Abstract
Archaeal histones wrap DNA into complexes, designated archaeal nucleosomes, that resemble the tetrasome core of a eukaryotic nucleosome. Therefore, all DNA interactions in vivo in Thermococcus kodakarensis, the most genetically versatile model species for archaeal research, must occur in the context of a histone-bound genome. Here we report the construction and properties of T. kodakarensis strains that have TK1413 or TK2289 deleted, the genes that encode HTkA and HTkB, respectively, the two archaeal histones present in this archaeon. All attempts to generate a strain with both TK1413 and TK2289 deleted were unsuccessful, arguing that a histone-mediated event(s) in T. kodakarensis is essential. The HTkA and HTkB amino acid sequences are 84% identical (56 of 67 residues) and 94% similar (63 of 67 residues), but despite this homology and their apparent redundancy in terms of supporting viability, the absence of HTkA and HTkB resulted in differences in growth and in quantitative and qualitative differences in genome transcription. A most surprising result was that the deletion of TK1413 (ΔhtkA) resulted in a T. kodakarensis strain that was no longer amenable to transformation, whereas the deletion of TK2289 (ΔhtkB) had no detrimental effects on transformation. Potential roles for the archaeal histones in regulating gene expression and for HTkA in DNA uptake and recombination are discussed.
INTRODUCTION
The histone fold apparently evolved before the archaeal and eukaryotic lineages diverged ∼1.5 to 1.9 billion years ago, and now almost all eukaryotes, Euryarchaea, Nanoarchaea, Thaumarchaea, and some Crenarchaea employ histone fold-based DNA binding to wrap and compact their genomic DNA (2, 9, 14, 21, 28, 30, 38). All transactions in these species that involve chromosomal DNA must therefore be considered in terms of histone-bound chromatin. The presence of histones in Archaea, but not in Bacteria, was a major distinction first recognized ∼20 years ago (26), and research since then has established the detailed structure of archaeal histones (3, 19); the composition and architecture of the archaeal nucleosome (11, 20, 22, 24, 30); and the consequences of archaeal histone binding on DNA topology, replication, and transcription in vitro (7, 37, 43, 44). Archaeal nucleosomes resemble the eukaryotic tetrasome, the structure at the center of the eukaryotic nucleosome formed by ∼90 bp of DNA wrapped around a histone (H3+H4)2 tetramer (20, 28, 30). Archaeal histones do not, however, have homologues of the N- and C-terminal amino acid extensions (21) that contain the targets for eukaryotic histone acetylation and methylation and thus provide the basis for epigenetic regulation. Consistent with this, scrutiny of archaeal genome sequences has failed to detect recognizable homologues of the eukaryotic histone modification systems, and to date, no archaeal histone modification has been described (5). It is therefore intriguing and important to determine if archaeal histones nevertheless participate in regulating genome functions. In this regard, in species with more than one archaeal histone, differences in their expression levels do correlate with differences in laboratory growth rates (4, 25), but how this regulation is achieved and its physiological consequences are unknown.
Archaeal histone binding to DNA in vitro introduces DNA topology constraints, prevents transcription initiation, and impedes transcript extension (7, 20, 43, 44). However, only with the development of genetic approaches has it become possible to investigate how these in vitro observations relate to in vivo functions. Inactivation of the single histone-encoding gene in Methanosarcina mazei was possible, but the resulting strain exhibited reduced growth, UV sensitivity, and global transcriptome changes (41). Similarly, inactivation of either of the two histone-encoding genes in the distantly related methanogen Methanococcus voltae was not lethal, but the resulting strains had proteome differences (10). Thermococcus kodakarensis is naturally competent for DNA uptake and chromosomal transformation (36), and with this advantage, T. kodakarensis has now been developed into the most genetically versatile model species for archaeal research (12). T. kodakarensis contains two very similar archaeal histones (Fig. 1) (6), now designated HTkA and HTkB (encoded by TK1413 and TK2289, respectively) and previously designated HPkA and HPkB when T. kodakarensis was classified as Pyrococcus kodakaraensis (11). To add to the database and provide additional research tools, we have constructed and determined the phenotypes of T. kodakarensis strains with TK1413 or TK2289 precisely deleted. Apparently, at least one archaeal histone is required for viability, as we were unable to construct a strain with both TK1413 and TK2289 deleted. As described below, based on differences in growth phenotypes and the results of microarray hybridization experiments, the absence of each histone results in substantial quantitative and qualitative differences in genome expression. An unanticipated and surprising discovery is that the deletion of TK1413 (htkA), but not TK2289 (htkB), resulted in a T. kodakarensis strain that is no longer naturally amenable to DNA transformation.
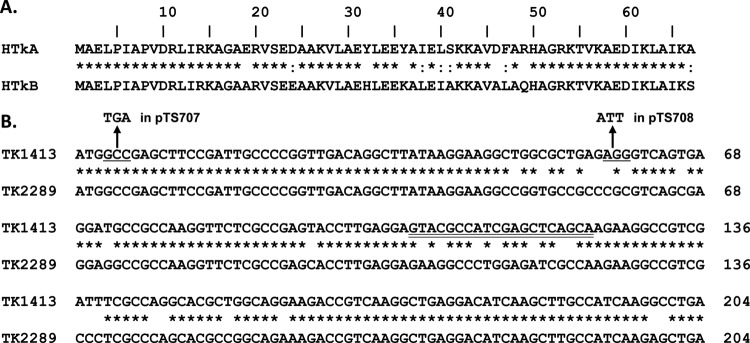
Fig 1.
Alignments of the T. kodakarensis histone sequences (6) using clustalW. Shown are alignments of the amino acid (A) and the encoding nucleic acid (B) sequences of HTkA (TK1413) and HTkB (TK2289). Between the sequences, identical amino acids and nucleotides are indicated by asterisks, and similar amino acids are indicated by colons. The codon changes introduced into TK1413 in plasmid pTS706 to generate plasmids pTS707 and pTS708 are indicated by underlining. An oligonucleotide with the sequence double underlined in TK1413 was synthesized, labeled with 32P, and used as the probe for Southern blotting (Fig. 2E).
MATERIALS AND METHODS
Plasmid and T. kodakarensis strain constructions.
The T. kodakarensis strains and plasmids used in this study are listed in Table 1. Standard molecular biology procedures were used for PCR amplifications, plasmid constructions, and Escherichia coli DH5α transformation and to select transformants and isolate plasmid preparations from E. coli. PCR amplicons generated from T. kodakarensis genomic DNA (primer sequences are available in Table S1 in the supplemental material) were cloned into plasmid pTS535 or pTS414, as previously described (32, 33).
Table 1.
