Abstract
Synthesis of integration-competent, double-stranded DNA from the (+)-RNA strand genome of retroviruses and long terminal repeat-containing retrotransposons reflects a multistep process catalyzed by the virus-encoded reverse transcriptase (RT). In conjunction with RNA- and DNA-templated DNA synthesis, a hydrolytic activity of the same enzyme (RNase H) is required to remove genomic RNA of the RNA/DNA replication intermediate. Together, these combined synthetic and degradative functions ensure correct selection, extension, and removal of the RNA primers of (−)- and (+)-strand DNA synthesis (tRNA and the polypurine tract, respectively). For HIV-1 RT, a quarter century of research has not only illuminated the biochemical properties, structure, and conformational dynamics of this highly versatile enzyme but has also witnessed drug discovery advances from the first Food and Drug Administration-approved anti-RT drug to recent use of RT inhibitors as potential colorectal microbicides. Salient features of HIV-1 RT and extension of these findings into programs of drug discovery are reviewed here.
Keywords: DNA Synthesis, Drug Discovery, Human Immunodeficiency Virus, Nucleic Acid Enzymology, Reverse Transcription, Ribonuclease H
HIV-1 DNA Synthesis
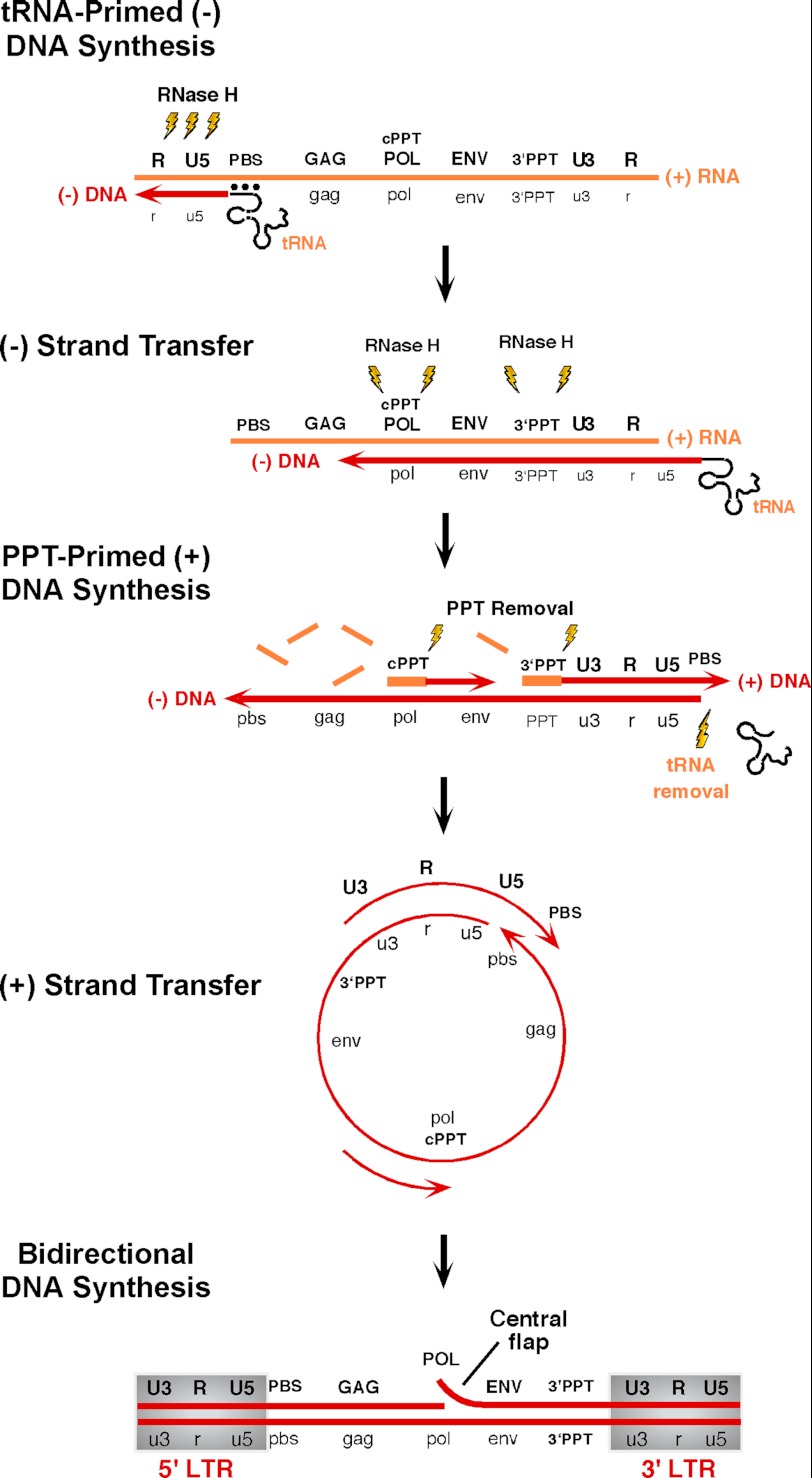
The individual steps of HIV-1 DNA synthesis, catalyzed by the multifunctional reverse transcriptase (RT),2 are summarized schematically in Fig. 1. (−)-Strand DNA synthesis, initiated from a cellular tRNA (tRNA3Lys) hybridized to the genome-encoded primer-binding site, continues to the 5′ terminus, creating (−)-strong-stop DNA. RNase H-mediated degradation of the resulting RNA/DNA hybrid promotes relocation of nascent (−)-DNA to the genome 3′ terminus by a strand transfer event that exploits sequence homology between the 5′ and 3′ termini. RNA-templated DNA synthesis continues, accompanied by RNase H-mediated degradation of the RNA genome, the exception to which are two short purine-rich segments (the 3′- and central polypurine tracts (PPTs)) from which (+)-strand DNA-dependent DNA synthesis is initiated. Newly synthesized (−)-strand DNA and 18 nucleotides of the covalently attached tRNA3Lys primer provide the template for 3′-PPT-primed (+)-strand DNA synthesis until the replication complex stalls at a position corresponding to the first modified tRNA base (A57). As a consequence, the C-terminal RNase H domain is positioned at the (−)-DNA/tRNA junction, and degradation of the tRNA “template” promotes a second or (+)-strand transfer event supported by homology between (−)- and (+)-strand DNA primer-binding sites. Although bidirectional DNA synthesis would be sufficient to complete DNA synthesis, HIV utilizes a second, central PPT primer, thereby producing a (+)-strand discontinuity (1). Following (+)-strand transfer, 3′-PPT-mediated DNA synthesis continues, displacing ∼100 nucleotides of central PPT-primed (+)-DNA, but abruptly ceases at the central termination sequence, a prominent feature of which is phased A-tracts that induce minor groove compression (1–3), creating the “central flap” (Fig. 1) (4–7). Although central flap function remains controversial (8, 9), its mutation or deletion has been shown to impair virus replication (4, 10, 11), and it improves transduction efficiency when incorporated into lentiviral vectors (12–16).
FIGURE 1.
Synthesis of integration-competent, double-stranded HIV-1 DNA from the (+)-strand RNA retroviral genome. See text for details. DNA and RNA are indicated in red and yellow, respectively. PBS, primer-binding site; cPPT, central PPT.
