Abstract
AMP phosphorylase (AMPpase), ribose-1,5-bisphosphate (R15P) isomerase, and type III ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) have been proposed to constitute a novel pathway involved in AMP metabolism in the Archaea. Here we performed a biochemical examination of AMPpase and R15P isomerase from Thermococcus kodakarensis. R15P isomerase was specific for the α-anomer of R15P and did not recognize other sugar compounds. We observed that activity was extremely low with the substrate R15P alone but was dramatically activated in the presence of AMP. Using AMP-activated R15P isomerase, we reevaluated the substrate specificity of AMPpase. AMPpase exhibited phosphorylase activity toward CMP and UMP in addition to AMP. The [S]-v plot (plot of velocity versus substrate concentration) of the enzyme toward AMP was sigmoidal, with an increase in activity observed at concentrations higher than approximately 3 mM. The behavior of the two enzymes toward AMP indicates that the pathway is intrinsically designed to prevent excess degradation of intracellular AMP. We further examined the formation of 3-phosphoglycerate from AMP, CMP, and UMP in T. kodakarensis cell extracts. 3-Phosphoglycerate generation was observed from AMP alone, and from CMP or UMP in the presence of dAMP, which also activates R15P isomerase. 3-Phosphoglycerate was not formed when 2-carboxyarabinitol 1,5-bisphosphate, a Rubisco inhibitor, was added. The results strongly suggest that these enzymes are actually involved in the conversion of nucleoside monophosphates to 3-phosphoglycerate in T. kodakarensis.
INTRODUCTION
Archaea comprise the third domain of life and exhibit unique metabolic features that are not found in bacteria and eukaryotes. The metabolic enzymes and pathways utilized for glycolysis and pentose biosynthesis in many archaea differ from the classical Embden-Meyerhof (EM)/Entner-Doudoroff (ED) pathways (24, 27, 33, 35) and the pentose phosphate pathway (11, 19, 28), respectively. A previous study of the hyperthermophilic archaeon Thermococcus kodakarensis KOD1 (3, 9) suggested the presence of a novel pathway involved in nucleic acid metabolism (25). The pathway consists of a type III ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) and two novel enzymes, AMP phosphorylase (AMPpase) and ribose-1,5-bisphosphate (R15P) isomerase. In the first reaction of this pathway, catalyzed by AMPpase, the adenine base of AMP is released and is replaced by a phosphate group to generate R15P. In the following R15P isomerase reaction, R15P is converted to ribulose 1,5-bisphosphate (RuBP). In the final reaction, Rubisco catalyzes the conversion of RuBP, CO2, and H2O to 2 molecules of 3-phosphoglycerate (3-PGA), which is an intermediate of central sugar metabolism. Genome sequences indicate that this pathway is distributed broadly among the Archaea, including all members of the Thermococcales, Archaeoglobales, Methanomicrobiales, and Methanosarcinales. The pathway is also found in several members of the Halobacteriales, Methanococcales, Desulfurococcales, and Thermoproteales. The pathway at present seems to be confined to the Archaea, since a complete set of genes cannot be found in any of the genomes of the Bacteria and Eucarya.
AMPpase is a unique enzyme in terms of its substrate specificity. It is the first enzyme identified to catalyze a phosphorolysis reaction on a nucleotide. This is distinct from the structurally related phosphorylases that have been characterized previously, which act specifically on nucleosides (6, 12, 20, 30). R15P isomerase catalyzes the isomerization of R15P, an aldopentose with a phosphorylated 1-hydroxy group. The reaction is of interest because opening of the furanose ring cannot precede isomerization due to the phosphate group on the C-1 carbon. The structure of R15P isomerase revealed that the enzyme displayed a hexameric assembly (18). Crystal structures bound to the substrate/product and mutational studies indicated that Cys133 and Asp202 act as active-site residues in the enzyme and that the reaction proceeds via a cis-phosphoenolate intermediate. R15P isomerase is structurally related to 5-methylthioribose-1-phosphate (MTR1P) isomerase. MTR1P isomerase functions in the methionine salvage pathway and catalyzes a similar reaction, the isomerization of MTR1P, an aldopentose with a phosphorylated 1-hydroxy group, to 5-methylthioribulose 1-phosphate (2, 26). Enzymatic (22) and structural (5, 29) examinations have been carried out on the enzymes from Bacillus subtilis and Saccharomyces cerevisiae. One of the reaction mechanisms that have been proposed for this enzyme also proceeds via a cis-phosphoenolate intermediate (29).
The enzymatic and structural features of type III Rubiscos have been reported previously (1, 7, 8, 15–17, 34). As mentioned above, we have recently determined the 3-dimensional structure of T. kodakarensis R15P isomerase (18). However, no detailed biochemical characterization of the two novel enzymes, AMPpase and R15P isomerase, has been reported. Since unregulated breakdown of AMP would be detrimental to the cells, one can suppose that the pathway should be regulated so that activity is rapidly shut down once intracellular levels of AMP become low. Here we performed the first detailed biochemical analysis of AMPpase and R15P isomerase. The enzymes from T. kodakarensis were examined by focusing mainly on their substrate specificity and kinetic behavior, as well as how their activities are regulated at the protein level in response to the presence of various metabolites. We have also confirmed the conversion of nucleoside monophosphates (NMPs) to 3-PGA in cell extracts, providing strong evidence that the pathway functions in T. kodakarensis.
MATERIALS AND METHODS
Strains and growth conditions.
Escherichia coli strains DH5α and BL21-CodonPlus (DE3)-RIL were used for plasmid construction and heterologous gene expression, respectively. These strains were cultivated at 37°C in Luria-Bertani medium containing 100 μg/ml ampicillin (23). T. kodakarensis KOD1 was cultivated under strictly anaerobic conditions at 85°C in a nutrient-rich growth medium based on artificial seawater (ASW) (21). Briefly, ASW-YT medium was composed of 0.8× ASW, 0.5% yeast extract, and 0.5% tryptone. Either 0.2% elemental sulfur (ASW-YT-S0), 0.5% sodium pyruvate (ASW-YT-Pyr), or 0.5% sodium pyruvate and a mixture of adenosine, guanosine, cytidine, and uridine at 2.5 mM (ASW-YT-Pyr-Nuc) were added.
Preparation of the recombinant proteins.
