Abstract
Leukocyte telomere length (LTL) is associated with a number of common age-related diseases and is a heritable trait. Previous genome-wide association studies (GWASs) identified two loci on chromosomes 3q26.2 (TERC) and 10q24.33 (OBFC1) that are associated with the inter-individual LTL variation. We performed a meta-analysis of 9190 individuals from six independent GWAS and validated our findings in 2226 individuals from four additional studies. We confirmed previously reported associations with OBFC1 (rs9419958 P = 9.1 × 10−11) and with the telomerase RNA component TERC (rs1317082, P = 1.1 × 10−8). We also identified two novel genomic regions associated with LTL variation that map near a conserved telomere maintenance complex component 1 (CTC1; rs3027234, P = 3.6 × 10−8) on chromosome17p13.1 and zinc finger protein 676 (ZNF676; rs412658, P = 3.3 × 10−8) on 19p12. The minor allele of rs3027234 was associated with both shorter LTL and lower expression of CTC1. Our findings are consistent with the recent observations that point mutations in CTC1 cause short telomeres in both Arabidopsis and humans affected by a rare Mendelian syndrome. Overall, our results provide novel insights into the genetic architecture of inter-individual LTL variation in the general population.
INTRODUCTION
Rare mutations in telomere maintenance genes―resulting in critically short leukocyte telomere length (LTL)―cause monogenic diseases with a spectrum of clinical manifestations. These include diseases such as dyskeratosis congenita, idiopathic pulmonary fibrosis and liver cirrhosis, the common manifestation of which is often aplastic anemia (1–4). In contrast, little is known about the consequences of common genetic variants that contribute to the considerable inter-individual variation in LTL in the general population (5). Patients with clinical manifestations of atherosclerosis and those at risk for this aging-related disease often display short LTL (6–9). Moreover, recent studies have reported LTL to be inversely related to survival in the elderly (10–12). Therefore, genetic variants that affect LTL dynamics (LTL at birth and age-dependent attrition) might play a role in the aging of the cardiovascular system and in longevity.
To date, genome-wide association studies (GWASs) have identified two main genetic variants associated with LTL variation (rs3772190 at 3q26.2 and rs4387287 at 10q24.33) (13–15). However, these findings explained only a very small part of the genetic variation in LTL (<1 and 2.26% for rs3772190 and rs4387287, respectively) (13,14). As has been demonstrated for other complex traits, the larger the sample size, the more likely it is to unveil additional LTL-regulating genes. This would bolster understanding of the role of LTL dynamics in aging, aging related-disease and longevity.
Here, we report the results of a GWAS meta-analysis, performed on a total of 9190 individuals from six independent observational studies. We have identified two novel LTL loci (on chromosome 17p13.1 and 19p12), highlighted a number of genes for which verification in larger studies is warranted and reaffirmed that LTL in humans is fashioned by a large number of common genetic variants with small individual effects.
RESULTS
We carried out a meta-analysis of GWAS of LTL in 9190 individuals of European ancestry from six collaborating studies: TwinsUK (n = 3222), the Family Heart Study (FamHS) (n = 2508), the Framingham Heart Study (FHS) (n = 1146), the Cardiovascular Health Study (CHS) (n = 1061), Hypertension Genetic Epidemiology Network Study (HyperGEN) (n = 920) and the Bogalusa Heart Study (BHS) (n = 333). Blood samples were collected in each cohort and LTL was measured by Southern blot (Table 1). Mean age ranged across the cohorts from 35 to 75 years and the mean LTL varied from 6.33 to 7.22 kb (Table 1).
Table 1.
Cohort characteristics for the telomere length GWAS analysis
| N | Mean, age (year range) | Women (%) | Body mass index (mean ± STD) | Telomere length (mean ± STD) | |
|---|---|---|---|---|---|
| Cohort | |||||
| Framingham Heart Study | 1146 | 59 (33–86) | 51 | 28.0 ± 5.0 | 6.96 ± 0.58 (Kb) |
| Family Heart Study | 2508 | 57 (30–93) | 54 | 28.9 ± 5.7 | 6.78 ± 0.67 (Kb) |
| Cardiovascular Health Study | 1061 | 75 (67–95) | 62 | 26.6 ± 4.4 | 6.33 ± 0.61 (Kb) |
| Bogalusa Heart Study | 333 | 35 (20–48)a | 42 | 28.0 ± 6.7a | 7.22 ± 0.70 (Kb) |
| HyperGEN | 920 | 50 (18–87) | 50 | 29.5 ± 6.3 | 6.78 ± 0.61 (Kb) |
| TwinsUK | 3222 | 48 (18–82) | 92 | 26.1 ± 4.9 | 6.97 ± 0.68 (Kb) |
| Replication | |||||
| Health ABC | 337 | 73.6 (69–80) | 49 | 27.7 ± 4.8 | 6.13 ± 0.51 (Kb) |
| Jerusalem LRC | 620 | 43.2 (41–46) | 33 | 27.2 ± 4.5 | 7.00 ± 0.63 (Kb) |
| ADELAHYDE/Nancy/ERA-France | 316 | 61.7 (25–84) | 51 | 27.4 ± 4.6 | 6.28 ± 0.74 (Kb) |
| Danish collection | 964 | 80.7 (58–101) | 77 | 24.3 ± 4.0 | 5.74 ± 0.67 (Kb) |
aThese values include two time points for each individual.
Each cohort underwent GWA for LTL on 2.5 million imputed single-nucleotide polymorphisms (SNPs) with adjustment for age, sex, BMI and smoking status (pack years). In the individual cohorts participating to this study, the GWAS inflation (λGC) ranged from 0.995 to 1.076, indicating that there was no significant population stratification or that it was very minor. In addition, there was no between-study heterogeneity as indicated by the P-values of the heterogeneity test ranging from 0.15 to 0.68 (Table 2). Finally, the quantile–quantile plot demonstrated that the statistical association for all SNPs included in the meta-analysis follows the distribution expected under the null hypothesis (Supplementary Material, Fig. S1).
Table 2.
Meta-analysis results for the most significant SNPs (P < 5 × 10−8) at each locus
| Most significant SNP | Chromosome | Location | minor/major allele | MAF | βa (SE) | P-value | Het Pb | Analysis | closest gene |
|---|---|---|---|---|---|---|---|---|---|
| rs9419958 | 10 | 105665936 | t/c | 0.1353 | 0.0829 (0.013) | 9.13E−11 | 0.15 | Discovery | OBFC1 |
| rs1317082 | 3 | 170980279 | g/a | 0.2875 | 0.0679 (0.011) | 1.14E−08 | 0.26 | Discovery | TERC |
| rs412658 | 19 | 22151280 | t/c | 0.3534 | 0.0559 (0.010) | 3.32E−08 | 0.65 | Discovery | ZNF676 |
| t/c | 0.3697 | 0.0241 (0.021) | 1.49E−01 | 0.69 | Replication | ||||
| t/c | 0.3564 | 0.0497 (0.009) | 9.75E−09 | 0.68 | Combined | ||||
| rs3027234 | 17 | 8076817 | t/c | 0.1678 | −0.0669 (0.012) | 3.58E−08 | 0.45 | Discovery | CTC1 |
| t/c | 0.2304 | −0.0226 (0.022) | 2.80E−01 | 0.78 | Replication | ||||
| t/c | 0.1794 | −0.0573 (0.011) | 2.29E−08 | 0.40 | Combined |
aEffect reported in Kbp relative to the minor allele.
bP-value of the heterogeneity test.
