Background: APPL2 is an endosomal Rab effector forming part of a signaling pathway linking cell surface and nucleus.
Results: Crystal and solution structures of APPL2 were solved, and Rab partners were identified.
Conclusion: APPL2 interacts tightly with Rab31, and APPL2 structures reveal unexpected domain motion that could have functional implications.
Significance: APPL2 dynamics and interactions may be crucial for its cell signaling role.
Keywords: Crystal Structure, Protein Structure, Protein-Protein Interactions, Rab Proteins, X-ray Scattering, BAR Domain Protein, SAXS
Abstract
The APPL1 and APPL2 proteins (APPL (adaptor protein, phosphotyrosine interaction, pleckstrin homology (PH) domain, and leucine zipper-containing protein)) are localized to their own endosomal subcompartment and interact with a wide range of proteins and small molecules at the cell surface and in the nucleus. They play important roles in signal transduction through their ability to act as Rab effectors. (Rabs are a family of Ras GTPases involved in membrane trafficking.) Both APPL1 and APPL2 comprise an N-terminal membrane-curving BAR (Bin-amphiphysin-Rvs) domain linked to a PH domain and a C-terminal phosphotyrosine-binding domain. The structure and interactions of APPL1 are well characterized, but little is known about APPL2. Here, we report the crystal structure and low resolution solution structure of the BARPH domains of APPL2. We identify a previously undetected hinge site for rotation between the two domains and speculate that this motion may regulate APPL2 functions. We also identified Rab binding partners of APPL2 and show that these differ from those of APPL1, suggesting that APPL-Rab interaction partners have co-evolved over time. Isothermal titration calorimetry data reveal the interaction between APPL2 and Rab31 has a Kd of 140 nm. Together with other biophysical data, we conclude the stoichiometry of the complex is 2:2.
Introduction
BAR domain proteins are crescent- or banana-shaped molecules that generate, sense, or maintain membrane curvature (1). Six subgroups of BAR domain proteins have been identified (2), and these proteins play diverse roles in fundamentally important membrane processes, including endocytosis (3–5).
APPL1 (DIP13α) and APPL2 (DIP13β) are BAR domain proteins and Rab5 effectors associated with a distinct subpopulation of early endosomes, termed APPL endosomes, that link cell surface signaling, endocytosis, and mitogenesis (6). Homo sapiens APPL1 (NP 036228.1) and APPL2 (NP 06064.2) share 52% sequence identity by ClustalW pair-wise alignment (7). APPL1 translocates from membranes to the nucleus in response to EGF binding or oxidative stress, where it interacts with a nucleosome remodeling and histone deacetylase complex (6).
APPL proteins are members of a subgroup of BAR domain proteins that combine BAR (bin-amphiphysin-Rvs167) and pleckstrin homology (PH)4 domains (8–10). The APPLs also encode a C-terminal phosphotyrosine binding (PTB) domain (see Fig. 1A). Homo sapiens APPL1 but not APPL2 has been structurally characterized. Thus, structures of the APPL1 BARPH domain (Refs. 11 and 12, and Riken Structural Genomics Institute (RSGI)5), the APPL1 BAR domain (Ref. 12, RSGI5) and the APPL1 PTB domain (RSGI5) are available in the Protein Data Bank (13). The APPL1 BAR domain adopts the typical crescent-shaped dimer of BAR proteins with the PH domains located at the distal ends of the dimer. There is no structural information on APPL2.
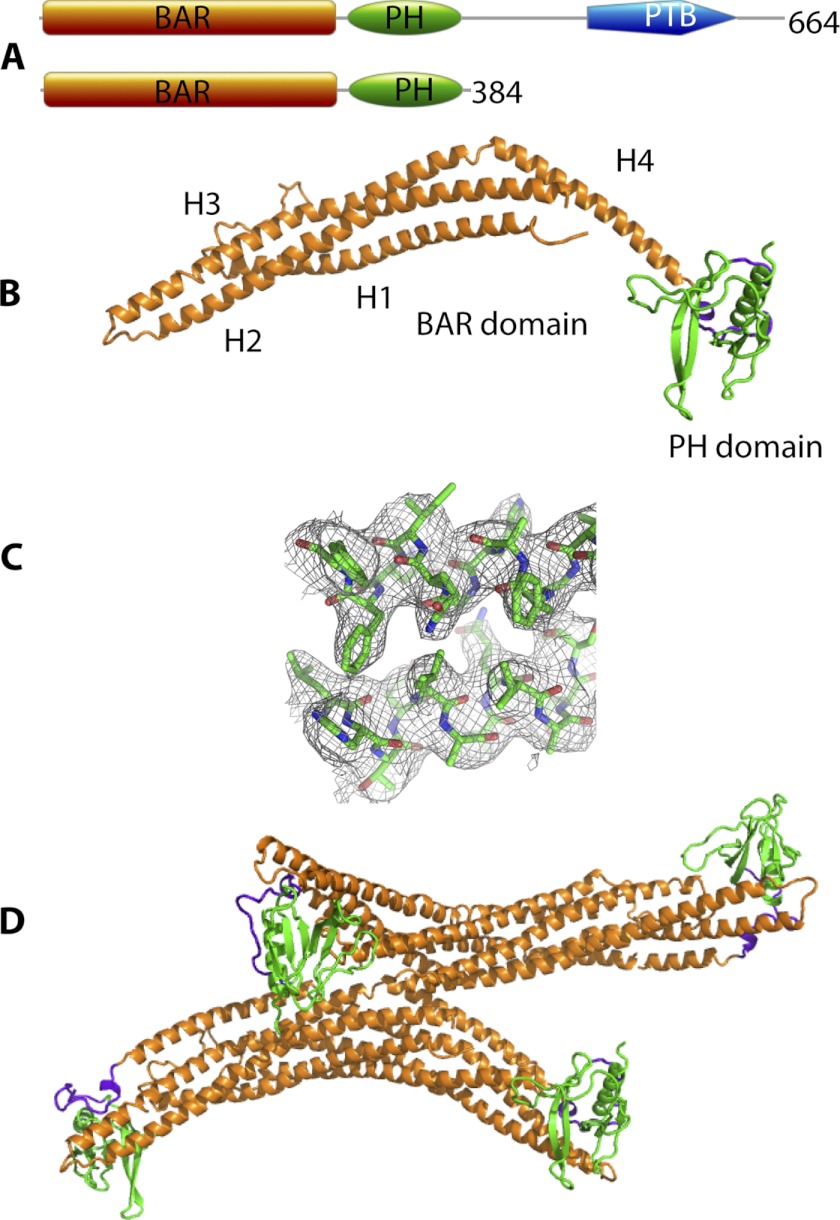
FIGURE 1.
Crystal structure of hAPPL2BARPH. A, domain structure of hAPPL2 and the hAPPL2BARPH construct used in this work. B, schematic representation of the crystal structure of hAPPL2BARPH chain A monomer showing the BAR domain (residues 18–257, orange), PH domain (residues 279–384, green), and linker (residues 258–278, purple). Helices (H1–H4) of the BAR domain are labeled. C, representative electron density of 2Fo − Fc map (contoured at 1σ) for the region around residues 109–118 (H2) and 200–210 (H3) of the BAR domain. D, schematic of the contents of the asymmetric unit of the crystal structure showing the arrangement of the two dimers.
Both APPL proteins target cell membranes (6), and the isolated PH and PTB domains individually also target membranes (14). Moreover, full-length APPL1 and APPL2 or their isolated PH and PTB domains bind phosphoinositides in vitro (11, 14) as do the APPL1 BAR and BARPH domains (11). Although the APPLs are Rab effectors, there is no structural information for an APPL-Rab complex, though a model of the APPL1-Rab5 complex has been proposed (12).
