Abstract
Retroviral integrases catalyze two reactions, 3′-processing of viral DNA ends, followed by integration of the processed ends into chromosomal DNA. X-ray crystal structures of integrase-DNA complexes from prototype foamy virus, a member of the Spumavirus genus of Retroviridae, have revealed the structural basis of integration and how clinically relevant integrase strand transfer inhibitors work. Underscoring the translational potential of targeting virus-host interactions, small molecules that bind at the host factor lens epithelium-derived growth factor/p75-binding site on HIV-1 integrase promote dimerization and inhibit integrase-viral DNA assembly and catalysis. Here, we review recent advances in our knowledge of HIV-1 DNA integration, as well as future research directions.
Keywords: AIDS, DNA Recombination, HIV-1, Integrase, Retrovirus
Introduction
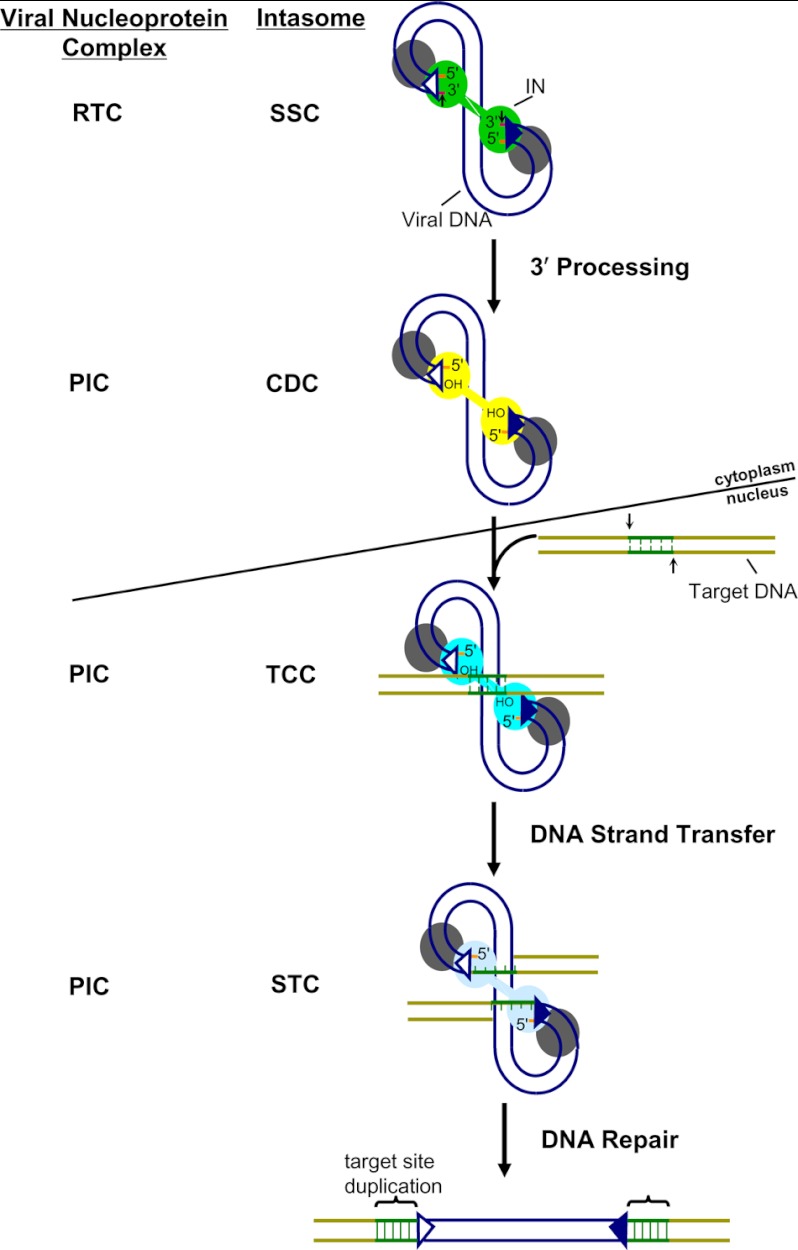
Retroviral replication proceeds through an obligate proviral or integrated DNA recombination intermediate. Integration provides a favorable environment for efficient gene expression, ensures inheritance of the virus in both daughter cells during mitosis, and forms the basis for latent HIV-1 reservoirs that persist in the face of highly active antiretroviral therapy. The early events of retroviral replication take place within the context of nucleoprotein complexes that are derived from the core of the infecting virus particle (1, 2). Within the confines of the reverse transcription complex (RTC),2 the reverse transcriptase enzyme copies single-stranded viral RNA into a linear double-stranded DNA molecule containing a copy of the LTR sequence at each end. The viral integrase (IN) engages the LTR ends prior to catalyzing two spatially and temporally distinct chemical reactions. Soon after their synthesis (3), IN site-specifically processes each LTR end adjacent to an invariant CA dinucleotide (4, 5), yielding CAOH 3′-hydroxyl groups that serve as the nucleophiles for the second reaction, DNA strand transfer. Because the viral replication intermediate at this point gains the ability to catalyze DNA strand transfer activity, 3′-processing operationally marks the transition of the RTC to the pre-integration complex (PIC) (Fig. 1). In the strand transfer step, IN utilizes the 3′-oxygen atoms to cut chromosomal DNA in a staggered fashion and simultaneously join the viral DNA ends to the 5′-phosphates of the target DNA (4, 6, 7). The resulting DNA recombination intermediate, with unjoined viral DNA 5′-ends, is repaired by host cell enzymes to yield the integrated provirus flanked by the duplication of the sequence of the staggered DNA cut (Fig. 1). The spumaviruses, which compose one of seven Retroviridae genera, are an exception to this generalized scheme, as DNA synthesis is largely complete prior to target cell infection (8). Research on the functionality of the active nucleoprotein complexes that mediate HIV-1 DNA integration has led to the development of antiviral inhibitors that target the IN and block integration.
FIGURE 1.
Retroviral nucleoprotein complexes and IN-mediated DNA cutting and joining reactions. The SSC substructure of the viral RTC forms when IN engages the LTR ends (U3 end of the upstream LTR (open triangle) and U5 end of the downstream LTR (closed triangle)) of the newly synthesized viral DNA. 3′-Processing (scissile phosphodiester bonds marked by short vertical arrows), which can occur in the cell cytoplasm (3), converts the RTC to the PIC, yielding the CDC with activated CAOH 3′ termini. The TCC is formed in the nucleus when the PIC engages chromosomal target DNA (dark green region delineates the 5-bp stagger cut made by HIV-1 IN). Integration of the viral LTR 3′-ends yields the STC, with viral DNA 5′-ends unjoined. Repair of the DNA recombination intermediate, which is mediated by host cell enzymes and likely requires active proteolysis of PIC components, yields the integrated provirus flanked by the duplication of the target DNA cut. Two of the four IN monomers within each complex are color-coded to match the inner monomers of the respective PFV intasome crystal structures as shown in Fig. 2.
