Abstract
The novel supervised learning method of vertex discriminant analysis (VDA) has been demonstrated for its good performance in multicategory classification. The current paper explores an elaboration of VDA for nonlinear discrimination. By incorporating reproducing kernels, VDA can be generalized from linear discrimination to nonlinear discrimination. Our numerical experiments show that the new reproducing kernel-based method leads to accurate classification for both linear and nonlinear cases.
Keywords: Gaussian reproducing kernels, nonlinear classifier, regular simplex, reproducing kernel Hilbert space
1. INTRODUCTION
Discriminant analysis seeks to build a separation rule for two or more classes of objects based on their corresponding predictive features. The procedure begins with a training data set {(xi, yi) : i = 1, 2, . . . ,n}, where xi ∈ ℝp is the feature vector of the ith observation, yi denotes the corresponding class indicator, n is the sample size, and p is the dimensionality of the feature space. Each observation pair (xi, yi) is assumed to be independently and identically distributed according to some unknown probability distribution function P(x, y). The goal is to establish a classification rule which can be used for future class prediction. The simplest case of discriminant analysis is binary classification, in which the class indicator is typically labeled as y = −1 or 1. The binary classification has been well studied. One of the most popular binary classification methods is the support vector machine (SVM) [1,2]. In essence, a binary SVM estimates a function ĥ(x) by maximizing the margin of separation between the two classes and uses its sign for future class prediction. Once ĥ(·) is obtained, for any future observation with feature vector x, its corresponding class indicator is predicted by ŷ = sign(ĥ(x)). The binary SVM has enjoyed great success in many different application areas such as cancer classification and digits recognition.
Our focus of the current paper is the more complicated multicategory classification. Because of its great success, the binary SVM has been extended to handle multicategory classification problems in several different ways. Although the one-versus-rest approach [3] or pairwise comparison coupled with the binary SVM is frequently used for multicategory classification, many authors have noticed the disadvantages of solving a series of binary problems [4,5]. Therefore, we focus on methods that consider all classes simultaneously. Examples of multicategory SVMs include, but are not limited to, refs [4,6,7]. In particular, the multicategory SVM of Crammer and Singer [7] was shown to be not Fisher consistent by Liu [8]. A loss function-based classification method is Fisher consistent if the theoretical minimizer of the conditional expectation of the loss function at any point of the feature space matches the Bayes rule. See ref. [8] for more detailed introduction to Fisher consistency. Wu and Liu [9] developed another SVM method, called the robust SVM, which uses the truncated hinge loss. This method is shown to be Fisher consistent for multicategory classification when an appropriate truncation is applied to the hinge loss function. Most of these aforementioned multicategory SVMs label the categorical responses as y ∈ {1, 2, . . . , k}, solve one complicated optimization problem to estimate k functions {ĥ1(x), ĥ2(x), . . . , ĥk(x)}, and make class prediction ŷ = argmaxj=1,2,...,k ĥj(x) for any new input vector x, where k denotes the number of classes.
Meanwhile, vertex discriminant analysis (VDA) is another supervised method of multicategory classification introduced by Lange and Wu [10]. The term ‘vertex’ comes from its class assignment in which each class is represented by a vertex of a regular simplex in an Euclidean space. The vertices are symmetrically arranged on the surface of the unit ball in ℝk−1. Classification is performed via the minimization of an objective function, which involves the ε-insensitive loss and penalties on the coefficients of the linear predictors. In the first paper of VDA [10], a ridge penalty is imposed on the coefficients to avoid overfitting. A primal MM (majorization–minimization) algorithm [11] is used to approximate the ε-insensitive loss function by a quadratic function and the surrogate function is iteratively minimized using reweighted least squares. The authors show that the VDA procedure shares many of the attractive properties of multicategory SVM with hinge loss, including simple class prediction, creation of dead regions where observations receive zero loss, and robustness to outliers. The authors also point out that the VDA method has the advantage of operating in ℝk−1 rather than in ℝk, therefore reducing p parameters. For example, for problems with k = 3 classes, the number of parameters is reduced from 3p to 2p, which is 33% less. If p is large, this will be a big saving. The virtues of assigning classes to vertices can be summarized as symmetry, elimination of excess parameters and constraints, and simplification of computation and model interpretation. Another advantage of VDA over SVM is the use of the ε-insensitive loss. Though both the hinge loss in SVM and the ε-insensitive loss in VDA penalize wrong predictions, hinge loss does not set penalty for predictions with wild values as long as they fall on the correct side of their class indicators. In addition, the ε-insensitive loss avoids the constraints evoked in hinge loss and this makes it possible to implement the rapid coordinate descent algorithm. It has been proved by Wu and Lange [12] that ε-insensitive loss is Fisher consistent. Wu and Lange [12] extended the original VDA method with the ridge penalty () to problems in high-dimensional feature spaces. By imposing singular penalties, namely lasso (‖β‖1) and Euclidean (‖β‖2) penalties, the extended VDA methods can handle the high-dimensional problems well even when the number of features p far exceeds the number of observations n. All these VDA’s are for linear classification.
Because of the attractive performance of the linear VDAs, we resume the study of VDA in the current paper and introduce a nonlinear VDA method. For problems with nonlinear classifiers, one solution is to add nonlinear terms in the feature space. However, this step would be arbitrary and subjective and some important nonlinear features might be missed. Alternatively one may begin with many nonlinear terms and use certain variable selection-capable penalties to select important nonlinear features. Yet a more natural way of incorporating nonlinear characteristics is desired. This has prompted us to redesign VDA. Here we propose an extension of the linear VDAs to reproducing kernel Hilbert spaces (RKHS). This research is motivated by the RKHS-based SVM [13], kernel logistic regression (KLR) and import vector machine (IVM) [14], and two-step kernel learning [15]. See the references therein for more related works on RKHS-based discriminant methods. In the settings with more predictors than cases, this kind of RKHS-based method is also able to reduce the number of parameters without eliminating predictor variables. We will see later that the number of parameters in the RKHS-based nonlinear VDA depends on the sample size instead of the dimensionality.
