SUMMARY
Oncogenic Myc alters mitochondrial metabolism, making it dependent on exogenous glutamine (Gln) for cell survival. Accordingly, Gln deprivation selectively induces apoptosis in MYC-overexpressing cells via unknown mechanisms. Using MYCN-amplified neuroblastoma as a model, we identify PUMA, NOXA and TRB3 as executors of Gln-starved cells. Gln depletion in MYC-transformed cells induces apoptosis through ATF4-dependent, but p53-independent, PUMA and NOXA induction. MYC-transformed cells depend on both glutamate-oxaloacetate transaminase and glutamate dehydrogenase to maintain Gln homeostasis and suppress apoptosis. Consequently, either ATF4 agonists or glutaminolysis inhibitors potently induce apoptosis in vitro and inhibit tumor growth in vivo. These results reveal mechanisms whereby Myc sensitizes cells to apoptosis and validate ATF4 agonists and inhibitors of Gln metabolism as potential Myc-selective cancer therapeutics.
INTRODUCTION
Neuroblastoma is one of the most frequent solid tumors detected in childhood, accounting for approximately 10-15% of all pediatric oncology deaths (Maris, 2010). Risk factors indicative of poor prognosis include age > 18 months at diagnosis, advanced stage, unfavorable histologic grade, and MYCN amplification (Maris, 2010). Recent studies demonstrate that mutations in the Anaplastic Lymphoma Kinase (ALK) gene are causal for most familial neuroblastoma and occur in ~10% of sporadic neuroblastomas, while polymorphisms in genes encoding BRCA1-associated RING Domain-1 (BARD1) and LIM domain only 1 (LMO1) influence disease susceptibility (Capasso et al., 2009; Mosse et al., 2008; Wang et al., 2011). Nevertheless, MYCN amplification remains the most important oncogenic driver and reliable prognostic factor, highly correlated with advanced disease stage and poor survival rate. MYCN amplification occurs in 20-25% of neuroblastomas overall and 40% of high-risk cases (Maris, 2010). Alternatively, elevated c-MYC expression correlates with poor prognosis in MYCN non-amplified neuroblastoma (Liu et al., 2008). High levels of Myc activity likely contribute to aggressive phenotypes by regulating and/or cooperating with other oncogenic pathways.
Neuroblastomas, like other solid tumors, require specific metabolic alterations to fuel their deregulated growth and invasion into surrounding tissues. Central metabolic pathways and energy production differ between normal and malignant cells in their regulation and dynamics. Fast-growing, poorly-differentiated tumor cells typically exhibit increased aerobic glycolysis, a phenomenon known as the “Warburg effect” (Vander Heiden et al., 2009). Meanwhile, cancer cells also depend on sustained mitochondrial activity, providing biosynthetic substrates to support enhanced proliferation and survival. Glucose and Gln are two of the most abundant nutrients consumed by neoplastic cells (DeBerardinis et al., 2008). In most human cancers, >80% of the absorbed glucose is catabolized into lactate. While glycolytic ATP generation maintains cellular bioenergetics, the remaining glucose enters the tricarboxylic acid cycle (TCA) cycle where it is converted to citrate. Citrate is then preferentially exported into the cytosol to support lipid synthesis. However, increased citrate efflux from mitochondria could deplete TCA cycle metabolites. To prevent this, Gln (another major substrate oxidized by tumor cells) replenishes a truncated TCA cycle through a process termed “anapleurosis” (DeBerardinis et al., 2008). Moreover, Gln metabolism maintains mitochondrial integrity and NADPH levels needed for redox homeostasis and macromolecular synthesis (DeBerardinis et al., 2008; Metallo et al., 2012; Mullen et al., 2012; Wise et al., 2011).
MYC oncogenes regulate multiple aspects of tumor metabolism, enabling cancer cells to avidly uptake both glucose and Gln (Dang, 2012). The MYC family contains three members, c-MYC, MYCN and MYCL. While c-MYC is broadly deregulated in many human tumors, MYCN expression is more restricted to neural tumors and MYCL is predominantly found in small cell lung cancer. Both c-Myc and N-Myc have been documented to enhance aerobic glycolysis by directly activating the transcription of glycolytic genes (Dang, 2012; Qing et al., 2010). Oncogenic c-Myc has been linked to increased glutaminolysis through coordinated transcriptional and posttranscriptional programs (Gao et al., 2009; Wise et al., 2008). For instance, c-Myc directly activates the transcription of SLC1A5 (solute carrier family 1, member 5, also known as ASCT2) and SLC38A5 (solute carrier family 38, member 5, also known as SN2), which encode two highly efficient Gln transporters (Wise et al., 2008). Furthermore, c-Myc indirectly stimulates glutaminase (encoded by GLS) expression through suppression of microRNAs miR-23a/b, which specifically target the GLS 3′ UTR and inhibit GLS mRNA translation (Gao et al., 2009). Thus, c-Myc coordinates the expression of multiple genes necessary for Gln metabolism, replenishing the TCA cycle and supplying essential intermediates for nucleic acid, amino acid, and glutathione biosynthesis.
MYCN amplification is strongly correlated with advanced stage neuroblastoma (Maris, 2010) and is used worldwide for patient risk-classification. MYCN-amplified neuroblastomas are frequently resistant to conventional therapeutic drugs due in part to defects in death-inducing signaling complex (DISC) components such as Caspase 8 (encoded by CASP8) (Teitz et al., 2000) and altered ABC transporter expression (Porro et al., 2010). Identification of effective druggable targets in neuroblastoma is therefore critical. Systemic inhibition in a Ras-mediated lung adenocarcinoma mouse model demonstrated the benefit of targeting MYC (a recipient of multiple oncogenic signals) as an efficient and tumor-specific cancer therapy (Soucek et al., 2008). However, small molecules targeting non-kinase oncoproteins like MYC have not been achieved thus far. MYCN-amplified neuroblastomas are highly vascular (Maris, 2010), making anti-angiogenic agents (e.g. those targeting VEGF) promising therapeutic approaches. Nevertheless, recent studies demonstrated that anti-angiogenics significantly increase invasion and metastasis in multiple tumor models (Ebos et al., 2009; Paez-Ribes et al., 2009), somewhat decreasing enthusiasm for this treatment of cancers like neuroblastoma.
Myc-mediated metabolic reprogramming triggers cellular dependency on exogenous Gln to sustain viability. Consequently, Gln depletion kills transformed cells in a Myc-dependent manner (Le et al., 2012; Wise et al., 2008; Yuneva et al., 2007). However, whether genes involved in Gln metabolism are deregulated in primary human tumors, and the mechanisms responsible for Gln deprivation-mediated cell death remain largely unknown. We therefore determined whether these cell death pathway(s) and/or alterations of Gln metabolism can be therapeutically exploited.
RESULTS
Gln Depletion Induces Neuroblastoma Cell Death in an N-Myc Dependent Manner
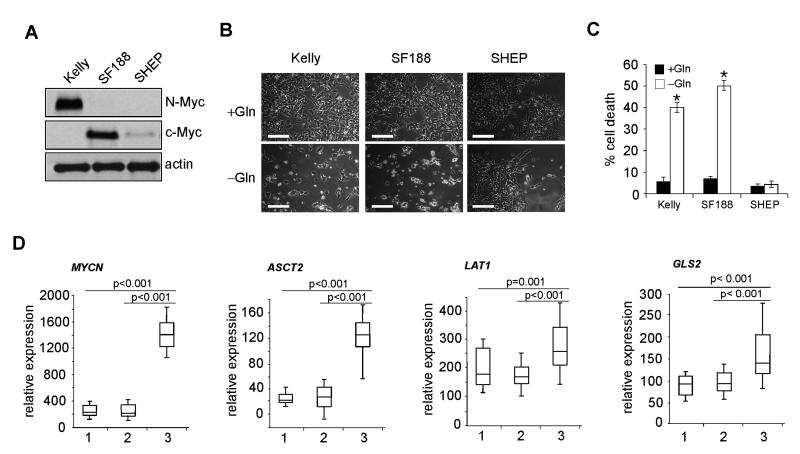
To evaluate the impact of MYC on cell death upon Gln starvation, we analyzed human tumor cell lines overexpressing N-Myc (Kelly, from MYCN-amplified neuroblastoma) or c-Myc (SF188, from glioma). SHEP cells (from MYCN non-amplified neuroblastoma) with low N- or c-Myc levels were used for comparison (Figure 1A). We then subjected these cells to Gln deprivation for 48 hr. As expected, Gln starvation induced significant cell death in Kelly and SF188 cells associated with Myc overexpression (Figures 1B and 1C), while SHEP cells exhibited minimal cell death under similar conditions (Figures 1B and 1C). Furthermore, direct N-Myc inhibition by specific siRNAs in Kelly cells alleviated cell death upon Gln loss (Figures S1A and S1B), confirming the essential role of Myc in this process. When tested in cultured cell lines, elevated c-Myc enhanced the transcription of genes involved in glutaminolysis (Wise et al., 2008). However, whether these in vitro observations are representative of what occurs in human tumors remained unknown. For this purpose, we evaluated 80 primary neuroblastomas of diverse risk-class with and without MYCN amplification. Of note, ASCT2, LAT1 (or SLC7A5, solute carrier family 7, member 5), LAT2 (or SLC7A6, solute carrier family 7, member 6), GLS2 (glutaminase 2), GOT2 (glutamate-oxaloacetate transaminase 2), and SLC1A7 (solute carrier family 1 glutamate transporter, member 7) mRNAs were significantly elevated in MYCN-amplified tumors when compared with non-amplified tumors (Figures 1D, Figure S1C, and data not shown). In contrast, GLS (also known as GLS1, glutaminase 1), GLUD1 (glutamate dehydrogenase), and SN1 (solute carrier family 38, member 3) expression was largely unchanged or even reduced (Figure S1C and data not shown), suggesting they are not N-Myc targets. Taken together, these results suggest that ASCT2, LAT1, LAT2, GLS2, GOT2 and SLC1A7 play a critical role in the regulation of Gln metabolism in MYCN-amplified neuroblastomas.
