Abstract
The lack of adipose triglyceride lipase (ATGL), a patatin-like phospholipase domain-containing enzyme that hydrolyzes fatty acids from triacylglycerol (TAG) stored in multiple tissues, causes the autosomal recessive disorder neutral lipid storage disease with myopathy (NLSD-M). In two families of Lebanese and Italian origin presenting with NLSD-M, we identified two new missense mutations in highly conserved regions of ATGL (p.Arg221Pro and p.Asn172Lys) and a novel nonsense mutation (p.Trp8X). The Lebanese patients harbor homozygous p.Arg221Pro, whereas the Italian patients are heterozygotes for p.Asn172Lys and the p.Trp8X mutation. The p.Trp8X mutation results in a complete absence of ATGL protein, while the p.Arg221Pro and p.Asn172Lys mutations result in proteins with minimal lipolytic activity. Although these mutations did not affect putative catalytic residues or the lipid droplet (LD)-binding domain of ATGL, cytosolic LDs accumulated in cultured skin fibroblasts from the patients. The missense mutations might destabilize a random coil (p.Asn172Lys) or a helix (p.Arg221Pro) structure within or proximal to the patatin domain of the lipase, thereby interfering with the enzyme activity, while leaving intact the residues required to localize the protein to LDs. Overexpressing wild-type ATGL in one patient's fibroblasts corrected the metabolic defect and effectively reduced the number and area of cellular LDs. Despite the poor lipase activity in vitro, the Lebanese siblings have a mild myopathy and not clinically evident myocardial dysfunction. The patients of Italian origin show a late-onset and slowly progressive skeletal myopathy. These findings suggest that a small amount of correctly localized lipase activity preserves cardiac function in NLSD-M.
INTRODUCTION
Human adipose triglyceride lipase (ATGL), a member of the patatin-like phospholipase domain-containing proteins (PNPLA) (1) that hydrolyze fatty acids from triacylglycerol (TAG) stored in multiple tissues, is a 504 amino acid protein containing the GXSXG serine hydrolase motif that is part of the catalytic dyad essential for lipase activity (2). ATGL also contains a C-terminus domain whose hydrophobic portion is required for binding to lipid droplets (LDs) (3). The expression of ATGL (also called PNPLA2, desnutrin, and calcium-independent phospholipase A2ζ) (1,4,5) is highest in brown and white adipose tissue, but is also present in heart, skeletal muscle, and most other tissues (1,5–7).
The absence of ATGL results in neutral lipid storage disease with myopathy (NLSD-M; MIM 610717) (8), an autosomal recessive disorder characterized by vacuolated granulocytes (Jordan's anomaly), cardiac and skeletal muscle myopathy, and LD storage in most tissues (9). Most of the reported ATGL mutations that cause NLSD-M result in truncated proteins with an unaltered catalytic site, but with defects in the C-terminus region which binds to LDs (8,10–12). These mutations indicate that preserving the lipase activity does not prevent NLSD-M if the enzyme cannot bind to LDs (3,12).
On the other hand, mutations located in the patatin/catalytic domain which severely impair the lipolytic capacity of the ATGL enzyme cause the complete expression of NLSD-M phenotype (8,13–16). A total loss of function is associated with a severe clinical phenotype in both humans and a rodent model (17). However, the clinical and biochemical impact of ATGL mutations that do not alter ATGL binding to LDs and also retain some lipase activity is less well characterized.
We now describe the clinical findings in four NLSD-M patients and report two novel missense mutations and a novel nonsense mutation in the ATGL gene. The nonsense mutation results in the lack of protein production, whereas the missense mutations severely diminish lipase activity, although the proteins retain their ability to bind to LDs.
RESULTS
Patient descriptions
Family 1 (Lebanese)
Patient 1A, reported in 1994 (18), is now 28 years old. Although he works as a caterer, he becomes exhausted after climbing three flights of stairs. He can run about 30–50 m. At 26 and 27 years of age, he reported chest discomfort. An echocardiogram showed hyperechogenic areas in the heart muscle. The electrocardiogram showed no arrhythmia. Plasma creatine kinase values were 4200 and 3800 IU/l without an increase of creatine kinase (CK)-MB, and troponin was negative. A stress test showed a maximum power of 125 watts (heart rate of 133 beats/min) with no signs of cardiac ischemia. The patient has been otherwise well except for episodes of pancreatitis in 2005 and in 2011 that required treatment in an intensive care unit. His leucocytes showed Jordan's anomaly (Fig. 1B).
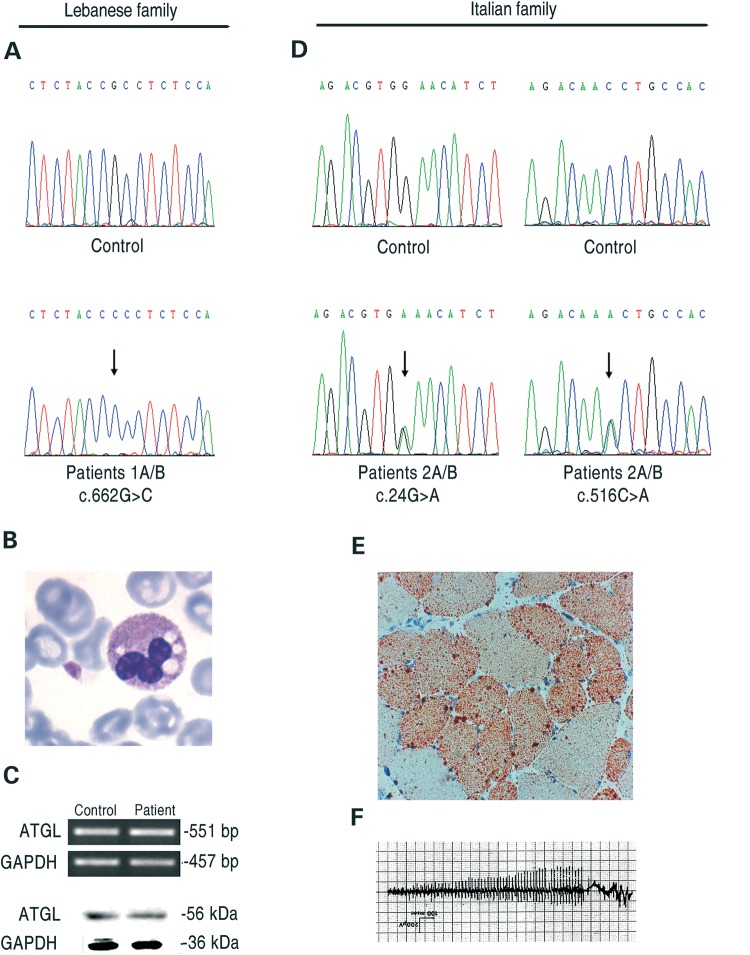
Figure 1.
Molecular and histochemical characterization of NLSD-M patients. (A) Electropherogram of the ATGL exon-5 sequence harboring the c.662G>C mutation in patients 1A/B; (B) microphotograph of LDs from patient 1B on buffy coats stained with May-Grünwald-Giemsa; (C) expression of ATGL in control and disease fibroblasts (patients 1A/B) at comparable passages as evaluated by RT-PCR and western blotting; (D) electropherograms of ATGL exon-2 and 5 sequences with the c. 24G>C and the c. 516C>A mutations, patients 2A/B; (E) light microscopy of frozen transverse section of a skeletal muscle biopsy from patient 2A stained with ORO; (F) electromyographic pattern of patient 2A showing myotonic discharges.