The rest of the paper is organized as follows. Section 2 precisely formulates the VDA model and reviews the previous work on the linear VDA methods. Section 3 introduces the new nonlinear VDA based on reproducing kernel Hilbert spaces. We extend the representer theorem accordingly and discuss computational algorithms in this section. Sections 4 and 5 report our numerical experiments of the nonlinear VDA on simulated and real data. The paper is concluded with a brief summary and discussion in Section 6.
2. LINEAR VERTEX DISCRIMINANT ANALYSIS
Vertex discriminant analysis (VDA) is a novel method of multicategory supervised learning [10]. It is based on linear discrimination among different categories. Discrimination is conducted via the minimization of the ε-insensitive loss plus a penalty. For the reason of symmetry, VDA uses the equidistant vertices of a regular simplex in an Euclidean space to indicate different classes. For k categories, one can choose the k vertices v1, . . . , vk of a regular simplex in ℝk−1. Among the many possible ways of constructing a regular simplex, Lange and Wu [10] give the following example
| (1) |
where
and ej is the jth coordinate vector in ℝk−1. The authors further prove that one can locate at most k equidistant points in ℝk−1. The k equidistant points will be used to label our class indicators. It is worthwhile to note that the MSVM method by Lee et al. [4] also uses a vertex-type symmetrical indicator, that is, (). Yet it is easy to see that the number of parameters in their method is k × p.
Given a loss function L(y, f), discriminant analysis seeks to minimize the average conditional loss . We use Y and X to denote the class indicator and feature vector, respectively, of a random case. The vector Y equals one of the vertices of the simplex depending on which class it belongs to, namely Y = vY. The linear VDA assumes the linear regression model f(x) = Ax + b, where A = (ajl) is a (k − 1) × p matrix of slopes and b = (bj) is a k − 1 column vector of intercepts. To avoid overfitting, penalties are imposed on the slope parameters ajl. In VDA, we take the loss function for case i to be g(yi − f(xi)), where g(z) is the ε-insensitive Euclidean distance
| (2) |
and ‖z‖2 is the Euclidean norm of vector z. Shown and proved in refs [10,12], the ε-insensitive Euclidean loss is Fisher consistent when , which is the largest possible value avoiding overlap of the ε-insensitive balls around the vertices of the regular simplex. We will use this optimal value throughout the paper. To understand the ε-insensitive loss function, one can imagine that there is a sphere surrounding each vertex with radius of ε. Inside each sphere, cases receive zero loss (dead region). Only the cases outside the spheres receive loss values greater than zero and they serve similarly as ‘support vectors’.
Classification proceeds by minimizing the objective function
| (3) |
where θ = (A, b) denotes the set of all unknown parameters, P(A) is the penalty on the matrix A of slope parameters, and λ ≥ 0 is a tuning constant. Since the loss function is convex, it is clearly advantageous to set P(A) to be convex as well. In the original VDAR method [10], the ridge penalty is employed. When λ > 0, a unique minimum to the objective function R(θ) is guaranteed because of its strict convexity. Once the estimates  and b̂ are obtained, we can assign a new case to the closest vertex, and hence the corresponding category. More explicitly, the categorical response is predicted by ŷ = argminj=1,2,...,k‖ vj − Âx − b̂‖2 for any new input feature x.
Motivated by modern genomics problems involving hundreds of thousands to millions of predictors, Wu and Lange [12] extended the problem scale the original VDA can handle. Instead of the ridge penalty, they impose a lasso penalty on individual parameters and an Euclidean penalty on grouped parameters. As both penalties are singular and hence capable of selecting important variables, the modified VDA methods (VDALE, VDAL, and VDAE) can handle the underdetermined problems where p ≫ n. The new VDA method this paper describes can easily avoid the high-dimensional problem without doing any variable selection. The number of parameters will be reduced from p × (k − 1) to n × (k − 1). In p ≫ n cases, this will be a huge saving.
3. REPRODUCING KERNEL HILBERT SPACE-BASED NONLINEAR VDA
One natural way of nonlinear extension is to use basis expansion. For example, one can add nonlinear terms like quadratic and cubic of the predictor variables. However, the choice of bases would be arbitrary and subjective. It might be difficult to identify important nonlinear characteristics. Alternatively, we introduce a reproducing kernel Hilbert space-based nonlinear VDA method.
For a bivariate kernel K(·, ·), we denote ℱK to be the reproducing kernel Hilbert space (RKHS) generated by K(·, ·). For a more detailed introduction to RKHS, interested readers may consult refs [16,17]. Similar to the RHKS-based SVM, we propose RKHS-based VDA by assuming that the classification function vector f(x) = (f1(x), f2(x), . . . , fk−1(x))T of the nonlinear VDA takes the form fj(x) = hj(x) + bj with hj (·) ∈ ℱK and bj ∈ ℝ. Then our RKHS-based VDA estimates f̂j (x), j = 1, 2, . . . , k − 1, can be obtained by minimizing the objective function
| (4) |
where is the squared norm of function hj in the RKHS ℱK and is also known as the roughness penalty. The minimization is performed with respect to hj ∈ ℱK and bj ∈ ℝ.
3.1. Representer Theorem of RKHS
The optimization of Eq. (4) is over an infinite-dimensional functional space and therefore it is a very challenging task, if possible at all. The following representer theorem proves that Eq. (4) can be converted into a finite-dimensional optimization problem, which is much easier to solve. The proof is a straightforward extension of refs [4,17] and we include it here for completeness.
PROPOSITION 1: Optimization of Eq. (4) with respect to fj(x) = hj (x) + bj with hj (·) ∈ ℱK and bj ∈ ℝ is equivalent to optimization of Eq. (4) with respect to .
