Abstract
Chromosomal aneuploidy, the gain or loss of whole chromosomes, is a hallmark of pathological conditions and a causal factor of birth defects and cancer. A number of studies indicate that aneuploid cells are present at a high frequency in the brain of mice and humans, suggesting that mosaic aneuploidies are compatible with normal brain function and prompting the question about their consequences. To explore the possible contribution of aneuploidy to functional decline and loss of cognitive functions during aging, we used a quantitative, dual-labeling interphase-fluorescence in situ hybridization approach to compare aneuploidy levels of chromosomes 1, 7, 14, 15, 16, 18, 19 and Y in the cerebral cortex of 4- and 28-month-old mice. We show that aneuploidy accumulates with age in a chromosome-specific manner, with chromosomes 7, 18 and Y most severely affected, i.e. up to 9.8% of non-neuronal brain nuclei in 28-month-old animals for chromosome 18. While at early age, both neuronal and glial cells are affected equally, the age-related increase was limited to the non-neuronal nuclei. No age-related increase in aneuploidy was observed in the cerebellum or in the spleen of the same animals. Extrapolating the average frequencies of aneuploidy from the average over 8 chromosomes to all 20 mouse chromosomes would indicate an almost 50% aneuploidy frequency in aged mouse brain. Such high levels of genome instability could well be a factor in age-related neurodegeneration.
INTRODUCTION
Aneuploidy, a chromosome content that deviates from a diploid genome, is a form of genomic instability commonly linked to pathological conditions (1). In somatic cells, aneuploidy is considered a cause of miscarriages (2) and also thought to contribute to cancer (3,4). A correlation between increased aneuploidy and aging was first described for humans, when recurrent losses of chromosomes X and Y were observed in lymphocytes and bone marrow (5–7). However, the lack of advanced cytogenetic methods essentially constrained efforts to systematically explore a possible causal relationship between aging and aneuploidy. Genomic variation in the form of mosaic aneuploidy has been described for the developing central nervous system of mammals. By using Spectral karyotyping to analyze metaphases isolated from murine embryonic neuroblasts, Rehen et al. (8) uncovered an unexpectedly high frequency of aneuploid cells, i.e. up to 33% for all chromosomes combined. A similarly high frequency of aneuploidy (30–35%) has been identified in the developing human nervous system (9). This finding has been interpreted as evidence for aneuploidy as a contributing factor to neuronal diversification in the developing brain (10). In human or mouse adult brain, in which only few chromosomes were analyzed by interphase fluorescence in situ hybridization (FISH), aneuploidy frequencies were also found to be very high, albeit much lower than in developmental mouse brain, i.e. ∼10% for all chromosomes combined (11–13). In this respect, it is conceivable that rather than contributing to neuronal diversification, most aneuploid cells are eliminated, for example, through the programmed cell death that occurs physiologically in the neuroproliferative zones of the brain (8,14). Indeed, considering the deleterious consequences of high levels of aneuploidy in other systems it is likely that aneuploid cells are eliminated or kept at bay in the adult brain, maybe under selective pressure.
As people age, their risk for neurodegenerative diseases increases and the accumulation of aneuploidy in the brain during aging could contribute to the impairment of cellular homeostasis, resulting in functional decline and neuropathies. In fact, aneuploidy was found to increase in the cerebral cortex of Alzheimer's disease (AD) and ataxia telangiectasia (AT) patients when compared with the cortex of normal controls. These findings suggest that aneuploidy could increase during aging of the brain and may contribute to neurodegeneration (12,15).
To obtain insight into the frequency and nature of chromosomal aneuploidy in the cerebral cortex during normal aging, we performed an extensive and detailed analysis at the single cell level of the frequency of whole chromosome gains and losses for seven randomly selected autosomes 1, 7, 14, 15, 16, 18, 19 and the chromosome Y. Our results indicate that aneuploidy does indeed increase with age in the cerebral cortex but not in the cerebellum or the spleen. Moreover, the observed increase in aneuploidy in the brain was found to be chromosome specific and to affect mainly non-neuronal cells.
RESULTS
Chromosome-specific aneuploidies are detected in the cortex of adult mice using a two-color single chromosome FISH approach
Standard FISH protocols to quantify low-frequency aneuploidy for single chromosomes in interphase cells are inaccurate because loss or gain of a signal loss or gain of a signal is frequently indistinguishable from the background. We therefore designed and extensively validated a two-color FISH approach that provides highly quantitative and reproducible aneuploidy data for individual chromosomes in interphase nuclei by assessing gain and loss based on the enumeration at two separate loci (Fig. 1A). In this study, we scored only those aneuploidies that were seen as gains or losses of both probes. While this approach may slightly underestimate the frequency of aneuploidy, it greatly reduces the number of false positives when only one chromosome-specific probe is used (Supplementary Material, Fig. S1).