Here, we describe the first structural information on APPL2, specifically, the BARPH domains of human APPL2 (hAPPL2BARPH). We report both the crystal structure and SAXS solution structure of APPL2, showing unexpectedly that there is flexibility in the arrangement of the BAR and PH domains. In addition, we identify by yeast two-hybrid screen that Rab31 is a binding partner for APPL2, and we establish the thermodynamics and stoichiometry of the interaction between hAPPL2BARPH and Rab31.
EXPERIMENTAL PROCEDURES
Cloning
For human APPL2 (Open Biosystems, accession no. BC033731; GenBank accession no. 55198) residues 2–384 were subcloned into the LIC vector pMCSG7 (15) encoding an N-terminal polyhistidine tag with a tobacco etch virus cleavage site. For Rab31, the same subcloning strategy was adopted using a codon-optimized construct consisting of residues 1–167 of hRab31 (GeneArt; GenBank accession no. U59877) subcloned into the LIC vector.
Protein Production
hAPPL2BARPH and Rab 31 were expressed in 500-ml cultures of Escherichia coli BL21(DE3)pLysS using auto induction (16). Cultures were grown at 30 °C and typically harvested after 17 h at an A600 of ∼8.0. Pellets were harvested by centrifugation, snap frozen in liquid nitrogen and stored at −80 °C.
The pellet was thawed in buffer A (25 mm Tris, pH 7.5, 150 mm NaCl, 1 mm-mercaptoethanol), with 0.5% Triton X-100, 0.30 units/ml DNase (Roche Applied Science), and protease inhibitor mixture (100 μm PMSF, 2.0 μm bestatin, 0.3 μm pepstatin A, 0.3 μm E-64, 0.08 μm aprotinin, and 1.0 μm leupepti-Astral) at 4 °C. The thawed pellet was sonicated on ice, and the cell debris was removed by centrifugation. The supernatant was then mixed with TALONTM resin (Clontech) pre-equilibrated in buffer A. This suspension was mixed with rotation for 1 h at 4 °C and left overnight on ice, before transferring to a gravity column. The bound lysate was washed with three bed volumes of buffer A, followed by three bed volumes of buffer A containing 10 mm imidazole. Protein was then eluted in three bed volumes of buffer A containing 300 mm imidazole. The eluant was concentrated and exchanged into buffer B (25 mm Tris, pH 7.5, containing 25 mm NaCl and 1 mm β-mercaptoethanol) using a 10-kDa concentrator (Amicon) before loading onto a MonoQ 5/50 GL column equilibrated in buffer B on an ÄKTA FPLC (GE Life Sciences). The column was washed with 10 ml of buffer B, and protein was eluted using a 70-ml 25–500 mm NaCl gradient in buffer B.
Pooled protein fractions were concentrated using a 10-kDa concentrator (Amicon) and applied to a size exclusion chromatography column (HiLoad 16/60 Superdex 200) equilibrated with 25 mm HEPES, pH 7.5, containing 150 mm NaCl and 1 mm DTT on an ÄKTA FPLC (GE Life Sciences). Fractions containing hAPPL2BARPH were pooled and dialyzed against 25 mm HEPES, pH 7.5, containing 150 mm NaCl , 1 mm DTT and 1 mm EDTA. Purified samples of hAPPL2BARPH were concentrated using a 10-kDa concentrator (Amicon) and stored in aliquots at −80 °C at an A280 of 80.0.
Rab31 was purified in a similar manner. However, the buffers used after the lysis and throughout the purification were at pH 8.5 and contained 5 mm MgCl2. TALONTM eluant was concentrated and exchanged into buffer C (25 mm Tris, pH 8.5, containing 25 mm NaCl, 1 mm β-mercaptoethanol, and 5 mm MgCl2) using a 10-kDa concentrator (Amicon) before loading onto a MonoQ 5/50 GL column equilibrated in buffer C on an ÄKTA FPLC (GE Life Sciences). The column was washed with 10 ml of buffer C, and protein was eluted using a 70-ml 25–500 mm NaCl gradient in buffer C. Size exclusion chromatography was performed with a HiLoad 16/60 Superdex 75 column (GE Life Sciences) equilibrated with 25 mm HEPES, pH 8.5, 150 mm NaCl, 1 mm DTT, and 5 mm MgCl2. Purified Rab 31 was stored frozen at −80 °C typically at 3–4 mg/ml.
MALDI-TOF mass spectrometry estimates of the masses of purified hAPPL2BARPH and Rab31 were 46.3 and 21.3 kDa, respectively. These are consistent with the theoretical masses of 46.1 and 21.3 kDa, respectively. Chemical cross-linking performed using bis(sulfosuccinimidyl) suberate suggested that hAPPL2BARPH is predominantly a dimer in solution (data not shown).
Crystal Structure Determination of hAPPL2BARPH
Crystals were grown at 20 °C in a 96-well hanging drop plate using ViewDrop II seals (TTP Labtech) where the well conditions were 20% (v/v) glycerol, 40 mm hexammine cobalt (III) chloride, and 2–4% (w/v) PEG 3350, or 1.5–3.0% (w/v) PEG 8000. The hanging drops were prepared using TTP Labtech's Mosquito robot and were composed of 200 nl of 15.0 mg/ml hAPPLBARPH and 200 nl of the well solution. The plates were imaged using a RockImager (Formulatrix). hAPPL2BARPH crystallized in the P212121 space group with two dimers in the asymmetric unit. The crystals contain 79% solvent and have a Matthews coefficient of 5.9 A3/Da (estimated using the CCP4 suite (21)).
Diffraction data were measured from each of two crystals, both harvested within 2 days of setting drops. One crystal grew in 2.55% (w/v) PEG 3350 and had dimensions of ∼100 × 100 × 100 μm3. The second crystal grew from 2.9% (w/v) PEG 8000 and had dimensions of ∼220 × 140 × 120 μm3. The cryo-protectant used for both crystals contained 5.0% PEG 8000, 20% glycerol, 40 mm hexammine cobalt (III) chloride, and 2.3 m trimethylamine oxide. Data used for phasing were collected on the first crystal and measured on a Rigaku FR-E Superbright X-ray generator with Osmic HiRes2 optics and a Rigaku R-Axis IV++ image plate detector. Data used for refinement were measured from the second crystal on the MX2 beamline at the Australian Synchrotron (17). Data were processed using HKL2000 (18), and statistics are shown in Table 1.
TABLE 1.
Data collection and refinement statistics for hAPPL2BARPH
Values in parentheses are outer shell values.
| hAPPL2BARPH | hAPPL2BARPH | |
|---|---|---|
| Data collection | R-AXIS (in-house) | MX2 |
| Wavelength (Å) | 1.5418 | 0.9537 |
| Space group | P212121 | P212121 |
| a, b, and c (Å); α, β, and γ (°) | 97.2, 207.0, 217.2; 90, 90, 90 | 97.9, 208.0, 218.4; 90, 90, 90 |
| Resolution range (Å) | 50–3.5 (3.58–3.50) | 50–3.5 (3.63–3.50) |
| No. of observations | 138,807 | 662,189 |
| Unique reflections | 53,994 | 55,792 |
| Redundancy | 2.6 (2.5) | 11.9 (11.7) |
| Completeness (%) | 96.0 (96.1) | 100 (100) |
| I/σI (all) | 5.9 (2.0) | 11.3 (2.2) |
| Rmerge (%)a | 14.7 (44.2) | 17.5 (>99) |
| Refinement | ||
| Resolution range (Å) | 46.9–3.5 | |
| No. of reflections | 54,413 | |
| Rworkb (%) | 20.9 | |
| Rfreec (%) | 25.6 | |
| No. of atoms | 23,872 | |
| Wilson B (Å2) | 98 | |
| Average B (Å2) | 128 | |
| r.m.s.d. in ideal | ||
| Bond lengths (Å) | 0.005 | |
| Bond angles (°) | 0.98 | |
| Ramachandran plotd | ||
| Favored (%) | 93.2 | |
| Outliers (%) | 1.1 | |
| Molprobity score | 2.3 (100th percentile) | |
a Rmerge = ΣhΣj|Ihj − 〈Ih〉|/ΣhΣj〈Ih〉.
b Rwork = Σh,k,l‖Fobs(h,k,l)| − |Fcalc(h,k,l)‖/Σh,k,l|Fobs(h,k,l)|.
c Rfree is Rwork calculated using 5.16% of reflections set aside from refinement.
d Values were calculated using Molprobity (21).