Figure 1. Gln Starvation Triggers Tumor Cell Death in a MYC-dependent Manner.
(A) Western blot analysis of Myc (N-Myc and c-Myc) expression in Kelly, SF188 and SHEP cells. β-actin was used as a loading control.
(B) Representative images of Kelly, SF188 and SHEP cells in the presence or absence of Gln for 48 hr. Scale bars represent 50 μm.
(C) Quantification of cell death by PI-Annexin V staining. Data are presented as an average of triplicates. Error bars represent standard deviation. *p< 0.001.
(D) Relative expression of MYCN, ASCT2, LAT1, and GLS2 in primary neuroblastoma tumors. 1: low-risk group (28 tumors); 2: MYCN non-amplified, high-risk group (32 tumors); 3: MYCN-amplified, high-risk group (20 tumors); defined using International Neuroblastoma Risk Group (INRG) criteria (Maris, 2010). Data are presented as box plots: the box represents 25th percentile through the 75th percentile (the line through the box denotes the median), and the whiskers above and below extend to the 90th and 10th percentiles respectively.
See also Figure S1.
Gln Depletion Induces Tumor Cell Death Largely Dependent on Bax and Caspase Activities
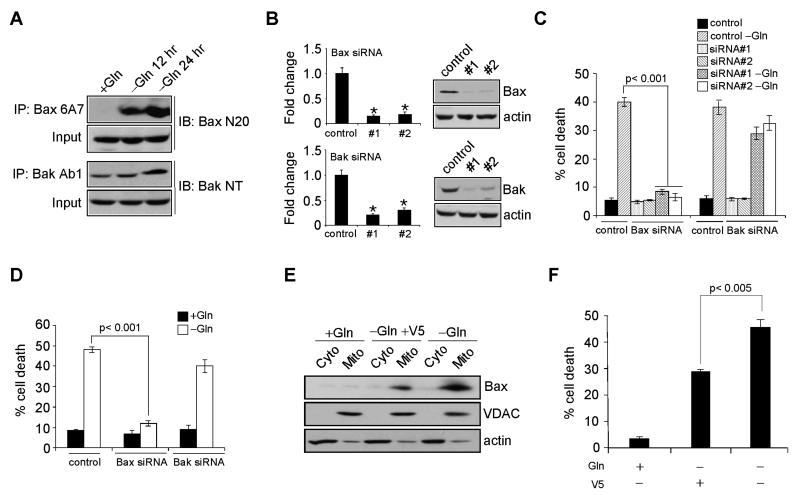
Studies using gene-targeted mice revealed that cellular apoptosis is frequently governed by two proapoptotic Bcl-2 family proteins, Bax and Bak (Wei et al., 2001). Bax and Bak exert similar function in most stress-induced apoptotic pathways and can functionally substitute for each other. To determine whether Gln deprivation induces apoptosis via Bax and/or Bak, we first examined Bax and Bak activation in Kelly cells using conformation-specific antibodies. Gln starvation activated both Bax and, to a lesser extent, Bak, as the 6A7 antibody for Bax and Ab1 antibody for Bak efficiently immunoprecipitated these proteins in comparison to non-starved cells (Figure 2A). Bax and Bak were then depleted by two distinct siRNAs each (Figure 2B), and cells were subjected to Gln starvation. Surprisingly, inhibition of Bax, but not Bak, significantly decreased N-Myc overexpressing Kelly cell death upon Gln deprivation (Figure 2C). This was recapitulated in c-Myc overexpressing SF188 cells (Figure 2D), suggesting a general mechanism predominantly dependent on Bax for Myc-mediated cell death during Gln limitation. Of note, previous data demonstrated Bax depletion cooperates with c-Myc during murine lymphomagenesis (Eischen et al., 2001). Since Bax activates downstream caspase-dependent and/or -independent pathways, we treated Kelly cells with escalating doses of z-VAD-fmk (z-VAD), a broad-spectrum caspase inhibitor, and found 50 μM of z-VAD completely inhibited Caspase-3 activation and Kelly cell death (data not shown). Therefore, a caspase-dependent pathway regulates Myc-mediated cell death during Gln starvation. Consistent with this finding, blocking Bax mitochondrial translocation using the cell membrane-permeable V5 peptide inhibitor (Sawada et al., 2003) (Figure 2E) partially rescued Gln deprivation-induced cell death (Figure 2F). Therefore, MYCN-amplified neuroblastomas provide an attractive model for studying the molecular mechanisms underlying the connection between Myc overexpression and Gln addiction.
Figure 2. Gln Depletion Triggers Bax-dependent, but Bak-independent Cell Death.
(A) Immunoprecipitation of active Bax and Bak using conformation-specific antibodies in Kelly cells subjected to Gln starvation at indicated time points.
(B) Protein levels of Bax and Bak upon siRNA knockdown in Kelly cells. Bar graphs show the quantification results. *p< 0.001.
(C-D) Viability of Kelly cells (C) and SF188 cells (D) upon indicated siRNA knockdown were examined by PI-Annexin V staining after 48 hr Gln starvation. Data are shown as an average of triplicates.
(E) Immunoblotting of Bax in the cytosolic (Cyto) and mitochondrial (Mito) fraction of NLF cells treated with V5 peptide in the presence or absence of Gln. VDAC and actin antibodies were used as controls for mitochondrial and cytosolic protein purification.
(F) Gln starvation-induced cell death with or without V5 peptide treatment was examined by PI-Annexin V staining. Shown are average results from three experiments.
All error bars represent standard deviation.
PUMA, NOXA, and TRB3 Promote Gln Deprivation-mediated Cell Death
Unlike Bak, Bax can be sequestered in the cytosol by specific interaction with the non-Bcl2 family protein Ku70 in an acetylation-dependent fashion (Figure S2A). Cellular stress causes Ku70 acetylation by CBP and PCAF, leading to subsequent Bax dissociation, Bax activation, and apoptosis (Sawada et al., 2003). We examined whether Gln deprivation affects Ku70 acetylation and Bax association. Kelly cell starvation did not result in detectable changes in either Ku70 acetylation or Ku70-Bax interaction, while treatment with the broad deacetylase inhibitor TSA increased Ku70 acetylation and disrupted Ku70-Bax binding (Figure S2B). Therefore, we concluded that Ku70 is not involved in Bax activation upon Gln withdrawal.