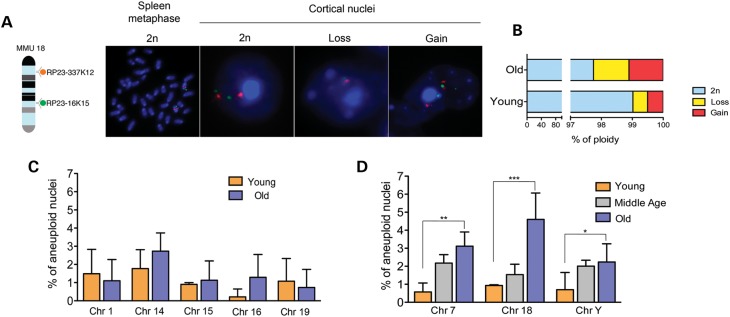
Figure 1.
Analysis of ploidy in cortical nuclei of young and old mice. (A) Two differently labeled BAC clones (red spectrum orange and green spectrum green) mapping to chromosome 18 at two distinct genomic loci enable the identification of diploid from aneuploid cells. A diploid cell contains two copies of each autosome, which are identified in our dual labeling system by two red and two green signals, thus allowing clear identification. Examples of diploid cells (2n), one in metaphases and one in interphase are shown. When a cell is aneuploid, its DNA content deviates from the expected two copies for single chromosomes. In our case, we observed a loss of chromosome 18, visualized by one red and one green signal (middle panel) and gain of chromosome 18, visualized by three red and three green signals (right panel). (B) The analysis of global ploidy on cortical nuclei for the eight chromosomes tested demonstrates that aneuploidy levels increase with age. (C) Individual aneuploidy frequencies for the chromosomes that did not show an age-related increase. (D) Statistically significant age-related increase in aneuploidy for chromosomes 7, 18 and Y (**P = 0.01, ***P < 0.0001, *P = 0.02, respectively).
Initially, we believed that the surprisingly high aneuploidy levels in the human and mouse brain observed by others could be artifacts associated with the methodology. However, applying our two-color FISH approach to populations of nuclei isolated by sucrose gradient centrifugation from the cerebral cortex of 4-month old male mice, we confirmed these high levels of aneuploidy. While thus far only very few chromosomes had been tested (mainly the sex chromosomes), we compared aneuploidy levels for eight arbitrarily selected chromosomes of different size and gene content (1,7,14,15,16,18 and 19) and a sex chromosome (Y) in six young mice. The isolation protocol used enriches for neuronal nuclei and immunostaining performed with anti-NeuN, a specific marker for neuronal nuclei, showed a population equally composed of neuronal and non-neuronal nuclei (Supplementary Material, Fig. S2). Hence, these results fully confirm the previously demonstrated high levels of aneuploidy already present in young animals (11), i.e. 1.0% per chromosome [95% confidence interval (CI): 0.7–1.3%; Fig. 1B, bottom graph].
Subsequent analysis of the cerebral cortex of eight old, i.e. 28 months, mice revealed a more than 2-fold, cumulative age-related elevation in the frequency of aneuploid cells across all chromosomes tested (2.3% per chromosome; 95% CI: 1.9–2.7%; P < 0.00001 when compared with young mice) (Fig. 1B, top graph). Chromosome gains and losses occurred at roughly similar frequency (Fig. 1B; Supplementary Material, Fig. S3). Interestingly, of the eight tested chromosomes, not all contributed equally to the elevated level of aneuploidy observed at old age. Individually, chromosomes 7, 18 and Y contributed most to the increased levels of aneuploidy at old age, with especially chromosome 18 severely affected. The average aneuploidy levels span from 0.9% (95% CI: 0.2–1.7%) in young to 4.5% (95% CI: 3.1–5.9%) in old mice for chromosome 18 (Fig. 1C and D).
Aneuploidy is not an early event
Since it is possible that the observed high levels of aneuploidy are an early phenomenon occurring during the first 10 months of life, we also studied mice of middle age (n = 3 animals at 15 months) to quantify the ploidy of the three most unstable chromosomes during aging (7,18 and Y). The results suggest that increases in aneuploidy for these chromosomes take place between adulthood and middle age, but continue also to accumulate after 15 months, with increases for chromosome 18 even accelerating towards old age. The average aneuploidy for chromosome 18 in this group spans from 0.9% (95% CI: 0.2–1.7%) in young mice to 1.5% (95% CI: 0.2–2.9%) in the middle age group up to 4.5% (95% CI: 3.1–5.9%) observed in old mice (P = 0.015 MA vs. O and Fig. 1D).
NeuN negative cells are the targets of aneuploidy in the old cortex
As it has been proposed that defects in chromosomal segregation are the main mechanisms responsible for the alteration of ploidy (16), we reasoned that mitotically active cells were the most likely targets for the accumulation of aneuploidy during aging. To address this issue, we investigated whether neuronal or non-neuronal cells were the source of aneuploidy in the old cortex (28-month-old mice, n = 3 animals analyzed). Using a flow cytometric approach based on the neuronal-specific marker, NeuN we enriched for the NeuN+ and NeuN− populations. Post-sorting quantification of NeuN-stained cells confirmed enrichment as being NeuN+ 99.5% neurons and NeuN− 99.5% glia (Fig. 2A). Subsequent FISH experiments with dual probes for chromosomes 1 and 18, the least and most susceptible to aneuploidy, revealed aneuploidy in both cellular fractions (Fig. 2B and C). However, only the non-neuronal cells contributed in a statistically significant manner to the age-related increase in chromosome 18 aneuploidy that was quantified at 2.1% (95%CI: 0.6–3.3%) and 9.8% (95%CI: 8.4–1.1%), respectively (P < 0.0001).