IN Domain Structure and Reaction Mechanism
Retroviral INs belong to a superfamily of proteins known as the retroviral IN superfamily, which contains other nucleic acid-metabolizing enzymes such as RNase H, RuvC, bacteriophage MuA transposase, and the nuclease component of the RNA-induced silencing complex Argonaute (9). Common features of these enzymes are the RNase H fold adopted by their catalytic domains and active sites composed of electronegative Asp and Glu side chains (9, 10). 3′-Processing and DNA strand transfer are in-line bimolecular nucleophilic substitution (SN2) reactions (7), and the catalytic site residues coordinate the positions of two Mg2+ ions to activate the attacking nucleophile (the oxygen atom of a water molecule for 3′-processing and the 3′-hydroxyl of viral DNA for strand transfer) and to destabilize the scissile phosphodiester bonds (Figs. 1 and 2) (11–13). Sequence database analysis has indicated the presence of a much larger DD(E/D) superfamily named for catalytic triad amino acid signatures, although in many cases, verification of active site domain RNase H folds and functional active site residues awaits validation (14).
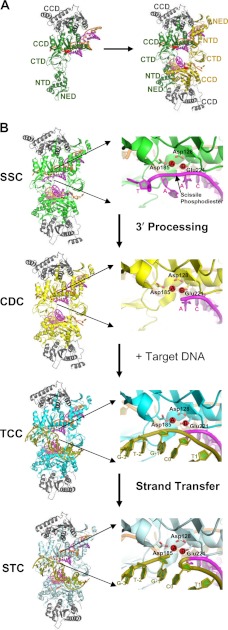
FIGURE 2.
PFV intasome structures. A, left, crystallographic asymmetric unit with the IN dimer bound to a single viral DNA end (Protein Data Bank (PDB) code 3OY9) (18, 68) and different IN domains labeled. The inner monomer is colored green, the outer monomer gray, and the DNA strands magenta (the transferred or to-be-integrated strand) and orange (the non-transferred strand). Right, two dimers come together to form the functional CDC (with the second inner monomer shown in gold). CTD, C-terminal domain; NED, N-terminal extension domain. B, left, structures of the PFV intasome complexes along the integration pathway (SSC, PDB code 4E7I; CDC, same as in A; TCC, PDB code 4E7K; and STC, PDB code 4E7L) (13). The coloring of the inner monomers matches the analogous complexes in Fig. 1. Right, enlargements of the active site of the upper inner monomer to the left, with Mn2+ ions A and B and the oxygen atoms of active site residues Asp-128, Asp-185, and Glu-221 colored red and the target DNA strand colored olive green (other coloring as described for A). The scissile viral DNA phosphodiester adjacent to the invariant CA dinucleotide and terminal residues of the transferred LTR strand in the active site regions of the SSC and CDC are labeled. The labeling of target DNA nucleotides in the TCC and STC active sites indicates the utilized oligonucleotide sequence and position of DNA strand transfer (26). Metal ion A in the SSC activates a water molecule to process the CA/AT phosphodiester bond, whereas metal ion B in the TCC activates the resulting CAOH 3′-hydroxyl for strand transfer into target DNA (13).
All retroviral IN proteins share three common domains, the N-terminal domain (NTD), catalytic core domain (CCD), and C-terminal domain (for a recent review on domains and IN structure, see Ref. 15). As mentioned, the CCD adopts an RNase H fold, harboring the D,DX35E catalytic triad common to all retroviral and retrotransposon INs, as well as certain bacterial transposase proteins (10, 16, 17). The NTD adopts a helix-turn-helix fold, utilizing conserved His and Cys residues to bind a single Zn2+ ion, whereas the positively charged C-terminal domain adopts an SH3 (Src homology 3) fold. Crystal structures of the prototype foamy virus (PFV) IN in complex with DNA revealed a fourth domain upstream from the NTD, termed the N-terminal extension domain (Fig. 2A) (18). Based on amino sequence alignment, the IN proteins from two additional genera, the gammaretroviruses and epsilonretroviruses, are predicted to harbor N-terminal extension domains.
Retroviral IN-DNA Nucleoprotein Complexes
The identification of specific nucleoprotein complexes that mediate the transposition of mobile DNA elements such as phage Mu and Tn10 (19) predated analogous work with retroviruses, so it is convenient to adopt DNA transposon terminology to describe the salient retroviral complexes. The stable synaptic complex (SSC) is formed upon IN binding to the LTR DNA ends, and 3′-processing yields the cleaved donor complex (CDC; donor refers to mobile element DNA, which, for retroviruses, is the viral DNA). Engagement of chromatin acceptor or target DNA by the CDC in the cell nucleus yields the target capture complex (TCC), and integration of the viral DNA 3′-ends yields the strand transfer complex (STC) (Fig. 1). The term “intasome,” which was originally used to describe the higher order nucleoprotein complex between bacteriophage λ integrase (Int; note phage λ and retroviruses integrate through fundamentally different mechanisms) (20) and λ substrate DNA (21), was first adopted by the retrovirus field to describe an assembly responsible for the approximate 250-bp functional footprints at the LTR end regions of Moloney murine leukemia virus and HIV-1 PICs (22, 23). The term has subsequently been used to describe the heart of the DNA recombination machine, composed of a tetramer of IN protein and two viral DNA ends (Fig. 2A, right) (18). The SSC, CDC, TCC, and STC accordingly represent different stages of the retroviral intasome that correspond to the distinguishable nucleoprotein complexes along the integration pathway (Figs. 1 and 2). The relationship between the relatively large footprints observed in HIV-1 PICs and smaller ∼16–32-bp regions of protection observed with purified IN and substrate DNA in vitro (24, 25) is unknown, although it seems likely that PIC-associated viral and/or cellular factors could impact the breadth of DNA protection.
X-ray crystal structures of functional PFV intasomes have yielded unprecedented details on the mechanism of retroviral DNA integration (13, 18, 26). The intasome is composed of a dimer of IN dimers, with each monomer within the dimer playing a distinct role (Fig. 2A). The inner monomer (colored green in Fig. 2A, left) adopts an extended conformation and makes all contacts with the viral LTR end. Only the CCD of the outer monomer is resolved in the electron density maps (18). The molecules within each dimer interact through an extensive CCD-CCD interface that is repeatedly observed in structures of one- and two-domain IN constructs in the absence of DNA (reviewed in Ref. 15). Consistent with results of in vitro integration assays with recombinant IN protein (27–30) and crystal structures of a lentiviral IN two-domain NTD-CCD construct (31), the NTD of each inner monomer engages the CCD of the opposed inner monomer with its bound viral DNA in trans (Fig. 2A, right) (18). Such trans-configurations, which can be found throughout the retroviral IN superfamily (32, 33), as well as in other DNA recombination systems (34), help to ensure coordinated activity by a multimer of recombinase protein on a pair of participating DNA strands. An outstanding question with the PFV structure is the role of the “missing” domains of the outer IN molecules. The visualized domains of the tetramer remain largely unchanged as the SSC progresses to the STC (Fig. 2B) (13, 26). Because the outer IN domains remain unresolved during this transition (Fig. 2B), it seems they are unlikely to play direct roles in DNA recombination and are more likely to help support the architecture of the inner pair of workhorse monomers.
Recombinant PFV IN was amenable to intasome structural biology due to its inherent solubility in relatively low ionic strength buffer and its favorable DNA recombination properties (18, 35, 36). The enzyme importantly integrates relatively short (≥16 bp) DNA oligonucleotide mimics of the U5 LTR end into both strands of target DNA separated by 4 bp, the spacing of the target site duplication observed during PFV infection (36). By contrast, HIV-1 IN is poorly soluble under conditions of limited ionic strength and struggles to integrate two LTR ends into target DNA in concerted fashion (37), characteristics that have severely limited its utility in intasome structural biology. Various parameters can increase the efficiency of concerted integration catalyzed by HIV-1 IN, including the length of the recombinant LTR substrate (38) or various IN- or DNA-binding cofactors such as lens epithelium-derived growth factor (LEDGF)/p75 (39) and HIV-1 nucleocapsid (40), respectively. Purification of HIV-1 IN under conditions that suppressed protein multimerization yielded monomers that supported efficient concerted integration activity of oligonucleotide substrate DNA (41), suggesting one possible way to improve the enzyme for intasome structural biology. LEDGF/p75 binding increases both the activity (39, 42, 43) and solubility (44) of HIV-1 IN, and a higher order complex comprising HIV-1 IN, LEDGF/p75, and 21-bp U5 LTR DNA was solved to ∼14–17 Å resolution by cryo-negative staining electron microscopy (43). The x-ray structure of the PFV intasome (18) has more recently spurred numerous attempts to model analogous HIV-1 structures (45–47).