T. kodakarensis strains and plasmids
| Strain or plasmid | Genotype or relevant feature | Reference |
|---|---|---|
| Strains | ||
| TS517 | ΔpyrF ΔtrpE::pyrF ΔTK0664 | 33 |
| LC124 | ΔpyrF ΔtrpE::pyrF ΔTK0664 ΔTK1413 | This study |
| LC125 | ΔpyrF ΔtrpE::pyrF ΔTK0664 ΔTK2289 | This study |
| Plasmids | ||
| pTS535 | pUC118 with MCS1-(PTK2279-TK0254 PhmtB-TK0664)-MCS2 | 33 |
| pLC113 | pTS535::TK1412-TK1413-TK0664-TK0254-TK1412-TK1414-TK1415 | This study |
| pLC114 | pTS535::TK2286-TK2287-TK2288-TK2289-TK0664-TK0254-TK2286-TK2287-TK2288-TK2290 | This study |
| pLC124 | pTS535::TK1412-TK0664-TK0254-TK1412-TK1414-TK1415 | This study |
| pLC125 | pTS535::TK2286-TK2287-TK2288-TK0664-TK0254-TK2286-TK2287-TK2288-TK2290 | This study |
| pUDHisD | pUC118 derivative; ΔhisD::trpE | 36 |
| pTS503 | pUC118::PhmtB-TK1827-trpE-TK1828 with TAG at codon 5 of TK1827 | 33 |
| pLC70 | PTK2279-TK0254 Pgdh-PF1848 | 32 |
| pLC71 | pLC70 Δp24 | 32 |
| pTS414 | pLC70::PhmtB-rpoL-HA | 32 |
| pTS706 | pLC70::PhmtB-TK1413 | This study |
| pTS707 | pLC70::PhmtB-TK1413 with TGA at codon 2 of TK1413 (A2STOP) | This study |
| pTS708 | pLC70::PhmtB-TK1413 with ATT at codon 20 of TK1413 (R20I) | This study |
Plasmids incapable of autonomous replication in T. kodakarensis were constructed and introduced into T. kodakarensis TS517 (ΔpyrF ΔtrpE::pyrF ΔTK0664) (32). The recombination of the plasmid into the chromosome resulted in transformants that were capable of growth on plates containing artificial seawater (ASW) medium supplemented with vitamins, elemental sulfur (S0), and 19 amino acids but lacking tryptophan (ASW+S0+AA−trp medium). Transformants (intermediate strains) (Fig. 2 and 3) in which two homologous recombinations had replaced the chromosomal DNA with the desired plasmid DNA, including the TK0254 (trpE) plus TK0664 (TK0254-TK0664) cassette, were identified via diagnostic PCRs. The expression of TK0664 resulted in sensitivity to 6-methylpurine (6MP). Dilutions of the intermediate strains, grown in medium containing ASW-yeast extract-tryptone plus sulfur (ASW-YT-S0 medium) were plated onto ASW+S0+AA medium containing 6MP. Spontaneously 6MP-resistant (6MPr) mutants were screened for the precise markerless deletion of TK1413 or TK2289. Two such 6MPr isolates were designated T. kodakarensis LC124 and LC125 (Table 1) and used in subsequent studies.
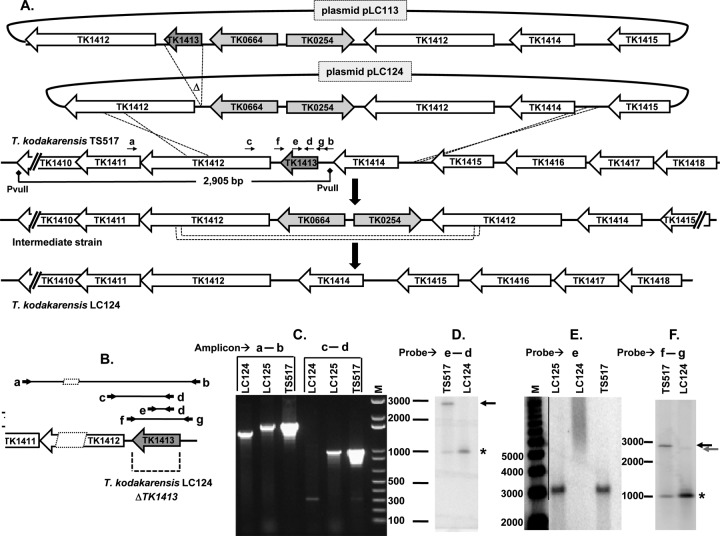
Fig 2.
Construction and confirmation of T. kodakarensis LC124 (ΔTK1413). (A) Plasmid pLC113 was generated from pTS535 (Table 1) by cloning amplicons from T. kodakarensis genomic DNA upstream and downstream of the TK0254-TK0664 cassette. Plasmid pLC124, generated by deletion of TK1413 from pLC113, was used as the donor DNA to transform T. kodakarensis TS517. The genome structure of the intermediate strain generated was confirmed, as illustrated, and recombination between the duplicated sequences deleted the cassette to yield T. kodakarensis LC124 with the genome as shown. The positions of primers (primers a to g [see Table S1 in the supplemental material for sequences]) used to generate diagnostic amplicons (C) and probes for Southern blotting (D to F) are indicated by arrows above the T. kodakarensis TS517 genome. The 2,905-bp PvuII restriction fragment to which the probes hybridized is indicated. (B) Expanded illustration of the primer locations (heavy arrows) and the amplicons generated. (C) Ethidium bromide-stained agarose gel for electrophoretic separation of the amplicons generated using the primer pairs a-b and c-d from T. kodakarensis LC124, LC125, and TS517 genomic DNAs. (D to F) Southern blots of PvuII-digested T. kodakarensis TS517, LC124, and LC125 DNAs probed with DIG-labeled amplicons (primer pairs c-d and f-g) or 32P-labeled oligonucleotide primer e. In panel D, the 2,905-bp PvuII fragment that contains TK1413 in T. kodakarensis TS517 is indicated by an arrow. This fragment is not present in T. kodakarensis LC124. The PvuII fragment present in both genomic DNAs which contains TK2289 and thus cross-hybridizes with the probe is noted by an asterisk. In panel F, as the probe includes sequences immediately adjacent to TK1413, it hybridized to the PvuII fragment in T. kodakarensis LC124 DNA that contains the ΔTK1413 deletion (gray arrow). As the probe contains the entire TK1413 sequence, it cross-hybridized to the ∼1-kbp PvuII fragment present which contains TK2289 (asterisk) in both T. kodakarensis TS517 and LC124 DNAs. M, molecular size marker (in base pairs).
Fig 3.
Construction and confirmation of T. kodakarensis LC125 (ΔTK2289). (A) Plasmid pLC114 was constructed by cloning amplicons from T. kodakarensis genomic DNA upstream and downstream of the TK0254-TK0664 cassette. Plasmid pLC125, generated by deletion of TK2289 from pLC114, was used as the donor DNA to transform T. kodakarensis TS517. The genome structure of the intermediate strain generated was confirmed, as illustrated, and recombination between the duplicated sequences deleted the cassette to yield T. kodakarensis LC125 with the genome as shown. The positions of primers (primers h to m [see Table S1 in the supplemental material]) used to generate the diagnostic amplicons (C and D) and the probe used for Southern blotting (E) are indicated by arrows above the T. kodakarensis TS517 genome. The 5,534- and 5,330-bp EcoRI restriction fragments to which the probe hybridized in digests of T. kodakarensis TS517 and LC125 genomic DNAs are indicated. (B) Expanded illustration of the primer locations (heavy arrows) and the amplicons generated. (C and D) Ethidium bromide-stained agarose gel electrophoretic separations of the amplicons generated with primer pairs h-i and j-I (see Table S1 in the supplemental material) from T. kodakarensis LC124, LC125, and TS517 genomic DNAs. (E) Southern blot of EcoRI-digested T. kodakarensis TS517 and LC125 DNAs. The amplicon generated by primer pair k-m was DIG labeled and used as the probe. The 5,534-bp EcoRI fragment present in T. kodakarensis TS517 DNA (black arrow) is shortened to 5,330-bp by the ΔTK2289 mutation, as indicated for T. kodakarensis LC125 DNA (gray arrow). The probe also cross-hybridized to an ∼11-kbp EcoRI fragment (*) present in both genomes that contains TK1413.
Plasmids capable of autonomous replication and expression of cloned genes in T. kodakarensis were generated from plasmid pLC70 (Table 1) (32), which, when introduced into T. kodakarensis strains, resulted in transformants that were both tryptophan prototrophs and resistant to mevinolin. Plasmid pTS706 was generated from pTS414 by replacing TK1167 (rpoL) with TK1413 (htkA). Plasmids pTS707 and pTS708 were generated by using the QuikChange procedure (Agilent Technologies, Santa Clara, CA) to introduce the desired sequence changes into TK1413 in pTS706 (Table 1 and Fig. 1).