We calculated that our study (n = 9190) had 80% power to detect a variant (minor allele frequency 10%) which has an effect on LTL of ±0.12 kb at a statistical threshold of P < 5 × 10−8 (Supplementary Material, Fig. S2).
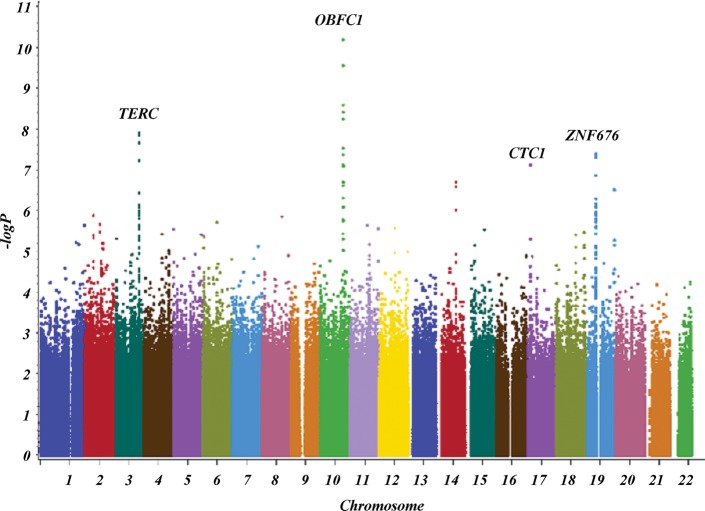
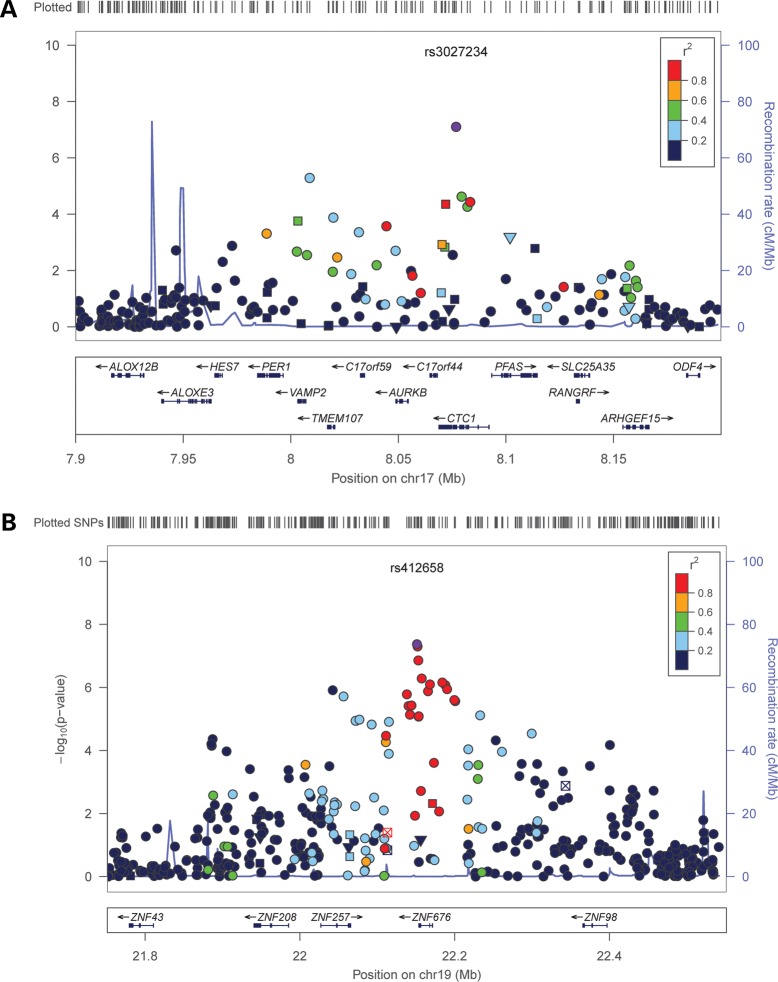
Our analysis confirmed the previously reported associations (13,14) of LTL with the oligonucleotide/oligosaccharide-binding fold containing one gene (OBFC1; rs9419958 P = 9.1 × 10−11) and the telomerase RNA component (TERC) gene (rs1317082, P = 1.1 × 10−8) (Fig. 1). We also identified two novel loci associated with LTL at the conventional genome-wide significance level (P < 5 × 10−8). These newly identified loci include variants in a conserved telomere maintenance complex component 1 (CTC1; rs3027234, P = 3.6 × 10−8) and in the zinc finger protein 676 (ZNF676; rs412658, P = 3.3 × 10−8) (Fig. 2).
Figure 1.
Manhattan plot of discovery meta-analysis.
Figure 2.
Regional plots of chromosome 17 (A) and 19 (B) loci. SNPs are plotted by position on chromosome against association (−log10 P-value) with LTL using discovery GWAS meta-analysis data. In each panel, the SNP with the strongest association is denoted with a purple dot: the P-value attached represents the final P-value obtained in the meta-analysis (Table 2). Estimated recombination rates (from HapMap-CEU) are plotted in blue to reflect the local LD structure on a secondary Y-axis. The SNPs surrounding the most significant SNP (purple dot) are color-coded (see legend) to reflect their LD with the lead SNP (using pair-wise r2-values from HapMap CEU). Genes position, as well as transcription direction, is shown below the plots (using data from the UCSC genome browser, genome.ucsc.edu).
A total of 2226 individuals from four independent studies were included in replication analyses of the novel two genome-wide significant SNPs (rs3027234 and rs412658) (Table 1). There was no evidence for heterogeneity of effect size among the replication studies (Table 2). In the replication sample set, the effect of the minor allele was in the same direction as in the discovery sample for both SNPs. In the combined meta-analysis of the discovery and replication samples, both rs3027234 and rs412658 (Table 2) showed a smaller P-value (rs412658, P = 9.7 × 10−9; rs3027234, P = 2.3 × 10−8) than in discovery.
Furthermore, we examined whether the effect of the top four SNPs associated with LTL variation identified in the discovery stage was modified by gender. Although we observed higher P-values for the female-specific analyses compared with those of the males, the effect size was not significantly different between the genders (P-values ranging from 0.07 to 0.89) (Supplementary Material, Table S2). However, in our study, the female to male ratio was nearly 2 to 1 (Supplementary Material, Table S2), which may compromise the power to find a significant gender interaction for ZNF676 and TERC SNPs.
To explore the potentially functional impacts of these two top SNPs, we used genome-wide expression data from the Multiple Tissue Human Expression Resource (MuTHER) (16) (http://www.muther.ac.uk/) based on ∼170 unselected twins sampled for skin, adipose tissue and lymphoblastoid cell lines (LCLs). We focused our analysis only on LCLs and found that the minor allele (T) of rs3027234 is highly associated with lower expression of CTC1 (P = 4.8 × 10−6). No expression data were present in the analyzed array for ZNF676.
Further, we used two independent pathway-based methods [STRING (17) and GRAIL (18)] to identify connections among our single-marker associations and link them with broader biological processes. Although both approaches are based on published data linking the gene products of our top hits in functional annotated pathways, they use substantially different methods.
