Abstract
Retinal ganglion cells (RGCs) transmit visual information topographically from the eye to the brain, creating a map of visual space in retino-recipient nuclei (retinotopy). This process is affected by retinal activity and by activity-independent molecular cues. Phr1, which encodes a presumed E3 ubiquitin ligase (PHR1), is required presynaptically for proper placement of RGC axons in the lateral geniculate nucleus and the superior colliculus, suggesting that increased levels of PHR1 target proteins may be instructive for retinotopic mapping of retinofugal projections. To identify potential target proteins, we conducted a proteomic analysis of optic nerve to identify differentially abundant proteins in the presence or absence of Phr1 in RGCs. 1D gel electrophoresis identified a specific band in controls that was absent in mutants. Targeted proteomic analysis of this band demonstrated the presence of PHR1. Additionally, we conducted an unbiased proteomic analysis that identified 30 proteins as being significantly different between the two genotypes. One of these, heterogeneous nuclear ribonucleoprotein M (hnRNP-M), regulates antero-posterior patterning in invertebrates and can function as a cell surface adhesion receptor in vertebrates. Thus we have demonstrated that network analysis of quantitative proteomic data is a useful approach for hypothesis generation and for identifying biologically relevant targets in genetically altered biological models.
Keywords: Phr1, Mycbp2, retinal ganglion cell, proteomics, hnRNP-M, retinotopy, ubiquitin ligase, label-free quantitative proteomics, LC-MS, network analysis
Introduction
Phr1 is a novel activity-independent regulator of retinal ganglion cell (RGC) axonal projections to their central targets in the brain (retinofugal projections; Figure S1). When Phr1 is conditionally knocked out in mouse retinal ganglion cells (Phr1-rko), RGC axons are mistargeted in the dorsal lateral geniculate nucleus (dLGN)1 and the superior colliculus (SC).2 The ipsilateral retinogeniculate projection in the dLGN is interrupted and shifted within the nucleus, reflecting disrupted retinogeniculate topographic mapping. Retinocollicular projections are similarly affected, particularly those originating in the dorsal nasal quadrant of the retina. These terminate in multiple locations in the mutant, as opposed to a single termination zone in normal animals. This phenotype is strikingly similar to that of the ephrin-A triple knock-out mouse3 in both the morphology of the mapping defect of retinofugal projections and in the fact that retinal activity is preserved and monocular segregation of inputs proceeds normally in both mutants. Despite these similarities, Phr1 mutant RGCs respond normally to ephrin-A in vivo,1 defining Phr1 as an activity and ephrinA-independent regulator of retinofugal mapping.
Phr1 is the ortholog of Protein Associated with Myc (PAM) in humans, highwire (hiw) in Drosophila, and regulator of presynaptic morphology 1 (rpm-1) in C. elegans, all of which share common domains that confer E3 dependent ubiquitin ligase function.4-7 E3 ubiquitin ligases identify proteins for proteasome degradation by covalently attaching ubiquitin to lysine residues on target molecules. Phr1 protein (PHR1) is a presumed E3 ubiquitin ligase, and may indirectly regulate retinofugal mapping through intracellular protein degradation of instructive molecular mediators. Thus, the topographic disruption that occurs in Phr1 knockout mice is likely due to increased levels of proteins which PHR1 normally targets for degradation.
Traditional protein analysis techniques have proven to be inadequate in the identification of targets of PHR1. First, available antibodies to murine PHR1 are of poor quality and are unreliable for even routine studies such as Western blotting and co-immunoprecipitation. Second, because PHR1 presumably acts as an E3 ubiquitin ligase, it has only a transient association with its target proteins, further limiting the utility of co-immunoprecipitation in identifying its potential targets, which are covalently modified and rapidly degraded. Additionally, because the phenotype of Phr1 mutants is specific to tissue-level axonal targeting, identifying targets of PHR1 in cell lines may not accurately reflect the same targets that regulate retinofugal mapping RGCs in vivo. In light of these limitations, proteomic analysis represents an ideal method to identify the targets of PHR1 in RGCs in situ, through identification of proteins that are increased in RGCs where Phr1 is knocked out in comparison to those with normal Phr1 function.
Proteomics-based approaches have their own limitations. Gel-based proteomic methods are tedious and biased to identify high-abundance proteins, which are often structural or ‘house-keeping’ proteins that are not apparently relevant to regulatory pathways and thus are unhelpful in the elucidation of the mechanisms under investigation. Quantitative proteomics using LC-MS and peptides as surrogates of relative protein concentrations has increased the efficiency and number of lower abundance proteins identified.8 However, this approach often generates extremely long lists of differentially expressed peptides and their corresponding proteins which must be organized and analyzed for target identification and hypothesis generation. Recently, network biological methods have been applied to large genomic datasets9 and increasingly used for the analysis of proteomics data.10, 11
In order to identify candidate proteins that are normally targeted by PHR1, an unbiased proteomics label-free quantitative approach was taken to identify proteins that are differentially expressed between wild-type (WT) and Phr1-rko optic nerve samples. Because Phr1 is conditionally deleted only in the retina of the Phr1-rko mouse, supporting oligodendrocytes, connective tissues, and endothelial cells are genetically identical in mutant and control optic nerve. Thus, differences in protein expression between mutant and control optic nerve samples can be attributed directly or indirectly to the loss of PHR1 from RGC axons. Discovery and directed proteomic analyses12 were used for analysis of complex mutant and control optic nerve samples. Using high-resolution label-free quantitative proteomics,13 peptide intensities from each LC-MS analysis were aligned and normalized from replicate endoprotease digestions of optic nerve samples from WT and Phr1-rko animals. Statistical differences in the relative abundances of the proteins were determined using ANOVA statistics of the protein grouped peptide intensity data.14 Thirty proteins were identified with significant differential expression between WT and Phr1-rko. Of these, twenty were increased in the mutant and therefore are potential targets of PHR1 in mouse optic nerve.
Materials and Methods
Mice
Phr1 retinal knock-out (Phr1-rko) mice have been described previously.1 Briefly, Phr1 floxed-allele mice (KOPMfl/fl)15 were mated with Math5/Cre16 mice to conditionally delete expression of Phr1 in retinal ganglion cells (RGCs). These are maintained and periodically back-crossed in the c57/Bl6 background.
Procurement of tissue
Care was taken during the dissection process to minimize contamination of the samples with human keratin. Dissections were carried out in a laminar flow hood under a microscope. All equipment and work surfaces were cleaned with ethanol and allowed to dry. Tools used were soaked in ethanol and allowed to dry under the hood. Gloves, masks, and hair bonnets were worn during dissections. Neonatal mice (postnatal day 2-13) were euthanized. The frontal cortex was exposed. For brain tissue samples, one half of the cortex was removed and placed in an Eppendorf tube and flash frozen in liquid nitrogen. For optic nerve samples, the frontal cortex was carefully elevated to reveal both the right and left optic nerves from where they exit the eye posterior to the globe and extending to the chiasm. Both nerves were carefully dissected free from the globes and removed together with the chiasm intact, placed in Eppendorf centrifuge tubes, flash frozen in liquid nitrogen, and stored at −80 °C until digestion. For our studies we prepared the following sets of tissue samples: i) 1D gel from WT whole brain lysates enriched for proteins >150 kDa to identify Phr1 protein, ii) 1D gel from optic nerve to identify Phr1 protein, iii) label-free proteomics on optic nerve samples to identify Phr1 protein, and iv) label-free quantitative proteomics on optic nerve, 4 WT vs. 4 Phr1-rko.
Protein extraction and digestion
Proteins were extracted using either SDS sample buffer for 1D PAGE (described below) or Rapigest17 (Waters Corporation, Millford, MA; 5% in 50 mM ammonium bicarbonate) for label-free quantitative LC-MS. Proteins were extracted by heating samples for two minutes at 55 °C, cooling to room temperature and vortexing until no obvious tissue pieces were visible. The samples were centrifuged at 1300 rpm for 10 minutes to remove insolubles, and the supernatants were collected.
1D SDS-PAGE and in situ endoprotease digestion
Optic nerve and brain samples were prepared to confirm the absence of PHR1 in Phr1-rko samples using 1D SDS-PAGE with mass spectrometric identification of proteins. For analytical SDS-PAGE, an aliquot of the sample was diluted with 5 μL of 4X sample buffer (Bio-Rad Laboratories, Hercules, CA) and 1μl 20X reductant (Bio-Rad). The fractionated sample was heated to 95 °C for 5 min, cooled to room temperature and spun down at 13000 rpm for 30-60 sec. 10 μl of the fraction samples containing 10-15 μg of protein were loaded on a 4-12% Criterion XT Bis-Tris gel with molecular weight markers (Bio-Rad) and run in MES buffer. Once the blue dye front had run off the gel, the gel was removed and placed in fixative solution for at least 1 h. The gel was then stained with SyproRuby for at least 2 h, destained for 30 min and scanned on the Typhoon 9400 using the following settings: 457 nm excitation, 610BP30 emission filter, and PMT voltage adjusted to stay just below saturation for the darkest band.