Results of chemical cross-linking (48), solution biochemistry (24, 25, 49, 50), and structure-based approaches (18, 43, 51) confirm that a tetramer of IN catalyzes DNA strand transfer activity, although somewhat less clear is the multimeric nature of the 3′-processing protagonist. Although a dimer of IN may suffice to process an individual LTR end in vitro (48, 52), some substitutions that affect the ability of HIV-1 IN to tetramerize compromise 3′-processing activity severely (31). Furthermore, the PFV IN tetramer efficiently processed a pair of LTR ends in crystallo (13). Consistent with the observation that the NTD works in trans with the CCD to catalyze 3′-processing activity (27, 28), Li et al. (24) demonstrated that a single HIV-1 DNA end can moreover be processed and integrated in the context of the intasomal tetramer in vitro. Such asymmetry may very well account for the different rates at which the U3 versus U5 ends of the HIV-1 LTR are processed during acute infection (3), as well as for the reasonable titers of single LTR end mutant viruses (53, 54), where IN-mediated integration of the wild-type end presumably templates subsequent host-mediated integration of the mutant viral DNA end (55, 56). Skalka and co-workers (57) have interestingly reported a novel “reaching dimer” structure for avian sarcoma virus IN protein based on small-angle x-ray scattering and chemical cross-linking that mimics the extended inner monomers of the PFV intasome. Although missing the canonical CCD-CCD interface, the reaching dimer nevertheless seemingly affords IN domain cis/trans-relationships consistent with solution measures of 3′-processing activity (27, 28). Additional work is required to determine whether the reaching dimer plays a physiological relevant role in catalyzing 3′-processing activity or in the assembly of the functional IN tetramer.
IN Strand Transfer Inhibitors
The establishment of DNA oligonucleotide-based HIV-1 IN 3′-processing and DNA strand transfer assays (37, 58) led to multiwell plate formats amenable for small-molecule library screening (59–61). As alluded to above, catalysis under these conditions occurs in large aggregate assemblies more so than discrete soluble complexes (35, 62). Early hits included polyanions, which could disrupt the binding of positively charged HIV-1 IN to DNA in a nonspecific manner and accordingly failed to effectively inhibit integration during HIV-1 infection or the activity of PICs extracted from infected cells (63). Reconfiguring the assay design to more faithfully recapitulate PIC biology by querying DNA strand transfer activity after prebinding IN to preprocessed LTR DNA (64) in large part overcame this undesirable outcome, leading to the identification of diketo acid (DKA) compounds that specifically inhibited strand transfer activity in vitro and during HIV-1 infection (65). Likely due to similar two-metal ion-dependent active sites, DKAs are also potent hepatitis C virus RNA polymerase inhibitors, and raltegravir (RAL), a pyrimidinone carboxamide, came from utilizing both enzymes in DKA optimization trials (66). RAL, the sole licensed HIV-1 IN inhibitor to date, has become a mainstay antiviral agent since its introduction into the clinic in October 2007.
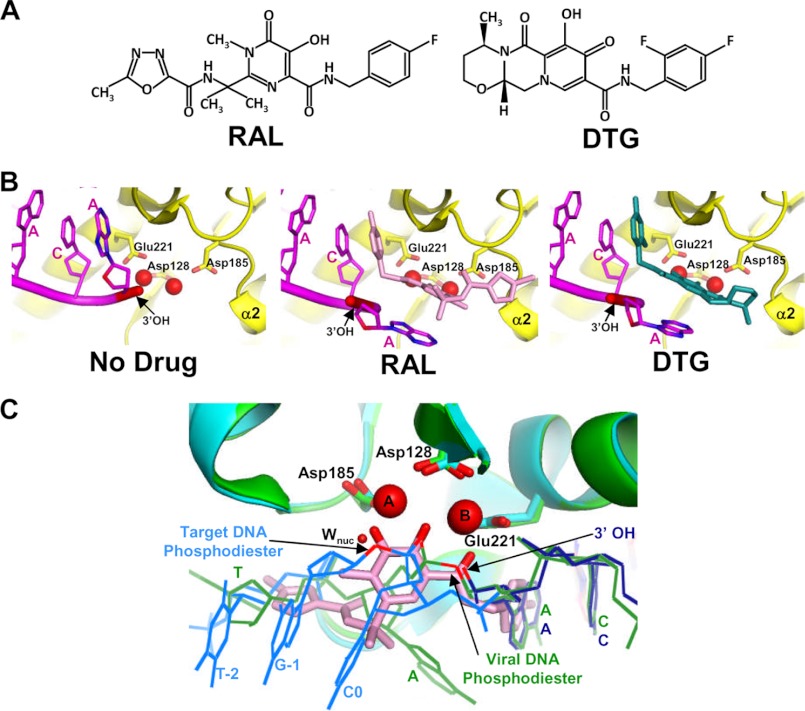
Potent IN strand transfer inhibitors (INSTIs) harbor two critical chemical moieties: heteroatom clusters reminiscent of the oxygen-based DKA pharmacophore and a halobenzyl side chain (Fig. 3A). The similar chemical nature of amino acid carboxylate side chains and DKAs led to the suggestion that the drug moieties engage bound metal ion at the IN active site (67), which was subsequently verified through co-crystallization of INSTIs with the PFV CDC (18, 68, 69). The crystal structures also revealed the role of the halobenzyl groups, which is to supplant the adenosine ring of the terminal deoxyadenylate residue at the processed LTR end and concordantly eject the nucleotide with its associated 3′-OH strand transfer nucleophile from the enzyme active site (Fig. 3B) (18, 68, 69). INSTI engagement of the IN active site within the PFV CDC clashes with the position of the scissile dinucleotide at the uncleaved LTR end in the analogous SSC structure (Fig. 3C), accounting for INSTI specificity to inhibit strand transfer activity over 3′-processing activity (13).
FIGURE 3.
Mechanism of action of INSTIs. A, chemical structures of RAL and DTG. B, comparison of drug-free (left; PDB code 3OY9), RAL-bound (middle; PDB code 3OYA), and DTG-bound (right; PDB code 3S3M) PFV intasomes shows the displacement of the terminal dA nucleotide (carbon, oxygen, and nitrogen atoms are colored magenta, red, and blue, respectively) and the reactive 3′-OH (colored red and marked by arrows) of the transferred DNA strand. C, IN active sites from the SSC (green; PDB code 4E7I), TCC (cyan; PDB code 4E7K), and RAL-bound (PDB code 3OYA; only the drug, in pink sticks, is shown) CDC are superimposed using the Cα atoms of active site residues Asp-128, Asp-185, and Glu-221. The unprocessed viral DNA end in the SSC is green, whereas the processed viral DNA and bound target DNA strands from the TCC structure are in dark blue and cyan, respectively. Mn2+ ions (labeled A and B), metal-chelating oxygens of RAL, bridging oxygen atoms of the scissile viral DNA and target DNA phosphodiester bonds (arrows), and 3′-processing (Wnuc, sphere) and DNA strand transfer (3′-OH, arrow) nucleophiles are colored red to highlight substrate mimicry of INSTIs during retroviral DNA integration (13).