Figure 2.
Identification of cell types targeted by aneuploidy in the old cortex. (A) Flow cytometric analysis of cortical nuclei with anti-NeuN antibody, detected with Alexa-488 secondary antibody. Isotype control and anti-NeuN specific staining on old cortex are shown. Isolated mouse cortical nuclei were gated based on forward scatter and side scatter (data not shown) and gated into NeuN+ DAPI+ and NeuN− DAPI+ populations. The gates were set based on the IgG1 isotype control. (B) Levels of aneuploidy obtained by interphase FISH analysis of chromosomes 1 and 18 performed on FACS enriched cortical neuronal and non- neuronal nuclei in 28-month-old mice. Chromosome 18 shows a statistically significantly higher level of aneuploidy than chromosome 1 in NeuN− cells (dark gray *P < 0.0001). (C) To avoid interference of the locus specific probe with the green fluorescence arising from NeuN staining (Alexa 488), the same BAC probe previously used has been labeled with spectrum Aqua. We chose yellow as pseudo-color to display the spectrum Aqua dye. Examples of NeuN- cortical nuclei aneuploid for chromosomes 1 and 18 performed with our two-color FISH approach are shown. The left panel shows a nucleus that carries three copies of chromosome 1, visualized by three red and three yellow signals (gain). The right panel shows that a nucleus carries one copy of chromosome 18, visualized by one red and one yellow signal (loss).
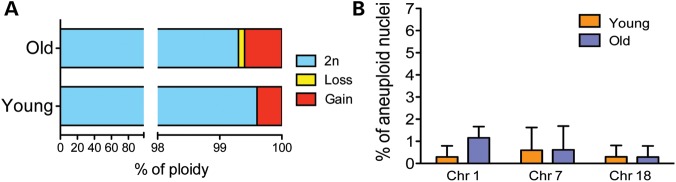
Lack of an age-related increase in aneuploidy in the cerebellum of adult mice
We next questioned if the accumulation of aneuploidy observed during aging is restricted to the cerebral cortex or a more widespread phenomenon, possibly targeting more regions of the adult brain. To begin addressing this question, we analyzed the cerebellum using tissue from the same mice for which we analyzed the cortex (Supplementary Material, Table S1). We selected the cerebellum because several lines of evidence show only minor morphological and gene expression changes in this area of the brain as a function of age (17,19). Nuclei from the cerebellum of three young and three old mice were analyzed with dual probes specific for chromosomes 1, 7 and 18. We chose these chromosomes based on the data obtained from the cortex where chromosomes 7 and 18 appeared the most susceptible to aneuploidy during aging and chromosome 1 the least. The results revealed a low level of aneuploidy in both age groups that is, overall, even lower than what was observed in the cerebral cortex (0.4%; 95% CI: 0–0.8% for young versus 0.7%; 95% CI: 0.2–1.2% for old mice old, Fig. 3). Even when the chromosome frequencies were analyzed for each single chromosome, the levels were low and no increase in aneuploidy levels was observed with age (Fig. 3).
Figure 3.
Analysis of ploidy in nuclei from the cerebellum of young and old mice. (A) The picture summarizes the analysis of global ploidy on cerebellum nuclei performed for chromosomes 1, 7 and 18, one of the least and of the most aneuploid chromosome founded previously in the cortex. The analysis demonstrates that the accumulation of aneuploidy during aging in the cerebellum for the chromosomes tested is absent. (B) Aneuploidy frequencies for single chromosomes 1,7 and 18 also show a lack of individual significant increase during aging.
Lack of an age-related increase in aneuploidy in spleen
We then asked the question if age-related chromosome-specific aneuploidy increases are restricted to the brain or a more general phenomenon also detectable in other aging tissues. To investigate this we analyzed the spleen, a mitotically active tissue, which allows for the analysis of both interphase cells and metaphase chromosomes. We first scored the number of chromosomes in metaphase spreads from young (n = 3) and old (n = 4) mice. This approach, although commonly used for assessing aneuploidy, is inaccurate in estimating aneuploidy and especially biased in the enumeration of chromosome loss. Despite the overall level of aneuploidy being higher than observed using interphase FISH, we could not detect significant changes in ploidy between the two groups (Fig. 4A). We then analyzed the interphase splenocytes of the two groups using our highly sensitive custom-designed FISH approach for chromosomes 1, 16, 18 and Y (Fig. 4B and C). The interphase FISH analysis confirmed the metaphase spread data revealing a similar profile of chromosome instability between the young and old groups and failing to detect accumulation of aneuploidy with age (0.6% per chromosome; 95% CI: 0.3–1.0% in young versus 0.8% per chromosome; 95% CI: 0.3–1.3 in the old spleen). We conclude that in the spleen, contrary to what we observed in the cerebral cortex, the cells are mainly diploid in both age groups.
Figure 4.