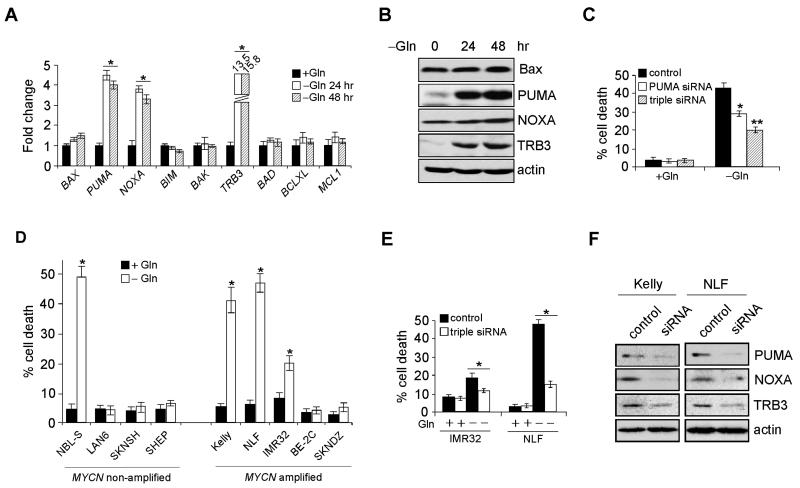
We then investigated expression of multiple factors inducing apoptosis upstream of Bax, and observed in Gln starved Kelly cells a significant increase in PUMA, NOXA and TRB3 (tribbles homolog 3) mRNA (Figure 3A) and protein (Figure 3B). TRB3 is a mammalian homologue of the Drosophila protein “tribbles”, a pseudo-kinase protein frequently induced by endoplasmic reticulum (ER) stress (Ohoka et al., 2005). Of note, TRB3 is activated by the ATF4/CHOP pathway, and in turn represses CHOP, possibly via direct CHOP interaction, blocking CHOP coactivator recruitment. Depletion of PUMA alone by specific siRNAs significantly inhibited Myc-mediated Kelly cell death upon Gln deprivation, and a triple depletion of PUMA, NOXA and TRB3 further reduced apoptosis (Figure 3C). We then extended our study to a number of additional neuroblastoma cell lines. A total of seventeen neuroblastoma lines, including nine MYCN-amplified and eight MYCN non-amplified lines were subjected to Gln starvation. Of note, 6/9 MYCN-amplified lines, but only one MYCN non-amplified line exhibited Gln dependence (Figure 3D and data not shown). The single outlier for Gln dependence absent MYCN amplification was NBL-S, a cell line with markedly deregulated N-Myc activity through protein stabilization (Liu et al., 2008). We examined N-Myc expression in two MYCN-amplified lines (NLF and IMR32) and NBL-S, and corroborated similar levels of deregulated protein (Figure S2C and Qing et al., 2010), underscoring the association between deregulated MYCN and Gln dependence. Moreover, significant PUMA, NOXA and TRB3 induction was observed in all three cell lines when Gln was absent (Figure S2D), and a combined inhibition of PUMA, NOXA and TRB3 reversed IMR32 and NLF cell death upon Gln removal (Figure 3E). Efficient siRNA-mediated depletion of PUMA, NOXA and TRB3 protein in both Kelly and NLF cells was confirmed by western blot (Figure 3F). Taken together, these results demonstrate that PUMA, NOXA and TRB3 work in concert to regulate Gln deprivation-mediated cell death in MYCN-amplified neuroblastomas.
Figure 3. PUMA, NOXA and TRB3, but Not Ku70, are Involved in Gln Deprivation-mediated Cell Death.
(A) RT-PCR analysis of genes involved in apoptosis. Data are shown as an average of triplicates.
(B) Western blot analysis of indicated protein levels in Gln-starved Kelly cells.
(C) Viability of Kelly cells transfected with indicated siRNAs in the presence or absence of Gln was analyzed by AnnexinV-PI staining. Data are shown as an average of triplicates.
(D) Examination of Gln starvation-induced cell death in five MYCN-amplified, and four MYCN non-amplified neuroblastoma cell lines. Cell death was measured by Annexin V-PI staining.
(E) Viability of IMR32 or NLF cells transfected with siRNAs targeting PUMA, NOXA, and TRB3 in the presence or absence of Gln was analyzed by AnnexinV-PI staining. Data are shown as an average of triplicates.
(F) Western blots confirming the effect of siRNA knockdown.
All error bars represent standard deviation. *p< 0.01. **p< 0.005. See also Figure S2.
PUMA, NOXA, and TRB3 are Regulated by a p53-independent, ATF4-dependent Mechanism
PUMA potently induces apoptosis and its overexpression is sufficient to cause cell death in numerous cell types, as it directly binds and antagonizes many anti-apoptotic Bcl2 family members, resulting in mitochondrial dysfunction and Bax activation (Letai et al., 2002). PUMA is normally expressed at low levels, but rapidly induced by p53 and other factors (Yu et al., 2008). Since Gln loss in Kelly cells specifically activates PUMA and NOXA (well-known p53 targets), we inhibited p53 by siRNA (Figure 4A), and then subjected Kelly cells to Gln starvation. Interestingly, p53 inhibition had no effect on PUMA, NOXA and TRB3 induction (Figure 4B), while the expression of another p53 target, TIGAR (p53-induced glycolysis and apoptosis regulator) (Bensaad et al., 2006), was significantly inhibited under the same conditions (Figure 4B). In addition to p53, other transcription factors have been implicated in context-dependent PUMA and/or NOXA induction, including Myc, FOXO3, SP1, and E2F1. Each factor was inhibited by specific siRNAs without a detectable effect on PUMA and NOXA activation (data not shown).
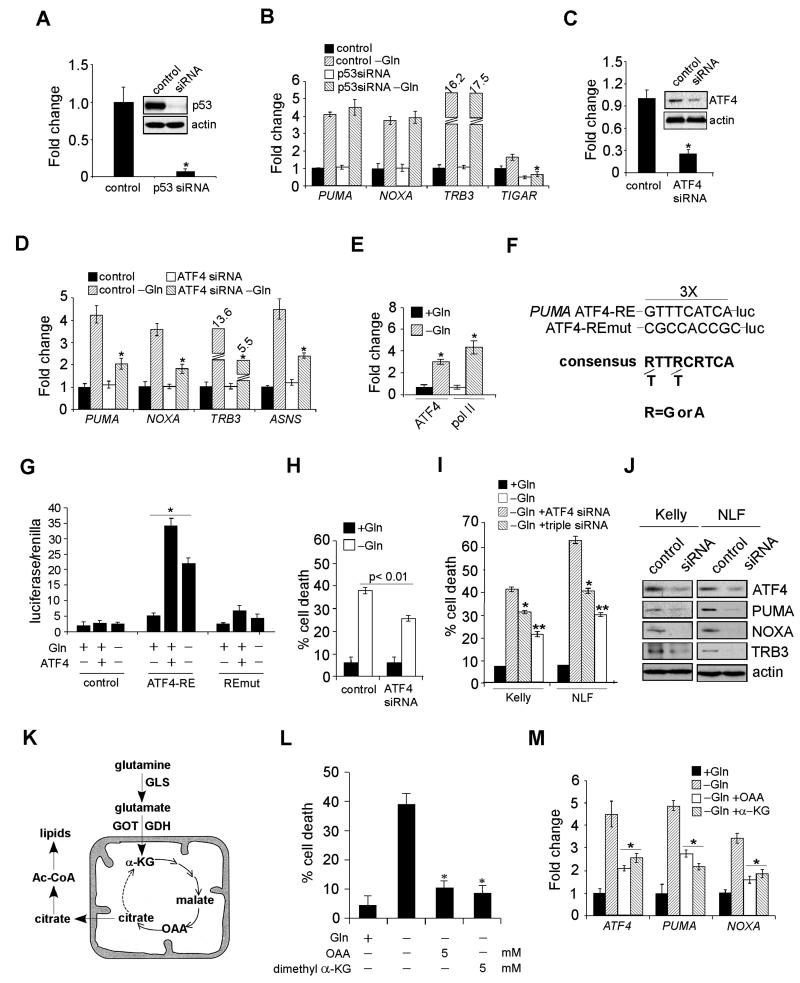
Figure 4. ATF4, but Not p53, is Responsible for PUMA, NOXA, and TRB3 Activation.
(A) Protein levels of p53 with or without siRNA knockdown.
(B) mRNA expression of indicated genes was examined by RT-PCR in Kelly cells transfected with a control siRNA or p53 siRNA in the presence or absence of Gln. Data are shown as an average of triplicates.
(C) Protein levels of ATF4 in Kelly cells with or without siRNA knockdown.
(D) Indicated gene expression was quantitated by RT-PCR in Kelly cells transfected with a control or ATF4 siRNA in the presence or absence of Gln. Data are shown as an average of triplicates.
(E) Specific chromatin binding of ATF4 evaluated by ChIP assay. Recruitment of Pol II was also assessed.
(F) Schematic representation of the consensus ATF4-binding site, the ATF4 response element (ATF4-RE) within the PUMA promoter and its mutant (ATF4-REmut).
(G) Luciferase assay was performed using control, ATF4-RE, and ATF4-REmut constructs with or without exogenous ATF4 expression or Gln starvation. Data are shown as an average of triplicates.
(H) Viability of Kelly cells transfected with a control or ATF4 siRNA in the presence or absence of Gln was examined by PI-Annexin V staining. Data are shown as an average of triplicates.
(I) Evaluation of Kelly and NLF cell death upon siRNA knockdown of ATF4 (using independent siRNAs from [C] and [H]), or a combination of PUMA, NOXA, and TRB3, in the absence of Gln.
(J) Western blots confirming the effect of siRNA knockdown in (I).
(K) Diagram depicting Gln metabolism in the TCA cycle. See text for more details.
(L) Evaluation of Gln-starved Kelly cell death upon the addition of OAA or α-KG.