With GRAIL, we investigated an excess of connectivity between the four established/novel loci (seed regions) and those loci that did not reach genome-wide significance, but showed suggestive associations (P < 5 × 10−5) with LTL variation (query regions). Among the 75 suggestively associated loci (Supplementary Material, Table S1), we observed five genes to be connected with those at the genome-wide significant loci at Pgrail < 0.01. These included TERT (Pgrail 3.2 × 10−5), ZNF256 (Pgrail 1.4 × 10−3), ZNF587 (Pgrail 4.5 × 10−3), WDR21A (Pgrail 7.3 × 10−3) and RPS29 (Pgrail 7.6 × 10−3).
We then used STRING to uncover additional protein interaction pathways among the genes within 500 Kb of the 79 independent SNPs with P < 5 × 10−5.
We observed two clusters (Supplementary Material, Fig. S3). The major cluster included TERT (rs10069690, P = 3.6 × 10−6), SMAD3 (rs17208967, P = 3.5 × 10−6), YWHAZ (rs884488, P = 3.6 × 10−5), PDK1 (rs4972842, P = 2.0 × 10−5), PIK3R1 (rs16897810, P = 1.3 × 10−5), GNG2 (rs7154422, P = 4.1 × 10−5), ADRA1B (rs10515803, P = 3.7 × 10−5), PROK2 (rs6790891, P = 3.2 × 10−5), CHRNA7 (rs11637923, P = 4.0 × 10−5), DLC1 (rs17254431, P = 2.3 × 10−5), LRP1B (rs11886422, P = 1.8 × 10−5) and GRM7 (rs9843107, P = 9.9 × 10−7). Of particular interest for LTL homeostasis was the interaction between TERT and YWHAZ (19), TERT and SMAD3 (20) and the interaction between CTC1 and OBFC1 (21), which formed the second protein–protein cluster.
DISCUSSION
In this meta-analysis of GWAS results from six observational studies, we confirmed associations with LTL at loci harboring OBFC1 and TERC, two intrinsic telomere maintenance genes. We, also, identified genome-wide significant associations of two new loci [a conserved telomere maintenance CTC1 and a zinc finger protein 676(ZNF676)] with LTL variation. Expression analysis showed that the rs3027234 minor allele was associated with both short LTL and reduced expression of the CTC1 gene. Based on its position on the sequence (rs3027234 is in introns 11–12, at 132 bp from exon 11) and the LD patterns of the region, we hypothesize that this variation (or another SNP in LD) may disrupt (or create a new) splicing site which results in the reduced gene expression.
The variance in telomere length explained by all four identified LTL loci was ∼1.6%. The minor allele of rs3027234 and the major allele of rs412658 were associated with a mean LTL that was, respectively, ∼57 and 49 bp shorter. To put this in context, in TwinsUK, LTL decreased by 27 bp on average per year (22); therefore, carriage of rs3027234 minor allele was associated with shorter LTL equivalent to 2.1 years of average age-related LTL attrition rate.
The genetic influence of telomere maintenance genes on telomere dynamics in the hematopoietic systems is clearly displayed in rare Mendelian diseases that result from major mutations in genes that encode the catalytic subunit of telomerase reverse transcriptase (TERT), TERC and other proteins that regulate telomerase and protect telomeres (1–4). The cardinal manifestation of these diseases is aplastic anemia, presumably because very short telomeres affect replicative capacity within the hematopoietic system. In addition, common variants near the TERT affect susceptibility to a variety of cancer subtypes (23–28), suggesting that more subtle variation in telomere maintenance increases cancer risk. At the same time, recent GWASs have also found that loci that contain OBFC1 and TERC explain some of the inter-individual variation in LTL in the general population (13,14). The protein encoded by OBFC1 is a member of the heterotrimer CST complex that was originally discovered in budding yeast (Cdc13, Stn1 and Ten1); this complex is conserved in plants and vertebrates (21,29). In human cells, CTC1 carries out the functions of cdc13 (30), while OBFC1 is equivalent to Stn1. Thus, the genes that encode the human heterotrimer CST complex are CTC1, OBFC1 and TEN1. The CST complex is distinguished from the shelterin complex, which consist of six proteins (TRF1, TRF2, TIN2, TPP1, RAP1 and POT1) (31). While the shelterin complex is distinctly involved in telomere maintenance through telomere protection, the CST complex is also involved in other tasks that are not entirely understood. Based on knock-downs of CTC1 and OBFC1, it appears that the complex is not only involved in telomere maintenance, perhaps by negatively regulating telomerase (30,32), but is also engaged in the functioning of the DNA replicative fork, which is not strictly related to telomere protection (29,33). In a recent study, point mutations in CTC1 were identified in Coats plus disorder, a rare recessive Mendelian syndrome (34). Individuals carrying mutations in CTC1 had a short LTL.
The findings that OBFC1 (14) and now CTC1 are associated with LTL in the general population underscore that the CST complex is indeed engaged in telomere maintenance in humans.
The mechanisms that link CTC1 to LTL evidently stem from the gene being a member of the CST complex, but the role of ZNF676 in LTL regulation is unknown. Theoretically, ZNF676 can modify LTL in two broad ways. First, by direct binding to DNA it might alter the expression (repression/activation) of genes engaged in telomere maintenance and through its interaction with RNAs/proteins alter the post-translational signaling of these genes (35,36). Second, there is evidence that the single-stranded telomeric DNA can fold into a structure known as a G-quartet or G-quadruplex (37,38), which at the 3′ end of the telomeres might inhibit telomere elongation by telomerase (37). Zinc finger proteins can bind specifically to and stabilize G-quadruplex DNA (39–41), including telomeric DNA (39,41).
Interestingly, we found that two SNPs that did not reach genome-wide significance, but showed suggestive levels of association (P < 5 × 10−5) with LTL variation, are in or near genes (TERT and SMAD3) pivotal for telomere regulation. Indeed, our results are consistent with findings that the expression of the human telomerase reverse transcriptase (hTERT) (a key element in telomerase activation, telomere maintenance and tumor development) is mediated by SMAD3 (20,42–44).
Leukocyte telomeres may become critically short during the long human lifespan because the hematopoietic system is the most proliferative tissue in the body. Thus, the premise that GWAS of LTL might ultimately provide a mechanistic insight into the role of genes in the telomere dynamics of hematopoietic stem cell has been borne out by this and recent investigations (13,14). That is because LTL dynamics principally reflect hematopoietic stem cell telomere dynamics (45,46). Based on the existing body of research, it is now clear that a large subset of genes contain common variants with very small effects on human LTL maintenance. These genes, as shown recently (47), might play a role in human longevity.
Finally, as women have longer LTL than men (48,49) and African Americans have longer LTL than whites of European ancestry (11,50,51), it will be important to test in future studies whether the functions of the LTL genes identified thus far by GWAS (and other LTL-regulating genes) are modified by sex or ethnicity and whether these genes interact with the environment. In this study, we examined the possibility that the loci identified may have a gender effect. Although we observed that gender had no effect on the function of the identified genes, we could not rule out any possible gender effect because the sample size of the males included in our analysis did not have enough power compared with females.