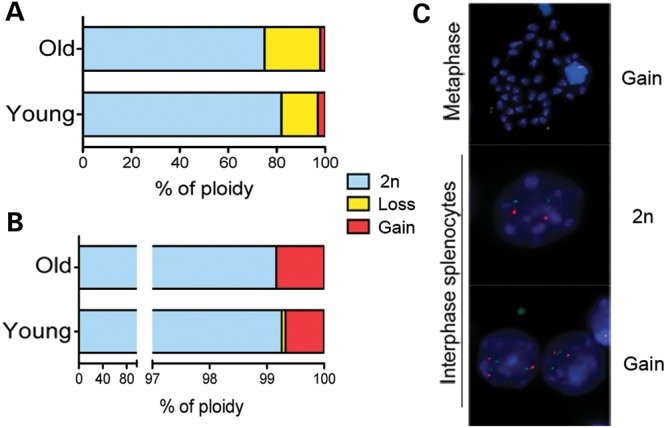
Analysis of ploidy in splenocytes of young and old mice. (A) Chromosomes count in metaphase spreads. (B) 2-color FISH analysis perfprmed on chromosomes 1, 16 and 18 on young FISH analysis on young and old splenocytes revealed that aneuploidy does not increase with age and has similar frequencies for all chromosomes analyzed (plotted average of the four chromosomes). (C) Examples of ploidy detected by two-color FISH analysis for chromosome 16 on splenocytes. FISH signals corresponding to 2n and 2n plus 1 extra chromosome 16 in interphase and metaphase cells are shown.
DISCUSSION
Based on a two-color interphase FISH strategy to examine cortical nuclei, we report for the first time a comprehensive analysis of the frequency of aneuploidy in the aging mouse brain across a number of chromosomes. Apart from confirming the previously reported high levels of aneuploidy in the brain of young adult mice (∼1% on average for a single chromosome), our extended analysis across eight chromosomes now shows that aneuploidy significantly increases with age, albeit the accumulation appeared restricted to the cortex, a part of the brain especially vulnerable to age-related changes (15,17). We also demonstrate that the age-related increase in aneuploidy is chromosome specific and that at least one other tissue, i.e. spleen, remains unaffected. The lack of a similar age-related increase in aneuploidy in the spleen indicates that chromosomal aneuploidy is also a tissue-specific phenomenon.
By combining the two-color FISH analysis with flow cytometric analysis and sorting (FACS), using a neuronal specific marker, our results indicate that age-related aneuploidy predominantly affects the non-neuronal cells (glia). Contrary to neurons, the glia preserves the ability to divide in adulthood. Acute glial proliferation occurs in response to trauma or injury, but gliosis also occurs in the cortex during aging where it provides a protective and nutritional function for neurons (18). Hence, these results strongly suggest that age-related aneuploidy is caused by mitotic errors in non-neuronal cells, with some level of aneuploidy at early age being the likely result of segregation errors during division of neuronal precursor stem cells during embryogenesis (10). Aneuploidy early in life could involve neurons remaining functionally active. Indeed, evidence has been provided that aneuploid neurons contribute to mosaic neural circuitries as part of the normal organization of the mammalian brain (20). The theory known as the ‘GIN'n'CIN hypothesis of brain aging’ (21) proposes that mature mosaic aneuploid neurons persist at low frequency in the adult brain and contribute to the functional decline and neurodegeneration observed later in life.
Our present data suggest that mosaic aneuploidy in the old brain is more complex than that previously predicted. The fraction of aneuploid neurons surviving embryonic selection into adulthood remains constant (∼1%), but coexist with a much higher level of aneuploid non-neuronal cells (∼9.8%) accumulated during aging. In this respect, our data suggest that also aneuploidy in non-neuronal cells may contribute significantly to genomic instability observed in pathological conditions associated with age (as further discussed below). The observed chromosome-specific aneuploidy frequencies preclude easy extrapolation from the 8 chromosomes analyzed in this study to all 20 mouse chromosomes. However, it should be noted that based on the calculated average of 2.3% per chromosome in aged mouse brain as many as 46% of cells may be affected. In our opinion, such exceptionally high levels of aneuploidy are unlikely to have no functional consequences.
We also observed chromosome and tissue specificity in the age-related increase in aneuploidy. The most likely explanation for this phenomenon, in our opinion, is selection against particular aneuploidies based on the function of the tissue affected. Normally, natural selection would work against chromosomal imbalances, albeit most strongly at early age. Aneuploidy at older age would probably be tolerated, provided its functional consequences are relatively minor. This could be the case for certain chromosomes, such as chromosome 18 in non-neuronal cells, aneuploidy of which could therefore rise to such high levels at very old age. The fact that mosaic aneuploidies are tolerated at such unexpectedly high levels in the old cerebral cortex raises several important questions that remain to be addressed. Aneuploidy is detrimental at the cellular level causing reduction of cell proliferation (22). However, fully differentiated cells have limited proliferation capabilities and, therefore, a growth disadvantage is most likely not the most deleterious consequence of increased aneuploidy in the brain. Changes in cellular metabolism, however, have been shown to occur in mouse embryonic fibroblasts as a consequence of single aneuploidies for chromosomes 1, 13, 16 and 19 (22). Williams et al. (22) suggest that cellular imbalance caused by aneuploidy and the resulting cellular stress could induce an increase in mutation rate, gene amplification and/or genomic instability. Indeed budding yeast strains that carry extra copies of single chromosomes exhibit increased genomic instability in the form of chromosome missegregation, increased mutation rate and recombination defects (23). Thus, accumulation of aneuploidy in the old brain could lead to an increase in genomically unstable cells with the potential of deleterious consequences. Of note, in humans, aging of the brain shows great individual variation and it is not inconceivable that high aneuploidy levels in some individuals may predispose to neurodegeneration, while lower levels in others would be more compatible with optimal brain function for extended periods of time.