Phasing was performed on the Rigaku-measured diffraction data set by performing molecular replacement with Phaser MR in the Phenix package (19). The APPL1BARPH crystal structure (PDB code 2Q13, with waters removed) (12) was used as the search model. Initial attempts to produce a phase solution using molecular replacement were frustrated by the expectation that the asymmetric unit was composed of eight or ten molecules, corresponding to solvent contents of 58.4 and 48.0%, respectively. A partial solution was eventually obtained that included two monomers, which were well placed in the electron density. The two monomers represented one subunit each of two dimers, allowing all four molecules in the asymmetric unit to be built in by comparison with the dimer structure of APPL1. The structure was refined with several rounds of manual rebuilding in COOT (20) and maximum likelihood refinement including TLS refinement in Phenix.refine (19). As recommended by Phenix developers, hydrogens were included, and the riding hydrogen model was used in refinement.
The final structure includes the following: chains A and B, residues 2–378 and 5–378, respectively, except residues 75–78 in loop 1 of chain A were not modeled; chains C and D, residues 2–378 and 5–377, respectively, except residues 76–78 in loop 1 of chain C were not modeled. No density was present for the polyhistidine tag at the N terminus, though for chains A and C the tobacco etch virus cleavage site (residues Asn-6 to Ala1) was modeled. The quality and geometry of the model were evaluated by Molprobity (21). Statistics for the final refined structure are provided in Table 1. Coordinates and structure factors have been deposited with the Protein Data Bank code 4H8S.
Yeast Two Hybrid Analysis
A Rab library, comprising constitutively active Rabs containing the catalytic glutamine to leucine mutation, was generated as described previously (22). Briefly, the cDNA encoding the Rab was cloned into the pGBKT7 vector (Clontech). hAPPL2 and hAPPL1 were cloned into the pGADT7 vector (Clontech). Plasmids for Rabs and APPLs were co-transformed into chemically competent Saccharomyces cerevisiae strain AH109 (Clontech) and selected for co-transformation by growth on −Leu/Trp plates. Co-transformants were spotted in serial dilution on −Leu/Trp plates (+His) as a control and −Leu/Trp/His (−His) plates to select for interaction.
SAXS Data Collection and Analysis
Data were collected on the SAXS-WAXS beamline at the Australian Synchrotron with a sample to detector distance set at 1567 mm and an X-ray wavelength of λ = 1.033 Å, allowing access to a q-range spanning ∼0.01–0.55 Å−1 (q = (4πsinθ)/λ, where θ is half of the scattering angle). Immediately prior to loading, all samples were centrifuged at 10,000 × g to remove large particles from the solution. To minimize the effects of radiation damage, 50 μl of each sample was flowed past the beam in 1.5-mm quartz capillaries (Hampton Research) at room temperature (22 °C). Data were collected at several concentrations for hAPPL2BARPH to assess the extent of concentration-dependent attractive or repulsive interactions. For each sample, five frames (each with a 2-s exposure time) were measured. Data reduction was carried out using SAXS15ID software (23), averaging all five measured frames and correcting for solvent scattering, sample transmission, detector sensitivity, and background radiation. Data were placed on an absolute scale by normalization against a water standard (24).
Data quality was assessed by inspection of the concentration dependence of the scattering data, linearity of the Guinier region of the data (qRg < 1.3), and the estimated molecular mass of the protein. At all concentrations, the Guinier plot (ln I(q) versus q2) was linear, and yielded I(0) values normalized by concentration that showed no systematic trend. Estimated molecular masses were determined (25), where the program MULCh (26) was used to calculate the contrast (Δρ) and partial specific volume (ν). Estimated molecular masses were close to the expected values. Taken together, these quality assessments indicate that the protein solutions are homogeneous, free of significant inter-particle interactions, and that the data are of high quality.
Modeling of hAPPL2BARPH was performed using BUNCH (27), which is designed to model the structure of a single polypeptide chain, typically where flexible or unstructured regions link structured domains. The program models regions of known structure as rigid units, and the remainder as chains of dummy residues, to generate an envelope that includes side chains. The position of the BAR (2–263) domain was fixed, and the position and orientation of the PH domain (272–384) was optimized. The N-terminal portion of the protein incorporating the His6 tag (24 residues) and linker region between the PH and BAR domains (264–271) were modeled as flexible loops. C2 symmetry of the dimer was enforced. The optimization was run 10 times, and the best model was chosen based on χ2. A summary of the SAXS data collection and analysis is given in Table 2.
TABLE 2.
SAXS data collection and analysis details
| Data collection | |
| Instrument | SAXS-WAXS (Australian Synchrotron) |
| Beam geometry | Point |
| Wavelength (Å) | 1.033 |
| q-range (Å−1) | 0.01–0.55 |
| Exposure time (s) | 10 (5 × 2-s exposures) |
| Protein concentration (mg/ml) | 0.75–2.00 |
| Temperature (°C) | 22 |
| Standard | Water |
| Structural parametersa | |
| I(0) (cm−1) (from p(r)) | 0.0504 ± 0.0004 |
| Rg (Å) (from p(r)) | 53.9 ± 0.5 |
| I(0) (cm−1) (from Guinier) | 0.0496 ± 0.0010 |
| Rg (Å) (from Guinier) | 51.3 ± 1.7 |
| Dmax (Å) | 180 ± 9 |
| Rg (Å) (crystal structure) | 51.7 |
| Dmax (Å) [crystal structure] | 168 |
| Molecular mass determination | |
| Partial specific volume (cm3 g−1) | 0.74 |
| Contrast, Δρ (1010 cm−2) | 2.85 |
| Molecular mass, Mr (from I(0)) | 91,000 ± 2000 |
| Molecular mass, Mr (crystal structure) | 92,600 |
| Software employed | |
| Primary data reduction | SAXS15ID |
| Data processing | PRIMUS and GNOM |
| Rigid body modeling | BUNCH |
| Three-dimensional graphics | PyMOL |
a Values displayed correspond to a sample with a protein concentration of 0.75 mg/ml. Values from Guinier analysis of other samples were as follows: I(0) = 0.0743 ± 0.0005 cm−1; Rg = 51.6 ± 0.8 Å; Mr = 91,400 ± 2000 (1.10 mg/ml); I(0) = 0.1340 ± 0.0008 cm−1; Rg = 51.7 ± 0.5 Å; Mr = 90,700 ± 2000 (2.00 mg/ml). There is no significant systematic change in Rg or Mr as concentration is varied, consistent with monodisperse solutions of noninteracting particles.
