Abstract
HIV entry involves binding of the trimeric viral envelope glycoprotein (Env) gp120/gp41 to cell surface receptors, which triggers conformational changes in Env that drive the membrane fusion reaction. The conformational landscape that the lipids and Env navigate en route to fusion has been examined by biophysical measurements on the microscale, whereas electron tomography, x-rays, and NMR have provided insights into the process on the nanoscale and atomic scale. However, the coupling between the lipid and protein pathways that give rise to fusion has not been resolved. Here, we discuss the known and unknown about the overall HIV Env-mediated fusion process.
Keywords: HIV, Membrane Biophysics, Membrane Fusion, Membrane Lipids, Membrane Proteins, Viral Protein, Viral Entry, Six-helix Bundle, HIV Env Trimer Structure and Function, HIV gp120 Structure
The Virus and Its Target
HIV virions exist as roughly spherical nanoparticles ∼100 nm in diameter and are coated by the viral envelope membrane (1). The viral membrane is a lipid bilayer ∼4 nm thick, interspersed with membrane-embedded glycoproteins. The HIV matrix protein Gag Pr55 drives viral assembly by recruiting all the building blocks, which include both viral and cellular components required for the formation of a fully infectious virion (2). A typical HIV viral membrane contains ∼300,000 lipids, with a rather unique distribution of lipids compared with the lipid composition of cells from which it is derived (3, 4). In addition to the virally encoded envelope glycoprotein gp120/gp41, the HIV membrane also incorporates a plethora of cell membrane proteins in the process of assembly and budding (5). The HIV envelope proteins are expressed in the endoplasmic reticulum as a precursor protein, gp160, which transits the Golgi apparatus, where glycosylation is completed (6, 7). The precursor is cleaved in the trans-Golgi by the cellular protease furin into two proteins, gp120 and gp41 (8). These proteins are present on the cell surface as the envelope complex, a mushroom-shaped trimer of heterodimers of gp120 and gp41, incorporated into the viral envelope through the transmembrane region of gp41 as virus particles bud from the cell surface (9).
The number of gp120/gp41 trimers per virion ranges between 10 and 100 depending on the isolate (10). HIV gp120 is ∼50% carbohydrate and is one of the most glycosylated proteins known (11). The number of accessory HIV membrane proteins has, in general, not been determined, with the exception of HLA class II in HIV-1MN derived from H9 cells, which has ∼50 native HLA class II complexes (12). The lipids and proteins are glycoconjugated, providing an additional shield for HIV against immune and environmental challenges. The viral genome is thus comfortably ensconced in a wrapper of lipid and protein covered by carbohydrate. However, mannose-type carbohydrates on HIV are recognized by DC-SIGN (dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin), which is a C-type lectin receptor present on dendritic cells (13). The dendritic cells internalize HIV-DC-SIGN complexes and migrate to the lymph nodes, where they interact with T cells. The HIV-DC-SIGN complexes are then recycled to the cell periphery, and HIV is transferred to CD4+ T cells via a virological synapse (14). To infect a CD4+ T cell, HIV has to transfer its genome across both viral and cell membranes, which are also highly resistant to environmental challenges. The double-crossing is achieved by fusion of the viral and cell membranes, which provides a large pore for the viral genome to gain access to the cell's cytoplasm. This process requires a partial opening up of the overall virus structure to expose bare lipid patches of ∼50–300 nm2 in area (15, 16).
Resting peripheral blood lymphocytes have typically 70,000 CD4, 2500 CXCR4, and 600 CCR5 molecules on their surface (17), and these numbers could change upon stimulation. Cell lines are often used as targets that overexpress orders of magnitude larger amounts of these molecules. The first step in HIV entry is docking, which involves a stepwise recognition of gp120 by CD4 and co-receptor (CR)2 on the target cells. Binding of gp120 to CD4 produces conformational changes in gp120 that lead to recognition of a CR, CXCR4 or CCR5. Although HIV fusion/entry has been studied extensively, we are still far from elucidating its mechanism. One problem is that little information is available on the proper time and length scales of the process. Data are available that can be used to explore only certain aspects of the process at different resolutions: atomic scale, nanoscale, and microscale (Figs. 1 and 2).
FIGURE 1.
HIV Env protein structure: atomic scale and nanoscale. A, x-ray crystallographic structure of gp120 with the V3 loop (Protein Data Bank code 2B4C) (19). The inner domain is in blue, the bridging sheet is in yellow, and the outer domain with the V3 loop is in red. B, crystallographic structure of the gp41 ectodomain in the 6HB conformation (Protein Data Bank code 2X7R) (30). The advantage of this recent structure is the inclusion of the FP proximal region and MPER segments. Although refolding of gp41 into this more stable conformation facilitates fusion, it is still unclear what stage of the process is driven by the released energy. C and D, cryo-electron microscopy tomogram density maps of HIV-1 Env gp120/gp41 trimers in the closed and soluble CD4-triggered open states (98), respectively. The fitted atomic coordinates of gp120 are shown in red, and those of soluble CD4 are shown in yellow. The same color code applies for the schematic insets, with the addition of cyan for gp41.
FIGURE 2.
Microscale determination of the HIV fusion cascade. Shown is a schematic representation of the experimentally determined sequence of events in HIV Env-mediated fusion determined on the micrometer scale. A, the viral membrane is labeled with a lipid dye (light blue). It contains Env trimers (gp120, dark blue; and gp41, red) and accessory proteins, such as HLA-DR (purple). It encapsulates the viral core, Vpr-BlaM (green), and small aqueous dyes (yellow). Although the target membrane may contain various attachment molecules (99), only the most important ones, CD4 (gray) and CR (CXCR4 or CCR5; red violet), are indicated. Specific lipids in the viral and target cell membranes are not indicated. B, docking of the virus via gp120 to the cell surface receptor gives rise to Env conformational changes (72, 73) and aggregation of Env proteins (1, 43). C, further conformational changes lead to lipid mixing (47) and redistribution of small aqueous dyes (53, 100). These data were gathered in HIV Env-mediated cell fusion experiments. The 6HB (orange) is on its way to formation at this stage. D, fusion pore expansion indicated by movement of larger molecules, Vpr-BlaM and GFP-Gag (green) (60, 61), as well as the viral core (brown), into the cytosol. At this point, viral membrane-embedded proteins redistribute over the cell surface (57).
HIV Env Structure on the Atomic Scale and Nanoscale
Because the gp120/gp41 trimer is unstable when separated from its membrane-anchored environment, subunits have been taken apart, and high resolution structures have been determined separately. The structure of gp120 is composed of two mixed α- and β-domains (Fig. 1A): the so-called “inner domain,” which interacts with the gp41 subunit, and the larger, less conserved “outer domain,” which is highly glycosylated and interacts with CD4 and the CR proteins of the target cells. The structure is unique, without obvious links to other proteins or subdomains (18). Due to problems in crystallizing the intact protein alone, the first structures obtained were mostly of the core domains bound to CD4 and an antibody, omitting the five variable loop regions. Subsequent studies were able to include the V3 loop, which determines CR selectivity (19); the V1/V2 loops (20); the interface with gp41 (21); and the unbound state in simian immunodeficiency virus (SIV) (22) and HIV (23).
