Abstract
We previously demonstrated that the histone deacetylase (HDAC) inhibitor, 4b, which preferentially targets HDAC1 and HDAC3, ameliorates Huntington's disease (HD)-related phenotypes in different HD model systems. In the current study, we investigated extensive behavioral and biological effects of 4b in N171-82Q transgenic mice and further explored potential molecular mechanisms of 4b action. We found that 4b significantly prevented body weight loss, improved several parameters of motor function and ameliorated Huntingtin (Htt)-elicited cognitive decline in N171-82Q transgenic mice. Pathways analysis of microarray data from the mouse brain revealed gene networks involving post-translational modification, including protein phosphorylation and ubiquitination pathways, associated with 4b drug treatment. Using real-time qPCR analysis, we validated differential regulation of several genes in these pathways by 4b, including Ube2K, Ubqln, Ube2e3, Usp28 and Sumo2, as well as several other related genes. Additionally, 4b elicited increases in the expression of genes encoding components of the inhibitor of kappaB kinase (IKK) complex. IKK activation has been linked to phosphorylation, acetylation and clearance of the Htt protein by the proteasome and the lysosome, and accordingly, we found elevated levels of phosphorylated endogenous wild-type (wt) Htt protein at serine 16 and threonine 3, and increased AcK9/pS13/pS16 immunoreactivity in cortical samples from 4b-treated mice. We further show that HDAC inhibitors prevent the formation of nuclear Htt aggregates in the brains of N171-82Q mice. Our findings suggest that one mechanism of 4b action is associated with the modulation of the ubiquitin–proteasomal and autophagy pathways, which could affect accumulation, stability and/or clearance of important disease-related proteins, such as Htt.
INTRODUCTION
Epigenetic dysregulation and transcriptional dysregulation have emerged as common pathological mechanisms in many neurological disorders, especially Huntington's disease (HD) (1–10). Accordingly, therapeutic approaches that target gene expression represent an encouraging new avenue of exploration for these disorders. Several studies have identified histone deacetylase (HDAC) inhibitors as candidate drugs for the treatment of different neurological disorders (11,12). Although early studies were performed using broad-spectrum HDAC inhibitors, it is becoming clear that inhibitors with isoform-selectivity may prove more beneficial for targeting neurological disease symptoms and minimizing harmful side effects (13). Class I-specific, benzamide-type HDAC inhibitors have been developed as a therapeutic approach for Friedreich's ataxia (14,15). We have previously studied one particular compound in this class, 4b, and have shown that it preferentially targets HDAC1 and HDAC3 enzymes and exhibits low in vitro and in vivo toxicity (16,17). Studies using HD model systems have revealed that 4b can ameliorate motor and behavioral symptoms and correct transcriptional abnormalities in R6/2 transgenic mice, when drugs were administered in drinking water (17), and can improve Htt-elicited phenotypes in HD Drosophila and in STHdhQ111 striatal cells (16). However, the mechanisms by which 4b, and related compound of this class, are imparting benefit in disease models are unclear.
In the current study, we tested the effects of different doses of HDACi 4b on a range of disease phenotypes in N171-82Q transgenic mice and further explored potential mechanisms of the action of HDACi 4b by performing network analysis of gene expression data and validating the expression changes of selected genes by real-time qPCR analysis. Our findings demonstrate that HDAC inhibition by 4b can affect protein post-translational modification processes at the level of gene expression, can increase phosphorylation and acetylation of endogenous Htt protein and can prevent formation of mutant Htt aggregates in the brain; these effects may contribute to its clinically beneficial properties observed in HD mice. Because post-translational modification of target proteins can modify their accumulation, stability and/or clearance, we suggest that HDAC inhibitors may be acting to modify the processing of important proteins in the cell, such as the Htt protein.
RESULTS
The effects of 4b on disease phenotypes in N171-82Q transgenic mice
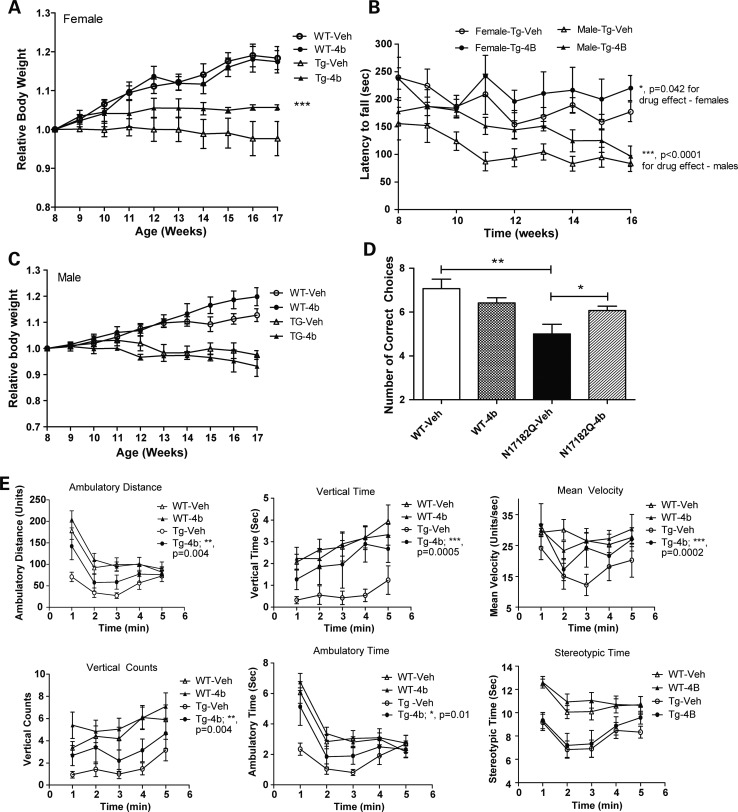
Groups of N171-82Q transgenic mice and wt littermate controls (both males and females) were treated with HDACi 4b (15, 50 or 100 mg/kg; s.c.; 2.5 injections per week; n = 12–20 per drug and vehicle groups) for 10 weeks, beginning at 8 weeks of age. The effects of HDACi 4b administration on several disease phenotypes, including body weight, rotarod performance, T-maze and open-field exploratory behavior, were measured. These results are summarized in Table 1. Consistent with our previous studies, we found that 4b significantly improved body weight of N171-82Q mice, at a dose of 100 mg/kg, although this effect was observed in females only (Fig. 1A). In male mice, 4b caused a small but significant increase in body weight of wt mice (Fig. 1B).
Table 1.
Summary of the HD phenotypes affected by 4b treatment at different doses
| Test | Sex | 100 mg/kg | 50 mg/kg | 15 mg/kg |
|---|---|---|---|---|
| Body weight | Male | NS | NS | NS |
| Female | **P < 0.001 | NS | NS | |
| Rotarod | Male | ***P < 0.0001 | ***P < 0.0001 | NS |
| Female | *P = 0.041 | *P = 0.042 | NS | |
| Brain weight | Male and female | ND | NS | ND |
| T-maze | Male and female | ND | *P < 0.05 | ND |
| Survival | Male and female | NS | NS | NS |
| Open field | ||||
| Ambulatory distance | Male and female | ND | **P = 0.004 | *P = 0.011 |
| Ambulatory time | Male and female | ND | P = 0.01 | *P = 0.017 |
| Stereotypic time | Male and female | ND | NS | *P = 0.025 |
| Mean velocity | Male and female | ND | ***P = 0.0002 | NS |
| Vertical counts | Male and female | ND | **P = 0.004 | NS |
| Vertical time | Male and female | ND | ***P = 0.0005 | NS |
All doses were administered subcutaneously. Statistical analyses for the different tests included two-way ANOVAs and Student's t-tests, as described in Materials and Methods. NS, non-significant; ND, not determined.
Figure 1.
The effects of 4b on the relative body weights of female (A) and male (C) wt and N171-82Q transgenic mice. The relative body weights were based on the weight at 8 weeks of age. Differences in drug- versus vehicle-treated mice were determined by two-way ANOVA (F(1,63) = 11.8; **P = 0.0011). (B) Rotarod performance of vehicle- and HDACi 4b-treated female and male N171-82Q transgenic mice from 8 to 16 weeks of age. Two-way ANOVA revealed significant differences between vehicle- and HDACi 4b-treated female transgenic mice (F(1,135) = 4.2; P = 0.040), as well as a significant effect of drug treatment in male N171-82Q transgenic mice (F(1,126) = 23.5; P < 0.0001). (D) Number of correct choice of first 10 trials in the alternating T-maze test. One-way ANOVA revealed significant differences between vehicle-treated wt and N171-82Q transgenic mice (**P < 0.001), and also a significant difference between vehicle-treated and HDACi 4b-treated N171-82Q mice (*P < 0.05). Bars represent mean score ± SEM (n = 6–7 per group). (E) Effects of HDACi 4b treatment on open-field activity of wt and N171-82Q transgenic mice (n = 15–20 mice per group) are shown over a 5 min test period. Two-way ANOVA revealed significant differences between HDACi 4b-treated and vehicle-treated N171-82Q transgenic mice in ambulatory distance (P = 0.004), vertical time (P = 0.0005), mean velocity (P = 0.0002), vertical counts (P = 0.004) and ambulatory time (P = 0.01). The summary of total effects of HDACi 4b treatment at different doses is shown in Table 1.