Isothermal Titration Calorimetry (ITC)
Rab31-Gpp(NH)p was prepared by incubating Rab31 (1.3 mg/ml) in 50 mm Hepes, pH 8.5, containing 150 mm NaCl, 5 mm MgCl2, and 10 mm EDTA with 6.3 mm Gpp(NH)p. After 4 h at 25 °C, MgCl2 was added to 10 mm, and the exchange solution was desalted into 50 mm Hepes, pH 8.5, containing 150 mm NaCl, 1 mm DTT, and 5 mm MgCl2. hAPPL2BARPH was also exchanged into 50 mm Hepes, pH 8.5 containing 150 mm NaCl, 5 mm MgCl2, and 1 mm DTT.
ITC experiments were carried out at 10 °C using a MicroCal AutoITC20 (GE Life Sciences). Origin software (version 7) was used to integrate the heat released and calculate the binding enthalpy (ΔH), equilibrium constant Ka (1/Kd), and stoichiometry (N), based on a “one set of sites” model fit to the data. Several other more complex binding models were trialed, but there was no improvement in the fit to the data. The Gibbs free energy (ΔG) was calculated using the following equation: ΔG = −RTln (Ka); binding entropy (ΔS) was calculated using ΔG = ΔH − TΔS.
The titration was performed by adding hAPPL2BARPH (160 μm) in 19 injections (2 μl) to 200 μl Rab31-Gpp(NH)p (12 μm). The sample was stirred at 1000 rpm for all measurements. The titration was performed in triplicate, and the reported parameters are the averaged values from the individual analyses.
Multiangle Laser Light Scattering (MALLS)
Rab31-Gpp(NH)p was prepared as for the ITC experiment. MALLS was performed as described previously (28, 29) using a miniDAWN Tristar laser light scattering photometer and Optilab DSP interferometric refractometer (Wyatt Technology). The size exclusion chromatography was performed using a SuperdexTM 200 10/300 (Pharmacia Biotech) at 25 °C in 50 mm Hepes, pH 8.5, containing 150 mm NaCl, 1 mm DTT, and 5 mm MgCl2. All samples were injected in a volume of 400 μl, and Mr estimates were determined using Debye fitting. Concentrations used were as follows: APPL2, 128 μm; APPL2, 24.6 μm and Rab31-Gpp(NH)p, 425.8 μm for the APPL2-Rab31-Gpp(NH)p complex.
RESULTS
Structure of hAPPL2BARPH
The crystal structure of hAPPL2BARPH was refined to 3.5 Å resolution (Table 1), revealing two dimers in the asymmetric unit (Fig. 1). Each dimer is crescent-shaped and formed by an anti-parallel arrangement of the helical BAR domains of each hAPPL2BARPH monomer. The apex of the crescent is shaped by a six-helix arrangement contributed by three helices (1, 2, and 3) from each of the BAR domains; this arrangement is typical of BAR domain dimers (2). Helix 4 is separated from helix 3 by two residues (residues Phe-218 and Ser-219) forming a kink that orients the long axes of the two helices ∼120° relative to each other. Helices 2 and 3 have minor kinks at residues 107–112 and 188–192, respectively, which also contribute to the overall shape of the hAPPL2BARPH dimer. The PH domain is a typical seven-stranded β-barrel capped by an α-helix at its C-terminal end (30). It is joined to helix 4 of the BAR domain by a linker region (Fig. 1A).
Two regions of the BAR domain interact with the PH domain of a dimer mate. Residues 148–153 in loop 2 of the BAR domain monomer interact with residues 334–339 of the PH domain in the loop between β-strand 5 and 6 of its monomer mate. On the other side of the same PH domain, residues 312–316 in the loop between β-strands 3 and 4 interact with amino acids 16–21 in the N-terminal region of the BAR domain of its monomer mate. Two alternate monomer conformations are present in the asymmetric unit because in chain C the orientation of the PH domain relative to the BAR domain differs from that of the other three chains. This is discussed further below.
Electrostatic Surface
The electrostatic charge mapped to the surface of hAPPL2BARPH and hAPPL1BARPH dimers is presented in Fig. 2. The concave surface has prominent basic patches at either end of the dimer at the BAR/PH domain interface. These patches are symmetrically arranged and are contributed by residues at the BAR and PH domain interface, from the BAR domain residues Arg-20, Lys-146, Arg-149, and Arg-171 of one monomer and from the PH domain of its dimer mate residues Arg-299, Arg-314, and Arg-338. The less prominent two patches near the center of the dimer are again symmetrically arranged and are contributed by Lys-63 and Lys-70 of one monomer and Arg-46 of its dimer mate.
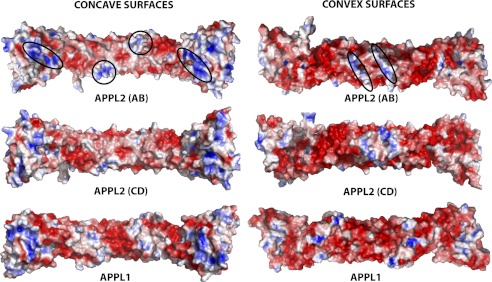
FIGURE 2.
Electrostatic surface charge of the hAPPL2BARPH and hAPPL1BARPH dimers. The proteins are arranged to display the concave and convex surfaces. The electrostatic surface charge colored red to blue from −15 to 15 KbT/ec (J/coulomb) (blue is positive, red is negative, and white is uncharged or hydrophobic) were generated using APBS (46) through PyMOL (47). On the concave surface of hAPPL2BARPH, the ellipses enclose residues Arg-20, Lys-146, Arg-149, and Arg-171 of chain A and residues Arg-299, Arg-314, and Arg-338 of chain B (left side of the molecule); and residues Arg-20, Lys-146, Arg-149, and Arg-171 of chain B and Arg-299, Arg-314, and Arg-338 of chain A (right side of the molecule), whereas the circles enclose residues Lys-63 and Lys-70, of chain B and Arg-46 of chain A (center bottom) and residues Lys-63 and Lys-70 of chain A and Arg-46 of chain B (center top). On the convex surface of hAPPL2BARPH, the ellipses enclose residues Lys-220 and Arg-221 of chain B and residues Lys-212 and Lys-213 of chain A (left center) and residues Lys-220 and Arg-221 of chain A and residues Lys-212 and Lys-213 of chain B (right center).
The electrostatic surface of the convex face of the hAPPL2BARPH and hAPPL1BARPH dimers is also presented in Fig. 2. A prominent basic patch is apparent at the apex of the hAPPL2BARPH dimer composed of residues Lys-212 and Lys-213 of one monomer and Lys-220 and Arg-221 of its dimer mate.
An end-on viw of the electrostatic surface of hAPPL2BARPH is shown in Fig. 3. Two basic patches are apparent. The more prominent one is formed by residues Lys-152, Lys-153, and Lys-154 in loop 2 of one BAR domain monomer and by residues Arg-338, Arg-339, Arg-360, and Lys-361 of the PH domain from the dimer mate and is part of the basic groove that essentially runs around the molecule at the BAR and PH domain interface. The less prominent patch in Fig. 2C is contributed from residues Arg-287 and Lys-289 of the PH domain. Minor differences in the distribution of surface charge were observed between the AB chain and CD chain monomers.