MATERIALS AND METHODS
Cohorts
A detailed description of demographic of the six cohorts participating in this GWAS consortium (the FHS, the FamHS, the CHS, the BHS, the HyperGEN and TwinsUK) can be found in Table 1. All studies received institutional review-board approvals and all participants provided written informed consent for participation in the parent study, including genetic analysis and the use of DNA for the measurement of LTL. Further details related to the discovery cohorts can be found elsewhere (50,52–57). The same covariates (age, age2 sex, smoking history) were included GWA analysis by all members of the consortium. Evidence of non-European ancestry was assessed in all cohorts by PCA comparison with HapMap populations.
LTL assay
LTL measurement was performed by Southern blot analysis of the terminal restriction fragments, generated by the restriction enzymes HinfI and RsaI after verification of DNA integrity (58). The overlay method was used for the FHS and the FamHS (49) and the standard method was used for the other cohorts. These two methods use the same measurement principles and are highly correlated. The mean length of terminal restriction fragments (expressed in kilobases) was used as a measure of LTL. Coefficients of variation ranged from 0.9 to 2.4% in replicate samples.
Genome-wide genotyping
For the FHS, genotyping was conducted on 9274 participants using the Affymetrix 500K mapping array and the Affymetrix 50K gene-focused molecular imprinted polymer array (SNP Health Association Resource at http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000007.v2.p1). Participants were excluded from association analyses if the call rate across all genotyped SNPs was <97%. A total of 503 551 SNPs available for analysis after exclusions and 1146 individuals met the eligibility criteria, based on LTL measurements and covariate information. The genotype data for 2.5 million SNPs were imputed using MACH (version 1.0.15) (59) software for autosomal SNPs and the reference panel for the imputation was the publicly available phased haplotypes from HapMap (release 22, build 36, CEU population). The final genotyping data for analysis were composed of 2 543 887 SNPs in HapMap using allele dosage (0–2) values. Family members were included. Thus, significance of the association between SNPs and LTL was tested using the linear mixed-effects regression model, which considers the random individual effects correlated within pedigree according to kinship relationships.
For the FamHS, participants were genotyped in three phases using the Illumina HumMap 550, Human 6100-Quadv1 or Human 1M-Duov3 arrays. Because telomere lengths were only measured on blood samples obtained at the second exam of FamHS, all of the subjects of European descent in this study were genotyped on the HumMap550 array. For the subjects of European descent, there were 530 092 SNP markers available for analysis in the FamHS. After exclusions for deviations from the Hardy–Weinberg equilibrium (P < 10−6), minor allele frequency <1% and markers not in the HapMap, 499 558 markers were used for imputation. MACH (version 1.0.15) (59) was used to impute up to 2 543 887 SNPs using the publicly available phased haplotypes from HapMap (release 22, build 26, CEU population) as a reference population. A random sample of unrelated 100 men and 100 women, excluding individuals with the highest rates of missing genotypes, was used to estimate parameters that were then applied to the remaining subjects. The GenAbel (60) package in R was used for analysis to allow for the pedigree structures to be modeled appropriately.
For the CHS, genotyping was performed using the Illumina 370CNV BeadChip system on 1080 samples from individuals of European ancestry were available with both LTL measurements and GWA analysis. Because all other cohorts included only Caucasian individuals, the African-American participants were excluded from this analysis (n = 19). Participants with a call rate ≤95% were excluded. The following exclusions were applied to identify a final set of 306 655 autosomal SNPs: call rate <97%, Hardy–Weinberg equilibrium P < 10−5, more than two duplicate errors or Mendelian inconsistencies (for reference Centre d'Etude du Polymorphisme Humain trios), heterozygote frequency = 0 or SNP not found in HapMap. Imputation was performed using BIMBAM (v0.99) (61) with reference to HapMap CEU using release 22, build 36 with one round of imputations and the default expectation–maximization warm-ups and runs. During the analysis, SNPs were excluded for variance on the allele dosage ≤0.01.
For the BHS, participants were genotyped on the Illumina Human610 Genotyping Beadchip. Genotypes were called using the BeadStudio clustering algorithm. Samples were filtered based on the 10th and 50th percentile GenCall score and overall call rates (<99%). Related individuals were identified using Identity-by-descent measures in PLINK (62). Individuals were filtered such that there were no pairs with pi-hat >0.10. This resulted in a total of 333 individuals. SNPs were filtered based on call rates (<90%). Cluster plots for SNPs with call rates between 90 and 95% or with cluster separation scores <0.30 were manually inspected. By genotyping 29 samples in duplicate (18 known replicates and 11 blind replicates), we observed reproducibility >99.99%. There were 626 145 genotyped SNPs used in the reproducibility analysis. Imputation was performed with MACH (v. 1.0.16) (59) using phased CEU haplotypes from HapMap Phase 2 (release 22) and 550 798 genotyped SNPs. A random sample of 200 individuals was used to estimate recombination and error rates before imputing the entire sample. This resulted in imputing 2 543 887 SNPs. By masking 0.1% of the genotypes before imputation, we observed allelic error rates of 1.6% and genotypic error rates of 3.1%. As LTL for BHS participants was measured at two time points, we modeled and tested associations between SNPs and the two LTL measures using a linear mixed model as implemented in the nlme R package (48). A covariance structure for the two LTL measurements was chosen by testing all spatial structures with the model, and the structure that resulted in the lowest Akaike Information Criterion score was used (in this study, the best covariance was exponential). If an SNP had been genotyped, the genotype value (0, 1 or 2) was used as a predictor, whereas if the SNP was imputed, the estimated dosage was used.
For HyperGEN, a subset of 1319 subjects of European descent (none from Framingham) were genotyped on an Affymetrix 5.0 array. After removing the HyperGEN subject overlap with the Family Heart Study, those without telomere lengths or those who did not meet quality control standards, 920 Caucasian subjects had complete data for inclusion in the meta-analysis. The total 358 327 SNPs were used for imputation using MACH (59) after excluding SNPs with Mendelian errors (P < 10−6) or minor allele frequencies <1%. The final number of imputed SNPs was 2 355 427. The GenABEL package (60) in R was used for analysis to allow for the pedigree structures to be modeled appropriately.
TwinsUK samples were typed with the Infinium 317K and 610K assay (Illumina, San Diego, USA) at two different centers, the Centre for Inherited Diseases Research (USA) and the Wellcome Trust Sanger Institute. We pooled the normalized intensity data and called genotypes on the basis of the Illluminus algorithm. No calls were assigned if the most likely call was less than a posterior probability of 0.95. Validation of pooling was done by visual inspection of 100 random, shared SNPs for overt batch effects; none was observed. We excluded SNPs that had a call rate <97% (SNPs with MAF ≥ 5%) or <99% (for 1% ≤MAF < 5%), Hardy–Weinberg P-values < 10−6 and minor allele frequencies <1%. We also removed subjects where genotyping failed for >2% of SNPs. The overall genotyping efficiency of the GWA was 98.7%. Imputation of genotypes was carried out using the software IMPUTE (63).
Because of the relatedness in the TwinsUK cohort, we utilized the GenABEL software package (60) which is designed for GWAS analysis of family-based data by incorporating a pair-wise kinship matrix calculated using genotyping data in the polygenic model to correct relatedness and hidden population stratification. The score test implemented in the software was used to test the association between a given SNP and LTL.
GWA meta-analysis
The meta-analysis was carried out independently by two analysts using METAL (64). The results were then compared for consistency.