Patient 1B, the unemployed 33-year-old sister of patient 1A, is physically limited. She can climb a single flight of stairs with difficulty and can walk slowly, but only for 30 min because she experiences leg cramps and shortness of breath. She also complains of painful cramps in her legs which occur without exercise and which require her to use a car with hand controls. She has muscle spasms of her hands and fingers, sometimes lasting for hours. Plasma CK values were 897 IU/l. Her gall bladder was removed at age 26 because of multiple gall stones. She has chronic diarrhea of unknown cause. She has not had pancreatitis or diabetes mellitus and has not been evaluated by a cardiologist. Her three pregnancies were complicated by prolonged labor and looping of the umbilical cords. Her leucocytes showed Jordan's anomaly.
Family 2 (Italian)
Patient 2A, an unemployed 65-year-old woman, has hypertension and hypothyroidism. She first noted leg myalgia and cramps at age 25. At 40 years of age, her electromyography (EMG) showed myotonic discharges (Fig. 1F). In 1995, the histological diagnosis was made: a muscle biopsy showed excess Oil Red O (ORO) staining (Fig. 1E), and transmission electron microscopy confirmed a lipid storage myopathy. Her leucocytes showed Jordan's anomaly. Serum and muscle carnitine levels and carnitine palmitoyltransferase activity were normal. Her CK was normal until she was 40 years old, but now ranges between 440 and 730 IU/l. Cardiac ultrasound and scintigraphy in 2005 showed mild hypertrophic cardiomyopathy. She has been subjected to regular clinical cardiological controls until 2011. Holter electrocardiogram (ECG) performed in 2011 showed slight tachycardia, signs of left ventricular pressure overload, no pauses and no significant alteration of S-T tract. She currently has severe weakness in her upper arms with deltoid strength of 1–2/5 and quadriceps 3–4/5 (Medical Research Council scale). She can rise from a chair with support, but is unable to abduct her arms and cannot cut her food or brush her hair. She climbs stairs by pulling herself up, using the bannister and with the help of an assistant. One of her two sons has an elevated plasma CK concentration.
Patient 2B, the 58-year-old brother of patient 2A, is a photographer who first presented with Jordan's anomaly. He enjoyed competitive sports until age 40 when he first experienced painful leg cramps. He has proximal lower limb weakness with quadriceps strength of 4.5/5 and gluteus maximus strength of 4/5 (Medical Research Council scale), a waddling gait and exertional dyspnea and fatigue after walking 1 km. He complains of painful cramps in his legs at rest, which are partially steroid responsive. Upper arm strength is normal. An EMG showed myotonic discharges. Cardiac ultrasound and scintigraphy in 2005 and 2008 showed left ventricular hypertrophy (1.7 mm), an aneurysm of the interventricular septum, an ejection fraction of 66% and mild perfusion defects after exercise. The ECG after effort was normal. In 2011 a transthoracic echocardiogram showed a normal ejection fraction, a ventricular septal defect without shunting and a mild increase in left atrial dimension. Twenty-four hour Holter monitoring did not record significant arrhythmias. Routine laboratory tests remain within the normal range, but plasma CK values range from 380 to 680 IU/L, and recent plasma cholesterol concentrations were 248 and 273 mg/dl.
Mutation in family 1
Analysis of the ATGL gene in fibroblasts from patient 1A (18) showed a new homozygous mutation, c.662G>C (p.Arg221Pro) (Fig. 1A). This mutation was not observed in >100 alleles from control subjects, and it was submitted to GenBank (accession number HQ651812). The same mutation was identified in DNA extracted from peripheral blood from patient 1B. Because ATGL mRNA and protein were normally expressed in the patient's fibroblasts (Fig. 1C), we aligned the protein sequences orthologous to human ATGL aa 211–231 (Hs) to that identified in the genomes of 10 mammals and observed that the p.Arg221Pro mutation is located in a highly conserved region of the ATGL protein (Supplementary Material, Fig. S1). Although the mutation is not within the patatin domain, we hypothesized that it might affect the function and/or location of ATGL.
In support of this interpretation, the TAG content of fibroblasts from patient 1A was 2-fold higher than that of control fibroblasts (control: 1.55 nmol TAG/mg protein; patient: 3.1 nmol TAG/mg protein) with no change in the cellular content of cholesterol, cholesterol esters or phospholipids (Fig. 2A). However, the combined content of diacylglycerol and monoacylglycerol was 20% lower (control 0.75 nmol/mg protein; patient: 0.6 nmol/mg protein). Analysis of the fatty acid composition of TAG showed that the mutant fibroblasts contained 1.45- to 2-fold more 16:0 (control: 10.5%; patient: 21.5%; p/c = 2), 16:1 (control: 3%; patient: 5.7%; p/c = 1.9), 18:0 (control: 12.7%; patient: 20.2%; p/c = 1.59) and 18:1 (control: 20.4%; patient: 29.5%; p/c = 1.45) than control cells and significantly lower amounts of 20–24 carbon, n-3 and n-6 fatty acid species (Fig. 2B).
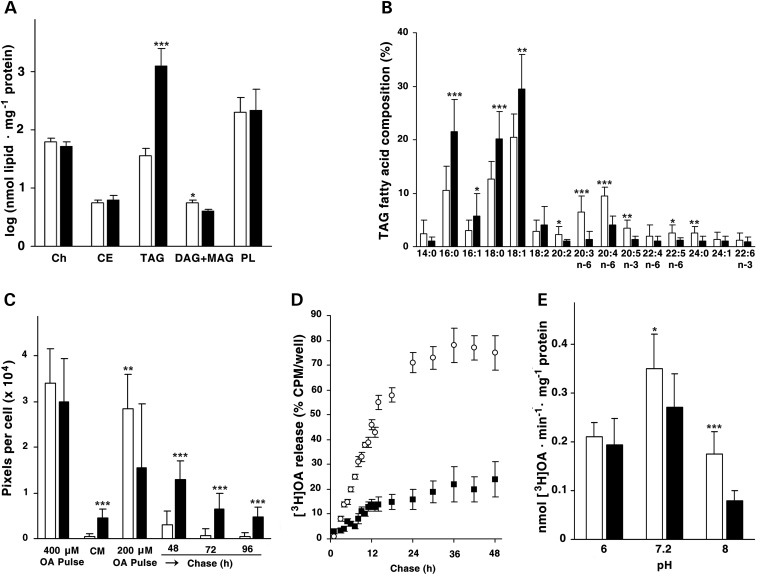
Figure 2.
Biochemical phenotype of the patient's 1A cultured skin fibroblasts. (A) Composition of the lipid fraction of confluent fibroblasts, determined in triplicate by TLC. *P < 0.05; ***P < 0.001. (B) Relative fatty acid composition of TAGs isolated from fibroblasts. The values are the mean of three independent GLC analyses. **P < 0.01. (C) OA pulse-chase experiments on patient and control fibroblasts. Fluorescence images (Supplementary Material, Fig. S2) were analyzed for pixel density per cell. (D) OA pulse-chase experiments on patient and control fibroblasts adding to the culture medium 200 µm OA complexed to BSA and enriched with [3H]OA (1.2 µCi/well). (E) Acylglycerol hydrolase activity of LDs isolated from control and patient fibroblasts was measured at three different pH values (6, 7.2 and 8). Detailed methods are reported in the Supplementary Material.