Lastly, we provide evidence that aneuploidy in the old brain may affect different areas. The cerebellum, contrary to the cerebral cortex, shows minor morphological changes as a function of age (19). At the molecular level, genome-wide expression analysis suggests that during aging many more genes undergo consistent expression changes in the cortex than in the cerebellum (17). Because alteration of gene expression has been shown to occur as a consequence of single chromosome aneuploidies (24), we can envision that the accumulation of aneuploidy observed during aging in the cortex may be responsible for changes in the transcriptome profile observed at old age (17). Consequently, the absence of gene expression changes in the cerebellum at old age may be due to the lack of aneuploid cells.
Because of the complexity of alteration in cellular functions resulting from aneuploidy, it is not surprising that aneuploidy has been found associated with age-dependent neurodegeneration. AD, Huntington's disease, AT and cortical basal ganglionic degeneration are progressive neurodegenerative disorders affecting the cerebral cortex. FISH studies of 10 human chromosomes in AT and AD patients revealed a significant increase in aneuploidy levels associated with these neuropathies (12,16). In the AT brain, Iourov et al. (25) observed a 2–5-fold increase in aneuploidy, randomly affecting the 10 chromosomes analyzed in 20–50% of the cells analyzed. The increased aneuploidy was found to be specific to the cerebellum and the authors showed chromosome-specific rearrangements (HSA 14), affecting primarily NeuN− cells occurring in an increased background of stochastic aneuploidy. In the AD brain, these same authors reported a dramatic, 10-fold increase in chromosome 21-specific aneuploidy. Other groups have reported mosaic aneuploidy for chromosome 21 in the brain of AD patients and a functional consequence of this chromosome-specific aneuploidy in AD is supported by the observation that trisomy 21/down syndrome patients develop early onset AD. The amyloid precursor protein (APP), the precursor molecule whose proteolysis generates the beta-amyloid peptide contained in the amyloid plaques found in the brain of AD patients, maps to human chromosome 21. It is possible that alteration in APP gene dosage and expression resulting from aneuploidy is an important causal contributor to AD. Taken together, these observations support the possibility that age-related accumulation of aneuploidy has deleterious consequences.
In principle, one could speculate that rather than being deleterious, the observed high levels of aneuploidy in the aged mouse brain could contribute to brain function, possibly in response to stress. In budding yeast, the inhibition of HSP90 leads to the production of a population of cells with an unbalanced karyotype. Long-term exposure to Hsp90 induces stress resulting in the accumulation of specific chromosomal aneuploidy that potentiates cellular adaptation (26). However, in mammals, it appears more likely that, similar to other forms of genomic instability, increased aneuploidy is part of the deteriorative nature of aging, possibly by predisposing the brain to neurodegenerative disease, which would be in keeping with the elevated levels of aneuploidy in the cerebral cortex of AD and AT patients (27).
To better understand the nature of the possible link between increased aneuploidy and age-related neurodegeneration, some fundamental questions remain to be addressed. Most notably, based on our findings, it is not the neuron that is the main target of age-related aneuploidy, but the glia compartment. Hence, a better understanding of the chromosome specificity of increased aneuploidy in this cellular compartment is necessary to understand functional relevance.
MATERIALS AND METHODS
Tissue isolation
We analyzed a total of 6 young (4 months), 3 middle aged (15 months) and 11 old (28 months) male C57BL6/J mice, obtained from the National Institute on Aging. The mice were anaesthetized with avertin (10 μl/g) and then sacrificed by cervical dislocation. The brain was quickly removed from the skull and the cerebral cortex or the cerebellum were dissected with the aid of a stereomicroscope. The cortex was placed in a steel mouse brain slicer and cut in coronal slices of 2.0 mm interval. The slices were placed in a petri-dish with ice-cold PBS. The cortex and the cerebellum were then processed for nuclei extraction [(28) and below]. The spleen of three young (4 months) and four old (28 months) mice was removed and placed into a homogenizer with 3 ml of RPMI medium 640 and processed for the metaphase preparation. All samples were de-identified for blinded analysis.
Extraction of cortical nuclei for direct FISH analysis
The entire procedure was performed on ice using pre-cooled buffers to preserve the integrity of the nuclei. The cortex was homogenized for 45 s in 5 ml of lysis buffer. The suspension was transferred to a clear ultra-centrifuge tube (14 × 89 mm, Beckman Coulter Inc., Brea, CA, USA) and layered on 9 ml of sucrose solution. The samples were placed into SWiT40 rotor buckets and ultra-centrifuged at 4°C for 2.5 h at 25 000 rpm. After the centrifugation, 1 ml of PBS was added to the pellet and kept on ice for 20 min. The nuclei were then dissolved through pipetting for 1 min, centrifuged at 4°C for 10 min at 160g and then fixed with ice-cold methanol/acetic acid (3:1) for 10′ on ice. The nuclei were stored in fixative solution at −20°C until they were dropped onto a clean slide in 48% humidity at 24°C for FISH analysis.