Recombinant His6-tagged R15P isomerase and Rubisco were produced in E. coli and were purified as described previously (18). The recombinant AMPpase was expressed as described elsewhere (25). In order to purify recombinant AMPpase, cells were harvested, resuspended in 50 mM Tris-HCl (pH 7.5), and sonicated. Soluble proteins were incubated for 10 min at 80°C and were centrifuged (at 20,000 × g for 30 min) to remove thermolabile proteins deriving from the host cells. The supernatant was applied to an anion-exchange column (Resource Q; GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom), and proteins were eluted with a linear gradient of NaCl (0 to 1.0 M) in 50 mM Tris-HCl (pH 7.5). After the fractions containing AMPpase were concentrated with an Amicon Ultra centrifugal filter unit (molecular weight cutoff [MWCO], 100,000; Millipore, Billerica, MA), the sample was applied to a gel filtration column (Superdex 200; GE Healthcare) with a mobile phase of 150 mM NaCl in 50 mM Tris-HCl (pH 7.5) at room temperature.
Enzymatic synthesis of ribose 1,5-bisphosphate with AMP phosphorylase.
Enzymatic preparation of the substrate R15P, used for activity measurements of R15P isomerase, was performed as follows. The AMPpase reaction was performed in a 500-μl mixture containing 560 nM purified AMPpase, 100 mM Tris-HCl (pH 7.5), 30 mM sodium phosphate (pH 7.5), and 30 mM AMP. After preincubation at 85°C for 3 min, the reaction was initiated by the addition of AMP. After a further 10-min incubation, the reaction was terminated by rapid cooling on ice for 5 min, and AMPpase was removed by ultrafiltration with an Amicon Ultra centrifugal filter unit (MWCO, 30,000; Millipore). The resulting R15P mixture was concentrated by vacuum drying and centrifugation and was used as the substrate for the R15P isomerase reaction. By use of high-performance liquid chromatography (HPLC) with a refractive index detector, the concentration of R15P was calculated to be approximately 5 mM. This enzymatically prepared substrate mixture is designated E-R15P.
Measurement of ribose-1,5-bisphosphate isomerase activity by use of coupling enzymes.
R15P isomerase activity was measured either with coupling enzymes or by HPLC (see below). The former procedure was carried out as follows. First, the R15P isomerase reaction was performed coupled with the carboxylase reaction of Rubisco. The reaction mixture (100 μl) was composed of 81 nM purified R15P isomerase, 1.0 μM purified Rubisco (90 mU), 100 mM NaHCO3, and 30 μl of E-R15P (1.5 mM) or 5 mM chemically synthesized R15P (C-R15P; Tokyo Chemical Industry Co., Tokyo, Japan) in 10 mM MgCl2 and 100 mM Bicine-NaOH (pH 8.3). NAD+, which had been included in the reaction mixture previously (25), was found to have no effect on activity (see below) and was excluded from the reaction mixture. When C-R15P was used as the substrate, AMP was added to the reaction mixture in order to activate the enzyme. After preincubation at 85°C for 3 min, the reaction was initiated by the addition of NaHCO3 and R15P. The reaction was carried out for 5 min at 85°C and was terminated by rapid cooling on ice for 5 min, and the enzymes were removed with an Amicon Ultra centrifugal filter unit (MWCO, 30,000). After appropriate dilution, the amount of 3-PGA synthesized by the reaction was determined by a second coupling reaction, described elsewhere (7). The second reaction mixture (100 μl) was composed of 5 mM ATP, 0.2 mM NADH, 8 mM MgCl2, 80 mM Bicine-NaOH (pH 8.3), 20 μl of coupling enzyme solution, and an aliquot of the R15P isomerase reaction mixture. The coupling enzyme solution contained 563 U ml−1 3-phosphoglycerate phosphokinase, 125 U ml−1 glyceraldehyde-3-phosphate dehydrogenase, 260 U ml−1 triose-phosphate isomerase, 22.5 U ml−1 glycerophosphate dehydrogenase, 5 mM reduced glutathione, 0.1 mM EDTA, and 20% glycerol in 50 mM Bicine-NaOH (pH 8.0). After preincubation at 25°C for 3 min, the reaction was initiated with the addition of the enzymes. The decrease in absorbance at 340 nm due to the consumption of NADH was measured.
The effects of various compounds on the R15P isomerase reaction were examined by adding one of the following compounds to the reaction mixture: 18 mM sodium phosphate, 0.4 mM adenine, 0.4 mM NAD+, or 3 mM AMP, CMP, GMP, UMP, TMP, ADP, ATP, adenosine, dAMP, 5′-methylthioadenosine (MTA), S-adenosylmethionine (SAM), S-adenosylhomocysteine (SAH), phosphoribosylpyrophosphate (PRPP), ribose 5-phosphate (R5P), fructose 1,6-bisphosphate (FBP), fructose 6-phosphate (F6P), 3-PGA, glucose, or pyruvate. We could not use the enzyme coupling method to measure the effects of ADP and 3-PGA, because ADP inhibits 3-phosphoglycerate phosphokinase, and 3-PGA is the product of the R15P isomerase/Rubisco reactions. In these cases, HPLC was applied for activity measurements (described below).
Measurement of ribose-1,5-bisphosphate isomerase activity using HPLC.
When activity was measured by HPLC, the reaction mixture (100 μl) was composed of 140 nM purified R15P isomerase, 3 mM AMP, and 5 mM C-R15P in 10 mM MgCl2 and 100 mM Bicine-NaOH (pH 8.3). After preincubation at 85°C for 3 min, the reaction was initiated by the addition of C-R15P, followed by incubation for 3, 5, or 7 min, and was terminated by rapid cooling on ice for 5 min. R15P isomerase was removed with an Amicon Ultra centrifugal filter unit (MWCO, 30,000). After the addition of an equal volume of 600 mM sodium phosphate (pH 4.4) to the filtrate in order to adjust the phosphate concentration to that of the HPLC mobile phase, the sample was analyzed on an amino column (Asahipak NH2P-50 4E column; Shodex, Tokyo, Japan) with 300 mM sodium phosphate buffer (pH 4.4) as the mobile phase. When the effects of ADP and 3-PGA on R15P isomerase activity were investigated, activity was calculated by measuring the decrease in R15P levels. Isomerase activity toward 40 mM PRPP, R5P, ribose, FBP, F6P, glucose 1,6-bisphosphate (G16P), glucose 6-phosphate (G6P), or glucose 1-phosphate (G1P) was examined by monitoring substrate consumption and/or product generation in the presence or absence of R15P isomerase. When activity toward PRPP was examined, a C18 column (Cosmosil 5C18-PAQ; Nacalai Tesque) was utilized with 50 mM NaH2PO4 (pH 4.3) as the mobile phase. Column temperatures were set at 40°C, and compounds were detected with a refractive index detector in all cases.
Measurement of AMP phosphorylase activity.