Rotarod performance was tested weekly in vehicle- and drug-treated mice from 8 to 16 weeks of age. Owing to the significant difference in rotarod performance between male and female transgenic mice [two-way analysis of variance (ANOVA); F(1,67) = 77.6; P < 0.0001], we analyzed the drug effect on males and females separately. 4b treatment (50 and 100 mg/kg) significantly improved rotarod performance of N171-82Q transgenic mice of both sexes, although greater effects were detected in male mice (35% improvement in mean performance for males: two-way ANOVA; F(1,126) = 23.5; P < 0.0001; versus 11.4% improvement in mean performance for females: F(1,135) = 4.2; P = 0.040) (Fig. 1C). 4b had no significant effects on rotarod performance of wt mice (data not shown).
4b treatment elicited significant improvement in several measures of open-field activity. These included ambulatory time, ambulatory distance, rearing activity and mean velocity (two-way ANOVA; P-values = 0.01–0.0001) (Fig. 1E). The greatest beneficial effects of 4b were on rearing behavior, in particular vertical time, whereby 4b-treated mice performed at the same level as wt mice (Fig. 1E).
We further tested 4b for its ability to improve cognitive deficits observed in N171-82Q transgenic mice, as measured by the alternating T-maze test. 4b-treated N171-82Q transgenic mice made significantly more correct choices for a food reward than vehicle-treated N171-82Q transgenic mice (Fig. 1D).
Gender differences in HDAC subtype gene expression
Given the sex differences observed by 4b treatment on body weight and rotarod, we measured the expression of HDAC subtypes targeted by 4b in treated male and female mice by real-time qPCR analysis. We found that Hdac1 mRNA expression was significantly higher in the cortex of male transgenic mice compared with female transgenic mice, with no changes in the expression of Hdac2 or Hdac3 (Supplementary Material, Fig. S1). No changes in the expression of HDAC subtypes were detected between male and female wt mice.
Pathways analysis of 4b-mediated transcriptome changes
From our previously published microarray data, we found that a 3-day treatment with 4b elicited a wide range of expression changes, including both up- and down-regulated genes [(17); GEO accession GSE26317]. Using the Ingenuity Pathways Analysis (IPA) software, functional analysis of the top gene expression changes from the brains of 4b-treated mice revealed significant associations with post-translational modification pathways, which included protein phosphorylation and ubiquitination (Table 2). Using IPA Network analysis, which identifies molecular relationships among genes or gene products, we identified several networks of highly connected genes related to post-translational modification processes that were regulated by 4b treatment. Strong connectivity among these genes was detected in different brain regions (cortex, striatum and cerebellum) and in both 4b-treated wt and transgenic mice (Table 3). Selected gene network connectivity maps from the cortex, striatum and cerebellum of 4b-treated mice are shown in Supplementary Material, Figure S2.
Table 2.
Post-translational modification categories associated with 4b treatment in the mouse brain
| Post-translational modification category | P-value | Number of molecules |
|---|---|---|
| Moiety attachment of protein | 6.86E − 05 | 63 |
| Phosphorylation of l-tyrosine | 1.15E − 03 | 12 |
| Phosphorylation of protein | 1.46E − 03 | 39 |
| Modification of protein | 2.14E − 03 | 70 |
| Phosphorylation of l-amino acid | 2.68E − 03 | 14 |
| Moiety attachment of amino acids | 1.39E − 02 | 15 |
| Ubiquitination of protein | 1.44E − 02 | 17 |
Post-translational modification categories were annotated as in the Ingenuity Systems database. The P-value was calculated using the right-tailed Fisher exact test, considering the number of molecules participating in a given function and the total number of molecules associated with that particular function in Ingenuity's knowledge base.
Table 3.
Network analysis of microarray data from brains of 4b-treated HD mice
| Top functions | Molecules in network | Score | Number of genes |
|---|---|---|---|
| 4b-treated N171-82Q cortex | |||
| Molecular transport, post-translational modification, protein folding | ARNTL2, C3orf17, CCNB2, CFTR, CHRNA5, COPE, CSMD3, DCUN1D5, GORAB, IDI1, MARK3, MESP2, NKD2, PAX3, POLH, PRKCI, PRKCZ, RAD51B, RCHY1, RLBP1, RNASE12, SCT, SLC26A9, SLC39A2, SLC44A3, SRR, TBX22, TCF3, TCF15, UBC, USP10, USP18, USP28, USP36, ZDHHC15 | 23 | 16 |
| Amino acid metabolism, energy production, post-translational modification | ATF6, ATF6B, BARX1, C11orf82, C11orf86, CYBASC3, ENO1, EPX, HMGCS2, HP, IFI6, IFI44L, IFNG, IL37, IL10RB, IL4I1, Irgm2, ISL1, KY, LCN2, LPO, MAFA, MANBA, MBTPS2, MBTPS, MERTK, PPARD, PPP1R1C, Runx1, SMAD3, TIA1, XAF1, ZNF8 | 17 | 13 |
| 4b-treated N171-82Q striatum | |||
| Post-translational modification, tissue morphology, cellular organization | AES, ARHGDIA, ARIH2, ASB6, ASB7, CIT, DVL2, GFI1B, HCN3, HCN4, LHX3, NDUFS2, NDUFS5, NELL2, PARK2, PITX2, RHOF, RNF7, RNF11, RNF103, SDHC, SNCAIP, TRIM23, TRIM29, UBE2, UBE2D3, UBE2R2, UBE4A, VANGL1, Wnt, WNT2B, ZNRD1 | 41 | 30 |
| Gene expression, RNA post-transcriptional modification, connective tissue development and function | ARL6IP4, CCNT1, CCNT2, CLDN8, CTDP1, DDX5, DOT1L, EPAS1, GTF2A1, GTF2H2, GTF3C5, HBZ, Histone h3, HOXA9, HOXD13, Ikb, IKBKG, IKK, ITGBL1, LRPPRC, P-TEFb, PADI3, POLR2B, PTCD2, TAF7, Taf, TAF11, TAF12, TBP, TFIIH, ZFP64, ZNF238 | 32 | 26 |
| 4b treated N171-82Q cerebellum | |||
| Post-translational modification, RNA post-transcriptional modification, developmental disorder | ADPGK, BLMH, CD2BP2, CENPB, DUB, FAM20C, GDF9, GRAP2, LSM10, PI3K, POLR3A, POLR3H, POP5, POP7, PRPS2, PRPSAP1, RAE1, RPP25, SART1, SF3B3, Shc, SMN1/SMN2, snRNP, SNRPB, SNRPN, SREK1, TBL3, USP1, USP8, USP11, USP16, USP18, USP53, WDR24 | 33 | 30 |
| Cell-to-cell signaling and interaction, small molecule biochemistry, post-translational modification | 7S NGF, 14-3-3, AKT1S1, AS3MT, C14orf133, CAB39, CHAT, DUOX2, JAM2, LGALS9B, MARK3, NIPSNAP1, P38 MAPK, PARD6G, PCSK2, PCSK7, PCSK9, Pcsk, PNMA3, RASGRF1, RGS7, RGS(2,4,7,16,18), SERCA, SIK3, SLC12A2, SLC18A3, SLC5A7, SOX11, TEAD2, VPS11, VPS33A, YAP1, YWHAH | 27 | 27 |
| 4b-treated wt cortex | |||
| Gene expression, protein synthesis, RNA post-transcriptional modification | AKT1S1, C2orf3, CBFB, CCND3, CRY2, Cyclin E, DGUOK, DST, DTNBP1, EIF4E, Eif4g, EPHA8, INTS6, KCNIP2, KCNIP4, MTORC1, NCOA3, NPAS2, p70 S6k, PER3, PFKP, PHF5A, PI3K RBP1, RPS6, RPS6KA, RPS6KB1, RPS6KB2, Rsk, SVIL, T3-TR-RXR, TSH, UCP3, UPP1 | 33 | 25 |
| Protein synthesis, RNA post-transcriptional modification, gene expression | AGTR1, AMHR2, ARRB2, BOP1, CEP250, CPA1, CSDA, DCD, EIF2AK3, ERF, FBXL5, HNRNPA0, LOC100505503/RPS17, MFAP1, MYC, NFYA, NKTR, NOLC1, PAPOLG, POLR1B, RAD51L3, RBM10, RPL6, RPL18, RPL19, RPL21, RPL30, RPL35A, SP6, SPI1, SPIN1, STAT1, TCOF1, TUBA8, TWISTNB | 18 | 17 |
| 4b-treated wt striatum | |||
| Amino acid metabolism, drug metabolism, post-translational modification | AHR, Ahr-Arnt, Arnt-Hif1a, ATRN, BTK, COPS5, CYSLTR2, E2F8, EIF2AK4, EWSR1, FZD9, JUN, KISS1R, KLC1, KLHL18, LOC100505793/SRSF10, LUC7L3, LYST, MBTPS2, MLLT1, MRPS31, MTMR9, PLXNA2, PPP1R10, PPP2CA, RAB37, RMND5B, SGOL2, SLC30A9, SMAD4, STK11IP, TNFRSF19, UGT1A3, UGT1A6, UQCRB | 14 | 13 |
| Post-translational modification, tissue development, cellular assembly and organization | C20orf94, CORO2A, CSRNP2, CUL1, DCTN4, DMRTC2, DYNC1I1, DYNC1I2, DYNC1LI1, DYNC1LI2, DYNC2H1, DYNLRB1, DYNLRB2, EIF5B, FBXL20, FBXW4, FNTA, GULP1, HIF1A, MAL, MLF1, NR3C1, NRN1, OTX2, PCDH10, PCNT, PTP4A2, RABGGTB, SEMA5B, TCTE3, TNFAIP8, UBXN7, WDR6, ZBTB3 | 14 | 14 |
| 4b-treated wt cerebellum | |||
| Post-translational modification, cell cycle, DNA replication, recombination and repair | ACTN3, ANAPC7, BAALC, BAP1, Cmya5, DGKZ, DMD, DUB, ENTPD3, FOXK1, HIST2H2AA3/HIST2H2AA4, IKK, KRI1, LRRC59, MAOB, MAP7, PLEKHA1, PSTPIP2, RCSD1, SCN3B, SGSH, SNTA1, SNTB2, SSSCA1, TP53BP1, TPM3, USP1, USP4, USP8, USP16, USP20, USP28, USP29, USP47, USP53 | 40 | 33 |
| Post-translational modification, DNA replication, recombination, and repair, protein degradation | API5, BRD4, CHFR, CPEB1, FBXO18, HTATSF1, IL12, Il18r, LRSAM1, MGAT4B, MKRN3, ADH dehydrogenase, NADH2 dehydrogenase, NDUFA7, NDUFA8, NDUFB8, NDUFC2, NDUFS7, P-TEFb, PIM2, PINK1, POLD2, POLDIP3, POLM, RFC5, RFWD3, RNF4, RNF8, UBE2E1, UBE2N, UFL1, XRCC4 | 28 | 27 |
Data were analyzed through the use of IPA (Ingenuity® Systems, www.ingenuity.com). The Network score is based on the hypergeometric distribution and is calculated with the right-tailed Fisher exact test. The score is the negative log of this P-value. The higher the score, the lower the probability of finding the observed number of Network-eligible molecules in a given network by random chance.