Biogenesis and Structural Organization of HIV-1 RT
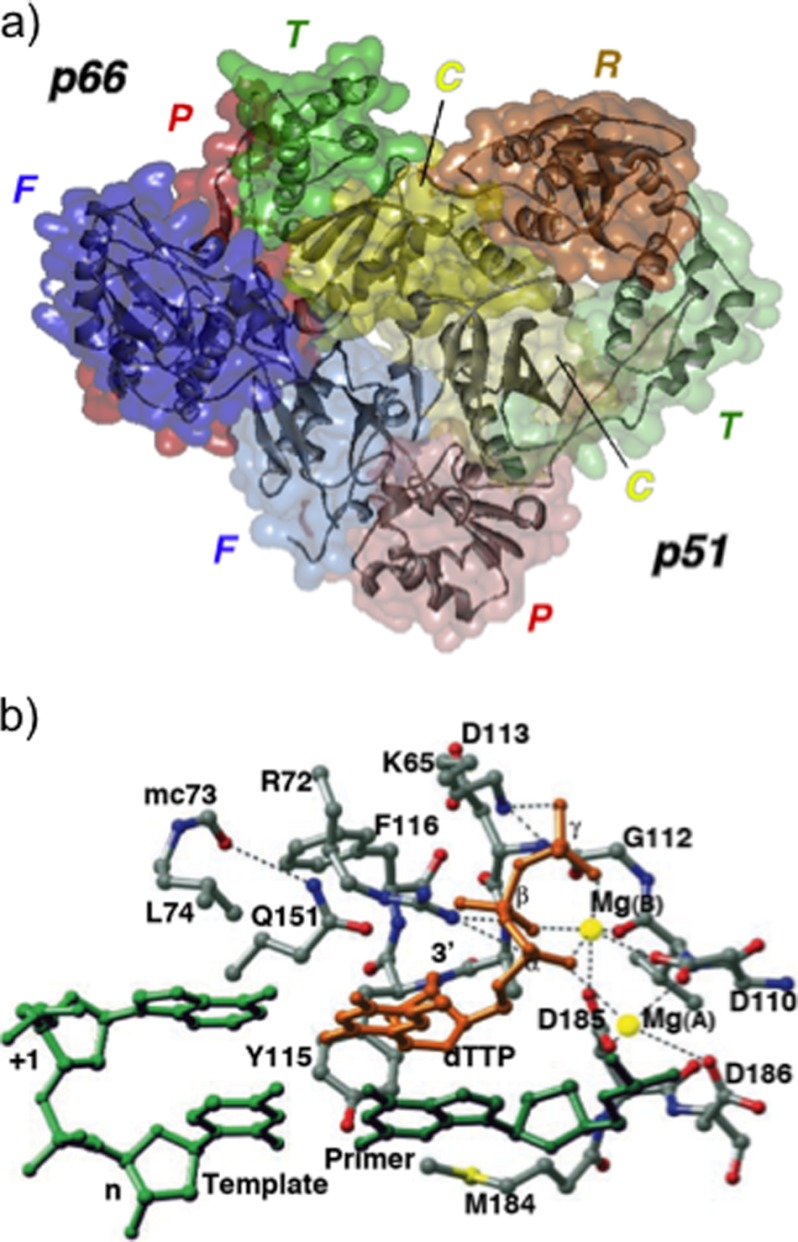
Although HIV-1 RT is encoded by a single open reading frame of the Gag/Pol precursor polyprotein, the biologically relevant enzyme is a heterodimer of 66 and 51 kDa polypeptides (p66/p51) (Fig. 2a) derived via cleavage of p66 by the virus-encoded protease between Phe-440 and Tyr-441 (17, 18). Both subunits thus contain four similar subdomains, designated fingers, palm, thumb, and connection, whereas p66 retains the C-terminal RNase domain (Fig. 2a) (19, 20). Despite identity in primary sequence, the p66 and p51 subdomains adopt significantly different folds, i.e. although the p66 DNA polymerase domain exhibits an open extended structure with a large active-site cleft, the equivalent region of p51 forms a closed compact structure incapable of participating in catalysis (19, 21). Both the DNA polymerase and RNase H activities are catalyzed by the p66 subunit, whereas the proposed roles for p51 include providing structural support to p66 (19, 20, 22, 23), facilitating p66 loading onto a template-primer (24), and stabilizing the appropriate p66 conformation during tRNA-primed initiation of reverse transcription (25, 26). In contrast to p66, which undergoes large-scale motions (especially the fingers and thumb subdomains), the p51 subunit is essentially rigid (27). Although the extreme p51 C terminus has not been resolved crystallographically, its contribution to maintaining RT architecture is supported by observations that reconstituted enzymes with short deletions show increased RNase H inhibitor sensitivity (28) and altered thermal stability.3
FIGURE 2.
a, structure of the p66/p51 HIV-1 RT heterodimer. For both subunits, the finger (F), palm (P), thumb (T), and connection (C) subdomains have been color-coded blue, red, green, and yellow, respectively, whereas the lighter shading has been used for the p51 subdomains. The p66 RNase H domain (R) has been colored in gold. b, DNA polymerase active site of HIV-1 RT. Template and primer strands are colored light and dark green, respectively, and dTTP is in gold. Mg2+ ions are represented as yellow spheres, and assigned hydrogen bonds and metal ligand interactions are indicated as dotted lines. This figure was adapted from Ref. 30 with permission.
The p66 nucleic acid-binding cleft is formed by its finger, palm, and thumb subdomains, and co-crystal structures of HIV-1 RT with duplex DNA and an RNA/DNA hybrid have identified numerous contacts with both strands of the template and primer (20, 29–31). Interactions are primarily between the sugar phosphate backbone and highly conserved motifs of the p66 DNA polymerase and RNase H domains. Superimposing x-ray structures of unliganded and nucleic acid-containing enzymes highlights considerable flexibility of the p66 thumb subdomain (32). Both x-ray crystallography and spin labeling studies of unliganded RT (33) depict the thumb as folded into the nucleic acid-binding cleft (34, 35), whereas nucleic acid binding produces large changes in orientation relative to the p66 palm, resulting in a more open conformation. α-Helix H of the thumb mediates extensive contacts with the primer strand in the minor groove of the DNA (36–38). Pro-227–His-235 comprises the β12-β13 hairpin, designated the DNA polymerase “primer grip.” This highly conserved motif (39) has been proposed to maintain the primer 3′-OH in an orientation appropriate for nucleophilic attack on the incoming dNTP (20). Important primer grip contacts involve the main chain atoms of Met-230 and Gly-231 with the primer terminal phosphate (29), and mutations of these residues induce pleiotropic effects, altering DNA polymerase and RNase H activities, as well as reducing dimer stability (36, 37, 40–46). Contact with the template strand is mediated by the “template grip,” comprising elements of the p66 palm (αB-β6 loop, β-strand 9, α-helix E, and β8-αE connecting loop) and fingers (β-strand 4) (20, 29). In contrast to original proposals (20), the co-crystal structure of Huang et al. (30) showed that the template overhang ahead of the polymerase active site was not co-linear with the duplex but was bent away and contacting the p66 fingers, revealing contacts with nucleobases +1, +2, and +3.
The DNA Polymerase Active Site
The palm subdomain of HIV-1 RT houses the DNA polymerase active site (Fig. 2b) characterized by the Asp-110, Asp-185, and Asp-186 catalytic triad, a common feature of nucleic acid-polymerizing enzymes (19, 20). Among polymerase families, palm subdomain architecture is also highly conserved, comprising a four- to six-stranded β-sheet flanked on one side by two α-helices (47). In nucleic acid-containing crystal structures, catalytic aspartates are close to the 3′ terminus of the primer. Asp-185 and Asp-186 are part of the conserved -Tyr-Met-Asp-Asp- motif, which adopts an unusual β-turn conformation (29, 35, 48), possibly to promote their positioning for catalysis, whereas the Tyr-183 phenoxy side chain is involved in hydrogen bonding with nucleobases at position −2 (29). Comparison of the crystal structures of DNA-bound RT with unliganded or non-nucleoside RT inhibitor (NNRTI)-bound enzymes reveals significant conformational differences for the -Tyr-Met-Asp-Asp- motif, implicating a high degree of structural flexibility (29).