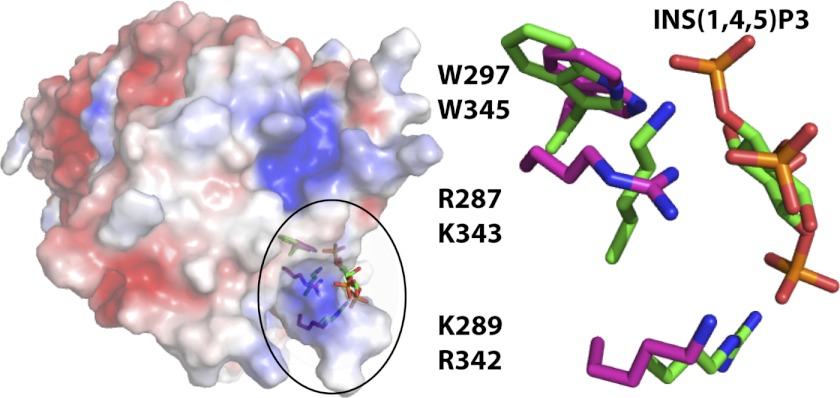
FIGURE 3.
A potential phospholipid binding site for the hAPPL2BARPH PH domain. The hAPPL2BARPH chain A monomer was superimposed with the ARHGAP9 PH domain bound to inositol (1,4,5)-trisphosphate (PDB code 2P0D). Shown is the ARHGAP9 bound conformation of the Ins(1,4,5)P3 and the electrostatic surface of hAPPL2BARPH chain A. The inset highlights the contact residues for ARHGAP9 PH domain (green) and the superimposed residues of hAPPL2BARPH (magenta) showing the correspondence between the two.
The hAPPL2BARPH dimer surface can be described as mostly electronegative with distinct electropositive ends and a central electropositive band that rings the BAR domain dimer. When the electrostatic surface of hAPPL2BARPH is compared with that of hAPPL1BARPH, very similar surface features are found with 19 of the 22 basic residues used in the description of the electrostatic surface of hAPPL2BARPH being conserved. Two of these exceptions Lys-220 and Lys-221 that form the basic patch on the convex surface of hAPPL2BARPH are not conserved in hAPPL1BARPH.
Potential Phosphoinositol Binding Site
The PH domain of APPL2 is reported to interact with phosphoinositides (11, 14). We investigated the potential binding site by comparing the structure of the hAPPL2BARPH PH domain with the crystal structure of the ARHGAP9 PH domain bound to inositol (1,4,5)-trisphosphate (Ins(1,4,5)P3) (PDB code 2P0D) (31). DaliLite (32) alignment of this structure with the PH domain of hAPPLPH gave a root mean square deviation (r.m.s.d.) of 2.0–2.2 Å for 89–92 Cα atoms and sequence identity of 20%. An overlay of the two structures suggested that similar to ARHGAP9, hAPPL2BARPH may bind Ins(1,4,5)P3 in a spectrin-like noncanonical manner. In this binding mode, the phosphoinositol is bound between strands β1 and β2 and the loop joining β5 and β6 of the hAPPL2BARPH PH domain, rather than between strands β1 and β2 and strands β3 and β4 of the PH domain as in the canonical binding mode (33). If the binding mode of Ins(1,4,5)P3 is conserved between the two proteins, then Ins(1,4,5)P3 would be located close to a complementary basic surface in hAPPL2BARPH (Fig. 3). Furthermore, there is an interesting correspondence between the contact residues for Ins(1,4,5)P3 in ARHGAP9 (31) and the hAPPL2BARPH residues in this region: ARHGAP9 has Arg-342, Lys-343, and Trp-345; hAPPL2BARPH has Lys-289, Arg-287, and Trp-297. Curiously, the equivalent residues of hAPPL2 are not related to those in ARHGAP9 by their position in the sequence, suggesting that if hAPPL2 does bind Ins(1,4,5)P3 in this manner, it may have evolved by convergent rather than divergent evolution.
Domain Flexibility
The APPL2 BAR and PH domains are linked by residues 258–278. The hAPPL2BARPH crystal structure reveals that these two domains move with respect to one another because the arrangement in chain C differs from the other three chains in the asymmetric unit (Fig. 4). Analysis of the PH domain rotation in the hAPPL2BARPH dimers was assessed using DynDom (34). This identified loop 254–263 254LLSVDESVYT/P264 as the bending region and found that the PH domain rotates by 13° with respect to the aligned BAR domains with minimal translation.
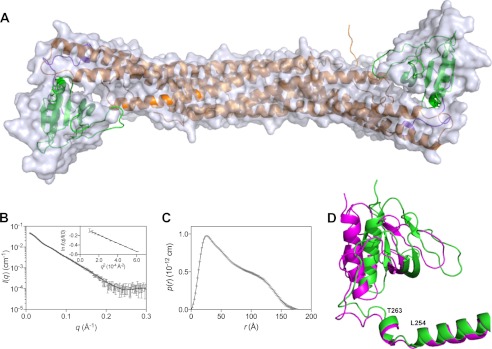
FIGURE 4.
Solution x-ray scattering data and model for hAPPL2BARPH. A, molecular surface of hAPPL2BARPH (obtained by optimizing against the SAXS data using the program BUNCH (27)), overlaid with the AB dimer crystal structure (normalized spatial discrepancy, 1.11). The model derived from the solution scattering data suggests a small outward movement of the PH domain relative to the BAR domain in both the AB and CD dimers of the crystal structure. B, scattering data for hAPPL2BARPH, and associated BUNCH model profile (solid black line, χ2 = 0.9). Inset, Guinier plot of the scattering data displaying a linear fit (R2 = 0.99), consistent with a monodisperse solution. C, pair-distance distribution function derived from the scattering data using GNOM (47) D, structural superimposition of the C chain monomer (green) with the D chain monomer (magenta) of the hAPPL2BARPH crystal structure showing the altered conformation of the PH domain of chain C (13° rotation relative to the aligned BAR domains). The two residues Leu-254 and Thr-263 at either end of the Dyndom-defined bending region are indicated. (these are part of the linker between the BAR and PH domains.) A and D were prepared using PyMOL (47).
We compared the structures of the four hAPPL2BARPH monomers with each other and with hAPPL1BARPH. Chains A, B, and D of hAPPL2BARPH are very similar to each other (r.m.s.d. of 1.1–1.3 Å for 369–373 Cα atoms). However, rotation of the PH domain relative to the BAR domain in chain C doubles the overall r.m.s.d. (2.5–2.8 Å for 370–378 Cα atoms), though the individual domains are structurally very similar (for the BAR domains, r.m.s.d. 0.7–1.2 for 256–260 Cα atoms; and for the PH domains, r.m.s.d. 1.0–1.2 Å for 102–103 Cα atoms).
SAXS Analysis of hAPPLBARPH
Structural modeling was performed against the solution scattering data using the program BUNCH. The PH domains sampled a range of dispositions relative to the BAR domains during the optimization, producing a model that provided an excellent representation of the data (χ2 = 0.9, cf. the crystal structures: χ2 (AB dimer), 2.6; χ2 (CD dimer), 2.7). The SAXS-derived solution structure of the hAPPL2BARPH dimer (Fig. 4a) is similar to the crystal structure, but the PH domain is shifted outward by a small amount. This movement is consistent with the experimental pair-distance distribution function, which indicated that the maximum dimension of the molecule in solution was ∼180 Å (cf. the maximum size of a dimer in the crystal structure is ∼170 Å).