Expression validation of ubiquitination-related genes
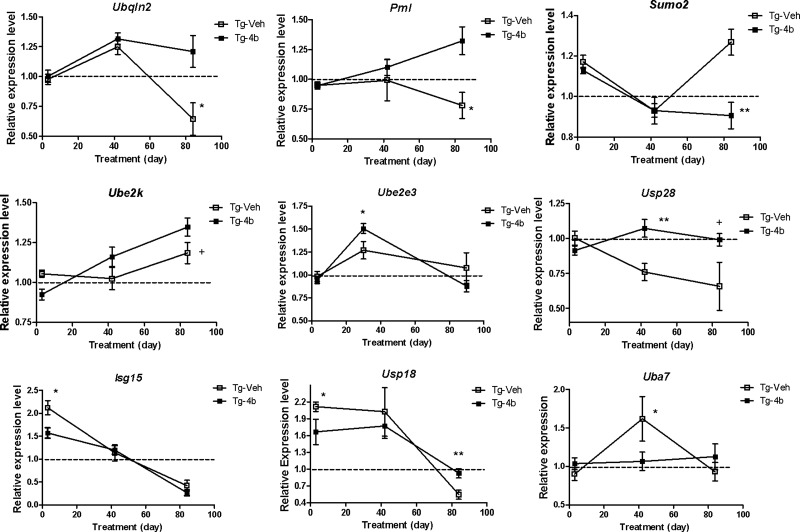
We validated expression differences of selected genes related to the ubiquitination pathway by real-time qPCR analysis at later time points of drug treatment, 6 and 12 weeks, in the striatum and cortex of N171-82Q transgenic mice. The most pronounced expression differences detected in the striatum were late in treatment (12 weeks of treatment; 20 weeks of age), in contrast to the cortex, where expression differences were most notable earlier in treatment (6 weeks of treatment, 14 weeks of age or less) (Figs 2 and 3). In the striatum, 4b caused significant ∼2-fold increases in the expression of Ubiquilin 2 (Ubqln2) and Promyelocytic leukemia (Pml) and a 1.5-fold increase in the expression of Ubiquitin-specific peptidase 28 (Usp28) in N171-82Q mice after 12 weeks of treatment (Fig. 2). Decreased expression in N171-82Q mice in response to 4b was detected for other genes, including Ubiquitin-like modifier activating enzyme 7 (Uba7; a.k.a. Ube1l) and SMT3 suppressor of mif two 3 homolog 2 (Sumo2) at 6 and 12 weeks of treatment, respectively. Changes in the expression of other ubiquitin-related genes in N171-82Q mice were more modest, yet in several cases, still statistically significant (Fig. 2).
Figure 2.
Real-time qPCR analysis showing the effects of 4b on the expression of the indicated ubiquitination-related genes in the striatum of N171-82Q transgenic mice. Values shown are the mean ± SEM expression values (n = 5–6 mice per group) compared with a wt expression level of 1, which is indicated by the dotted line. Significant differences were determined by Student's t-tests (unpaired; one-tailed) at the indicated treatment time points. *P < 0.05; **P < 0.01; +P < 0.08.
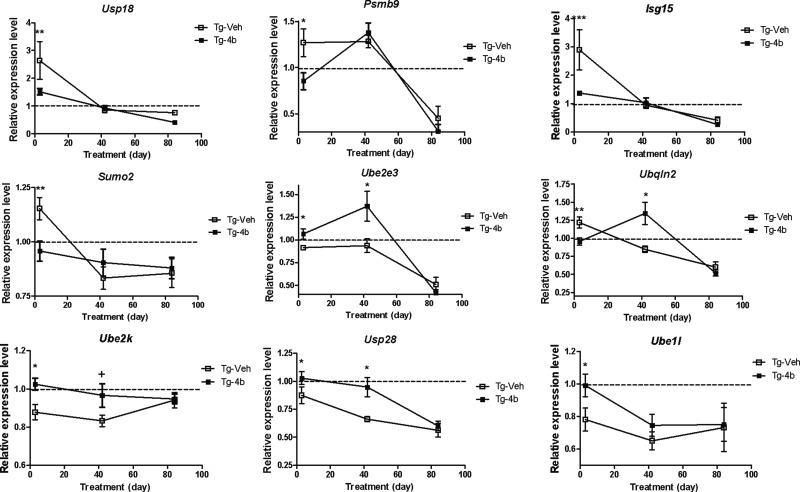
Figure 3.
Real-time qPCR analysis showing the effects of 4b on the expression of the indicated ubiquitination-related genes in the cortex of N171-82Q transgenic mice. Values shown are the mean ± SEM expression values (n = 5–6 mice per group) compared with a wt expression level of 1, which is indicated by the dotted line. Significant differences were determined by Student's t-tests (unpaired; one-tailed) at the indicated treatment time points. *P < 0.05; **P < 0.01; ***P < 0.001; +P < 0.08.
In the cortex, 4b normalized Htt-induced deficits in the expression of Ubiquitin-conjugating enzyme E2K (Ube2k; a.k.a. Hip2), Uba7 and Usp28, at 3 days and 6 weeks of treatment but not at the late stages of disease (Fig. 3). 4b also normalized Htt-elicited expression increases after only 3 days of treatment for Usp18, Psmb9, SMT3 suppressor of mif two 3 homolog 2 (Sumo2) and ISG15 ubiquitin-like modifier (Isg15) (Fig. 3).