For identification of proteins in 1D gels, the robotically-excised18 gel cores (1.8 mm, diameter) were digested in situ using trypsin according to a modified19 previously-described method.20 The peptide pools were dissolved in aqueous 1% formic acid in 1% acetonitrile and analyzed using LC-MS as described.
Preparation of peptides from optic nerve lysates
Label-free quantitative proteomics was performed in a single block analysis with replicate optic nerve preparations from 8 animals (n=4 WT, n=4 Phr1-rko). The homogenized optic nerves were precipitated using the vendor protocol for the 2D clean-up kit (GE Healthcare, Pittsburgh, PA). The protein pellets were solubilized in 20 μL of Tris buffer (100 mM, pH 8.5) containing 8 M urea. The protein disulfide bonds were reduced with 1 mM TCEP (2 μL of a 50 mM solution) (TCEP bond breaker, 0.5 M solution, Thermo Fisher, Waltham, MA) and placed at room temperature for 30 min. Alkylation of the cysteine residues was performed using iodoacetamide (2.2 μL of a 100 mM solution). After 30 min at room temperature in the dark, the reaction was quenched with 10 mM DTT at room temperature for 15 min. The reduced and alkylated proteins (~30 μL) were digested in 8 M urea with 1 μg of endoproteinase Lys-C (2 μl of a 0.5 μg/LL stock; Roche, Basel, Switzerland) after an overnight incubation at 37 °C. The samples were diluted 1:4 with 100 mM Tris, pH 8.5, trypsin (Sigma) was added (~1:4 enzyme ratio), and the incubation was continued for 24 h at 37 °C. The digests were acidified with aqueous 5% formic acid (3.3 μL) (Fluka, St. Louis, MO). The peptides were extracted with a conditioned Nutip carbon tip (Glygen, Columbia, MD). The tips were prepared by repetitive pipetting with 25μL (x 3) of the peptide elution solvent (60% acetonitrile in 1% formic acid) and then equilibrated with 10 washes (25μL) of extraction solvent (1% formic acid). The samples were loaded with 50 pipetting cycles. The tips were then washed four times with extraction solution. The peptides were recovered by 20 pipetting cycles with 25μL of elution solution, followed by four washes (20 μL each) of elution solution. The extraction and wash solutions were combined in an autosampler vial (SunSri, Rockwood, TN) and dried in a Speed Vac (Thermo-Savant). The vial caps for the AS2 autosampler were from National Scientific (Rockwood, TN).
Mass Spectrometry
Nano-LC-FTMS was carried out using hybrid linear ion trap Fourier transform mass spectrometers (LTQ XL Orbitrap or LTQ FT Ultra, ThermoFisher, San Jose, CA). An autosampler (AS1, Eksigent, Dublin, CA) was used to load 5 μL of sample from a 10 μL loop at 1 μL/min onto a Picofrit Column (75 μm ID, ~15 cm long; New Objective, Woburn, MA) packed with reversed phase C18 beads (5 μm diameter, 200 Å pores; Michrom Bioresources, Auburn, CA). The peptides were separated using 1-3 hour gradients from 2-60% organic phase provided by a nanoflow HPLC (260 nL/min; Nano1D Plus, Eksigent). The organic phase was acetonitrile (Riedel-de Haen) and the aqueous phase was water (Riedel-de Haen) both modified with 0.1% formic acid (Sigma Aldrich, St. Louis, MO). Both control and mutant samples were run on the same column separated by two full gradient blank injections between samples to minimize carryover. Peptides eluted from the column were sprayed (PicoView PV550; New Objective) directly into the mass spectrometer. The nanospray source was operated between 1.8 and 2.3 kV and was tuned using a 25% organic isocratic flow containing angiotensin. Full MS scans (MS1) were acquired at 100,000 (LTQ-FT Ultra) or 60,000 (Orbitrap) resolution with MS1 target values of 2×106 (LTQ-FT Ultra) and 5×105 (Orbitrap). For data-dependent analysis, preview mode was enabled and the six most abundant peaks were selected for wideband collisionally activated dissociation. The LTQ MSn target value was 3×104, with an activation Q of 0.250, a reaction time of 30 ms, and a normalized collision energy of 30. Dynamic exclusion was enabled. For directed analysis, preview mode and dynamic exclusion were not enabled. Parent ion lists derived from label free analysis (described below) were processed with Xcalibur software (ThermoElectron) and ion injection times were adjusted based on peak intensities (i.e., injection times were increased for low abundance features). All data were acquired in the profile mode.
Proteomics data processing
The LC-MS data were processed as shown in Figure S2. Protein identifications from 5 optic nerve experiments were accomplished using MASCOT Distiller (Matrix Science, version 2.3.0.2 to generate files *.mgf) for database searching with MASCOT (Matrix Science, version 2.2.04) and a UniProt Mouse protein database downloaded 20110502 (72,503 entries). LC-MS file processing by Distiller was performed as previously described.13 The searches were conducted using trypsin as the specified protease, allowing 4 missed cleavages. Carbamidomethyl cysteine was specified as a fixed modification, and methionine oxidation was specified as a variable modification. All specific parameters are given in Figure S2. Database searching with four missed cleavages was performed to assess the fidelity of endoprotease digestion for quantitative proteomics analysis. Eighty three percent (36,960) of the 44,676 peptides had no missed cleavages and 15.3% (6,857) peptides had a single missed cleavage with < 2% comprising the remainder. The protein annotations were qualified in Scaffold (version 3.00.03; Proteome Software, Portland, OR, www.proteomesoftware.com) using the Protein and Peptide Prophet algorithms.21, 22 Protein identifications were accepted using protein and peptide probabilities of 95% and 50%, respectively. The distribution of Mascot ion scores is shown in Figure S3 with > 99% of the peptides having a score of > 30. The proteins, peptides, ion scores and mass spectrometric data for all LC-MS analysis are summarized in Table S1. The qualified proteins were subjected to knowledge-based network and pathway analyses using Ingenuity Pathway analysis software (IPA, Ingenuity® Systems, www.ingenuity.com) to determine literature based protein associations (Table S2).
For identification of PHR1 in gel plugs, .raw files were processed and searched using Sorcerer-SEQUEST (Sage-N Research, Sorcerer Version 3.5 release, Sorcerer Software 4.0.4 build). Database searching was performed against a custom in house database (232 entries). Fragment ion tolerance was 1.00 Da; all other parameters were identical to those detailed above. Peptides assigned to PHR1 at greater than 95% probability were manually validated; theoretical values for parent and fragment ions were calculated using the MS-Product utility within Protein Prospector (http://prospector.ucsf.edu/prospector/mshome.htm).
Label-free quantification was performed using high resolution MS1 data with an experimental design of four biological replicates from each animal type (WT vs KO), as summarized in Figure S2. The eight LC-MS analyses were performed in the same analytical block with randomized sample queuing over ~42 continuous hours of LC-MS analysis. To minimize carryover between LC-MS analyses, the nano-column was washed with two gradients after injection of solvent A. The eight unprocessed LC-MS files were imported into Rosetta Elucidator™ software and the ion chromatograms were aligned by m/z and time as features and normalized.23 After peptide annotation and qualification of peptides, as described above within Rosetta Elucidator™ software, the intensities of the aligned, annotated ion chromatograms (peak heights) were used to determine the relative protein abundances within the DAnTE-R suite of quantitative proteomic algorithms.14,24 The peptides were grouped by gene name from the Uniprot database ‘protein descriptor’ field (Table S3) and the fold-changes and p values for differential protein abundances between WT and KO groups were determined using ANOVA statistics as applied to proteomics data.25 The fold changes and p-values are summarized in Table S4.
Western Analysis
Total protein from mutant and control optic nerve was measured by Bradford assay and samples were normalized for total protein prior to loading. Proteins were separated by 4-12% Novex Bis-Tris Mini Gel (Invitrogen) and transferred to PVDF membranes. Membranes were blocked in 1X Tris-buffered saline with Tween-20 (TBST), 5% milk for 1 h at room temperature (RT). Anti-PAM (Santa Cruz; 1:250) and anti-hnRNP-M (Santa Cruz; 1:200) primary antibodies were diluted in 1X TBST, 5% milk either for 1 h at RT or overnight at 4 °C. Secondary antibody-HRP conjugates were blotted in 1X TBST, 5% milk for 1 hr at RT and the signals detected with Luminata Crescendo western HRP Substrate (Millipore). Drosophila larval brains were prepared as described previously.26 Briefly, Drosophila wild-type or hiw mutant larval brains were crushed by pestle in ice cold homogenization buffer to prevent degradation. Samples were boiled 3 minutes and centrifuged. Total protein was run on a gel and probed with anti Rump primary antibody27 (1:1000) or anti-β-tubulin (Developmental Studies Hybridoma Bank; 1:500) as a loading control. ImageJ software was used to quantify the intensity of bands on the Western blots. Paired t-test was performed to compare hnRNP-M band intensities or the ratio of the intensity of anti rump bands to that of β-tubulin bands to determine the statistical significance of the differences between genotypes.