When fibroblasts were incubated for 18 h with 400 µm oleic acid (OA), the amount of accumulated lipid was equivalent in the patient and control cells. At 200 µm, although the patient's cells accumulated less lipid during the incubation, loss of lipid was almost completely abrogated during the chase, so that after 96 h, the patient's cells contained 9.6-fold more TAG than the control cells (Fig. 2C; Supplementary Material, Fig. S2). Adding triacsin C to the cells that had been labeled with [3H] OA showed that these differences in lipid accumulation had resulted from diminished lipid hydrolysis. Because triacsin C is an inhibitor of several acyl-CoA synthetases, it blocks the synthesis of acyl-CoA used for re-esterification and promotes the release of hydrolyzed fatty acids into the media. When triacsin C was present for 48 h, normal fibroblasts released 3.2-fold more [3H] OA than did the patient's fibroblasts (Fig. 2D). The patient's fibroblasts showed a 60% lower TAG lipase specific activity at pH 8, but not at pH 7.2 or pH 6 (Fig. 2E).
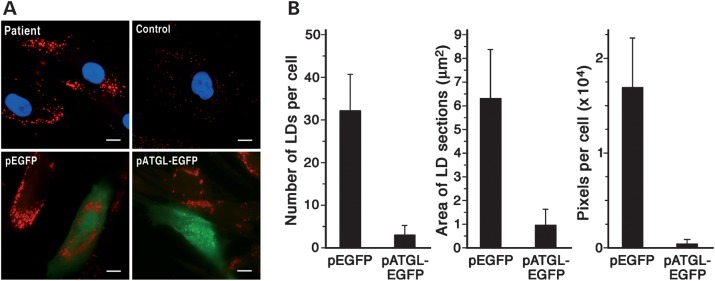
To confirm that the retention of LDs and lower TAG lipase activity had resulted from the mutant ATGL (Arg221Pro), we transfected patient A1's fibroblasts with either EGFP or ATGL-EGFP (Fig. 3A). Overexpressing the normal ATGL decreased the number of ORO-stained LDs by 90.6% and decreased their area by 84% (Fig. 3B).
Figure 3.
Correction of the mutant cell phenotype by over expressing wild-type ATGL. (A) Upper images are from fibroblasts from patient 1A and a control cultured at comparable passages in MEM plus Earle's salts containing FBS (10% v/v). Fixed cells (paraformaldehyde, 4% w/v) were stained with ORO and nuclei counterstained with DAPI (blue). Lower images are from the patient's fibroblasts (passage 8) that were cultured in the same medium as above and transiently transfected with either pEGFP or pATGL-EGFP. After 24 h, the cells were fixed and stained with ORO. Fluorescence of EGFP and ATGL-EGFP fusion proteins is in green. Images are representative of four independent transfections. Scale bar: 10 µm. (B) ORO-stained LDs of fibroblasts from patient 1A transfected with either pEGFP or pATGL-EGFP were analyzed for number, dimension (cross-section area) and pixels per cell. The values represent the means of 70–90 cells. Experiments were run in duplicate.
Mutations in family 2
The Italian patients 2A and 2B were compound heterozygotes for two novel mutations, c.24G>C (p.Trp8X) and c.516C>A (p.Asn172Lys) (Fig. 1D). These mutations were not observed in >100 alleles from control subjects, and they were submitted to GenBank (accession numbers: JF279441 and JF279442). Because the p.Trp8X causes the production of an ATGL peptide of only eight amino acids, the allele carrying this mutation can be considered a ‘null allele’. The second variation is a missense mutation, p.Asn172Lys. This mutation is located in the terminal region of the patatin-like domain. Alignment of the protein sequences orthologous to human ATGL, aa 162–182, revealed that the p.Asn172Lys mutation is located in a highly conserved region of the patatin domain of the ATGL protein (Supplementary Material, Fig. S1).
Functional study of missense ATGL mutations
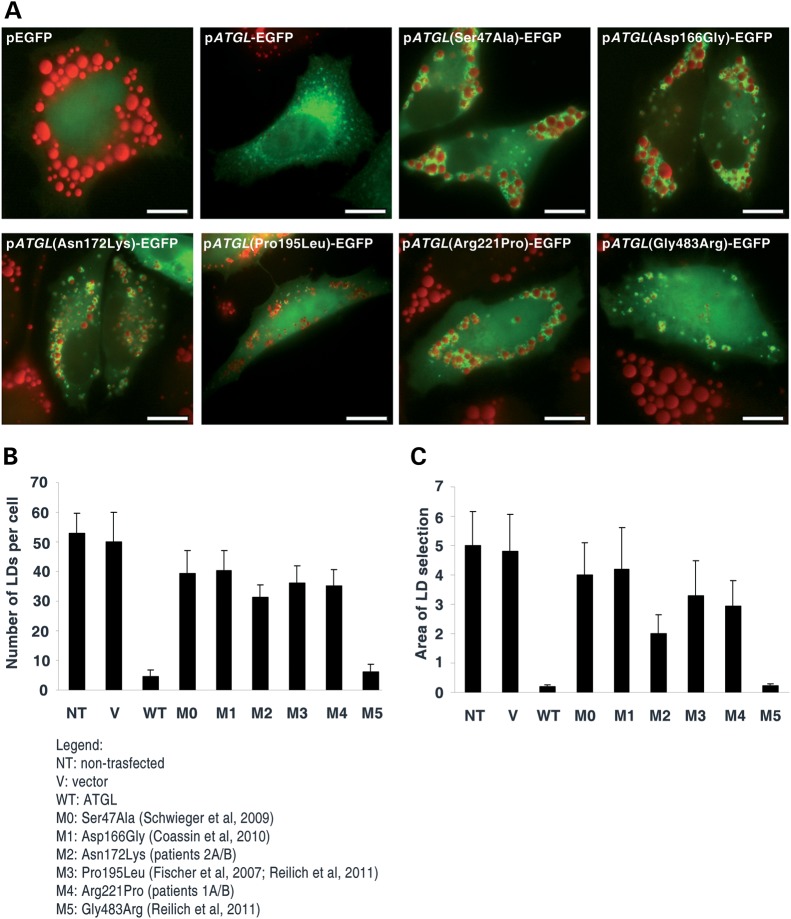
To verify the pathogenic effect of ATGL allelic variations identified in our patients, we constructed different recombinant plasmids, each with EGFP at the N-terminus. The plasmids contained either human ATGL cDNA (pATGL-EGFP), human ATGL with the p.Arg221Pro mutation (pATGL(Arg221Pro)-EGFP) and human ATGL with the p.Asn172Lys mutation (pATGL(Asn172Lys)-EGFP). For comparison, we constructed human ATGL with p.Ser47Ala and p.Asp166Gly mutations (pATGL(Ser47Ala)-EGFP), (pATGL(ASp166Gly)-EGFP). Ser 47 and Asp 66 are invariant residues in the core of the catalytic domain and essential for ATGL enzyme activity (19). Finally, we performed mutagenesis for all ATGL missense mutations previously reported for NLSD-M patients (p.Pro195L, p.Gly483Arg).