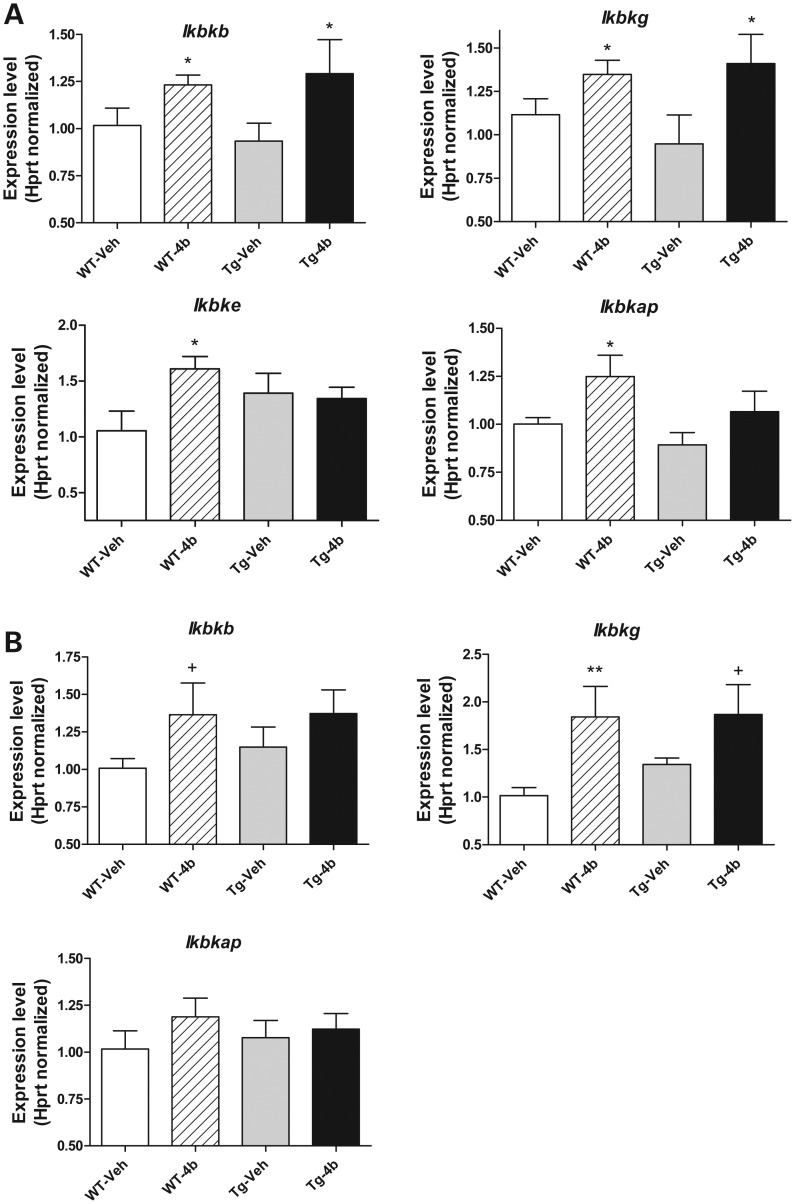
Validation of IKK-related genes
Although several different types of protein kinase genes were found to be altered by 4b in our microarray studies, we focused our follow-up studies on the IKK complex pathway, which has been previously associated with modulating the toxicity of Htt protein and the induction of cellular autophagy (18–22). We measured the expression levels of four IKK-related genes in response to 6 weeks of 4b treatment: Inhibitor of kappaB kinase beta (Ikbkb), Inhibitor of kappaB kinase gamma (Ikbkg), Inhibitor of kappaB kinase epsilon (Ikbke), Inhibitor of kappa light polypeptide enhancer in B cells, kinase complex-associated protein (Ikbkap). We found that 4b elevated the expression of all four genes in the cortex from wt mice and Ikbkb and Ikbkg in the striatum of wt mice (Fig. 4). These latter two genes, Ikbkb and Ikbkg, were elevated in the cortex of N171-82Q transgenic mice, and only Ikbkg in the striatum of N171-82Q transgenic mice in response to 4b treatment (Fig. 4). Ikbke was not detected at consistently detectable levels in the striatum and hence, could not be assessed. As a correlate, we measured the expression of selected genes related to autophagy and found that 4b treatment significantly elevated the expression of the Autophagy related 4c (Atg4c) gene in both the cortex and striatum of wt and/or N171-82Q transgenic mice, decreased the expression of Autophagy related 9b (Atg9b) in the striatum of N171-82Q mice and decreased the expression of BCL2/adenovirus E1B interacting protein 3 (Bnip3) in the striatum of wt and N171-82Q mice (Supplementary Material, Fig. S3). This is consistent with our microarray data, which also revealed increased expression of Map1lc3a (LC3), a well-known autophagy marker, in the cortex of 4b-treated mice (data not shown).
Figure 4.
Real-time qPCR analysis showing the effects of 4b treatment on the IKK-related genes in the cortex (A) and striatum (B) of wt and N171-82Q transgenic mice. Values shown are the mean ± SEM expression values (n = 5–6 mice per group). Significant differences were determined by Student's t-tests (unpaired; one-tailed) at the indicated treatment time points. *P < 0.05; **P < 0.01; +P < 0.08.
Post-translational modification of Htt protein
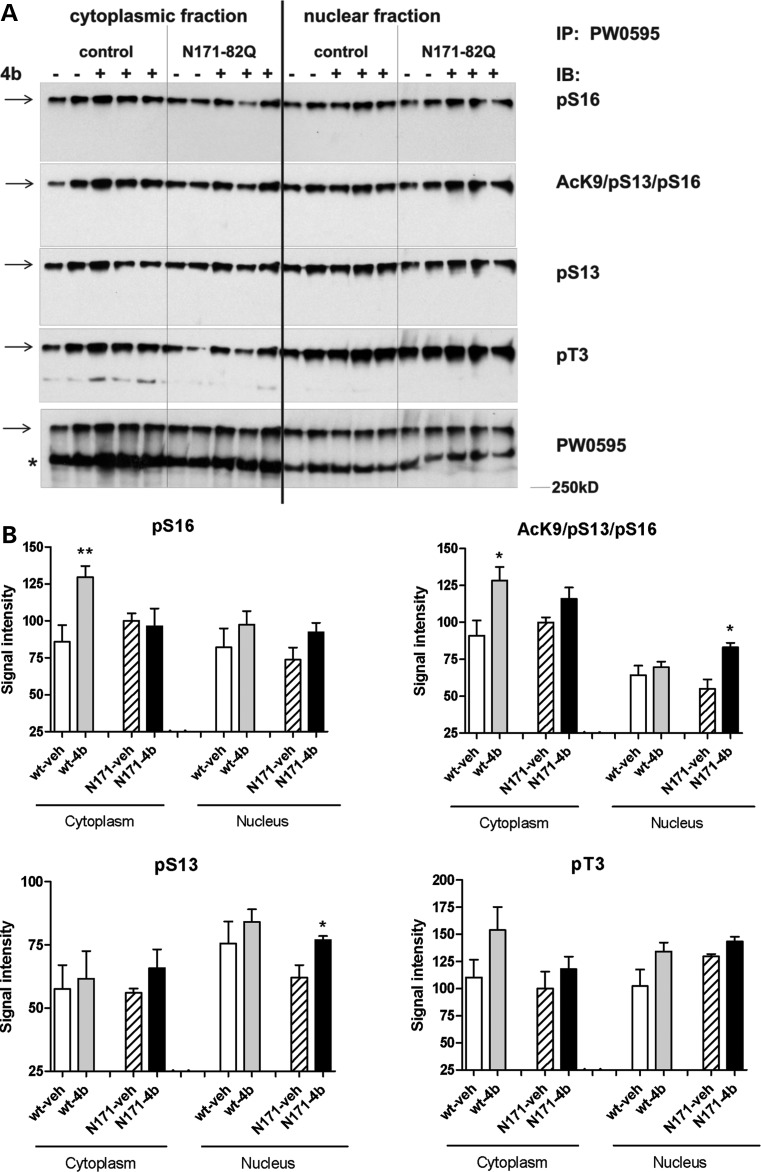
Previous studies have shown that IKK activation leads to increased phosphorylation and acetylation of Htt protein and clearance by the proteasome and lysosome, thereby modulating Htt-mediated toxicity (19–22). Therefore, we measured the effects of 4b on phosphorylation of Htt protein at serine residues S13 and S16, and threonine T3, using western blot analysis. We first immunoprecipitated total Htt from cytoplasmic and nuclear fractions of cortex from vehicle- and 4b-treated mice, using a novel method that involved an anti-Htt antibody (PW0595) covalently linked to magnetic beads using a hydrazide linkage. Immunoprecipitated fractions were then probed with antibodies generated against the specific modifications of Htt. Immunoprecipitated phosphorylated and acetylated Htt runs on SDS–PAGE as a high molecular weight relatively insoluble species and can be resolved only when a loading buffer rich in beta-mercaptoethanol and SDS is used. This Htt species can be transferred to membrane for western analysis if extra methanol and SDS are added to the transfer buffer (23). In general, phosphorylation of Htt at all three sites was elevated in response to 4b treatment in cytoplasmic and/or nuclear fraction, although only pS16 in the cytoplasm and pS13 in the nucleus reached statistical significance (Fig. 5). Phosphorylation of S13/S16 has been shown to promote acetylation of Htt at K9 (20); hence, we further measured the effects of 4b treatment on acetylated Htt at lysine K9, a form of Htt that co-exists with phosphorylation of S13/S16. A significant increase in acK9/pS13/pS16 reactivity was detected in cytoplasmic fractions of the cortex from 4b-treated wt mice and in nuclear fractions of N171-82Q mice compared with vehicle-treated wt and N171-82Q mice, respectively (Fig. 5).
Figure 5.
Modifications of Htt protein by 4b treatment in the cortex of wt (control) and N171-82Q transgenic mice. (A) Cytosolic and nuclear protein fractions from cortex of vehicle- and 4b-treated mice were probed with antibodies recognizing different phosphorylation and acetylation states of Htt protein. Homogenates were immunoprecipitated (IP) using PW0595 anti-Htt antibody as described in Materials and Methods. Immunoblots (IB) were run using the indicated specific antibodies. (B) Quantitation of band intensity was done for the high molecular weight modified Htt species designated by the arrow, which runs larger than the standard full-length 350 kDa Htt species (*), shown in (A). Bar graphs represent data from n = 6–7 samples for pT3, pS16 and AcK9/pS13/pS16 in the cytoplasmic fractions and n = 2–3 samples for all nuclear fractions and the pS13 cytoplasmic fraction. Statistical significance was determined using Student's t-tests comparing 4b- and vehicle-treated samples. *P < 0.05; **P < 0.01.