The phosphorylase reaction of nucleoside monophosphates (NMPs) was performed in a mixture (100 μl) containing 100 mM Tris-HCl (pH 7.5), 20 mM sodium phosphate (pH 7.5), 190 nM purified AMPpase, and 20 mM NMP. After preincubation at 85°C for 3 min, the reaction was initiated by the addition of an NMP. The reaction was carried out at 85°C for 5 min and was terminated by rapid cooling on ice for 5 min, and AMPpase was removed with an Amicon Ultra centrifugal filter unit (MWCO, 30,000). The R15P generated in the AMPpase reaction was then converted to 3-PGA by the isomerase activity of R15P isomerase and the carboxylase activity of Rubisco. The reaction mixture (100 μl) was composed of 1.3 μM purified R15P isomerase (170 mU), 1.0 μM purified Rubisco (90 mU), 10 mM AMP, 100 mM NaHCO3, and 10 μl of the AMPpase reaction mixture. After preincubation at 85°C for 3 min, the reaction was initiated by the addition of NaHCO3 and the AMPpase reaction mixture. The reaction was carried out for 10 min at 85°C and was terminated by rapid cooling on ice for 5 min, and the enzymes were removed with an Amicon Ultra centrifugal filter unit (MWCO, 30,000). The 3-PGA generated in this reaction was quantified by the second coupling reaction, described above.
In order to examine the substrate specificity of AMPpase, phosphorylase activities toward the following substrates were measured by HPLC: 20 mM dNMP, adenosine, cytidine, uridine, ADP, ATP, SAM, SAH, PRPP, or R5P or 2 mM MTA. The reactions were carried out for 3, 5, or 7 min. Compounds were detected with a refractive index detector and/or a UV detector (A254). The phosphorylase activity was quantified by substrate consumption and/or nucleobase release.
Examination of 3-phosphoglycerate synthesis in cell extracts.
Cell extracts (CFE) of T. kodakarensis KOD1 were prepared as follows. Cells cultivated in ASW-YT-S0 medium for 17 h were harvested by centrifugation (5,000 × g, 15 min, 4°C) and were lysed in 50 mM Tris-HCl (pH 7.5) containing 0.1% Triton X-100 at a volume of 1/500 of the culture. After mixing with a vortex for 30 min, the supernatant after centrifugation (20,000 × g, 30 min, 4°C) was used as the CFE. Examination of 3-PGA synthesis with the CFE was performed in a mixture (100 μl) containing 100 mM Bicine-NaOH (pH 8.3), 10 mM MgCl2, the CFE (corresponding to 100 μg protein), 20 mM sodium phosphate (pH 7.5), 100 mM NaHCO3, and 20 mM NMP. When necessary, 20 mM dAMP (to activate R15P isomerase) and/or 20 mM 2-carboxyarabinitol 1,5-bisphosphate (CABP) (to inhibit Rubisco) was also added to the reaction mixture. After preincubation at 85°C for 3 min, the reaction was initiated by the addition of NaHCO3 and an NMP. The reaction was carried out at 85°C for 30 min and was terminated by rapid cooling on ice for 5 min, and proteins were removed with an Amicon Ultra centrifugal filter unit (MWCO, 3,000). The 3-PGA generated in this reaction was quantified by a second coupling reaction, described above.
Western blot analysis.
Cell extracts from T. kodakarensis, grown in ASW-YT-Pyr or ASW-YT-Pyr-Nuc, were prepared as described above, and proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12.5% acrylamide) and were electroblotted onto a Hybond-P membrane (GE Healthcare). After blocking with blocking reagents (GE Healthcare), membranes were hybridized with a rabbit antiserum containing polyclonal anti-AMPpase, anti-R15P isomerase, or anti-Rubisco antibodies, washed, hybridized with horseradish peroxidase (HRP)-conjugated recombinant protein G (dilution, 1:100,000; Zymed Laboratories, South San Francisco, CA), and washed again. For signal detection, the ECL Advance Western blotting detection system (GE Healthcare) and a LumiVision PRO 400EX image analyzer (Aisin, Kariya, Japan) were used.
RESULTS
Identification of compounds that activate ribose-1,5-bisphosphate isomerase.
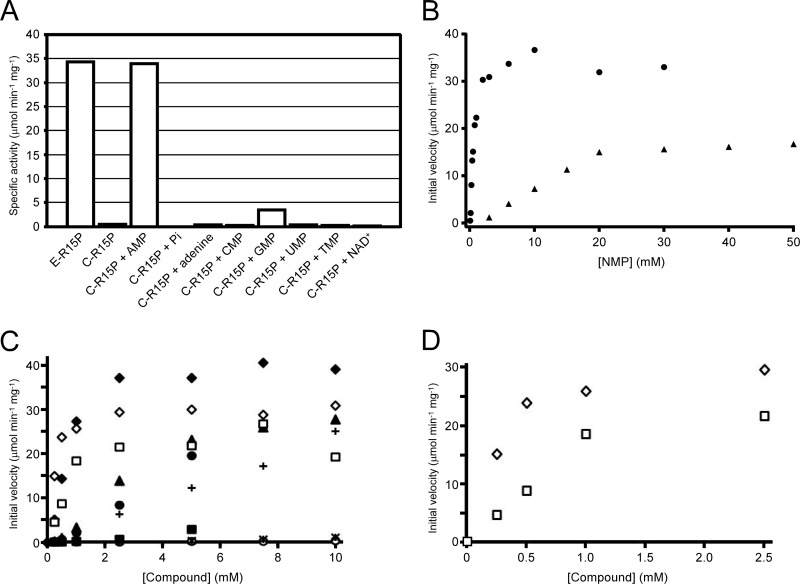
In previous studies, the substrate (R15P) used in measuring R15P isomerase activity was prepared enzymatically from AMP and sodium phosphate using AMPpase (18, 25). R15P isomerase displayed a specific activity of about 35 μmol min−1 mg−1, a level similar to those observed in the previous studies, when the AMPpase reaction mixture, which includes R15P as well as AMP, adenine, and phosphate (referred to here as E-R15P), was used as the substrate (Fig. 1A). In order to remove the effects of the other components in the enzymatically prepared substrate, we used chemically synthesized R15P (C-R15P) as the substrate here. Intriguingly, we could observe only very low levels of R15P isomerase activity by using C-R15P. This suggested that R15P isomerase was activated by a component(s) present in the AMPpase reaction mixture. R15P isomerase activity with C-R15P was thus measured in the presence of individual components of the AMPpase reaction mixture: AMP, sodium phosphate, or adenine. In the presence of AMP, R15P isomerase displayed activity with C-R15P at a level comparable to that observed with E-R15P. When R15P was excluded from the reaction mixture in the presence of AMP, RuBP synthesis was not observed. Furthermore, HPLC analyses of the R15P isomerase reaction with AMP revealed a time-dependent decrease in the R15P level, whereas consumption of AMP was not detected (see Fig. S1 in the supplemental material). These results indicated that AMP itself was not a substrate of R15P isomerase but an activator. On the other hand, the addition of sodium phosphate or adenine had no effect on R15P isomerase activity. We further examined the possibility that AMP activated Rubisco, which was utilized as a coupling enzyme in the R15P isomerase activity measurements, consequently enhancing the conversion of R15P to 3-PGA. We found that the addition of AMP had no effect on Rubisco activity (data not shown). These results clearly indicate that AMP dramatically activates R15P isomerase, with an increase of >40-fold in activity levels. All further analyses using R15P were performed with chemically synthesized R15P.