When HeLa cells were transiently transfected with either pATGL-EGFP or mutated ATGL recombinant plasmids and then incubated with 400 µm OA bound to bovine serum albumin (BSA), both wild-type ATGL and all ATGL mutant proteins were observed on LDs (Fig. 4A). Compared with the vector control, transfection of pATGL-EGFP reduced the number and area of the LDs 91% and 96.25%, respectively (Fig. 4B and C, WT). As expected, the two transfected mutant ATGL plasmids carrying p.Ser47Ala and p.Asp166Gly mutations had little effect, suggesting that the TAG lipase activity associated with these variants was almost absent (Fig. 4B and C, M0 and M1). Considering ATGL(Ser47Ala) and ATGL(Asp166Gly) proteins as totally inactive (virtually no normal activity), transfection of the mutant proteins ATGL(Asn172Lys) and ATGL(Arg221Pro) suggested that a residual enzymatic activity might be present. With ATGL(Asn172Lys), the number of LDs per cell is 21.63% lower and the area of LD sections is 51.23% lower. With ATGL(Arg221Pro), the number of LDs per cell is 8.8% lower and the area of LD sections is 28.78% lower (Fig. 4B and C, M2 and M4). The pATGL(Gly483Arg) mutant protein is almost completely active and reduces the number and the area of LDs 84.93% and 94.63%, respectively (Fig 4B and C, M5). In contrast, the activity of pATGL(Pro195Leu) was severely impaired, similar to that of pATGL(Arg221Pro) (Fig. 4B and C, M3).
Figure 4.
Transient transfection of ATGL mutant proteins in HeLa cells. (A) HeLa cells, loaded for 18 h with OA (400 µm) complexed to BSA (6:1 molar ratio), were transiently transfected with one of the following plasmids: pEGFP, pATGL-EGFP, pATGL(Ser47Ala)-EGFP, pATGL(Asp166Gly)-pEGFP, pATGL(Asn172Lys)-EGFP, pATGL(Pro195Leu)-EGFP, pATGL(Arg221Pro)-EGFP and pATGL(Gly483Arg)-EGFP. After 24 h, the cells were fixed and stained with ORO. Fluorescence of EGFP and ATGL-EGFP fusion proteins is in green. Scale bar: 10 µm. (B and C) ORO-stained LDs of Hela cells non-transfected and transfected with pEGFP (V), pATGL-EGFP (WT), pATGL(Ser47Ala)-EGFP (M0), pATGL(Asp166Gly)-pEGFP (M1), pATGL(Asn172Lys)-EGFP (M2), pATGL(Pro195Leu)-EGFP (M3), pATGL(Arg221Pro)-EGFP (M4) and pATGL(Gly483Arg)-EGFP (M5) plasmids. The cells were analyzed for number (b) and dimension (c) (cross-section area) of LDs per cell. The values represent the means of 80–95 cells for each transfected mutant. Experiments were run in triplicate.
DISCUSSION
NLSD results from deficiencies of either ATGL or CGI-58 (9), a 39 kDa protein that associates with LDs and is both a coactivator of ATGL (20) and a lysophosphatidic acid acyltransferase (21,22). Patients with deficiencies of either CGI-58 or ATGL have vacuolated granulocytes and excess TAG storage in LDs in multiple tissues. Clinically, the primary difference between the two deficiencies is that a lack of CGI-58 results in ichthyosis, whereas a lack of ATGL causes skeletal and cardiac myopathy. Atgl null mice accumulate excess TAG in virtually all tissues, and in contrast to the disorder in humans, have larger adipose depots than wild-type controls (17). The heart is severely affected in Atgl−/− mice, with TAG comprising as much as 30% of its weight. The resulting cardiac dysfunction causes death as early as 12 weeks of age (23,24). Increased glucose use in the Atgl−/− mice suggested that the low rates of lipolysis in adipose stores were insufficient to supply fatty acids for normal β-oxidation (23), or that the rate of fatty acid oxidation is low (25). It is now apparent that the murine cardiomyopathy is caused, in part, by a lack of peroxisome proliferator-activated receptor-α (PPARα) activation of β-oxidation target genes and that PPARα agonists can partially correct the cardiomyopathy (26).
Human ATGL is a 504 amino acid protein with a patatin domain in its N-terminal region (residues 1–251) which contains a catalytic dyad that includes the active site serine (Ser47) within a canonical GXSXG site and the critical aspartic acid residue (Asp166) (9). Within the C-terminal half of the protein, a hydrophobic region (315–360) is required for binding to LDs.
To date, 15 different mutations of the ATGL gene have been reported in 18 patients. In the present manuscript, we have clinically and genetically described an additional four patients with three new mutations. In order to highlight the complex effects that different ATGL gene variations may have on the lipase activity, we have performed a functional characterization of the novel, as well as the previously reported ATGL missense mutations.
The p.Arg221Pro and p.Asn172Lys mutations identified in our NLSD-M patients are located in a highly conserved region within the α/β/α sandwich structure, and both include and extend from the patatin domain. Our data suggest that these amino acid changes might perturb the conformation of the catalytic site and affect lipase activity, whereas binding to the LD would remain unaffected.
The p.Arg221Pro mutation was homozygous in patients 1A and 1B, whereas the p.Asn172Lys was heterozygous in patients 2A and 2B who also had the p.Trp8X mutation. Since the c.24G>C ATGL allele (p.Trp8X) can be considered to be a null allele, only the contribution of p.Asn172Lys ATGL protein may be critically evaluated for the clinical phenotype. In the literature, a very young patient carrying a p.Asp166Gly mutation in homozygous status was reported (14). The p.Asp166Gly mutation disrupts the catalytic dyad, comprised of Ser47 and Asp166, and abolishes lipase activity, despite binding to LDs. The patient carrying the p.Asp166Gly mutation presented with a severe cardiomyopathy and is currently awaiting a cardiac transplant, confirming the highly deleterious effect of a total deficiency in ATGL activity. Our functional studies using recombinant ATGL(Asp166Gly) protein confirm that the lipase activity is completely abrogated, similar to the ATGL(Ser47Ala) protein (Fig. 4B, M0 and M1). In the ATGL gene, only one other missense mutation, c.584C>T, has been reported; this mutation leads to an amino acid change within the α/β-fold at position 195 (p.Pro195Leu). Although this change does not directly affect the catalytic site, the p.Pro195Leu protein has defective lipase activity, but interacts with CGI-58 and, like the ATGL from our patients, is located on LDs (3). The c.584C>T (p.Pro195Leu) mutation was heterozygous in the reported NLSD-M patient (8), with a second allele harboring a c.808delC mutation (p.Leu319X). This patient presented with a mild phenotype (mild myopathy and hepatomegaly); the presence of cardiomyopathy was not ascertained (see Supplementary Material). Because the second allele retains enzyme activity (p.Leu319X), it may be able to partially compensate for the catalytically inactive p.Pro195Leu. Recently, the c.584C>T (p.Pro195Leu) mutation was identified in homozygous status in a patient with distal muscle weakness, elevated plasma CK and a cardiomyopathy (27). In accordance with the clinical phenotype, our data on mutant transfection show that p.Pro195Leu ATGL protein activity is highly impaired and resembles that of p.Ser47Ala (Fig. 4, M3). In the same patient, the p.Gly483Arg mutation was also identified, but this variation is most probably a benign polymorphism (Fig. 4, M5).