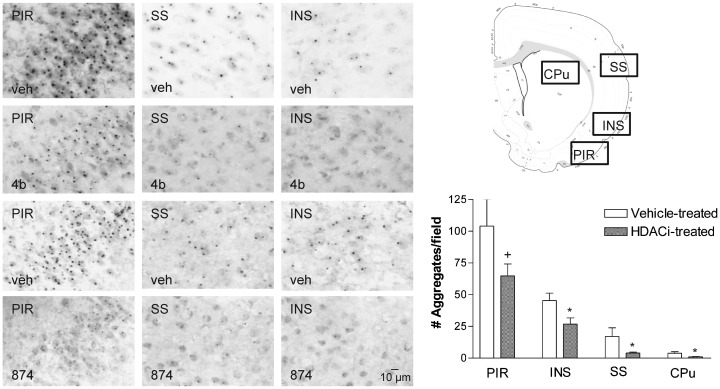
We next investigated the effects of 4b on mutant Htt aggregates in different brain regions of N171-82Q transgenic mice, using immunohistochemistry with the widely used anti-Htt antibody, EM48. We found that brains from 4b-treated N171-82Q transgenic mice contained less Htt aggregates in the cortex and striatum compared with vehicle-treated mice, although the striatum exhibited substantially fewer aggregates than cortical regions in general (Fig. 6). To validate this effect, we also measured mutant Htt aggregates in the brains of mice treated with another HDAC inhibitor, 874, which is similar to 4b in structure and in HDAC subtype selectivity [targeting primarily HDACs 1 and 3; (16)]. Structurally, HDACi 874 differs from 4b in replacement of the ‘left’ amide in the linker region with an ether and contains a different cap group, as described previously (16). Quantification of neurons containing Htt protein aggregates revealed significantly fewer EM48-positive aggregates in the cortex and striatum of 874-treated brains compared with vehicle (Fig. 6). In addition to the numbers of aggregates being lower, the size of the aggregates in HDAC inhibitor-treated HD mice was also smaller (Fig. 6).
Figure 6.
The effect of 4b and 874 on Htt aggregates in different brain regions from N171-82Q transgenic mice. Immunohistochemistry was performed on free-floating sections (25 µm) from vehicle-, 4b- and 874-treated mice (6 weeks of treatment), using the anti-Htt antibody EM48 (1:500 dilution). Aggregates were quantified by counting the number of EM48-positive aggregates per 150 μm × 150 μm field at 64× magnification. Micrographs from caudate putamen are not shown due to very low numbers of aggregates. Bar graphs depict quantification of Htt aggregates from 874-treated N171-82Q transgenic mice (n = 6 per group). PIR, piriform cortex; INS, insular cortex; SS, somatosensory cortex; CPu, caudate putamen. Asterisks denote statistically significant values using Student's t-test, unpaired, two-tailed: *P < 0.05; **P < 0.01.
DISCUSSION
In this study, we report beneficial effects of the benzamide-type HDAC inhibitor, HDACi 4b, on disease phenotypes in N171-82Q transgenic mice, which include significantly improved movement and motor function, cognitive behavior and delayed weight loss over vehicle-treated animals. We linked these beneficial effects to changes in the expression of genes related to post-translational modification providing a potential mechanism of action for 4b. Post-translational modification of target proteins can regulate their stability and clearance from cells; hence, we hypothesize that 4b, and other related HDAC inhibitors, could affect a wide range of important target proteins, such as Htt, by altering these processes.
In our behavioral studies, we found that 4b was most effective at a dose of 50 mg/kg (s.c.), although some benefits were seen in selected tasks at a dose of 15 mg/kg. The greatest beneficial effects of 4b, in magnitude, were detected on rearing behavior, whereby 4b-treated mice performed at the same level as wt mice, although 4b also significantly improved measures of ambulatory distance and increased average velocity in the open field. We found rotarod behavior to be highly variable in the N171-82Q mice. First, significant differences were detected in performance between males and females, similar to data reported in a previous study (24). And, although 4b was effective at improved behavior in both sexes, the effect on males was much greater than that observed in females. 4b also showed a differential sex effect on body weight, whereby the positive effects of drug treatment were observed only in female N171-82Q transgenic mice. We have previously shown that 4b selectively targets HDACs 1 and 3 (16). Herein, we found that Hdac1 mRNA expression was significantly elevated in male compared with female N171-82Q transgenic mice, although Hdac2 and Hdac3 mRNA expression were not significantly different between genders or genotypes. This could suggest that different HDAC enzymes are preferentially associated with different behaviors. For example, HDAC1 activity may be linked to metabolism and ensuing body weight loss in N171-82Q mice, whereby male mice, which show elevated HDAC1 expression, may be insensitive to compounds that inhibit HDAC1 in this measure. These findings further suggest that all symptoms in HD may not be driven by identical pathological mechanisms.
In addition to improving motor behavior, 4b improved performance in the T-maze forced alternation task, which assesses cognitive ability. 4b-treated mice showed a higher number of correct choices in alternating arm choices for a food reward. The blood–plasma ratio for 4b is 0.45 (16), reflecting its ability to penetrate the brain; nonetheless, it is important to measure CNS-driven phenotypes, given that 4b might also be acting on peripheral tissues (i.e. skeletal muscle) to improve motor phenotypes.
4b and related HDAC inhibitors have been shown to elevate histone acetylation, thereby altering chromatin structure to promote gene transcription (14,15,17). Hence, we explored potential mechanisms of action associated with altered gene transcription. First, we performed functional and network analyses of our previously generated microarray gene expression data, followed by validation of expression changes for selected genes after chronic 4b administration. Functional and network analyses implicated post-translational modification, notably ubiquitination and phosphorylation, as significantly associated with short-term 4b treatment. Both of these processes can affect the removal of misfolded or damaged proteins in the cell, which is critical for optimal cell functioning. In the cytosol and the nucleus, a major proteolytic pathway to recycle misfolded or damaged proteins is the ubiquitin–proteasome system, although phosphorylation of proteins is also known to regulate protein degradation.
Of the ubiquitin–proteasome pathway genes that we investigated, we found that 4b increased the expression of two genes, Ube2k and Ube2e3, encoding ubiquitin-conjugating enzymes, which play vital roles in the ubiquitin–proteasome system. The ubiquitin–proteasome system is the main intracellular pathway for regulated protein turnover. This system is essential for maintaining cellular homeostasis and for regulating fundamental cellular events, such as cell division, apoptosis and neuronal functioning (25,26). It is thought that dysregulated ubiquitination of target proteins could play a role in neurodegenerative disorders, many of which are pathologically characterized by the presence of ubiquitin-positive protein aggregates, suggesting impairment of the ubiquitin–proteasome system (25,27–30).
Another important ubiquitination player identified to be regulated by 4b in this study is Ubqln2, a member of the ubiquitin-like protein family of ubiquilins, which regulates the ubiquitin–proteasomal system by delivering ubiquinated proteins to the proteasome. We found that 4b significantly upregulated Ubqln2 in both the cortex and the striatum, but at different time points (6 and 12 weeks of treatment, respectively). Interestingly, mutations in the Ubqln2 gene have been linked to amyotrophic lateral sclerosis, and functional analysis demonstrated that such mutations were associated with an impairment of protein degradation (31).
4b also elicited decreases in the expression of ubiquitin-related genes, notably Sumo2, which encodes a member of the SUMO (small ubiquitin-like modifier) protein family. Similar to ubiquitin, the SUMOs are protein modifiers that are covalently attached to target proteins as part of a post-translational modification process. However, unlike ubiquitin, which generally targets proteins for degradation, SUMO proteins are involved in a variety of cellular processes, such as nuclear transport, transcriptional regulation, apoptosis as well as protein stability (32). Sumo2 was found to be upregulated in N171-82Q mice in both the cortex and the striatum, where 4b treatment decreased Sumo2 expression back down to wt levels.
Our qPCR findings are consistent with the notion that 4b treatment acts to promote ubiquitin-mediated protein degradation by either elevating the expression of genes associated with protein degradation or decreasing the expression of genes promoting protein stability. Although the specific proteins targeted by these systems are not known, one important candidate protein is Htt itself. One consequence of elevated Ube2k or Ube2e3 expression could be an increase in the ubiquitination of Htt protein, which has been linked to reduced toxicity of mutant Htt protein, most likely through increased clearance by the proteasome (33–35). In the Htt protein, SUMOylation and ubiquitination can each modify target lysines at amino acids 6, 9 and 15 (36). Although genetically reduced neuronal ubiquitination has been shown to increase truncated mutant Htt toxicity in a Drosophila HD model, less neuronal SUMOylation reduces toxicity (36), similar to our qPCR findings that 4b reduces Sumo2 expression but increases the ubiquitin machinery in parallel with its neuroprotective effects in the N171-82Q mouse HD model.
We also investigated the effects of 4b on the expression of genes related to protein phosphorylation. Protein phosphorylation is an abundant and important regulatory mechanism that plays a significant role in a wide range of cellular processes. Phosphorylation is also known to regulate protein degradation, alter subcellular localization of proteins and lead to changes in other protein modifications such as ubiquitination, SUMOylation and acetylation. In particular, the IKK pathway has been linked to autophagic protein degradation, independent of its known role to activate nuclear factor-kappaB (18). Further, IKK activation has been shown to induce phosphorylation and acetylation of the Htt protein, leading to its clearance by the proteasome and lysosome (20). We found that 4b treatment significantly increased the expression of several IKK gene family members, suggesting that it may be acting on autophagic processes. Additionally, 4b was found to increase the expression of the Atg4c and Map1lc3a genes in the mouse cortex, providing additional support for this notion. These findings are consistent with previous studies showing that two HDAC inhibitors, butyrate and suberoylanilide hydroxamic acid, can activate autophagy (37,38), by a mechanism involving increased expression of the autophagic factor LC3, and inhibition of the nutrient-sensing kinase mammalian target of rapamycin (38).