Proof: For any function fj(x) = hj (x) + bj with hj (·) ∈ ℱK, one may decompose hj (x) into , where cij ∈ ℝ, and ϕj (·) ∈ ℱK is a function orthogonal to the function space spanned by {K(xi, ·), i = 1, 2, . . . , n}.
As K(·, ·) is a reproducing kernel, we have 〈hj, K(xi, ·)〉ℱK = hj (xi ) for i = 1, 2, . . . , n and j = 1, 2, . . . , k − 1, because of the reproducing property, where 〈·, ·〉ℱK denotes the inner product in the RKHS ℱK. Consequently, we have
This implies that the data fit term does not depend on ϕj (·) for j = 1, 2, . . . , k − 1.
In addition, we have . Thus, the additional terms ϕj’s do not reduce the data fit term but cause an increase in the roughness penalty term. Hence, the minimizer of Eq. (4) must satisfy ϕj(·) = 0, j = 1, 2, . . . , k − 1. This completes the proof.
3.2. RKHS-Based VDA
Given a bivariate kernel K(·, ·), according to Proposition 1, the RKHS-based VDA boils down to the estimation of the function vector f(x) = (f1(x), . . . , fk−1(x))T with components . Note that the squared norm of is given by . Thus, the coefficients cij and bj can be estimated by solving
Denote K to be an n × n matrix with the (i, j)th element K(xi, xj) and C to be a n × (k − 1) matrix with element cij . The roughness penalty of the nonlinear function fj(x) is given by , where cj is the j th column of C. Therefore, the roughness of the function vector f(x) is tr(CT KC). In a more compact format, the objective function to be minimized can be rewritten as
| (5) |
where θ denotes the set of all unknown parameters cij and bj. The loss function g(·) is the ε-insensitive Euclidean loss defined in Eq. (2).
3.3. Computational Algorithm
The minimization of Eq. (5) can be implemented by coupling the MM algorithm [11,12,18] and weighted least squares estimation [10,19]. The MM algorithm is a more general form of the EM algorithm [20]. For a minimization problem, the first M means ‘majorization’, which creates a surrogate function to be minimized in the second M (‘minimization’) step. The majorization step is equivalent to, but more general than, the Expectation step of the EM algorithm in the missing data framework. For more details, please refer to refs [11,12]. The ε-insensitive loss function (2) can be majorized at all points except the two kinks at ±ε. One possible majorizer is given by Lange and Wu [10]
| (6) |
| (7) |
for residuals ri = yi − f(xi), weights
argument shifts
and constant f depending on the residuals at iteration m but not on the current values of A and b. As pointed out by Lange and Wu [10], the majorization Eq. (6) is advantageous as it separates the parameter for each dimension of ℝk−1. The minimization step of the MM algorithm therefore reduces to solving k − 1 least squares problems of size n rather than one large one of (k − 1) × n. The pseudocode for the RKHS-based VDA method is summarized below.
Another possible computing algorithm is to follow Wu and Lange [12] and use the modified ε-insensitive loss and the coordinate descent algorithm. Experimented by the authors, the latter approach does not have much computational advantage to the above algorithm. The modified VDA methods in ref. [12] perform variable selection when training the classifier. When the solution is sparse, the updates of most parameters are skipped. Therefore, coordinate descent saves computational times as only nonzero parameters get updated. Yet, in the RKHS-based VDA method, the roughness penalty tr(CTKC) does not perform variable selection, so no updates can be skipped. More explicitly, in every step, the estimates ĉij are nonzero. Hence, coordinate descent cannot speed up the computation. According to our limited numerical experiences, the weighted least squares estimation is more preferred.
Table 8.
| Pseudocode for the RKHS-based VDA method | |
|---|---|
| 1. | Set the iteration counter m = 0, and initialize C(0) = 0 and b (0) = 0; |
| 2. | Select a kernel function K(·, ·) and calculate the n × n matrix K; |
| 3. | Define yi = vj if the ith subject belongs to category j, where vj is defined in Eq. (1); |
| 4. | Majorize the regularized loss function as indicated in equation (6) with ith current residual |
| 5. | Minimize the quadratic surrogate function by weighted LSE and determine C (m+1) and b (m+1) by solving k − 1 sets of linear equations; |
| 6. | If ‖C (m+1) − C (m)‖ < γ and |R(θ(m+1)) − R(θ(m))| < γ both hold for γ = 10−4, then stop; |
| 7. | Otherwise repeat steps 4 through 6. |
4. SIMULATION EXAMPLES
To evaluate the performance of the RKHS-based VDA method (VDAK, with ‘K’ standing for kernel), we perform simulation analyses on both linear and nonlinear classification problems. Comparisons to its competitors such as linear discriminant analysis (LDA), quadratic discriminant analysis (QDA), k-nearest neighbors (KNN), one-versus-rest SVM (OVR), classification and regression tree (CART), random forest (RandForest), and RKHS-based multicategory truncated hinge loss SVM (MTSVM) are included. The best performer in each setting is highlighted in boldface.
In each example, for any method with regularization parameters to be tuned (e.g., λ in VDAK), we select the tuning parameter by minimizing the classification error on a separate tuning dataset with 200 observations based on a grid search. The classifiers with the optimal tuning parameters are then applied to a testing dataset with 20 000 observations. For methods that are not needed to be tuned (e.g., LDA and QDA), we combine the training and the tuning datasets to train the classifiers, which will then be tested on the 20 000 testing samples. We fix k = 2 or search for the optimal k value for the k-nearest neighbors method. Fifty random replications are performed in each example.
Note that both MTSVM and the RKHS-based VDA need to specify the kernel functions. There are many kernel functions available. The problem of how to choose the kernel function is not the focus of this paper. In our numerical studies, we fix the Gaussian kernel
because of its great flexibility, where σ is the data width parameter to be specified. Borrowing the idea of Brown et al. [21], we set σ to be the median pairwise Euclidean distance between classes. Specifically, it is defined as the median of ‖xi − xj‖2 for any i and j such that yi and yj do not belong to the same class.