Southern blots.
Genomic DNA preparation, restriction enzyme digestions, digoxigenin (DIG)-labeled probe preparation, DNA transfer, and the development of blots using anti-DIG antibodies coupled to alkaline phosphatase were performed as described previously (23, 31). An oligonucleotide (sequence double underlined in Fig. 1B) was synthesized, labeled with 32P by incubation with T4 polynucleotide kinase and [γ-32P]ATP, and used as the probe for Southern blotting.
Growth curves.
Cultures of T. kodakarensis TS517, LC124, and LC125 were grown in ASW-YT-S0 medium under an atmosphere of 95% N2 plus 5% H2 at 85°C. Changes in the optical density at 600 nm (OD600) were measured by using a Genesys 20 spectrophotometer (Thermo Scientific, Waltham, MA).
RNA extraction and microarray analyses.
RNA extractions and microarray hybridizations were performed as previously described (16). T. kodakarensis cultures were grown at 85°C in medium containing ASW-yeast extract-tryptone plus sodium pyruvate (ASW-YT-Pyr medium), cells were harvested during exponential growth (OD660 ≈ 0.2), and RNA preparations were isolated by using RNeasy Midi kits (Qiagen). The microarrays (Array Tko2) were manufactured by TaKaRa Bio (Otsu, Japan) and contained target sequences corresponding to all 2,306 open reading frames (ORFs) annotated in the T. kodakarensis KOD1 genome (6). Two copies of each sequence were present at different locations on each microarray, providing two data sets for each microarray hybridization experiment. The data files contain the individual signal intensity ratios for each target sequence plus the average ratio and standard deviation (SD) values.
RESULTS
T. kodakarensis is viable with either TK1413 (htkA) or TK2289 (htkB) deleted.
Plasmids pLC124 and pLC125 were constructed such that transformation and recombination into the T. kodakarensis TS517 genome resulted in the deletion of the target gene (TK1413 or TK2289), insertion of the two-gene (TK0254-TK0664) cassette, and a duplication of genes that flanked the inserted cassette (Fig. 2 and 3). Representative transformants, selected by growth on ASW medium lacking tryptophan, were confirmed by diagnostic PCRs (see below) to have the desired genome organization. As these intermediate strains lacked TK1413 or TK2289, these results established that neither histone was essential for T. kodakarensis growth under laboratory conditions. Plating onto ASW+S0+AA medium containing 10 μM 6MP selected spontaneously 6MPr clones. Diagnostic PCRs (see below) and sequencing confirmed the absence of the TK0254-TK0664 cassette and the precise and markerless deletion of TK1413 or TK2289 in two such isolates, designated T. kodakarensis LC124 and LC125, respectively (Table 1), which were used in subsequent studies.
Differences in transformation and recombination frequencies.
Construction of T. kodakarensis LC125 (ΔTK2289) was performed according to routine procedures (12, 33). Transformation of T. kodakarensis TS517 with 1 μg of pLC125 DNA generated >103 tryptophan-independent transformants, and genomic screening confirmed the desired cassette integration in all transformants thus evaluated. Spontaneous 6MPr isolates were readily and abundantly obtained with the expected loss of the TK0254-TK0664 cassette and the presence of the ΔTK2289 markerless deletion. In contrast, transformation of T. kodakarensis TS517 with 1 μg of pLC124 resulted in ∼100-fold-fewer tryptophan-independent transformants, and for ∼90% of these transformants, genomic screening revealed that a single recombination event had integrated the entire pLC124 sequence into the T. kodakarensis genome adjacent to TK1413. In the remaining ∼10% of transformants, either the pLC124 sequence was integrated at a site remote from TK1413 or the cells contained two genomes, one with the desired TK0254-TK0664 integration and thus with TK1413 deleted and a second that had not undergone recombination and had retained TK1413. By subjecting such isolates to repeated single-colony isolations, clones were obtained in which genome segregation had occurred and which contained only the genome with the TK0254-TK0664 cassette and the ΔTK1413 deletion. Plating onto 6MP-containing medium then selected for 6MPr clones that had lost the TK0254-TK0664 cassette, but such mutants occurred at a 100- to 1,000-fold-lower frequency than routinely observed. Apparently, spontaneous recombinations between the duplicated sequences flanking the TK0254-TK0664 cassette occurred at a much-reduced frequency in the absence of HTkA.
PCR and Southern blot confirmation of the genome structures of T. kodakarensis LC124 and LC125.
The genome structures of T. kodakarensis LC124 (ΔTK1413) and T. kodakarensis LC125 (ΔTK2289) were confirmed by three different diagnostic PCRs and by Southern blot hybridizations (Fig. 2 and 3). PCRs with primers that hybridized on either side of TK1413 or TK2289 confirmed the deletion of the target gene from its wild-type location and eliminated the presence of a second genome that retained the target gene at the wild-type locus (Fig. 2C and 3C). The failure to obtain an amplicon in PCRs using one primer that hybridized within TK1413 or TK2289 and a second primer that hybridized to a sequence adjacent to TK1413 or TK2289 further confirmed the deletion of the target gene from the wild-type locus (Fig. 2C). The failure to obtain an amplicon with a primer pair that would generate an amplicon from within TK1413 or TK2289 eliminated the possibility of the retention of TK1413 or TK2289 at any location within the T. kodakarensis genome.
Genomic DNA preparations from T. kodakarensis TS517, LC124, and LC125 were also subjected to Southern blot analyses with probes that hybridized to sequences within TK1413 and TK2289 and/or to genomic sequences directly adjacent to these genes. PvuII digestion of T. kodakarensis TS517 DNA generated a 2,905-bp fragment that contained TK1413 (Fig. 2A). An amplicon probe generated from within TK1413 hybridized to this PvuII fragment in T. kodakarensis TS517 DNA (Fig. 2D, black arrow), but this fragment was not present in PvuII digests of T. kodakarensis LC124 DNA. With the sequence similarities of TK1413 and TK2289 (Fig. 1B), this probe also hybridized to an ∼1-kbp PvuII fragment that contained TK2289 (Fig. 2D, asterisk), which was present in both T. kodakarensis TS517 and LC124 DNAs but not in T. kodakarensis LC125 DNA (not shown). Southern blotting using the oligonucleotide underlined in the TK1413 sequence shown in Fig. 1B as the probe (designated primer e [see Table S1 in the supplemental material]), chosen as likely to minimize cross-hybridization with TK2289, confirmed that TK1413 was absent from T. kodakarensis LC124 but present on the 2,905-bp PvuII fragment in T. kodakarensis LC125 and TS517 DNAs (Fig. 2E). As indicated in Fig. 3A, EcoRI digestion of T. kodakarensis TS517 DNA generated a 5,534-bp restriction fragment that contained TK2289 (Fig. 3E, black arrow), and this fragment was reduced to 5,330 bp in T. kodakarensis LC125 DNA by the ΔTK2289 mutation (Fig. 3E, gray arrow). Both of these fragments hybridized to the amplicon probe generated with primer pair k-m that contains TK2289 and sequences directly adjacent to TK2289. With the histone sequence conservation (Fig. 1A), this probe also hybridized to an ∼11-kbp EcoRI fragment present in both T. kodakarensis TS517 and LC125 which contains TK1413 (Fig. 3E, asterisk).
Failure to generate a strain with both TK1413 and TK2289 deleted.