In the main meta-analysis, we combined the individual P-values obtained for each cohort using Fisher's method (65), which allows P-values and direction of effect in each cohort to be combined taking cohort size into account, independently of β-estimates. Individual cohort results were corrected for residual inflation of the test statistics using lambda of genomic control (GC) estimates (66). The GC values were estimated for each study using all analyzed SNPs.
The level of conventional genome-wide significance level was established at P < 5 × 10−8. A P-value between 5 × 10−8 and 5 × 10−7 was considered strongly suggestive of association.
The inverse variance weighted method was used to combine the cohort-specific β-estimates to evaluate the effect size only for the markers that reached a genome-wide significance level.
The estimate of the variance explained by the top four SNPs at each locus were calculated in a two-stage approach using GCTA (v 1.0) (67). TwinsUK (n = 3222) genotypes and LTL information were used for this analysis. In the first stage, we estimated the genetic relationship matrix for all the genotyped SNPs in order to account for family structure in the second stage. In the second stage, we performed a restricted maximum likelihood analysis to estimate the variance explained by the top four SNPs. In this stage, age and sex were also included as covariates.
Replication cohorts
Samples from the Jerusalem LRC Longitudinal Study (N = 620) (68), Health ABC study (N = 337) (69), from ADELAHYDE-Nancy study and ERA-France study (N = 316) (70,71) and from a Danish collection (N = 964) (47) were included in the replication stage. A detailed description for each cohort can be found elsewhere (47,68,69). All studies received institutional review-board approvals and all participants provided written informed consent, including genetic analysis and the use of DNA for the measurement of LTL.
LTL for all the samples was measured by Southern blot analysis of the terminal restriction fragments, as in the discovery data set (47,58).
A linear regression analysis adjusting for same covariates (age, age2 sex, smoking history) included in the discovery analysis was performed in the replication sample to test association between the two novel SNPs and LTL variation.
Power calculations
Power calculations were performed using QUANTO (v 1.2.4) (72). We calculated the power in our combined discovery cohorts (n = 9190) to detect effects of various sizes at a genome-wide significance level (P < 5 × 10−8) for variants with allele frequencies ranging from 0.1 to 0.5.
Pathway analysis
Grail
We used the GRAIL (18) online tool to evaluate whether genome-wide loci associated with LTL were enriched for connectivity between genes representing particular pathways. The method performs a text-based analysis looking at abstracts in PubMed (we used abstracts prior to December 2006 to avoid confounding from GWAS results). We investigated connectivity between our genome-wide significant signals (seed regions) and those which did not reach genome-wide significance but were suggestively associated with LTL (P < 5 × 10−5) (query regions) as previously described (73).
We identified 75 query regions defined by all SNPs with a P < 5 × 10−5. We also pruned SNPs with r2 > 0.05 in each region represented by more than one SNP using SNAP pairwise LD online tool (74).
String
We used the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) (www.string-db.org) (17) to perform a more in-depth pathway analysis. The interactions explored by STRING include direct (physical) and indirect (functional) associations. We examined potential interactions among all the gene proteins in a range of 500 kb from the 79 independent SNPs with a P < 5 × 10−5 utilized in the GRAIL analysis.
Expression analysis
We used the genome-wide expression data from the LCLs from the Multiple Tissue Human Expression Resource (MuTHER) (16). The expression values were derived from a subset of twins from TwinsUK, which were also included in the association analysis. The analysis was performed only on rs3027234 and the expression levels of CTC1 because there was no expression data for ZNF676. The analysis was performed using MERLIN (75), taking in account the family structure. For this analysis, the significance was defined as P < 0.05, as only one independent test was performed.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
TwinsUK. The study was funded by the Wellcome Trust; European Community's Seventh Framework Programme (FP7/2007-2013), ENGAGE project grant agreement (HEALTH-F4-2007-201413). The study also receives support from the Dept of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy's & St Thomas’ NHS Foundation Trust in partnership with King's College London. T.D.S. is an NIHR senior Investigator and is holder of an ERC Advanced Principal Investigator award. Genotyping was performed by The Wellcome Trust Sanger Institute, support of the National Eye Institute via an NIH/CIDR genotyping project.
The Bogalusa Heart Study. This study was supported by grants HD-061437 and HD-062783 from the National Institute of Child Health and Human Development, and AG-16592 from the National Institute on Aging.
The Framingham Heart Study. Supported by NIH contract N01-HC-25195. This project was supported in part by intramural funding from the National Heart, Lung, and Blood Institute and the Center for Population Studies of the NHLBI.
CHS. This CHS research was supported by NHLBI grant 1 R01 HL80698-01 and contracts N01-HC-85239, N01-HC-85079 through N01-HC-85086; N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133, HHSN268201200036C and NHLBI grants HL080295, HL087652, HL105756 with additional contribution from NINDS. Additional support was provided through AG-023629, AG-15928, AG-20098 and AG-027058 from the NIA. See also http://www.chs-nhlbi.org/pi.htm. DNA handling and genotyping were supported in part by National Center of Advancing Translational Technologies CTSI grant UL1TR000124 and National Institute of Diabetes and Digestive and Kidney Diseases grant DK063491 to the Southern California Diabetes Endocrinology Research Center and Cedars-Sinai Board of Governors’ Chair in Medical Genetics (J.I.R.).
The Center of Human Development and Aging. A.A. grant support: NIH: Human Telomere Genetics R01AG20132; Telomeres & Vascular Aging, R01AG21593; Leukocyte Telomere Dynamics, Gender, Menopause, Insulin Resistance, R01AG030678.
Jerusalem LRC Longitudinal Study The study was funded by the US-Israel Binational Science Foundation and the Israel Science Foundation.
ADELAHYDE-Nancy study and ERA-France study. The study received support from the French Fondation pour la Recherche Médicale (FRM DCV2007-0409250) and the Plan Pluriformation (PPF815 PSVT-2005). Special thanks to Ms Cynthia Thiriot (INSERM U961, Nancy France) for her contribution to the geotyping of French cohorts.
HyperGEN. HyperGEN was supported by cooperative agreements HL54471, HL54472, HL54473, HL54495, HL54496, HL54509, HL54515 and grant HL055673. HyperGEN investigators and institutions can be found at http://www.biostat.wustl.edu/hypergen/hypergen.shtml.
Health ABC. This research was supported by NIA contracts N01AG62101, N01AG62103 and N01AG62106. The GWAS was funded by NIA grant 1R01AG032098-01A1 to Wake Forest University Health Sciences and genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN268200782096C. This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging. Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust.