Probe Synthesis
We purchased the hnRNP-M plasmid (Open Biosystems). The plasmid was grown overnight in original organisms on ampicillin resistant plates at 37 °C. The plasmid was then amplified using the Wizard® Plus Midipreps DNA Purification System (Promega). Gene fragments from verified plasmids were then linearized by direct amplification by sequence- or vector-specific PCR (gene, accession number, forward primer, reverse primer): HNRNPM, BC065172.1, AATTAACCCTCACTAAAGGGTGGCTG, GTAATACGACTCACTATAGGCATGCG. Digoxigenin (DIG) labeled anti-sense and sense RNA probes were made using PCR products as template and T7, T3, or SP6 RNA polymerases (Roche). cRNA probes were purified using Quick Spin columns (Roche) and quantified by spectrophotometry. Probes were used at a concentration of 1-2 μg/ml. Sense counterparts of all probes were tested to ensure probe specificity.
In situ hybridization
The protocol for in situ hybridization of the retina and optic nerve was modified from VanDunk et al.28 Briefly, retinas are harvested and fixed in 4% PFA, cryoprotected in 30% sucrose solution overnight and sectioned on a cryostat. Sections were post fixed in 4% PFA, permeabilized with proteinase K, washed in 0.1 M triethanolamine-HCl with 0.25% acetic anhydride, and blocked in hybridization buffer. They were then incubated in DIG labeled sense or anti sense cRNA for hnRNP-M overnight at 65°C. Sections were washed, blocked with normal horse serum, and incubated in alkaline phosphatase-labeled anti-DIG secondary antibody (Roche; 1:2000) overnight. Labeling was visualized using nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (Roche). Staining was stopped after visual inspection and slides coverslipped in Vectashield Mounting Medium (Vector Laboratories).
RGC explant culture
Retinal explant culture has been described previously.1 E16 retinas were plated ganglion cell-side down on culture dishes that had be previously coated in Poly-L-Ornithine and laminin. Cultures were grown at 37 °C for 3-4 days in Neurobasal with B27 (Invitrogen), enriched with BDNF (50 ng/ml, 0.1%) and CTNF (1 ng/ml, 0.2%, Peprotech), and Forsklin (0.1%, Sigma).
Immunohistochemistry/immunocytochemistry
The protocol for immunohistochemistry and double labeling in retina is described previously.1 Briefly, retinas were harvested and fixed in paraformaldehyde (PFA). 20 μm cryo sections were obtained and stained with combinations of primary antibodies to label hnRNP-M (Santa Cruz Biotech; 1:500) and brn-3b (Abcam; 1:500) (RGCs). Fluorescence-conjugated secondary antibodies (Cy3 or Alexa 488, Jackson ImmunoResearch; 1:1000) were used to visualize the distribution of primary antibodies. Confocal microscopy (Vision Core) was used to acquire images of the double-labeled sections.
Explant Immunocytochemistry
The mouse primary antibody to the external N-terminal portion of rat hnRNP-M (Upstate Cell Signaling Solutions; 1:500) was diluted in Neurobasal growth media and applied to live RGC explant cultures for 30 minutes at 37 °C/5% CO2. The cultures were carefully rinsed with fresh Neurobasal media to remove unbound antibody and then fixed in 4% PFA for 30 minutes at room temperature. Cy3 conjugated secondary antibody (Jackson ImmunoResearch; 1:1000) was then applied to fixed explant cultures to visualize the primary antibody and to determine background labeling (secondary only control).
Results
Phr1 protein is identified in WT but not Phr1-rko optic nerve
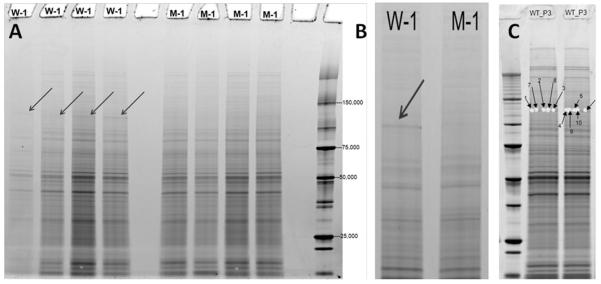
A constitutive Phr1 mutant mouse created by Cre mediated excision of Phr1 floxed exons 8 and 9 with beta-actin Cre results in an in-frame deletion in the Phr1 gene that is phenotypically indistinguishable from complete Phr1 deletion.15 Because existing antibodies to PHR1 are unreliable for detecting wild-type Phr1 protein, antibodies have limited utility in demonstrating whether Cre mediated excision results in a Phr1 protein null. Initial visual examination of a 1D gel of Phr1-mutant and control optic nerves identified a band in the control sample between 100 and 150 kDa that was notably absent in the mutant sample (Figure 1). Proteomic analysis of the excised samples from this band demonstrated the presence of PHR1 in control optic nerve samples from postnatal day 3 and 13 mice (Scaffold Protein Identification Probability = 100%). Based upon the genomic sequence, the expected size of PHR1 is ~460 kDa, thus this band likely represents a portion of Phr1 protein breakdown product in the reduced optic nerve samples. Attempts to identify PHR1 and to confirm the identity of this band as PHR1 in optic nerve samples using antibody staining were unsuccessful: no labeling was detected in wild-type optic nerve samples using commercially available anti-PAM antibodies. However, because PHR1 is highly enriched in brain tissue, we attempted to use these antibodies to identify PHR1 in brain samples. Multiple bands were identified with this antibody, including bands >250 kDa. A band of similar size to that identified as PHR1 in WT optic nerve on 1D gel (100-150 kDa) was detected by PHR1 antibody staining in reduced, but not in non-reduced, mouse brain tissue samples (data not shown). Additionally, PHR1 was detected by proteomic analysis of 1D gel samples enriched for proteins greater than 150 kDa in WT mouse whole brain lysate (Protein Prophet Identification Probability = 100%; Figure S4). The identification of Phr1 product in control, but not mutant, optic nerve is consistent with the interpretation that Cre mediated excision of the floxed Phr1 allele in the Phr1-rko mutant results in significant disruption of protein expression.
Figure 1.
Gel electrophoresis of Phr1-mutant and control optic nerves. (A) Arrows indicate a band between 100 and 150 kDa which appears to be enriched in wild-type (W-1) compared to mutant (M-1) optic nerves. (B) Higher magnification view of band of interest. (C) Samples were cut out of the identified band from wild-type optic nerve from postnatal day 3 and 13 mice. The interpreted MS2 spectra that support the identification of PHR1 in these samples are shown in Figure S4.
Identification of optic nerve proteins
We identified 887 unique proteins in our unbiased analysis of mammalian optic nerve (Table S1). Ingenuity pathway analysis of these proteins (Table S2) organized these proteins into 25 networks (Table S5; Figure S5). The top ranked canonical pathways included EIF2 signaling, regulation of eIF4 and p70S6K signaling, mTOR and the protein ubiquitination pathway (p-values and other canonical pathways are summarized in Table S6).
Identification of potential regulators of topographic mapping
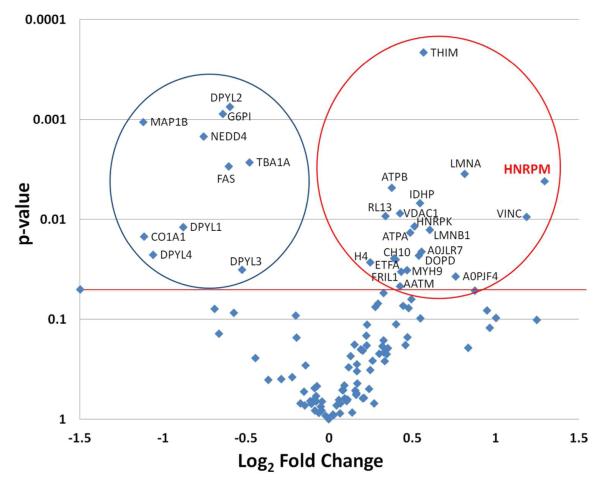
To begin to understand how Phr1 regulates topographic mapping, we sought to identify downstream effector proteins by differentially comparing protein levels in WT and Phr1-rko optic nerve. We designed a label-free quantitative proteomics experiment in which four biological replicates (i.e. four individual optic nerves) from WT and Phr1-rko each were analyzed using high-resolution LC-MS. ANOVA analysis was performed on the log2 transformed peptide intensity data from replicate optic nerve preparations for WT and Phr1-rko (Table S3). Of the proteins that were identified with two or more unique peptides sequences and not containing Met residues, we found 30 proteins with statistically different levels between WT and Phr1-rko optic nerve (p<0.05; Figure 2). Twenty of these proteins were up-regulated in the Phr1-rko, making them potential targets for the PHR1 E3 ubiquitin ligase in RGC axons. Some of these have a nuclear localization, and therefore are most likely originating from the supporting cells in the optic nerve (Table 1, bold). Because the supporting cells are genetically normal in the mutant, these differences likely reflect cell non-autonomous effects secondary to the loss of Phr1 protein in the RGC axons in the optic nerve. Because the cell bodies of RGCs are in the retina, not the optic nerve, proteins which are potential candidates for regulators of RGC topographic mapping are likely to have a cytoplasmic or plasma membrane localization (Table 1, not bold). One of these, heterogeneous nuclear ribonucleoprotein M (hnRNP-M), which showed the greatest fold change in our sample, has been shown to function as a cell surface receptor in some mammalian cells, making it a particularly promising candidate regulator of topographic mapping.29, 30
Figure 2.