The incoming dNTP is tightly coordinated by p66 finger residues Lys-65 and Arg-72, the main chain NH groups of Asp-113 and Ala-114, and two divalent metal ions, whereas its ribose is accommodated by a pocket lined by Asp-113, Tyr-115, and Phe-116 on one side and Glu-151 and Arg-72 on the other. Additional dNTP contacts involve base pairing and base stacking with the template overhang (30). The fidelity of dNTP insertion is critically influenced by interactions of the γ-phosphate with Lys-65 (49), whereas mutagenesis studies have designated Tyr-115 as the “steric gate,” suggesting that it aids in discriminating between deoxy- and ribonucleoside triphosphates (50, 51).
The RNase H Domain and Catalytic Mechanism
RNase H-mediated hydrolysis is divalent metal-dependent, with a preference for Mg2+. Although models for one-metal (52) and two-metal (53) assisted catalysis have been proposed, structures of Bacillus halodurans (54, 55) and human RNase H (56) bound to an RNA/DNA hybrid have confirmed the original two-metal hypothesis by Steitz and Steitz (53) (Fig. 3). Metal ion coordination is also substrate-dependent, i.e. at physiologically relevant concentrations, productive binding occurs only in the presence of substrate (57). Based on crystallographic studies (54–57), the RNase H catalytic cycle can be summarized in the following steps.
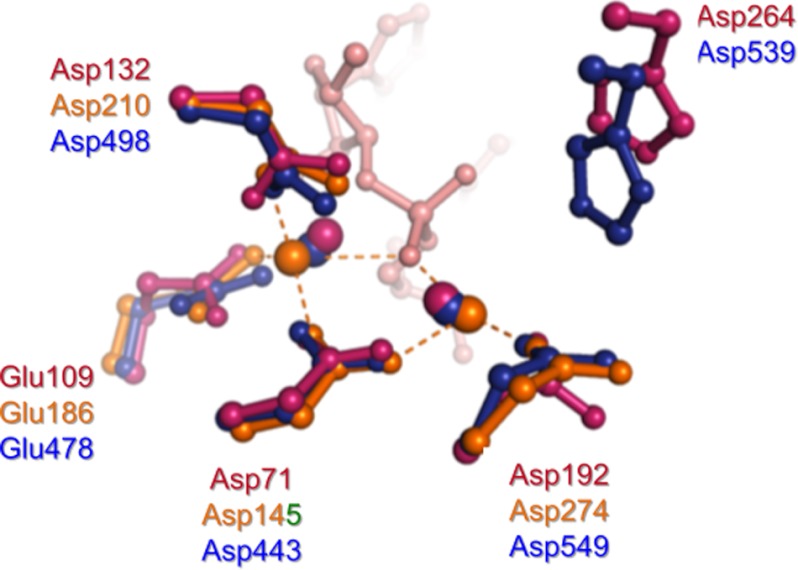
FIGURE 3.
Superposition of active-site residues from the substrate complexes of B. halodurans RNase H1 (magenta) and human RNase H1 (orange) and the complex of HIV-1 RNase H containing the natural product hydroxytropolone β-thujaplicinol (blue) (89). Metal ions (Mg2+ for B. halodurans RNase H1, Ca2+ for human RNase H1, and Mn2+ for HIV-1 RNase H) are depicted as spheres. The RNA strand from the B. halodurans RNase H1-substrate complex is shown in pink. The inhibitor from the HIV-1 RNase H co-crystal structure has been omitted for clarity.
1) In a “resting” state, divalent metal ions A and B are separated by ∼4 Å, whereas during catalysis, their position and separation vary according to the coordination environment. 2) Similar to nucleic acid phosphoryl transfer reactions, hydrolysis proceeds via an SN2 mechanism, involving a pentacoordinated intermediate and resulting in inversion of configuration at the phosphorus. 3) Metal ion A (complexed by conserved carboxylates Asp-443 and Asp-549) coordinates a water molecule, reducing its pKa and aligning this for in-line nucleophilic attack on the scissile phosphodiester bond. 4) In turn, metal ion B (coordinated by Asp-443, Glu-478, and Asp-498) is correctly positioned to stabilize the transition state, facilitating leaving of the 3′-oxyanion group.
Because metal ion B undergoes a change from an irregular five-ligand coordination and non-ideal geometry in the substrate and intermediate complexes to a regular octahedral geometry in the enzyme-product complex, this likely lowers the energy barrier for product formation (57). Crystallographic studies have revealed extensive contacts between HIV-1 RT and nucleic acid immediately ahead of the RNase H active site (31). This motif, which interacts with the DNA primer 4–9 nucleotides upstream of the scissile bond of the RNA/DNA hybrid, is collectively designated the “RNase H primer grip” (31). Amino acids of the RNase H primer grip include p66 residues Gly-359, Ala-360, His-361, Thr-473, Asn-474, Gln-475, Lys-475, Tyr-501, Ile-505, and Lys-359 and p51 residue Glu-396. Through interactions with DNA of the RNA/DNA hybrid, the RNase H primer grip is believed to impose the appropriate trajectory on the RNA strand for catalysis when it enters the RNase H active site. RNase H primer grip residues are conserved among retroviral RTs and Escherichia coli RNase H1, and both in vitro and in vivo site-directed mutagenesis studies have demonstrated their importance with respect to cleavage specificity (58, 59).
The B. halodurans RNase H-RNA/DNA co-crystal structure (54) has demonstrated that the RNA and DNA strands adopt A- and B-form geometry, respectively. Unfortunately, a structure of HIV-1 RT-associated RNase H with the RNA/DNA hybrid positioned in the active site to promote catalysis remains elusive. Structures of HIV-1 RT containing either duplex DNA (30) or a PPT-derived RNA/DNA hybrid (31) indicate that substrate is positioned for primer extension by the polymerase catalytic center and extends into the RNase H domain but does not reach its active site. Modeling studies based on RNase H-RNA/DNA co-crystals (56) propose that the RNA/DNA hybrid cannot be correctly positioned at both active sites simultaneously, necessitating a conformational change to permit “toggling” between catalytic centers. Although this is a plausible model, Beilhartz et al. (60) have shown that RNase H activity persists in the presence of the pyrophosphate analog foscarnet, which “traps” the nucleic acid substrate in the pre-translocated conformation (31). If, indeed, there is a mechanism of substrate toggling, it remains to be established whether this is specific to sequences resembling the PPT that are refractory to hydrolysis.