T. kodakarensis LC124 (ΔTK1413) and T. kodakarensis LC125 (ΔTK2289) retained the ΔpyrF ΔtrpE::pyrF ΔTK0664 mutations (Table 1) and thus are tryptophan auxotrophs and should be amenable to a second round of gene deletion by a repetition of the tryptophan selection-6MPr counterselection protocol (12, 33). However, despite repeated attempts, we were unable to generate a T. kodakarensis strain with both TK1413 and TK2289 deleted. Transformation of T. kodakarensis LC125 with plasmid pLC124 DNA (Fig. 2) did generate tryptophan-independent transformants, but TK1413 was still present in all of the >100 transformants screened. The TK0254-TK0664 cassette was integrated either via a single entire-plasmid insertion event or via nonhomologous recombination at a remote site. All attempts to transform T. kodakarensis LC124 with pLC125 DNA were unsuccessful, and this led to the discovery that the deletion of TK1413 resulted in a T. kodakarensis strain that was no longer amenable to transformation.
Deletion of TK2289 (htkB) reduces growth.
As shown in Fig. 4, when grown in nutrient-rich ASW-YT-S0 medium, cultures of T. kodakarensis LC125 (ΔTK2289) reached only ∼75% of the final cell density of T. kodakarensis TS517 cultures, whereas T. kodakarensis LC124 (ΔTK1413) cultures grew to almost the same final cell density as T. kodakarensis TS517. Despite this difference in growth in liquid culture, the three strains formed colonies on Gelrite-solidified medium with the same plating efficiency. Inactivation of the histone-encoding gene in M. mazei resulted in a strain with increased UV sensitivity (41), but there were no detectable differences in the UV sensitivities of T. kodakarensis TS517, LC124, and LC125 (results not shown).
Fig 4.
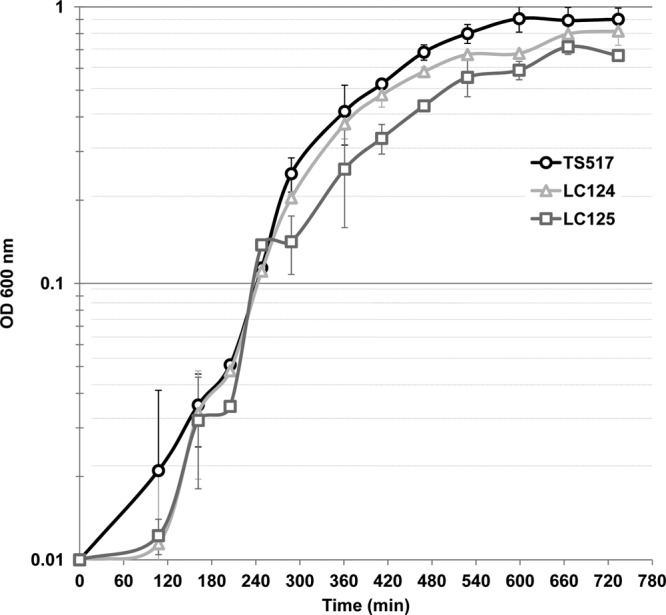
Comparison of the growth profiles of T. kodakarensis TS517, LC124, and LC125 cultures in ASW-YT-S0 medium at 85°C monitored by measurements of the increases in the optical density at 600 nm. The curves show the average values, with errors, obtained in three independent experiments from a total of 9 cultures of each strain.
Loss of HTkA or HTkB results in quantitative and qualitative changes in the T. kodakarensis transcriptome.
The consequences of the loss of HTkA and HTkB on the abundance of transcripts in vivo were documented and quantified by comparing the steady-state levels of transcripts in T. kodakarensis TS517 versus LC124 and in T. kodakarensis TS517 versus LC125. Fluorophore-labeled cDNAs, generated from RNA preparations isolated from exponentially growing cultures, were incubated with T. kodakarensis microarrays that carried duplicated spots of sequences generated from all 2,306 ORFs annotated in the T. kodakarensis genome (6). Reproducible and meaningful results were obtained for transcripts of 2,138 ORFs (∼96%) in comparisons between T. kodakarensis TS517 and LC124 and for 2,153 ORFs (∼97%) in comparisons between T. kodakarensis TS517 and LC125 (Table 2). Deletion of TK1413 resulted in a ≥2-fold change in the abundances of transcripts of 65 ORFs (∼3% of transcripts measured), with transcripts of 30 ORFs increasing and transcripts of 35 ORFs decreasing in abundance in the absence of HTkA. Deletion of TK2289 resulted in a ≥2-fold change in the abundances of transcripts of 87 ORFs (∼4% of transcripts measured), with transcripts of 56 ORFs increasing and transcripts of 31 ORFs decreasing in abundance in the absence of HTkB.
Table 2.
Microarray comparisons of RNA abundances in T. kodakarensis LC124 versus T. kodakarensis TS517 and in T. kodakarensis LC125 versus TS517a
| Comparison and gene | Predicted or known function | Fold difference |
|---|---|---|
| LC124 vs TS517 | ||
| Upregulated by deletion of TK1413 (histone A) | ||
| TK2195 | Aspartate carbamoyltransferase, regulatory subunit | 3.5 |
| TK2196 | Aspartate carbamoyltransferase, catalytic subunit | 3.1 |
| TK1927 | Hypothetical membrane protein | 3.4 |
| TK0190 | GMP synthase, glutamine amidotransferase component | 2.2 |
| TK0191 | Predicted nucleic acid-binding protein | 1.1 |
| TK0192 | Hypothetical protein | 2.1 |
| TK0193 | GMP synthase, PP-loop-ATPase component | 3.3 |
| TK0194 | Inosine-5′-monophosphate dehydrogenase | 2.9 |
| TK0157 | Xanthine/uracil permease | 3.1 |
| TK0475 | d-Arabino 3-hexulose-6-phosphate formaldehyde lyase, fused to phosphohexuloisomerase | 2.9 |
| TK0268 | 2-Dehydro-3-deoxyphosphoheptonate aldolase | 2.8 |
| TK0269 | Transketolase, C-terminal section | 1.3 |
| TK0270 | Transketolase, N-terminal section | 2.6 |
| TK0271 | Hypothetical protein | 2.0 |
| TK1893 | Glycerate kinase-related protein | 2.7 |
| TK0835 | Phosphoribosylaminoimidazole carboxylase, ATPase subunit | 2.6 |
| TK0204 | Phosphoribosylamine-glycine ligase | 2.5 |
| TK0203 | Hypothetical protein | 2.2 |
| TK1895 | Purine-nucleoside phosphorylase | 2.4 |
| TK0010 | Hypothetical protein | 2.4 |
| TK2147 | Methyl-accepting chemotaxis protein | 2.2 |
| TK1002 | Adenylosuccinate synthase | 2.2 |
| TK1287 | Uracil phosphoribosyltransferase | 2.2 |
| TK2138 | Orotate phosphoribosyltransferase | 2.1 |
| TK1737 | Xanthine/guanine phosphoribosyltransferase | 2.1 |
| TK0766 | Hypothetical membrane protein | 2.1 |
| TK1605 | Hydrolase, metallo-beta-lactamase superfamily | 2.1 |
| TK1961 | Replication factor A complex, RPA41 subunit | 2.1 |
| TK1459 | Hypothetical protein | 2.0 |
| TK0252 | Indole-3-glycerol phosphate synthase | 2.0 |
| Downregulated by deletion of TK1413 (histone A) | ||
| TK0161 | ABC-type multidrug transport system, ATPase component | 3.0 |
| TK0162 | Hypothetical membrane protein | 2.0 |
| TK0163 | Hypothetical membrane protein | 2.3 |
| TK0164 | S-layer-like array protein | 3.8 |
| TK0165 | Hypothetical protein | 2.0 |
| TK0166 | Hypothetical protein | 5.3 |
| TK1413 | Archaeal histone A | 3.6 |
| TK0982 | Hypothetical membrane protein | 3.5 |
| TK0465 | Acetyl-CoA synthetase I, beta subunit | 2.3 |
| TK0466 | Hypothetical protein, conserved | 1.2 |
| TK0467 | Hypothetical protein | 3.4 |
| TK0468 | Hypothetical protein | 2.1 |
| TK1025 | Hypothetical protein | 3.4 |
| TK2114 | Hypothetical protein | 3.3 |
| TK1463 | Hypothetical protein | 3.0 |
| TK0589 | Hypothetical protein | 2.8 |
| TK1229 | Hypothetical protein | 2.8 |
| TK1316 | Predicted membrane protease subunit, stomatin/prohibitin homologue | 2.7 |