(M) RT-PCR analysis of indicated genes in Kelly cells cultured in Gln-free or replete medium, or Gln-free medium supplemented with OAA or α-KG. Data are shown as an average of triplicates.
All error bars represent standard deviation. *p< 0.01. **p< 0.005. See also Figure S3.
To identify PUMA regulatory factor(s), we performed bioinformatic analyses, and identified a conserved ATF4 binding site within the PUMA promoter region (Figure S3A). ATF4 knockdown (Figure 4C) potently inhibited PUMA, NOXA and TRB3 activation in Gln-starved Kelly, IMR32, and NLF neuroblastoma cells (Figures 4D and S3B). Chromatin immunoprecipitation (ChIP) analysis demonstrated that significant ATF4 pools were recruited to the PUMA promoter along with enhanced RNA polymerase II (Pol II) occupancy in Gln-starved Kelly cells (Figure 4E). We created a luciferase reporter construct using a pGL3 plasmid containing the putative ATF4-binding site (in triplicate) 5′ of the SV40 promoter (ATF4-RE/luc, Figure 4F). As a control, we generated an additional construct harboring mutations in the ATF4-binding motif (ATF4-REmut/luc, Figure 4F). Compared with empty pGL3 vector and ATF4-REmut/luc, ATF4-RE luciferase activity was significantly increased by exogenous ATF4 or Gln starvation (Figure 4G). Interestingly, we identified a different ATF4 response element within the NOXA promoter (Figure S3C). Both ChIP and luciferase assays confirmed ATF4 directly activated NOXA transcription through binding to this site (Figures S3D-F). Furthermore, two distinct ATF4 siRNAs significantly reduced Gln starvation-induced death in Kelly and NLF cells (Figures 4H and 4I), which was further alleviated by combinatorial inhibition of PUMA, NOXA and TRB3 (Figures 4I and 4J). To control for off-target effects, independent siRNAs for PUMA, NOXA, and TRB3 were used in these assays (Figures 4I and 4J, compared to Figures 3C, 3E and 3F). In contrast, ATF4 depletion in SF188 glioma cells had little effect on Gln starvation-induced cell death (Figures S3G and S3H), possibly due to a different tumorigenic background. A recent study indicated that ATF4 directly activates CHOP expression, which subsequently stimulates PUMA and apoptosis (Galehdar et al., 2010). However, CHOP inhibition in Gln-starved Kelly cells failed to impact PUMA and NOXA activation (Figure S3I), or concomitant cell death (Figure S3J), suggesting CHOP was not involved in this process. Taken together, we determined that ATF4 functions as a transcription factor directly regulating PUMA,NOXA, and TRB3 expression in MYCN transformed cells.
Gln can be converted by two deamination reactions into α-ketoglutarate (α-KG) to replenish a functional TCA cycle (Figure 4K). Because the TCA cycle provides a “hub” for multiple metabolic pathways, loss of cycle intermediates could have various consequences, including cell death (DeBerardinis et al., 2008). Indeed, oxaloacetate (OAA), a cell membrane-permeable TCA cycle intermediate, or dimethyl α-KG, a cell membrane-permeable α-KG analog, prevented apoptosis of Gln-starved cells (Figure 4L). We reasoned that if ATF4 mediated PUMA stimulation is critical to cell death, then, OAA or α-KG should decrease PUMA and NOXA expression when Gln is absent. As expected, supplement of OAA or α-KG (4 mM) efficiently inhibited PUMA and NOXA induction (Figure 4M). Of note, both PUMA and NOXA inhibition correlated with that of ATF4 (Figure 4M), raising the possibility that enhanced ATF4 transcription occurs following Gln withdrawal. Moreover, Kelly cell treatment with actinomycin D, which abrogates ATF4 transcriptional induction, significantly reduced ATF4 protein levels when Gln was absent (Figure S3K). ATF4 transcriptional control is largely unknown, except for stress-regulated transcription factor p8 which induces ATF4 during cannabinoid-mediated apoptosis (Carracedo et al., 2006). Although p8 expression was significantly increased at both the mRNA and protein levels in Gln starved cells, p8 inhibition had no detectable effect on ATF4 (Figure S3L). Since ATF4 is activated by Gln starvation, we investigated whether ATF4 is regulated by signaling pathways promoting Gln metabolism, e.g. AMPK-mTOR (Nicklin et al., 2009) or Myc (Gao et al., 2009; Wise et al., 2008; Yuneva et al., 2007). However, siRNA knockdown of AMPKα1 and α2 catalytic subunits or N-Myc had no significant impact on ATF4 mRNA levels (Figure S3M). Finally, bioinformatic analysis of ATF4 regulatory regions identified ten potential transcription factors involved in ATF4 induction. However, their RNAi-mediated inhibition failed to decrease ATF4 induction in Gln-starved cells (data not shown), suggesting other unidentified factor(s) are involved in this process.
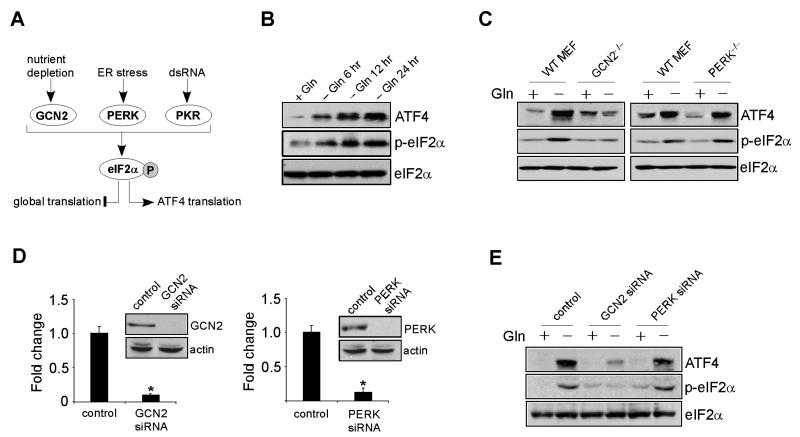
ATF4 Translation Depends on the GCN2 kinase, but not PERK, in the Absence of Gln
Translation of ATF4 mRNA is also regulated by stress signals (Kilberg et al., 2009), transduced by multiple eIF2α kinases, including GCN2 (general control nonrepressed-2), PERK (protein kinase-like ER kinase) and PKR (double-strand RNA activated protein kinase). Amino acid deprivation (usually essential ones like leucine and lysine) triggers the amino acid response (AAR) signal transduction pathway via GCN2, while ER disruption and viral infection activate PERK and PKR, respectively. Mechanistically, these kinases promote eIF2α phosphorylation at Ser 51. Phospho-eIF2α then binds eIF2B in a non-functional complex that suppresses global protein synthesis, while promoting increased translation of select mRNAs, including ATF4 (Figure 5A). We first examined eIF2α phosphorylation in Kelly cells at different time points. As expected, Gln starvation gradually elevated eIF2α phosphorylation concomitant with a dramatic increase in total ATF4 protein (Figure 5B), suggesting that ATF4 translation is also enhanced in Gln-starved Kelly cells. Of note, this stress response is tightly linked to MYC overexpression. Although ATF4, PUMA, NOXA, and TRB3 transcripts were somewhat increased in three MYCN non-amplified cell lines (SHEP, SKNAS, and EBC1) upon Gln depletion (Figure S4A), their protein levels did not change in all cell lines tested (Figure S4B). Consistent with this result, MYCN non-amplified neuroblastoma cells failed to engage the eIF2α-ATF4 pathway, as shown by similar levels of p-eIF2α in both conditions (Figure S4B).
Figure 5. GCN2-eIF2α Pathway Activates ATF4 Translation in the Absence of Gln.
(A) Diagram depicting pathways involved in selective ATF4 translation. See text for more details.
(B) Western blot analysis of ATF4 and phosphorylated eIF2α (p-eIF2α) in Kelly cells, in the presence or absence of Gln. Total eIF2α was used as a loading control.
(C) Immunoblotting of ATF4 and phosphorylated eIF2α (p-eIF2α) in wild-type (WT) MEFs and GCN2- or PERK-deficient (KO) MEFs, with or without Gln starvation.
(D) Protein levels of GCN2 and PERK upon indicated siRNA knockdown in Kelly cells. Error bars represent standard deviation. *p< 0.005.
(E) Protein levels of ATF4 and phosphorylated eIF2α (p-eIF2α) were detected in Kelly cells transfected with a control or GCN2 or PERK siRNA, in the presence or absence of Gln.
See also Figure S4.