Conformational Dynamics of Reverse Transcription
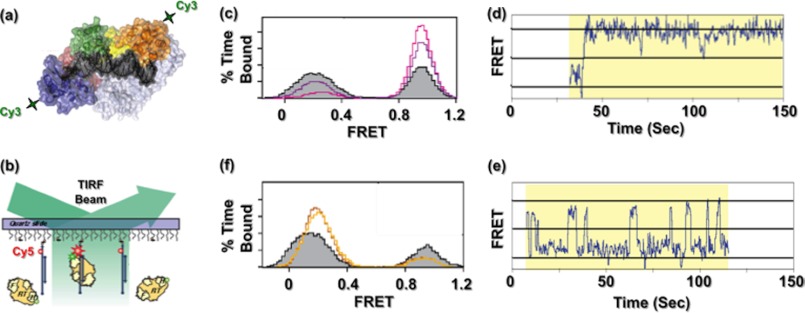
X-ray crystallography has provided incisive insights into structures of the unliganded HIV-1 RT (34) and co-crystals with nucleic acid (20, 30) and inhibitors of both DNA polymerase and RNase H function (19, 61–64). However, crystallographic analysis does not explain how RT assumes alternative conformations required to catalyze HIV-1 DNA synthesis. We therefore investigated HIV-1 RT-nucleic acid complexes by single-molecule spectroscopy (SMS), placing a fluorescent dye on the p66 N or C terminus of reconstituted p66/p51 and a second on a surface-immobilized nucleic acid duplex (Fig. 4, a and b) (65). Although the FRET signal obtained from RT bound to a 50-nucleotide DNA template/19-nucleotide DNA primer confirmed placement of the DNA polymerase catalytic center over the primer 3′ terminus, reversal of the FRET signal in the presence of an RNA primer of identical sequence indicated a binding mode with the C-terminal RNase H domain now over the primer terminus. Substituting chimeric RNA/DNA primers whose RNA component progressively increased from the 5′ terminus indicated that introducing two ribonucleotides (i.e. 2 RNA/17 DNA) was sufficient to initiate redistribution in enzyme reorientation and that the process was virtually complete on a duplex whose primer contained five 5′-ribonucleotides. The structure of HIV-1 RT and a PPT-containing RNA/DNA hybrid (31) shows that p66 residues Glu-89, Gln-91, Ser-280 (substituted for Cys-280 in Ref. 31), and Arg-284 contact the ribose 2′-OH at several positions near the RNA 5′ terminus. Because equivalent contacts are absent with duplex DNA (20, 30), additional hydrogen bonding afforded by the RNA strand may stabilize the inverted orientation. Applying SMS to PPT-containing RNA/DNA hybrids indicated that the presence of the incoming dNTP favored a polymerization-competent binding mode, whereas the NNRTI nevirapine (NVP; 11-cyclopropyl-4-methyl-5,11-dihydro-6H-dipyrido[3,2-b:2′,3′-e][1,4]diazepin-6-one) increased the frequency with which RT assumed the opposite orientation (Fig. 4, c–f). Because NNRTIs potently and preferentially inhibit initiation of HIV-1 (+)-strand DNA synthesis (66), SMS suggests that this might be due in part to their ability to induce enzyme binding in a polymerase-incompetent mode. Consistent with the notion that NNRTIs occupy a hydrophobic pocket at the base of the p66 thumb, thereby “loosening” the grip on nucleic acid, SMS also demonstrated increased “sliding” of HIV-1 RT on the template in the presence of NVP (67). Although an unresolved feature of this and related studies (68) was the ability of HIV-1 RT to adopt alternative orientations (collectively referred to as “flipping”) without dissociating from its nucleic acid substrate, our work demonstrated the value of SMS in dissecting the intricate events of reverse transcription.
FIGURE 4.
Examining HIV-1 RT dynamics by SMS. a, HIV-1 RT is site-specifically labeled with Cy3 at either the N-terminal finger subdomain or the C-terminal RNase H domain. b, Cy3-labeled RT interacts with surface-immobilized DNA containing the FRET acceptor Cy5, and fluorescence of individual substrates is followed by total internal reflection fluorescence (TIRF) microscopy. c–f, HIV-1 RT adopts alternative orientations on the PPT. c, ternary complex formation promotes binding to the primer terminus in a polymerization orientation. Shown is a FRET histogram of RT bound to the PPT (filled gray trace) in the presence of 10 μm (purple trace) and 1 mm (cyan trace) dTTP. d, FRET time trace of HIV-1 RT bound to a chain-terminated PPT substrate in the presence of dNTPs, establishing a stable ternary complex. e, FRET time trace of HIV-1 RT bound to the PPT substrate in the presence of NVP, indicating multiple transitions or flipping between high and low FRET states. f, NNRTI binding promotes RT binding to the PPT 3′ terminus in an RNase H orientation. Shown are histograms of RT bound in the absence (filled gray trace) and presence of 10 μm (red trace) and 100 μm (orange trace) NVP. This figure is adapted from Ref. 65.
DNA Polymerase and RNase H Inhibitor Development
Since the Food and Drug Administration approval of azidothymidine (1-[(2R,4S,5S)-4-azido-5-(hydroxymethyl)oxolan-2-yl]-5-methylpyrimidine-2,4-dione) in 1987, several nucleoside RT inhibitors (NRTIs) have been approved for clinical use, including lamivudine (2′,3′-dideoxy-3′-thiacytidine), emtricitabine (4-amino-5-fluoro-1-[(2S,5R)-2-(hydroxymethyl)-1,3-oxathiolan-5-yl]-1,2-dihydropyrimidin-2-one), abacavir({(1S,4R)-4-[2-amino-6-(cyclopropylamino)purin-9-yl]cyclopent-2-en-1-yl}methanol), didanosine (9-[(2R,5S)-5-(hydroxymethyl)tetrahydrofuran-2-yl]-3H-purin-6(9H)-one), and stavudine (1-[(2R,5S)-5-(hydroxymethyl)-2,5-dihydrofuran-2-yl]-5-methylpyrimidine-2,4(1H,3H)-dione). NRTIs contain modifications to their sugar moiety, the nucleobase, or both (69). Upon phosphorylation to the triphosphate derivative by cellular kinases, NRTIs are incorporated into nascent DNA, whereas their general lack of a 3′-OH prevents incorporation of the subsequent dNTP. Nucleotide RT inhibitors, e.g. tenofovir (TFV; [(2R)-1-(6-aminopurin-9-yl)propan-2-yl]oxymethylphosphonic acid), which was also approved in 1988 for treatment of hepatitis B, function by an equivalent chain-terminating mechanism but harbor a phosphonate group, requiring only two phosphorylation steps to their active derivative. In contrast, NNRTIs, e.g. NVP and efavirenz, impose allosteric control of DNA synthesis by occupying a site at the base of the p66 thumb that “locks” this subdomain in a configuration incompatible with catalysis (19, 70). Subsequent SMS analysis indicated an additional property of NNRTIs of inducing dislocation of HIV-1 RT from the polymerization target site and sliding on the nucleic acid duplex. Finally, indolopyridones (INDOPYs) represent a new class of RT inhibitors with a unique mechanism. The prototype compound, INDOPY-1 (which is active against NNRTI-resistant HIV), binds and stabilizes RT-DNA/DNA complexes, trapping them in a post-translocational state. INDOPY-1 binding depends on the chemical nature of the ultimate base pair at the primer 3′ terminus rather than the chemical nature of the templated base engaged in classic base pairing (71, 72). Although the clinical benefits of RT inhibitors in reducing rates of HIV transmission are clear, the rapid emergence of drug resistance continues to pose a challenge (73). On a more positive note, however, the availability of a wealth of high resolution co-crystal structures has spawned the development of second generation NNRTIs that interact with highly conserved residues immediately adjacent to the allosteric binding pocket. A more comprehensive synopsis of current anti-RT drugs and their use in combination antiretroviral therapy is provided in Ref. 69.
Despite documentation that loss of virus infectivity following selective inhibition of RNase H function (74) identified this activity as a potential therapeutic target, RNase H inhibitors have failed to advance toward clinical trials. The retroviral enzyme belongs to a superfamily of nucleotidyltransferases, raising concerns of toxicity due to lack of specificity. In particular, inactivating the eukaryotic RNase H counterpart is associated with a lethal embryonic defect in mice due to failure to accumulate mitochondrial DNA (75). These issues notwithstanding, a considerable body of biochemical and structural data for small molecule RNase H inhibitors has accumulated. As might be predicted, chelating the divalent metal essential for catalysis is a shared feature of these inhibitors. For example, the pharmacophore of the N-hydroxyimides described by Klumpp and Mirzadegan (76) contains a 3-oxygen motif and was originally developed as an inhibitor of influenza virus endonuclease, which shares the two-metal ion mechanism of catalysis (77, 78). A second class includes the natural product hydroxytropolones β-thujaplicinol and manicol, the former of which specifically inhibited HIV-1 RNase H with ∼30- and ∼250-fold enhanced specificity with respect to human and bacterial RNases H, respectively (79). Originally developed as metal-dependent inhibitors of HIV-1 integrase (80, 81), compounds containing a diketo acid moiety are moderately effective RNase H inhibitors, among which RDS 1643 inhibited RNase H activity and prevented HIV replication in cell culture with an EC50 of 14 μm (82).