| TK0773 | Predicted ATP-dependent endonuclease | 2.7 |
| TK0751 | Hypothetical protein | 2.7 |
| TK1405 | Phosphoenolpyruvate carboxykinase | 2.6 |
| TK2053 | ABC-type multidrug transport system, ATPase component | 2.2 |
| TK2079 | Probable formate transporter | 2.2 |
| TK0443 | Hypothetical membrane protein | 2.2 |
| TK0719 | ABC-type molybdate transport system, ATPase component | 2.2 |
| TK0878 | Translation initiation factor eIF-5A | 2.1 |
| TK0844 | Tungsten-containing oxidoreductase | 2.1 |
| TK1481 | NADH:polysulfide oxidoreductase | 2.0 |
| TK1335 | Hypothetical protein | 2.0 |
| TK1780 | Hypothetical protein | 2.0 |
| TK0675 | Hypothetical protein | 2.0 |
| TK1949 | Hypothetical protein | 2.0 |
| TK1123 | 2-Oxoacid:ferredoxin oxidoreductases, gamma subunit | 2.0 |
| TK1334 | Dephospho-CoA kinase | 2.0 |
| TK1060 | Hypothetical protein | 2.0 |
| LC125 vs TS517 | ||
| Upregulated by deletion of TK2289 (histone B) | ||
| TK0264 | Archaeal shikimate kinase | 7.8 |
| TK2195 | Aspartate carbamoyltransferase, regulatory subunit | 6.5 |
| TK2196 | Aspartate carbamoyltransferase, catalytic subunit | 4.3 |
| TK0766 | Hypothetical membrane protein | 5.4 |
| TK0157 | Xanthine/uracil permease | 4.7 |
| TK0202 | Phosphoribosylformylglycinamidine synthase, PurS component | 2.6 |
| TK0203 | Hypothetical protein | 3.6 |
| TK0204 | Phosphoribosylamine-glycine ligase | 4.0 |
| TK0190 | GMP synthase, glutamine amidotransferase component | 2.5 |
| TK0191 | Predicted nucleic acid-binding protein | 1.6 |
| TK0192 | Hypothetical protein | 2.4 |
| TK0193 | GMP synthase, PP-loop-ATPase component | 4.0 |
| TK0194 | Inosine-5′-monophosphate dehydrogenase | 3.1 |
| TK0835 | Phosphoribosylaminoimidazole carboxylase, ATPase subunit | 3.4 |
| TK0207 | Formate-dependent phosphoribosylglycinamide formyltransferase | 2.9 |
| TK0603 | Hypothetical protein | 2.9 |
| TK0604 | Hypothetical protein | 2.8 |
| TK1090 | Hypothetical membrane protein | 2.7 |
| TK0561 | Adenylosuccinate lyase | 2.7 |
| TK1352 | Hypothetical protein | 2.6 |
| TK1583 | Hypothetical protein | 2.6 |
| TK0418 | Hypothetical protein | 2.5 |
| TK1287 | Uracil phosphoribosyltransferase | 2.5 |
| TK0260 | Probable aromatic aminotransferase | 2.4 |
| TK1895 | Purine-nucleoside phosphorylase | 2.4 |
| TK0084 | Hypothetical protein | 2.3 |
| TK0612 | Hypothetical protein | 2.3 |
| TK1923 | Hypothetical membrane protein | 2.3 |
| TK2180 | Hypothetical protein | 2.3 |
| TK1002 | Adenylosuccinate synthase | 2.3 |
| TK0199 | Predicted permease | 2.2 |
| TK0475 | d-Arabino 3-hexulose-6-phosphate formaldehyde lyase, fused to phosphohexuloisomerase | 2.2 |
| TK0838 | Thioredoxin | 2.2 |
| TK0983 | Hypothetical protein | 2.2 |
| TK1822 | Predicted ATPase | 2.2 |
| TK0762 | Glycosyltransferase | 2.2 |
| TK2138 | Orotate phosphoribosyltransferase | 2.2 |
| TK0531 | Hypothetical protein | 2.2 |
| TK1841 | Predicted site-specific integrase-resolvase | 2.2 |
| TK0836 | Phosphoribosylaminoimidazole carboxylase, catalytic subunit | 2.2 |
| TK0407 | Hypothetical protein | 2.2 |
| TK2182 | Hypothetical protein | 2.2 |
| TK0737 | Hypothetical protein | 2.2 |
| TK0210 | Phosphoribosylaminoimidazole-succinocarboxamide synthase | 2.1 |
| TK1557 | Predicted dehydrogenase | 2.1 |
| TK0197 | Phosphoribosylformylglycinamidine synthase II | 2.1 |
| TK1591 | Predicted transcription regulator | 2.1 |
| TK1869 | Probable phosphate transport system regulator | 2.1 |
| TK0120 | Proline dehydrogenase, gamma subunit (4Fe-4S cluster-binding component) | 2.1 |
| TK0625 | Multisubunit sodium/hydrogen antiporter, MnhD subunit | 2.1 |
| TK0906 | Endonuclease V (deoxyinosine 3′-endonuclease) | 2.1 |
| TK0396 | Hypothetical protein | 2.1 |
| TK0091 | Hypothetical protein | 2.1 |
| TK1838 | Hypothetical membrane protein | 2.0 |
| TK1444 | Homoserine kinase | 2.0 |
| TK1289 | Sodium-driven multidrug efflux pump protein | 2.0 |
| Downregulated by deletion of TK2289 (histone B) | ||
| TK2289 | Archaeal histone B | 9.8 |
| TK0179 | Hypothetical protein | 2.3 |
| TK0180 | Acetyl-CoA acetyltransferase | 3.6 |
| TK0181 | 3-Hydroxy-3-methylglutaryl-CoA synthase | 3.4 |
| TK2076 | Formate:ferredoxin oxidoreductase, alpha subunit | 3.2 |
| TK2077 | Formate:ferredoxin oxidoreductase, 4Fe-4S cluster-binding beta subunit | 2.9 |
| TK2078 | Formate:ferredoxin oxidoreductase, 4Fe-4S cluster-binding gamma subunit | 2.8 |
| TK2079 | Probable formate transporter | 3.1 |
| TK1830 | Probable alpha-amylase | 3.2 |
| TK1413 | Archaeal histone A | 2.7 |
| TK1405 | Phosphoenolpyruvate carboxykinase | 2.6 |
| TK1431 | Glutamate dehydrogenase | 2.6 |
| TK1046 | Hypothetical protein | 2.4 |
| TK1624 | Methylmalonyl-CoA decarboxylase, gamma subunit | 2.3 |
| TK0560 | Archaeal chromatin protein, Alba | 2.2 |
| TK1245 | Hypothetical protein | 2.2 |
| TK2048 | Hypothetical protein | 2.2 |
| TK0136 | Indolepyruvate:ferredoxin oxidoreductase, alpha subunit | 2.1 |
| TK0307 | SSU ribosomal protein S10P | 2.1 |
| TK2268 | Aspartate aminotransferase | 2.1 |
| TK0352 | Thymidine phosphorylase | 2.1 |
| TK2035 | Glycine cleavage system protein T (aminomethyltransferase) | 2.1 |
| TK1539 | LSU ribosomal protein L2P | 2.1 |
| TK0878 | Translation initiation factor eIF-5A | 2.1 |
| TK1500 | SSU ribosomal protein S9P | 2.1 |
| TK0465 | Acetyl-CoA synthetase I, beta subunit | 2.0 |
| TK1534 | Protein translation factor SUI1 homologue | 2.0 |
| TK1295 | Predicted thiol protease | 2.0 |
| TK0308 | Translation elongation factor EF-1, alpha subunit | 2.0 |
| TK1469 | Hydrolase, metallo-beta-lactamase superfamily | 2.0 |
| TK1174 | Predicted acetyltransferase, isoleucine patch superfamily | 2.0 |
Shown are average values from two independent experiments for all transcripts for which expression levels increased or decreased by at least 2-fold in T. kodakarensis LC124 or LC125 compared with their abundances in T. kodakarensis TS517. The microarrays had amplicons from all 2,306 T. kodakarensis ORFs (6) spotted twice at different locations. Genes in operons that exhibited significant changes in expression levels are listed even when the expression levels of some genes did not change by 2-fold. Shading highlights operons and genes with the same transcription responses to the absence of HTkA or HTkB. CoA, coenzyme A; SSU, small subunit; LSU, large subunit.