Volcano plot of differentially expressed proteins in mutant vs. control mouse. ANOVA analysis was performed on the log2 transformed peptide intensity data from replicate optic nerve preparations for WT (n=4) and the Phr1-rko mutant (n=4) using the DAnTE-R software. The aligned intensities for the qualified peptides are given in Table S3. The protein level ANOVA comparisons were performed after using the peptide ‘R-Rollup’ utility in DAnTE-R. Only the proteins that contained a minimum of two unique peptide sequences were quantified using the R-Rollup option and these are listed in Table S4. Proteins with significantly different expression are shown above the thick line. The circle on the left encompasses the proteins (n=10) with significant down regulation in the Phr1-rko mutant compared to WT. Those within the circle on the right (n=20) are proteins that were significantly increased in the mutant versus control mice, including the protein hnRNP-M. The results of the ANOVA including significantly and not significantly changed proteins are given in Table S4.
Table 1.
List of 30 significantly up- or down-regulated proteins in Phr-rko compared to control.
| Protein | Protein Description | Fold change |
p-value | Location | Role |
|---|---|---|---|---|---|
| HNRPM_MOUSE | heterogeneous nuclear ribonucleoprotein M |
2.45 | <0.005 | Nucleus; Plasma membrane |
Other |
| VINC_MOUSE | Vinculin | 2.27 | 0.01 | Plasma membrane |
Enzyme |
| LMNA_MOUSE | lamin A/C | 1.76 | <0.005 | Nucleus | Other |
| A0PJF4_MOUSE | AHNAK nucleoprotein | 1.69 | 0.04 | Nucleus | Other |
| LMNB1_MOUSE | lamin B1 | 1.52 | 0.01 | Nucleus; cytoplasm |
Other |
| THIM_MOUSE | acetyl-CoA acyltransferase 2 | 1.48 | <0.005 | Cytoplasm | Enzyme |
| A0JLR7_MOUSE | AHNAK nucleoprotein | 1.47 | 0.02 | Nucleus | Other |
| IDHP_MOUSE | isocitrate dehydrogenase 2 (NADP+), mitochondrial |
1.46 | 0.01 | Cytoplasm | Enzyme |
| DOPD_MOUSE | D-dopachrome tautomerase | 1.45 | 0.02 | Cytoplasm | Enzyme |
| HNRPK_MOUSE | heterogeneous nuclear ribonucleoprotein K |
1.43 | 0.01 | Nucleus | Other |
| ATPA_MOUSE | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle |
1.40 | 0.01 | Cytoplasm | Transporter |
| MYH9_MOUSE | myosin, heavy chain 9, non- muscle |
1.38 | 0.03 | Cytoplasm | Enzyme |
| FRIL1_MOUSE | ferritin, light polypeptide | 1.35 | 0.03 | ? | ? |
| VDAC1_MOUSE | voltage-dependent anion channel 1 |
1.34 | 0.01 | Cytoplasm | Ion channel |
| AATM_MOUSE | glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2) |
1.34 | 0.05 | Cytoplasm | Enzyme |
| CH10_MOUSE | heat shock 10 kDa protein 1 (chaperonin 10) |
1.32 | 0.03 | Cytoplasm | Enzyme |
| ETFA_MOUSE | electron-transfer-flavoprotein, alpha polypeptide |
1.31 | 0.02 | Cytoplasm | Transporter |
| ATPB_MOUSE | ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
1.30 | <0.005 | Cytoplasm | Transporter |
| RL13_MOUSE | ribosomal protein L13 pseudogene 12 |
1.26 | 0.01 | ? | ? |
| H4_MOUSE | histone H4 | 1.19 | 0.03 | unknown | group |
| MAP1B_MOUSE | microtubule-associated protein 1B |
−2.18 | <0.005 | ? | ? |
| CO1A1_MOUSE | collagen, type I, alpha 1 | −2.17 | 0.01 | Extracellular space |
Other |
| DPYL4_MOUSE | dihydropyrimidinase-like 4 | −2.09 | 0.02 | Cytoplasm | Enzyme |
| DPYL1_MOUSE | collapsin response mediator protein 1 |
−1.84 | 0.01 | Cytoplas | Enzyme |
| NEDD4_MOUSE | neural precursor cell expressed, developmentally down regulated 4 |
−1.69 | <0.005 | Cytoplasm | Enzyme |
| G6PI_MOUSE | glucose-6-phosphate isomerase |
−1.56 | <0.005 | Extracellular space |
Enzyme |
| FAS_MOUSE | fatty acid synthase | −1.52 | <0.005 | Cytoplasm | Enzyme |
| DPYL2_MOUSE | dihydropyrimidinase-like-2 | −1.52 | <0.005 | Cytoplasm | Enzyme |
| DPYL3_MOUSE | dihydropyrimidinase-like 3 | −1.44 | 0.03 | Cytoplasm | Enzyme |
| TBA1A_MOUSE | tubulin, alpha 1a | −1.40 | <0.005 | Cytoplasm | Other |
Shaded rows indicate proteins with only nuclear localization that likely reflect cell non-autonomous effects on the supporting cells contacting Phr1-rko mutant axons within the optic nerve.
Western analysis of hnRNP-M
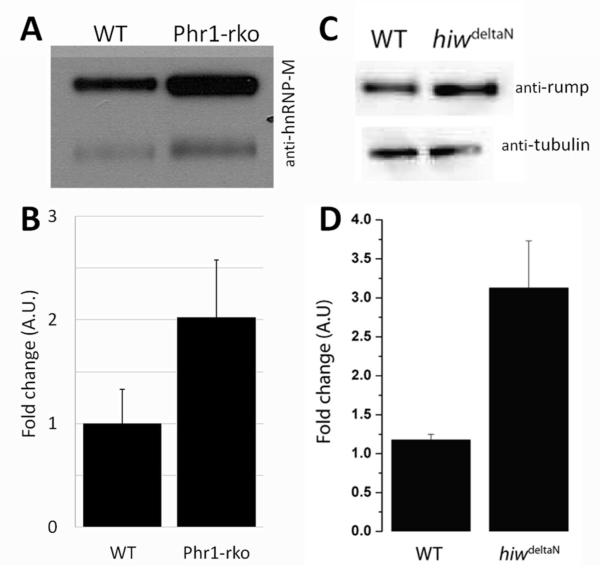
To confirm that hnRNP-M is enriched in Phr1-rko optic nerve compared to WT optic nerve, Western analysis was performed on separate optic nerve samples. Samples were normalized for total protein as measured by the Bradford assay prior to loading. Two bands were consistently labeled by the anti-hnRNP-M antibody at ~68 and ~72 kDa in both the WT and Phr1-rko samples (Figure 3A). We found that hnRNP-M was increased on average 2-fold in the Phr1-rko relative to the control in both bands (Figure 3B; p=0.04). This finding confirms the results from proteomic analysis that hnRNP-M is increased in Phr1-rko optic nerve, suggesting that hnRNP-M may be regulated by PHR1 in RGC axons in vivo.
Figure 3. hnRNP-M is upregulated in the absence of Phr1.
(A) Western analysis of WT and Phr1-rko optic nerve samples normalized for total protein by Bradford assay and probed with anti hnRNP-M antibody demonstrates 2 bands with increased levels in Phr1-rko optic nerve relative to WT. (B) Quantification of this change demonstrates a 2-fold enrichment of hnRNP-M in Phr1-rko optic nerve (n=5). (C) A similar increase in the hnRNP-M ortholog Rump was demonstrated in Drosophila hiw mutant brain suggesting that this relationship is evolutionarily conserved. Tubulin is shown as a loading control. (D) Quantitative assessment of this change in Drosophila shows roughly a 3-fold enrichment in the absence of hiw (n=3).
The Phr1/hnRNP-M effector pathway is evolutionarily conserved in Drosophila and in mice
There is a Drosophila ortholog of hnRNP-M, termed rumpelstiltskin (Rump).27 We sought to determine whether the Rump levels may be similarly regulated by hiw, the Drosophila ortholog of Phr1. We compared Rump levels in fly larval brains of WT and hiw null mutants by Western analysis and found that there was a significant ~3-fold increase in Rump levels in hiw mutant flies (Figure 3C, D). This finding suggests that the PHR1/hnRNP-M effector pathway is evolutionarily conserved in flies and in mice.
hnRNP-M is expressed in RGCs in vivo, localizes to RGC axons in vitro
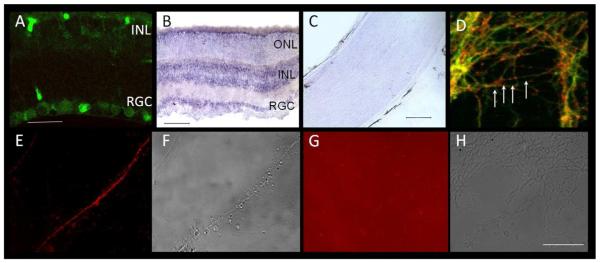
We sought to determine whether hnRNP-M is expressed in retinal ganglion cells. Anti-hnRNP-M labeling of retina slices demonstrated the presence of hnRNP-M in retinal ganglion cell bodies (Figure 4A). To determine whether the source of hnRNP-M in optic nerve is RGC axons and/or supporting optic nerve cells, we conducted in situ hybridization of retina and optic nerve. We found high levels of hnRNP-M mRNA in RGCs and the inner nuclear layer of the retina, but not in the optic nerve in WT or mutant animals (Figure 4B, C). Next, we sought to assess whether hnRNP-M protein localized to RGC axons in addition to RGC cell bodies. In order to visualize individual RGC axons, we cultured retinal explants in vitro. We found punctate hnRNP-M labeling along RGC axons in culture (Figure 4D). In rat Kupffer cells, hnRNP-M localizes to the cell surface and functions as a receptor for a cancer cell adhesion protein, carcinoembryonic antigen.30, 31 To determine whether hnRNP-M is present on the cell surface in mouse RGC axons, we labeled live cultured RGCs with an antibody against the N-terminus of rat hnRNP-M that recognizes the external portion of the protein in Kupffer cells (CEA L).31 We found that this antibody bound to live axons, confirming localization of hnRNP-M to the cell surface in mouse RGC axons in vitro (Figure 4E-H).