On the basis of structural information on pyrimidinol carboxylic RNase H inhibitors, Lansdon et al. (63) have suggested that the relatively open inhibitor-binding site is unfavorable and provides a major obstacle to compound optimization. In light of the potential drawbacks of active-site inhibitors, targeting a region outside the active site might offer the possibility of allosteric inhibition of RNase H activity, akin to NNRTI-mediated restriction of thumb movement, and should be further explored. Compounds fulfilling this requirement include N-acylhydrazones such as dihydroxy benzoyl naphthyl hydrazone ((E)-3,4-dihydroxy-N′-((2-methoxynaphthalen-1-yl)methylene)benzohydrazide). Surprisingly, while demonstrating selectivity for RNase H function, x-ray crystallography has shown that dihydroxy benzoyl naphthyl hydrazone binds ∼50 Å from the active site, interacting with Trp-229 of the primer grip and Asp-186 of the DNA polymerase active site (61). Thienopyrimidinones represent a second class of allosteric inhibitor, demonstrated by both protein footprinting and site-directed mutagenesis to bind at the interface between the p51 thumb subdomain and p66 RNase H domain (28, 84, 85). Because interactions of p51 thumb residues Cys-280–Thr-290 and p66 RNase H residues Pro-537–Glu-546 constitute ∼33% of the buried surface at the dimer interface (86) thienopyrimidinones are unlikely to interact with active-site residues but rather induce a change in active-site geometry that is inconsistent with catalysis. A more thorough summary of HIV-1 RNase H inhibitors can be found in Ref. 87.
The Final Chapter? RT Inhibitors as Microbicides
Although considerable structural and biochemical data are now available for HIV-1 RT, it is essential to recognize this in the context of developing new and improved antiretroviral agents to reduce viral burden and rates of HIV transmission. Antiretroviral agents targeting specific enzyme functions of the HIV replication cycle, and in particular RT, have recently emerged as promising vaginal and rectal microbicides (88). Prominent among these is the nucleotide RT inhibitor TFV, which functions through a chain-terminating mechanism. In clinical trials, TFV was demonstrated to be safe and well tolerated in a study on HIV-negative women with a vaginal gel applied during 24 weeks. Repeated application of TFV intravaginal gel was well tolerated, produced low plasma levels, and, importantly, did not select for resistance-conferring mutations. NNRTIs such as dapivirine (DPV; 4-({4-[(2,4,6-trimethylphenyl)amino]pyrimidin-2-yl}amino)benzonitrile) have also displayed promising virucidal properties. When applied intravaginally, DPV is absorbed by the outer mucosal layers while plasma concentrations reportedly remained low. Long-term constant release of DPV has been obtained from a variety of intravaginal rings. Although encouraging, HIV microbicide development (reviewed in Ref. 88) still faces considerable formidable challenges, including conclusive demonstration of efficacy in non-human primates, selection of drug-resistant virus in clinical settings, cultural acceptability, and affordability. These issues notwithstanding, advances in HIV RT research over that last 25 years, which have ranged from expressing active recombinant enzyme for high throughput screening to the potential of introducing vaginal and rectal microbicides in resource-limited settings, should be considered a bench-to-bedside success and a model for development of future antiviral agents.
Acknowledgment
I thank M. Nowotny (International Institute of Molecular and Cell Biology, Warsaw, Poland) for providing Fig. 3.
This work was supported, in whole or in part by the National Institutes of Health Intramural Research Program of the National Cancer Institute, Department of Health and Human Services. This is the second article in the Thematic Minireview Series on Understanding Human Immunodeficiency Virus-Host Interactions at the Biochemical Level.
S. Chung and S. F. Le Grice, unpublished data.
- RT
- reverse transcriptase
- PPT
- polypurine tract
- NNRTI
- non-nucleoside RT inhibitor
- SMS
- single-molecule spectroscopy
- NVP
- nevirapine
- NRTI
- nucleoside RT inhibitor
- TFV
- tenofovir
- INDOPY
- indolopyridone
- DPV
- dapivirine.
REFERENCES
- 1. Lavigne M., Roux P., Buc H., Schaeffer F. (1997) DNA curvature controls termination of plus strand DNA synthesis at the centre of HIV-1 genome. J. Mol. Biol. 266, 507–524 [DOI] [PubMed] [Google Scholar]
- 2. Lavigne M., Buc H. (1999) Compression of the DNA minor groove is responsible for termination of DNA synthesis by HIV-1 reverse transcriptase. J. Mol. Biol. 285, 977–995 [DOI] [PubMed] [Google Scholar]
- 3. Lavigne M., Polomack L., Buc H. (2001) DNA synthesis by HIV-1 reverse transcriptase at the central termination site. A kinetic study. J. Biol. Chem. 276, 31429–31438 [DOI] [PubMed] [Google Scholar]
- 4. Charneau P., Alizon M., Clavel F. (1992) A second origin of DNA plus-strand synthesis is required for optimal human immunodeficiency virus replication. J. Virol. 66, 2814–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Charneau P., Clavel F. (1991) A single-stranded gap in human immunodeficiency virus unintegrated linear DNA defined by a central copy of the polypurine tract. J. Virol. 65, 2415–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hungnes O., Tjotta E., Grinde B. (1991) The plus strand is discontinuous in a subpopulation of unintegrated HIV-1 DNA. Arch. Virol. 116, 133–141 [DOI] [PubMed] [Google Scholar]
- 7. Hungnes O., Tjøtta E., Grinde B. (1992) Mutations in the central polypurine tract of HIV-1 result in delayed replication. Virology 190, 440–442 [DOI] [PubMed] [Google Scholar]
- 8. Dvorin J. D., Bell P., Maul G. G., Yamashita M., Emerman M., Malim M. H. (2002) Reassessment of the roles of integrase and the central DNA flap in human immunodeficiency virus type 1 nuclear import. J. Virol. 76, 12087–12096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marsden M. D., Zack J. A. (2007) Human immunodeficiency virus bearing a disrupted central DNA flap is pathogenic in vivo. J. Virol. 81, 6146–6150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Charneau P., Mirambeau G., Roux P., Paulous S., Buc H., Clavel F. (1994) HIV-1 reverse transcription. A termination step at the center of the genome. J. Mol. Biol. 241, 651–662 [DOI] [PubMed] [Google Scholar]
- 11. De Rijck J., Debyser Z. (2006) The central DNA flap of the human immunodeficiency virus type 1 is important for viral replication. Biochem. Biophys. Res. Commun. 349, 1100–1110 [DOI] [PubMed] [Google Scholar]
- 12. Ao Z., Yao X., Cohen E. A. (2004) Assessment of the role of the central DNA flap in human immunodeficiency virus type 1 replication by using a single-cycle replication system. J. Virol. 78, 3170–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dardalhon V., Herpers B., Noraz N., Pflumio F., Guetard D., Leveau C., Dubart-Kupperschmitt A., Charneau P., Taylor N. (2001) Lentivirus-mediated gene transfer in primary T cells is enhanced by a central DNA flap. Gene Ther. 8, 190–198 [DOI] [PubMed] [Google Scholar]