4.1. Simulation Example 1
The first simulation example is modified from Example 1 of Wang and Shen [22]. In this example, we consider two different settings with either k = 4 or k = 8 classes, n = 300 training observations, and p = 2 predictors. Data are generated in two steps. We first generate the categorical response randomly with the same probability in each category. If observation i belongs to class c ∈ {1, . . . , k}, the corresponding j th predictor xij is generated by
where a1 = d · cos[2(c − 1)π/k] and a2 = d · sin[2(c − 1) π/k].
The uij’s are independent standard normal. The constant d determines the degree of overlap of the classes and thus controls the difficulty of classification. The various combinations of k ∈ {4, 8} and d ∈ {1, 3} generate four datasets with different difficulty levels. Table 1 reports testing errors averaged over 50 random replicates for different methods. Numbers in parentheses are the corresponding standard deviations.
Table 1.
Comparison of VDAK to competitive methods in simulation Example 1 with p = 2. Average testing errors (%) on a sample of 20 000 testing observations based on 5 0 random replicates are reported. The corresponding standard deviations are given in parentheses.
| Method | k = 4, d = 1 | k = 4, d = 3 | k = 8, d = 1 | k = 8, d = 3 |
|---|---|---|---|---|
| LDA | 22.83 (0.58) | 0.20 (0.03) | 54.14 (0.64) | 5.11 (0.18) |
| QDA | 23.35 (0.76) | 0.24 (0.05) | 54.75 (0.84) | 5.52 (0.29) |
| KNN (k = 2) | 25.90 (0.73) | 0.18 (0.04) | 57.06 (0.54) | 6.44 (0.54) |
| KNN (optimal k) | 20.63 (1.29) | 0.19 (0.06) | 52.59 (1.97) | 5.68 (0.61) |
| OVR | 19.97 (0.34) | 0.16 (0.03) | 57.02 (2.28) | 5.98 (0.35) |
| CART | 28.41 (6.54) | 0.73 (0.43) | 67.71 (2.68) | 16.37 (7.98) |
| RandForest | 19.92 (0.71) | 0.25 (0.08) | 51.58 (0.69) | 3.82 (0.45) |
| MTSVM | 19.69 (0.77) | 0.24 (5e−4) | 51.61 (2.28) | 5.29 (0.81) |
| VDAK | 19.85 (0.95) | 0.15 (0.06) | 50.65 (1.51) | 5.09 (0.47) |
Table 1 shows that the RKHS-based VDA outperforms LDA, QDA, KNN, OVR, and CART across all the four settings. It has the smallest testing errors in two out of the four settings. In the other two settings, the RKHS-based VDA ranks the second and the margin to the best method is quite small.
4.2. Simulation Example 2
This simulation example aims to demonstrate the capability of nonlinear discrimination of the RKHS-based VDA. It is similar to Example 5.3 of Zhang et al. [23], except that noise predictors are excluded here. The predictor variables x1 and x2 are uniformly distributed over [−3, 3] and [−6, 6], respectively. The categorical response is generated using the conditional probabilities pk(x) = P(class = k|x = x) ∝ exp(fk(x)), k = 1, 2, 3, where
