et al. 2008. The actinobacterial mce4 locus encodes a steroid transporter. J. Biol. Chem. 283:35368–35374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pandey AK, Sassetti CM. 2008. Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl. Acad. Sci. U. S. A. 105:4376–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Petrusma M, Dijkhuizen L, van der Geize R. 2009. Rhodococcus rhodochrous DSM 43269 3-ketosteroid 9-alpha-hydroxylase, a two-component iron-sulfur-containing monooxygenase with subtle steroid substrate specificity. Appl. Environ. Microbiol. 75:5300–5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Petrusma M, Hessels G, Dijkhuizen L, van der Geize R. 2011. Multiplicity of 3-ketosteroid-9 alpha-hydroxylase enzymes in Rhodococcus rhodochrous DSM43269 for specific degradation of different classes of steroids. J. Bacteriol. 193:3931–3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rosloniec KZ, et al. 2009. Cytochrome P450 125 (CYP125) catalyses C26-hydroxylation to initiate sterol side-chain degradation in Rhodococcus jostii RHA1. Mol. Microbiol. 74:1031–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Seto M, et al. 1995. A novel transformation of polychlorinated biphenyls by Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 61:3353–3358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Swain K, Casabon I, Eltis LD, Mohn WW. 2012. Two transporters essential for reassimilation of novel cholate metabolites by Rhodococcus jostii RHA1. J. Bacteriol. 194:6720–6727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tenneson ME, Bilton RF, Mason AN. 1978. The degradation of taurocholic acid and glycocholic acid by Pseudomonas spp. NCIB10590. Biochem. Soc. Trans. 6:975–979 [DOI] [PubMed] [Google Scholar]
- 28. Tenneson ME, Bilton RF, Mason AN. 1978. A scheme for the microbial degradation of lithocholic acid involving testosterone as an intermediate. Biochem. Soc. Trans. 6:428–430 [DOI] [PubMed] [Google Scholar]
- 29. van der Geize R, Dijkhuizen L. 2004. Harnessing the catabolic diversity of rhodococci for environmental and biotechnological applications. Curr. Opin. Microbiol. 7:255–261 [DOI] [PubMed] [Google Scholar]
- 30. van der Geize R, Grommen AWF, Hessels GI, Jacobs AAC, Dijkhuizen L. 2011. The steroid catabolic pathway of the intracellular pathogen Rhodococcus equi is important for pathogenesis and a target for vaccine development. PLoS Pathog. 7:e1002181 doi:10.1371/journal.ppat.1002181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. van der Geize R, Hessels GI, van Gerwen R, van der Meijden P, Dijkhuizen L. 2001. Unmarked gene deletion mutagenesis of kstD, encoding 3-ketosteroid Delta(1)-dehydrogenase, in Rhodococcus erythropolis SQ1 using sacB as counter-selectable marker. FEMS Microbiol. Lett. 205:197–202 [DOI] [PubMed] [Google Scholar]
- 32. van der Geize R, et al. 2007. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc. Natl. Acad. Sci. U. S. A. 104:1947–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wilkinson CJ, et al. 2002. Increasing the efficiency of heterologous promoters in actinomycetes. J. Mol. Microbiol. Biotechnol. 4:417–426 [PubMed] [Google Scholar]
- 34. Yam KC, et al. 2009. Studies of a ring-cleaving dioxygenase illuminate the role of cholesterol metabolism in the pathogenesis of Mycobacterium tuberculosis. PLoS Pathog. 5:e1000344 doi:10.1371/journal.ppat.1000344 [DOI] [PMC free article] [PubMed] [Google Scholar]