Fig 1.
Effects of various compounds on the activity of R15P isomerase. (A) R15P isomerase activity was measured by the enzyme coupling method with enzymatically prepared or chemically synthesized R15P (E-R15P or C-R15P, respectively). With C-R15P, the effects of the following compounds were examined: 3 mM AMP, 18 mM sodium phosphate (Pi), 0.4 mM adenine, 3 mM CMP, 3 mM GMP, 3 mM UMP, 3 mM TMP, or 0.4 mM NAD+. (B) Activation of R15P isomerase in the presence of varying concentrations of AMP or GMP. The initial velocities of R15P isomerase measured with coupling enzymes in the presence of AMP (circles) or GMP (triangles) are shown. (C) Activation of R15P isomerase in the presence of various compounds with an adenosyl moiety. The initial velocities of R15P isomerase were measured with coupling enzymes at varying concentrations of the following compounds: AMP (filled diamonds), dAMP (filled triangles), methylthioadenosine (filled circles), adenosine (plus signs), S-adenosylhomocysteine (filled squares), S-adenosylmethionine (asterisks), or ATP (open circles). AMP (open diamonds) and ADP (open squares) data are the results of measurements by HPLC. (D) Results of AMP and ADP measurements by HPLC in panel C, up to 2.5 mM. In all experiments, C-R15P was used at a concentration of 5 mM.
Considering the effects of AMP, we next examined the effects of other nucleotides on R15P isomerase activity. Among CMP, GMP, UMP, TMP, and NAD+, we observed a slight increase in R15P isomerase activity in the presence of GMP. In order to compare the extents of activation brought about by AMP and GMP, R15P isomerase activity was measured in the presence of various concentrations of AMP or GMP. As shown in Fig. 1B, higher degrees of activation were observed for AMP than for GMP at lower concentrations, suggesting that AMP is the major activator of R15P isomerase in vivo.
We further explored the possibilities of other compounds acting as activators of R15P isomerase. Various compounds were added at a concentration of 3 mM, and in order to expand the range of compounds that we could examine, we also performed activity measurements with HPLC when necessary. This allowed us to analyze the effects of compounds such as ADP and 3-PGA, which would be difficult with the enzyme coupling assays. The enzyme coupling assay and the HPLC analysis revealed that the addition of 3 mM ADP resulted in increases in R15P isomerase activity comparable to those observed with AMP. Other compounds with an adenosyl moiety also exhibited activating effects, but at higher concentrations. On the other hand, R15P isomerase was not activated by R5P, PRPP, FBP, F6P, glucose, pyruvate, or 3-PGA. These results suggested a tendency for compounds with an adenosyl moiety to activate R15P isomerase. To compare the extents of activation, R15P isomerase activity was measured in the presence of varying concentrations of these compounds. As shown in Fig. 1C, the effects of SAM, SAH, and ATP were relatively small, while significant activation was detected in the presence of dAMP, adenosine, or MTA. However, in terms of the degree of activation and the concentration range within which activation was observed, AMP had the most dramatic effect. At low concentrations of 250 μM, which may better represent physiological concentrations, activity with AMP was more than 3-fold higher than that observed with ADP. The dissociation constant values of the activators calculated from the HPLC measurements for AMP and ADP (Fig. 1D) were 217 μM (KAMP) and 667 μM (KADP). The effects of other compounds at this concentration were negligible.
Kinetic analysis and substrate specificity of ribose-1,5-bisphosphate isomerase.
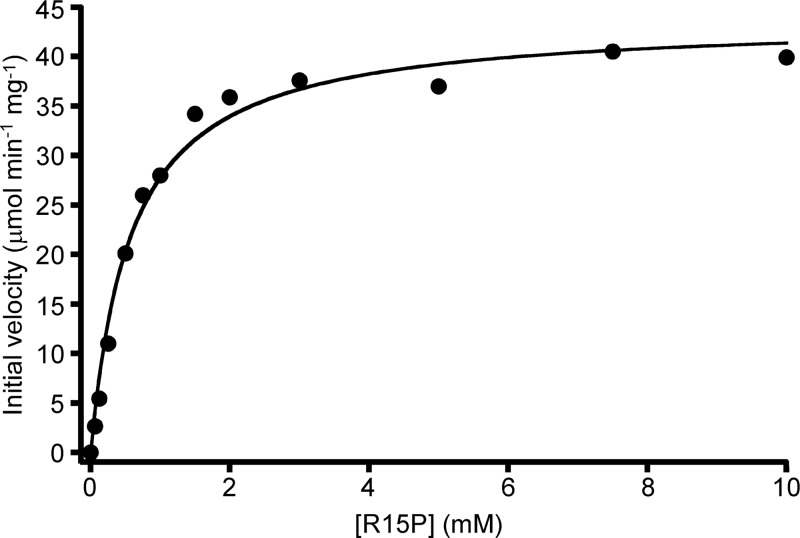
Kinetic analysis of R15P isomerase was carried out in the presence of 10 mM AMP by using the enzyme coupling method (Fig. 2). The isomerase reaction from R15P to RuBP followed Michaelis-Menten kinetics, with a Km of 0.6 ± 0.1 mM for R15P and a kcat of 29.2 ± 0.7 s−1 at 85°C (Table 1).
Fig 2.
Kinetic analysis of R15P isomerase. Initial velocities of R15P isomerase were measured with coupling enzymes in the presence of varying concentrations of C-R15P. AMP (10 mM) was present for all measurements.
Table 1.