Supplementary Material
REFERENCES
- 1.Armanios M.Y., Chen J.J., Cogan J.D., Alder J.K., Ingersoll R.G., Markin C., Lawson W.E., Xie M., Vulto I., Phillips J.A., 3rd, et al. Telomerase mutations in families with idiopathic pulmonary fibrosis. N. Engl. J. Med. 2007;356:1317–1326. doi: 10.1056/NEJMoa066157. doi:10.1056/NEJMoa066157. [DOI] [PubMed] [Google Scholar]
- 2.Calado R.T., Young N.S. Telomere diseases. N. Engl. J. Med. 2009;361:2353–2365. doi: 10.1056/NEJMra0903373. doi:10.1056/NEJMra0903373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parry E.M., Alder J.K., Qi X., Chen J.J., Armanios M. Syndrome complex of bone marrow failure and pulmonary fibrosis predicts germline defects in telomerase. Blood. 2011;117:5607–5611. doi: 10.1182/blood-2010-11-322149. doi:10.1182/blood-2010-11-322149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsakiri K.D., Cronkhite J.T., Kuan P.J., Xing C., Raghu G., Weissler J.C., Rosenblatt R.L., Shay J.W., Garcia C.K. Adult-onset pulmonary fibrosis caused by mutations in telomerase. Proc. Natl Acad. Sci. USA. 2007;104:7552–7557. doi: 10.1073/pnas.0701009104. doi:10.1073/pnas.0701009104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aviv A. Genetics of leukocyte telomere length and its role in atherosclerosis. Mutat. Res. 2012;730:68–74. doi: 10.1016/j.mrfmmm.2011.05.001. doi:10.1016/j.mrfmmm.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benetos A., Gardner J.P., Zureik M., Labat C., Xiaobin L., Adamopoulos C., Temmar M., Bean K.E., Thomas F., Aviv A. Short telomeres are associated with increased carotid atherosclerosis in hypertensive subjects. Hypertension. 2004;43:182–185. doi: 10.1161/01.HYP.0000113081.42868.f4. doi:10.1161/01.HYP.0000113081.42868.f4. [DOI] [PubMed] [Google Scholar]
- 7.Brouilette S.W., Moore J.S., McMahon A.D., Thompson J.R., Ford I., Shepherd J., Packard C.J., Samani N.J. Telomere length, risk of coronary heart disease, and statin treatment in the West of Scotland Primary Prevention Study: a nested case-control study. Lancet. 2007;369:107–114. doi: 10.1016/S0140-6736(07)60071-3. doi:10.1016/S0140-6736(07)60071-3. [DOI] [PubMed] [Google Scholar]
- 8.O'Donnell C.J., Demissie S., Kimura M., Levy D., Gardner J.P., White C., D'Agostino R.B., Wolf P.A., Polak J., Cupples L.A., et al. Leukocyte telomere length and carotid artery intimal medial thickness: the Framingham Heart Study. Arterioscler. Thromb. Vasc. Biol. 2008;28:1165–1171. doi: 10.1161/ATVBAHA.107.154849. doi:10.1161/ATVBAHA.107.154849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samani N.J., van der Harst P. Biological ageing and cardiovascular disease. Heart. 2008;94:537–539. doi: 10.1136/hrt.2007.136010. doi:10.1136/hrt.2007.136010. [DOI] [PubMed] [Google Scholar]
- 10.Bakaysa S.L., Mucci L.A., Slagboom P.E., Boomsma D.I., McClearn G.E., Johansson B., Pedersen N.L. Telomere length predicts survival independent of genetic influences. Aging Cell. 2007;6:769–774. doi: 10.1111/j.1474-9726.2007.00340.x. doi:10.1111/j.1474-9726.2007.00340.x. [DOI] [PubMed] [Google Scholar]
- 11.Fitzpatrick A.L., Kronmal R.A., Kimura M., Gardner J.P., Psaty B.M., Jenny N.S., Tracy R.P., Hardikar S., Aviv A. Leukocyte telomere length and mortality in the Cardiovascular Health Study. J. Gerontol. A Biol. Sci. Med. Sci. 2011;66:421–429. doi: 10.1093/gerona/glq224. doi:10.1093/gerona/glq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimura M., Hjelmborg J.V., Gardner J.P., Bathum L., Brimacombe M., Lu X., Christiansen L., Vaupel J.W., Aviv A., Christensen K. Telomere length and mortality: a study of leukocytes in elderly Danish twins. Am. J. Epidemiol. 2008;167:799–806. doi: 10.1093/aje/kwm380. doi:10.1093/aje/kwm380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Codd V., Mangino M., van der Harst P., Braund P.S., Kaiser M., Beveridge A.J., Rafelt S., Moore J., Nelson C., Soranzo N., et al. Common variants near TERC are associated with mean telomere length. Nat. Genet. 2010;42:197–199. doi: 10.1038/ng.532. doi:10.1038/ng.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levy D., Neuhausen S.L., Hunt S.C., Kimura M., Hwang S.J., Chen W., Bis J.C., Fitzpatrick A.L., Smith E., Johnson A.D., et al. Genome-wide association identifies OBFC1 as a locus involved in human leukocyte telomere biology. Proc. Natl Acad. Sci. USA. 2010;107:9293–9298. doi: 10.1073/pnas.0911494107. doi:10.1073/pnas.0911494107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mangino M., Richards J.B., Soranzo N., Zhai G., Aviv A., Valdes A.M., Samani N.J., Deloukas P., Spector T.D. A genome-wide association study identifies a novel locus on chromosome 18q12.2 influencing white cell telomere length. J. Med. Genet. 2009;46:451–454. doi: 10.1136/jmg.2008.064956. doi:10.1136/jmg.2008.064956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nica A.C., Parts L., Glass D., Nisbet J., Barrett A., Sekowska M., Travers M., Potter S., Grundberg E., Small K., et al. The architecture of gene regulatory variation across multiple human tissues: the MuTHER study. PLoS Genet. 2011;7:e1002003. doi: 10.1371/journal.pgen.1002003. doi:10.1371/journal.pgen.1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P., et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. doi:10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raychaudhuri S., Plenge R.M., Rossin E.J., Ng A.C., Purcell S.M., Sklar P., Scolnick E.M., Xavier R.J., Altshuler D., Daly M.J. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. doi:10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seimiya H., Sawada H., Muramatsu Y., Shimizu M., Ohko K., Yamane K., Tsuruo T. Involvement of 14-3-3 proteins in nuclear localization of telomerase. EMBO J. 2000;19:2652–2661. doi: 10.1093/emboj/19.11.2652. doi:10.1093/emboj/19.11.2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H., Xu D., Li J., Berndt M.C., Liu J.P. Transforming growth factor beta suppresses human telomerase reverse transcriptase (hTERT) by Smad3 interactions with c-Myc and the hTERT gene. J. Biol. Chem. 2006;281:25588–25600. doi: 10.1074/jbc.M602381200. doi:10.1074/jbc.M602381200. [DOI] [PubMed] [Google Scholar]
- 21.Miyake Y., Nakamura M., Nabetani A., Shimamura S., Tamura M., Yonehara S., Saito M., Ishikawa F. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol. Cell. 2009;36:193–206. doi: 10.1016/j.molcel.2009.08.009. doi:10.1016/j.molcel.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 22.Valdes A.M., Andrew T., Gardner J.P., Kimura M., Oelsner E., Cherkas L.F., Aviv A., Spector T.D. Obesity, cigarette smoking, and telomere length in women. Lancet. 2005;366:662–664. doi: 10.1016/S0140-6736(05)66630-5. doi:10.1016/S0140-6736(05)66630-5. [DOI] [PubMed] [Google Scholar]