- 14. Sirven A., Pflumio F., Zennou V., Titeux M., Vainchenker W., Coulombel L., Dubart-Kupperschmitt A., Charneau P. (2000) The human immunodeficiency virus type 1 central DNA flap is a crucial determinant for lentiviral vector nuclear import and gene transduction of human hematopoietic stem cells. Blood 96, 4103–4110 [PubMed] [Google Scholar]
- 15. Van Maele B., De Rijck J., De Clercq E., Debyser Z. (2003) Impact of the central polypurine tract on the kinetics of human immunodeficiency virus type 1 vector transduction. J. Virol. 77, 4685–4694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zennou V., Serguera C., Sarkis C., Colin P., Perret E., Mallet J., Charneau P. (2001) The HIV-1 DNA flap stimulates HIV vector-mediated cell transduction in the brain. Nat. Biotechnol. 19, 446–450 [DOI] [PubMed] [Google Scholar]
- 17. di Marzo Veronese F., Copeland T. D., DeVico A. L., Rahman R., Oroszlan S., Gallo R. C., Sarngadharan M. G. (1986) Characterization of highly immunogenic p66/p51 as the reverse transcriptase of HTLV-III/LAV. Science 231, 1289–1291 [DOI] [PubMed] [Google Scholar]
- 18. Lowe D. M., Aitken A., Bradley C., Darby G. K., Larder B. A., Powell K. L., Purifoy D. J., Tisdale M., Stammers D. K. (1988) HIV-1 reverse transcriptase: crystallization and analysis of domain structure by limited proteolysis. Biochemistry 27, 8884–8889 [DOI] [PubMed] [Google Scholar]
- 19. Kohlstaedt L. A., Wang J., Friedman J. M., Rice P. A., Steitz T. A. (1992) Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 256, 1783–1790 [DOI] [PubMed] [Google Scholar]
- 20. Jacobo-Molina A., Ding J., Nanni R. G., Clark A. D., Jr., Lu X., Tantillo C., Williams R. L., Kamer G., Ferris A. L., Clark P., Hizi A., Hughesi S. H., Arnold E. (1993) Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc. Natl. Acad. Sci. U.S.A. 90, 6320–6324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang J., Smerdon S. J., Jäger J., Kohlstaedt L. A., Rice P. A., Friedman J. M., Steitz T. A. (1994) Structural basis of asymmetry in the human immunodeficiency virus type 1 reverse transcriptase heterodimer. Proc. Natl. Acad. Sci. U.S.A. 91, 7242–7246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Le Grice S. F., Naas T., Wohlgensinger B., Schatz O. (1991) Subunit-selective mutagenesis indicates minimal polymerase activity in heterodimer-associated p51 HIV-1 reverse transcriptase. EMBO J. 10, 3905–3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Amacker M., Hübscher U. (1998) Chimeric HIV-1 and feline immunodeficiency virus reverse transcriptases: critical role of the p51 subunit in the structural integrity of heterodimeric lentiviral DNA polymerases. J. Mol. Biol. 278, 757–765 [DOI] [PubMed] [Google Scholar]
- 24. Harris D., Lee R., Misra H. S., Pandey P. K., Pandey V. N. (1998) The p51 subunit of human immunodeficiency virus type 1 reverse transcriptase is essential in loading the p66 subunit on the template primer. Biochemistry 37, 5903–5908 [DOI] [PubMed] [Google Scholar]
- 25. Jacques P. S., Wöhrl B. M., Howard K. J., Le Grice S. F. (1994) Modulation of HIV-1 reverse transcriptase function in “selectively deleted” p66/p51 heterodimers. J. Biol. Chem. 269, 1388–1393 [PubMed] [Google Scholar]
- 26. Arts E. J., Ghosh M., Jacques P. S., Ehresmann B., Le Grice S. F. (1996) Restoration of tRNA3Lys-primed (−)-strand DNA synthesis to an HIV-1 reverse transcriptase mutant with extended tRNAs. Implications for retroviral replication. J. Biol. Chem. 271, 9054–9061 [DOI] [PubMed] [Google Scholar]
- 27. Bahar I., Erman B., Jernigan R. L., Atilgan A. R., Covell D. G. (1999) Collective motions in HIV-1 reverse transcriptase: examination of flexibility and enzyme function. J. Mol. Biol. 285, 1023–1037 [DOI] [PubMed] [Google Scholar]
- 28. Chung S., Miller J. T., Johnson B. C., Hughes S. H., Le Grice S. F. (2012) Mutagenesis of human immunodeficiency virus reverse transcriptase p51 subunit defines residues contributing to vinylogous urea inhibition of ribonuclease H activity. J. Biol. Chem. 287, 4066–4075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ding J., Das K., Hsiou Y., Sarafianos S. G., Clark A. D., Jr., Jacobo-Molina A., Tantillo C., Hughes S. H., Arnold E. (1998) Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 Å resolution. J. Mol. Biol. 284, 1095–1111 [DOI] [PubMed] [Google Scholar]
- 30. Huang H., Chopra R., Verdine G. L., Harrison S. C. (1998) Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282, 1669–1675 [DOI] [PubMed] [Google Scholar]
- 31. Sarafianos S. G., Das K., Tantillo C., Clark A. D., Jr., Ding J., Whitcomb J. M., Boyer P. L., Hughes S. H., Arnold E. (2001) Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 20, 1449–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Carvalho A. T., Fernandes P. A., Ramos M. J. (2006) Molecular dynamics model of unliganded HIV-1 reverse transcriptase. Med. Chem. 2, 491–498 [DOI] [PubMed] [Google Scholar]
- 33. Kensch O., Restle T., Wöhrl B. M., Goody R. S., Steinhoff H. J. (2000) Temperature-dependent equilibrium between the open and closed conformation of the p66 subunit of HIV-1 reverse transcriptase revealed by site-directed spin labelling. J. Mol. Biol. 301, 1029–1039 [DOI] [PubMed] [Google Scholar]
- 34. Rodgers D. W., Gamblin S. J., Harris B. A., Ray S., Culp J. S., Hellmig B., Woolf D. J., Debouck C., Harrison S. C. (1995) The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. U.S.A. 92, 1222–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hsiou Y., Ding J., Das K., Clark A. D., Jr., Hughes S. H., Arnold E. (1996) Structure of unliganded HIV-1 reverse transcriptase at 2.7 Å resolution: implications of conformational changes for polymerization and inhibition mechanisms. Structure 4, 853–860 [DOI] [PubMed] [Google Scholar]
- 36. Beard W. A., Stahl S. J., Kim H. R., Bebenek K., Kumar A., Strub M. P., Becerra S. P., Kunkel T. A., Wilson S. H. (1994) Structure/function studies of human immunodeficiency virus type 1 reverse transcriptase. Alanine scanning mutagenesis of an α-helix in the thumb subdomain. J. Biol. Chem. 269, 28091–28097 [PubMed] [Google Scholar]
- 37. Bebenek K., Beard W. A., Casas-Finet J. R., Kim H. R., Darden T. A., Wilson S. H., Kunkel T. A. (1995) Reduced frameshift fidelity and processivity of HIV-1 reverse transcriptase mutants containing alanine substitutions in helix H of the thumb subdomain. J. Biol. Chem. 270, 19516–19523 [DOI] [PubMed] [Google Scholar]
- 38. Bebenek K., Beard W. A., Darden T. A., Li L., Prasad R., Luton B. A., Gorenstein D. G., Wilson S. H., Kunkel T. A. (1997) A minor groove binding track in reverse transcriptase. Nat. Struct. Biol. 4, 194–197 [DOI] [PubMed] [Google Scholar]