Kinetic parameters of R15P isomerase and AMPpasea
| Enzyme | Substrate | Kinetic parameter |
||
|---|---|---|---|---|
| Km (mM) | kcat (s−1) | kcat/Km (s−1 mM−1) | ||
| R15P isomerase | R15P | 0.6 ± 0.1 | 29.2 ± 0.7 | 48.7 |
| AMPpase | CMP | 6.2 ± 0.5 | 39.1 ± 1.2 | 6.3 |
| UMP | 4.4 ± 0.5 | 10.5 ± 0.4 | 2.4 | |
| GMP | 18.5 ± 1.3 | 2.7 ± 0.1 | 0.1 | |
| Pi | 2.8 ± 0.1 | 15.0 ± 0.2 | 5.4 | |
The kinetic parameters of the R15P isomerase reaction were examined in the presence of 10 mM AMP. The kinetic parameters of the AMPpase reaction toward NMPs and Pi were investigated in the presence of 20 mM Pi and 20 mM AMP, respectively. The kinetic parameters of the AMPpase reaction toward AMP and dCMP were not determined, because kinetic equations that fit our data well could not be identified.
We next examined the substrate specificity of R15P isomerase. Among the two anomers of R15P, we observed previously that the enzyme utilizes only the α-anomer compound (18). Here we investigated whether R15P isomerase can recognize other phosphorylated sugars. Activities toward the following substrates were investigated by HPLC: R5P, ribose, G16P, G6P, G1P, FBP, F6P, and PRPP. We did not observe any decrease in the levels of substrates or synthesis of products dependent on R15P isomerase with any of the compounds except PRPP. Although a decrease in PRPP levels and an increase in RuBP levels were observed, the decrease in PRPP levels was not dependent on the R15P isomerase enzyme. We presume that PRPP displayed thermal degradation during the incubation at 85°C, resulting in the generation of R15P, the substrate of R15P isomerase. These results indicated that among the compounds examined, R15P isomerase can utilize only α-R15P, implying that the substrate specificity of this enzyme is strict.
Substrate specificity of AMP phosphorylase.
AMPpase was previously reported to catalyze an AMP-specific phosphorylase reaction generating R15P and adenine. Relevant levels of phosphorylase activity could not be observed with other nucleoside monophosphates, such as UMP, GMP, CMP, and TMP (25). However, because the activity measurements were performed with coupling enzymes that included R15P isomerase, we realized that phosphorylase activities toward nucleoside monophosphates other than AMP might have been overlooked or underestimated due to the low activity levels of R15P isomerase in the absence of AMP. Therefore, we reevaluated the substrate specificity of AMPpase by using the coupling enzymes, but under the condition that R15P isomerase was activated with saturating concentrations of AMP. As a result, we found that AMPpase exhibited phosphorylase activity not only toward AMP but also toward CMP and UMP, while only low phosphorylase activity was observed with GMP and IMP (Table 2).
Table 2.
Substrate specificity of AMPpase toward NMPs and dNMPs
| Substrate | Phosphorylase activity (μmol min−1 mg−1)a determined by the following method: |
|
|---|---|---|
| Coupling enzymes | HPLC | |
| AMP | 15.9 ± 0.7 | — |
| CMP | 37.5 ± 1.4 | 35.2 ± 0.7 |
| UMP | 10.8 ± 0.5 | — |
| GMP | 1.8 ± 0.2 | — |
| IMP | 0.3 ± 0.1 | — |
| dAMP | — | 1.5 ± 0.02 |
| dCMP | — | 15.7 ± 0.5 |
| dUMP | — | 0.4 ± 0.02 |
| dGMP | — | 0.4 ± 0.03 |
Activities were investigated in the presence of 20 mM NMP and 20 mM Pi. —, not performed.
In order to investigate whether AMPpase could catalyze the phosphorolysis of other compounds related to nucleotides/nucleosides, activity measurements with HPLC were performed on the following compounds: dAMP, dCMP, dGMP, dUMP, TMP, adenosine, cytidine, uridine, ADP, ATP, MTA, SAM, SAH, PRPP, and R5P. We observed significant levels of cytosine released from dCMP during the reaction. On the other hand, the use of dAMP, dGMP, or dUMP resulted in the generation of only trace amounts of their corresponding nucleobases, adenine, guanine, or uracil, respectively (Table 2). No nucleobase synthesis and no substrate consumption were observed when dCMP, dAMP, dGMP, and dUMP were incubated in the absence of the enzyme or phosphate, indicating that the nucleobases were not the product of thermal degradation but were dependent on the enzyme activity of AMPpase. We did not detect enzyme activities toward TMP, adenosine, cytidine, uridine, ADP, ATP, MTA, SAM, SAH, PRPP, or R5P.
Kinetic analysis of AMP phosphorylase.
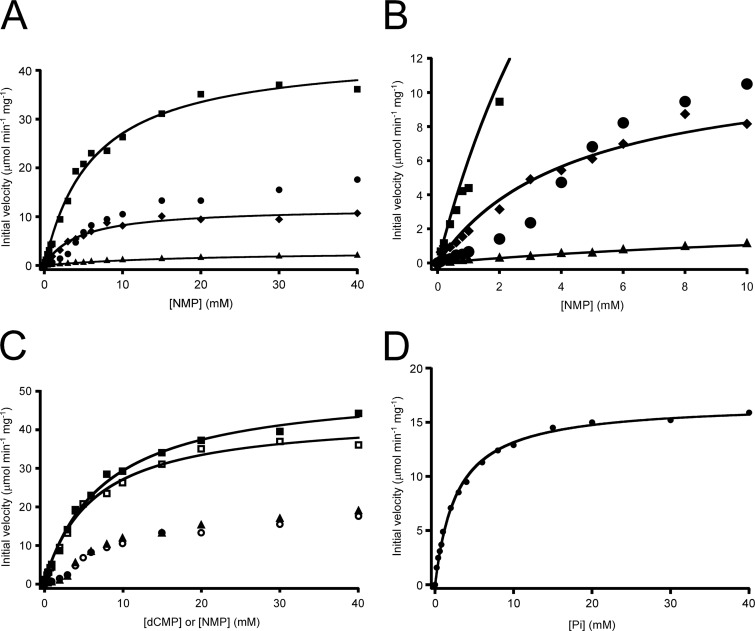
Kinetic analyses of the activities of AMPpase toward AMP, CMP, GMP, and UMP were carried out (Fig. 3A). Toward CMP, GMP, and UMP, the plots of velocity versus substrate concentration ([S]-v plots) followed Michaelis-Menten kinetics with the kinetic parameters shown in Table 1. Although activity toward GMP was relatively low, activities with CMP and UMP were as high as or higher than that observed with AMP. These results confirmed that AMPpase could convert not only AMP but also CMP and UMP. It should be noted that the [S]-v plot with AMP as the substrate displayed a sigmoidal curve, indicating regulation of AMPpase by AMP (Fig. 3B). A number of kinetic models, including cooperativity, were considered, but no equation that fit our data well could be identified. Kinetic analysis of AMPpase activity toward dCMP was also carried out by HPLC (Fig. 3C). The [S]-v plot was very similar to that obtained with AMP, suggesting a similar mode of regulation. In order to confirm that the activity levels obtained by measurements with HPLC and coupling enzymes could be accurately compared with one another, we performed a kinetic analysis on CMP again, using HPLC. No large differences were observed between the two procedures (Fig. 3C).