- 23.Haiman C.A., Chen G.K., Vachon C.M., Canzian F., Dunning A., Millikan R.C., Wang X., Ademuyiwa F., Ahmed S., Ambrosone C.B., et al. A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor-negative breast cancer. Nat. Genet. 2011;43:1210–1214. doi: 10.1038/ng.985. doi:10.1038/ng.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kote-Jarai Z., Olama A.A., Giles G.G., Severi G., Schleutker J., Weischer M., Campa D., Riboli E., Key T., Gronberg H., et al. Seven prostate cancer susceptibility loci identified by a multi-stage genome-wide association study. Nat. Genet. 2011;43:785–791. doi: 10.1038/ng.882. doi:10.1038/ng.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Law M.H., Montgomery G.W., Brown K.M., Martin N.G., Mann G.J., Hayward N.K., MacGregor S. Meta-analysis combining new and existing data sets confirms that the TERT-CLPTM1L locus influences melanoma risk. J. Invest. Dermatol. 2011;132:485–487. doi: 10.1038/jid.2011.322. doi:10.1038/jid.2011.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rafnar T., Sulem P., Stacey S.N., Geller F., Gudmundsson J., Sigurdsson A., Jakobsdottir M., Helgadottir H., Thorlacius S., Aben K.K., et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat. Genet. 2009;41:221–227. doi: 10.1038/ng.296. doi:10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stacey S.N., Sulem P., Masson G., Gudjonsson S.A., Thorleifsson G., Jakobsdottir M., Sigurdsson A., Gudbjartsson D.F., Sigurgeirsson B., Benediktsdottir K.R., et al. New common variants affecting susceptibility to basal cell carcinoma. Nat. Genet. 2009;41:909–914. doi: 10.1038/ng.412. doi:10.1038/ng.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turnbull C., Rapley E.A., Seal S., Pernet D., Renwick A., Hughes D., Ricketts M., Linger R., Nsengimana J., Deloukas P., et al. Variants near DMRT1, TERT and ATF7IP are associated with testicular germ cell cancer. Nat. Genet. 2010;42:604–607. doi: 10.1038/ng.607. doi:10.1038/ng.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giraud-Panis M.J., Teixeira M.T., Geli V., Gilson E. CST meets shelterin to keep telomeres in check. Mol. Cell. 2010;39:665–676. doi: 10.1016/j.molcel.2010.08.024. doi:10.1016/j.molcel.2010.08.024. [DOI] [PubMed] [Google Scholar]
- 30.Wan M., Qin J., Songyang Z., Liu D. OB fold-containing protein 1 (OBFC1), a human homolog of yeast Stn1, associates with TPP1 and is implicated in telomere length regulation. J. Biol. Chem. 2009;284:26725–26731. doi: 10.1074/jbc.M109.021105. doi:10.1074/jbc.M109.021105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palm W., de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. doi:10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 32.Li S., Makovets S., Matsuguchi T., Blethrow J.D., Shokat K.M., Blackburn E.H. Cdk1-dependent phosphorylation of Cdc13 coordinates telomere elongation during cell-cycle progression. Cell. 2009;136:50–61. doi: 10.1016/j.cell.2008.11.027. doi:10.1016/j.cell.2008.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price C.M., Boltz K.A., Chaiken M.F., Stewart J.A., Beilstein M.A., Shippen D.E. Evolution of CST function in telomere maintenance. Cell Cycle. 2010;9:3157–3165. doi: 10.4161/cc.9.16.12547. doi:10.4161/cc.9.16.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anderson B.H., Kasher P.R., Mayer J., Szynkiewicz M., Jenkinson E.M., Bhaskar S.S., Urquhart J.E., Daly S.B., Dickerson J.E., O'Sullivan J., et al. Mutations in CTC1, encoding conserved telomere maintenance component 1, cause Coats plus. Nat. Genet. 2012;44:338–342. doi: 10.1038/ng.1084. doi:10.1038/ng.1084. [DOI] [PubMed] [Google Scholar]
- 35.Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu. Rev. Biochem. 2010;79:213–231. doi: 10.1146/annurev-biochem-010909-095056. doi:10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- 36.Matthews J.M., Sunde M. Zinc fingers—folds for many occasions. IUBMB Life. 2002;54:351–355. doi: 10.1080/15216540216035. doi:10.1080/15216540216035. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q., Liu J.Q., Chen Z., Zheng K.W., Chen C.Y., Hao Y.H., Tan Z. G-quadruplex formation at the 3′ end of telomere DNA inhibits its extension by telomerase, polymerase and unwinding by helicase. Nucleic Acids Res. 2011;39:6229–6237. doi: 10.1093/nar/gkr164. doi:10.1093/nar/gkr164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zahler A.M., Williamson J.R., Cech T.R., Prescott D.M. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. doi:10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 39.Isalan M., Patel S.D., Balasubramanian S., Choo Y. Selection of zinc fingers that bind single-stranded telomeric DNA in the G-quadruplex conformation. Biochemistry. 2001;40:830–836. doi: 10.1021/bi001728v. doi:10.1021/bi001728v. [DOI] [PubMed] [Google Scholar]
- 40.Ladame S., Schouten J.A., Roldan J., Redman J.E., Neidle S., Balasubramanian S. Exploring the recognition of quadruplex DNA by an engineered Cys2-His2 zinc finger protein. Biochemistry. 2006;45:1393–1399. doi: 10.1021/bi050229x. doi:10.1021/bi050229x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu H., Wang X., Fu M., Ren J., Qu X. Chiral metallo-supramolecular complexes selectively recognize human telomeric G-quadruplex DNA. Nucleic Acids Res. 2008;36:5695–5703. doi: 10.1093/nar/gkn569. doi:10.1093/nar/gkn569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cassar L., Li H., Jiang F.X., Liu J.P. TGF-beta induces telomerase-dependent pancreatic tumor cell cycle arrest. Mol. Cell. Endocrinol. 2010;320:97–105. doi: 10.1016/j.mce.2010.02.002. doi:10.1016/j.mce.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 43.Hu B., Tack D.C., Liu T., Wu Z., Ullenbruch M.R., Phan S.H. Role of Smad3 in the regulation of rat telomerase reverse transcriptase by TGFbeta. Oncogene. 2006;25:1030–1041. doi: 10.1038/sj.onc.1209140. doi:10.1038/sj.onc.1209140. [DOI] [PubMed] [Google Scholar]
- 44.Li H., Liu J.P. Mechanisms of action of TGF-beta in cancer: evidence for Smad3 as a repressor of the hTERT gene. Ann. N. Y. Acad. Sci. 2007;1114:56–68. doi: 10.1196/annals.1396.016. doi:10.1196/annals.1396.016. [DOI] [PubMed] [Google Scholar]
- 45.Kimura M., Gazitt Y., Cao X., Zhao X., Lansdorp P.M., Aviv A. Synchrony of telomere length among hematopoietic cells. Exp. Hematol. 2010;38:854–859. doi: 10.1016/j.exphem.2010.06.010. doi:10.1016/j.exphem.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sidorov I., Kimura M., Yashin A., Aviv A. Leukocyte telomere dynamics and human hematopoietic stem cell kinetics during somatic growth. Exp. Hematol. 2009;37:514–524. doi: 10.1016/j.exphem.2008.11.009. doi:10.1016/j.exphem.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 47.Soerensen M., Thinggaard M., Nygaard M., Dato S., Tan Q., Hjelmborg J., Andersen-Ranberg K., Stevnsner T., Bohr V.A., Kimura M., et al. Genetic variation in TERT and TERC and human leukocyte telomere length and longevity: a cross-sectional and longitudinal analysis. Aging Cell. 2011 doi: 10.1111/j.1474-9726.2011.00775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nawrot T.S., Staessen J.A., Gardner J.P., Aviv A. Telomere length and possible link to X chromosome. Lancet. 2004;363:507–510. doi: 10.1016/S0140-6736(04)15535-9. doi:10.1016/S0140-6736(04)15535-9. [DOI] [PubMed] [Google Scholar]