- 39. Xiong Y., Eickbush T. H. (1990) Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 9, 3353–3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wisniewski M., Palaniappan C., Fu Z., Le Grice S. F., Fay P., Bambara R. A. (1999) Mutations in the primer grip region of HIV reverse transcriptase can increase replication fidelity. J. Biol. Chem. 274, 28175–28184 [DOI] [PubMed] [Google Scholar]
- 41. Wöhrl B. M., Krebs R., Thrall S. H., Le Grice S. F., Scheidig A. J., Goody R. S. (1997) Kinetic analysis of four HIV-1 reverse transcriptase enzymes mutated in the primer grip region of p66. Implications for DNA synthesis and dimerization. J. Biol. Chem. 272, 17581–17587 [DOI] [PubMed] [Google Scholar]
- 42. Powell M. D., Ghosh M., Jacques P. S., Howard K. J., Le Grice S. F., Levin J. G. (1997) Alanine-scanning mutations in the “primer grip” of p66 HIV-1 reverse transcriptase result in selective loss of RNA priming activity. J. Biol. Chem. 272, 13262–13269 [DOI] [PubMed] [Google Scholar]
- 43. Palaniappan C., Wisniewski M., Jacques P. S., Le Grice S. F., Fay P. J., Bambara R. A. (1997) Mutations within the primer grip region of HIV-1 reverse transcriptase result in loss of RNase H function. J. Biol. Chem. 272, 11157–11164 [DOI] [PubMed] [Google Scholar]
- 44. Ghosh M., Williams J., Powell M. D., Levin J. G., Le Grice S. F. (1997) Mutating a conserved motif of the HIV-1 reverse transcriptase palm subdomain alters primer utilization. Biochemistry 36, 5758–5768 [DOI] [PubMed] [Google Scholar]
- 45. Ghosh M., Jacques P. S., Rodgers D. W., Ottman M., Darlix J. L., Le Grice S. F. (1996) Alterations to the primer grip of p66 HIV-1 reverse transcriptase and their consequences for template-primer utilization. Biochemistry 35, 8553–8562 [DOI] [PubMed] [Google Scholar]
- 46. Jacques P. S., Wöhrl B. M., Ottmann M., Darlix J. L., Le Grice S. F. (1994) Mutating the “primer grip” of p66 HIV-1 reverse transcriptase implicates tryptophan 229 in template-primer utilization. J. Biol. Chem. 269, 26472–26478 [PubMed] [Google Scholar]
- 47. Brautigam C. A., Steitz T. A. (1998) Structural and functional insights provided by crystal structures of DNA polymerases and their substrate complexes. Curr. Opin. Struct. Biol. 8, 54–63 [DOI] [PubMed] [Google Scholar]
- 48. Esnouf R., Ren J., Ross C., Jones Y., Stammers D., Stuart D. (1995) Mechanism of inhibition of HIV-1 reverse transcriptase by non-nucleoside inhibitors. Nat. Struct. Biol. 2, 303–308 [DOI] [PubMed] [Google Scholar]
- 49. Garforth S. J., Kim T. W., Parniak M. A., Kool E. T., Prasad V. R. (2007) Site-directed mutagenesis in the fingers subdomain of HIV-1 reverse transcriptase reveals a specific role for the β3-β4 hairpin loop in dNTP selection. J. Mol. Biol. 365, 38–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Martín-Hernández A. M., Domingo E., Menéndez-Arias L. (1996) Human immunodeficiency virus type 1 reverse transcriptase: role of Tyr115 in deoxynucleotide binding and misinsertion fidelity of DNA synthesis. EMBO J. 15, 4434–4442 [PMC free article] [PubMed] [Google Scholar]
- 51. Cases-Gonzalez C. E., Gutierrez-Rivas M., Menéndez-Arias L. (2000) Coupling ribose selection to fidelity of DNA synthesis. The role of Tyr-115 of human immunodeficiency virus type 1 reverse transcriptase. J. Biol. Chem. 275, 19759–19767 [DOI] [PubMed] [Google Scholar]
- 52. Cowan J. A., Ohyama T., Howard K., Rausch J. W., Cowan S. M., Le Grice S. F. (2000) Metal-ion stoichiometry of the HIV-1 RT ribonuclease H domain: evidence for two mutually exclusive sites leads to new mechanistic insights on metal-mediated hydrolysis in nucleic acid biochemistry. J. Biol. Inorg. Chem. 5, 67–74 [DOI] [PubMed] [Google Scholar]
- 53. Steitz T. A., Steitz J. A. (1993) A general two-metal-ion mechanism for catalytic RNA. Proc. Natl. Acad. Sci. U.S.A. 90, 6498–6502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nowotny M., Gaidamakov S. A., Crouch R. J., Yang W. (2005) Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell 121, 1005–1016 [DOI] [PubMed] [Google Scholar]
- 55. Nowotny M., Yang W. (2006) Stepwise analyses of metal ions in RNase H catalysis from substrate destabilization to product release. EMBO J. 25, 1924–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nowotny M., Gaidamakov S. A., Ghirlando R., Cerritelli S. M., Crouch R. J., Yang W. (2007) Structure of human RNase H1 complexed with an RNA/DNA hybrid: insight into HIV reverse transcription. Mol. Cell 28, 264–276 [DOI] [PubMed] [Google Scholar]
- 57. Yang W., Lee J. Y., Nowotny M. (2006) Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell 22, 5–13 [DOI] [PubMed] [Google Scholar]
- 58. Julias J. G., McWilliams M. J., Sarafianos S. G., Alvord W. G., Arnold E., Hughes S. H. (2003) Mutation of amino acids in the connection domain of human immunodeficiency virus type 1 reverse transcriptase that contact the template-primer affects RNase H activity. J. Virol. 77, 8548–8554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rausch J. W., Lener D., Miller J. T., Julias J. G., Hughes S. H., Le Grice S. F. (2002) Altering the RNase H primer grip of human immunodeficiency virus reverse transcriptase modifies cleavage specificity. Biochemistry 41, 4856–4865 [DOI] [PubMed] [Google Scholar]
- 60. Beilhartz G. L., Wendeler M., Baichoo N., Rausch J., Le Grice S., Götte M. (2009) HIV-1 reverse transcriptase can simultaneously engage its DNA/RNA substrate at both DNA polymerase and RNase H active sites: implications for RNase H inhibition. J. Mol. Biol. 388, 462–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Himmel D. M., Sarafianos S. G., Dharmasena S., Hossain M. M., McCoy-Simandle K., Ilina T., Clark A. D., Jr., Knight J. L., Julias J. G., Clark P. K., Krogh-Jespersen K., Levy R. M., Hughes S. H., Parniak M. A., Arnold E. (2006) HIV-1 reverse transcriptase structure with RNase H inhibitor dihydroxy benzoyl naphthyl hydrazone bound at a novel site. ACS Chem. Biol. 1, 702–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chung S., Himmel D. M., Jiang J. K., Wojtak K., Bauman J. D., Rausch J. W., Wilson J. A., Beutler J. A., Thomas C. J., Arnold E., Le Grice S. F. (2011) Synthesis, activity, and structural analysis of novel α-hydroxytropolone inhibitors of human immunodeficiency virus reverse transcriptase-associated ribonuclease H. J. Med. Chem. 54, 4462–4473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lansdon E. B., Liu Q., Leavitt S. A., Balakrishnan M., Perry J. K., Lancaster-Moyer C., Kutty N., Liu X., Squires N. H., Watkins W. J., Kirschberg T. A. (2011) Structural and binding analysis of pyrimidinol carboxylic acid and N-hydroxy quinazolinedione HIV-1 RNase H inhibitors. Antimicrob. Agents Chemother. 55, 2905–2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Su H. P., Yan Y., Prasad G. S., Smith R. F., Daniels C. L., Abeywickrema P. D., Reid J. C., Loughran H. M., Kornienko M., Sharma S., Grobler J. A., Xu B., Sardana V., Allison T. J., Williams P. D., Darke P. L., Hazuda D. J., Munshi S. (2010) Structural basis for the inhibition of RNase H activity of HIV-1 reverse transcriptase by RNase H active site-directed inhibitors. J. Virol. 84, 7625–7633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Abbondanzieri E. A., Bokinsky G., Rausch J. W., Zhang J. X., Le Grice S. F., Zhuang X. (2008) Dynamic binding orientations direct activity of HIV reverse transcriptase. Nature 453, 184–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Grobler J. A., Dornadula G., Rice M. R., Simcoe A. L., Hazuda D. J., Miller M. D. (2007) HIV-1 reverse transcriptase plus-strand initiation exhibits preferential sensitivity to non-nucleoside reverse transcriptase inhibitors in vitro. J. Biol. Chem. 282, 8005–8010 [DOI] [PubMed] [Google Scholar]