One highly conserved function of MYC family members across species is to activate ribosome biogenesis and mRNA translation essential for cell growth (Dang, 2012). Gln is utilized in O-linked β-N-acetylglucosamine (O-GlcNAc) modification of nascent polypeptides, a posttranslational modification critical for transcription factor function (Butkinaree et al., 2010). Theoretically, Gln deprivation results in protein mis-folding and ER accumulation, leading to ER stress pathway activation. In addition, oncogenic Myc converts the nonessential amino acid Gln into an essential one triggering cellular addiction to Gln (Wise et al., 2008; Yuneva et al., 2007), and AAR stress pathway activation. Indeed, the mRNA levels of ER stress-inducing genes ASNS (asparagine synthetase), GRP78 (glucose-regulated protein 78), and XBP-1 (X-box binding protein 1) were significantly increased in Gln-starved Kelly cells (data not shown). Whereas glucose starvation in MEFs induces ATF4 translation via both GCN2 and PERK, Gln depletion affects ATF4 levels in a GCN2-dependent, but PERK-independent manner (Ye et al., 2010). To confirm this result, wild-type (WT), Gcn2−/−, and Perk−/− MEFs were examined. Indeed, lack of GCN2, but not PERK, abolished ATF4 translation in response to Gln depletion, as increased ATF4 abundance was selectively lost in Gcn2−/−, but not Perk−/− cells (Figure 5C). In parallel, eIF2α phosphorylation was exclusively abrogated in Gcn2−/− fibroblasts (Figure 5C). We then depleted GCN2 and PERK in Kelly cells (Figure 5D), and demonstrated that GCN2 inhibition selectively abrogated eIF2α phosphorylation in association with decreased ATF4 translation (Figure 5E).
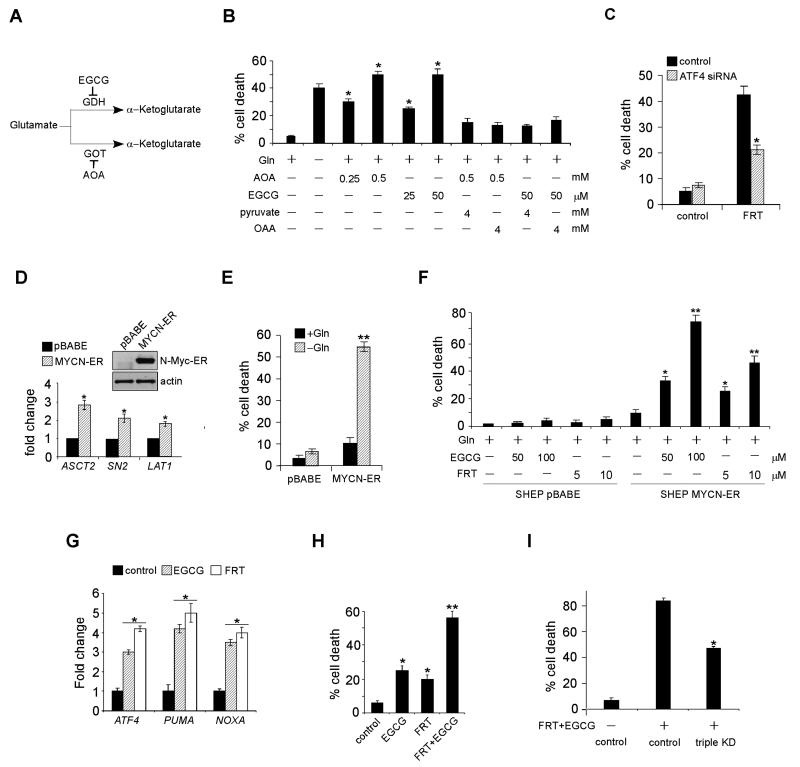
Either Pharmacological Intervention of Gln Metabolism or ATF4 Stimulation Induces Cell Death in vitro and Inhibits Tumorigenesis in vivo
Since Gln is critical for maintaining TCA cycle homeostasis and providing precursors for protein, nucleotide and lipid synthesis, we tested if pharmacological inhibition of Gln metabolism induces apoptosis. Gln is metabolized to α-ketoglutarate (α-KG), a critical TCA cycle intermediate, through two deamination steps: glutaminase converts Gln to glutamate and then glutamate dehydrogenase (GDH) or transaminases transfer the amino group to α-keto acids to generate amino acids such as alanine and aspartate (Figure 6A). Both amino oxyacetate (AOA), a chemical inhibitor of glutamate-dependent transaminases (Wise et al., 2008), and epigallocatechin gallate (EGCG), which inhibits GDH (Li et al., 2007), induced cell death in a dose-dependent manner in Kelly cells (Figure 6B). Both AOA and EGCG effectively induced apoptosis at comparable levels to that resulting from Gln depletion. The effect of AOA and EGCG was specific to Gln metabolism, as supplementation of OAA or cell permeable pyruvate significantly inhibited the toxic effects of both chemical inhibitors (Figure 6B). To highlight the importance of enzymes involved in glutaminolysis in maintaining cell viability, GDH inhibition was also achieved by siRNA knockdown (Figure S5A). Indeed, GLUD1 (encoding GDH) mRNA degradation (Figure S5A) triggered dramatic cell death independent of Gln status, while control cells underwent apoptosis only in the absence of Gln (Figure S5B).
Figure 6. Pharmacological Inhibition of Gln Metabolism or ATF4 Hyperactivation Triggers Dramatic Cell Death In Vitro.
(A) Diagram showing enzymes involved in glutamate metabolism. See text for details.
(B) Kelly cell death was examined by PI-Annexin V staining upon EGCG or AOA treatment in the presence of Gln. Where indicated, various metabolites were also supplemented.
(C) Kelly cell death was measured upon fenretinide treatment, with or without ATF4 knockdown.
(D) Induction of MYCN-ER and its target genes in SHEP cells. Data are shown as an average of triplicates.
(E) SHEP cell death with or without MYCN-ER induction was quantified by PI-Annexin V staining, in the presence or absence of Gln.
(F) SHEP cells with or without MYCN-ER induction were cultured in Gln-replete medium, and their viability was measured upon indicated drug treatments. Data are shown as an average of triplicates.
(G) RT-PCR analysis of ATF4, PUMA, and NOXA mRNAs in Kelly cells treated without or with EGCG (50 μM) or fenretinide (5 μM). Data are shown as an average of triplicates.
(H) Kelly cell death was quantitated by Annexin V-PI staining upon different drug treatments. Where indicated, 25 μM EGCG and/or 3 μM fenretinide were used, and data are shown as an average of triplicates.
(I) Evaluation of Kelly cell death upon PUMA/NOXA/TRB3 triple knockdown, in the presence or absence of fenretinide/EGCG. Experiments were repeated three times.
All error bars represent standard deviation. *p< 0.01. **p< 0.005. See also Figure S5.
Since Gln starvation resulted in ATF4 accumulation, PUMA/NOXA/TRB3 induction, and apoptosis, we evaluated the effect of ATF4 agonists. We chose fenretinide (FRT, also known as “4-hydroxyphenyl-retinamide”), a chemotherapeutic agent, for the following reasons: 1) preliminary data obtained from clinical trials demonstrate that FRT is well tolerated in humans (Villablanca et al., 2006); and 2) FRT induces a signaling cascade activating ATF4 and cell death (Corazzari et al., 2007). Indeed, Kelly or NLF cells incubated with FRT exhibited increased eIF2α phosphorylation (Figure S5C), indicating that FRT treatment leads to ER stress in neuroblastoma cells, promoting ATF4 translation. In addition, FRT administration mimics the effect of Gln withdrawal in a number of other ways, including resistance to Bak inhibition (Figure S5D), and partial rescue by ATF4 knockdown (Figure 6C), reinforcing the idea that FRT functions through the ATF4 pathway to induce cell death in neuroblastoma.
To confirm whether the effects of EGCG and FRT are dependent on N-Myc overexpression, we stably expressed MYCN-ER in SHEP, a MYCN non-amplified cell line exhibiting undetectable N-Myc expression (Figure 1A). Induction of MYCN-ER by 4-Hydroxytamoxifen in SHEP cells significantly increased the expression of genes involved in Gln metabolism (Figure 6D) and sensitized these cells to Gln deprivation (Figure 6E). More importantly, both EGCG and FRT induced dramatic cell death in N-Myc overexpressing SHEP cells (SHEP MYCN-ER) while causing minimal cell death in control SHEP cells (Figure 6F), suggesting both chemicals constitute a synthetic lethal interaction with MYC transformation in the context of neuroblastoma.