HIV gp41 can be divided into three major domains: the extracellular domain or ectodomain, the transmembrane domain, and the cytoplasmic domain (cytoplasmic tail (CT)). The major functions of the gp41 protein are mediated by the extracellular domain, which can be further subdivided into the following five functional regions: the fusion peptide (FP) followed by the N-terminal heptad repeat (NHR), the loop region, the C-terminal heptad repeat (CHR), and finally, the membrane-proximal external region (MPER) (24). The structure of the fusion peptide depends critically on the lipid environment in which the structure is determined (25). It is generally assumed that the FP inserts into the target and/or viral membrane following the triggering of gp120-gp41 complexes by host cell CD4 and CRs. However, FP insertion has not be demonstrated despite tremendous efforts that involve photolabeling, protein purification, fragmentation, and sequence analysis (26). Three NHR helices form a coiled-coil core, and the three CHR helices dock in the three hydrophobic grooves around the core, forming the iconic six-helix bundle (6HB) structure (27–29). This final conformation in the fusion process is the only atomic level (x-ray and NMR) information we have of gp41 to date. A recent crystallographic study has expanded our understanding by including the FP proximal region and MPER segments with the classic 6HB structure of gp41 (Fig. 1B) (30). These extra segments occur as helical extensions from the core coiled-coil structure but still interact to enhance the thermodynamic stability. An interesting bend at the C terminus of the MPER segments exposes conserved hydrophobic residues that may insert into the outer layer of the viral membrane to increase the curvature.
The unusually long CT of HIV-1 gp41 contains highly conserved lentiviral lytic peptides, which are amphipathic α-helical domains embedded in the viral membrane (26). Truncation of the HIV-1 CT has been shown to result in increased fusion efficiency while leading to the enhanced exposure of gp120 conformational epitopes (31). The HIV-1 CT therefore seems to be involved in an “inside-out” mechanism of regulation of the envelope function.
Binding of the gp120 subunits of Env spikes to CD4 from the target cell initiates a complex series of conformational changes. The relatively strong binding to CD4 (32) causes a rearrangement of the inner and outer domains of gp120, resulting in the joining of two β-hairpins to form the bridging sheet domain (18, 22). Studies of the intact trimer spike using cryo-electron tomography indicate that CD4 binding also initiates a dramatic change in quaternary structure, where the gp120 domains rotate and move away from the central stalk, exposing the V1/V2 and V3 loops to the target membrane (33), where they presumably are better able to interact with the CRs (Fig. 1, C and D). Similarly, the binding also induces a hinge-like change in the position of the two outer domains of CD4 (D1D2) relative to the inner ones (D3D4), which acts to pull the Env spike closer to the target membrane. The opening up of the gp120 domains from the top of the stalk also allows for rearrangement of the gp41 domains in preparation for catalyzing fusion.
Microscale Determination of the HIV Fusion Cascade
Viral fusion involves the merging of the lipid bilayer membranes of virus and cell, as well as the mixing of aqueous compartments encapsulated by these membranes (34). For this to happen, the lipids must transiently leave their lamellar orientations. Fig. 3 shows these rearrangements in the framework of the “stalk-pore” model, which has been very well grounded in theory (35) and experiment (36). HIV membranes are remarkably enriched in cholesterol, ceramide, GM3, and dihydrosphingomyelin (3, 4). Their highly liquid-ordered state (37) would provide resistance to membrane bending and stalk formation. On the other hand, HIV membranes are also enriched in phosphoinositides (4), which, when situated on the inner monolayer, may provide positive curvature required for fusion pore formation (38).
FIGURE 3.

Lipid rearrangements in fusion. The starting point of the fusion process is the formation of a contact site (i) between two apposed membranes (“docking”). Fusion is initiated in the contact site when two apposed membranes each locally protrude as “nipples” toward each other (ii) (15). In pure lipid bilayers, the required energy for structural changes comes from thermal fluctuations, whereas in viral fusion, (part of) this energy comes from work exerted by viral envelope proteins on the lipid bilayer (16). The point-like membrane protrusions minimize the energy of the hydration repulsion between the proximal leaflets of the membranes coming into immediate contact. Nipple formation results in transient displacements of polar headgroups from each other, yielding small hydrophobic patches at the tip of the nipple. Because hydrophobic surfaces attract each other, cis-leaflets can merge to create a hemifusion stalk (iii), with proximal leaflets fused and distal leaflets unfused. On the nanoscale, stalk structures have been determined in pure lipids by x-ray diffraction (36). Stalk expansion yields the hemifusion diaphragm (iv). A fusion pore forms either in the hemifusion diaphragm bilayer or directly from the stalk (v). Dashed lines show the boundaries of the hydrophobic surfaces of monolayers. This figure was reprinted by permission from Ref. 38.
HIV Env-induced membrane fusion is a multistep “dance macabre,” in which the leading partner is the protein, and the following partner is the lipid. However, engagement of an inappropriate lipid partner on the target cell may lead to early abortion of the process. Such inappropriate partners have been identified in the form of overexpressed glycosphingolipids (39), ceramides (40), or other products of the sphingomyelin metabolism (41) and, more recently, dihydrosphingomyelin (42). Interestingly, the major effect of the inappropriate partner is bringing CD4 lateral movement to a grinding halt (39, 41), whereas the CR goes merrily on with the dance. From a cell biological point of view, it would be interesting to figure out why certain lipids have such a selective effect on the lateral movement of cell surface receptors. Non-participation of CD4 in the dance macabre presumably leads to defective viral junction formation (43), and consequently, fusion is blocked.
To study the kinetics of viral fusion, lipid dyes have been incorporated into the viral membrane (Fig. 2) in self-quenching concentrations, with dequenching as a result of fusion measured both in bulk at high time resolution and in single virions at high spatial resolution (44–46). The kinetics of HIV/SIV Env-mediated membrane fusion has been studied extensively using envelope glycoproteins expressed in cells interacting with target cells bearing CD4 and the appropriate CR in dye redistribution assays (47). Interestingly, the rates of HIV cell fusion and Env-mediated cell fusion are roughly similar (48). Although there are subtle differences between fusion of intact virus with cells and Env-mediated cell-cell fusion, the basic mechanisms that underpin both phenomena are the same.
The rearrangements that lipids undergo during fusion have been recapitulated in viral envelope protein-mediated cell fusion experiments, particularly in the case of influenza HA. The groundbreaking paper by White and colleagues (49) showing that lipid-anchored influenza HA promotes hemifusion firmly established this intermediate in the consciousness of fusion aficionados. In other studies, small pores have been seen before lipid redistribution (50), a process referred to as restricted hemifusion (51). Hemifusion in the HIV Env-mediated cascade was revealed by the observation that the entry inhibitor T20 had different sensitivities for lipid versus contents mixing (47, 52). Moreover, a mutant in the MPER exhibited a phenotype that allowed small molecules to pass but not pore expansion, leading to syncytia or nucleocapsid release (53). Additionally, HIV Env proteins with mutations in the loop region were found to exhibit hemifusion phenotypes (54, 55). The involvement of the loop region in hemifusion was further supported by a study with homologous peptides (56). According to one HIV pathogenesis hypothesis, hemifusion is considered to be dangerous in that HIV-infected CD4+ cells expressing HIV Env act like vampires that inflict apoptosis upon innocent CD4+ bystanders by a “kiss and run” process that is blocked by fusion inhibitors, such as enfuvirtide (24).