- 67. Liu S., Abbondanzieri E. A., Rausch J. W., Le Grice S. F., Zhuang X. (2008) Slide into action: dynamic shuttling of HIV reverse transcriptase on nucleic acid substrates. Science 322, 1092–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Liu S., Harada B. T., Miller J. T., Le Grice S. F., Zhuang X. (2010) Initiation complex dynamics direct the transitions between distinct phases of early HIV reverse transcription. Nat. Struct. Mol. Biol. 17, 1453–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mehellou Y., De Clercq E. (2010) Twenty-six years of anti-HIV drug discovery: where do we stand and where do we go? J. Med. Chem. 53, 521–538 [DOI] [PubMed] [Google Scholar]
- 70. Morningstar M. L., Roth T., Farnsworth D. W., Smith M. K., Watson K., Buckheit R. W., Jr., Das K., Zhang W., Arnold E., Julias J. G., Hughes S. H., Michejda C. J. (2007) Synthesis, biological activity, and crystal structure of potent nonnucleoside inhibitors of HIV-1 reverse transcriptase that retain activity against mutant forms of the enzyme. J. Med. Chem. 50, 4003–4015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ehteshami M., Scarth B. J., Tchesnokov E. P., Dash C., Le Grice S. F., Hallenberger S., Jochmans D., Götte M. (2008) Mutations M184V and Y115F in HIV-1 reverse transcriptase discriminate against “nucleotide-competing reverse transcriptase inhibitors.” J. Biol. Chem. 283, 29904–29911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jochmans D., Deval J., Kesteleyn B., Van Marck H., Bettens E., De Baere I., Dehertogh P., Ivens T., Van Ginderen M., Van Schoubroeck B., Ehteshami M., Wigerinck P., Götte M., Hertogs K. (2006) Indolopyridones inhibit human immunodeficiency virus reverse transcriptase with a novel mechanism of action. J. Virol. 80, 12283–12292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Johnson V. A., Brun-Vézinet F., Clotet B., Günthard H. F., Kuritzkes D. R., Pillay D., Schapiro J. M., Richman D. D. (2010) Update of the drug resistance mutations in HIV-1: December 2010. Top. HIV Med. 18, 156–163 [PubMed] [Google Scholar]
- 74. Tisdale M., Schulze T., Larder B. A., Moelling K. (1991) Mutations within the RNase H domain of human immunodeficiency virus type 1 reverse transcriptase abolish virus infectivity. J. Gen. Virol. 72, 59–66 [DOI] [PubMed] [Google Scholar]
- 75. Cerritelli S. M., Frolova E. G., Feng C., Grinberg A., Love P. E., Crouch R. J. (2003) Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol. Cell 11, 807–815 [DOI] [PubMed] [Google Scholar]
- 76. Klumpp K., Mirzadegan T. (2006) Recent progress in the design of small molecule inhibitors of HIV RNase H. Curr. Pharm. Des. 12, 1909–1922 [DOI] [PubMed] [Google Scholar]
- 77. Parkes K. E., Ermert P., Fässler J., Ives J., Martin J. A., Merrett J. H., Obrecht D., Williams G., Klumpp K. (2003) Use of a pharmacophore model to discover a new class of influenza endonuclease inhibitors. J. Med. Chem. 46, 1153–1164 [DOI] [PubMed] [Google Scholar]
- 78. Doan L., Handa B., Roberts N. A., Klumpp K. (1999) Metal ion catalysis of RNA cleavage by the influenza virus endonuclease. Biochemistry 38, 5612–5619 [DOI] [PubMed] [Google Scholar]
- 79. Budihas S. R., Gorshkova I., Gaidamakov S., Wamiru A., Bona M. K., Parniak M. A., Crouch R. J., McMahon J. B., Beutler J. A., Le Grice S. F. (2005) Selective inhibition of HIV-1 reverse transcriptase-associated ribonuclease H activity by hydroxylated tropolones. Nucleic Acids Res. 33, 1249–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Grobler J. A., Stillmock K., Hu B., Witmer M., Felock P., Espeseth A. S., Wolfe A., Egbertson M., Bourgeois M., Melamed J., Wai J. S., Young S., Vacca J., Hazuda D. J. (2002) Diketo acid inhibitor mechanism and HIV-1 integrase: implications for metal binding in the active site of phosphotransferase enzymes. Proc. Natl. Acad. Sci. U.S.A. 99, 6661–6666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hazuda D. J., Felock P., Witmer M., Wolfe A., Stillmock K., Grobler J. A., Espeseth A., Gabryelski L., Schleif W., Blau C., Miller M. D. (2000) Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287, 646–650 [DOI] [PubMed] [Google Scholar]
- 82. Tramontano E., Esposito F., Badas R., Di Santo R., Costi R., La Colla P. (2005) 6-[1-(4-Fluorophenyl)methyl-1H-pyrrol-2-yl]-2,4-dioxo-5-hexenoic acid ethyl ester, a novel diketo acid derivative that selectively inhibits the HIV-1 viral replication in cell culture and the ribonuclease H activity in vitro. Antiviral Res. 65, 117–124 [DOI] [PubMed] [Google Scholar]
- 83.Deleted in proof
- 84. Wendeler M., Lee H. F., Bermingham A., Miller J. T., Chertov O., Bona M. K., Baichoo N. S., Ehteshami M., Beutler J., O'Keefe B. R., Götte M., Kvaratskhelia M., Le Grice S. (2008) Vinylogous ureas as a novel class of inhibitors of reverse transcriptase-associated ribonuclease H activity. ACS Chem. Biol. 3, 635–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Chung S., Wendeler M., Rausch J. W., Beilhartz G., Gotte M., O'Keefe B. R., Bermingham A., Beutler J. A., Liu S., Zhuang X., Le Grice S. F. (2010) Structure-activity analysis of vinylogous urea inhibitors of human immunodeficiency virus-encoded ribonuclease H. Antimicrob. Agents Chemother. 54, 3913–3921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Srivastava S., Sluis-Cremer N., Tachedjian G. (2006) Dimerization of human immunodeficiency virus type 1 reverse transcriptase as an antiviral target. Curr. Pharm. Des. 12, 1879–1894 [DOI] [PubMed] [Google Scholar]
- 87. Tramontano E., Di Santo R. (2010) HIV-1 RT-associated RNase H function inhibitors: recent advances in drug development. Curr. Med. Chem. 17, 2837–2853 [DOI] [PubMed] [Google Scholar]
- 88. Lewi P., Heeres J., Ariën K., Venkatraj M., Joossens J., Van der Veken P., Augustyns K., Vanham G. (2012) Reverse transcriptase inhibitors as microbicides. Curr. HIV Res. 10, 27–35 [DOI] [PubMed] [Google Scholar]
- 89. Himmel D. M., Maegley K. A., Pauly T. A., Bauman J. D., Das K., Dharia C., Clark A. D., Jr., Ryan K., Hickey M. J., Love R. A., Hughes S. H., Bergqvist S., Arnold E. (2009) Structure of HIV-1 reverse transcriptase with the inhibitor β-thujaplicinol bound at the RNase H active site. Structure 17, 1625–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]