The patients whose mutations are reported here are limited physically by their skeletal myopathy but cardiac involvement appears to be relatively mild. The functional data obtained from transient transfection clearly show that the p.Arg221Pro and p.Asn172Lys ATGL proteins localize correctly to the LD surface (Fig. 4A). Moreover, the p.Arg221Pro protein shows less activity in comparison with p.Asn172Lys ATGL (Fig 4B, M2 and M4). Although they are likely to contribute only minimal lipase activity in vivo, it appears that the small amount of residual enzymatic function has protected patients 2A and 2B, who are now 58 and 66 years old, from the early onset of heart failure and arrhythmias. The latter observation appears particularly relevant also in consideration that they were older than NLSD-M patients reported so far (27), thus suggesting a slow progression of cardiomyopathy in these patients.
Compared with control fibroblasts, the fibroblasts from NLSD-M patients 1A and 1B contain higher amounts of the saturated and monounsaturated long-chain fatty acids, palmitic, stearic and OA. In contrast, the content of very-long-chain polyunsaturated fatty acids is lower in NLSD-M fibroblasts than in controls. It is not known whether these differences in fatty acid species are present in tissues of patients with NLSD-M or whether they contribute to the clinical phenotype.
The prognosis of patients 1A and 1B remains uncertain because (i) the p.Arg221Pro ATGL protein retains only a small amount of catalytic activity (Figs 2 and 4, M4), and (ii) among the 12 NLSD-M patients with cardiomyopathy described to date, 11 cases were clinically silent until the patients were 30–40 years old. As previously reported for another young patient (15), our Lebanese patients pose the crucial clinical dilemma of prognosis. The clinical course of these patients will be important to follow in order to determine whether their minimal ATGL lipase activity is sufficient to prevent heart failure. NLSD-I patients, also known as Chanarin Dorfman patients, do not develop cardiomyopathy. In these patients, ATGL is normally expressed but its enzymatic activity is low, since the coactivator, CGI-58, does not function or is absent. Our data suggest that low, but correctly localized ATGL lipase activity in human cardiomyocytes might preserve NLSD-M patients from severe cardiac dysfunction.
In patients similar to ours, it could be helpful to increase ATGL mRNA expression or to reduce the amount of the ATGL inhibitor protein G0S2 in order to improve ATGL protein synthesis. Although dexamethasone can up-regulate ATGL mRNA expression in vitro, it has little effect on cells from NLSD-M patients (27). Beta-adrenergic agonists, however, may inhibit G0S2 protein expression, thereby improving ATGL enzymatic activity (28). Overexpression of ATGL improved the disease phenotype in fibroblasts from our patients; thus, enzyme replacement therapy may be therapeutic. Supplying exogenous human recombinant enzyme improves the clinical outcome of several genetic diseases, including Fabry disease and glycogen storage disease 2 (29,30).
Recently, it was demonstrated that the low abundance of mRNA for PPARα target genes and the cardiac dysfunction of Atgl−/− mice can be corrected by the synthetic PPARα agonist Wy14 643 which increases energy supply to the heart through increased rates of FA β-oxidation (26). In vitro experimental studies on human cardiomyocytes are needed in order to verify the potential therapeutic effects of PPARα and/or beta-adrenergic agonists and to develop therapeutic protocols for NLSD-M patients.
Although all reported NLSD-M patients suffer from muscle weakness and skeletal muscle myopathy, the phenotypic severity appears to be highly variable. Cardiomyopathy was lethal in some patients or necessitated cardiac transplantation in young patients, but older NLSD-M patients have been described with less severe cardiac involvement (27). In the present study, we provide biochemical evidence that might help to explain the variable degrees of myopathy and cardiomyopathy in NLSD-M. Additional larger clinical studies are warranted to definitively elucidate genotype–phenotype correlations of NLSD-M mutations. Such studies may improve disease prognosis and aid in developing personalized approaches in treating patients with ATGL deficiency.
MATERIALS AND METHODS
Subjects
Human skin fibroblasts from a previously described patient (18), two previously unreported patients and a control subject were obtained from skin biopsies and cultured in Earle's minimum essential media (MEM) with 10% fetal bovine serum (FBS), 100 IU/ml penicillin and 100 μg/ml streptomycin at 37°C in a 5% CO2 incubator. Informed consent was obtained from the study participants.
Amplification and sequencing of ATGL genomic DNA fragments
Genomic DNA was extracted using a Puregene DNA isolation kit (Gentra Systems, Minneapolis), according to the manufacturer's instructions. To amplify the sequences of ATGL exons (GeneBank NM02376), we used eight primer pairs. PCR was performed with 200 ng of genomic DNA. Primer sequences and PCR amplification conditions for the analysis of ATGL coding regions were previously reported (8). All PCR products were purified (NucleoSpin Extract II; M-Medical) and sequenced on 3730 DNA analyzers by the BigDye® Terminator V1.1 Cycle sequencing kit (Applied Biosystems, Foster City, CA, USA).
RT-PCR analysis of ATGL mRNA expression in fibroblasts
Total RNA (1 μg) isolated from fibroblasts with TRIzol (Invitrogen, Carslbad, CA, USA) was converted to cDNA by RT-PCR using random hexamers (0.5 μg), 400 units of MMLV-RT, 1.6 mm total dNTPs, 20 units of Rnasin and 0.4 mm dithiothreitol, in a 50 μl reaction solution containing 10× RT Buffer. Forty nanograms of cDNA were used to perform PCR amplification using ATGL-3F (5′-TCTAAAGAGGCCCGGAAGCG-3′) and ATGL-6R (5′-TTCAGGAGGCGTTCCGCTG-3′) primers designed to produce a 551 bp fragment of the ATGL transcript (Accession No. AY894804.1 GI:58759050). PCR conditions for ATGL3F/6R primers were as follows: denaturation at 95°C for 3 min, annealing at 57°C for 30 s and extension at 72°C for 30 s for the first round, denaturation at 94°C for 30 s, annealing at 57°C for 30 s and extension at 72°C for 30 s for 28 cycles; denaturation at 94°C for 30 s, annealing at 57°C for 30 s and terminal extension at 72°C for 3 min for the last cycle. GAPDH, glyceraldehyde-3-phosphate dehydrogenase, gene expression was used as the internal control (31). The PCR products were electrophoresed on a 2% agarose gel containing ethidium bromide.
Western blot analysis of ATGL protein expression in fibroblasts
Cell extracts were prepared from confluent cultures grown in serum-containing medium. After extensive washing with cold PBS, cells were recovered by scraping with a rubber policeman in 1 ml of 0.05% (wt/vol) sodium dodecyl sulfate (SDS). The total protein concentration of cell extracts was quantified by a Coomassie (Bradford) protein assay kit (PIERCE). Laemmli's polyacrylamide gel electrophoresis, in the presence of SDS and under reducing conditions, was carried out using a 12% vertical slab gel. Proteins (25 μg/well) were loaded, then immunoblotted using a mouse polyclonal antibody (dil. 1:5000) raised against full-length recombinant ATGL protein (ABNOVA, Taiwan corporation) and a mouse monoclonal antibody (dil: 1:7000) to GAPDH (Ambion, Austin, TX, USA). Specifically bound immunoglobulins were detected using the SuperSignal West Pico Complete Mouse Detection Kit (PIERCE) containing ImmunoPure Peroxidase Conjugated Goat anti-Mouse IgG (dil 1:20.000).