Resistance to RAL arises through one of three predominant pathways that include changes at HIV-1 IN amino acid residues Tyr-143, Gln-148, and Asn-155 (70). The amino acid sequences of the HIV-1 and PFV IN CCDs are 22% identical, and crystal structures of PFV intasomes harboring wild-type or drug resistance changes, with or without bound drug, have helped explain the structural basis of HIV-1 drug resistance (18, 68, 69). RAL in particular is unlikely to directly contact the side chains of resistance substitutions that occur at position 148 or 155 (45). Instead, the substitutions appear to subtly affect local active site geometry, such that binding of the drug to mutant enzymes requires significant energetically unfavorable rearrangement of the IN CCD (68). Consistent with this interpretation, resistance changes correlate with significant increases in drug dissociation rates from these enzymes (71). The PIC harbors a unique enzymatic antiviral target: formed through IN processing of the LTR ends in the cytoplasm, the functional CDC must persist for sufficient time to accommodate PIC nuclear import, chromatin targeting, and IN strand transfer activity (Fig. 1), which would require a few hours to days, depending on the activation state of the target cell. When dissociation constants are significantly longer than the PIC half-life, CDC-specific drugs like INSTIs theoretically afford a “one-shot kill” mechanism (72).
3′-Processing and integration of modeled PFV LTR DNA have recently been followed in crystallo, allowing unprecedented visualization of the reaction mechanisms (Fig. 2B) (13). Overlaying the position of RAL in the PFV CDC co-crystal structure with the respective 3′-processing and strand transfer nucleophiles and scissile phosphodiester bonds from the SSC and TCC structures moreover revealed impressive similarities in positions of salient oxygen atoms (Fig. 3C). The oxygen of the heteroatom cluster distal from the halobenzyl group mimics the attacking water nucleophile during processing (labeled Wnuc in Fig. 3C) and a bridging oxygen atom of the target phosphodiester bond during strand transfer. Vice versa, the proximal oxygen mimics the positions of the attacking 3′-OH nucleophile for DNA strand transfer and a bridging oxygen atom of the scissile phosphodiester bond during processing (Fig. 3C). These data are fully consistent with the reversible “ping-pong” role played by the two metal ions during RNase H catalysis (73) and furthermore suggest novel ways by which INSTIs could be modified to increase drug potency (13).
Dolutegravir (DTG) (Fig. 3A) is a promising second-generation INSTI that retains activity against many RAL-resistant viruses (69). Co-crystallization with the PFV CDC revealed an extended linker region between the metal-binding heteroatoms and the halobenzyl arm that allowed the drug to intimately contact the viral LTR end and IN β4-α2 loop region (Fig. 3B) (69), likely contributing to the slower dissociation of DTG versus RAL from HIV-1 IN-LTR complexes in vitro (71). The relative difficulty in selecting for DTG-resistant viruses ex vivo (47) suggests that the generation of resistance in vivo might occur less frequently than what has been seen with RAL. If so, such differences should be forthcoming over the next year or so as DTG segues into the AIDS clinic.
IN-binding Host Factors and Allosteric IN Inhibitors
Numerous host factors have been described to bind to HIV-1 IN (see Refs. 74 and 75 for recent reviews). Of these, LEDGF/p75 has been determined to play a vital role in viral DNA integration (reviewed in Refs. 76 and 77).
The different genera of retroviruses differentially target particular aspects of host cell chromatin during viral DNA integration (see Ref. 78 for review). Lentiviruses like HIV-1 preferentially integrate along the bodies of active genes, although integration negatively correlates with the uppermost levels of transcriptional activity (79). Moloney murine leukemia virus, a gammaretrovirus, also favors genes but, by contrast to HIV-1, favors promoters over internal regions (80). Although the mechanisms that underlie different retroviral preferences for particular chromatin features during integration are largely unknown, the lentiviral preference to integrate along the bodies of active genes is largely determined by LEDGF/p75 (76, 77).
LEDGF/p75 primarily acts as a bifunctional molecular tether: N-terminal elements that harbor a PWWP domain and a pair of AT-hook DNA-binding motifs confer constitutive chromatin-binding activity (81, 82), whereas a C-terminally located IN-binding domain (IBD) engages lentiviral IN protein (83, 84). The overall efficiency of HIV-1 integration is reduced by as much as 10-fold in cells depleted for LEDGF/p75 content by RNA interference (85, 86) or gene knock-out (87, 88). Although LEDGF/p75 co-fractionates with PICs extracted from HIV-1-infected cells (89), PICs made in knock-out cells display normal levels of DNA strand transfer activity in vitro (88). LEDGF/p75 therefore plays no apparent role in the regulation of IN catalysis prior to chromatin engagement. Concordantly, order-of-addition experiments reveal that effective IN activity requires LTR substrate binding prior to LEDGF/p75 binding in vitro (46, 90). It therefore seems that HIV-1 PICs most likely engage LEDGF/p75 as a chromatin component, where it preferentially associates along gene bodies (91). LEDGF/p75-bound IN is then activated for strand transfer in the local vicinity. The mechanism that underlies the chromatin distribution of LEDGF/p75 is unknown but predictably relies on engagement of specific histone modifications by the N-terminal PWWP domain (92, 93).