Identification of Rabs That Interact with APPL2
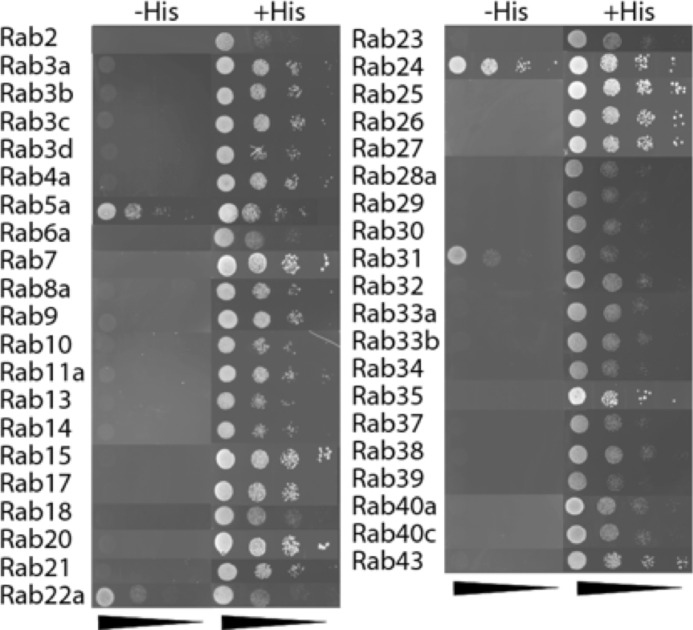
APPL1 and APPL2 have been reported to interact with Rab5 (6, 12). APPL1 has also been shown to interact with Rab21 (12). We hypothesized that APPL2 may also bind to other Rab partners. We therefore performed a yeast two-hybrid screen with APPL2 and a comprehensive Rab library, comprising 46 Rab GTPases containing at least one member of each subfamily (12). The Rabs used in this analysis were variants that stabilize their GTP-bound form and hence are constitutively active. As a control, we also performed the screen using APPL1 as the bait and, in agreement with published literature (12), we identified Rabs 5 and 21 as APPL1-interacting proteins (data not shown). For APPL2, the yeast two-hybrid screen revealed interactions with Rabs 5, 22a, 24, and 31 (Fig. 5). APPL2 was previously shown to interact with Rab5 (6, 14), but Rabs 22a, 24, and 31 are novel APPL2 interactors.
FIGURE 5.
hAPPL2 interacts with Rab5a, Rab22a, Rab24, and Rab31. Yeast two-hybrid screen was performed with hAPPL2 and the catalytically active Rab library. Co-transformants were spotted in serial dilutions on +His plates as a control and on −His plates to select for interactions. Representative images of three independent experiments are shown.
Analysis of hAPPL2BARPH Interaction with Rab31
To follow up on the yeast two-hybrid results, we cloned and expressed Rab31 and generated the Rab31 complex with the non-hydrolysable GTP analog (Gpp(NH)p). We evaluated the interaction between hAPPL2BARPH and Rab31-Gpp(NH)p using ITC and found that two Rab31 molecules bind one hAPPL2BARPH dimer (n = 0.98 ± 0.01) with a binding affinity, Kd, of 140 ± 30 nm. Thermodynamic analysis of the interaction indicated that binding is entropically driven with ΔH of 8.85 ± 0.30 kcal/mol and ΔS of 62.5 ± 0.7 cal/mol/degree, suggesting that the binding of Rab31-Gpp(NH)p to hAPPL2BARPH involves the displacement of structured water into bulk solvent.
We also performed MALLS experiments to confirm the stoichiometry of the complex. MALLS data yielded molecular mass estimates of 129.5 kDa ± 0.8% for the complex (theoretical mass of 114 kDa for 2:1 complex; 135.4 kDa of for 2:2 complex), 95.1 kDa ± 2.0% for hAPPL2BARPH (theoretical mass of 92.1 for dimer), and 24.9 kDa ± 8.0% (theoretical monomer mass, 21.9 kDa) for Rab31-Gpp(NH)p. These results are consistent with the formation of a complex comprising an hAPPL2BARPH dimer and two Rab31-Gpp(NH)p monomers in agreement with the ITC results.
DISCUSSION
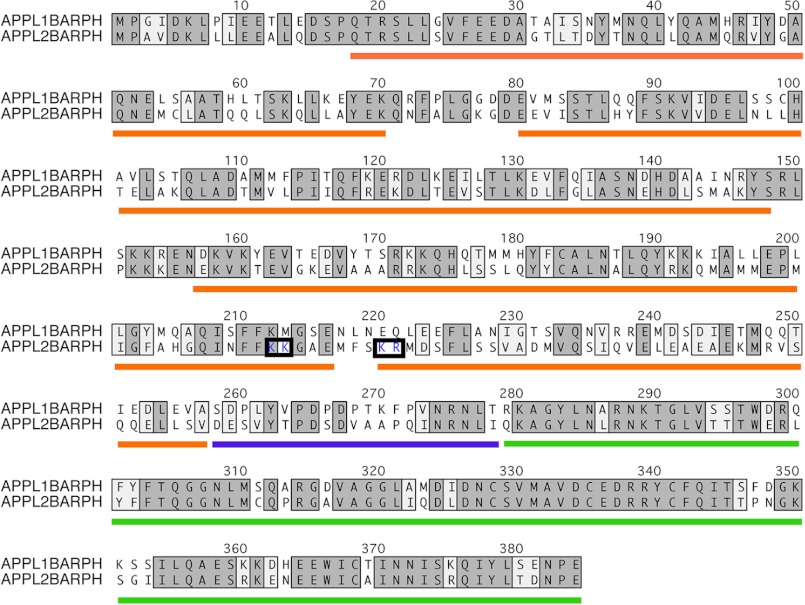
BAR domain-containing proteins are reported to possess a charge dipole across the BAR domain dimer with one face, usually the concave face, thought to be important for their ability to bind to negatively charged membrane lipids (1). The electrostatic surface of the hAPPL2BARPH dimer does reveal a concave surface that is less electronegative than its convex surface with patches of electropositive residues at the center and the ends of the banana-shaped molecule. A striking difference between the electrostatic surfaces of hAPPL1 and hAPPL2 dimers is that APPL2 has an additional basic patch on the apex of the convex surface formed by residues Lys-220 and Arg-221 of one monomer and Lys-212 and Lys-213 of the second monomer: the equivalent residues in hAPPL1 are Lys-Met and Glu-Gln, respectively (Fig. 6).
FIGURE 6.
Pair-wise alignment of full-length hAPPL1 and hAPPL2. Equivalent residues are highlighted in gray. The four helices of the BAR domain are indicated by orange underline, the PH domain by green underline, and the linker region by purple underline. The four basic residues that form the basic patch at the apex of the convex surface in APPL2BARPH are boxed and shown in blue (see Fig. 2 APPL2 (AB) convex surface). It is evident from this alignment that the PH domain is more highly conserved than the BAR domain and that helix 4 is the least conserved of the four BAR domain helices.
The crystal structure of hAPPL2BARPH incorporates two dimers, whereas APPL1 crystal structures all have one dimer in the asymmetric unit. The APPL2 structure reveals that the PH domain is able to rotate with respect to the BAR domain, and that this can occur unilaterally. SAXS studies have shown the BAR-PX domain of SNX9 is flexible (35), however, while our SAXS data from hAPPL2BARPH are consistent with a small movement of the PH domain (Fig. 4), it does not conclusively demonstrate flexibility between the BAR and PH domains of APPL2.
Many PH domains bind phosphoinositides (36), and although debate exists about the phospholipid binding ability of the PH domain of APPL1 (11, 12), the isolated PH domains of APPL1 and APPL2 have both been shown to bind phosphoinositides (14). Both APPL PH domains lack the high affinity and high specificity binding motif for phosphoinositides, KXn(K/R)XR (37). However, we found that the APPL2 PH domain residues Arg-287, Lys-289, and Trp-297 superimpose with the noncanonical inositol (1,4,5)-trisphosphate binding residues of ARHGAP9 (PDB code 2P0D) (31), suggesting that this alternate binding mode may be used. Equivalent residues are also present in hAPPL1, although are not all modeled in the available crystal structures. Nevertheless, it seems possible that both APPL proteins may bind phosphoinositides using this noncanonical binding site.