Fig 3.
Kinetic analyses of AMPpase. (A) Kinetic analysis of AMPpase activities toward NMPs. Initial velocities of AMPpase were measured with coupling enzymes in the presence of varying concentrations of AMP (circles), CMP (squares), GMP (triangles), or UMP (diamonds). The concentration of Pi was constant at 20 mM. (B) Enlarged view of the results shown in panel A. (C) Kinetic analysis of AMPpase activity toward dCMP. Initial velocities of AMPpase were measured in the presence of varying concentrations of dCMP (triangles), AMP (circles), or CMP (squares). The results for CMP were obtained by both the enzyme coupling method (open symbols) and HPLC (filled symbols). The concentration of Pi was constant at 20 mM. (D) Dependence of AMPpase activity on Pi concentrations. Initial velocities were measured in the presence of varying concentrations of Pi and 20 mM AMP.
Kinetic analysis of AMPpase toward its other substrate, Pi (Fig. 3D), revealed that this phosphorylase reaction followed Michaelis-Menten kinetics, with a Km of 2.8 ± 0.1 mM and a kcat of 15.0 ± 0.2 s−1 at 85°C (Table 1).
Conversion of nucleoside monophosphates to 3-phosphoglycerate in cell extracts.
We indicated previously that the recombinant AMPpase, R15P isomerase, and Rubisco could catalyze sequential reactions converting AMP to 3-PGA in vitro (25). In order to gain insight on whether this was also the case in vivo, we investigated whether NMPs were converted to 3-PGA in cell extracts (CFE). When AMP was used as the substrate, we clearly observed the generation of 3-PGA in the CFE (Table 3). We could not observe 3-PGA synthesis when CMP or UMP was added alone. We presumed that this was due to the fact that R15P isomerase was not activated. We then added dAMP to the reaction mixture, since this compound activates R15P isomerase but does not serve as a substrate to generate 3-PGA (Table 3). With the addition of dAMP, we detected significant levels of 3-PGA synthesis from CMP and UMP. Since the amounts of 3-PGA generated with CMP and UMP were lower than that observed for AMP, we investigated whether activation of R15P isomerase by dAMP was not as effective as that by AMP. We added R15P directly to the CFE and measured 3-PGA synthesis in the presence or absence of dAMP. The addition of dAMP led to a dramatic increase in 3-PGA formation, from 12.1 to 202 μM. The fact that the addition of dAMP enhanced 3-PGA generation strongly suggests that the conversion of AMP, CMP, and UMP to 3-PGA is brought about by the three enzymes AMPpase, R15P isomerase, and Rubisco. In order to gain further support, CABP, an inhibitor of Rubisco, was added to the reaction mixture. In the presence of CABP, the significant levels of 3-PGA observed with AMP, CMP, and UMP in the experiments described above could not be detected (Table 3), providing further indications that AMP, CMP, and UMP are converted to 3-PGA via the archaeal AMP metabolic pathway in T. kodakarensis.
Table 3.
3-Phosphoglycerate formation from NMPs in T. kodakarensis cell extracts
| Substratea | Additional componenta |
3-PGA concn (μM)b | |
|---|---|---|---|
| dAMP | CABP | ||
| − | − | − | <10 |
| − | + | − | <10 |
| − | − | + | <10 |
| RuBP | − | − | 1,240 |
| − | + | 15.1 | |
| AMP | − | − | 94.3 |
| − | + | <10 | |
| CMP | − | − | <10 |
| + | − | 42.4 | |
| + | + | <10 | |
| UMP | − | − | <10 |
| + | − | 14.4 | |
| + | + | <10 | |
| R15P | − | − | 12.1 |
| − | + | <10 | |
| + | − | 202 | |
The substrate, dAMP, and CABP were each used at a concentration of 20 mM. −, absence; +, presence.
Detected after a 30-min reaction with the substrates and the additional components along with 100 mM NaHCO3 and 20 mM Pi in the cell extract (100 μg protein).
Protein levels of AMPpase, R15P isomerase, and Rubisco.
To investigate whether the three enzymes AMPpase, R15P isomerase, and Rubisco are subject to regulation at the transcriptional/translational level in addition to their responses to AMP at the activity level, we raised polyclonal antibodies against each protein and performed Western blot analyses. Since we had observed previously that nucleosides are assimilated well in T. kodakarensis (19), we grew the cells in ASW-YT-Pyr medium with or without a mixture of nucleosides (Fig. 4). We observed increases in the protein levels of R15P isomerase and Rubisco in the cells cultivated with nucleosides. In contrast, protein levels of AMPpase were more or less equivalent in cells grown in the presence or absence of nucleosides. This result suggests that the archaeal AMP metabolic pathway, in addition to its response at the activity level to AMP, also responds to nucleosides or related metabolites at the transcriptional and/or translational level.
Fig 4.

Protein levels of AMPpase, R15P isomerase, and Rubisco in T. kodakarensis cells. Western blot analyses using an antiserum containing polyclonal anti-AMPpase, anti-R15P isomerase, or anti-Rubisco antibodies were performed against extracts of T. kodakarensis cells grown in ASW-YT-Pyr or ASW-YT-Pyr-Nuc medium.