We generate n = 200 samples to train the classifier.
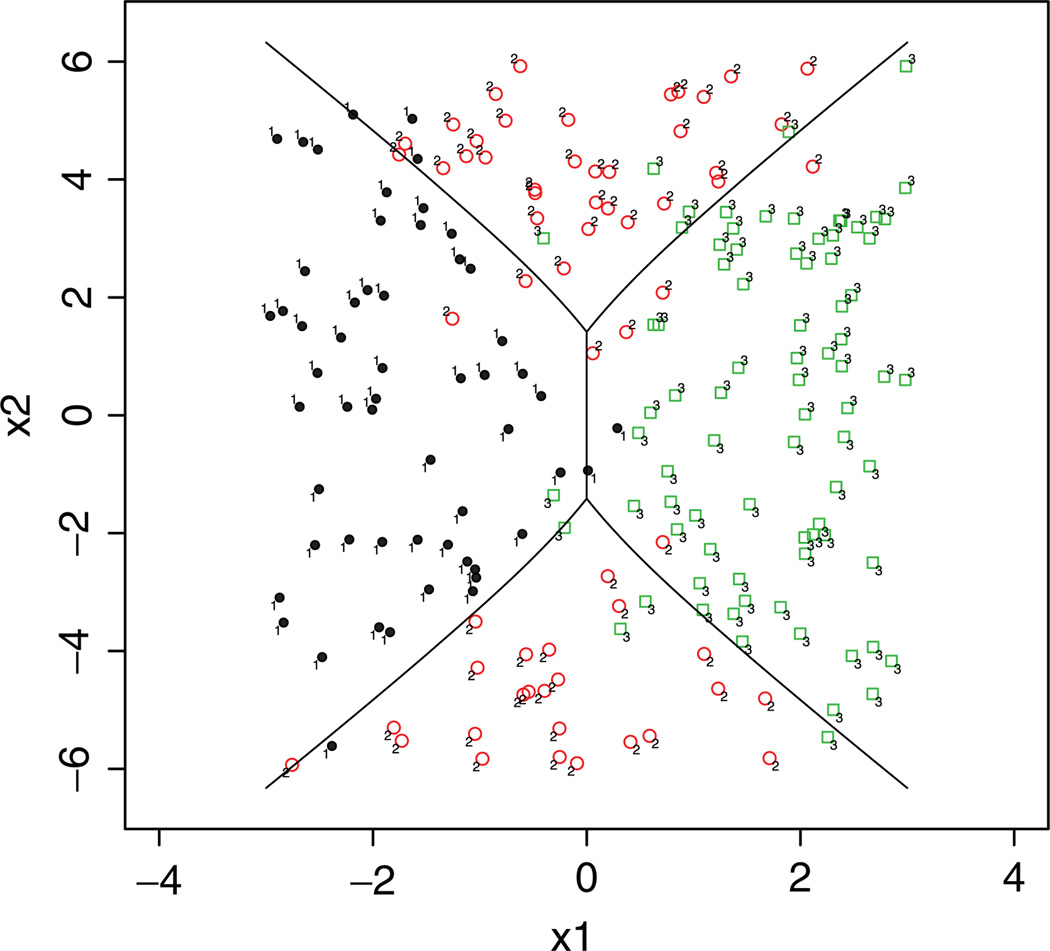
Figure 1 displays the Bayes boundaries and the observations from three classes. Obviously, it is a nonlinear classifier. We compare the RKHS-based VDA method with the original VDAR method [10] and other competing methods in Table 2. To test the nonlinear classification performance, we first apply the original VDAR method with predictors x1 and x2 only. As expected, without including the second order polynomial terms, VDAR fails to capture the nonlinear properties and gives high testing errors. In this simulation example, in addition to predictors x1 and x2 themselves, their corresponding second order terms ,, and x1x2 are relevant to classification. By adding the second order terms , and x1, x2 in the basis functions, VDAR outperforms all the other methods since it corresponds to an oracle model specification. The RKHS-based VDA ranks the second. However, we argue that the advantage of the RKHS-based VDA with the Gaussian kernel is the flexibility and the margin won by VDAR with the oracle model specification is really negligible. Clearly, if the important basis functions are missing, the original VDAR might result in bad classification. We know in this simulation example which high order terms to add. However, in practice, it is very challenging, if possible at all, to know which basis functions to include.
Figure 1.
Bayes boundaries (solid lines) for the nonlinear classifier in Example 2. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Table 2.
Average testing errors (%) of VDAK and competitive methods in Example 2 based on 50 random replicates. The corresponding standard deviations for these averages appear in parentheses.
| Method | Testing error (%) |
|---|---|
| Bayes | 12.0 |
| LDA | 27.94 (0.11) |
| QDA | 22.42 (0.62) |
| KNN (k = 2) | 18.73 (0.53) |
| KNN (optimal k) | 16.21 (1.11) |
| OVR | 14.05 (0.55) |
| CART | 20.07 (2.01) |
| RandForest | 15.60 (0.51) |
| MTSVM | 14.37 (0.80) |
| VDAR with main effects only | 29.46 (1.13) |
| VDAR with main and second order effects | 13.50 (0.64) |
| VDAK | 13.89 (0.77) |
4.3. Simulation Example 3: Gaussian Clouds
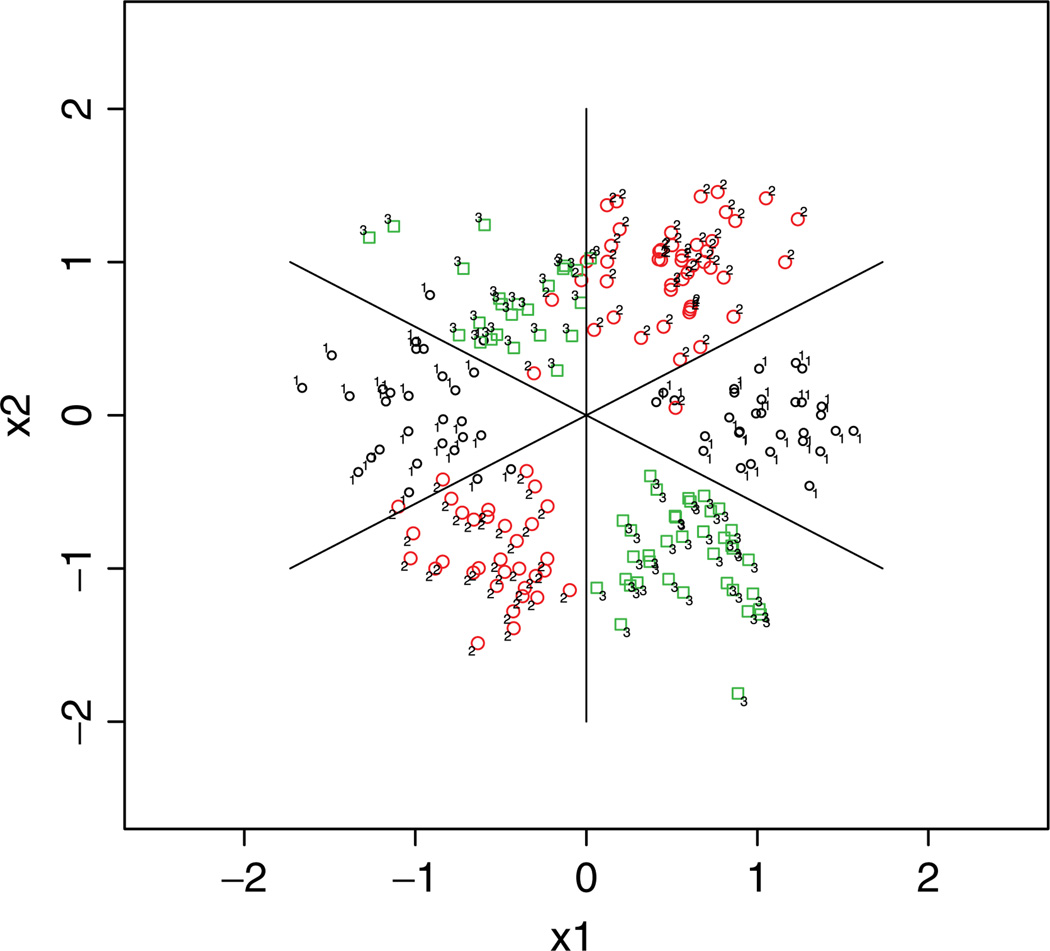
In this simulation, we let k = 3, n = 200, and p = 2. The predictors xij are generated from six Gaussian clouds centered at (a1, a2) such that
Each Gaussian cloud is generated with the variance–covariance matrix being σ2I2, where I2 is a 2 × 2 identity matrix. The variance σ2 determines the degree of overlap of the classes and hence classification difficulty. The Bayes boundaries are plotted in Fig. 2.