In addition, administration of either EGCG or FRT to Kelly cells significantly elevated ATF4, PUMA and NOXA expression (Figure 6G), suggesting that either direct inhibition of Gln metabolism or activation of upstream signaling achieved a similar phenotype to Gln starvation. When combined with EGCG, FRT treatment resulted in enhanced cell death (Figure 6H), indicating their combination could achieve anti-tumor activity without significant toxicity. Interestingly, other glutaminolysis enzymes, especially those up-regulated in high-risk neuroblastomas (Figures 1D and S1C), may also become feasible drug targets, given that they underlie neuroblastoma Gln addiction (Figures S5E and S5F). Finally, to highlight the importance of PUMA, NOXA, and TRB3 as the major cell death mediators upon drug treatment, we constructed a PUMA/NOXA/TRB3 stable knockdown variant of the NLF cell line. In contrast to control cells, NLF cells with PUMA, NOXA, and TRB3 triple inhibition exhibited strong resistance to Gln starvation or FRT/EGCG treatment (Figures S5G, S5H and 6I).
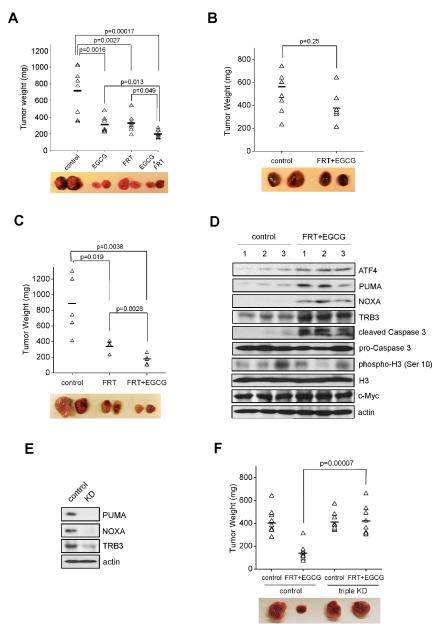
Based on these in vitro results, there appears to be a functional interplay between Gln metabolism, ATF4 activity and the Myc-driven oncogenic phenotype of MYCN-amplified neuroblastomas, which can be pharmacologically targeted by FRT and/or EGCG. However, whether these signaling events occur in vivo, or whether similar therapeutic approaches can be applied to other tumors remained to be determined. To investigate this issue, we established subcutaneous xenografts in nude mice using Kelly cells. Mice with palpable tumors were randomized into 4 groups receiving different drug treatments. Consistent with the in vitro findings, administration of EGCG or FRT significantly suppressed tumor progression, and combined treatment resulted in more marked tumor inhibition (Figure 7A). SKNAS, a MYCN non-amplified neuroblastoma cell line, was found to be highly resistant to FRT and EGCG (Figure 7B). Xenograft assays were performed using P493B Burkitt’s lymphoma cells that overexpress c-Myc, and are addicted to Gln (Figures S6A and S6B). P493B tumors were also sensitive to FRT and/or EGCG administration, to a similar extent as Kelly xenografts (Figure 7C). Importantly, P493B tumors treated with FRT/EGCG exhibited increased expression of ATF4, PUMA, NOXA, and TRB3 (Figure 7D), indicating that the ATF4-PUMA/NOXA/TRB3 pathway was activated by these drugs in xenograft lymphomas. In addition, we generated PUMA/NOXA/TRB3 triple knockdown P493B cells (Figure 7E), and found that P493 cells with PUMA/NOXA/TRB3 depletion exhibited substantial resistance to FRT/EGCG treatments in vivo (Figure 7F), as did PUMA/NOXA/TRB3-depleted NLF cells (Figure S6C). We also probed responses to FRT, AOA, EGCG or Gln deprivation in A549 lung cancer cells driven by an oncogenic KRAS mutation. Interestingly, A549 cells were resistant to all of these conditions (Figure S6D), and no significant increases in ATF4, PUMA, NOXA, and TRB3 protein levels were observed (Figure S6E). Moreover, FRT/EGCG treatment failed to inhibit A549-initiated xenograft tumor growth (Figure S6F), further demonstrating that ATF4-regulated Gln dependence is tightly linked to MYC overexpression.
Figure 7. Pharmacological Intervention of Gln Metabolism or ATF4 Stimulation Significantly Inhibits MYC-mediated Xenograft Tumor Growth.
(A) Xenograft tumor growth assay was performed using Kelly cells with EGCG and/or fenretinide administration. Representative pictures of subcutaneous tumors under different treatments are shown.
(B-C) SKNAS cells (B) and P493B lymphoma cells overexpressing c-Myc (C) were subjected to the same xenograft experiments as performed in (A).
(D) Western blot analysis of P493B-inititated xenograft tumor lysates for indicated proteins.
(E) ShRNA viruses targeting PUMA, NOXA, and TRB3 were transduced into P493B cells and knockdown efficiencies were evaluated by Western blots.
(F) Xenografts of P493B cells with or without PUMA/NOXA/TRB3 triple knock-down in the presence or absence of fenretinide/EGCG.
The horizontal lines in (A-C) and (F) represent the average tumor weights for each group. See also Figure S6.
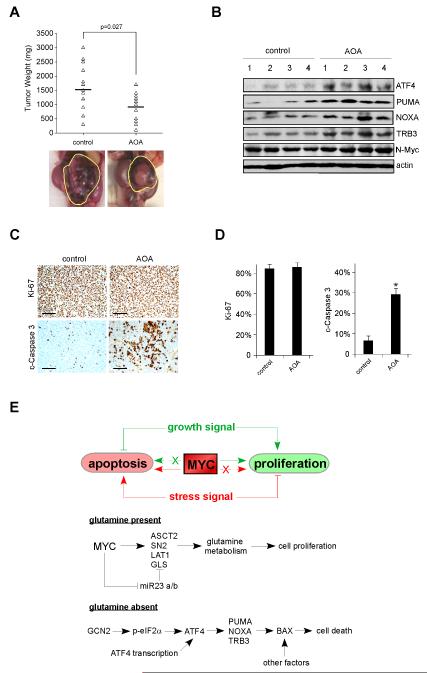
Finally, we tested this approach using the TH-MYCN transgenic mouse model in which spontaneous neuroblastomas arise in autochthonous tumor sites due to enforced MYCN expression in neural crest tissues. Homozygous TH-MYCN mice were treated with AOA or vehicle control at the time a palpable tumor was documented. As a result, AOA therapy led to inhibition of tumor growth by comparison to control mice (Figure 8A), and tumors treated with AOA exhibited induction of ATF4, PUMA, NOXA, and TRB3 (Figure 8B). The anti-tumor effects of these drugs (AOA/FRT/EGCG) are not due to decreased tumor cell proliferation (evaluated by phosphorylated H3 immunoblotting in Figure 7D or Ki-67 staining in Figures 8C and 8D) or MYC expression (Figures 7D, 8B, S7A-C), but instead due to significantly increased intratumoral apoptosis as quantified by cleaved Caspase 3 levels (Figures 7D, 8C and 8D). All of these results support the clinical potential of glutaminolysis inhibitors as cancer therapeutics against MYC driven tumors.
Figure 8. Transaminase Inhibitor Amino Oxyacetate (AOA) Reduces Autochthonous Neuroblastoma Growth in the TH-MYCN Transgenic Mouse Model.
(A) Tumor bearing homozygous TH-MYCN mice were i.p. injected daily with PBS or 10 mg/kg AOA as described in the Experimental Procedures. 8 Days later, tumors were isolated and weighed. The horizontal lines represent the average tumor weights for each group. Pictures shown are of representative neuroblastomas.
(B) Tumors harvested from homozygous TH-MYCN mice as described in (A) were lysed and subjected to Western blot analysis using indicated antibodies.
(C) Ki-67 and cleaved Caspase 3 (c-Caspase 3) staining were performed on paraffin-embedded tumor tissue sections derived from (A). Representative staining micrographs were shown. Scale bars represent 100 μm.
(D) Quantification of the results in (C), n=8 and *p< 0.001. Error bars represent standard deviation.
(E) Model depicting the action of Gln in MYC-overexpressing tumors. See text for additional details.
See also Figure S7.
DISCUSSION
Cancer cells exhibit increased metabolic autonomy in comparison to normal cells, importing and metabolizing nutrients required to support their growth and proliferation (Vander Heiden et al., 2009). The important role played by bioenergetics and cellular metabolism in the process of cancer development is receiving renewed attention. In order to achieve rampant proliferation, tumor cells must duplicate their entire biomass, including nucleic acids, proteins and lipids, and assemble these components in daughter cells (DeBerardinis et al., 2008). Therefore, in addition to enhanced energy generation, tumors alter their metabolism to more efficiently generate macromolecules. In these processes, Gln plays an important role. Recently, oncogenic MYC has been shown to reprogram glutaminolysis to support biosynthetic activities through transcriptional and posttranscriptional stimulation of genes involved in Gln metabolism (Gao et al., 2009; Le et al., 2012; Wise et al., 2008). However, precisely how Gln deprivation leads to apoptosis in MYC-transformed cells was unknown. Using neuroblastoma as a model system, we identified a pathway linking ATF4 to PUMA/NOXA/TRB3 activation and cell death upon Gln starvation, and propose the model shown in Figure 8E. Our results are highly consistent with the notion that Myc functions as a double-edged sword in regulating cellular activities; that is, oncogenic Myc promotes proliferation or apoptosis depending on upstream signals and enforced dependencies. In Gln replete conditions, Myc induces genes (e.g. ASCT2, GLS1, LAT1) involved in Gln metabolism to support increased biosynthetic activities. It should be noted that Myc also regulates gene networks activating glucose metabolism, mitochondrial biogenesis and ribosome biogenesis (Dang, 2012). In concert, these lead to a robust growth phenotype. When Gln is depleted, cells initiate a distinct network including ATF4 activation. Mechanistically, two pathways mediate ATF4 stimulation upon Gln deprivation: increased transcription through a currently unknown mechanism and enhanced translation via GCN2-eIF2α. ATF4 then activates PUMA and other genes (e.g. NOXA, TRB3) involved in the execution of cell death, sensitizing tumor cells to Myc-mediated apoptosis.