Cloning the ATGL cDNA and generation of site-directed mutagenesis plasmids
ATGL cDNA was amplified by PCR using DNA clone ID 4875483 (Open Biosystem) as template. The PCR product was cloned into pEGFP-N1 from Clonetech to produce pEGFP-N1-ATGL, expressing ATGL with GFP at the C-terminus. Point mutations were performed using the QuikChange XL site-directed mutagenesis kit (Stratagene). Mutations in ATGL cDNA were introduced using the following primers: Ser47Ala: forward 5′-CACATCTACGGCGCCGCGGCCGGGGCGCTCACGG-3′ and reverse 5′-CCGTGAGCGCCCCGGCCGCGGCGCCGTAGATGTG-3′; Asp166Gly forward 5′-TGCGCTACGTGGGTGGTGGCATTTC-3′ and reverse 5′-GAAATGCCACCACCCACGTAGCGCA; Asn172Lys: forward 5′-GGCATTTCAGACAAACTGCCACTCTATGAGCT-3′ and reverse 5′-AGCTCATAGAGTGGCAGTTTGTCTGAAATGCC-3′; Pro195Leu forward 5′-GAGTGACATCTGTCTGCAGGACAGCTCCAC-3′ and reverse 5′-GTGGAGCTGTCCTGCAGACAGATGTCACTC-3′; Arg221Pro: forward 5′-CCTGCGCAACCTCTACCCCCTCTCCAAGGCCCTC-3′ and reverse 5′-GAGGGCCTTGGAGAGGGGGTAGAGGTTGCGCAGG-3′ Gly483Arg forward 5′-ACCAGCTGGCCCGGCCTGCCC-3′ and reverse 5′-GGGCAGGCCGGGCCAGCTGGT-3′. All constructs were verified by DNA sequencing.
Expression of recombinant ATGL plasmids in HeLa cells
HeLa cells were cultured on glass coverslips in Dulbecco's modified Eagle's medium supplemented with 10% FBS and allowed to adhere overnight. To increase TAG synthesis and storage, 400 μm oleate complexed to BSA (6:1 molar ratio) was added to the medium and incubated overnight. The cells were then transiently transfected with recombinant pEGFP ATGL plasmids [pEGFP, pATGL-EGFP, pATGL(Ser47Ala)-EGFP, pATGL(Asp166Gly)-EGFP, pATGL(Asn172Lys)-EGFP, pATGL(Pro195Leu)-EGFP, pATGL(Arg221Pro)-EGFP and pATGL(Gly483Arg)-EGFP] using the siPORT XP-1 transfection reagent, according to the manufacturer's protocol (Ambion). After 24 h, the cells were fixed and stained with ORO, and LDs from immunofluorescent images were quantified.
TLC analysis
Total cellular lipid extracts (8:4:3 v/v chloroform–methanol–water solution) were dried in a centrifugal vacuum concentrator (SPD111V; Thermo Electron Corp., Madison, WI, USA) at 30°C then re-dissolved in 20 μl of chloroform/methanol (2:1 v/v). Aliquots were applied onto silica gel thin-layer chromatography (TLC) plates (60 G, 100 × 200 × 0.25 mm; Merck AG, Darmstadt, Germany) and separated in a solvent system of hexane/diethyl ether/formic acid (80:20:2; v/v). The developed plates were heated at 110°C for 30 min, then cooled at RT, sprayed with 50% (v/v) sulfuric acid solution in ethanol and redried at 170°C for 20 min. Qualitative and quantitative analyses of individual lipid classes were made through comparison with standards (cholesterol oleate, triolein, 1,3-dioleoyl-sn-glycerol, 1,2-dioleoyl-sn-glycerol, sn-1- and 2-monooleoyl glycerol and 3-sn-phosphatidic acid sodium salt; Sigma-Aldrich).
Fatty acid composition of TAG
Purified TAG for gas–liquid chromatography (GLC) analysis was obtained from the TLC plates. TAG bands were detected by exposing the developed TLC plates to iodine vapor. After iodine sublimation, the TAG spots were scraped from the silica gel plates, and TAG was extracted with methanol. After evaporating the solvent, the TAG was transmethylated at 50°C for 1 h with BF3/methanol (40% w/v). After evaporation, fatty acid methyl esters were re-dissolved in hexane and analyzed with a Hewlett-Packard model 5700A (Round Rock, TX, USA) gas chromatograph equipped with a SP-1000 column (30 m × 0.25 mm ID; Supelco Inc., Bellafonte, Pennsylvania , USA) and a flame ionization detector. Lipid standards were obtained from Sulpelco Inc. FFA C23:0 was used as the internal standard for TAG quantification.
Oleic acid pulse-chase experiments on disease and control fibroblasts
Oleic acid (O-1630, Sigma) was dissolved in a 2.1 mm solution of FFA-free BSA, previously prepared in 0.1 m Tris (pH 8), to a final concentration of 12.5 mm. After the fatty acid was completely dissolved, the solution was heated to 37°C, then cooled, filtered on 0.20 μm Minisart and stored in aliquots at 4°C.
Metabolic pulse experiment with 400 μm oleic acid
Fibroblasts (200 000 per dish) were seeded on coverslips in a serum-free Earle's MEM culture medium and allowed to adhere for 7 h. Then the cell culture medium was supplemented with the 12.5 mm oleate/BSA solution at a final concentration of 400 μm (pulse medium). After an overnight incubation, the medium was removed, and the cells were washed with Dulbecco's phosphate-buffered saline (D-PBS) and stained with Nile Red (NR) dye (NR, 9-diethylamino-5H-benzo[alpha]phenoxazine-5-one; Sigma-Aldrich).
Metabolic pulse-chase experiment with 200 μm oleic acid
For the oleic acid (OA) pulse-chase experiment, control and patient fibroblasts were seeded on glass coverslips and grown in a serum-free Earle's MEM culture medium to 90% confluence, then incubated for 24 h in a culture medium containing OA/BSA (200 μm). The day after, in some (1/4) of the dishes, the medium containing OA/BSA was removed, the cells were washed twice with D-PBS, fixed on the coverslips with 4% paraformaldehyde, and labeled with NR dye. Fibroblasts in the remaining dishes were washed twice with D-PBS, and then incubated for 48, 72 or 96 h in the serum-free Earle's MEM culture medium containing fatty acid-free BSA (2% w/v) to enhance cellular lipolysis (chase). BSA was present in the medium to capture OA released from cells. At the end of each chase (48, 72 and 96 h), the medium was removed, and the cells were washed twice with D-PBS and stained with NR. Quantitative analysis of fatty acid stores used ‘WCIF ImageJ 1.35j’ software that allowed evaluation of pixels per cell related to NR emission.
Metabolic pulse-chase experiment with 200 μm OA enriched with [3H]OA
Confluent fibroblasts (1.5 × 105 cells/well) were incubated with OA (200 µm) complexed to BSA and enriched with [3H]OA (1.2 µCi/well). After 18 h, the cells were washed, and the medium was replaced by MEM plus Earle's salts containing fatty acid-free BSA (3% w/v) and triacsin C (5 µm). [3H]OA release into the medium was determined in aliquots of the water phase by liquid scintillation counting.
Acylglycerol hydrolase activity of lipid droplets isolated from fibroblasts
Isolation of the LD fraction was performed as described by Fujimoto et al. (32). The acylglycerol hydrolase activity of LDs isolated from control and patient fibroblasts was measured at three different pH values (6, 7.2 and 8) in 40 mm potassium phosphate buffer containing fatty acid-free BSA (2% w/v) and using [3H]trioleoylglycerol (40 000 cpm/nmol) as the substrate. After an incubation of 30 min at 37°C, the reaction was terminated by extraction of the alkalinized mixture with an organic phase. Free [3H]OA was determined in aliquots of the water phase by liquid scintillation counting.