Figure 2.
Bayes boundaries (solid lines) for the nonlinear classifier generated around a circle in Example 3 with σ2 = 0.52. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Testing errors of the RKHS-based VDA and other methods are summarized in Table 3. The RKHS-based VDA captures the nonlinear characteristics of the classifier without specifying the basis functions and performs better than most of the competing methods.
Table 3.
Average testing errors (%) of VDAK and competitive methods in Example 3 based on 50 random replicates. The corresponding standard deviations for these averages appear in parentheses.
| Method | σ= 0.5 | σ= 0.3 |
|---|---|---|
| Bayes | 31.12 | 9.59 |
| LDA | 66.29 (5.55) | 65.89 (8.26) |
| QDA | 32.42 (0.41) | 10.01 (0.20) |
| KNN (k=2) | 42.71 (0.69) | 15.67 (0.52) |
| KNN (optimal k) | 35.80 (1.96) | 11.23 (0.68) |
| OVR | 32.31 (0.51) | 10.02 (0.21) |
| CART | 42.46 (3.38) | 15.47 (1.54) |
| RandForest | 36.79 (0.65) | 12.55 (0.55) |
| MTSVM | 37.69 (9.98) | 10.99 (0.95) |
| VDAR with main effects only | 61.21 (4.15) | 55.98 (6.17) |
| VDAR with main and second order effects | 33.91 (1.06) | 11.08 (0.86) |
| VDAK | 33.46 (0.99) | 10.62 (0.49) |
4.4. Simulation Example 4: Nested Circles
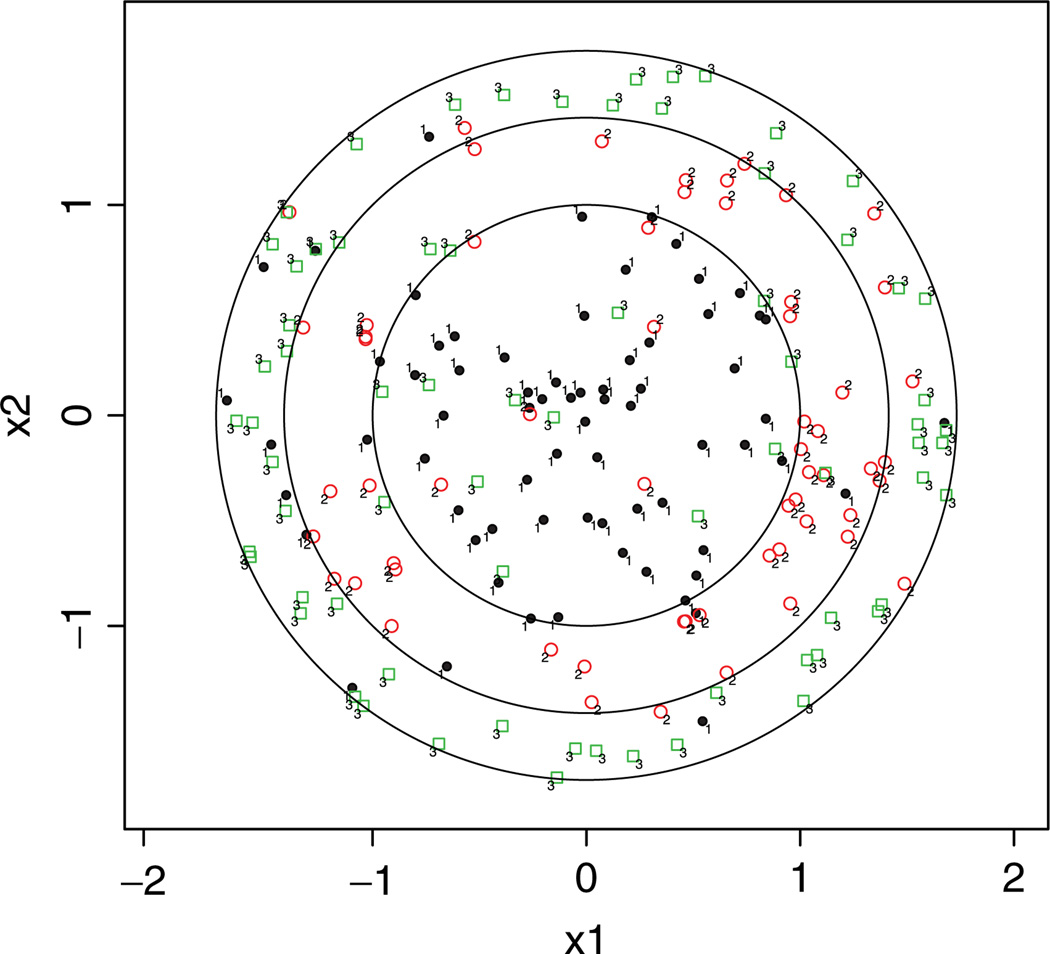
Generate (x1, x2) uniformly in the disc . For any individual sample with predictor (x1, x2), we generate an intermediate response , where ⌊r⌋ + 1 denotes the largest integer that is less than r. Next we apply a random flipping to generate the response y, namely y = ỹ with probability p and y ∈ {1, 2, . . . , k} \ ỹ with probability 1 − p. Here y ∈ {1, 2, . . . , k} \ ỹ means that y has an equal probability of (1 − p)/(k − 1) to be in any class other than class ỹ. In this way, p is the Bayes error and controls the difficulty of the problem. The Bayes boundaries and observations from three classes are plotted in Fig. 3.