Like Myc, ATF4 also exhibits a dual role in regulating cellular activities. ATF4’s typical role as a protective factor is well documented (Ameri et al., 2008). Genome-wide profiling in ATF4 wild-type and deficient MEFs revealed that ATF4 regulates amino acid metabolism and resistance to oxidative stress (Ameri et al., 2008). Indeed, Atf4−/− fibroblasts are prone to death in response to stress, including oxidative stress and amino acid deprivation. Nevertheless, numerous reports also describe a pro-death role for ATF4 in neurons (Carracedo et al., 2006; Lange et al., 2008; Ohoka et al., 2005). In contrast to fibroblasts, ATF4 is a pro-apoptotic factor in neurons both in vitro and in vivo (Lange et al., 2008). A subset of ATF4-regulated genes, including TRB3 (Ohoka et al., 2005), promote neuronal apoptosis, suggesting that context-dependent ATF4 regulation may account for the divergent phenotypes observed. ATF4 activation also results in tumor cell death under stress conditions (Ameri et al., 2008; Carracedo et al., 2006), suggesting that ATF4 agonists constitute a potential therapeutic strategy for inhibiting tumor growth. The different metabolic demands of MYCN-amplified and non-amplified neuroblastomas might explain distinct roles for ATF4 in divergent cell types. Given that ATF4 plays opposing roles (pro-survival vs pro-death) in different tumors, caution is necessary when considering ATF4 agonists or antagonists as potential therapeutics for cancer treatment.
MYC deregulation frequently occurs in human cancers, and has been estimated to contribute to at least 40% of all human cancers (Dang, 2012). In multiple models, MYC is continuously required for tumor maintenance (Shachaf et al., 2004; Soucek et al., 2008), suggesting that directly targeting Myc is an effective therapeutic strategy. However, attempts to chemically disrupt its function have met with limited success, possibly due to the inherent difficulty of inhibiting transcription factors with small molecules. Here, we demonstrate and validate an alternative, pharmacologic approach exploiting the enforced Gln addiction present in Myc-overexpressing tumor cells. Previous treatments inducing Gln deficiency or interfering with its metabolism (e.g. administration of 6-diazo-5-oxo-l-norleucine [DON] and acivicin) showed great promise in animal models (Ahluwalia et al., 1990), but were unacceptably toxic in humans and eventually abandoned. In screens for compounds suppressing Rho GTPase activation by oncogenic Dbl (for Diffuse B cell lymphoma), recent work identified a tetrahydrobenzo derivative (“no 968”), which blocks Rho GTPase-mediated transformation and tumor growth via mitochondrial glutaminase inhibition (Wang et al., 2010). Consistent with this finding, administration of BPTES, another glutaminase inhibitor, significantly decreased xenograft tumor growth initiated by c-Myc-transformed lymphoma cells (Le et al., 2012). Nevertheless, the efficacy and toxicity of either compound 968 or BPTES in treating human patients remain largely unknown. We show here that combination of FRT (4-hydroxyphenyl-retinamide) and EGCG is effective when administered both in vitro and in vivo in killing neuroblastoma and lymphoma cells. Both drugs are well tolerated in humans (Khan et al., 2008; Villablanca et al., 2006), and may have clinical utility for human cancers overexpressing the MYC oncogene. While FRT alone may not elicit robust anti-tumor responses, it could be combined with EGCG as a strategy to move forward in patient care. Moreover, the development of drugs targeting Gln metabolism in neuroblastoma should occur with some urgency, as patients with MYCN amplifications are likely to respond.
Experimental Procedures
Cell Culture
Cells were maintained in DMEM medium supplemented with 10% fetal bovine serum (FBS), 2 mM Gln, and 1× penicillin and streptomycin. To deplete Gln, cells were cultured in Gln-free DMEM medium supplemented with 10% dialyzed FBS. Where indicated, Gln was added back at a final concentration of 2 mM.
siRNA Knockdown, RNA and Protein Analysis
All the assays were performed as described in Qing et al., 2010. Details regarding the applied siRNAs, primers and antibodies are provided in Supplemental Experimental Procedures.
Cell Death Assay
Cells were harvested by combining floating cells in the medium and adherent cells detached by 0.25% trypsin, and cell pellets were washed once with cold PBS. Apoptosis was analyzed using the Annexin V-FITC Apoptosis Kit (BioVision), and data are presented as an average of triplicates.
Luciferase Reporter Assay
0.5 μg empty pGL3 luciferase vector or pGL3 expressing ATF4-RE (or indicated mutants) were transiently co-transfected in triplicate into HEK293T cells using Fugene 6 (Roche Molecular Biochemicals) with 0.1 μg Renilla luciferase reporter. Where indicated, 0.5 μg pCMV-ATF4 plasmid was included. Luciferase activities were measured 16-20 hr later using Dual Luciferase Kit (Promega). Firefly luciferase activities were normalized to Renilla luciferase control values and shown as an average of triplicates.
Human Subjects
Primary human neuroblastoma samples were collected and handled at the Children’s Hospital of Philadelphia with the approval of their institutional review board (IRB) committees. The related procedures were performed following ethical and legal standards regarding human subjects, and informed consent was obtained.
Animal Studies
All animal experiments were approved by the Animal Care and Use Committee at the University of Pennsylvania. For xenograft experiments, female BALB/C nude mice (Charles River) were injected subcutaneously on both flanks with three million Kelly, NLF, SKNAS, A549 or P493B cells diluted in 100 μl DMEM mixed with equal volume of matrigel (BD Bioscience). Once palpable tumors were established, mice were randomly divided into several groups receiving different treatments. Fenretinide (1.5 mg/kg) was intravenously injected every three days, and EGCG (50 mg/kg) was intraperitoneally (i.p.) injected daily. Tumor weight was measured at the time of sacrifice. For experiments using the transgenic mouse model, thirty homozygous TH-MYCN mice bearing palpable intra-abdominal tumors (ultrasound verified) were randomly divided into two groups, and i.p. injected daily with PBS or 10 mg/kg AOA, respectively. Eight days later the mice were sacrificed and tumors were isolated and weighed. To prepare tumor lysates for Western blot analysis, tumors were snap-frozen in liquid nitrogen, grounded with a mortar, and then lysed. To prepare tumor tissue sections for Ki-67 and cleaved Caspase 3 staining, fresh tumors were fixed in 10% formalin or 4% formaldehyde, and then dehydrated with a series of ethanol solutions (75%-95%-100%). Immunohistochemistry analysis of Ki-67 and cleaved Caspase 3 were performed by the histology core at the Children’s Hospital of Philadelphia. ImageJ software was used to quantify the staining results.
Supplementary Material
SIGNIFICANCE.
Myc-transformed cells depend on elevated glutaminolysis for survival, but little is known about the molecular pathways that trigger apoptosis upon Gln withdrawal. Here we describe a regulatory mechanism in which ATF4 induces expression of the pro-apoptotic proteins PUMA, NOXA, and TRB3 to promote apoptosis in Gln-deprived, Myc-overexpressing neuroblastoma cells. Moreover, xenograft and autochthonous murine neuroblastoma tumor growth is inhibited by small molecules that either enhance ATF4 activity or block glutaminolysis. Importantly, we demonstrate that MYCN-amplified human neuroblastomas selectively overexpress high-affinity Gln transporters and other glutaminolytic enzymes, which correlate with poor prognosis. These data suggest that drugs that inhibit glutaminolysis, or elevate ATF4 function, may be effective therapeutic strategies for treating the ≥40% of human cancers that overexpress Myc.
HIGHLIGHTS.