Transient transfection of wild-type ATGL cDNA in disease fibroblasts
Patient's fibroblasts were cultured on glass coverslips until 80% of confluence and transiently transfected with either pEGFP or ATGL-EGFP plasmid with the siPORT XP-1 transfection reagent (Ambion). After 24 h, the cells were fixed and stained with ORO, and LDs were quantified from immunofluorescent images.
Immunofluorescence microscopy
NR staining solution was freshly prepared in DPBS (1:100 v/v) from a saturated solution (1 mg/ml) in dimethyl sulfoxide. The cells were fixed on a glass slide using 4% paraformaldehyde. After rinses with distilled water, slides were incubated with NR solution for 20 min in the dark.
A 1.66× solution of ORO was prepared as reported by Koopman et al. (33). The cells fixed on glass coverslips were incubated with 2 ml of fresh 1X ORO solution for 30 min under darkened conditions.
After washing with 2 ml of distilled water, fibroblasts and HeLa transfected cells labeled with lipophilic dyes (ORO and NR) were mounted on glass microscope slides with Vectashield mounting medium (Vector Laboratories, Burlingame, CA, USA) and examined with a Leica MB5000B microscope equipped with 20×, 40× and 100× Fluorart oil immersion objectives. Excitation fluorescence filters used for ORO and NR images were TX2-FITC (540–580 nm) and I3-TRITC (450–490 nm), respectively. Fluorescence images were captured using a Leica DFC480 R2 digital camera and the Leica Application Suite (LAS) software that allows one to change a large number of measurement parameters. The values chosen for the exposure time, saturation, gamma and gain parameters were, respectively, 4 s, 1.2, 1.43, 1× for ORO stained fibroblasts and 1 s, 1.1, 1.21, 1.1× for NR stained fibroblasts. In HeLa transfected cells stained with ORO, the values for the exposure time, saturation, gamma and gain parameters were, respectively, 12 s, 1.20, 1.43, 5×.
Fluorescent images captured by immunofluorescent microscopy were analyzed using the public domain Java image processing program ‘WCIF ImageJ 1.35j’ (developed by W. Rasband, NIH, Bethesda, MD, USA). This software allows us to isolate components having the same wavelength and to evaluate and quantify several parameters like area, numbers of selected units (LDs, in this case) and pixels per cell; 15 inches2 was chosen as threshold value for LD area in order to discard fluorescent emissions due to impurity.
Statistical analysis
The statistical analysis of quantitative data on LDs identified from cells (fibroblasts and HeLa transfected cells) by image analysis of immunofluorescence experiments as well as those obtained from TLC, TLC–GLC, the pulse-chase experiments and the acylglycerol hydrolase activity assay was made using SPSS v.19 package (SPSS, Chicago, IL, USA). The values were compared with Student's unpaired t-test, χ2-squared test and linear regression analysis using Spearman's software. A P-value of ≤0.05 was considered to be statistically significant.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by grants from the NIH (DK56598 and DK59935), the Cariplo Foundation (Milan, Italy) and Guido Berlucchi & C. Spa (Brescia, Italy).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to GianPaolo Martelli for management consulting and to Silvia Cristini and Dario Degiorgio for generous scientific assistance.
Conflict of Interest statement. None declared.
References
- 1.Zimmermann R., Strauss J.G., Haemmerle G., Schoiswohl G., Birner-Gruenberger R., Riederer M., Lass A., Neuberger G., Eisenhaber F., Hermetter A., Zechner R. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306:1383–1386. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
- 2.Kurat C.F., Natter K., Petschnigg J., Wolinski H., Scheuringer K., Scholz H., Zimmermann R., Leber R., Zechner R., Kohlwein S.D. Obese yeast: triglyceride lipolysis is functionally conserved from mammals to yeast. J. Biol. Chem. 2006;281:491–500. doi: 10.1074/jbc.M508414200. [DOI] [PubMed] [Google Scholar]
- 3.Schweiger M., Schoiswohl G., Lass A., Radner F.P., Haemmerle G., Malli R., Graier W., Cornaciu I., Oberer M., Salvayre R., et al. The C-terminal region of human adipose triglyceride lipase affects enzyme activity and lipid droplet binding. J. Biol. Chem. 2008;283:17211–17220. doi: 10.1074/jbc.M710566200. [DOI] [PubMed] [Google Scholar]
- 4.Jenkins C.M., Mancuso D.J., Yan W., Sims H.F., Gibson B., Gross R.W. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J. Biol. Chem. 2004;279:48968–48975. doi: 10.1074/jbc.M407841200. [DOI] [PubMed] [Google Scholar]
- 5.Villena J.A., Roy S., Sarkadi-Nagy E., Kim K.H., Sul H.S. Desnutrin, an adipocyte gene encoding a novel patatin domain-containing protein, is induced by fasting and glucocorticoids: ectopic expression of desnutrin increases triglyceride hydrolysis. J. Biol. Chem. 2004;279:47066–47075. doi: 10.1074/jbc.M403855200. [DOI] [PubMed] [Google Scholar]
- 6.Lake A.C., Sun Y., Li J.L., Kim J.E., Johnson J.W., Li D., Revett T., Shih H.H., Liu W., Paulsen J.E., Gimeno R.E. Expression, regulation, and triglyceride hydrolase activity of adiponutrin family members. J. Lipid Res. 2005;46:2477–2487. doi: 10.1194/jlr.M500290-JLR200. [DOI] [PubMed] [Google Scholar]
- 7.Kershaw E.E., Hamm J.K., Verhagen L.A., Peroni O., Katic M., Flier J.S. Adipose triglyceride lipase: function, regulation by insulin, and comparison with adiponutrin. Diabetes. 2006;55:148–157. [PMC free article] [PubMed] [Google Scholar]
- 8.Fischer J., Lefèvre C., Morava E., Mussini J.M., Laforêt P., Negre-Salvayre A., Lathrop M., Salvayre R. The gene encoding adipose triglyceride lipase (PNPLA2) is mutated in neutral lipid storage disease with myopathy. Nat. Genet. 2007;39:28–30. doi: 10.1038/ng1951. [DOI] [PubMed] [Google Scholar]
- 9.Schweiger M., Lass A., Zimmermann R., Eichmann T.O., Zechner R. Neutral lipid storage disease: genetic disorders caused by mutations in adipose triglyceride lipase/PNPLA2 or CGI-58/ ABHD5. Am. J. Physiol. Endocrinol. Metab. 2009;297:289–296. doi: 10.1152/ajpendo.00099.2009. [DOI] [PubMed] [Google Scholar]
- 10.Campagna F., Nanni L., Quagliarini F., Pennisi E., Michailidis C., Pierelli F., Bruno C., Casali C., DiMauro S., Arca M. Novel mutations in the adipose triglyceride lipase gene causing neutral lipid storage disease with myopathy. Biochem. Biophys. Res. Commun. 2008;377:843–846. doi: 10.1016/j.bbrc.2008.10.081. [DOI] [PubMed] [Google Scholar]