- 49.Vasan R.S., Demissie S., Kimura M., Cupples L.A., Rifai N., White C., Wang T.J., Gardner J.P., Cao X., Benjamin E.J., et al. Association of leukocyte telomere length with circulating biomarkers of the renin-angiotensin-aldosterone system: the Framingham Heart Study. Circulation. 2008;117:1138–1144. doi: 10.1161/CIRCULATIONAHA.107.731794. doi:10.1161/CIRCULATIONAHA.107.731794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hunt S.C., Chen W., Gardner J.P., Kimura M., Srinivasan S.R., Eckfeldt J.H., Berenson G.S., Aviv A. Leukocyte telomeres are longer in African Americans than in whites: the National Heart, Lung, and Blood Institute Family Heart Study and the Bogalusa Heart Study. Aging Cell. 2008;7:451–458. doi: 10.1111/j.1474-9726.2008.00397.x. doi:10.1111/j.1474-9726.2008.00397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu H., Wang X., Gutin B., Davis C.L., Keeton D., Thomas J., Stallmann-Jorgensen I., Mooken G., Bundy V., Snieder H., et al. Leukocyte telomere length in healthy Caucasian and African-American adolescents: relationships with race, sex, adiposity, adipokines, and physical activity. J. Pediatr. 2011;158:215–220. doi: 10.1016/j.jpeds.2010.08.007. doi:10.1016/j.jpeds.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feinleib M., Kannel W.B., Garrison R.J., McNamara P.M., Castelli W.P. The Framingham Offspring Study. Design and preliminary data. Prev. Med. 1975;4:518–525. doi: 10.1016/0091-7435(75)90037-7. doi:10.1016/0091-7435(75)90037-7. [DOI] [PubMed] [Google Scholar]
- 53.Fried L.P., Borhani N.O., Enright P., Furberg C.D., Gardin J.M., Kronmal R.A., Kuller L.H., Manolio T.A., Mittelmark M.B., Newman A., et al. The Cardiovascular Health Study: design and rationale. Ann. Epidemiol. 1991;1:263–276. doi: 10.1016/1047-2797(91)90005-w. doi:10.1016/1047-2797(91)90005-W. [DOI] [PubMed] [Google Scholar]
- 54.Higgins M., Province M., Heiss G., Eckfeldt J., Ellison R.C., Folsom A.R., Rao D.C., Sprafka J.M., Williams R. NHLBI Family Heart Study: objectives and design. Am. J. Epidemiol. 1996;143:1219–1228. doi: 10.1093/oxfordjournals.aje.a008709. doi:10.1093/oxfordjournals.aje.a008709. [DOI] [PubMed] [Google Scholar]
- 55.Moayyeri A., Hammond C.J., Valdes A.M., Spector T.D. Cohort Profile: TwinsUK and Healthy Ageing Twin Study. Int. J. Epidemiol. 2012 doi: 10.1093/ije/dyr207. doi:10.1093/ije/dyr207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tell G.S., Fried L.P., Hermanson B., Manolio T.A., Newman A.B., Borhani N.O. Recruitment of adults 65 years and older as participants in the Cardiovascular Health Study. Ann. Epidemiol. 1993;3:358–366. doi: 10.1016/1047-2797(93)90062-9. doi:10.1016/1047-2797(93)90062-9. [DOI] [PubMed] [Google Scholar]
- 57.Williams R.R., Rao D.C., Ellison R.C., Arnett D.K., Heiss G., Oberman A., Eckfeldt J.H., Leppert M.F., Province M.A., Mockrin S.C., et al. NHLBI family blood pressure program: methodology and recruitment in the HyperGEN network. Hypertension genetic epidemiology network. Ann. Epidemiol. 2000;10:389–400. doi: 10.1016/s1047-2797(00)00063-6. doi:10.1016/S1047-2797(00)00063-6. [DOI] [PubMed] [Google Scholar]
- 58.Kimura M., Stone R.C., Hunt S.C., Skurnick J., Lu X., Cao X., Harley C.B., Aviv A. Measurement of telomere length by the Southern blot analysis of terminal restriction fragment lengths. Nat. Protoc. 2010;5:1596–1607. doi: 10.1038/nprot.2010.124. doi:10.1038/nprot.2010.124. [DOI] [PubMed] [Google Scholar]
- 59.Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. doi:10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aulchenko Y.S., Ripke S., Isaacs A., van Duijn C.M. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. doi:10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 61.Scheet P., Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 2006;78:629–644. doi: 10.1086/502802. doi:10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. doi:10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. doi:10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. doi:10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fisher R.A., Immer F.R., Tedin O. The genetical interpretation of statistics of the third degree in the study of quantitative inheritance. Genetics. 1932;17:107–124. doi: 10.1093/genetics/17.2.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Devlin B., Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. doi:10.1111/j.0006-341X.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 67.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. doi:10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kark J.D., Goldberger N., Kimura M., Sinnreich R., Aviv A. Energy intake and leukocyte telomere length in young adults. Am. J. Clin. Nutr. 2012;95:479–487. doi: 10.3945/ajcn.111.024521. doi:10.3945/ajcn.111.024521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thorpe R.J., Jr, Koster A., Bosma H., Harris T.B., Simonsick E.M., van Eijk J.T., Kempen G.I., Newman A.B., Satterfield S., Rubin S.M., et al. Racial differences in mortality in older adults: factors beyond socioeconomic status. Ann. Behav. Med. 2012;43:29–38. doi: 10.1007/s12160-011-9335-4. doi:10.1007/s12160-011-9335-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Benetos A., Adamopoulos C., Bureau J.M., Temmar M., Labat C., Bean K., Thomas F., Pannier B., Asmar R., Zureik M., et al. Determinants of accelerated progression of arterial stiffness in normotensive subjects and in treated hypertensive subjects over a 6-year period. Circulation. 2002;105:1202–1207. doi: 10.1161/hc1002.105135. doi:10.1161/hc1002.105135. [DOI] [PubMed] [Google Scholar]
- 71.Kearney-Schwartz A., Rossignol P., Bracard S., Felblinger J., Fay R., Boivin J.M., Lecompte T., Lacolley P., Benetos A., Zannad F. Vascular structure and function is correlated to cognitive performance and white matter hyperintensities in older hypertensive patients with subjective memory complaints. Stroke. 2009;40:1229–1236. doi: 10.1161/STROKEAHA.108.532853. doi:10.1161/STROKEAHA.108.532853. [DOI] [PubMed] [Google Scholar]
- 72.Gauderman W.J. Candidate gene association analysis for a quantitative trait, using parent-offspring trios. Genet. Epidemiol. 2003;25:327–338. doi: 10.1002/gepi.10262. doi:10.1002/gepi.10262. [DOI] [PubMed] [Google Scholar]
- 73.Raychaudhuri S., Thomson B.P., Remmers E.F., Eyre S., Hinks A., Guiducci C., Catanese J.J., Xie G., Stahl E.A., Chen R., et al. Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat. Genet. 2009;41:1313–1318. doi: 10.1038/ng.479. doi:10.1038/ng.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Johnson A.D., Handsaker R.E., Pulit S.L., Nizzari M.M., O'Donnell C.J., de Bakker P.I. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. doi:10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Abecasis G.R., Cherny S.S., Cookson W.O., Cardon L.R. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 2002;30:97–101. doi: 10.1038/ng786. doi:10.1038/ng786. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.