Figure 3.
Bayes boundaries of the nested circle data in Example 4. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
We generate n = 200 training samples. Each entry of Table 4 represents an average of testing errors over 50 repetitions. Again, the RKHS-based VDA captures the nonlinear characteristics of the classifier without specifying the basis functions. It performs better than most of the competing methods.
Table 4.
Average testing errors (%) of VDAK and competitive methods in Example 4 based on 50 random replicates. The corresponding standard deviations for these averages appear in parentheses.
| Method | Testing error (%) |
|---|---|
| Bayes | 19.89 |
| LDA | 49.10 (0.03) |
| QDA | 34.36 (1.94) |
| KNN (k= 2) | 37.89 (1.08) |
| KNN (optimal k) | 33.99 (1.85) |
| OVR | 34.79 (1.04) |
| CART | 33.28 (2.67) |
| RandForest | 29.51 (0.89) |
| MTSVM | 27.30 (1.90) |
| VDAK | 28.06 (2.40) |
4.5. Simulation Example 5: Waveform
The waveform data of Breiman et al. [24] is a standard benchmark featured in Example 12.7.1 of ref. [25]. There are k = 3 categories and p = 21 predictors. The j th predictor of observation i is generated by
where j = 1, . . . , 21, Ui is a uniform deviate on [0,1], Zij is a standard normal deviate, and
The training set contains 300 cases. Average testing errors and the standard deviations over 50 simulations are summarized in Table 5. The RKHS-based VDA achieves low testing error rates.
Table 5.
Average testing errors (%) of VDAK and competitive methods in Example 5 based on 50 random replicates. The corresponding standard deviations for these averages appear in parentheses.
| Method | Testing error (%) |
|---|---|
| LDA | 16.93 (0.74) |
| QDA | 18.81 (0.39) |
| KNN (k = 2) | 23.86 (0.60) |
| KNN (optimal k) | 18.94 (1.54) |
| OVR | 15.76 (0.44) |
| CART | 30.40 (1.29) |
| RandForest | 15.71 (0.34) |
| MTSVM | 16.18 (1.20) |
| VDAK | 14.47 (0.72) |
In summary, the five simulation examples have demonstrated that the RKHS-based VDA performs competitively when the true classifier is either linear or nonlinear. When the true classifier is nonlinear, the flexibility of the RKHS-based VDA is especially obvious as it does not require to speculate the basis functions.
5. REAL DATA EXAMPLES
5.1. Four UCI Data Examples
To test the performance of our model on real data, we first analyze four datasets (wine, glass, zoo, and lymphography) from the University of California Irvine (UCI) machine learning repository (http://archive.ics.uci.edu/ml/) [26]. Table 6 compares the performance of VDA to its competitors. For all four datasets, the error rates in the table are average testing error rates based on threefold cross-validation.
Table 6.
Average cross-validated error rates for empirical examples from the UCI data repository. The triples beneath each data set give in order the number of categories k, the number of cases n, and the number of predictors p.
| Method | Wine (3,178,13) |
Glass (6,214,10) |
Zoo (7,101,16) |
Lymphog- raphy (4,148,18) |
|---|---|---|---|---|
| LDA | 3.45 | 34.76 | NA | 19.44 |
| QDA | 12.07 | NA | NA | NA |
| KNN (k = 2) | 5.75 | 31.90 | 12.12 | 27.08 |
| KNN (optimal k) | 4.60 | 25.23 | 8.08 | 73.61 |
| OVR | 1.72 | 32.86 | 6.06 | 16.67 |
| CART | 12.07 | 37.14 | 18.18 | 26.39 |
| RandForest | 2.30 | 21.90 | 5.05 | 16.67 |
| MTSVM | 2.30 | 37.62 | 5.05 | 26.39 |
| VDAK | 1.69 | 34.58 | 3.98 | 14.89 |
The wine dataset has 178 cases spread over three categories. It aims to classify the place of origin of the sampled wines based on the 13 continuous chemical measurements. The glass data consist of 6 categories, 214 cases, and 10 continuous predictors. The zoo data [27] involves animal classification and have 101 cases over 7 categories: mammal, bird, fish, reptile, amphibian, insect, and invertebrate. Each case is scored on 16 features, 15 of which are binary and 1 of which (number of legs) is numeric. The lymphography domain data of Zwitter and Soklic [28] consist of 148 cases spread over four categories: normal finding, metastases, malignant lymphoma, and fibrosis. Nine out of the 18 predictors are binary.
The results displayed in Table 6 are encouraging for the RKHS-based VDA even though random forest prediction outperforms it on the glass example. LDA do not work for the zoo data because the within-class covariance matrix is close to singular. QDA does not work for glass, zoo, and lymphography data because of the small sample size of certain classes.
5.2. Vowel Data Example
The vowel data [29] involve speaker recognition of the eleven steady state vowels of British English. The data are available online at http://www-stat.stanford.edu/~tibs/ElemStatLearn/. The training sample consists of 528 cases with 11 classes (state vowels) and 10 features (lpc derived log area ratios). The testing set consists of 462 cases. We first standardize each feature to have mean zero and standard deviation one. Tuning parameters are selected using a five-fold cross-validation over a grid search for any that needs to be tuned (i.e., MTSVM and VDAK). Gaussian kernels are also used with the data width parameter being the median pairwise Euclidean distance. The results reported in Table 7 show our RKHS-based VDA method has the smallest testing error. The 58.2% correct classification rate also compares favorably with the results reported in Tony Robinson’s Ph.D. thesis cited in the website of Hastie et al.’s [29] book (http://www-stat.stanford.edu/~tibs/ElemStatLearn/).
Table 7.