In HD, post-translational modifications to the Htt protein have been implicated in disease pathogenesis (22,36,39–41). Given the role of IKK to induce phosphorylation and acetylation of the Htt protein (20), we tested whether 4b could affect post-translational modification of Htt in the brain. We found elevated levels of endogenous phosphorylated Htt protein at serine 16 and threonine 3, and increased AcK9/pS13/pS16 immunoreactivity in cortical samples from 4b-treated mice. Phosphorylation and acetylation of Htt at these residues have been linked to autophagic clearance of Htt by the lysosome (20); hence, these findings suggest that 4b may be acting, in part, to alter clearance of Htt by the lysosomal pathway. The exact role of how modified endogenous Htt protein can affect the mutant protein and subsequent effects on pathology are unclear. We were unable to detect mutant Htt protein in our western blot assays, due to the insolubility of the transgenic Htt protein in this context; however, our immunohistochemistry experiments show that HDAC inhibitors have dramatic effects on the formation of mutant Htt aggregates. Both 4b- and 874-treated N171-82Q mice showed significantly lower numbers of Htt aggregates, suggesting that HDACis can prevent the formation of Htt aggregates or promote the clearance of Htt aggregates once they are formed. Although it is unclear whether aggregates are the toxic Htt species, their production requires high concentrations of misfolded Htt; hence, their elimination has been shown to be correlated with reductions in pathogenic Htt protein (42). These results provide further evidence for our hypothesis that 4b and related compounds prevent or reduce Htt aggregate formation by promoting degradation and/or clearance of misfolded Htt fragments, and that this action involves the activation of the ubiquitin–proteasomal and/or autophagy systems.
Our findings demonstrate that HDAC inhibition by 4b affects protein post-translational modification processes at the level of gene expression and that these effects may contribute to its clinically beneficial properties observed in HD mouse models. Because post-translational modification of target proteins can modify their clearance, we suggest that 4b, as well as other HDAC inhibitors, may be acting to modify the stability, processing and/or clearance of important proteins, including Htt. This could occur via the activation of autophagy or the ubiquitin–proteasome systems, the major protein degradation pathways in eukaryotic cells, or a combination of both in light of recent studies suggesting active crosstalk between the two systems (43).
MATERIALS AND METHODS
Mice
A B6C3-Tg(HD82Gln)81Dbo/J (N171-HD82Q) line (Jackson Laboratories) has been maintained at The Scripps Research Institute by breeding male heterozygous N171-HD82Q mice with F1 hybrids of the same background. At the age of 3–4 weeks, mice were genotyped according to the Jackson Laboratories protocol to determine hemizygosity for the HD transgene. The CAG repeat lengths in these mice were verified by commercial genotyping (Laragen, Los Angeles, CA, USA) and found to be 82 ± 1 CAGs for the major transgene species. The lifespan of the N171-82Q HD is ∼17–20 weeks with HD-like symptoms beginning at 10–12 weeks of age.
Drug treatment
HDACi 4b [N1-(2-aminophenyl)-N7-phenylheptanediamide] was synthesized by Scynexis Corporation (Durham, NC, USA) as described previously (14). HDACi 874 was provided by Repligen Corporation (Waltham, MA, USA). N171-82Q transgenic mice were housed and maintained on a normal 12 h light/dark cycle with lights on at 6:00 a.m. and free access to food and water. Groups of mice (n = 12–20 per genotype and drug treatment) were administered HDACi 4b (15, 50 and 100 mg/kg) for 10–12 weeks by s.c. injection (2.5 injections/week) beginning at 8 weeks of age. HDACi 4b was dissolved with 75% polyethylene glycol 200; control mice received ddH2O containing an equal volume of drug vehicle. Body weights were recorded two times per week. Mice were killed 6 h after the final injection, brains removed and striata and cortices dissected out for gene expression assays. All procedures were in strict accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals.
Motor behavioral assessments
Rotarod
An AccuRotor rotarod (AccuScan Instruments) was used to measure motor coordination and balance. Mice were tested on during the light phase of the 12 h light/dark cycle by using an accelerating rotation paradigm. Mice were placed on a rotarod accelerating from 0 to 20 r.p.m. over 300 s, and latency to fall was recorded. The best of three trials was used for analysis. Mice were trained on the rotarod before the start of drug treatment (8 weeks of age) to establish a behavioral baseline. At baseline, there were no significant differences in rotarod performance between wt and N17182Q transgenic mice.
Open-field test
Open-field exploration was measured in a square Plexiglas chamber (27.3 cm × 27.3 cm × 20.3 cm) (Med Associates, Inc., St Albans, VT, USA). Several behavioral parameters (ambulatory distance, ambulatory time, stereotypic time, vertical time, vertical counts and mean velocity) were recorded during a 10 min observation period.
T-maze forced alternation
The forced alternation T-maze test was performed using methods similar to those described in Deacon and Rawlins (44). The T-maze is made up of transparent Plexiglas with a central arm (75 cm long × 12 cm wide × 20 cm high) and two lateral arms (32 cm long × 12 cm wide × 20 cm high) positioned at a 90° angle relative to the central arm. Mice were habituated to the food reward (Froot loops) in their home cage, and then on three successive days before the start of testing, animals were habituated to the T-maze with food rewards at the ends of each arm. Before the forced alternation training and free choice testing, mice were fasted with 1.5 g of food per mouse overnight. For the T-maze training, each mouse was put into the initial ‘T’ arm facing the wall and allowed to enter only the left arm with the food reward at the end of the arm. After eating the food reward, the mouse begins a second trial in which only the right arm is open with a food reward at the end. Each mouse was trained for 10 trials for three consecutive days. Each trial lasted no longer than 2 min. For the free choice test, both left and right arms were baited, providing an initial free choice for either arm. As the mouse entered an arm, a sliding door closed behind the mouse, forcing it to choose that arm to retrieve the food reward. The chosen arm at first entry was noted, and the percentage of alternation over the next nine trials was determined. This percentage can be used as an index of working memory performance (45). Total trials to criterion (trials to criterion day 1 + trials to criterion day 2) were used for statistical analysis. Statistical analyses were performed using analysis of variance (ANOVA) (GraphPad Prism, San Diego, CA, USA).
Real-time PCR analysis
Real-time PCR experiments were performed using the ABI StepOne Detection System (Applied Biosystems, Foster City, CA, USA) as described previously (33). Amplification was performed on a cDNA amount equivalent to 25 ng of total RNA with 1× SYBR® Green Universal PCR Master Mix (Applied Biosystems) containing deoxyribonucleotide triphosphates, MgCl2, AmpliTaq Gold DNA polymerase and forward and reverse primers. PCR reactions were performed on groups of n = 4–6 mice per condition. Specific primers for each sequence were designed using the Primer Express 1.5 software, and their specificity for binding to the desired sequences was searched against NCBI database (Supplementary Material, Table S2). Standard curves were generated for each gene of interest, using serial dilutions of mouse cDNAs. Experimental samples and no-template controls were all run in duplicate. The PCR cycling parameters were 50°C for 2 min, 95°C for 10 min, and 40 cycles of 94°C for 15 s, 60°C for 1 min. The amount of cDNA in each sample was calculated using the SDS2.1 software by the comparative threshold cycle (Ct) method and expressed as 2Ct, using hypoxanthine guanine phosphoribosyl transferase as an internal control. Statistical significance was determined using Student's t-tests (unpaired; one-tailed). One-tailed tests were used given that the direction of the expression changes was predicted a priori from the microarray data. All statistical tests were performed using the GraphPad software (GraphPad Prism).
Network and functional pathways analysis
The functional relevance of microarray data generated from different brain regions from 4b-treated HD mice (GEO accession no. GSE26317) was analyzed using IPA (Ingenuity® Systems, www.ingenuity.com). For the functional analyses, the P-values were calculated using the right-tailed Fisher exact test, considering the number of molecules participating in a given function and the total number of molecules associated with that function in Ingenuity's knowledge base. Gene Network analysis was used to identify the extent of gene connectivity among those genes changed by 4b treatment. Each identified network was given a ‘score’, which is based on the hypergeometric distribution and was calculated using the right-tailed Fisher exact test. The score is the −log of this P-value, with the higher the score corresponding to a lower the probability of finding the observed number of Network-eligible molecules in a given network by random chance.
Western blot analysis of Htt protein
Cellular fractionation
Cortical samples were homogenized on ice with a glass-on-glass dounce in 500 μl of homogenizing buffer [0.25 m sucrose, 15 mm Tris–HCl, pH 8.0, 60 mm KCl, 5 mm EDTA, 1 mm EGTA, 1 mm PMSF, 5 mm NaF, containing an EDTA-free mini protease inhibitor pellet (Roche), a Phos-Stop pellet (Roche), 20 mm N-ethylmaleimide, 5 mm butyrate, 5 mm nicotinamide]. The intact nuclei were isolated by centrifuging at 800g for 15 min, and this first supernatant was centrifuged for 10 min at 16 000g, sonicated for 1 × 10 s on ice and designated the cytoplasmic fraction. The initial 800g pellet was washed two times in 1 ml of buffer at 800g for 5 min and resuspended in 500 μl of TPER (Thermo) containing phosphatase inhibitors 2 and 3 (Sigma), and sonicated 3 × 10 s on ice. This fraction was designated the nuclear fraction. Proteins were quantified by the Bradford assay.