A photosensitized labeling methodology provided a reliable time course of fusion of HIV and SIV with biological membranes (57). The assay reports the redistribution of a protein (HLA-DR) from the viral membrane to the cell membrane (Fig. 2D). The most striking result is that SIV reaches maximum fusion at 37 °C, with a t½ of ∼20 min. This indicates that the CD4- and CR-induced triggering events leading to HIV/SIV Env-mediated fusion are stochastic, leading to much slower fusion kinetics compared with the low pH fusion induced by influenza HA and vesicular stomatitis virus, whose pH-triggered fusion events are more synchronized. The lack of synchrony in the activation of HIV/SIV Env proteins therefore provides an opportunity for the C-terminal peptide inhibitors to bind to the prehairpin grooves, which become transiently exposed following CD4-induced triggering of HIV-1 Env. Indeed, there is a strong correlation between HIV/SIV fusion kinetics and potency of fusion inhibitors, such as enfuvirtide (58). Another fusion assay was later developed that relies on the co-incorporation within the virus of an enzyme linked to the HIV accessory protein Vpr (59). The loading of target cells with fluorescent probes attached to a substrate of this enzyme gives the means to follow fusion specifically, as the activity of the enzyme is observed only within the cytoplasm of the target cell, does not necessitate uncoating, and occurs with small fusion pores (60). Kinetic information can be obtained by stopping the fusion process through nonpermissive temperatures or the use of inhibitors at different time points, confirming the slow fusion process of HIV.
Dye redistribution from single HIV pseudotyped virions engineered to incorporate GFP-Gag does show reliable fusion (61), although the kinetics cannot be assessed. In an interesting twist, Melikyan and co-workers (62) found that hemifusion occurred at the plasma membrane but that pore expansion occurred following endocytosis in the endosome. Although the endocytic pathway was revealed by observation on the micrometer scale (62, 63), it was not supported by examination on the nanoscale in primary lymphocytes (64). Inhibitors of dynamin- and clathrin-dependent endocytosis (62, 65) have been invoked to argue for HIV entry via endocytosis, but such inhibitors have multifaceted effects on cellular processes, including cell-cell fusion (66). Endosomal acidification inhibitors could prevent degradation and allow endocytic fusion at neutral pH, but enhanced endocytosis causes lysosomal degradation of HIV (67, 68). Interestingly “fusion from without” has been used as a measure of virus-cell fusion, which is followed by Env-induced cell fusion (69). However, if the initial process (virus-cell fusion) were to occur in the endosome, Env would have to traffic back to the cell surface to cause cell fusion. This process needs to be examined. A kinetic argument in favor of the endocytic pathway is the observation that entry inhibitor arrest precedes cold arrest in HIV entry (62). This observation does need to be confirmed for Env-induced cell fusion. Finally, observations of differential uptake (70) and infection (71) of vesicular stomatitis virus (which does enter via the pH-dependent endocytic pathway) versus HIV seem to be at odds. Regardless of where entry eventually occurs, which may depend on the type of target cell HIV infects, the mechanics of fusion will likely be the same.
Conformational Changes of HIV Env Proteins in the Course of HIV Env-mediated Fusion
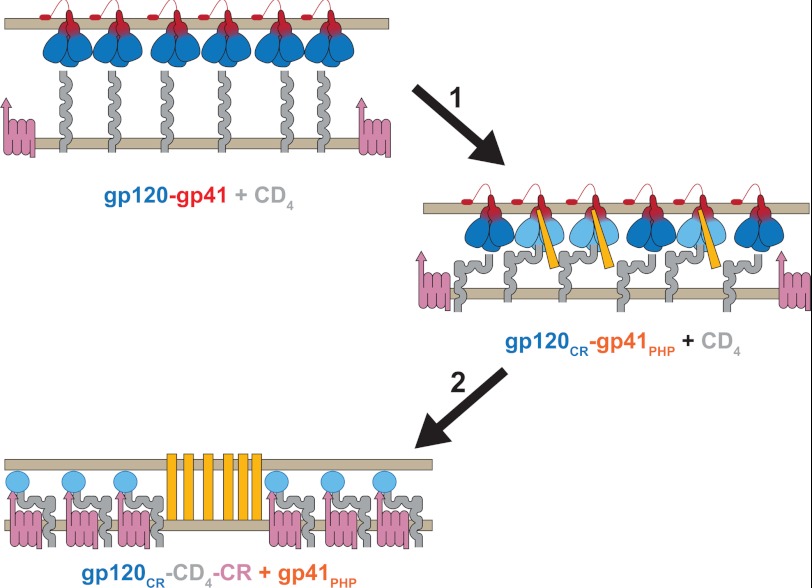
The complex choreography of the protein in its pathway to fusion is being unraveled by structural studies on the Env proteins on the nanometer scale level for the trimer and on the atomic level for the subunits. In addition to structural information, a wealth of HIV Env-mediated fusion data, including inhibition by peptides that mimic the sequences of the N- and C-terminal helical regions and fusion kinetics (48), has provided information on this complex choreography. In the absence of complete structural information, some of the details of the HIV-1 Env-mediated fusion reaction have been inferred from immunochemical, biochemical, and mutagenic analyses. Conformational changes in gp120-gp41 expressed on cells have been monitored as a function of time by analytical and quantitative video microscopy following interactions of Env-expressing cells with target cells using nonspecific probes that report on hydrophobicity changes (72, 73), as well as reagents that are specific for the different conformations of triggered HIV-1 gp120/gp41 (74, 75). After the interaction with target cell surface-bound CD4, a substantial increase in immunoreactivity to certain antibodies is observed, reflecting conformational changes in gp120, resulting in exposure of the CR-binding site (gp120CR), and in gp41, resulting in the prehairpin conformation (gp41PHP) (Fig. 4, step 1) (76). The increased reactivity has been observed in the absence of gp120 dissociation (77), indicating that subunit dissociation is not absolutely required for these CD4-induced conformational changes to occur. However, engagement of the gp120-gp41PHP-CD4 complex with CR triggers a further barrage of conformational changes, resulting in dissociation of gp120 and further release of gp41PHP (Fig. 4, step 2). These interactions lead to viral junction formation (43), which presumably involves annular aggregation of gp120-gp41-CD4-CR complexes into an “entry claw” observed by electron microscopy (1).
FIGURE 4.