Mean five-fold cross-validated testing error rates (%) for empirical examples of vowel data.
| Method | Testing error |
|---|---|
| LDA | 54.76 |
| QDA | 58.44 |
| KNN (k = 2) | 50.65 |
| OVR | 50.43 |
| CART | 74.68 |
| MTSVM | 45.67 |
| VDAR with main effects only | 67.53 |
| VDAK | 41.77 |
6. DISCUSSION
Encouraged by the good performance of VDAR and its modifications for high-dimensional features, we extend VDA from linear classification to nonlinear classification in this paper. The new RKHS-based VDA does not require the specification of basis functions. This great flexibility, plus the virtues inherited from its predecessors, leads to the top performance of RKHS-based VDA for both linear and nonlinear classifiers in our numerical experiments.
One point is worth making that no variable selection is conducted in the current RKHS-based nonlinear method. It will be interesting to investigate the possibility of achieving variable selection for RKHS-based methods.
ACKNOWLEDGMENTS
The authors thank the Editor, Associate Editor, and referees for their constructive comments that helped substantially to improve the article. T. T. Wu’s research is supported in part by NSF grant CCF-0926194. Y. Wu’s research is partially supported by NSF grants DMS-090 5561 and DMS-1055210, NIH/NCI grant R01-CA149569. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
REFERENCES
- 1.Vapnik V. Statistical Learning Theory. New York: Wiley; 1998. [Google Scholar]
- 2.Cristianini N, Shawe-Taylor J. An Introduction to Support Vector Machines and Other Kernel-based Learning Method. Cambridge: Cambridge University Press: 2000. [Google Scholar]
- 3.Rifkin R, Klautau A. In defense of one-vs-all classification. J Mach Learn Res. 2004;5:101–141. [Google Scholar]
- 4.Lee Y, Lin Y, Wahba G. Multicategory support vector machines: theory and application to the classification of microarray data and satellite radiance data. J Am Stat Assoc. 2004;99:67–81. [Google Scholar]
- 5.Kressel U. Advances in Kernel Methods: Support Vector Learning. Cambridge, MA: MIT Press: 1999. Pairwise classification and support vector machines. (chapter 15) [Google Scholar]
- 6.Weston J, Watkins C. Support vector machines for multi-class pattern recognition; Proceedings of the Seventh European Symposium On Artificial Neural Networks; 1999. [Google Scholar]
- 7.Crammer K, Singer Y. On the algorithmic implementation of multiclass kernel-based vector machines. J Mach Learn Res. 2001;2:265–292. [Google Scholar]
- 8.Liu Y. Fisher consistency of multicategory support vector machines. Eleventh International Conference on Artificial Intelligence and Statistics. 2007;2:289–296. [Google Scholar]
- 9.Wu Y, Liu Y. Robust truncated-hinge-loss support vector machines. J Am Stat Assoc. 2007;102:974–983. [Google Scholar]
- 10.Lange K, Wu TT. An MM algorithm for multicategory vertex discriminant analysis. J Comput Graph Stat. 2008;17:527–544. [Google Scholar]
- 11.Lange K. Optimization. New York: Springer-Verlag; 2004. [Google Scholar]
- 12.Wu TT, Lange K. Multicategory vertex discriminant analysis for high-dimensional data. Ann Appl Stat. 2010;4:1698–1721. doi: 10.1214/09-AOAS279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wahba G. Support vector machines, reproducing kernel Hilbert spaces, and randomized GAC. Cambridge, MA: MIT Press; 1999. [Google Scholar]
- 14.Zhu J, Hastie T. Kernel logistic regression and the import vector machine. J Comput Graph Stat. 2005;14:185–205. [Google Scholar]
- 15.Wang L, Zhu J. Financial market forecasting using a two-step kernel learning method for the support vector regression. Ann Oper Res. 2010;174:103–120. [Google Scholar]
- 16.Kimeldorf G, Wahba G. Some results on Tchebycheffian spline functions. J Math Anal Appl. 1971;33:82–95. [Google Scholar]
- 17.Wahba G. Spline Models for Observational Data. Philadelphia, PA: Society for Industrial and Applied Mathematics; 1990. [Google Scholar]
- 18.Hunter DR, Lange K. A tutorial on MM algorithms. Am Stat. 2004;58:30–37. [Google Scholar]
- 19.Golub GH, Van Loan CF. Matrix Computations. 3rd ed. Baltimore, MD: Johns Hopkins University Press; 1996. [Google Scholar]
- 20.Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. J R Stat Soc Ser B. 1977;39:1–38. [Google Scholar]
- 21.Brown MPS, Grundy WN, Lin D, Cristianini N, Sugnet CW, Furey TS, Ares M, Haussler D. Knowledgebased analysis of microarray gene expression data by using support vector machines. Proc Natl Acad Sci. 2000;97:262–267. doi: 10.1073/pnas.97.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L, Shen X. On L 1-norm multiclass support vector machines. J Am Stat Assoc. 2007:583–594. [Google Scholar]
- 23.Zhang HH, Liu Y, Wu Y, Zhu J. Variable selection for the multicategory SVM via adaptive sup-norm regularization. Elec J Stat. 2008;2:149–167. [Google Scholar]
- 24.Breiman L, Friedman J, Olshen R, Stone C. Classification and Regression Trees. New York: Chapman & Hall/ CRC; 1984. [Google Scholar]
- 25.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. New York: Springer-Verlag; 2001. [Google Scholar]
- 26.Frank A, Asuncion A. UCI Machine Learning Repository. Irvine, CA: University of California, School of Information and Computer Science: 2010. [Google Scholar]
- 27.Forsyth RS. PC/BEAGLE User’s Guide. 1990. [Google Scholar]
- 28.Zwitter M, Soklic M. Lymphography Domain. 1988. http://archive.ics.uci.edu/ml/datasets/Lymphography. [Google Scholar]
- 29.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. 2nd ed. New York: Springer-Verlag; 2008. [Google Scholar]