- 11.Hirano K., Ikeda Y., Zaima N., Sakata Y., Matsumiya G. Triglyceride deposit cardiomyovasculopathy. N. Engl. J. Med. 2008;359:2396–2398. doi: 10.1056/NEJMc0805305. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi K., Inoguchi T., Maeda Y., Nakashima N., Kuwano A., Eto E., Ueno N., Sasaki S., Sawada F., Fujii M., et al. The lack of the C-terminal domain of adipose triglyceride lipase causes neutral lipid storage disease through impaired interactions with lipid droplets. J. Clin. Endocrinol. Metab. 2008;93:2877–2884. doi: 10.1210/jc.2007-2247. [DOI] [PubMed] [Google Scholar]
- 13.Akiyama M., Sakai K., Ogawa M., McMillan J.R., Sawamura D., Shimizu H. Novel duplication mutation in the patatin domain of adipose triglyceride lipase (PNPLA2) in neutral lipid storage disease with severe myopathy. Muscle Nerve. 2007;36:856–859. doi: 10.1002/mus.20869. [DOI] [PubMed] [Google Scholar]
- 14.Coassin S., Schweiger M., Kloss-Brandstätter A., Lamina C., Haun M., Erhart G., Paulweber B., Rahman Y., Olpin S., Wolinski H., et al. Investigation and functional characterization of rare genetic variants in the adipose triglyceride lipase in a large healthy working population. PLoS Genet. 2010;6:e1001239. doi: 10.1371/journal.pgen.1001239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akman H.O., Davidzon G., Tanji K., Macdermott E.J., Larsen L., Davidson M.M., Haller R.G., Szczepaniak L.S., Lehman T.J., Hirano M., DiMauro S. Neutral lipid storage disease with subclinical myopathy due to a retrotransposal insertion in the PNPLA2 gene. Neuromuscul. Disord. 2010;20:397–402. doi: 10.1016/j.nmd.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J., Hong D., Wang Z., Yuan Y. A novel PNPLA2 mutation causes neutral lipid storage disease with myopathy (NLSDM) presenting muscular dystrophic features with lipid storage and rimmed vacuoles. Clin. Neuropathol. 2010;29:351–356. doi: 10.5414/npp29351. [DOI] [PubMed] [Google Scholar]
- 17.Haemmerle G., Lass A., Zimmermann R., Gorkiewicz G., Meyer C., Rozman J., Heldmaier G., Maier R., Theussl C., Eder S., et al. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science. 2006;312:734–737. doi: 10.1126/science.1123965. [DOI] [PubMed] [Google Scholar]
- 18.Wessalowski R., Schroten H., Neuen-Jacob E., Reichmann H., Melnik B.C., Lenard H.G., Voit T. Multisystem triglyceride storage disorder without ichthyosis in two siblings. Acta Paediatr. 1994;83:93–98. doi: 10.1111/j.1651-2227.1994.tb12960.x. [DOI] [PubMed] [Google Scholar]
- 19.Zechner R., Kienesberger P., Haemmerle G., Zimmermann R., Lass A. Adipose triglyceride lipase and the lipolytic catabolism of cellular fat stores. J. Lipid Res. 2009;50:3–21. doi: 10.1194/jlr.R800031-JLR200. [DOI] [PubMed] [Google Scholar]
- 20.Lass A., Zimmermann R., Haemmerle G., Riederer M., Schoiswohl G., Schweiger M., Kienesberger P., Strauss J.G., Gorkiewicz G., Zechner R. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell. Metab. 2006;3:309–319. doi: 10.1016/j.cmet.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 21.Ghosh A.K., Ramakrishnan G., Chandramohan C., Rajasekharan R. CGI-58, the causative gene for Chanarin-Dorfman syndrome, mediates acylation of lysophosphatidic acid. J. Biol. Chem. 2008;283:24525–24533. doi: 10.1074/jbc.M801783200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montero-Moran G., Caviglia J.M., McMahon D., Rothenberg A., Subramanian V., Xu Z., Lara-Gonzalez S., Storch J., Carman G.M., Brasaemle D.L. CGI-58/ABHD5 is a coenzyme A-dependent lysophosphatidic acid acyltransferase. J. Lipid Res. 2010;51:709–719. doi: 10.1194/jlr.M001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schoiswohl G., Schweiger M., Schreiber R., Gorkiewicz G., Preiss-Landl K., Taschler U., Zierler K.A., Radner F.P., Eichmann T.O., Kienesberger P.C., et al. Adipose triglyceride lipase plays a key role in the supply of the working muscle with fatty acids. J. Lipid Res. 2010;51:490–499. doi: 10.1194/jlr.M001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huijsman E., van de Par C., Economou C., van der Poel C., Lynch G.S., Schoiswohl G., Haemmerle G., Zechner R., Watt M.J. Adipose triacylglycerol lipase deletion alters whole body energy metabolism and impairs exercise performance in mice. Am. J. Physiol. Endocrinol. Metab. 2009;297:E505–E513. doi: 10.1152/ajpendo.00190.2009. [DOI] [PubMed] [Google Scholar]
- 25.Wölkart G., Schrammel A., Dörffel K., Haemmerle G., Zechner R., Mayer B. Cardiac dysfunction in adipose triglyceride lipase deficiency: treatment with a PPARα agonist. Br. J. Pharmacol. 2012;165:380–389. doi: 10.1111/j.1476-5381.2011.01490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haemmerle G., Moustafa T., Woelkart G., Büttner S., Schmidt A., van de Weijer T., Hesselink M., Jaeger D., Kienesberger P.C., Zierler K., et al. ATGL-mediated fat catabolism regulates cardiac mitochondrial function via PPAR-α and PGC-1. Nat. Med. 2011;17:1076–1085. doi: 10.1038/nm.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reilich P., Horvath R., Krause S., Schramm N., Turnbull D.M., Trenell M., Hollingsworth G., Gorman G.S., Hans V.H., Reimann J., et al. The phenotypic spectrum of neutral lipid storage myopathy due to mutations in the PNPLA2 gene. J. Neurol. 2011;258:1987–1997. doi: 10.1007/s00415-011-6055-4. [DOI] [PubMed] [Google Scholar]
- 28.Lu X., Yang X., Liu J. Differential control of ATGL-mediated lipid droplet degradation by CGI-58 and G0S2. Cell Cycle. 2010;9:2719–2725. doi: 10.4161/cc.9.14.12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kampmann C., Kalkum G., Beck M., Whybra C. Successful long-term enzyme replacement therapy in a young adult with Fabry disease. Clin. Genet. 2012 doi: 10.1111/j.1399-0004.2012.01916.x. doi:10.1111/j.1399-0004.2012.01916.x. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 30.Richard E., Douillard-Guilloux G., Caillaud C. New insights into therapeutic options for Pompe disease. IUBMB Life. 2011;63:979–986. doi: 10.1002/iub.529. [DOI] [PubMed] [Google Scholar]
- 31.Tavian D., Salvi A., De Petro G., Barlati S. Stable expression of antisense urokinase mRNA inhibits the proliferation and invasion of human hepatocellular carcinoma cells. Cancer Gene Ther. 2003;10:112–120. doi: 10.1038/sj.cgt.7700533. [DOI] [PubMed] [Google Scholar]
- 32.Fujimoto Y., Itabe H., Sakai J., Makita M., Noda J., Mori M., Higashi Y., Kojima S., Takano T. Identification of major proteins in the lipid droplet-enriched fractio isolated from the human hepatocyte cell line HuH7. Biochim. Biophys. Acta. 2004;1644:47–59. doi: 10.1016/j.bbamcr.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 33.Koopman R., Schaart G., Hesselink M.K. Optimisation of oil red O staining permits combination with immunofluorescence and automated quantification of lipids. Histochem. Cell Biol. 2001;116:63–68. doi: 10.1007/s004180100297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.