Figure 4. hnRNP-M is expressed in RGCs.
(A) RGCs and cells in the INL label with anti-hnRNP-M antibody. (B) In situ hybridization demonstrates strong labeling of hnRNP-M mRNA in RGCs and the INL (dark purple) in the retina, consistent with antibody labeling. (C) In situ hybridization in optic nerve does not detect hnRNP-M mRNA in cell bodies endogenous to the nerve. (D) In RGC explant cultures in vitro, hnRNP-M (red) localizes to axons of RGCs (green) in a punctate pattern (arrows). (E) Labeling unfixed, live cultures with an anti hnRNP-M antibody to an epitope that corresponds to the ligand binding region of rat hnRNP-M demonstrates that hnRNP-M is expressed in RGC axons and localizes to the cell surface (red). (F) Non-fluorescent image of (E). (G, H) Secondary-only control confirms that the fluorescence labeling in E is due to anti hnRNP-M binding the cell surface. INL=inner nuclear layer, RGC=retinal ganglion cell layer, ONL=outer nuclear layer. Scale bars as follows: A=20 μm; B=100 μm; C, D-H=50 μm.
Discussion
Phr1 is a presumed E3 ubiquitin ligase
Phr1 and its orthologs (highwire [hiw] in Drosophila, rpm-1 in C. elegans, Mycbp2/PAM in humans, and esrom in zebrafish) encode large, multi-domain proteins that have been linked to several crucial processes in neural development.1, 2, 5-7, 15, 32,35 These proteins share common functional domains including B-box and RING zinc finger clusters that confer E3 dependent ubiquitin ligase function.5, 7, 36, 37 The Drosophila and C. elegans orthologs function as E3 ubiquitin ligases with defined molecular targets. Indeed, ubiquitin ligase activity has also been described for the mammalian Phr1 orthologs in rat spinal cord38 and zebrafish visual system (esrom).39
The Phr1 effector pathway in mammals is not known
In Drosophila hiw mutants, the molecular cascade associated with the one phenotype, synaptic overgrowth at the neuromuscular junction (NMJ), has been clearly defined mechanistically.40 Highwire functions as a ubiquitin ligase that modulates levels of Wallenda, a mitogen-activated protein kinase kinase kinase (MAP3K). In the absence of Highwire, Wallenda is not sufficiently downregulated via proteasome degradation, causing Wallenda levels to rise in the cell. This results in excess phosphorylation of Jun NH2-terminal kinase (JNK) that in turn activates D-fos mediated synaptic overgrowth.40 In C. elegans, RPM-1 targets dual leucine kinase (DLK), the ortholog of wallenda (wnd).37 However, the mechanism of action of PHR1 in vertebrate species is less clear. Wallenda is orthologous to DLK in the mouse, but unlike in the fly, DLK is not upregulated in Phr1 mutants, and loss of DLK in the mouse fails to rescue Phr1 dependent pathfinding defects.15 Inhibitors of p38MAPK activity have been shown to reverse morphological defects of Phr1 mutant spinal motor axons in vitro, suggesting p38MAPK as a potential target of PHR1.34 In contrast, in esrom zebrafish mutants, abnormal pathfinding and microtubule looping of retinal ganglion cell axons in vivo is unaffected by p38MAPK inhibitors. Phospho-JNK was increased in the esrom mutant, but pharmacologic blockers of JNK upregulation failed to rescue the morphological defects, indicating that Esrom acts independently of both p38MAPK and JNK.35 These conflicting results may reflect species differences, cell type-specific differences, or a complex system that has multiple components that are differentially regulated in vivo and in vitro. What is clear is that the hiw-wnd pathway, while necessary and sufficient for the NMJ overgrowth phenotype, can not account for all hiw/Phr1 phenotypes.
Loss of Phr1 disrupts retinofugal mapping
Phr1 is required presynaptically for the proper localization of retinogeniculate1 and retinocollicular2 projections. The loss of Phr1 from RGCs does not affect axon path-finding to correct target nuclei, spontaneous retinal activity, or monocular segregation of retinofugal projections. It is, however, required for retinotopic mapping of retinofugal projections within their central targets. Other molecules known to affect retinotopic mapping independently of activity include the Eph/ephrin family of receptors41 and Bone Morphogenic Proteins (BMPs)42. Because PHR1 is a presumed E3 ubiquitin ligase, this suggests intracellular protein degradation as a novel mechanism for the regulation of retinofugal mapping. Yet how Phr1 regulates retinotopic mapping is not known.
Proteomic analysis is a valuable hypothesis-generating method when traditional methods are not reliable
Identification of PHR1 and its targets by traditional protein analysis techniques has proven difficult. There is no commercially available source of reliable and consistent antibodies to PHR1, and attempts to generate our own antibody were unsuccessful. For this reason, co-immunoprecipiation of Phr1 orthologs in Drosophila and C. elegans required epitope-tagging of the ~5000 amino acid protein to isolate it and its associated proteins.43,44 Additionally, the targets of ubiquitin ligases do not tend to bind tightly to these enzymes, but instead are covalently modified and rapidly degraded. As a consequence, these studies in flies and worms yielded other members of the ubiquitination complex such as the fbox protein Fsn and RAE1, rather than potential targets of Phr1 orthologs.43,45,46 An alternate strategy, examination of differentially ubiquitinated proteins in the presence or absence of E3 ligases, is also of limited utility in our system. Proteasome inhibitors can reduce the degradation of ubiquitinated proteins to aid in their identification in cultured cells, but because of their toxicity and systemic effects, proteasome inhibitors are impractical for use in a conditional knock-out in vivo. Thus, to identify differentially abundant proteins in RGC axons lacking Phr1 we used an alternate approach: we conducted an unbiased proteomic analysis to identify alterations in protein levels in Phr1 mutant optic nerve as compared to controls.
Potential Phr1 targets can be identified by proteomic analysis
We reasoned that the topographic disruption that occurs in the Phr1 mutant mouse could occur as a consequence of increased levels of instructive proteins that would have undergone ubiquitination and proteolysis in the presence of the Phr1 ligase. Using an unbiased proteomics approach we compared expression patterns of proteins in normal and Phr1-rko optic nerve samples to identify potential protein candidates that were differentially expressed in the presence or absence of Phr1. To increase the yield of potential targets that are biologically relevant to retinofugal mapping specifically, we utilized the Phr1 conditional mutant mouse. Because Cre mediated excision of Phr1 occurs only in the retina in these animals, only the retinal ganglion cell axons in the optic nerve lack Phr1 function while all other supporting cells are genetically normal. Thus differences in protein expression between Phr1-rko and control optic nerves should be a consequence of either cell autonomous deletion of Phr1 in RGC axons or a cell non autonomous effect of Phr1 loss in RGCs upon genetically normal cells in the optic nerve such as supporting oligodendrocytes, connective tissues, or endothelial cells. Because the mapping phenotype becomes apparent during the first few weeks of postnatal life in mice, we used optic nerve samples from neonatal mice to increase the chance of identifying developmentally relevant PHR1 targets.
Proteomic analysis reveals Phr1 protein products in wild-type but not Phr1-rko optic nerve
We first sought to characterize PHR1 protein expression in the Phr1 conditional mutant. However, currently available antibodies to Phr1 orthologs are unreliable in mouse tissue samples. Thus, traditional Western blot analysis is insufficient for this task.15 The conditional allele of Phr1 is rendered non-functional by Cre mediated excision of floxed exons 8 and 9. These mice have a phenotype that is indistinguishable from Phr1 null mice with a piebald deletion on chromosome 14 that includes the Phr1 gene.15 The location of the flox sites, however, results in an in-frame deletion that could potentially result in non-functional protein production. 1D gel analysis demonstrated a band with a molecular weight between 100-150 kDa in WT optic nerve that was lacking from the Phr1-rko optic nerve. Our proteomic analysis identified PHR1 in samples excised from these bands in wild-type animals, demonstrating that Phr1 protein localizes to retinal ganglion cell axons in WT animals. Loss of the PHR1 band in the Phr1-rko suggests substantial disruption of the protein following Cre mediated excision of exons 8 and 9. The demonstration of the presence of Phr1 protein in control optic nerve is an important step in characterizing the function of Phr1 in topographic mapping of retinal axons to their central targets in the brain. This localization may indicate that Phr1 plays a role as an E3 ubiquitin ligase targeting proteins for degradation within retinal axons themselves during the process of axonal targeting. This finding also demonstrates that proteomic analysis of targeted samples from 1D (or 2D) gel analysis could be of further utility in characterizing the Phr1 mutant and determining additional localizations and targets of PHR1.