BAR domain-containing and PH domain-containing proteins are binding partners for small GTPases (10, 38), and both APPLs are Rab effectors (6). In addition, APPL1 has been shown to bind directly to Rab5 and Rab21 (12). However, relatively little is known of the interacting partners for APPL2. We therefore undertook a global screen for its Rab partners and found interactions with Rabs 5, 22a, 24, and 31. APPL2 did not bind the APPL1 partners Rab21 and none of the novel APPL2-interacting Rabs were found to interact with APPL1. Thus, our yeast two-hybrid results show that APPL1 and APPL2 differ in their Rab binding spectrum. We confirmed by ITC, MALLS in conjunction with size exclusion chromatography that Rab31 does indeed interact with hAPPL2BARPH, with both ITC and MALLS indicating the formation of a 2:2 complex.
Rab31 is also called Rab22b due to its close similarity with Rab22a. Intriguingly, all three novel APPL2-binding Rabs are members of the same branch of the Rab superfamily, with Rabs 22a and 31 more closely related than Rabs 5 and 21. Rab24 is less closely related but still resides in the same branch (39). Our results imply that during evolution the Rabs and their cognate effector proteins APPL1 and APPL2 co-evolved.
Our structural studies shed light on two potential sequence motifs of BARPH proteins (Fig. 6). First, a candidate amphipathic helix has been identified at the N terminus of the BAR domains of the BARPH proteins centaurin-β2 and oligophrenin-1 and have been suggested as possible membrane insertion motifs (40). The sequence 14SPRFRAALEEVEGDVAEL31 in centaurin-β2 aligns with residues 16SPQTRSLLSVFEEDAGTL33 of hAPPL2. These residues form an N-terminal helical region in the crystal structure of hAPPL2BARPH that interacts with a PH domain loop located between strands β3 and β4. Second, a putative nuclear localization sequence has been identified in the APPL2 sequence 151PKKKENE157 (6). Again, this motif is part of the BAR domain (loop 2), which interacts with a PH domain loop located between strands β5 and β6 in the hAPPL2BARPH crystal structure. Taken together, these data support a model where the PH domain may mask interactions with these sequence motifs. We speculate that dynamic motion of the PH domain in solution or in response to specific stimuli may alter the accessibility of these motifs, switching on or off their specific functions. B-factor analysis of the linker region between the two domains (residues 258–278) supports the notion that this region is flexible. Thus, for chain A, the average B value (all atoms) for this linker is 126 Å2 compared with 109 Å2 for the entire chain; values for chain B are 159 Å2 linker, 116 Å2 overall; and for chain D are 161 Å2 for linker, 119 Å2 overall. Curiously, the increased B-value for the linker region does not hold true for chain C in which the PH domain rotation is present: the average B value for the linker region in chain C is 110 Å2, which is similar to the overall value for the chain of 114 Å2. We note that rotation of the PH domain in chain C does not fully expose the NLS and putative membrane insertion regions, although the SAXS data suggest the PH domain can adopt multiple conformations in solution. The possibility that PH domain rotation might enable these regions to be revealed, and whether this occurs in response to specific signals, will therefore require further study.
The interaction between APPL2 and Rab31 is of considerable interest in view of the known roles of these proteins in the regulation of metabolism. Adiponectin is an adipokine, which is secreted from adipocytes to fine control insulin regulation of metabolism in other organs similar to the liver and muscle (41). APPL2 is a negative regulator of adiponectin receptor signaling, whereas APPL1 has been reported to act as a positive regulator of adiponectin action (42, 43). For example, knockdown of APPL2 in C2C12 myocytes enhances adiponectin-stimulated glucose uptake and fatty acid oxidation (42). Moreover, overexpression of Rab31 or its GTP exchange factor, Gapex-5, blocks glucose uptake in adipocytes (44). This suggests that APPL2 and Rab31 may act as a complex to negatively regulate the adiponectin signaling pathway to control glucose uptake in fat and muscle cells. If this model is correct, this would suggest that different APPL isoforms may coordinate opposing trafficking steps by invoking the activity of different Rab proteins that are the master controllers of these pathways. Consistent with this, Rab5/APPL1 controls endocytosis, whereas Rab31 is thought to control TGN/endosomal trafficking (45).
Acknowledgments
We thank Brett Collins for advice on ITC data measurement and analysis. We are grateful to the Australian Synchrotron for access to the MX2 and SAXS/WAXS beamlines and thank the beamline staff for help and advice. We also acknowledge access to the University of Queensland ROCX Diffraction Facility and thank Karl Byriel for providing research support.
This work was supported by the Australian National Health and Medical Research Council (NHMRC) Program Grant 535921. The scattering data were collected with support from Australian Synchrotron Grant AS111/SAXSFI/3171.
The atomic coordinates and structure factors (code 4H8S) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Riken Structural Genomics Initiative, unpublished data.
- PH
- pleckstrin homology
- PTB
- phosphotyrosine binding
- hAPPL2
- human APPL2
- PDB
- Protein Data Bank
- ITC
- isothermal titration calorimetry
- MALLS
- multiangle laser light scattering
- Gpp(NH)p
- guanosine 5′-(β,γ-imido)triphosphate
- SAXS
- small-angle X-ray scattering.
REFERENCES
- 1. Peter B. J., Kent H. M., Mills I. G., Vallis Y., Butler P. J., Evans P. R., McMahon H. T. (2004) BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303, 495–499 [DOI] [PubMed] [Google Scholar]
- 2. Qualmann B., Koch D., Kessels M. M. (2011) Let's go bananas: revisiting the endocytic BAR code. EMBO J. 30, 3501–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tarricone C., Xiao B., Justin N., Walker P. A., Rittinger K., Gamblin S. J., Smerdon S. J. (2001) The structural basis of Arfaptin-mediated cross-talk between Rac and Arf signalling pathways. Nature 411, 215–219 [DOI] [PubMed] [Google Scholar]
- 4. Nakano-Kobayashi A., Kasri N. N., Newey S. E., Van Aelst L. (2009) The Rho-linked mental retardation protein OPHN1 controls synaptic vesicle endocytosis via endophilin A1. Curr. Biol. 19, 1133–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Smaczynska-de Rooij I. I., Allwood E. G., Mishra R., Booth W. I., Aghamohammadzadeh S., Goldberg M. W., Ayscough K. R. (2012) Yeast dynamin Vps1 and amphiphysin Rvs167 function together during endocytosis. Traffic 13, 317–328 [DOI] [PubMed] [Google Scholar]
- 6. Miaczynska M., Christoforidis S., Giner A., Shevchenko A., Uttenweiler-Joseph S., Habermann B., Wilm M., Parton R. G., Zerial M. (2004) APPL proteins link Rab5 to nuclear signal transduction via an endosomal compartment. Cell 116, 445–456 [DOI] [PubMed] [Google Scholar]
- 7. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 8. Dowler S., Currie R. A., Campbell D. G., Deak M., Kular G., Downes C. P., Alessi D. R. (2000) Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem. J. 351, 19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Billuart P., Bienvenu T., Ronce N., des Portes V., Vinet M. C., Zemni R., Roest Crollius H., Carrié A., Fauchereau F., Cherry M., Briault S., Hamel B., Fryns J. P., Beldjord C., Kahn A., Moraine C., Chelly J. (1998) Oligophrenin-1 encodes a rhoGAP protein involved in X-linked mental retardation. Nature 392, 923–926 [DOI] [PubMed] [Google Scholar]