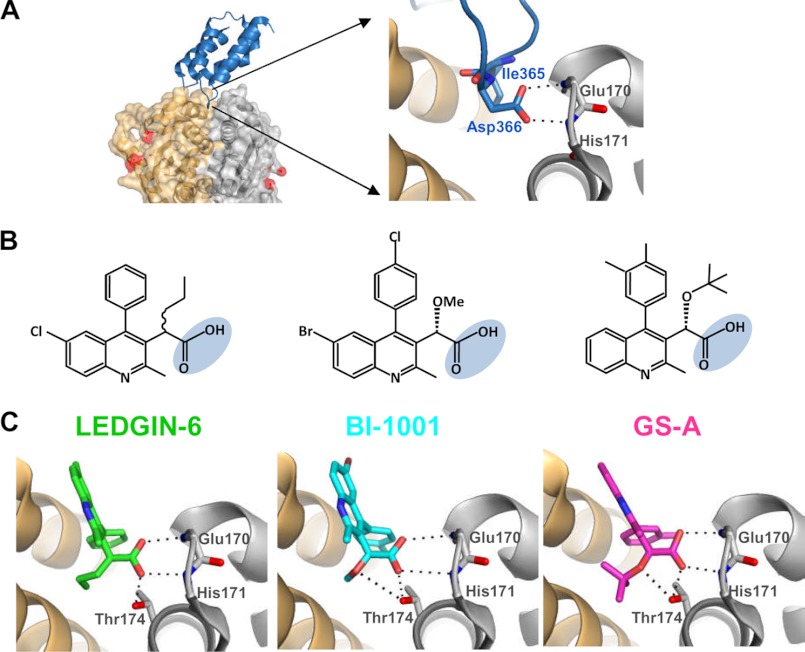
The LEDGF/p75 IBD is a PHAT (pseudo-HEAT repeat analogous topology) domain composed of two copies of a helix-hairpin-helix HEAT (huntingtin/elongation factor 3/subunit A of protein phosphatase 2A/yeast PI3K Tor1) repeat (94) and Ile-365 and Asp-366 at the tip of the N-terminal hairpin nestle into a cleft at the IN CCD-CCD interface (Fig. 4A) (95). Through an impressive example of structure-based drug design, Debyser and co-workers (96) identified a series of 2-(quinolin-3-yl)acetic acid derivatives that mimicked the positions of Ile-365 and Asp-366 in silico and inhibited IN-LEDGF/p75 binding in vitro and HIV-1 replication in cell culture. Although one of their more potent compounds, LEDGIN-6, did not inhibit IN 3′-processing activity in vitro, Boehringer Ingelheim identified a remarkably similar series of compounds using a high throughput screen for HIV-1 IN 3′-processing activity (Fig. 4B) (97). Due to this apparent discrepancy, LEDGIN-6 was compared with BI-1001 in a series of assays, which revealed that the compounds similarly inhibit IN 3′-processing and DNA strand transfer activities in the absence of added LEDGF/p75 protein (98). Concurrent work using more potent derivatives, including GS-A from Gilead Sciences, Inc. (Fig. 4B), verified host factor-independent inhibition of IN catalytic function (99, 100). Because the compounds engage the IN CCD at the host factor-binding cleft (Fig. 4C) distal from the active site (Fig. 4A), they are bona fide allosteric inhibitors of enzyme activity. The compounds apparently stabilize an inactive dimeric form of IN, thereby preventing proper assembly of the HIV-1 SSC in vitro (98–100). They also display steep dose-response curves during virus infection, which is indicative of a cooperative antiviral mechanism (98). X-ray structures of allosteric IN inhibitor (ALLINI)-CCD co-crystals moreover indicate that antiviral potency is dictated by the strength of the interaction (Fig. 4C), suggesting that further drug modifications could increase efficacy. Although the underlying mechanisms of cooperative drug behavior during HIV-1 infection are not entirely clear, inhibition of both intasome assembly and intasome-LEDGF/p75 interaction are reasonable possibilities (98–100). Virus produced in the presence of drug is unable to initiate a new round of infection (100),3 indicating that inhibition of steps in the HIV-1 life cycle that precede integration might contribute to the multimode mechanism of action of these intriguing compounds.
FIGURE 4.
ALLINI structures and binding mechanisms. A, the x-ray co-crystal structure of the HIV-1 IN-LEDGF/p75 complex (left; PDB code 2B4J) shows one IBD molecule (blue) bound at the interface of two IN CCD monomers (gold and silver), with IN active site residues colored red and the interhelical loop of LEDGF/p75 penetrating into the cavity at the dimer interface. The side chains of LEDGF/p75 contact residues Ile-365 and Asp-366 and main chain atoms of Glu-170 and His-171 of IN are shown as sticks, with oxygen and nitrogen atoms colored red and blue, respectively, and H-bonds drawn as dotted lines (right) (95). B, chemical structures of LEDGIN-6 (left), BI-1001 (middle), and GS-A (right), with the carboxyl group that mimics LEDGF/p75 hot spot residue Asp-366 highlighted. C, binding of LEDGIN-6 (left; PDB code 3LPU), BI-1001 (middle; PDB code 4DMN), and GS-A (right; PDB code 4E1M) at the HIV-1 IN CCD-CCD interface. Oxygen and nitrogen atoms of the compounds and Glu-170, His-171 (for simplicity, only main chain atoms of these residues are shown), and Thr-174 are colored red and blue, respectively. H-bonds between the drugs and IN are shown as dotted lines. Approximate values for 50% inhibition of HIV-1 replication in cell culture are 12, 6, and 0.1 μm for LEDGIN-6, BI-1001 (98), and GS-A (99), respectively.
Conclusions
The retroviral integration field has witnessed remarkable advances in recent years. Co-crystal structures of PFV intasomes have revealed unprecedented details of IN-DNA interactions and reaction mechanisms along the complete DNA recombination pathway (Fig. 2) (13, 18, 26) and have helped elucidate the structural basis of HIV-1 drug resistance to clinical INSTIs (18, 68). Attempts to solve crystal structures of analogous IN-DNA complexes from other retroviral genera, spurred on by the successes with PFV, are under way in a number of laboratories. DTG, a second-generation INSTI that retains activity against first-generation RAL-resistant viruses (69), is nearing its introduction into the clinic. Selection of HIV-1 strains resistant to pipeline ALLINIs can occur readily ex vivo (96, 99), perhaps tempering excitement for this new class of IN inhibitors. Their cooperative mode of action (98), which indicates antiviral activity against more than one step in the replication cycle, nevertheless highlights ALLINIs as unique HIV-1 intasome structural probes with potential relevance for the treatment of HIV/AIDS.
Acknowledgments
We thank Robert Craigie and Peter Cherepanov for critical review of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants AI039394 and AI070042. This is the third article in the Thematic Minireview Series on Understanding Human Immunodeficiency Virus-Host Interactions at the Biochemical Level.
K. Jurado, Y. Koh, and A. Engelman, unpublished data.
- RTC
- reverse transcription complex
- IN
- integrase
- PIC
- pre-integration complex
- NTD
- N-terminal domain
- CCD
- catalytic core domain
- PFV
- prototype foamy virus
- SSC
- stable synaptic complex
- CDC
- cleaved donor complex
- TCC
- target capture complex
- STC
- strand transfer complex
- LEDGF
- lens epithelium-derived growth factor
- DKA
- diketo acid
- RAL
- raltegravir
- INSTI
- IN strand transfer inhibitor
- DTG
- dolutegravir
- IBD
- IN-binding domain
- ALLINI
- allosteric IN inhibitor
- PDB
- Protein Data Bank.