DISCUSSION
In this study, we performed the first detailed biochemical characterization of two novel enzymes functioning in an archaeal AMP metabolic pathway, AMPpase and R15P isomerase. Examination of the substrate specificity of AMPpase indicated that this enzyme utilized not only AMP but also CMP, UMP, and dCMP. However, since intracellular concentrations of dNMPs are in general much lower than those of NMPs (32), it is likely that the major substrates of AMPpase in T. kodakarensis cells are the ribonucleotides AMP, CMP, and UMP. We found that 3-PGA was synthesized from AMP, CMP, and UMP in T. kodakarensis cell extracts, suggesting that the three enzymes AMPpase, R15P isomerase, and Rubisco most likely function as a metabolic pathway in vivo. Kinetic analysis of AMPpase activity toward CMP, UMP, and Pi revealed that the [S]-v plots for these substrates followed Michaelis-Menten kinetics. In contrast, the [S]-v plot for AMP displayed a sigmoidal curve, indicating that this enzyme is regulated by AMP. This behavior toward AMP may act as a regulation mechanism to prevent excess degradation of AMP in T. kodakarensis cells. There may also be an unidentified mechanism to regulate the degradation of CMP and UMP, a hypothesis supported by the observation that the levels of 3-PGA synthesized from CMP and UMP in CFE were lower than that from AMP. We cannot rule out the possibility that CMP and UMP are rapidly consumed in other pathways present in CFE, but we think this unlikely, since we added the substrates at high concentrations. On the other hand, the equilibrium constant of the AMPpase reaction ([R15P] [adenine]/[AMP] [Pi]) has been determined previously as 6.02 × 10−3 ± 0.46 × 10−3 (25), indicating that the reaction, from a thermodynamic point of view, favors AMP synthesis. This is also the case for other nucleoside phosphorylases (4, 13, 14, 20). Since the reaction toward other NMPs can also be expected to favor NMP synthesis, AMPpase may be involved in nucleotide interconversion between NMPs via R15P and nucleobases, regulating the intracellular NMP ratio when the in vivo concentration of AMP is low and R15P is not actively consumed by R15P isomerase. Although nucleoside interconversion has been found in Bacteria and Eucarya (10, 31), nucleotide interconversion has not been reported until now, and this may represent a novel regulation mechanism in nucleotide metabolism. The fact that the protein levels of AMPpase were constitutive and did not respond to the addition of nucleosides in the medium may be related to this mechanism.
Through enzymatic characterization of R15P isomerase, we clarified that, among various compounds we used, R15P isomerase was activated more than 40-fold in the presence of 1 mM AMP. This property of R15P isomerase suggests that the flux of the AMP metabolic pathway responds to the intracellular concentrations of AMP. The regulation of R15P isomerase activity responding to AMP basically answers our question as to how excess degradation of intracellular AMP by the AMP degradation pathway is prevented. The recently reported crystal structure of R15P isomerase did not include AMP (18). However, this enzyme form without AMP displays activity, although it is low, as shown in this study and by the fact that cocrystallization of the wild-type enzyme with the substrate (R15P) led to a protein structure bound to the product (RuBP). Further studies will be necessary to determine how AMP binding affects enzyme structure and activity. MTR1P isomerase, a protein homologous to R15P isomerase, displays a 3-dimensional structure resembling the dimer unit of R15P isomerase (5, 18, 29). This enzyme has not been examined for activation by metabolites (22), and the question of whether this enzyme is also regulated similarly or not is of interest.
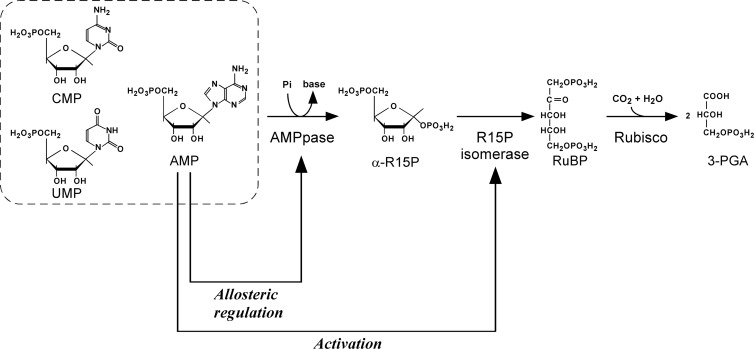
Based on the novel insight obtained in this study, our present understanding of the archaeal AMP metabolic pathway and its regulation is illustrated in Fig. 5. AMP phosphorylase and R15P isomerase catalyze the reversible reactions between AMP (or CMP or UMP) and RuBP. The equilibrium of the AMP phosphorylase reaction is greatly skewed toward nucleoside monophosphate formation, but the irreversible reaction catalyzed by Rubisco results in the unidirectional formation of 3-PGA by this pathway. Activation of R15P isomerase by AMP stimulates the conversion between AMP and RuBP, and this activation, together with Rubisco, promotes 3-PGA formation. A decrease in AMP concentration reduces the activity levels of R15P isomerase, thus preventing excess degradation of AMP. Low levels of AMP would also prevent the breakdown of CMP or UMP via the regulation of R15P isomerase. We have shown previously that T. kodakarensis can assimilate exogenous nucleosides (19). In this study, we have shown that R15P isomerase and Rubisco are upregulated by the addition of nucleosides. Therefore, the pathway may be involved in this nucleoside assimilation/degradation by directing the ribose moieties of nucleosides to central carbon metabolism (3-PGA) via AMP, CMP, or UMP. This is possible either through conventional routes that involve nucleoside phosphorylase, phosphopentomutase, PRPP synthetase, and nucleobase phosphoribosyltransferase or through a direct conversion via nucleoside kinases. The pathway may also be involved in metabolic conversions other than exogenous nucleoside/nucleotide breakdown, and the presence of unidentified metabolic pathways that are linked to these three enzymes via the nucleoside monophosphates should be examined.
Fig 5.
Substrate specificities and regulatory properties of AMPpase, R15P isomerase, and Rubisco in T. kodakarensis.
ACKNOWLEDGMENTS
This work was partially supported by the Japan Science and Technology Agency (to H.A., T.I., and K.M.), the Ministry of Education, Culture, Sports, Science, and Technology (Targeted Proteins Research Program; to K.M., T.I., and H.A.), and the Japan Society for the Promotion of Science (KAKENHI 22750150; to T.S.).