Flow cytometric analysis and sorting (FACS)
For the FACS analysis, we performed the extraction of cortical nuclei as described above; however, after the nuclei were dissolved in PBS, we proceeded as follows: to improve the permeability of the sample, the cortical nuclei were incubated for 10 min on ice in 1 ml of PBS 1x + 0.6% Triton X-100. The dilutions of primary and secondary antibodies were performed with a buffer solution composed by 2% of fetal bovine serum and 0.3% of Triton X-100 in PBS. The sample was centrifuged for 10 min at 4°C at 160g and the supernatant removed. The pellet was resuspended in 100 μl of primary antibody solution and incubated at 4°C for 20 min (mouse anti-NeuN antibody 1:100, Millipore, Billerica, MA, USA). A fraction of nuclei was incubated with the corresponding isotype control antibody (anti- IgG1, Millipore). After adding 2 ml of wash buffer, the nuclei were re-centrifuged. Finally, the pellet was resuspended in 100 μl of secondary antibody solution and DAPI (Alexa fluor 488, Invitrogen, Carlsbad, CA, USA) and incubated for 20 min on ice. After a third centrifugation, the pellet was resuspended in 500 μl of PBS 1× and 0.1% FCS and filtered. Stained nuclei were FACS sorted using BD FACSAria II and data analyzed using FloJo software (29).
Metaphase preparation from mouse spleen
Spleens were dissected from 4 and 28 months old mice and single cell suspensions were obtained with the aid of a Potter–Elvehjem tissue homogenizer. To promote cell proliferation, splenocytes were incubated for 48 h at 37°C with concavalin A (5 μg/μl), LPS (25 μg/ml) and 0.5% of β-mercaptoethanol. Splenocytes undergoing active cell division were arrested in metaphase by exposure to colcemid (1 h at 37°C, KaryoMax 10 μg/ml, Invitrogen). The cell suspension was centrifuged for 10 min at 300g and incubated with hypotonic KCl solution (0.075 m KCl pre-warmed at 37°C, ThermoFisher). The cells were then fixed and washed four times with methanol/acetic acid solution (3:1). 40 μl of the cell suspension was dropped onto a clean slide, in 48% humidity and 24°C, and then stored at 37°C until analyzed. Chromosomes were counted in 25 metaphases for each condition for a total 100 spread/mouse analyzed.
Fluorescence in situ hybridization
FISH was performed using two locus-specific probes for each chromosome that map to different regions: the BAC clones RP23-34K7 (1qA1) and RP23-102B11 (1qA2) for chromosome 1; the BAC clones RP23-336P12 (7qA1) and RP23-316N16 (7qF5) for chromosome 7; the BAC clones RP23-348I17 (14qA3) and RP23-185C10 (14qE1) for chromosome 14; the BAC clones RP23-4E14 (15qB2) and RP23-23L17 (15qD1) for chromosome 15; the BAC clones RP24-329N7 (16qA1) and RP24-266L1 (16qB2) for chromosome 16; the BAC clones RP23-332K12 (18qA2) and RP23-16K15 (18qE1) for chromosome 18 (Fig. 1); the BAC clones RP24-375I7 (19qB) and RP24-214I3 (19qD1) for chromosome 19; the BAC clones RP24-275C21 (Y1qA1) and RP24-332J21 (YqA1) for chromosome Y (S1). The probes were labeled by nick translation using spectrum orange-dUTP (Invitrogen) and DY-505-aadUTP (Dyomics, Jena, GE, USA); in the case of FACS/FISH combined techniques, the nick translation was performed with spectrum orange-dUTP (Invitrogen) and DY-415-aadUTP (Dyomics). The slides with cortical nuclei or spleen metaphases were denatured with 50%FA/2× SSC at 80°C for 1.5 min and then dehydrated with serial ethanol washing steps (ice cold 70, 90 and 100% for 3 min each). Probes were denaturated in the hybridization solution (50% dextran sulfate/2× SSC) at 85°C for 5 min, applied to the slides and incubated overnight at 37°C in a humidified chamber. The slides were then washed three times for 5 min with 50% formamide/2× SSC,1× SSC and 4× SSC/0.1%Tween. Slides were dehydrated with serial ethanol washing steps (see above) and mounted with ProLong Gold antifade reagent with DAPI (Invitrogen) for imaging.
Image acquisition
FISH images were acquired with a manual inverted fluorescence microscope (Axiovert 200, Zeiss) with fine focusing oil immersion lens (×40, NA 1.3 oil and ×60, NA 1.35 oil). Multiple focal planes were acquired for each channel to ensure that signals on different focal planes were included. The resulting fluorescence emissions were collected using 425–475 nm (for DAPI), 546–600 nm (for spectrum orange), 422–449 nm (for spectrum aqua) and 500–550 nm (for AlexaFluor488) filters. The microscope was equipped with a Camera Hall 100 and the Applied Spectral Imaging software.