HIV Env conformational changes during fusion. Shown is a schematic representation of experimentally determined conformational changes during the initial stages of HIV Env-mediated fusion. HIV Env trimers (gp120, blue; and gp41, red) on the viral membrane (top) are poised to interact with CD4 (gray) on the target membrane (bottom). Binding of gp120 to CD4 (step 1) leads to conformational changes in gp120 that involve exposure of its CR-binding site (gp120CR; light blue) and formation of the gp41 prehairpin (gp41PHP; orange). Upon engagement of CR (red violet; step 2), gp120 dissociates into subunits (blue oval), allowing gp41PHP to engage with the target membrane (27, 28) and/or viral membrane (16). The mechanism of coupling between HIV Env refolding and lipid rearrangements in Fig. 3 leading to fusion is unresolved.
Coupling between Refolding of HIV Env and Lipid Rearrangements
Figs. 3 and 4 show pathways the lipids and viral Env, respectively, take en route to fusion. Although these pathways have been fairly well grounded in theory and experiment, the coupling between the two has not been resolved. The classical “harpoon model” (27, 28) posits that the triggering of Env by CD4 and CR results in gp41 prehairpin formation and the displacement of FP in the direction of the target membrane into which it inserts. The CHR region is then thought to jackknife and pack against the inner NHR coiled-coil core, forming the 6HB, which brings the attached target cell and viral membranes together. However, no mechanism is provided for membrane bending and fusion. An alternative model posits that both the FP and transmembrane domain are anchored in the viral membrane, and refolding into 6HB exerts a force that bends the viral membrane into a saddle-like shape (16), from whence the process indicated in Fig. 3 can ensue. There is no experimental evidence for either model because it still remains to be shown that the FP inserts into the target and/or viral membrane. The energy required for formation of the stalk (Fig. 3) ranges from 25 to 50 kcal/mol at body temperature depending on the lipid compositions of the membrane layers. How many Env proteins are involved in the fusion process and how much energy is released by the refolding of a single protein trimer are still hotly debated. Data have been generated from in vitro infectivity assays using pseudotyped virions that express mixed trimers consisting of functional and nonfunctional envelope proteins (78). However, mathematical models are required to interpret such data in terms of Env stoichiometry. Although the original model posited that only one Env protein is required, subsequent analyses came up with five (79) or eight (80) Env proteins. The energy released from 6HB formation has been addressed mostly by thermodynamic studies (e.g. thermal denaturation and isothermal titration calorimetry) and theoretical calculations. Considerable work has gone into studying the stability of the 6HB conformation of gp41 in pursuit of developing fusion-inhibiting drugs for HIV (81). Unfortunately, obtaining an exact answer is complicated by many factors, such as that melting of the 6HB is not necessarily a simple two-state process, that the experimental environments of the protein are different from in vivo, and that studies tend to utilize only fragments of the protein. Indeed, Buzon et al. (30) demonstrated that including the FP proximal region and MPER segments in the gp41 6HB complex significantly enhances the stability. Simple back of the (viral) envelope calculations on the energy required for stalk formation versus recoup from 6HB formation indicate that activation of a single trimer would be more than sufficient to drive merging of the membranes (82). However, even if one trimer is capable of doing the heavy lifting, it still has to work against the other five or more proteins that are keeping the membranes apart in their prehairpin state. In an interesting twist, Melikyan and co-workers (83) showed that although 6HB formation is indeed required for the fusion process, some (if not all) of this occurs after the fusion pore has formed. This suggests that we must consider other facilitators of fusion pore expansion, such as earlier conformational changes following activation that destabilize the outer leaflets of the viral and target membranes. This is consistent with the recent crystal structure of gp41 determined by Buzon et al. (30), which shows the MPER segment ideally positioned for shallow insertion into the viral membrane.
Fusion Pore Opening
Although the cost of forming a stalk and fusion pore appears to be manageable and accommodated by conformational changes of one or more gp41 trimers, this only leads to a relatively small opening. Whether the pore stabilizes and dilates depends on the balance between membrane tension that drives dilation and line tension that drives pore closure (84). Pore dilation appears to be the most energy-demanding step of fusion (85). So, even if several Env glycoproteins form a prefusion complex, it remains uncertain how these proteins create a pore large enough to permit the release of the HIV nucleocapsid (50 nm). Therefore, it has been proposed that cellular processes, such as signaling and actin rearrangement, might be co-opted by the virus to help enlarge a fusion pore. Upon the discovery of CXCR4 and CCR5 as the CRs for HIV entry (86), questions have risen regarding the relevance of their signaling capability in the infection process. At the same time, results indicated that actin dynamics may have a role to play in HIV entry (87). However, it is only recently that we start getting initial insights into the mechanism of this process. gp120 induces a capping of CD4 and CR on the surfaces of target cells at the site of interaction with the virus. This capping is facilitated by the gp120 activation of RhoA, which in turn phosphorylates cofilin (88). This transient inhibition of the actin depolymerization factor cofilin contributes to a capping mediated by the physical and functional adapter filamin A, which has the ability to bind to CD4, CXCR4, and actin. These cortical actin filaments can then be attached to the plasma membrane through the ERM (ezrin/radixin/moesin) cytoskeletal proteins. Indeed, gp120 has been shown to induce the phosphorylation of ERM proteins, leading to the recruitment of moesin to the rich actin cap (89). Although early deactivation of cofilin is helpful to facilitate the gathering of the different participants in the fusion event, its reactivation is necessary to clear the cortical actin, which then represents a barrier for the penetration of the viral material within the cells (90). The participants in this complex and dynamic orchestration are still being unraveled, as the modulation of the process by Arf6 (91), LIMK1 (92), and syntenin-1 (93) has recently been evidenced. Harmon et al. (94) went a step farther in the characterization of the process by identifying the stage at which the blockage occurs. They showed that Abl and the Wave2 signaling complex were involved at a post-hemifusion step. Indeed, the blockage induced by silencing of those components could be relieved by the addition of chlorpromazine or trifluoroperazine, agents that are able to induce positive curvature and that are believed to selectively destabilize a hemifusion diagram (95). An important mediator of these processes might be found at the lipid level with phosphatidylinositol 4,5-bisphosphate (PIP2) (89, 93). PIP2 is indeed an important modulator of membrane remodeling (96) and has been implicated in the control of syncytial formation (97). PIP2 can often be the target of proteins such as epsins, Arf proteins, and some BAR domain proteins, which have the ability to sense and induce membrane curvature. The cell therefore harbors an arsenal of membrane-shaping proteins to carry out its compartment biogenesis that could be hijacked by the virus to facilitate the highly energy-demanding pore expansion that is needed for the proper delivery of its genetic cargo.
Conclusion
The high resolution determination of the structure of the gp41 core from HIV-1 has provided a well defined landmark in the terrain HIV Env navigates following CD4- and CR-induced conformational changes. Further elucidation has been accomplished by experimental data on lipids and proteins involved in this process on the microscale, nanoscale, and atomic scale and by computational analysis. The obligatory steps taken by lipid and proteins in this process are delineated in Figs. 3 and 4. However, several critical pieces of the puzzle, which include fusion peptide insertion, oligomerization, and pore expansion, are still missing. These uncertainties underscore the incompleteness of any present model of HIV entry. We need to develop more advanced biochemical (photolabeling, mass spectroscopy) and biophysical (force measurements, time-resolved nanoscale imaging) techniques to resolve these issues. Despite many efforts in the areas of drug delivery and gene therapy, we have not been able to engineer an efficient fusion machine. Following Richard Feynmans' dictum “what I cannot create, I do not understand,” we are not there yet in the case of HIV Env-mediated fusion.