REFERENCES
- 1. Bowerman B., Brown P. O., Bishop J. M., Varmus H. E. (1989) A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 3, 469–478 [DOI] [PubMed] [Google Scholar]
- 2. Fassati A., Goff S. P. (2001) Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J. Virol. 75, 3626–3635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Miller M. D., Farnet C. M., Bushman F. D. (1997) Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 71, 5382–5390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fujiwara T., Mizuuchi K. (1988) Retroviral DNA integration: structure of an integration intermediate. Cell 54, 497–504 [DOI] [PubMed] [Google Scholar]
- 5. Roth M. J., Schwartzberg P. L., Goff S. P. (1989) Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence on IN function and terminal DNA sequence. Cell 58, 47–54 [DOI] [PubMed] [Google Scholar]
- 6. Brown P. O., Bowerman B., Varmus H. E., Bishop J. M. (1989) Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc. Natl. Acad. Sci. U.S.A. 86, 2525–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Engelman A., Mizuuchi K., Craigie R. (1991) HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell 67, 1211–1221 [DOI] [PubMed] [Google Scholar]
- 8. Yu S. F., Baldwin D. N., Gwynn S. R., Yendapalli S., Linial M. L. (1996) Human foamy virus replication: a pathway distinct from that of retroviruses and hepadnaviruses. Science 271, 1579–1582 [DOI] [PubMed] [Google Scholar]
- 9. Nowotny M. (2009) Retroviral integrase superfamily: the structural perspective. EMBO Rep. 10, 144–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dyda F., Hickman A. B., Jenkins T. M., Engelman A., Craigie R., Davies D. R. (1994) Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science 266, 1981–1986 [DOI] [PubMed] [Google Scholar]
- 11. Mizuuchi K. (1992) Polynucleotidyl transfer reactions in transpositional DNA recombination. J. Biol. Chem. 267, 21273–21276 [PubMed] [Google Scholar]
- 12. Yang W., Lee J. Y., Nowotny M. (2006) Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell 22, 5–13 [DOI] [PubMed] [Google Scholar]
- 13. Hare S., Maertens G. N., Cherepanov P. (2012) 3′-Processing and strand transfer catalysed by retroviral integrase in crystallo. EMBO J. 31, 3020–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yuan Y. W., Wessler S. R. (2011) The catalytic domain of all eukaryotic cut-and-paste transposase superfamilies. Proc. Natl. Acad. Sci. U.S.A. 108, 7884–7889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li X., Krishnan L., Cherepanov P., Engelman A. (2011) Structural biology of retroviral DNA integration. Virology 411, 194–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Engelman A., Craigie R. (1992) Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J. Virol. 66, 6361–6369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kulkosky J., Jones K. S., Katz R. A., Mack J. P., Skalka A. M. (1992) Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol. Cell. Biol. 12, 2331–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hare S., Gupta S. S., Valkov E., Engelman A., Cherepanov P. (2010) Retroviral intasome assembly and inhibition of DNA strand transfer. Nature 464, 232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chaconas G. (1999) Studies on a “jumping gene machine”: higher-order nucleoprotein complexes in Mu DNA transposition. Biochem. Cell Biol. 77, 487–491 [PubMed] [Google Scholar]
- 20. Mizuuchi K., Adzuma K. (1991) Inversion of the phosphate chirality at the target site of Mu DNA strand transfer: evidence for a one-step transesterification mechanism. Cell 66, 129–140 [DOI] [PubMed] [Google Scholar]
- 21. Richet E., Abcarian P., Nash H. A. (1986) The interaction of recombination proteins with supercoiled DNA: defining the role of supercoiling in lambda integrative recombination. Cell 46, 1011–1021 [DOI] [PubMed] [Google Scholar]
- 22. Wei S. Q., Mizuuchi K., Craigie R. (1997) A large nucleoprotein assembly at the ends of the viral DNA mediates retroviral DNA integration. EMBO J. 16, 7511–7520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen H., Wei S. Q., Engelman A. (1999) Multiple integrase functions are required to form the native structure of the human immunodeficiency virus type 1 intasome. J. Biol. Chem. 274, 17358–17364 [DOI] [PubMed] [Google Scholar]
- 24. Li M., Mizuuchi M., Burke T. R., Jr., Craigie R. (2006) Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J. 25, 1295–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bera S., Pandey K. K., Vora A. C., Grandgenett D. P. (2009) Molecular interactions between HIV-1 integrase and the two viral DNA ends within the synaptic complex that mediates concerted integration. J. Mol. Biol. 389, 183–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maertens G. N., Hare S., Cherepanov P. (2010) The mechanism of retroviral integration through X-ray structures of its key intermediates. Nature 468, 326–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Engelman A., Bushman F. D., Craigie R. (1993) Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J. 12, 3269–3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van Gent D. C., Vink C., Groeneger A. A., Plasterk R. H. (1993) Complementation between HIV integrase proteins mutated in different domains. EMBO J. 12, 3261–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pahl A., Flügel R. M. (1995) Characterization of the human spuma retrovirus integrase by site-directed mutagenesis, by complementation analysis, and by swapping the zinc finger domain of HIV-1. J. Biol. Chem. 270, 2957–2966 [DOI] [PubMed] [Google Scholar]
- 30. Jonsson C. B., Donzella G. A., Gaucan E., Smith C. M., Roth M. J. (1996) Functional domains of Moloney murine leukemia virus integrase defined by mutation and complementation analysis. J. Virol. 70, 4585–4597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hare S., Di Nunzio F., Labeja A., Wang J., Engelman A., Cherepanov P. (2009) Structural basis for functional tetramerization of lentiviral integrase. PLoS Pathog. 5, e1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Savilahti H., Mizuuchi K. (1996) Mu transpositional recombination: donor DNA cleavage and strand transfer in trans by the Mu transposase. Cell 85, 271–280 [DOI] [PubMed] [Google Scholar]
- 33. Swanson P. C. (2001) The DDE motif in RAG-1 is contributed in trans to a single active site that catalyzes the nicking and transesterification steps of V(D)J recombination. Mol. Cell. Biol. 21, 449–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen J. W., Lee J., Jayaram M. (1992) DNA cleavage in trans by the active site tyrosine during Flp recombination: switching protein partners before exchanging strands. Cell 69, 647–658 [DOI] [PubMed] [Google Scholar]
- 35. Delelis O., Carayon K., Guiot E., Leh H., Tauc P., Brochon J. C., Mouscadet J. F., Deprez E. (2008) Insight into the integrase-DNA recognition mechanism. A specific DNA-binding mode revealed by an enzymatically labeled integrase. J. Biol. Chem. 283, 27838–27849 [DOI] [PubMed] [Google Scholar]
- 36. Valkov E., Gupta S. S., Hare S., Helander A., Roversi P., McClure M., Cherepanov P. (2009) Functional and structural characterization of the integrase from the prototype foamy virus. Nucleic Acids Res. 37, 243–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bushman F. D., Craigie R. (1991) Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc. Natl. Acad. Sci. U.S.A. 88, 1339–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li M., Craigie R. (2005) Processing of viral DNA ends channels the HIV-1 integration reaction to concerted integration. J. Biol. Chem. 280, 29334–29339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hare S., Shun M. C., Gupta S. S., Valkov E., Engelman A., Cherepanov P. (2009) A novel co-crystal structure affords the design of gain-of-function lentiviral integrase mutants in the presence of modified PSIP1/LEDGF/p75. PLoS Pathog. 5, e1000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Carteau S., Gorelick R. J., Bushman F. D. (1999) Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J. Virol. 73, 6670–6679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pandey K. K., Bera S., Grandgenett D. P. (2011) The HIV-1 integrase monomer induces a specific interaction with LTR DNA for concerted integration. Biochemistry 50, 9788–9796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cherepanov P., Maertens G., Proost P., Devreese B., Van Beeumen J., Engelborghs Y., De Clercq E., Debyser Z. (2003) HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 278, 372–381 [DOI] [PubMed] [Google Scholar]