Data analysis of interphase cells
Images representing a minimum of 150 cells for each hybridization were randomly acquired and saved as .tiff composite files. An average 105–116 cells were visually inspected for each chromosome and FISH signals manually counted for both cortex and spleen. A minimum of 400 interphase cells to a maximum of 800 was counted for each cortex analyzed. The variation in the number of nuclei enumerated for each sample was dictated by the number of chromosomes tested for each mouse. A total of 3578 interphase nuclei were screened for the young mice group, 998 nuclei for middle age and 4787 for the old mice group to determine the frequency of ploidy in the cortex. For the cerebellum, we scored 982 nuclei for the young mice and 1020 for the old group. For the spleen, a total of 1492 interphase nuclei were screened in the young mice group and 1325 for the old mice group. For the analysis of spleen metaphases, we scored 371 spreads for the young group and 272 for the old one.
The detailed number of nuclei scored for each chromosome tested and the mice analyzed are listed in Supplementary Material, Tables S1–S3.
Statistical analysis
Frequencies of aneuploidy and corresponding 95% CIs were calculated. The aneuploidy frequencies were compared between different groups by Fisher's exact test. The 95% CI values for the spleen experiments are provided in the text. The 95% CIs are provided in the text and also summarized in Supplementary Material, Tables S1–S3.
SUPPLEMENTARY MATERIAL
FUNDING
The work of the authors was supported by grants from the NIH (AG17242 and AG032117 to J.V.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Guillermo O. Simkin and Dr Cristina Panaroni for technical support with the FACS analysis and the Molecular Cytogenetic Core at Albert Einstein College of Medicine for help with the FISH analysis.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Iourov I.Y., Vorsanova S.G., Yurov Y.B. Chromosomal variation in mammalian neuronal cells: known facts and attractive hypotheses. Int. Rev. Cytol. 2006;249:143–191. doi: 10.1016/S0074-7696(06)49003-3. doi:10.1016/S0074-7696(06)49003-3. [DOI] [PubMed] [Google Scholar]
- 2.Suzumori N., Sugiura-Ogasawara M. Genetic factors as a cause of miscarriage. Curr. Med. Chem. 2010;17:3431–3437. doi: 10.2174/092986710793176302. [DOI] [PubMed] [Google Scholar]
- 3.Kops G.J., Weaver B.A., Cleveland D.W. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat. Rev. Cancer. 2005;5:773–785. doi: 10.1038/nrc1714. doi:10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- 4.Weaver B.A., Silk A.D., Montagna C., Verdier-Pinard P., Cleveland D.W. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. doi:10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Jacobs P.A., Court Brown W.M., Doll R. Distribution of human chromosome counts in relation to age. Nature. 1961;191:1178–1180. doi: 10.1038/1911178a0. doi:10.1038/1911178a0. [DOI] [PubMed] [Google Scholar]
- 6.Pierre R.V., Hoagland H.C. Age-associated aneuploidy: loss of Y chromosome from human bone marrow cells with aging. Cancer. 1972;30:889–894. doi: 10.1002/1097-0142(197210)30:4<889::aid-cncr2820300405>3.0.co;2-1. doi:10.1002/1097-0142(197210)30:4<889::AID-CNCR2820300405>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 7.Guttenbach M., Koschorz B., Bernthaler U., Grimm T., Schmid M. Sex chromosome loss and aging: in situ hybridization studies on human interphase nuclei. Am. J. Hum. Genet. 1995;57:1143–1150. [PMC free article] [PubMed] [Google Scholar]
- 8.Rehen S.K., McConnell M.J., Kaushal D., Kingsbury M.A., Yang A.H., Chun J. Chromosomal variation in neurons of the developing and adult mammalian nervous system. Proc. Natl Acad. Sci. USA. 2001;98:13361–13366. doi: 10.1073/pnas.231487398. doi:10.1073/pnas.231487398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yurov Y.B., Iourov I.Y., Vorsanova S.G., Liehr T., Kolotii A.D., Kutsev S.I., Pellestor F., Beresheva A.K., Demidova I.A., Kravets V.S., et al. Aneuploidy and confined chromosomal mosaicism in the developing human brain. PLoS ONE. 2007;2:e558. doi: 10.1371/journal.pone.0000558. doi:10.1371/journal.pone.0000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muotri A.R., Gage F.H. Generation of neuronal variability and complexity. Nature. 2006;441:1087–1093. doi: 10.1038/nature04959. doi:10.1038/nature04959. [DOI] [PubMed] [Google Scholar]
- 11.Westra J.W., Peterson S.E., Yung Y.C., Mutoh T., Barral S., Chun J. Aneuploid mosaicism in the developing and adult cerebellar cortex. J. Comp. Neurol. 2008;507:1944–1951. doi: 10.1002/cne.21648. doi:10.1002/cne.21648. [DOI] [PubMed] [Google Scholar]