-
▶
MYCN-amplified neuroblastomas overexpress genes critical for glutamine metabolism
-
▶
PUMA/NOXA/TRB3 are executers of Myc-mediated cell death upon glutamine deprivation
-
▶
MYCN transgenic mice treated with glutaminolysis inhibitors develop smaller tumors
-
▶
ATF4 agonists or glutaminolysis inhibitors are potential cancer therapeutics
ACKNOWLEDGMENTS
We thank Dr. Chi Dang for providing the P493B lymphoma cells overexpressing c-Myc, and members of the Simon laboratory for helpful discussions. This work was supported by the Howard Hughes Medical Institute, NIH Grant CA104838 (to M.C. Simon), NIH grant CA097323 (to J.M. Maris), NIH grant CA97323 (to M.D. Hogarty), NIH F32 Training Grant 1F32CA137988 and the National Natural Science Foundation grant 81171928 from China (to G.L. Qing). M.C. Simon is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahluwalia GS, Grem JL, Hao Z, Cooney DA. Metabolism and action of amino acid analog anticancer agents. Pharmacology & therapeutics. 1990;46:243–271. doi: 10.1016/0163-7258(90)90094-i. [DOI] [PubMed] [Google Scholar]
- Ameri K, Harris AL. Activating transcription factor 4. The international journal of biochemistry & cell biology. 2008;40:14–21. doi: 10.1016/j.biocel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Butkinaree C, Park K, Hart GW. O-linked beta-N-acetylglucosamine (O-GlcNAc): Extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochimica et biophysica acta. 2010;1800:96–106. doi: 10.1016/j.bbagen.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capasso M, Devoto M, Hou C, Asgharzadeh S, Glessner JT, Attiyeh EF, Mosse YP, Kim C, Diskin SJ, Cole KA, et al. Common variations in BARD1 influence susceptibility to high-risk neuroblastoma. Nature genetics. 2009;41:718–723. doi: 10.1038/ng.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carracedo A, Lorente M, Egia A, Blazquez C, Garcia S, Giroux V, Malicet C, Villuendas R, Gironella M, Gonzalez-Feria L, et al. The stress-regulated protein p8 mediates cannabinoid-induced apoptosis of tumor cells. Cancer cell. 2006;9:301–312. doi: 10.1016/j.ccr.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Corazzari M, Lovat PE, Armstrong JL, Fimia GM, Hill DS, Birch-Machin M, Redfern CP, Piacentini M. Targeting homeostatic mechanisms of endoplasmic reticulum stress to increase susceptibility of cancer cells to fenretinide-induced apoptosis: the role of stress proteins ERdj5 and ERp57. British journal of cancer. 2007;96:1062–1071. doi: 10.1038/sj.bjc.6603672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell metabolism. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Ebos JM, Lee CR, Cruz-Munoz W, Bjarnason GA, Christensen JG, Kerbel RS. Accelerated metastasis after short-term treatment with a potent inhibitor of tumor angiogenesis. Cancer cell. 2009;15:232–239. doi: 10.1016/j.ccr.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eischen CM, Roussel MF, Korsmeyer SJ, Cleveland JL. Bax loss impairs Myc-induced apoptosis and circumvents the selection of p53 mutations during Myc-mediated lymphomagenesis. Molecular and cellular biology. 2001;21:7653–7662. doi: 10.1128/MCB.21.22.7653-7662.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galehdar Z, Swan P, Fuerth B, Callaghan SM, Park DS, Cregan SP. Neuronal apoptosis induced by endoplasmic reticulum stress is regulated by ATF4-CHOP-mediated induction of the Bcl-2 homology 3-only member PUMA. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:16938–16948. doi: 10.1523/JNEUROSCI.1598-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, Dang CV. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan N, Mukhtar H. Multitargeted therapy of cancer by green tea polyphenols. Cancer letters. 2008;269:269–280. doi: 10.1016/j.canlet.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilberg MS, Shan J, Su N. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends in endocrinology and metabolism: TEM. 2009;20:436–443. doi: 10.1016/j.tem.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange PS, Chavez JC, Pinto JT, Coppola G, Sun CW, Townes TM, Geschwind DH, Ratan RR. ATF4 is an oxidative stress-inducible, prodeath transcription factor in neurons in vitro and in vivo. The Journal of experimental medicine. 2008;205:1227–1242. doi: 10.1084/jem.20071460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, Tsukamoto T, Rojas CJ, Slusher BS, Zhang H, et al. Glucose-Independent Glutamine Metabolism via TCA Cycling for Proliferation and Survival in B Cells. Cell metabolism. 2012;15:110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letai A, Bassik MC, Walensky LD, Sorcinelli MD, Weiler S, Korsmeyer SJ. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer cell. 2002;2:183–192. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- Li M, Allen A, Smith TJ. High throughput screening reveals several new classes of glutamate dehydrogenase inhibitors. Biochemistry. 2007;46:15089–15102. doi: 10.1021/bi7018783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Mazanek P, Dam V, Wang Q, Zhao H, Guo R, Jagannathan J, Cnaan A, Maris JM, Hogarty MD. Deregulated Wnt/beta-catenin program in high-risk neuroblastomas without MYCN amplification. Oncogene. 2008;27:1478–1488. doi: 10.1038/sj.onc.1210769. [DOI] [PubMed] [Google Scholar]
- Maris JM. Recent advances in neuroblastoma. The New England journal of medicine. 2010;362:2202–2211. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosse YP, Laudenslager M, Longo L, Cole KA, Wood A, Attiyeh EF, Laquaglia MJ, Sennett R, Lynch JE, Perri P, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–935. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, Yang Y, Linehan WM, Chandel NS, DeBerardinis RJ. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohoka N, Yoshii S, Hattori T, Onozaki K, Hayashi H. TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. The EMBO journal. 2005;24:1243–1255. doi: 10.1038/sj.emboj.7600596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez-Ribes M, Allen E, Hudock J, Takeda T, Okuyama H, Vinals F, Inoue M, Bergers G, Hanahan D, Casanovas O. Antiangiogenic therapy elicits malignant progression of tumors to increased local invasion and distant metastasis. Cancer cell. 2009;15:220–231. doi: 10.1016/j.ccr.2009.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porro A, Haber M, Diolaiti D, Iraci N, Henderson M, Gherardi S, Valli E, Munoz MA, Xue C, Flemming C, et al. Direct and coordinate regulation of ATP-binding cassette transporter genes by Myc factors generates specific transcription signatures that significantly affect the chemoresistance phenotype of cancer cells. The Journal of biological chemistry. 2010;285:19532–19543. doi: 10.1074/jbc.M109.078584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing G, Skuli N, Mayes PA, Pawel B, Martinez D, Maris JM, Simon MC. Combinatorial regulation of neuroblastoma tumor progression by N-Myc and hypoxia inducible factor HIF-1alpha. Cancer research. 2010;70:10351–10361. doi: 10.1158/0008-5472.CAN-10-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada M, Sun W, Hayes P, Leskov K, Boothman DA, Matsuyama S. Ku70 suppresses the apoptotic translocation of Bax to mitochondria. Nature cell biology. 2003;5:320–329. doi: 10.1038/ncb950. [DOI] [PubMed] [Google Scholar]
- Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, Bachmann MH, Borowsky AD, Ruebner B, Cardiff RD, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitz T, Wei T, Valentine MB, Vanin EF, Grenet J, Valentine VA, Behm FG, Look AT, Lahti JM, Kidd VJ. Caspase 8 is deleted or silenced preferentially in childhood neuroblastomas with amplification of MYCN. Nature medicine. 2000;6:529–535. doi: 10.1038/75007. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villablanca JG, Krailo MD, Ames MM, Reid JM, Reaman GH, Reynolds CP. Phase I trial of oral fenretinide in children with high-risk solid tumors: a report from the Children’s Oncology Group (CCG 09709) Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2006;24:3423–3430. doi: 10.1200/JCO.2005.03.9271. [DOI] [PubMed] [Google Scholar]
- Wang JB, Erickson JW, Fuji R, Ramachandran S, Gao P, Dinavahi R, Wilson KF, Ambrosio AL, Dias SM, Dang CV, Cerione RA. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer cell. 2010;18:207–219. doi: 10.1016/j.ccr.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Diskin SJ, Zhang H, Attiyeh EF, Winter C, Hou C, Schnepp RW, Diamond M, Bosse K, Mayes PA, et al. Integrative genomics identifies LMO1 as a neuroblastoma oncogene. Nature. 2011;469:216–220. doi: 10.1038/nature09609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ, Roth KA, MacGregor GR, Thompson CB, Korsmeyer SJ. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, Thompson CB. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM, Platt JM, DeMatteo RG, Simon MC, Thompson CB. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, De Panis DN, Bobrovnikova-Marjon E, Diehl JA, Ron D, Koumenis C. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. The EMBO journal. 2010;29:2082–2096. doi: 10.1038/emboj.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Zhang L. PUMA, a potent killer with or without p53. Oncogene. 2008;27(Suppl 1):S71–83. doi: 10.1038/onc.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. The Journal of cell biology. 2007;178:93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.