- 43. Michel F., Crucifix C., Granger F., Eiler S., Mouscadet J. F., Korolev S., Agapkina J., Ziganshin R., Gottikh M., Nazabal A., Emiliani S., Benarous R., Moras D., Schultz P., Ruff M. (2009) Structural basis for HIV-1 DNA integration in the human genome, role of the LEDGF/P75 cofactor. EMBO J. 28, 980–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Busschots K., Vercammen J., Emiliani S., Benarous R., Engelborghs Y., Christ F., Debyser Z. (2005) The interaction of LEDGF/p75 with integrase is lentivirus-specific and promotes DNA binding. J. Biol. Chem. 280, 17841–17847 [DOI] [PubMed] [Google Scholar]
- 45. Krishnan L., Li X., Naraharisetty H. L., Hare S., Cherepanov P., Engelman A. (2010) Structure-based modeling of the functional HIV-1 intasome and its inhibition. Proc. Natl. Acad. Sci. U.S.A. 107, 15910–15915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kessl J. J., Li M., Ignatov M., Shkriabai N., Eidahl J. O., Feng L., Musier-Forsyth K., Craigie R., Kvaratskhelia M. (2011) FRET analysis reveals distinct conformations of IN tetramers in the presence of viral DNA or LEDGF/p75. Nucleic Acids Res. 39, 9009–9022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Quashie P. K., Mesplède T., Han Y. S., Oliveira M., Singhroy D. N., Fujiwara T., Underwood M. R., Wainberg M. A. (2012) Characterization of the R263K mutation in HIV-1 integrase that confers low-level resistance to the second-generation integrase strand transfer inhibitor dolutegravir. J. Virol. 86, 2696–2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Faure A., Calmels C., Desjobert C., Castroviejo M., Caumont-Sarcos A., Tarrago-Litvak L., Litvak S., Parissi V. (2005) HIV-1 integrase crosslinked oligomers are active in vitro. Nucleic Acids Res. 33, 977–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bao K. K., Wang H., Miller J. K., Erie D. A., Skalka A. M., Wong I. (2003) Functional oligomeric state of avian sarcoma virus integrase. J. Biol. Chem. 278, 1323–1327 [DOI] [PubMed] [Google Scholar]
- 50. Pandey K. K., Sinha S., Grandgenett D. P. (2007) Transcriptional coactivator LEDGF/p75 modulates human immunodeficiency virus type 1 integrase-mediated concerted integration. J. Virol. 81, 3969–3979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kotova S., Li M., Dimitriadis E. K., Craigie R. (2010) Nucleoprotein intermediates in HIV-1 DNA integration visualized by atomic force microscopy. J. Mol. Biol. 399, 491–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Guiot E., Carayon K., Delelis O., Simon F., Tauc P., Zubin E., Gottikh M., Mouscadet J. F., Brochon J. C., Deprez E. (2006) Relationship between the oligomeric status of HIV-1 integrase on DNA and enzymatic activity. J. Biol. Chem. 281, 22707–22719 [DOI] [PubMed] [Google Scholar]
- 53. Masuda T., Kuroda M. J., Harada S. (1998) Specific and independent recognition of U3 and U5 att sites by human immunodeficiency virus type 1 integrase in vivo. J. Virol. 72, 8396–8402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Brown H. E., Chen H., Engelman A. (1999) Structure-based mutagenesis of the human immunodeficiency virus type 1 DNA attachment site: effects on integration and cDNA synthesis. J. Virol. 73, 9011–9020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Oh J., Chang K. W., Hughes S. H. (2006) Mutations in the U5 sequences adjacent to the primer binding site do not affect tRNA cleavage by Rous sarcoma virus RNase H but do cause aberrant integrations in vivo. J. Virol. 80, 451–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Oh J., Chang K. W., Alvord W. G., Hughes S. H. (2006) Alternate polypurine tracts affect Rous sarcoma virus integration in vivo. J. Virol. 80, 10281–10284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bojja R. S., Andrake M. D., Weigand S., Merkel G., Yarychkivska O., Henderson A., Kummerling M., Skalka A. M. (2011) Architecture of a full-length retroviral integrase monomer and dimer, revealed by small angle x-ray scattering and chemical cross-linking. J. Biol. Chem. 286, 17047–17059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sherman P. A., Fyfe J. A. (1990) Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc. Natl. Acad. Sci. U.S.A. 87, 5119–5123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Craigie R., Mizuuchi K., Bushman F. D., Engelman A. (1991) A rapid in vitro assay for HIV DNA integration. Nucleic Acids Res. 19, 2729–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hazuda D. J., Hastings J. C., Wolfe A. L., Emini E. A. (1994) A novel assay for the DNA strand-transfer reaction of HIV-1 integrase. Nucleic Acids Res. 22, 1121–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vink C., Banks M., Bethell R., Plasterk R. H. (1994) A high-throughput, non-radioactive microtiter plate assay for activity of the human immunodeficiency virus integrase protein. Nucleic Acids Res. 22, 2176–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. van Gent D. C., Elgersma Y., Bolk M. W., Vink C., Plasterk R. H. (1991) DNA binding properties of the integrase proteins of human immunodeficiency viruses types 1 and 2. Nucleic Acids Res. 19, 3821–3827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Farnet C. M., Wang B., Lipford J. R., Bushman F. D. (1996) Differential inhibition of HIV-1 preintegration complexes and purified integrase protein by small molecules. Proc. Natl. Acad. Sci. U.S.A. 93, 9742–9747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hazuda D. J., Felock P. J., Hastings J. C., Pramanik B., Wolfe A. L. (1997) Differential divalent cation requirements uncouple the assembly and catalytic reactions of human immunodeficiency virus type 1 integrase. J. Virol. 71, 7005–7011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hazuda D. J., Felock P., Witmer M., Wolfe A., Stillmock K., Grobler J. A., Espeseth A., Gabryelski L., Schleif W., Blau C., Miller M. D. (2000) Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287, 646–650 [DOI] [PubMed] [Google Scholar]
- 66. Summa V., Petrocchi A., Bonelli F., Crescenzi B., Donghi M., Ferrara M., Fiore F., Gardelli C., Gonzalez Paz O., Hazuda D. J., Jones P., Kinzel O., Laufer R., Monteagudo E., Muraglia E., Nizi E., Orvieto F., Pace P., Pescatore G., Scarpelli R., Stillmock K., Witmer M. V., Rowley M. (2008) Discovery of raltegravir, a potent, selective orally bioavailable HIV-integrase inhibitor for the treatment of HIV-AIDS infection. J. Med. Chem. 51, 5843–5855 [DOI] [PubMed] [Google Scholar]
- 67. Grobler J. A., Stillmock K., Hu B., Witmer M., Felock P., Espeseth A. S., Wolfe A., Egbertson M., Bourgeois M., Melamed J., Wai J. S., Young S., Vacca J., Hazuda D. J. (2002) Diketo acid inhibitor mechanism and HIV-1 integrase: implications for metal binding in the active site of phosphotransferase enzymes. Proc. Natl. Acad. Sci. U.S.A. 99, 6661–6666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hare S., Vos A. M., Clayton R. F., Thuring J. W., Cummings M. D., Cherepanov P. (2010) Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance. Proc. Natl. Acad. Sci. U.S.A. 107, 20057–20062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hare S., Smith S. J., Métifiot M., Jaxa-Chamiec A., Pommier Y., Hughes S. H., Cherepanov P. (2011) Structural and functional analyses of the second-generation integrase strand transfer inhibitor dolutegravir (S/GSK1349572). Mol. Pharmacol. 80, 565–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cooper D. A., Steigbigel R. T., Gatell J. M., Rockstroh J. K., Katlama C., Yeni P., Lazzarin A., Clotet B., Kumar P. N., Eron J. E., Schechter M., Markowitz M., Loutfy M. R., Lennox J. L., Zhao J., Chen J., Ryan D. M., Rhodes R. R., Killar J. A., Gilde L. R., Strohmaier K. M., Meibohm A. R., Miller M. D., Hazuda D. J., Nessly M. L., DiNubile M. J., Isaacs R. D., Teppler H., Nguyen B. Y., and BENCHMRK Study Teams (2008) Subgroup and resistance analyses of raltegravir for resistant HIV-1 infection. N. Engl. J. Med. 359, 355–365 [DOI] [PubMed] [Google Scholar]