Proteomic and bioinformatic analysis of Phr1-rko optic nerve identified 30 differentially expressed proteins
We identified 887 unique proteins in our unbiased analysis of mammalian optic nerve (Table S1). Out of this group, 30 were found to be significantly differentially expressed between WT and Phr1-rko optic nerves (Figure 2; Table 1). Using ANOVA statistics to compare replicate analyses between the sample types, we concluded that these changes in protein abundances reflect significant biological differences between the WT and Phr1-rko optic nerve. It is important to note that both samples were almost identical in genetic makeup with the exception of the loss of PHR1 from the RGC axons traversing the optic nerve. In general, we expected molecules that were direct targets of the E3 ligase PHR1 to be increased in response to its loss from RGCs (20 proteins), while decreases (10 proteins) may reflect indirect regulation via intermediates. Additionally, changes in protein concentration in optic nerve between mutant and control animals may reflect either cell autonomous or cell non autonomous changes. The 30 proteins identified as being present in significantly different abundance between the mutant and control were then characterized as nuclear, cytoplasmic, or extracellular based on prior reports in the literature. Because the nuclei of RGCs are situated in the retina, not the optic nerve, we believe changes in the concentration of nuclear proteins are likely to reflect cell non-autonomous changes in neighboring cells due to abnormal RGC axons in their proximity. Changes in cytoplasmic or membrane protein concentrations, however, are more likely to reflect cell autonomous changes due to the absence of Phr-1 activity in RGC axons. Thus we stratified our list of potential PHR1 target candidates by those most likely to be cell-autonomous to retinal ganglion cells and focused our attention on these candidates for further investigation.
Increased expression of heterogeneous nuclear ribonucleoprotein M (hnRNP-M) in the absence of Phr-1 is conserved across species
The proteomics analysis revealed a 2.45-fold increase in the expression of hnRNP-M in the Phr1-rko mouse as compared to controls. Western blot of optic nerve samples of neonatal and juvenile mice confirmed an increase in hnRNP-M protein in Phr1-rkos as compared to wild-type controls. The Drosophila ortholog of hnRNP-M, rumpelstiltskin (rump), regulates antero-posterior patterning in fly larvae by sequestering instructive mRNAs at one pole of the developing embryo.27 The role of rump in antero-posterior patterning is a provocative association given the disruption of anterior-posterior mapping of RGCs in the superior colliculus of Phr1 mutant mice,2 making hnRNP-M an attractive candidate for mediating Phr1 dependent mapping. To determine if hnRNP-M regulation by Phr1 was generalizable across species, we studied the relationship between rump and hiw, the Drosophila ortholog of Phr1, in Drosophila brain. We found that, as in mice, loss of the E3 ubiquitin ligase hiw results in increased levels of the hnRNP-M ortholog Rump. Thus the regulation of Rump/hnRNP-M is a fundamental feature of Phr1/hiw and is likely important for its functional regulation of circuit formation across species. This finding supports the model that hnRNP-M is a biologically relevant target of Phr1.
HnRNP-M is expressed on RGC axons, suggesting a putative role as a cell surface receptor
The protein hnRNP-M is classically characterized as a nuclear protein with critical functions in posttranslational mRNA splicing. HnRNP-M has also been shown in mammalian systems to be at the cell-surface and in the cytoplasm.29-31 In rat Kupffer cells, hnRNP-M has been identified as the carcinoembryonic antigen receptor and is important for cell adhesion and anchoring.30, 31 Furthermore, in human tissues, hnRNP-M has been cloned as a thyroglobin receptor.29 Similarly, we have shown by antibody staining that hnRNP-M is expressed in RGCs in vivo and localizes to the cell surface of RGC axons in vitro. Taken together, these findings raise the possibility that hnRNP-M could regulate axonal targeting of retinal axons via cell surface receptor signaling in a fashion similar to that of the Eph/ephrin family of receptors.
Proteomic analysis revealed other provocative candidate molecular mediators of Phr-1 dependent retinofugal mapping
The microtubule associated protein MAP1B was found to be significantly reduced in Phr1-rko optic nerve. This finding is also intriguing because Phr1 mutant phenotypes have similarity to that of MAP1B-/- knockout mice. Cultured motor and sensory nerve axons that lack Phr1 function have an abnormal “kinked” axonal morphology as a consequence of disruption of the normal microtubule cytoskeleton during axonal outgrowth.34 Similarly, the axons of cultured sensory neurons from MAP1B-/- knock-out mice are described as “frizzy/wavy.”47 Furthermore, both Phr1−/−15 and MAP1B−/−48 mice share similar neuro-developmental phenotypes including agenesis of the corpus callosum and disruption of major fiber tracts in the brain that may reflect a causal role for MAP1B in some Phr1 phenotypes.
MAP1B is normally expressed at high levels during neuronal development but declines substantially with maturation in the central nervous system.49 Our optic nerve samples were from very young postnatal animals (P3). Thus the reduction of MAP1B in Phr1-rko optic nerve in this study might be the result of either a failure to maintain developmentally appropriate levels of MAP1B, or possibly that the loss of Phr1 leads to precocious maturation with a corresponding reduction of MAP1B. How Phr1 regulates MAP1B is not clear. Because MAP1B levels decline, rather than increase, in the absence of the presumed E3 ubiquitin ligase Phr1 it may be regulated indirectly via yet undefined mediators. For example, over-expression of gigaxonin, which is mutated in the human neurodegenerative disorder Giant Axonal Neuropathy and functions as substrate-specific adaptor for the ubiquitin activating enzyme E1, results in a reduction of MAP1B levels that can be rescued by proteasome inhibitors.50
Another protein that was increased in mutant optic nerve relative to controls, Lamin B1, was initially presumed to be the consequence of cell non-autonomous effects of mutant axons on genetically normal supporting cells due to its known localization to the nuclear membrane. Recently, however, Lamin B has been shown to be axonally synthesized in Xenopus RGC axons in vitro, and loss of Lamin B results in axonal degeneration.51 How increased expression of Lamin B1 may be related to axonal mistargeting is not clear, but the finding of extranuclear Lamin B expression in RGC axons in another vertebrate system warrants consideration of this protein as a potential candidate effector molecule in Phr1-mediated mapping.
Determining which of these candidate molecules plays a role in Phr1 mediated retinofugal mapping will be challenging. The great strength of our proteomic approach, conducting comparative proteomic analysis using an in vivo conditional mutant approach, is also the greatest challenge to identifying the causative factors for our phenotype: no in vitro system exists to screen candidate molecules. Because the Phr1-rko phenotype is a tissue-level axonal targeting defect, candidates must be manipulated in RGCs in vivo early in development and their mapping assessed after maturation. This could be done by generating transgenic mice with conditionally expressed constructs that are Cre dependent, thereby limiting their tissue expression (i.e., RGCs using Math5-Cre). While this approach most closely mimics the expression we observe in the Phr1-rko for comparison, it is both time consuming and expensive as a screening strategy. Electroporation of our candidate constructs into RGCs is an alternate approach, but because transfection of RGCs is only possible while they are dividing, electroporation must be done in utero before RGC precursors exit the cell cycle and differentiate beginning at E13.52 Petros and colleagues have shown that in utero electroporation of EphB1 in the developing eye disrupts normal chiasmal crossing of ipsilateral RGC axons and their subsequent mapping in the dLGN,53 suggesting that this may be an effective strategy to explore candidate molecules that mediate RGC mapping in vivo.