Acknowledgments
We thank Sriram Subramaniam, Leonid Chernomordik, Misha Kozlov, Gregory Melikyan, Stephen Harrison, Adrian Bax, Hans-George Kräusslich, David Ott, and Eric Freed for helpful answers to questions that arose during the preparation of the manuscript.
This was supported, in whole or in part, by National Institutes of Health Contract HHSN261200800001E from NCI; the National Institutes of Health Intramural Research Program of NCI, Center for Cancer Research, Frederick National Lab; and grants from the National Institutes of Health Intramural AIDS Targeted Antiviral Program (IATAP) and the NIAID Intramural Biodefense Research Program. This is the first article in the Thematic Minireview Series on Understanding Human Immunodeficiency Virus-Host Interactions at the Biochemical Level.
- CR
- co-receptor
- SIV
- simian immunodeficiency virus
- CT
- cytoplasmic tail
- FP
- fusion peptide
- NHR
- N-terminal heptad repeat
- CHR
- C-terminal heptad repeat
- MPER
- membrane-proximal external region
- 6HB
- six-helix bundle
- PIP2
- phosphatidylinositol 4,5-bisphosphate.
REFERENCES
- 1. Sougrat R., Bartesaghi A., Lifson J. D., Bennett A. E., Bess J. W., Zabransky D. J., Subramaniam S. (2007) Electron tomography of the contact between T cells and SIV/HIV-1: implications for viral entry. PLoS Pathog. 3, e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huang M., Orenstein J. M., Martin M. A., Freed E. O. (1995) p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69, 6810–6818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brügger B., Glass B., Haberkant P., Leibrecht I., Wieland F. T., Kräusslich H. G. (2006) The HIV lipidome: a raft with an unusual composition. Proc. Natl. Acad. Sci. U.S.A. 103, 2641–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chan R., Uchil P. D., Jin J., Shui G., Ott D. E., Mothes W., Wenk M. R. (2008) Retroviruses human immunodeficiency virus and murine leukemia virus are enriched in phosphoinositides. J. Virol. 82, 11228–11238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Esser M. T., Graham D. R., Coren L. V., Trubey C. M., Bess J. W., Jr., Arthur L. O., Ott D. E., Lifson J. D. (2001) Differential incorporation of CD45, CD80 (B7-1), CD86 (B7-2), and major histocompatibility complex class I and II molecules into human immunodeficiency virus type 1 virions and microvesicles: implications for viral pathogenesis and immune regulation. J. Virol. 75, 6173–6182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Willey R. L., Bonifacino J. S., Potts B. J., Martin M. A., Klausner R. D. (1988) Biosynthesis, cleavage, and degradation of the human immunodeficiency virus-1 envelope glycoprotein gp160. Proc. Natl. Acad. Sci. U.S.A. 85, 9580–9584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Walker B. D., Kowalski M., Goh W. C., Kozarsky K., Krieger M., Rosen C., Rohrschneider L., Haseltine W. A., Sodroski J. (1987) Inhibition of human immunodeficiency virus syncytium formation and virus replication by castanospermine. Proc. Natl. Acad. Sci. U.S.A. 84, 8120–8124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hallenberger S., Bosch V., Angliker H., Shaw E., Klenk H. D., Garten W. (1992) Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature 360, 358–361 [DOI] [PubMed] [Google Scholar]
- 9. White T. A., Bartesaghi A., Borgnia M. J., Meyerson J. R., de la Cruz M. J., Bess J. W., Nandwani R., Hoxie J. A., Lifson J. D., Milne J. L., Subramaniam S. (2010) Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure. PLoS Pathog. 6, e1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chertova E., Bess J. W., Jr., Crise B. J., Sowder R. C., 2nd, Schaden T. M., Hilburn J. M., Hoxie J. A., Benveniste R. E., Lifson J. D., Henderson L. E., Arthur L. O. (2002) Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. J. Virol. 76, 5315–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Leonard C. K., Spellman M. W., Riddle L., Harris R. J., Thomas J. N., Gregory T. J. (1990) Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J. Biol. Chem. 265, 10373–10382 [PubMed] [Google Scholar]
- 12. Trubey C. M., Chertova E., Coren L. V., Hilburn J. M., Hixson C. V., Nagashima K., Lifson J. D., Ott D. E. (2003) Quantitation of HLA class II protein incorporated into human immunodeficiency type 1 virions purified by anti-CD45 immunoaffinity depletion of microvesicles. J. Virol. 77, 12699–12709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Geijtenbeek T. B., Kwon D. S., Torensma R., van Vliet S. J., van Duijnhoven G. C., Middel J., Cornelissen I. L., Nottet H. S., KewalRamani V. N., Littman D. R., Figdor C. G., van Kooyk Y. (2000) DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 100, 587–597 [DOI] [PubMed] [Google Scholar]
- 14. McDonald D., Wu L., Bohks S. M., KewalRamani V. N., Unutmaz D., Hope T. J. (2003) Recruitment of HIV and its receptors to dendritic cell-T cell junctions. Science 300, 1295–1297 [DOI] [PubMed] [Google Scholar]
- 15. Kuzmin P. I., Zimmerberg J., Chizmadzhev Y. A., Cohen F. S. (2001) A quantitative model for membrane fusion based on low-energy intermediates. Proc. Natl. Acad. Sci. U.S.A. 98, 7235–7240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kozlov M. M., Chernomordik L. V. (1998) A mechanism of protein-mediated fusion: coupling between refolding of the influenza hemagglutinin and lipid rearrangements. Biophys. J. 75, 1384–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Viard M., Parolini I., Sargiacomo M., Fecchi K., Ramoni C., Ablan S., Ruscetti F. W., Wang J. M., Blumenthal R. (2002) Role of cholesterol in human immunodeficiency virus type 1 envelope protein-mediated fusion with host cells. J. Virol. 76, 11584–11595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kwong P. D., Wyatt R., Robinson J., Sweet R. W., Sodroski J., Hendrickson W. A. (1998) Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393, 648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang C. C., Tang M., Zhang M. Y., Majeed S., Montabana E., Stanfield R. L., Dimitrov D. S., Korber B., Sodroski J., Wilson I. A., Wyatt R., Kwong P. D. (2005) Structure of a V3-containing HIV-1 gp120 core. Science 310, 1025–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McLellan J. S., Pancera M., Carrico C., Gorman J., Julien J. P., Khayat R., Louder R., Pejchal R., Sastry M., Dai K., O'Dell S., Patel N., Shahzad-ul-Hussan S., Yang Y., Zhang B., Zhou T., Zhu J., Boyington J. C., Chuang G. Y., Diwanji D., Georgiev I., Kwon Y. D., Lee D., Louder M. K., Moquin S., Schmidt S. D., Yang Z. Y., Bonsignori M., Crump J. A., Kapiga S. H., Sam N. E., Haynes B. F., Burton D. R., Koff W. C., Walker L. M., Phogat S., Wyatt R., Orwenyo J., Wang L. X., Arthos J., Bewley C. A., Mascola J. R., Nabel G. J., Schief W. R., Ward A. B., Wilson I. A., Kwong P. D. (2011) Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480, 336–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pancera M., Majeed S., Ban Y. E., Chen L., Huang C. C., Kong L., Kwon Y. D., Stuckey J., Zhou T., Robinson J. E., Schief W. R., Sodroski J., Wyatt R., Kwong P. D. (2010) Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc. Natl. Acad. Sci. U.S.A. 107, 1166–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen B., Vogan E. M., Gong H., Skehel J. J., Wiley D. C., Harrison S. C. (2005) Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433, 834–841 [DOI] [PubMed] [Google Scholar]