Thirteen of the ORFs that were transcribed at higher levels in the absence of HTkA appear to be components of four operons (Table 2). Transcripts of three of these operons were also more abundant in the absence of HTkB, consistent with both histones normally decreasing transcription levels or the stability of these transcripts in vivo. Increases in the abundances of eight monocistronic transcripts also occurred in the absence of either HTkA or HTkB, but there were also increases and decreases in operon and single-gene transcript abundances that occurred specifically in the absence of HTkA or HTkB (Table 2). Notably, in the absence of HTkA, the level of transcription of an operon (TK0161 to TK0166) that is predicted to encode several membrane and cell surface components (6) was reduced, possibly playing a role in the loss of transformability of T. kodakarensis LC124. Similarly, in the absence of HTkB, the decreased transcription levels of several genes predicted to encode translation factors and ribosomal proteins seem notable as a potential factor in the reduced growth of T. kodakarensis LC125 cultures.
The transcript levels of several ORFs encoding proteins associated with purine metabolism increased in the absence of either histone, and the transcript level of one ORF (TK1591), predicted to encode a transcription regulator, was increased in the absence of HTkB but not in the absence of HTkA.
The deletion of TK1413 had no detectable effect on the transcripts of TK2289, but intriguingly, the deletion of TK2289 resulted in decreased abundances of TK1413 transcripts and of transcripts of TK0560, which encodes the unrelated chromatin protein Alba (29). The decrease in the abundance of TK1413 transcripts, however, must not reduce HTkA production below that needed for viability.
T. kodakarensis LC124 has lost competency for transformation.
The development of T. kodakarensis as a model system was founded on this species being naturally competent for DNA uptake and transformation (36). It was therefore important to explore the observation that transformation of T. kodakarensis TS517 with plasmid pLC124 DNA resulted in ∼103-fewer tryptophan-independent transformants than routinely observed with similar donor DNAs, including pLC125, that direct the integration of the TK0254-TK0664 cassette at other chromosomal locations (Table 3). Despite the low transformation frequency, T. kodakarensis LC124 (ΔTK1413) was constructed, but all subsequent attempts to obtain chromosomal transformants of T. kodakarensis LC124 were unsuccessful. Several different donor DNAs were used that generated large numbers of tryptophan-independent transformants by using the identical protocol and reagents used for T. kodakarensis TS517 and LC125 (12). One possibility was that T. kodakarensis LC124 had acquired an unusual sensitivity to some step in the protocol, but this was not the case. T. kodakarensis LC124 cells exposed to the transformation protocol formed colonies on nonselective media with the same plating efficiency as T. kodakarensis TS517 and LC125.
Table 3.
Transformation of T. kodakarensis TS517, LC124, and LC125 with replicative and nonreplicative plasmids
| Plasmid | Autonomous replicationa | Gene targetb | Avg no. of transformants recovered ± SDc |
||
|---|---|---|---|---|---|
| TS517 | LC124 (ΔTK1413) | LC125 (ΔTK2289) | |||
| pUDHisD | − | TK0244 | 1 × 104 ± 0.5 × 104 | 0 | 1 × 104 ± 0.5 × 104 |
| pLC124 | − | TK1413 | 1 × 101 ± 1 × 101 | NA | 1 × 101 ± 1 × 101 |
| pLC125 | − | TK2289 | 1 × 104 ± 0.5 × 104 | 0 | NA |
| pTS503 | − | TK1827 | 1 × 104 ± 0.5 × 104 | 0 | 1 × 104 ± 0.5 × 104 |
| pLC70 | + | 2 × 104 ± 1 × 104 | 1 ± 1 | 2 × 104 ± 1 × 104 | |
| pLC71 | + | 2 × 104 ± 1 × 104 | 1 ± 1 | 2 × 104 ± 1 × 104 | |
| pTS414 | + | 2 × 104 ± 1 × 104 | 1 ± 1 | 2 × 104 ± 1 × 104 | |
| pTS706 | + | 0 | 0 | 0 | |
| pTS707 | + | 2 × 104 ± 1 × 104 | 1 ± 1 | 2 × 104 ± 1 × 104 | |
| pTS708 | + | 0 | 0 | 0 | |
Plasmid capable (+) or incapable (−) of autonomous replication in T. kodakarensis.
Gene that the plasmid was constructed to delete.
Transformants recovered after incubation of the T. kodakarensis strain (1012 cells) with 1 μg of the plasmid DNA. NA, not applicable (gene target was already deleted).
T. kodakarensis strains can also be transformed by autonomously replicating plasmids (pLC70 and derivatives [32]) (Table 1). These plasmids are maintained in the T. kodakarensis cytoplasm by a replication machinery derived from plasmid pTN1 that was isolated from Thermococcus nautilus (40). Routinely, such plasmid transformations result in ∼2-fold more transformants per μg of donor DNA than chromosomal transformations (Table 3). Transformation of T. kodakarensis LC125 with pLC70 and pLC71 generated transformants at the same frequency as transformation of T. kodakarensis TS517. In contrast, transformation of T. kodakarensis LC124 with pLC70 and pLC71 resulted in ∼104-fold-fewer transformants (Table 3). However, once established in T. kodakarensis LC124, these plasmids were maintained as cytoplasmic replicons with the same copy number and stability as in T. kodakarensis TS517 and LC125.
Constitutive plasmid expression of HTkA is toxic.
Plasmid pTS706 was constructed to determine if the transformation defect of T. kodakarensis LC124 could be suppressed by plasmid expression of HTkA. Plasmid pTS706 was derived from pLC70 (Table 1) and therefore should replicate autonomously in T. kodakarensis and had TK1413 positioned appropriately downstream from PhmtB, a constitutive promoter that has been used for the expression of many genes in T. kodakarensis (31–33, 35, 42). Despite several attempts, incubation of T. kodakarensis LC124 with pTS706 never resulted in selectable transformants. Surprisingly, this was also the case in all attempts to transform T. kodakarensis TS517 and LC125 with pTS706 DNA. To determine if it was HTkA synthesis that was inhibiting the growth of transformants, the second codon (GCC) of TK1413 was changed to a translation stop codon (TGA). When the resulting plasmid, pTS707 (Table 1), was used as the donor DNA, transformants were obtained with T. kodakarensis TS517, LC125, and LC124, although transformants of T. kodakarensis LC124 were obtained at a much lower frequency than transformants of T. kodakarensis TS517 and LC125 (Table 3). DNA binding by archaeal histones is dependent on a universally conserved arginine residue (R20 in TK1413) (Fig. 1), and variants with an isoleucine at this location do not bind DNA (27, 39). Plasmid pTS708 was therefore generated with codon 20 of TK1413 changed from AGG to ATT to synthesize the HTkA(R20I) variant, but incubation of T. kodakarensis TS517, LC124, and LCI25 with pTS708 DNA failed to produce transformants. Apparently, therefore, the ectopic expression of an HTkA variant incapable of DNA binding was still toxic, although it remains possible that HTkA(R20I) monomers might assemble into toxic DNA-binding heterodimers with HTkB monomers synthesized from the chromosomal copy of TK2289.