Conclusions
Using an unbiased proteomic approach we have generated the most exhaustive proteomic analysis of mouse optic nerve to our knowledge. The proteins identified in this extensive study are not only relevant to the current study of the development of the normal retinofugal projection, but also possibly informative for studies of abnormal retinofugal development,54 other optic nerve disorders such as glaucoma and multiple sclerosis, or even optic nerve regeneration. Our approach demonstrates that proteomic analysis with bioinformatic analysis is a highly specific approach for identifying candidate targets in effector pathways. We found 30 proteins, including the provocative heterogeneous nuclear ribonucleoprotein M, to be differentially expressed in Phr1-mutant optic nerve, thus identifying them as potential mediators of disrupted topographic mapping in the Phr1-rko mutant mouse.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank A. Joseph Bloom and Cassandra VanDunk for technical advice and support, and Alan Davis for expert proteomics technical assistance and LC-MS analysis at the Washington University Proteomics Core Laboratory (PCL). The Proteomics Core Laboratory (PCL) is supported with grants from the National Center for Research Resources (NCRR 5P41RR000954-35), the National Institute of General Medical Science (8 P41 GM103422-35), and the National Center for Research Resources (UL1 RR024992), a component of the National Institutes of Health (NIH) and the NIH Roadmap for Medical Research. The PCL is also supported by the NIH Neuroscience Blueprint Core Grant P30 NS057105 to Washington University, and the Silvio O. Conte Digestive Diseases Core Center at Washington University School of Medicine and Barnes-Jewish Hospital in St. Louis, Missouri, which is supported in part by Center Support Grant P30 DK52574. Funding was provided by the National Institutes of Health/ National Institute of Diabetes and Digestive and Kidney Diseases (NIH/NIDDK T35 DK074375) to ARL, and the National Heart, Lung and Blood Institute (NIH/NHLBI; T35 HL007815) to JC. This work was also supported by awards to the Department of Ophthalmology and Visual Sciences at Washington University from a Research to Prevent Blindness, Inc. Unrestricted grant, the NIH Vision Core Grant P30 EY0268, the National Eye Institute (NEI; K12 EY016336) and the Horncrest Foundation to SC and NEI (K12 EY016336-ARRA) to SC and HR, and NIDA (DA020812) to AD. The DAnTE-R software was written by Tom Taverner (t.taverner@gmail.com) and Ashoka Polpitiya for the U.S. Department of Energy (PNNL, Richland, WA, USA, http://omics.pnl.gov/software) and the authors are grateful for their assistance in the deployment of the software at Washington University School of Medicine.
Footnotes
SUPPORTING INFORMATION Figure S1. Presynaptic deletion of the E3 ubiquitin ligase Phr1 results in mistargeting of retinofugal projections. Figure S2. Data processing for quantitative, label-free proteomics analysis of mouse optic nerves. Figure S3. Distribution of Mascot Ion Scores for all 44,676 peptides identified. Figure S4. Tandem mass spectra from differential gel band (MW ~130 kDa, Figure 1) of peptides consistent with the indicated sequences from mouse Phr1. Figure S5. Ingenuity Interaction Networks. Table S1. Mass spectrometry and database search results. Table S2. Proteins and annotation key for networks and pathways. Table S3. Aligned, amplitude normalized, annotated gene grouped peptide intensities from LC-MS analysis of WT and Phr1 mutant. Table S4. Relative quantification and statistical significance of protein abundances between WT and Phr1 mutant. Table S5. Ingenuity Interaction Networks. Table S6. Ingenuity Canonical Pathways. This material is available free of charge via the Internet at http://pubs.acs.org. The authors declare no competing financial interest.
REFERENCES
- 1.Culican SM, Bloom AJ, Weiner JA, DiAntonio A. Phr1 regulates retinogeniculate targeting independent of activity and ephrin-A signalling. Molecular and Cellular Neuroscience. 2009;41(3):304–312. doi: 10.1016/j.mcn.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vo BQ, Bloom AJ, Culican SM. Phr1 is required for proper retinocollicular targeting of nasal-dorsal retinal ganglion cells. Vis Neurosci. 2011;28:175–81. doi: 10.1017/S0952523810000386. [DOI] [PubMed] [Google Scholar]
- 3.Pfeiffenberger C, Cutforth T, Woods G, Yamada J, Renteria RC, Copenhagen DR, Flanagan JG, Feldheim DA. Ephrin-As and neural activity are required for eye-specific patterning during retinogeniculate mapping. Nat Neurosci. 2005;8(8):1022–7. doi: 10.1038/nn1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo Q, Xie J, Dang CV, Liu ET, Bishop JM. Identification of a large Myc-binding protein that contains RCC1-like repeats. Proc Natl Acad Sci U S A. 1998;95(16):9172–7. doi: 10.1073/pnas.95.16.9172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wan HI, DiAntonio A, Fetter RD, Bergstrom K, Strauss R, Goodman CS. Highwire regulates synaptic growth in Drosophila. Neuron. 2000;26(2):313–29. doi: 10.1016/s0896-6273(00)81166-6. [DOI] [PubMed] [Google Scholar]
- 6.Zhen M, Huang X, Bamber B, Jin Y. Regulation of presynaptic terminal organization by C. elegans RPM-1, a putative guanine nucleotide exchanger with a RING-H2 finger domain. Neuron. 2000;26(2):331–43. doi: 10.1016/s0896-6273(00)81167-8. [DOI] [PubMed] [Google Scholar]
- 7.Burgess RW, Peterson KA, Johnson MJ, Roix JJ, Welsh IC, O’Brien TP. Evidence for a conserved function in synapse formation reveals Phr1 as a candidate gene for respiratory failure in newborn mice. Mol Cell Biol. 2004;24(3):1096–105. doi: 10.1128/MCB.24.3.1096-1105.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elliott MH, Smith DS, Parker CE, Borchers C. Current trends in quantitative proteomics. Journal of Mass Spectrometry. 2009;44(12):1637–1660. doi: 10.1002/jms.1692. [DOI] [PubMed] [Google Scholar]
- 9.Dehmer M, Emmert-Streib F, Graber A, Salvador A. Applied Statistics for network biology. In: Dehmer M, Emmert-Streib F, editors. Quantitative and Network Biology. Wiley and Blackwell; 2011. [Google Scholar]
- 10.Zubarev RA, Nielsen ML, Fung EM, Savitski MM, Kel-Margoulis O, Wingender E, Kel A. Identification of dominant signaling pathways from proteomics expression data. Journal of Proteomics. 2008;71(1):89–96. doi: 10.1016/j.jprot.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 11.Lee S, Kim P-J, Jeong H. Global organization of protein complexome in the yeast Saccharomyces cerevisiae. BMC Systems Biology. 2011;5(1):126. doi: 10.1186/1752-0509-5-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Domon B, Aebersold R. Options and considerations when selecting a quantitative proteomics strategy. Nat Biotech. 2010;28(7):710–721. doi: 10.1038/nbt.1661. [DOI] [PubMed] [Google Scholar]
- 13.Nittis T, Guittat L, LeDuc RD, Dao B, Duxin JP, Rohrs H, Townsend RR, Stewart SA. Revealing Novel Telomere Proteins Using in Vivo Cross-linking, Tandem Affinity Purification, and Label-free Quantitative LC-FTICR-MS. Molecular & Cellular Proteomics. 2010;9(6):1144–1156. doi: 10.1074/mcp.M900490-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karpievitch Y, Stanley J, Taverner T, Huang J, Adkins JN, Ansong C, Heffron F, Metz TO, Qian W-J, Yoon H, Smith RD, Dabney AR. A statistical framework for protein quantitation in bottom-up MS-based proteomics. Bioinformatics. 2009;25(16):2028–2034. doi: 10.1093/bioinformatics/btp362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bloom AJ, Miller BR, Sanes JR, DiAntonio A. The requirement for Phr1 in CNS axon tract formation reveals the corticostriatal boundary as a choice point for cortical axons. Genes Dev. 2007;21(20):2593–606. doi: 10.1101/gad.1592107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Z, Ding K, Pan L, Deng M, Gan L. Math5 determines the competence state of retinal ganglion cell progenitors. Dev Biol. 2003;264(1):240–54. doi: 10.1016/j.ydbio.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 17.Zybailov B, Coleman MK, Florens L, Washburn MP. Correlation of Relative Abundance Ratios Derived from Peptide Ion Chromatograms and Spectrum Counting for Quantitative Proteomic Analysis Using Stable Isotope Labeling. Analytical Chemistry. 2005;77(19):6218–6224. doi: 10.1021/ac050846r. [DOI] [PubMed] [Google Scholar]
- 18.Bredemeyer A, Lewis R, Malone JP, Davis A, Gross J, Townsend RR, Ley T. A proteomic approach for the discovery of protease substrates. Proc Natl Acad Sci U S A. 2004;101(32):11785–90. doi: 10.1073/pnas.0402353101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rader J, Malone JP, Gross J, Gilmore P, Brooks R, Nguyen L, Crimmins D, Feng S, Wright JT,N, Zighelboim I, Funk M, Huettner P, Ladenson J, Gius D, Townsend RR. A unified sample preparation protocol for proteomic and genomic profiling of cervical swabs to identify biomarkers for cervical cancer screening. Proteomics Clin Appl. 2008;2(12):1658–69. doi: 10.1002/prca.200780146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Havlis J, Thomas H, Sebela M, Shevchenko A. Fast-response proteomics by accelerated in-gel digestion of proteins. Anal Chem. 2003;75(6):1300–6. doi: 10.1021/ac026136s. [DOI] [PubMed] [Google Scholar]