- 23. Kwon Y. D., Finzi A., Wu X., Dogo-Isonagie C., Lee L. K., Moore L. R., Schmidt S. D., Stuckey J., Yang Y., Zhou T., Zhu J., Vicic D. A., Debnath A. K., Shapiro L., Bewley C. A., Mascola J. R., Sodroski J. G., Kwong P. D. (2012) Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc. Natl. Acad. Sci. U.S.A. 109, 5663–5668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Garg H., Viard M., Jacobs A., Blumenthal R. (2011) Targeting HIV-1 gp41-induced fusion and pathogenesis for antiviral therapy. Curr. Top. Med. Chem. 11, 2947–2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lai A. L., Moorthy A. E., Li Y., Tamm L. K. (2012) Fusion activity of HIV gp41 fusion domain is related to its secondary structure and depth of membrane insertion in a cholesterol-dependent fashion. J. Mol. Biol. 418, 3–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Viard M., Ablan S. D., Zhou M., Veenstra T. D., Freed E. O., Raviv Y., Blumenthal R. (2008) Photoinduced reactivity of the HIV-1 envelope glycoprotein with a membrane-embedded probe reveals insertion of portions of the HIV-1 gp41 cytoplasmic tail into the viral membrane. Biochemistry 47, 1977–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chan D. C., Fass D., Berger J. M., Kim P. S. (1997) Core structure of gp41 from the HIV envelope glycoprotein. Cell 89, 263–273 [DOI] [PubMed] [Google Scholar]
- 28. Weissenhorn W., Dessen A., Harrison S. C., Skehel J. J., Wiley D. C. (1997) Atomic structure of the ectodomain from HIV-1 gp41. Nature 387, 426–430 [DOI] [PubMed] [Google Scholar]
- 29. Caffrey M., Cai M., Kaufman J., Stahl S. J., Wingfield P. T., Covell D. G., Gronenborn A. M., Clore G. M. (1998) Three-dimensional solution structure of the 44-kDa ectodomain of SIV gp41. EMBO J. 17, 4572–4584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Buzon V., Natrajan G., Schibli D., Campelo F., Kozlov M. M., Weissenhorn W. (2010) Crystal Structure of HIV-1 gp41 including both fusion peptide- and membrane-proximal external regions. PLoS Pathog. 6, e1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wyss S., Dimitrov A. S., Baribaud F., Edwards T. G., Blumenthal R., Hoxie J. A. (2005) Regulation of human immunodeficiency virus type 1 envelope glycoprotein fusion by a membrane-interactive domain in the gp41 cytoplasmic tail. J. Virol. 79, 12231–12241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kwong P. D., Doyle M. L., Casper D. J., Cicala C., Leavitt S. A., Majeed S., Steenbeke T. D., Venturi M., Chaiken I., Fung M., Katinger H., Parren P. W., Robinson J., Van Ryk D., Wang L., Burton D. R., Freire E., Wyatt R., Sodroski J., Hendrickson W. A., Arthos J. (2002) HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420, 678–682 [DOI] [PubMed] [Google Scholar]
- 33. Liu J., Bartesaghi A., Borgnia M. J., Sapiro G., Subramaniam S. (2008) Molecular architecture of native HIV-1 gp120 trimers. Nature 455, 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Blumenthal R., Clague M. J., Durell S. R., Epand R. M. (2003) Membrane fusion. Chem. Rev. 103, 53–69 [DOI] [PubMed] [Google Scholar]
- 35. Kozlovsky Y., Kozlov M. M. (2002) Stalk model of membrane fusion: solution of energy crisis. Biophys. J. 82, 882–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang L., Huang H. W. (2002) Observation of a membrane fusion intermediate structure. Science 297, 1877–1879 [DOI] [PubMed] [Google Scholar]
- 37. Lorizate M., Brügger B., Akiyama H., Glass B., Müller B., Anderluh G., Wieland F. T., Kräusslich H. G. (2009) Probing HIV-1 membrane liquid order by Laurdan staining reveals producer cell-dependent differences. J. Biol. Chem. 284, 22238–22247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chernomordik L. V., Kozlov M. M. (2008) Mechanics of membrane fusion. Nat. Struct. Mol. Biol. 15, 675–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rawat S. S., Gallo S. A., Eaton J., Martin T. D., Ablan S., KewalRamani V. N., Wang J. M., Blumenthal R., Puri A. (2004) Elevated expression of GM3 in receptor-bearing targets confers resistance to human immunodeficiency virus type 1 fusion. J. Virol. 78, 7360–7368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Finnegan C. M., Rawat S. S., Puri A., Wang J. M., Ruscetti F. W., Blumenthal R. (2004) Ceramide, a target for antiretroviral therapy. Proc. Natl. Acad. Sci. U.S.A. 101, 15452–15457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Finnegan C. M., Rawat S. S., Cho E. H., Guiffre D. L., Lockett S., Merrill A. H., Jr., Blumenthal R. (2007) Sphingomyelinase restricts the lateral diffusion of CD4 and inhibits human immunodeficiency virus fusion. J. Virol. 81, 5294–5304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vieira C. R., Munoz-Olaya J. M., Sot J., Jiménez-Baranda S., Izquierdo-Useros N., Abad J. L., Apellániz B., Delgado R., Martinez-Picado J., Alonso A., Casas J., Nieva J. L., Fabriás G., Mañes S., Goñi F. M. (2010) Dihydrosphingomyelin impairs HIV-1 infection by rigidifying liquid-ordered membrane domains. Chem. Biol. 17, 766–775 [DOI] [PubMed] [Google Scholar]
- 43. Dobrowsky T. M., Daniels B. R., Siliciano R. F., Sun S. X., Wirtz D. (2010) Organization of cellular receptors into a nanoscale junction during HIV-1 adhesion. PLoS Comput. Biol. 6, e1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Blumenthal R., Gallo S. A., Viard M., Raviv Y., Puri A. (2002) Fluorescent lipid probes in the study of viral membrane fusion. Chem. Phys. Lipids 116, 39–55 [DOI] [PubMed] [Google Scholar]
- 45. Lowy R. J., Sarkar D. P., Chen Y., Blumenthal R. (1990) Single influenza virus-cell fusion events observed by fluorescent video microscopy. Proc. Natl. Acad. Sci. U.S.A. 87, 1850–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lowy R. J., Sarkar D. P., Whitnall M. H., Blumenthal R. (1995) Differences in dispersion of influenza virus lipids and proteins during fusion. Exp. Cell Res. 216, 411–421 [DOI] [PubMed] [Google Scholar]