Immunoprecipitation
An anti-Htt antibody (PW0595) was covalently linked to magnetic beads using a hydrazide linkage as instructed by the vendor, using the BcMag Hydrazide-terminated Magnetic Beads and Kit (Bioclone) as follows: 25 μl of PW0595 anti-Htt antibody (Enzo) was added to 275 μl of 1× coupling buffer (Bioclone) with 600 μg of NaIO4 in a brown tube and rotated 30 min in the dark. To stop the reaction and remove unreacted NaIO4 by desalting and buffer exchange, the sample was run through a mini Sephadex G-25 spin column (Roche), pre-equilibrated in coupling buffer. An amount of 300 μl of sample was applied to the column and it was spun for 3 min at 750g, and the eluate collected. BcMag Hydrazide-terminated Magnetic Beads were washed in coupling buffer three times, and the antibody sample was added to 500 μl of beads and incubated in a total volume of 1 ml of coupling buffer overnight rotating in the dark at room temperature. The beads were then washed three times using a magnetic rack in 1× wash buffer (Bioclone), three times in PBS and stored at 4°C in 500 μl of PBS. Ten microliters of these PW0595-linked beads were used for each IP in a total volume of 500 μl of TPER plus phosphatase inhibitors 2 and 3 (Sigma). An amount of 300–1000 μg of cytoplasmic lysate or 500 μg of nuclear lysate was incubated overnight rotating at 4°C and washed four times with 500 μl of (i) 0.5× STEN, (ii) STEN containing 0.1% SDS, (iii) 0.8× STEN containing 1 m NaCl, and (iv) 1× STEN (300 mm NaCl, 100 mm Tris–HCl, pH 7.6, 4 mm EDTA, 0.4% NP-40). The beads were resuspended in 35 μl of a fresh or frozen loading buffer (2.083 ml of 1.5 m Tris–HCl, pH 6.8, 1 g of SDS, 5 ml of glycerol, 2.5 ml of beta-mercaptoethanol and 10 mg of bromophenol blue, then up to 10 ml with water) and boiled for 10 min.
SDS–PAGE and western transfer analysis
The samples were loaded on a hand-poured SDS–PAGE 8% gel upper layer and a 15% lower gel with a DATD stacking gel of 0.5 cm thickness. The stacking gel was composed of 30% total acrylamide, which contains 4.5% DATD (N,N′-diallyl-tartardiamide, Aldrich 156868-25) as a cross-linker. The stacking gel contained 1.25 ml of 1 m Tris, pH 6.8, 1.00 ml of 30%/4.5% DATD acrylamide stock, 50 μl of 20% SDS and 7.65 ml of water. An amount of 50 μl of 10% ammonium persulfate and 10 μl of TEMED were added to the stacking gel solution—the stacking gel was poured on the top of the separating gel. A 20-lane comb was inserted and the stack allowed to polymerase for 1 h. Western blotting was done using 16% methanol and 0.05% SDS in the standard Tris/glycine 1× buffer to PVDF (Millipore) membrane (overnight transfer at 4°C). The membranes were blocked in TBS T20 Starting Block Buffer (Thermo) and then incubated overnight rotating at 4°C with antibodies against different Htt modifications: anti-phosphorylated S16 (pS16, 1:1000) (20); anti-phosphorylated S13 (pS13, 1:1000) (20); anti-phosphorylated T3 (pT3, 1:1000) (46); anti-acetylated and phosphorylated Htt (AcK9/pS13/pS16, 1:1000) (20). The anti-Htt antibody, PW0595 (1:5000), was used to detect total Htt protein. Western blots were quantified using the Scion software (Scion Software Solutions).
Immunohistochemical analyses
Immunohistochemistry experiments were performed on free-floating brain sections (25 µm) from vehicle and 4b- and 874-treated N171-82Q transgenic mice, as described previously (47), using the anti-Htt, EM-48 (1:500 dilution). The immunoreaction was detected with the Vectastain ABC kit (Vector Laboratory, Inc., Burlingame, CA, USA) according to the instructions of the manufacturer. Enzymatic development was performed in 0.05% diaminobenzene in PBS containing 0.003% hydrogen peroxide for 3–5 min. For 4b-treated mice, sections were counterstained with Richardson's stain to visualize nuclei. Quantification of Htt aggregates was performed by counting the number of visible punctate EM48 staining per field of vision using a 64× oil objective. For each brain region, we analyzed five sections per mouse, from the following numbers of mice: vehicle-treated group 1, n = 6; 874-treated mice, n = 6; vehicle-treated group 2, n = 5; 4b-treated mice, n = 2. Significant differences in aggregate number due to 874 treatment were determined using Student's t-test (unpaired; two-tailed) (GraphPad Prism).
SUPPLEMENTARY MATERIAL
FUNDING
These studies were funded by National Institutes of Health grants NS072453, NS052789 and U01NS063953, the Hereditary Disease Foundation and the CHDI Foundation.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr Leslie Thompson (University of California, Irvine) for the kind gift of anti-pSer13, anti-pSer16 and anti-AcetylK9/pSer13/pSer16 antibodies, to Dr Larry Marsh (University of California, Irvine) for the anti-pT3 antibody, to Dr Joel Gottesfeld (The Scripps Research Institute) for the HDAC inhibitor 4b, and to Dr James Rusche and Dr Vincent Jacques (Repligen Corporation) for the gift of the HDAC inhibitor 874. We would like to thank Dr Simonetta Sipione (University of Alberta, Canada) for helpful discussion regarding protein sample loading buffer and western transfer conditions to detect high molecular weight post-translationally modified Huntingtin.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Cha J.H. Transcriptional dysregulation in Huntington's disease. Trends Neurosci. 2000;23:387–392. doi: 10.1016/s0166-2236(00)01609-x. [DOI] [PubMed] [Google Scholar]
- 2.Sugars K.L., Rubinsztein D.C. Transcriptional abnormalities in Huntington disease. Trends Genet. 2003;19:233–238. doi: 10.1016/S0168-9525(03)00074-X. [DOI] [PubMed] [Google Scholar]
- 3.Okazawa H. Polyglutamine diseases: a transcription disorder. Cell Mol. Life Sci. 2003;60:1427–1439. doi: 10.1007/s00018-003-3013-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas E.A. Striatal specificity of gene expression dysregulation in Huntington's disease. J. Neurosci. Res. 2006;84:1151–1164. doi: 10.1002/jnr.21046. [DOI] [PubMed] [Google Scholar]
- 5.Freiman R.N., Tjian R. Neurodegeneration. A glutamine-rich trail leads to transcription factors. Science. 2002;296:2149–2150. doi: 10.1126/science.1073845. [DOI] [PubMed] [Google Scholar]
- 6.Zhai W., Jeong H., Cui L., Krainc D., Tjian R. In vitro analysis of huntingtin-mediated transcriptional repression reveals multiple transcription factor targets. Cell. 2005;123:1241–1253. doi: 10.1016/j.cell.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 7.Steffan J.S., Bodai L., Pallos J., Poelman M., McCampbell A., Apostol B.L., Kazantsev A., Schmidt E., Zhu Y.Z., Greenwald M., et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 8.Ferrante R.J., Ryu H., Kubilus J.K., D'Mello S., Sugars K.L., Lee J., Lu P., Smith K., Browne S., Beal M.F., et al. Chemotherapy for the brain: the antitumor antibiotic mithramycin prolongs survival in a mouse model of Huntington's disease. J. Neurosci. 2004;24:10335–10342. doi: 10.1523/JNEUROSCI.2599-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ryu H., Lee J., Hagerty S.W., Soh B.Y., McAlpin S.E., Cormier K.A., Smith K.M., Ferrante R.J. ESET/SETDB1 gene expression and histone H3 (K9) trimethylation in Huntington's disease. Proc. Natl Acad. Sci. USA. 2006;103:19176–19181. doi: 10.1073/pnas.0606373103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stack E.C., Del Signore S.J., Luthi-Carter R., Soh B.Y., Goldstein D.R., Matson S., Goodrich S., Markey A.L., Cormier K., Hagerty S.W., et al. Modulation of nucleosome dynamics in Huntington's disease. Hum. Mol. Genet. 2007;16:1164–1175. doi: 10.1093/hmg/ddm064. [DOI] [PubMed] [Google Scholar]
- 11.Kazantsev A.G., Thompson L.M. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat. Rev. Drug Discov. 2008;7:854–868. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- 12.Hahnen E., Hauke J., Trankle C., Eyupoglu I.Y., Wirth B., Blumcke I. Histone deacetylase inhibitors: possible implications for neurodegenerative disorders. Expert Opin. Investig. Drugs. 2008;17:169–184. doi: 10.1517/13543784.17.2.169. [DOI] [PubMed] [Google Scholar]