- 71. Hightower K. E., Wang R., Deanda F., Johns B. A., Weaver K., Shen Y., Tomberlin G. H., Carter H. L., 3rd, Broderick T., Sigethy S., Seki T., Kobayashi M., Underwood M. R. (2011) Dolutegravir (S/GSK1349572) exhibits significantly slower dissociation than raltegravir and elvitegravir from wild-type and integrase inhibitor-resistant HIV-1 integrase-DNA complexes. Antimicrob. Agents Chemother. 55, 4552–4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Grobler J. A., McKenna P. M., Ly S., Stillmock K. A., Bahnck C. M., Danovich R. M., Dornadula G., Hazuda D. J., Miller M. D. (2009) HIV integrase inhibitor dissociation rates correlate with efficacy in vitro. Antiviral Ther. 14, Suppl. 1, A25 [Google Scholar]
- 73. Nowotny M., Gaidamakov S. A., Crouch R. J., Yang W. (2005) Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell 121, 1005–1016 [DOI] [PubMed] [Google Scholar]
- 74. Turlure F., Devroe E., Silver P. A., Engelman A. (2004) Human cell proteins and human immunodeficiency virus DNA integration. Front. Biosci. 9, 3187–3208 [DOI] [PubMed] [Google Scholar]
- 75. Van Maele B., Busschots K., Vandekerckhove L., Christ F., Debyser Z. (2006) Cellular co-factors of HIV-1 integration. Trends Biochem. Sci. 31, 98–105 [DOI] [PubMed] [Google Scholar]
- 76. Engelman A., Cherepanov P. (2008) The lentiviral integrase binding protein LEDGF/p75 and HIV-1 replication. PLoS Pathog. 4, e1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Poeschla E. M. (2008) Integrase, LEDGF/p75 and HIV replication. Cell Mol. Life Sci. 65, 1403–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bushman F., Lewinski M., Ciuffi A., Barr S., Leipzig J., Hannenhalli S., Hoffmann C. (2005) Genome-wide analysis of retroviral DNA integration. Nat. Rev. Microbiol. 3, 848–858 [DOI] [PubMed] [Google Scholar]
- 79. Schröder A. R., Shinn P., Chen H., Berry C., Ecker J. R., Bushman F. (2002) HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110, 521–529 [DOI] [PubMed] [Google Scholar]
- 80. Wu X., Li Y., Crise B., Burgess S. M. (2003) Transcription start regions in the human genome are favored targets for MLV integration. Science 300, 1749–1751 [DOI] [PubMed] [Google Scholar]
- 81. Llano M., Vanegas M., Hutchins N., Thompson D., Delgado S., Poeschla E. M. (2006) Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. J. Mol. Biol. 360, 760–773 [DOI] [PubMed] [Google Scholar]
- 82. Turlure F., Maertens G., Rahman S., Cherepanov P., Engelman A. (2006) A tripartite DNA-binding element, comprised of the nuclear localization signal and two AT-hook motifs, mediates the association of LEDGF/p75 with chromatin in vivo. Nucleic Acids Res. 34, 1653–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Cherepanov P., Devroe E., Silver P. A., Engelman A. (2004) Identification of an evolutionarily conserved domain in LEDGF/p75 that binds HIV-1 integrase. J. Biol. Chem. 279, 48883–48892 [DOI] [PubMed] [Google Scholar]
- 84. Cherepanov P. (2007) LEDGF/p75 interacts with divergent lentiviral integrases and modulates their enzymatic activity in vitro. Nucleic Acids Res. 35, 113–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Llano M., Saenz D. T., Meehan A., Wongthida P., Peretz M., Walker W. H., Teo W., Poeschla E. M. (2006) An essential role for LEDGF/p75 in HIV integration. Science 314, 461–464 [DOI] [PubMed] [Google Scholar]
- 86. Vandekerckhove L., Christ F., Van Maele B., De Rijck J., Gijsbers R., Van den Haute C., Witvrouw M., Debyser Z. (2006) Transient and stable knockdown of the integrase cofactor LEDGF/p75 reveals its role in the replication cycle of human immunodeficiency virus. J. Virol. 80, 1886–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Marshall H. M., Ronen K., Berry C., Llano M., Sutherland H., Saenz D., Bickmore W., Poeschla E., Bushman F. D. (2007) Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PLoS ONE 2, e1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Shun M. C., Raghavendra N. K., Vandegraaff N., Daigle J. E., Hughes S., Kellam P., Cherepanov P., Engelman A. (2007) LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 21, 1767–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Llano M., Vanegas M., Fregoso O., Saenz D., Chung S., Peretz M., Poeschla E. M. (2004) LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J. Virol. 78, 9524–9537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Raghavendra N. K., Engelman A. (2007) LEDGF/p75 interferes with the formation of synaptic nucleoprotein complexes that catalyze full-site HIV-1 DNA integration in vitro: implications for the mechanism of viral cDNA integration. Virology 360, 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. De Rijck J., Bartholomeeusen K., Ceulemans H., Debyser Z., Gijsbers R. (2010) High-resolution profiling of the LEDGF/p75 chromatin interaction in the ENCODE region. Nucleic Acids Res. 38, 6135–6147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Shun M. C., Botbol Y., Li X., Di Nunzio F., Daigle J. E., Yan N., Lieberman J., Lavigne M., Engelman A. (2008) Identification and characterization of PWWP domain residues critical for LEDGF/p75 chromatin binding and human immunodeficiency virus type 1 infectivity. J. Virol. 82, 11555–11567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Pradeepa M. M., Sutherland H. G., Ule J., Grimes G. R., Bickmore W. A. (2012) Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLoS Genet. 8, e1002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Cherepanov P., Sun Z. Y., Rahman S., Maertens G., Wagner G., Engelman A. (2005) Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75. Nat. Struct. Mol. Biol. 12, 526–532 [DOI] [PubMed] [Google Scholar]
- 95. Cherepanov P., Ambrosio A. L., Rahman S., Ellenberger T., Engelman A. (2005) Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc. Natl. Acad. Sci. U.S.A. 102, 17308–17313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Christ F., Voet A., Marchand A., Nicolet S., Desimmie B. A., Marchand D., Bardiot D., Van der Veken N. J., Van Remoortel B., Strelkov S. V., De Maeyer M., Chaltin P., Debyser Z. (2010) Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication. Nat. Chem. Biol. 6, 442–448 [DOI] [PubMed] [Google Scholar]
- 97. Fenwick C. W., Tremblay S., Wardrop E., Bethell R., Coulomb R., Elston R., Faucher A. M., Mason S., Simoneau B., Tsantrizos Y., Yoakim C. (2011) Resistance studies with HIV-1 non-catalytic site integrase inhibitors. Antiviral Ther. 16, Suppl. 1, A9 [Google Scholar]
- 98. Kessl J. J., Jena N., Koh Y., Taskent-Sezgin H., Slaughter A., Feng L., de Silva S., Wu L., Le Grice S. F., Engelman A., Fuchs J. R., Kvaratskhelia M. (2012) Multimode, cooperative mechanism of action of allosteric HIV-1 integrase inhibitors. J. Biol. Chem. 287, 16801–16811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Tsiang M., Jones G. S., Niedziela-Majka A., Kan E., Lansdon E. B., Huang W., Hung M., Samuel D., Novikov N., Xu Y., Mitchell M., Guo H., Babaoglu K., Liu X., Geleziunas R., Sakowicz R. (2012) New class of HIV-1 integrase (IN) inhibitors with a dual mode of action. J. Biol. Chem. 287, 21189–21203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Christ F., Shaw S., Demeulemeester J., Desimmie B. A., Marchand A., Butler S., Smets W., Chaltin P., Westby M., Debyser Z., Pickford C. (2012) Small molecule inhibitors of the LEDGF/p75 binding site of integrase (LEDGINs) block HIV replication and modulate integrase multimerization. Antimicrob. Agents Chemother. 56, 4365–4374 [DOI] [PMC free article] [PubMed] [Google Scholar]