- 21.Keller A, Nesvizhskii A, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Analytical Chemistry. 2002;74(20):5383–92. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 22.Nesvizhskii A, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Analytical Chemistry. 2003;75(17):4646–58. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 23.Neubert H, Bonnert TP, Rumpel K, Hunt BT, Henle ES, James IT. Label-Free Detection of Differential Protein Expression by LC/MALDI Mass Spectrometry. Journal of Proteome Research. 2008;7(6):2270–2279. doi: 10.1021/pr700705u. [DOI] [PubMed] [Google Scholar]
- 24.Polpitiya AD, Qian W-J, Jaitly N, Petyuk VA, Adkins JN, Camp DG, Anderson GA, Smith RD. DAnTE: a statistical tool for quantitative analysis of -omics data. Bioinformatics. 2008;24(13):1556–1558. doi: 10.1093/bioinformatics/btn217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oberg AL, Vitek O. Statistical Design of Quantitative Mass Spectrometry-Based Proteomic Experiments. Journal of Proteome Research. 2009;8(5):2144–2156. doi: 10.1021/pr8010099. [DOI] [PubMed] [Google Scholar]
- 26.Wairkar YP, Fradkin LG, Noordermeer JN, DiAntonio A. Synaptic Defects in a Drosophila Model of Congenital Muscular Dystrophy. The Journal of Neuroscience. 2008;28(14):3781–3789. doi: 10.1523/JNEUROSCI.0478-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jain RA, Gavis ER. The Drosophila hnRNP-M homolog Rumpelstiltskin regulates nanos mRNA localization. Development. 2008;135(5):973–982. doi: 10.1242/dev.015438. [DOI] [PubMed] [Google Scholar]
- 28.VanDunk C, Hunter LA, Gray PA. Development, Maturation, and Necessity of Transcription Factors in the Mouse Suprachiasmatic Nucleus. The Journal of Neuroscience. 2012;31(17):6457–6467. doi: 10.1523/JNEUROSCI.5385-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blanck O, Perrin C, Mziaut H, Darbon H, Mattei MG, Miquelis R. Molecular Cloning, cDNA Analysis, and Localization of a Monomer of the N-Acetylglucosamine-Specific Receptor of the Thyroid, NAGR1, to Chromosome 19p13.3-13.2. Genomics. 1994;21(1):18–26. doi: 10.1006/geno.1994.1219. [DOI] [PubMed] [Google Scholar]
- 30.Bajenova OV, Zimmer R, Stolper E, Salisbury-Rowswell J, Nanji A, Thomas P. Heterogeneous RNA-binding Protein M4 Is a Receptor for Carcinoembryonic Antigen in Kupffer Cells. Journal of Biological Chemistry. 2001;276(33):31067–31073. doi: 10.1074/jbc.M104093200. [DOI] [PubMed] [Google Scholar]
- 31.Bajenova OV, Stolper E, Gapon S, Sudina N, Zimmer R, Thomas P. Surface expression of heterogeneous nuclear RNA binding protein M4 on Kupffer cell relates to its function as a carcinoembryonic antigen receptor. Exp Cell Res. 2003;291(1):228–41. doi: 10.1016/s0014-4827(03)00373-2. [DOI] [PubMed] [Google Scholar]
- 32.DiAntonio A, Haghighi AP, Portman SL, Lee JD, Amaranto AM, Goodman CS. Ubiquitination-dependent mechanisms regulate synaptic growth and function. Nature. 2001;412(6845):449–52. doi: 10.1038/35086595. [DOI] [PubMed] [Google Scholar]
- 33.Schaefer AM, Hadwiger GD, Nonet ML. rpm-1, a conserved neuronal gene that regulates targeting and synaptogenesis in C. elegans. Neuron. 2000;26(2):345–56. doi: 10.1016/s0896-6273(00)81168-x. [DOI] [PubMed] [Google Scholar]
- 34.Lewcock JW, Genoud N, Lettieri K, Pfaff SL. The ubiquitin ligase Phr1 regulates axon outgrowth through modulation of microtubule dynamics. Neuron. 2007;56(4):604–20. doi: 10.1016/j.neuron.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 35.Hendricks M, Jesuthasan S. PHR Regulates Growth Cone Pausing at Intermediate Targets through Microtubule Disassembly. J. Neurosci. 2009;29(20):6593–6598. doi: 10.1523/JNEUROSCI.1115-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han S, Witt RM, Santos TM, Polizzano C, Sabatini BL, Ramesh V. Pam (Protein associated with Myc) functions as an E3 Ubiquitin ligase and regulates TSC/mTOR signaling. Cellular Signalling. 2008;20(6):1084–1091. doi: 10.1016/j.cellsig.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakata K, Abrams B, Grill B, Goncharov A, Huang X, Chisholm AD, Jin Y. Regulation of a DLK-1 and p38 MAP kinase pathway by the ubiquitin ligase RPM-1 is required for presynaptic development. Cell. 2005;120(3):407–20. doi: 10.1016/j.cell.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 38.Pierre SC, Hausler J, Birod K, Geisslinger G, Scholich K. PAM mediates sustained inhibition of cAMP signaling by sphingosine-1-phosphate. EMBO J. 2004;23(15):3031–40. doi: 10.1038/sj.emboj.7600321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D’Souza J, Hendricks M, Le Guyader S, Subburaju S, Grunewald B, Scholich K, Jesuthasan S. Formation of the retinotectal projection requires Esrom, an ortholog of PAM (protein associated with Myc) Development. 2005;132(2):247–56. doi: 10.1242/dev.01578. [DOI] [PubMed] [Google Scholar]
- 40.Collins CA, Wairkar YP, Johnson SL, DiAntonio A. Highwire restrains synaptic growth by attenuating a MAP kinase signal. Neuron. 2006;51(1):57–69. doi: 10.1016/j.neuron.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 41.McLaughlin T, O’Leary DDM. MOLECULAR GRADIENTS AND DEVELOPMENT OF RETINOTOPIC MAPS. Annual Review of Neuroscience. 2005;28(1):327–355. doi: 10.1146/annurev.neuro.28.061604.135714. [DOI] [PubMed] [Google Scholar]
- 42.Plas DT, Dhande OS, Lopez JE, Murali D, Thaller C, Henkemeyer M, Furuta Y, Overbeek P, Crair MC. Bone morphogenetic proteins, eye patterning, and retinocollicular map formation in the mouse. J Neurosci. 2008;28(28):7057–67. doi: 10.1523/JNEUROSCI.3598-06.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu C, Daniels R, DiAntonio A. DFsn collaborates with Highwire to down-regulate the Wallenda/DLK kinase and restrain synaptic terminal growth. Neural Development. 2007;2(1):16. doi: 10.1186/1749-8104-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grill B, Bienvenut WV, Brown HM, Ackley BD, Quadroni M, Jin Y. C. elegans RPM-1 regulates axon termination and synaptogenesis through the Rab GEF GLO-4 and the Rab GTPase GLO-1. Neuron. 2007;55(4):587–601. doi: 10.1016/j.neuron.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 45.Tian X, Li J, Valakh V, DiAntonio A, Wu C. Drosophila Rae1 controls the abundance of the ubiquitin ligase Highwire in post-mitotic neurons. Nat Neurosci. 2011;14(10):1267–1275. doi: 10.1038/nn.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grill B, Chen L, Tulgren ED, Baker ST, Bienvenut W, Anderson M, Quadroni M, Jin Y, Garner CC. RAE-1, a Novel PHR Binding Protein, Is Required for Axon Termination and Synapse Formation in Caenorhabditis elegans. The Journal of Neuroscience. 2012;32(8):2628–2636. doi: 10.1523/JNEUROSCI.2901-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bouquet C. l., Soares S, von Boxberg Y, Ravaille-Veron M. l., Propst F, Nothias F. Microtubule-Associated Protein 1B Controls Directionality of Growth Cone Migration and Axonal Branching in Regeneration of Adult Dorsal Root Ganglia Neurons. The Journal of Neuroscience. 2004;24(32):7204–7213. doi: 10.1523/JNEUROSCI.2254-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meixner A, Haverkamp S, Wässle H, FÃ1/2hrer S, Thalhammer J, Kropf N, Bittner RE, Lassmann H, Wiche G, Propst F. Map1b Is Required for Axon Guidance and Is Involved in the Development of the Central and Peripheral Nervous System. The Journal of Cell Biology. 2000;151(6):1169–1178. doi: 10.1083/jcb.151.6.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma D, Nothias F, Boyne LJ, Fischer I. Differential regulation of microtubule-associated protein 1B (MAP1B) in rat CNS and PNS during development. Journal of Neuroscience Research. 1997;49(3):319–332. doi: 10.1002/(sici)1097-4547(19970801)49:3<319::aid-jnr7>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 50.Allen E, Ding J, Wang W, Pramanik S, Chou J, Yau V, Yang Y. Gigaxonin-controlled degradation of MAP1B light chain is critical to neuronal survival. Nature. 2005;438(7065):224–228. doi: 10.1038/nature04256. [DOI] [PubMed] [Google Scholar]
- 51.Yoon Byung C., Jung H, Dwivedy A, O’Hare Catherine M., Zivraj Krishna H., Holt Christine E. Local Translation of Extranuclear Lamin B Promotes Axon Maintenance. Cell. 2012;148(4):752–764. doi: 10.1016/j.cell.2011.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(1):16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Petros TJ, Shrestha BR, Mason C. Specificity and Sufficiency of EphB1 in Driving the Ipsilateral Retinal Projection. The Journal of Neuroscience. 2009;29(11):3463–3474. doi: 10.1523/JNEUROSCI.5655-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blank M, Fuerst PG, Stevens B, Nouri N, Kirkby L, Warrier D, Barres BA, Feller MB, Huberman AD, Burgess RW, Garner CC. The Down Syndrome Critical Region Regulates Retinogeniculate Refinement. The Journal of Neuroscience. 2011;31(15):5764–5776. doi: 10.1523/JNEUROSCI.6015-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.