- 47. Muñoz-Barroso I., Durell S., Sakaguchi K., Appella E., Blumenthal R. (1998) Dilation of the human immunodeficiency virus-1 envelope glycoprotein fusion pore revealed by the inhibitory action of a synthetic peptide from gp41. J. Cell Biol. 140, 315–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gallo S. A., Finnegan C. M., Viard M., Raviv Y., Dimitrov A., Rawat S. S., Puri A., Durell S., Blumenthal R. (2003) The HIV Env-mediated fusion reaction. Biochim. Biophys. Acta 1614, 36–50 [DOI] [PubMed] [Google Scholar]
- 49. Kemble G. W., Danieli T., White J. M. (1994) Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell 76, 383–391 [DOI] [PubMed] [Google Scholar]
- 50. Zimmerberg J., Blumenthal R., Sarkar D. P., Curran M., Morris S. J. (1994) Restricted movement of lipid and aqueous dyes through pores formed by influenza hemagglutinin during cell fusion. J. Cell Biol. 127, 1885–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chernomordik L. V., Frolov V. A., Leikina E., Bronk P., Zimmerberg J. (1998) The pathway of membrane fusion catalyzed by influenza hemagglutinin: restriction of lipids, hemifusion, and lipidic fusion pore formation. J. Cell Biol. 140, 1369–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kliger Y., Gallo S. A., Peisajovich S. G., Munoz-Barroso I., Avkin S., Blumenthal R., Shai Y. (2001) Mode of action of an antiviral peptide from HIV-1. Inhibition at a post-lipid mixing stage. J. Biol. Chem. 276, 1391–1397 [DOI] [PubMed] [Google Scholar]
- 53. Muñoz-Barroso I., Salzwedel K., Hunter E., Blumenthal R. (1999) Role of the membrane-proximal domain in the initial stages of human immunodeficiency virus type 1 envelope glycoprotein-mediated membrane fusion. J. Virol. 73, 6089–6092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bär S., Alizon M. (2004) Role of the ectodomain of the gp41 transmembrane envelope protein of human immunodeficiency virus type 1 in late steps of the membrane fusion process. J. Virol. 78, 811–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Garg H., Joshi A., Freed E. O., Blumenthal R. (2007) Site-specific mutations in HIV-1 gp41 reveal a correlation between HIV-1-mediated bystander apoptosis and fusion/hemifusion. J. Biol. Chem. 282, 16899–16906 [DOI] [PubMed] [Google Scholar]
- 56. Ashkenazi A., Viard M., Wexler-Cohen Y., Blumenthal R., Shai Y. (2011) Viral envelope protein folding and membrane hemifusion are enhanced by the conserved loop region of HIV-1 gp41. FASEB J. 25, 2156–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Raviv Y., Viard M., Bess J., Jr., Blumenthal R. (2002) Quantitative measurement of fusion of HIV-1 and SIV with cultured cells using photosensitized labeling. Virology 293, 243–251 [DOI] [PubMed] [Google Scholar]
- 58. Reeves J. D., Gallo S. A., Ahmad N., Miamidian J. L., Harvey P. E., Sharron M., Pohlmann S., Sfakianos J. N., Derdeyn C. A., Blumenthal R., Hunter E., Doms R. W. (2002) Sensitivity of HIV-1 to entry inhibitors correlates with envelope/co-receptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. U.S.A. 99, 16249–16254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cavrois M., De Noronha C., Greene W. C. (2002) A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat. Biotechnol. 20, 1151–1154 [DOI] [PubMed] [Google Scholar]
- 60. Cavrois M., Neidleman J., Yonemoto W., Fenard D., Greene W. C. (2004) HIV-1 virion fusion assay: uncoating not required and no effect of Nef on fusion. Virology 328, 36–44 [DOI] [PubMed] [Google Scholar]
- 61. Markosyan R. M., Cohen F. S., Melikyan G. B. (2005) Time-resolved imaging of HIV-1 Env-mediated lipid and content mixing between a single virion and cell membrane. Mol. Biol. Cell 16, 5502–5513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Miyauchi K., Kim Y., Latinovic O., Morozov V., Melikyan G. B. (2009) HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell 137, 433–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dale B. M., McNerney G. P., Thompson D. L., Hubner W., de Los Reyes K., Chuang F. Y., Huser T., Chen B. K. (2011) Cell-to-cell transfer of HIV-1 via virological synapses leads to endosomal virion maturation that activates viral membrane fusion. Cell Host Microbe 10, 551–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Felts R. L., Narayan K., Estes J. D., Shi D., Trubey C. M., Fu J., Hartnell L. M., Ruthel G. T., Schneider D. K., Nagashima K., Bess J. W., Jr., Bavari S., Lowekamp B. C., Bliss D., Lifson J. D., Subramaniam S. (2010) 3D visualization of HIV transfer at the virological synapse between dendritic cells and T cells. Proc. Natl. Acad. Sci. U.S.A. 107, 13336–13341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Daecke J., Fackler O. T., Dittmar M. T., Kräusslich H. G. (2005) Involvement of clathrin-mediated endocytosis in human immunodeficiency virus type 1 entry. J. Virol. 79, 1581–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lai W., Huang L., Ho P., Montefiori D., Chen C. H. (2011) The role of dynamin in HIV type 1 Env-mediated cell-cell fusion. AIDS Res. Hum. Retroviruses 27, 1013–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schaeffer E., Soros V. B., Greene W. C. (2004) Compensatory link between fusion and endocytosis of human immunodeficiency virus type 1 in human CD4 T lymphocytes. J. Virol. 78, 1375–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Finnegan C. M., Blumenthal R. (2006) Fenretinide inhibits HIV infection by promoting viral endocytosis. Antiviral Res. 69, 116–123 [DOI] [PubMed] [Google Scholar]
- 69. Clavel F., Charneau P. (1994) Fusion from without directed by human immunodeficiency virus particles. J. Virol. 68, 1179–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pace M. J., Agosto L., O'Doherty U. (2011) R5 HIV Env and vesicular stomatitis virus G protein cooperate to mediate fusion to naive CD4+ T cells. J. Virol. 85, 644–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yu D., Wang W., Yoder A., Spear M., Wu Y. (2009) The HIV envelope but not VSV glycoprotein is capable of mediating HIV latent infection of resting CD4 T cells. PLoS Pathog. 5, e1000633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jones P. L., Korte T., Blumenthal R. (1998) Conformational changes in cell surface HIV-1 envelope glycoproteins are triggered by cooperation between cell surface CD4 and co-receptors. J. Biol. Chem. 273, 404–409 [DOI] [PubMed] [Google Scholar]
- 73. Chien M. P., Jiang S., Chang D. K. (2008) The function of co-receptor as a basis for the kinetic dissection of HIV type 1 envelope protein-mediated cell fusion. FASEB J. 22, 1179–1192 [DOI] [PubMed] [Google Scholar]