DISCUSSION
The results obtained demonstrate that the loss of either HTkA or HTkB can be accommodated by T. kodakarensis, but as it was impossible to generate a strain with both TK1413 and TK2298 deleted, the presence of at least one archaeal histone seems to be essential for viability. Given that homodimers of either HTkA or HTkB are sufficient for viability, the histone requirement must be only for functions common to both histones. The results obtained, however, also argue that HTkA and HTkB have unique functions. In the absence of HTkA but not HTkB, T. kodakarensis is no longer amenable to transformation, whereas the loss of HTkB but not HTkA reduced growth in liquid culture. As discussed below, these different phenotypes may correlate with differences in HTkA- versus HTkB-dependent gene expression.
Comparisons of the abundances of transcripts in T. kodakarensis LC124 (ΔTK1413) and T. kodakarensis LC125 (ΔTK2289) with those in the parental strain T. kodakarensis TS517 revealed that HTkA and HTkB participate in both transcription activation and repression in vivo (Table 2). For most ORFs for which an increase in the transcription level occurred, it occurred in the absence of either HTkA or HTkB, consistent with both histones normally negatively regulating the transcriptions of these genes. Based on in vitro studies, promoter binding by HTkA and HTkB most likely limits access of the transcription apparatus and thus limits the initiation of the transcription of these genes (7, 43, 44). By using high-resolution nucleosome position technology (1), and with the availability of T. kodakarensis LC124 and LC125, this prediction can now be tested. In the absence of HTkA and HTkB, reductions occurred in the abundances of transcripts of ∼30 ORFs, with little overlap in the genes thus regulated by HTkA versus HTkB. Transcription of these ORFs is most likely stimulated in vivo by HTkA and/or HTkB, either directly through histone interactions with the transcription machinery or by histone construction of a chromatin configuration(s) that promotes local transcription (43).
The most striking discovery of this research is that deletion of TK1413 results in a T. kodakarensis strain that is no longer amenable to transformation. Apparently, therefore, HTkA but not HTkB plays a role in transformation, possibly directly in DNA integration and/or indirectly through the expressions of other genes required for transformation. A direct role for HTkA in recombination would be consistent with the difficulty encountered in the construction of T. kodakarensis LC124. Recombination deleting the TK0254-TK0664 cassette from the genome of the intermediate strain, a strain which lacks TK1413, occurred at a frequency ∼103-fold lower than usually observed (12, 32). Also consistent with a direct role for HTkA in recombination, all attempts to integrate selectable genes by single- or double-crossover recombination into the genome of T. kodakarensis LC124 failed. However, as the level of transformation of T. kodakarensis LC124 with autonomously replicating plasmids is also severely reduced, deletion of TK1413 may also negatively affect other steps required for transformation. Possibly, the DNA uptake mechanism generates DNA fragments that require HTkA for protection (8, 42), or the deletion of TK1413 increases the expression level of a nuclease or decreases the synthesis of components of the DNA uptake system. Hinting at the latter possibility, transcripts of an operon (TK0161 to TK0166) that is predicted to encode membrane- and surface-located proteins are less abundant in T. kodakarensis LC124 (ΔTK1413) than in the parental T. kodakarensis strain TS517 (Table 2). In contrast, the loss of HTkB (ΔTK2289) had no detectable effects on the transcripts of this operon.
Attempts to confirm, by complementation, that it was the absence of HTkA that was responsible for the loss of transformation, by expression of TK1413 from a replicating plasmid (pTS706), led to the discovery that such HTkA synthesis was toxic to T. kodakarensis. Regardless of the recipient strain, with or without a chromosomal copy of TK1413 and/or TK2289, incubation of T. kodakarensis strains with pTS706 never resulted in transformants. Transformants were readily obtained with pTS707, which has a translation-terminating codon at position 2 of TK1413, consistent with the toxicity of pTS706 being a consequence of plasmid-directed synthesis of HTkA. As transformation with pTS708 was also impossible, synthesis of HTkA(R20I) was also toxic, and homodimers of HTkA(R20I) would not bind DNA (27, 39). Plasmid expression of HTkA(R20I), however, might still confer toxicity through DNA binding if these monomers assembled into HTkA(R20I)+HTkB heterodimers (20).
T. kodakarensis grows rapidly to high cell densities, tolerates air exposure, and has a high plating efficiency and natural competency, and many genetic tools are now available (12). Given these advantages, T. kodakarensis is now used widely as a model system for archaeal research in programs that investigate topics ranging from DNA replication (17, 18, 37) to hyperthermophily (13) to biofuel production (15, 34). The discovery that HTkA but not HTkB plays a critical role in genetic manipulations of T. kodakarensis adds a surprising feature that is likely relevant to many T. kodakarensis-based investigations.
ACKNOWLEDGMENTS
This work was supported by NIH grants GM098176 (to J.N.R. and T.J.S.) and GM100329 (to T.J.S.) and by the Japan Society for the Promotion of Science under a grant-in-aid for scientific research (to T.K.).