- 12.Iourov I.Y., Vorsanova S.G., Liehr T., Yurov Y.B. Aneuploidy in the normal, Alzheimer's disease and ataxia-telangiectasia brain: differential expression and pathological meaning. Neurobiol. Dis. 2009;34:212–220. doi: 10.1016/j.nbd.2009.01.003. doi:10.1016/j.nbd.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 13.Rehen S.K., Yung Y.C., McCreight M.P., Kaushal D., Yang A.H., Almeida B.S., Kingsbury M.A., Cabral K.M., McConnell M.J., Anliker B., et al. Constitutional aneuploidy in the normal human brain. J. Neurosci. 2005;25:2176–2180. doi: 10.1523/JNEUROSCI.4560-04.2005. doi:10.1523/JNEUROSCI.4560-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blaschke A.J., Weiner J.A., Chun J. Programmed cell death is a universal feature of embryonic and postnatal neuroproliferative regions throughout the central nervous system. J. Comp. Neurol. 1998;396:39–50. doi: 10.1002/(sici)1096-9861(19980622)396:1<39::aid-cne4>3.0.co;2-j. doi:10.1002/(SICI)1096-9861(19980622)396:1<39::AID-CNE4>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 15.Iourov I.Y., Vorsanov S.G., Liehr T., Kolotii A.D., Soloviev I.V., Vostrikov V.M., Uranova N.A., Yurov Y.B. Association of genome instability and neurodegeneration in the cerebral cortex and hippocampus in Alzheimer's disease brain: evidences for a new pathogenetic mechanism of the disease. Int. J. Psychophysiol. 2008;69:288. doi:10.1016/j.ijpsycho.2008.05.262. [Google Scholar]
- 16.Faggioli F., Vijg J., Montagna C. Chromosomal aneuploidy in the aging brain. Mech. Ageing Dev. 2011;132:429–436. doi: 10.1016/j.mad.2011.04.008. doi:10.1016/j.mad.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fraser H.B., Khaitovich P., Plotkin J.B., Paabo S., Eisen M.B. Aging and gene expression in the primate brain. PLoS Biol. 2005;3:e274. doi: 10.1371/journal.pbio.0030274. doi:10.1371/journal.pbio.0030274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartman T.K., Wengenack T.M., Poduslo J.F., van Deursen J.M. Mutant mice with small amounts of BubR1 display accelerated age-related gliosis. Neurobiol. Aging. 2007;28:921–927. doi: 10.1016/j.neurobiolaging.2006.05.012. doi:10.1016/j.neurobiolaging.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 19.Andersen B.B., Gundersen H.J., Pakkenberg B. Aging of the human cerebellum: a stereological study. J. Comp. Neurol. 2003;466:356–365. doi: 10.1002/cne.10884. doi:10.1002/cne.10884. [DOI] [PubMed] [Google Scholar]
- 20.Kingsbury M.A., Friedman B., McConnell M.J., Rehen S.K., Yang A.H., Kaushal D., Chun J. Aneuploid neurons are functionally active and integrated into brain circuitry. Proc. Natl Acad. Sci. USA. 2005;102:6143–6147. doi: 10.1073/pnas.0408171102. doi:10.1073/pnas.0408171102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yurov Y.B., Vorsanova S.G., Iourov I.Y. GIN'n'CIN hypothesis of brain aging: deciphering the role of somatic genetic instabilities and neural aneuploidy during ontogeny. Mol. Cytogenet. 2009;2:23. doi: 10.1186/1755-8166-2-23. doi:10.1186/1755-8166-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams B.R., Prabhu V.R., Hunter K.E., Glazier C.M., Whittaker C.A., Housman D.E., Amon A. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. doi:10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheltzer J.M., Blank H.M., Pfau S.J., Tange Y., George B.M., Humpton T.J., Brito I.L., Hiraoka Y., Niwa O., Amon A. Aneuploidy drives genomic instability in yeast. Science. 2011;333:1026–1030. doi: 10.1126/science.1206412. doi:10.1126/science.1206412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Upender M.B., Habermann J.K., McShane L.M., Korn E.L., Barrett J.C., Difilippantonio M.J., Ried T. Chromosome transfer induced aneuploidy results in complex dysregulation of the cellular transcriptome in immortalized and cancer cells. Cancer Res. 2004;64:6941–6949. doi: 10.1158/0008-5472.CAN-04-0474. doi:10.1158/0008-5472.CAN-04-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iourov I.Y., Vorsanova S.G., Liehr T., Kolotii A.D., Yurov Y.B. Increased chromosome instability dramatically disrupts neural genome integrity and mediates cerebellar degeneration in the ataxia-telangiectasia brain. Hum. Mol. Genet. 2009;18:2656–2669. doi: 10.1093/hmg/ddp207. doi:10.1093/hmg/ddp207. [DOI] [PubMed] [Google Scholar]
- 26.Chen G., Bradford W.D., Seidel C.W., Li R. Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature. 2012;482:246–250. doi: 10.1038/nature10795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zekanowski C., Wojda U. Aneuploidy, chromosomal missegregation, and cell cycle reentry in Alzheimer's disease. Acta Neurobiol. Exp. (Wars) 2009;69:232–253. doi: 10.55782/ane-2009-1748. [DOI] [PubMed] [Google Scholar]
- 28.Matevossian A., Akbarian S. Neuronal nuclei isolation from human postmortem brain tissue. J. Vis. Exp. 2008 doi: 10.3791/914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shankaran M., King C., Lee J., Busch R., Wolff M., Hellerstein M.K. Discovery of novel hippocampal neurogenic agents by using an in vivo stable isotope labeling technique. J. Pharmacol. Exp. Ther. 2006;319:1172–1181. doi: 10.1124/jpet.106.110510. doi:10.1124/jpet.106.110510. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.