- 74. Dimitrov A. S., Louis J. M., Bewley C. A., Clore G. M., Blumenthal R. (2005) Conformational changes in HIV-1 gp41 in the course of HIV-1 envelope glycoprotein-mediated fusion and inactivation. Biochemistry 44, 12471–12479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Dimitrov A. S., Jacobs A., Finnegan C. M., Stiegler G., Katinger H., Blumenthal R. (2007) Exposure of the membrane-proximal external region of HIV-1 gp41 in the course of HIV-1 envelope glycoprotein-mediated fusion. Biochemistry 46, 1398–1401 [DOI] [PubMed] [Google Scholar]
- 76. Jacobs A., Quraishi O., Huang X., Bousquet-Gagnon N., Nault G., Francella N., Alvord W. G., Pham N., Soucy C., Robitaille M., Bridon D., Blumenthal R. (2007) A covalent inhibitor targeting an intermediate conformation of the fusogenic subunit of the HIV-1 envelope complex. J. Biol. Chem. 282, 32406–32413 [DOI] [PubMed] [Google Scholar]
- 77. Finnegan C. M., Berg W., Lewis G. K., DeVico A. L. (2002) Antigenic properties of the human immunodeficiency virus transmembrane glycoprotein during cell-cell fusion. J. Virol. 76, 12123–12134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yang X., Kurteva S., Ren X., Lee S., Sodroski J. (2005) Stoichiometry of envelope glycoprotein trimers in the entry of human immunodeficiency virus type 1. J. Virol. 79, 12132–12147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Klasse P. J. (2007) Modeling how many envelope glycoprotein trimers per virion participate in human immunodeficiency virus infectivity and its neutralization by antibody. Virology. 369, 245–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Magnus C., Rusert P., Bonhoeffer S., Trkola A., Regoes R. R. (2009) Estimating the stoichiometry of human immunodeficiency virus entry. J. Virol. 83, 1523–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Eckert D. M., Kim P. S. (2001) Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70, 777–810 [DOI] [PubMed] [Google Scholar]
- 82. Harrison S. C. (2008) Viral membrane fusion. Nat. Struct. Mol. Biol. 15, 690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Markosyan R. M., Cohen F. S., Melikyan G. B. (2003) HIV-1 envelope proteins complete their folding into six-helix bundles immediately after fusion pore formation. Mol. Biol. Cell 14, 926–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Karatekin E., Sandre O., Guitouni H., Borghi N., Puech P. H., Brochard-Wyart F. (2003) Cascades of transient pores in giant vesicles: line tension and transport. Biophys. J. 84, 1734–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kozlov M. M., Chernomordik L. V. (2002) The protein coat in membrane fusion: lessons from fission. Traffic. 3, 256–267 [DOI] [PubMed] [Google Scholar]
- 86. Berger E. A. (1997) HIV entry and tropism: the chemokine receptor connection. AIDS 11, S3–S16 [PubMed] [Google Scholar]
- 87. Iyengar S., Hildreth J. E., Schwartz D. H. (1998) Actin-dependent receptor colocalization required for human immunodeficiency virus entry into host cells. J. Virol. 72, 5251–5255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Jiménez-Baranda S., Gómez-Moutón C., Rojas A., Martínez-Prats L., Mira E., Ana Lacalle R., Valencia A., Dimitrov D. S., Viola A., Delgado R., Martínez-A C., Mañes S. (2007) Filamin-A regulates actin-dependent clustering of HIV receptors. Nat. Cell Biol. 9, 838–846 [DOI] [PubMed] [Google Scholar]
- 89. Barrero-Villar M., Cabrero J. R., Gordón-Alonso M., Barroso-González J., Alvarez-Losada S., Muñoz-Fernández M. A., Sánchez-Madrid F., Valenzuela-Fernández A. (2009) Moesin is required for HIV-1-induced CD4-CXCR4 interaction, F-actin redistribution, membrane fusion, and viral infection in lymphocytes. J. Cell Sci. 122, 103–113 [DOI] [PubMed] [Google Scholar]
- 90. Yoder A., Yu D., Dong L., Iyer S. R., Xu X., Kelly J., Liu J., Wang W., Vorster P. J., Agulto L., Stephany D. A., Cooper J. N., Marsh J. W., Wu Y. (2008) HIV envelope-CXCR4 signaling activates cofilin to overcome cortical actin restriction in resting CD4 T cells. Cell 134, 782–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. García-Expósito L., Barroso-González J., Puigdomènech I., Machado J. D., Blanco J., Valenzuela-Fernández A. (2011) HIV-1 requires Arf6-mediated membrane dynamics to efficiently enter and infect T lymphocytes. Mol. Biol. Cell 22, 1148–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Vorster P. J., Guo J., Yoder A., Wang W., Zheng Y., Xu X., Yu D., Spear M., Wu Y. (2011) LIM kinase 1 modulates cortical actin and CXCR4 cycling and is activated by HIV-1 to initiate viral infection. J. Biol. Chem. 286, 12554–12564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Gordón-Alonso M., Rocha-Perugini V., Álvarez S., Moreno-Gonzalo O., Ursa A., López-Martín S., Izquierdo-Useros N., Martínez-Picado J., Muñoz-Fernández M. Á., Yáñez-Mó M., Sánchez-Madrid F. (2012) The PDZ-adaptor protein syntenin-1 regulates HIV-1 entry. Mol. Biol. Cell 23, 2253–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Harmon B., Campbell N., Ratner L. (2010) Role of Abl kinase and the Wave2 signaling complex in HIV-1 entry at a post-hemifusion step. PLoS Pathog. 6, e1000956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Melikyan G. B., Brener S. A., Ok D. C., Cohen F. S. (1997) Inner but not outer membrane leaflets control the transition from glycosylphosphatidylinositol-anchored influenza hemagglutinin-induced hemifusion to full fusion. J. Cell Biol. 136, 995–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Takenawa T. (2010) Phosphoinositide-binding interface proteins involved in shaping cell membranes. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 86, 509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Richard J. P., Leikina E., Langen R., Henne W. M., Popova M., Balla T., McMahon H. T., Kozlov M. M., Chernomordik L. V. (2011) Intracellular curvature-generating proteins in cell-to-cell fusion. Biochem. J. 440, 185–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Tran E. E., Borgnia M. J., Kuybeda O., Schauder D. M., Bartesaghi A., Frank G. A., Sapiro G., Milne J. L., Subramaniam S. (2012) Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog. 8, e1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ugolini S., Mondor I., Sattentau Q. J. (1999) HIV-1 attachment: another look. Trends Microbiol. 7, 144–149 [DOI] [PubMed] [Google Scholar]
- 100. Melikyan G. B., Markosyan R. M., Hemmati H., Delmedico M. K., Lambert D. M., Cohen F. S. (2000) Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151, 413–423 [DOI] [PMC free article] [PubMed] [Google Scholar]