Footnotes
Published ahead of print 12 October 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Brogaard K, Xi L, Wang JP, Widom J. 2012. A map of nucleosome positions in yeast at base-pair resolution. Nature 486:496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Čuboňová L, Sandman K, Hallam SJ, DeLong EF, Reeve JN. 2005. Histones in Crenarchaea. J. Bacteriol. 187:5482–5485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Decanniere K, Babu AM, Sandman K, Reeve JN, Heinemann U. 2000. Crystal structures of recombinant histones HMfA and HMfB from the hyperthermophilic archaeon Methanothermus fervidus. J. Mol. Biol. 303:35–47 [DOI] [PubMed] [Google Scholar]
- 4. Dinger ME, Baillie GJ, Musgrave DR. 2000. Growth phase-dependent expression and degradation of histones in the thermophilic archaeon Thermococcus zilligii. Mol. Microbiol. 36:876–885 [DOI] [PubMed] [Google Scholar]
- 5. Forbes AJ, et al. 2004. Targeted analysis and discovery of posttranslational modifications in proteins from methanogenic archaea by top-down MS. Proc. Natl. Acad. Sci. U. S. A. 101:2678–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fukui T, et al. 2005. Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 15:352–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Geiduschek EP, Ouhammouch M. 2005. Archaeal transcription and its regulators. Mol. Microbiol. 56:1397–1407 [DOI] [PubMed] [Google Scholar]
- 8. Grayling RA, Bailey KA, Reeve JN. 1997. DNA binding and nuclease protection by the HMf histones from the hyperthermophilic archaeon Methanothermus fervidus. Extremophiles 1:79–88 [DOI] [PubMed] [Google Scholar]
- 9. Hadjithomas M, Moudrianakis EN. 2011. Experimental evidence for the role of domain swapping in the evolution of the histone fold. Proc. Natl. Acad. Sci. U. S. A. 108:13462–13467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Heinicke I, Müller J, Pittelkow M, Klein A. 2004. Mutational analysis of genes encoding chromatin proteins in the archaeon Methanococcus voltae indicates their involvement in the regulation of gene expression. Mol. Genet. Genomics 272:76–87 [DOI] [PubMed] [Google Scholar]
- 11. Higashibata H, Fujiwara S, Takagi M, Imanaka T. 1999. Analysis of DNA compaction profile and intracellular contents of archaeal histones from Pyrococcus kodakaraensis KOD1. Biochem. Biophys. Res. Commun. 258:416–424 [DOI] [PubMed] [Google Scholar]
- 12. Hileman TH, Santangelo TJ. 2012. Genetics techniques for Thermococcus kodakarensis. Front. Microbiol. 3:195 doi:10.3389/fmicb.2012.00195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Imanaka T. 2011. Molecular bases of thermophily in hyperthermophiles. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 87:587–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jahn U, Aigner J, Längst G, Reeve JN, Huber H. 2009. Nanoarchaeal origin of histone H3? J. Bacteriol. 191:1092–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kanai T, et al. 2005. Continuous hydrogen production by the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Biotechnol. 116:271–282 [DOI] [PubMed] [Google Scholar]
- 16. Kanai T, Takedomi S, Fujiwara S, Atomi H, Imanaka T. 2010. Identification of the Phr-dependent heat shock regulon in the hyperthermophilic archaeon, Thermococcus kodakaraensis. J. Biochem. 147:361–370 [DOI] [PubMed] [Google Scholar]
- 17. Li Z, et al. 2011. A novel DNA nuclease is stimulated by association with the GINS complex. Nucleic Acids Res. 39:6114–6123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Z, Santangelo TJ, Cuboňová L, Reeve JN, Kelman Z. 2010. Affinity purification of an archaeal DNA replication protein network. mBio 1(5):e00221–10 doi:10.1128/mBio.00221-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li T, Sun F, Ji X, Feng Y, Rao Z. 2003. Structure based hyperthermostability of archaeal histone HPhA from Pyrococcus horikoshii. J. Mol. Biol. 325:1031–1037 [DOI] [PubMed] [Google Scholar]
- 20. Marc F, Sandman K, Lurz R, Reeve JN. 2002. Archaeal histone tetramer formation determines DNA affinity, and the direction of DNA supercoiling. J. Biol. Chem. 277:30879–30886 [DOI] [PubMed] [Google Scholar]
- 21. Mariño-Ramírez L, et al. 2011. The Histone Database: an integrated resource for histones and histone fold-containing proteins. Database 2011:bar048 doi:10.1093/database/bar048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maruyama H, et al. 2011. Histone and TK0471/TrmBL2 form a novel heterogeneous genome architecture in the hyperthermophilic archaeon Thermococcus kodakarensis. Mol. Biol. Cell 22:386–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pan M, Santangelo TJ, Li Z, Reeve JN, Kelman Z. 2011. Thermococcus kodakarensis encodes three MCM homologs but only one is essential. Nucleic Acids Res. 39:9671–9680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pereira S, Grayling RA, Lurz R, Reeve JN. 1997. Archaeal nucleosomes. Proc. Natl. Acad. Sci. U. S. A. 94:12633–12637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sandman K, Grayling RA, Dobrinski B, Lurz R, Reeve JN. 1994. Growth phase dependent synthesis of histones in the archaeon Methanothermus fervidus. Proc. Natl. Acad. Sci. U. S. A. 91:12624–12628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sandman K, Krzycki JA, Dobrinski B, Lurz R, Reeve JN. 1990. DNA binding protein HMf, from the hyperthermophilic archaebacterium Methanothermus fervidus, is most closely related to histones. Proc. Natl. Acad. Sci. U. S. A. 87:5788–5791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sandman K, Louvel H, Samson RY, Pereira SL, Reeve JN. 2008. Archaeal chromatin proteins histone HMtB and Alba have lost DNA-binding ability in laboratory strains of Methanothermobacter thermautotrophicus. Extremophiles 12:811–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sandman K, Reeve JN. 2000. Structure and functional relationships of archaeal and eukaryal histones and nucleosomes. Arch. Microbiol. 173:165–169 [DOI] [PubMed] [Google Scholar]
- 29. Sandman K, Reeve JN. 2005. Archaeal chromatin proteins: different structures but common function? Curr. Opin. Microbiol. 8:656–661 [DOI] [PubMed] [Google Scholar]
- 30. Sandman K, Reeve JN. 2006. Archaeal histones and the origin of the histone fold. Curr. Opin. Microbiol. 9:520–525 [DOI] [PubMed] [Google Scholar]
- 31. Santangelo TJ, Cuboňová L, James CL, Reeve JN. 2007. TFB1 or TFB2 is sufficient for Thermococcus kodakarensis viability and basal transcription in vitro. J. Mol. Biol. 367:344–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Santangelo TJ, Cuboňová L, Reeve JN. 2008. Shuttle vector expression in Thermococcus kodakaraensis: contributions of cis elements to protein synthesis in a hyperthermophilic archaeon. Appl. Environ. Microbiol. 74:3099–3104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Santangelo TJ, Cuboňová L, Reeve JN. 2010. Thermococcus kodakarensis genetics: TK1827-encoded beta-glycosidase, new positive-selection protocol, and targeted and repetitive deletion technology. Appl. Environ. Microbiol. 74:1044–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Santangelo TJ, Cuboňová L, Reeve JN. 2011. Deletion of alternative pathways for reductant recycling in Thermococcus kodakarensis increases hydrogen production. Mol. Microbiol. 81:897–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Santangelo TJ, Reeve JN. 2010. Deletion of switch 3 results in an archaeal RNA polymerase that is defective in transcript elongation. J. Biol. Chem. 285:23908–23915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sato T, Fukui T, Atomi H, Imanaka T. 2003. Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 185:210–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shin J-H, Santangelo TJ, Xie Y, Reeve JN, Kelman Z. 2007. Archaeal MCM helicase can unwind DNA bound by archaeal histones and transcription factors. J. Biol. Chem. 282:4908–4915 [DOI] [PubMed] [Google Scholar]
- 38. Slesarev AI, Belova GI, Kozyavkin SA, Lake JA. 1998. Evidence for an early prokaryotic origin of histones H2A and H4 prior to the emergence of eukaryotes. Nucleic Acids Res. 26:427–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Soares DJ, Sandman K, Reeve JN. 2000. Mutational analysis of archaeal histone-DNA interactions. J. Mol. Biol. 297:39–47 [DOI] [PubMed] [Google Scholar]
- 40. Soler N, et al. 2007. The rolling-circle plasmid pTN1 from the hyperthermophilic archaeon Thermococcus nautilus. Mol. Microbiol. 66:357–370 [DOI] [PubMed] [Google Scholar]
- 41. Weidenbach K, et al. 2008. Deletion of the archaeal histone in Methanosarcina mazei Gö1 results in reduced growth and genomic transcription. Mol. Microbiol. 67:662–671 [DOI] [PubMed] [Google Scholar]
- 42. Weng L, Liu D, Li Y, Cao S, Feng Y. 2004. An archaeal histone-like protein as an efficient DNA carrier in gene transfer. Biochim. Biophys. Acta 1702:209–216 [DOI] [PubMed] [Google Scholar]
- 43. Wilkinson SP, Ouhammouch M, Geiduschek EP. 2010. Transcriptional activation in the context of repression mediated by archaeal histones. Proc. Natl. Acad. Sci. U. S. A. 107:6777–6781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xie Y, Reeve JN. 2004. Transcription by an archaeal RNA polymerase is slowed but not blocked by an archaeal nucleosome. J. Bacteriol. 186:3492–3498 [DOI] [PMC free article] [PubMed] [Google Scholar]