- 13.Balasubramanian S., Verner E., Buggy J.J. Isoform-specific histone deacetylase inhibitors: the next step. Cancer Lett. 2009;280:211–221. doi: 10.1016/j.canlet.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 14.Herman D., Jenssen K., Burnett R., Soragni E., Perlman S.L., Gottesfeld J.M. Histone deacetylase inhibitors reverse gene silencing in Friedreich's ataxia. Nat. Chem. Biol. 2006;2:551–558. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- 15.Chou C.J., Herman D.M., Gottesfeld J.M. Pimelic diphenylamide 106 is a slow, tight-binding inhibitor of class I histone deacetylases. J. Biol. Chem. 2008;283:35402–35409. doi: 10.1074/jbc.M807045200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jia H., Pallos J., Jacques J., Lau A., Tang B., Cooper A., Syed A., Purcell J., Chen Y., Sharma S., et al. Histone deacetylase (HDAC) inhibitors targeting HDAC3 and HDAC1 ameliorate polyglutamine-elicited phenotypes in model systems of Huntington's disease. Neurobiol. Dis. 2012;46:351–361. doi: 10.1016/j.nbd.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas E.A., Coppola G., Desplats P.A., Tang B., Soragni E., Burnett R., Gao F., Fitzgerald K.M., Borok J.F., Herman D., et al. The HDAC inhibitor, 4b, ameliorates the disease phenotype and transcriptional abnormalities in Huntington's disease transgenic mice. Proc. Natl Acad. Sci. USA. 2008;105:15564–15569. doi: 10.1073/pnas.0804249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Criollo A., Senovilla L., Authier H., Maiuri M.C., Morselli E., Vitale I., Kepp O., Tasdemir E., Galluzzi L., Shen S., et al. The IKK complex contributes to the induction of autophagy. EMBO J. 2010;29:619–631. doi: 10.1038/emboj.2009.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khoshnan A., Ko J., Watkin E.E., Paige L.A., Reinhart P.H., Patterson P.H. Activation of the IkappaB kinase complex and nuclear factor-kappaB contributes to mutant huntingtin neurotoxicity. J. Neurosci. 2004;24:7999–8008. doi: 10.1523/JNEUROSCI.2675-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson L.M., Aiken C.T., Kaltenbach L.S., Agrawal N., Illes K., Khoshnan A., Martinez-Vincente M., Arrasate M., O'Rourke J.G., Khashwji H., et al. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. J. Cell Biol. 2009;187:1083–1099. doi: 10.1083/jcb.200909067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu X., Greiner E.R., Mishra R., Kodali R., Osmand A., Finkbeiner S., Steffan J.S., Thompson L.M., Wetzel R., Yang X.W. Serines 13 and 16 are critical determinants of full-length human mutant huntingtin induced disease pathogenesis in HD mice. Neuron. 2009;64:828–840. doi: 10.1016/j.neuron.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steffan J.S. Does Huntingtin play a role in selective macroautophagy. Cell Cycle. 2010;9:3401–3413. doi: 10.4161/cc.9.17.12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Pardo A., Maglione V., Alpaugh M., Horkey M., Atwal R.S., Sassone J., Ciammola A., Steffan J.S., Fouad K., Truant R., et al. Ganglioside GM1 induces phosphorylation of mutant huntingtin and restores normal motor behavior in Huntington disease mice. Proc. Natl Acad. Sci. USA. 2012;109:3528–3533. doi: 10.1073/pnas.1114502109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orr A.L., Huang S., Roberts M.A., Reed J.C., Li S., Li X.J. Sex-dependent effect of BAG1 in ameliorating motor deficits of Huntington disease transgenic mice. J. Biol. Chem. 2008;283:16027–16036. doi: 10.1074/jbc.M710606200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bence N.F., Sampat R.M., Kopito R.R. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 26.Finkbeiner S., Mitra S. The ubiquitin-proteasome pathway in Huntington's disease. Sci. World J. 2008;8:421–433. doi: 10.1100/tsw.2008.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Pril R., Fischer D.F., Maat-Schieman M.L., Hobo B., de Vos R.A., Brunt E.R., Hol E.M., Roos R.A., van Leeuwen F.W. Accumulation of aberrant ubiquitin induces aggregate formation and cell death in polyglutamine diseases. Hum. Mol. Genet. 2004;13:1803–1813. doi: 10.1093/hmg/ddh188. [DOI] [PubMed] [Google Scholar]
- 28.Bennett E.J., Shaler T.A., Woodman B., Ryu K.Y., Zaitseva T.S., Becker C.H., Bates G.P., Schulman H., Kopito R.R. Global changes to the ubiquitin system in Huntington's disease. Nature. 2007;448:704–708. doi: 10.1038/nature06022. [DOI] [PubMed] [Google Scholar]
- 29.Diaz-Hernandez M., Hernandez F., Martin-Aparicio E., Gomez-Ramos P., Moran M.A., Castano J.G., Ferrer I., Avila J., Lucas J.J. Neuronal induction of the immunoproteasome in Huntington's disease. J. Neurosci. 2003;23:11653–11661. doi: 10.1523/JNEUROSCI.23-37-11653.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J., Wang C.E., Orr A., Tydlacka S., Li S.H., Li X.J. Impaired ubiquitin-proteasome system activity in the synapses of Huntington's disease mice. J. Cell Biol. 2008;180:1177–1189. doi: 10.1083/jcb.200709080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng H.X., Chen W., Hong S.T., Boycott K.M., Gorrie G.H., Siddique N., Yang Y., Fecto F., Shi Y., Zhai H., et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–215. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilkinson K.A., Nakamura Y., Henley J.M. Targets and consequences of protein SUMOylation in neurons. Brain Res. Rev. 2010;64:195–212. doi: 10.1016/j.brainresrev.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jana N.R., Dikshit P., Goswami A., Kotliarova S., Murata S., Tanaka K., Nukina N. Co-chaperone CHIP associates with expanded polyglutamine protein and promotes their degradation by proteasomes. J. Biol. Chem. 2005;280:11635–11640. doi: 10.1074/jbc.M412042200. [DOI] [PubMed] [Google Scholar]
- 34.Kalchman M.A., Graham R.K., Xia G., Koide H.B., Hodgson J.G., Graham K.C., Goldberg Y.P., Gietz R.D., Pickart C.M., Hayden M.R. Huntingtin is ubiquitinated and interacts with a specific ubiquitin-conjugating enzyme. J. Biol. Chem. 1996;271:19385–19394. doi: 10.1074/jbc.271.32.19385. [DOI] [PubMed] [Google Scholar]
- 35.de Pril R., Fischer D.F., Roos R.A., van Leeuwen F.W. Ubiquitin-conjugating enzyme E2–25K increases aggregate formation and cell death in polyglutamine diseases. Mol. Cell Neurosci. 2007;34:10–19. doi: 10.1016/j.mcn.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 36.Steffan J.S., Agrawal N., Pallos J., Rockabrand E., Trotman L.C., Slepko N., Illes K., Lukacsovich T., Zhu Y.Z., Cattaneo E., et al. SUMO modification of Huntingtin and Huntington's disease pathology. Science. 2004;304:100–104. doi: 10.1126/science.1092194. [DOI] [PubMed] [Google Scholar]
- 37.Shao Y., Gao Z., Marks P.A., Jiang X. Apoptotic and autophagic cell death induced by histone deacetylase inhibitors. Proc. Natl Acad. Sci. USA. 2004;101:18030–18035. doi: 10.1073/pnas.0408345102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gammoh N., Lam D., Puente C., Ganley I., Marks P.A., Jiang X. Role of autophagy in histone deacetylase inhibitor-induced apoptotic and nonapoptotic cell death. Proc. Natl Acad. Sci. USA. 2012;109:6561–6565. doi: 10.1073/pnas.1204429109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeong H., Then F., Melia T.J., Jr, Mazzulli J.R., Cui L., Savas J.N., Voisine C., Paganetti P., Tanese N., Hart A.C., et al. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell. 2009;137:60–72. doi: 10.1016/j.cell.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anne S.L., Saudou F., Humbert S. Phosphorylation of huntingtin by cyclin-dependent kinase 5 is induced by DNA damage and regulates wild-type and mutant huntingtin toxicity in neurons. J. Neurosci. 2007;27:7318–7328. doi: 10.1523/JNEUROSCI.1831-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Humbert S., Bryson E.A., Cordelieres F.P., Connors N.C., Datta S.R., Finkbeiner S., Greenberg M.E., Saudou F. The IGF-1/Akt pathway is neuroprotective in Huntington's disease and involves Huntingtin phosphorylation by Akt. Dev. Cell. 2002;2:831–837. doi: 10.1016/s1534-5807(02)00188-0. [DOI] [PubMed] [Google Scholar]
- 42.Rubinsztein D.C. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 43.Kraft C., Peter M., Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat. Cell Biol. 2010;12:836–841. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 44.Deacon R.M., Rawlins J.N. T-maze alternation in the rodent. Nat. Protoc. 2006;1:7–12. doi: 10.1038/nprot.2006.2. [DOI] [PubMed] [Google Scholar]
- 45.Bontempi B., Whelan K.T., Risbrough V.B., Lloyd G.K., Menzaghi F. Cognitive enhancing properties and tolerability of cholinergic agents in mice: a comparative study of nicotine, donepezil, and SIB-1553A, a subtype-selective ligand for nicotinic acetylcholine receptors. Neuropsychopharmacology. 2003;28:1235–1246. doi: 10.1038/sj.npp.1300150. [DOI] [PubMed] [Google Scholar]
- 46.Aiken C.T., Steffan J.S., Guerrero C.M., Khashwji H., Lukacsovich T., Simmons D., Purcell J.M., Menhaji K., Zhu Y.Z., Green K., et al. Phosphorylation of threonine 3: implications for Huntingtin aggregation and neurotoxicity. J. Biol. Chem. 2009;284:29427–29436. doi: 10.1074/jbc.M109.013193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas E.A., Danielson P.E., Nelson P.A., Pribyl T.M., Hilbush B.S., Hasel K.W., Sutcliffe J.G. Clozapine increases apolipoprotein D expression in rodent brain: towards a mechanism for neuroleptic pharmacotherapy. J. Neurochem. 2001;76:789–796. doi: 10.1046/j.1471-4159.2001.00027.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.