- 10. Habermann B. (2004) The BAR-domain family of proteins: a case of bending and binding? EMBO Rep. 5, 250–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li J., Mao X., Dong L. Q., Liu F., Tong L. (2007) Crystal structures of the BAR-PH and PTB domains of human APPL1. Structure 15, 525–533 [DOI] [PubMed] [Google Scholar]
- 12. Zhu G., Chen J., Liu J., Brunzelle J. S., Huang B., Wakeham N., Terzyan S., Li X., Rao Z., Li G., Zhang X. C. (2007) Structure of the APPL1 BAR-PH domain and characterization of its interaction with Rab5. EMBO J. 26, 3484–3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. (2000) The Protein Data Bank. Nucleic Acids Res. 28, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chial H. J., Wu R., Ustach C. V., McPhail L. C., Mobley W. C., Chen Y. Q. (2008) Membrane targeting by APPL1 and APPL2: dynamic scaffolds that oligomerize and bind phosphoinositides. Traffic 9, 215–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stols L., Gu M., Dieckman L., Raffen R., Collart F. R., Donnelly M. I. (2002) A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr. Purif. 25, 8–15 [DOI] [PubMed] [Google Scholar]
- 16. Studier F. W. (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 17. McPhillips T. M., McPhillips S. E., Chiu H. J., Cohen A. E., Deacon A. M., Ellis P. J., Garman E., Gonzalez A., Sauter N. K., Phizackerley R. P., Soltis S. M., Kuhn P. (2002) Blu-Ice and the Distributed Control System: software for data acquisition and instrument control at macromolecular crystallography beamlines. J. Synch. Rad. 9, 401–406 [DOI] [PubMed] [Google Scholar]
- 18. Otwinowski Z., Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 19. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 21. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Westlake C. J., Junutula J. R., Simon G. C., Pilli M., Prekeris R., Scheller R. H., Jackson P. K., Eldridge A. G. (2007) Identification of Rab11 as a small GTPase binding protein for the Evi5 oncogene. Proc. Natl. Acad. Sci. U.S.A. 104, 1236–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cookson D. J. (2007) SAXS15ID - Software for Acquiring, Processing and Viewing SAXS/WAXS Image Data at ChemMatCARS Argonne National Laboratory, Argonne, IL [Google Scholar]
- 24. Orthaber D., Bergmann A., Glatter O. (2000) SAXS experiments on absolute scale with Kratky systems using water as a secondary standard. J. Appl. Cryst. 33, 218–225 [Google Scholar]
- 25. Jacques D. A., Trewhella J. (2010) Small-angle scattering for structural biology–Expanding the frontier while avoiding the pitfalls. Protein Sci. 19, 642–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Whitten A. E., Cai S. Z., Trewhella J. (2008) MULCh: modules for the analysis of small-angle neutron contrast variation data from biomolecular assemblies. J. Appl. Crystallogr. 41, 222–226 [Google Scholar]
- 27. Petoukhov M. V., Svergun D. I. (2005) Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys. J. 89, 1237–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rowland S. L., Burkholder W. F., Cunningham K. A., Maciejewski M. W., Grossman A. D., King G. F. (2004) Structure and mechanism of action of Sda, an inhibitor of the histidine kinases that regulate initiation of sporulation in Bacillus subtilis. Molecular cell 13, 689–701 [DOI] [PubMed] [Google Scholar]
- 29. Folta-Stogniew E., Williams K. R. (1999) Determination of molecular masses of proteins in solution: Implementation of an HPLC size exclusion chromatography and laser light scattering service in a core laboratory. J. Biomol. Tech. 10, 51–63 [PMC free article] [PubMed] [Google Scholar]
- 30. Ferguson K. M., Lemmon M. A., Schlessinger J., Sigler P. B. (1995) Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain. Cell 83, 1037–1046 [DOI] [PubMed] [Google Scholar]
- 31. Ceccarelli D. F., Blasutig I. M., Goudreault M., Li Z., Ruston J., Pawson T., Sicheri F. (2007) Non-canonical interaction of phosphoinositides with pleckstrin homology domains of Tiam1 and ArhGAP9. J. Biol. Chem. 282, 13864–13874 [DOI] [PubMed] [Google Scholar]
- 32. Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008) Searching protein structure databases with DaliLite v.3. Bioinformatics 24, 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lemmon M. A. (2008) Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9, 99–111 [DOI] [PubMed] [Google Scholar]
- 34. Hayward S., Berendsen H. J. (1998) Systematic analysis of domain motions in proteins from conformational change: new results on citrate synthase and T4 lysozyme. Proteins 30, 144–154 [PubMed] [Google Scholar]
- 35. Wang Q., Kaan H. Y., Hooda R. N., Goh S. L., Sondermann H. (2008) Structure and plasticity of endophilin and Sorting Nexin 9. Structure 16, 1574–1587 [DOI] [PubMed] [Google Scholar]
- 36. Kutateladze T. G. (2010) Translation of the phosphoinositide code by PI effectors. Nat. Chem. Biol. 6, 507–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Isakoff S. J., Cardozo T., Andreev J., Li Z., Ferguson K. M., Abagyan R., Lemmon M. A., Aronheim A., Skolnik E. Y. (1998) Identification and analysis of PH domain-containing targets of phosphatidylinositol 3-kinase using a novel in vivo assay in yeast. EMBO J. 17, 5374–5387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bunney T. D., Opaleye O., Roe S. M., Vatter P., Baxendale R. W., Walliser C., Everett K. L., Josephs M. B., Christow C., Rodrigues-Lima F., Gierschik P., Pearl L. H., Katan M. (2009) Structural insights into formation of an active signaling complex between Rac and phospholipase C γ 2. Mol. Cell 34, 223–233 [DOI] [PubMed] [Google Scholar]
- 39. Colicelli J. (2004) Human RAS superfamily proteins and related GTPases. Sci. STKE 2004, RE13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bhatia V. K., Madsen K. L., Bolinger P. Y., Kunding A., Hedegård P., Gether U., Stamou D. (2009) Amphipathic motifs in BAR domains are essential for membrane curvature sensing. EMBO J. 28, 3303–3314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dyck D. J. (2009) Adipokines as regulators of muscle metabolism and insulin sensitivity. Appl. Physiol. Nutr. Metab. 34, 396–402 [DOI] [PubMed] [Google Scholar]
- 42. Wang C., Xin X., Xiang R., Ramos F. J., Liu M., Lee H. J., Chen H., Mao X., Kikani C. K., Liu F., Dong L. Q. (2009) Yin-Yang regulation of adiponectin signaling by APPL isoforms in muscle cells. J. Biol. Chem. 284, 31608–31615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mao X., Kikani C. K., Riojas R. A., Langlais P., Wang L., Ramos F. J., Fang Q., Christ-Roberts C. Y., Hong J. Y., Kim R. Y., Liu F., Dong L. Q. (2006) APPL1 binds to adiponectin receptors and mediates adiponectin signalling and function. Nat. Cell Biol. 8, 516–523 [DOI] [PubMed] [Google Scholar]
- 44. Lodhi I. J., Chiang S. H., Chang L., Vollenweider D., Watson R. T., Inoue M., Pessin J. E., Saltiel A. R. (2007) Gapex-5, a Rab31 guanine nucleotide exchange factor that regulates Glut4 trafficking in adipocytes. Cell Metab. 5, 59–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rodriguez-Gabin A. G., Cammer M., Almazan G., Charron M., Larocca J. N. (2001) Role of rRAB22b, an oligodendrocyte protein, in regulation of transport of vesicles from trans Golgi to endocytic compartments. J. Neurosci. Res. 66, 1149–1160 [DOI] [PubMed] [Google Scholar]
- 46. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA [Google Scholar]