Footnotes
Published ahead of print 12 October 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Alonso H, Blayney MJ, Beck JL, Whitney SM. 2009. Substrate-induced assembly of Methanococcoides burtonii d-ribulose-1,5-bisphosphate carboxylase/oxygenase dimers into decamers. J. Biol. Chem. 284:33876–33882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ashida H, et al. 2003. A functional link between RuBisCO-like protein of Bacillus and photosynthetic RuBisCO. Science 302:286–290 [DOI] [PubMed] [Google Scholar]
- 3. Atomi H, Fukui T, Kanai T, Morikawa M, Imanaka T. 2004. Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea 1:263–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bose R, Yamada EW. 1974. Uridine phosphorylase, molecular properties and mechanism of catalysis. Biochemistry 13:2051–2056 [DOI] [PubMed] [Google Scholar]
- 5. Bumann M, et al. 2004. Crystal structure of yeast Ypr118w, a methylthioribose-1-phosphate isomerase related to regulatory eIF2B subunits. J. Biol. Chem. 279:37087–37094 [DOI] [PubMed] [Google Scholar]
- 6. Desgranges C, Razaka G, Rabaud M, Bricaud H. 1981. Catabolism of thymidine in human blood platelets: purification and properties of thymidine phosphorylase. Biochim. Biophys. Acta 654:211–218 [DOI] [PubMed] [Google Scholar]
- 7. Ezaki S, Maeda N, Kishimoto T, Atomi H, Imanaka T. 1999. Presence of a structurally novel type ribulose-bisphosphate carboxylase/oxygenase in the hyperthermophilic archaeon, Pyrococcus kodakaraensis KOD1. J. Biol. Chem. 274:5078–5082 [DOI] [PubMed] [Google Scholar]
- 8. Finn MW, Tabita FR. 2003. Synthesis of catalytically active form III ribulose 1,5-bisphosphate carboxylase/oxygenase in archaea. J. Bacteriol. 185:3049–3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fukui T, et al. 2005. Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 15:352–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Giorgelli F, et al. 1997. Recycling of α-d-ribose 1-phosphate for nucleoside interconversion. Biochim. Biophys. Acta 1335:16–22 [PubMed] [Google Scholar]
- 11. Grochowski LL, Xu H, White RH. 2005. Ribose-5-phosphate biosynthesis in Methanocaldococcus jannaschii occurs in the absence of a pentose-phosphate pathway. J. Bacteriol. 187:7382–7389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hamamoto T, Noguchi T, Midorikawa Y. 1996. Purification and characterization of purine nucleoside phosphorylase and pyrimidine nucleoside phosphorylase from Bacillus stearothermophilus TH 6-2. Biosci. Biotechnol. Biochem. 60:1179–1180 [DOI] [PubMed] [Google Scholar]
- 13. Heppel LA, Hilmoe RJ. 1952. Phosphorolysis and hydrolysis of purine ribosides by enzymes from yeast. J. Biol. Chem. 198:683–694 [PubMed] [Google Scholar]
- 14. Jensen KF, Nygaard P. 1975. Purine nucleoside phosphorylase from Escherichia coli and Salmonella typhimurium. Purification and some properties. Eur. J. Biochem. 51:253–265 [DOI] [PubMed] [Google Scholar]
- 15. Kitano K, et al. 2001. Crystal structure of a novel-type archaeal Rubisco with pentagonal symmetry. Structure 9:473–481 [DOI] [PubMed] [Google Scholar]
- 16. Kreel NE, Tabita FR. 2007. Substitutions at methionine 295 of Archaeoglobus fulgidus ribulose-1,5-bisphosphate carboxylase/oxygenase affect oxygen binding and CO2/O2 specificity. J. Biol. Chem. 282:1341–1351 [DOI] [PubMed] [Google Scholar]
- 17. Maeda N, Kanai T, Atomi H, Imanaka T. 2002. The unique pentagonal structure of an archaeal Rubisco is essential for its high thermostability. J. Biol. Chem. 277:31656–31662 [DOI] [PubMed] [Google Scholar]
- 18. Nakamura A, et al. 2012. Dynamic, ligand-dependent conformational change triggers reaction of ribose-1,5-bisphosphate isomerase from Thermococcus kodakarensis KOD1. J. Biol. Chem. 287:20784–20796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Orita I, et al. 2006. The ribulose monophosphate pathway substitutes for the missing pentose phosphate pathway in the archaeon Thermococcus kodakaraensis. J. Bacteriol. 188:4698–4704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Panova NG, et al. 2007. Substrate specificity of Escherichia coli thymidine phosphorylase. Biokhimiia 72:21–28 [DOI] [PubMed] [Google Scholar]
- 21. Robb FT, Place AR. 1995. Media for thermophiles, p 167–168 In Robb FT, Place AR. (ed), Archaea: a laboratory manual—thermophiles. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 22. Saito Y, et al. 2007. Enzymatic characterization of 5-methylthioribose 1-phosphate isomerase from Bacillus subtilis. Biosci. Biotechnol. Biochem. 71:2021–2028 [DOI] [PubMed] [Google Scholar]
- 23. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 24. Sato T, Atomi H. 2011. Novel metabolic pathways in Archaea. Curr. Opin. Microbiol. 14:307–314 [DOI] [PubMed] [Google Scholar]
- 25. Sato T, Atomi H, Imanaka T. 2007. Archaeal type III RuBisCOs function in a pathway for AMP metabolism. Science 315:1003–1006 [DOI] [PubMed] [Google Scholar]
- 26. Sekowska A, Danchin A. 2002. The methionine salvage pathway in Bacillus subtilis. BMC Microbiol. 2:8 doi:10.1186/1471-2180-2-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Siebers B, Schönheit P. 2005. Unusual pathways and enzymes of central carbohydrate metabolism in Archaea. Curr. Opin. Microbiol. 8:695–705 [DOI] [PubMed] [Google Scholar]
- 28. Soderberg T. 2005. Biosynthesis of ribose-5-phosphate and erythrose-4-phosphate in archaea: a phylogenetic analysis of archaeal genomes. Archaea 1:347–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tamura H, et al. 2008. Crystal structure of 5-methylthioribose 1-phosphate isomerase product complex from Bacillus subtilis: implications for catalytic mechanism. Protein Sci. 17:126–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tomoike F, Nakagawa N, Kuramitsu S, Masui R. 2011. A single amino acid limits the substrate specificity of Thermus thermophilus uridine-cytidine kinase to cytidine. Biochemistry 50:4597–4607 [DOI] [PubMed] [Google Scholar]
- 31. Tozzi MG, Camici M, Mascia L, Sgarrella F, Ipata PL. 2006. Pentose phosphates in nucleoside interconversion and catabolism. FEBS J. 273:1089–1101 [DOI] [PubMed] [Google Scholar]
- 32. Traut TW. 1994. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 140:1–22 [DOI] [PubMed] [Google Scholar]
- 33. Verhees CH, et al. 2003. The unique features of glycolytic pathways in Archaea. Biochem. J. 375:231–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Watson GM, Yu JP, Tabita FR. 1999. Unusual ribulose 1,5-bisphosphate carboxylase/oxygenase of anoxic Archaea. J. Bacteriol. 181:1569–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zaparty M, Tjaden B, Hensel R, Siebers B. 2008. The central carbohydrate metabolism of the hyperthermophilic crenarchaeote Thermoproteus tenax: pathways and insights into their regulation. Arch. Microbiol. 190:231–245 [DOI] [PubMed] [Google Scholar]