Background: Lower level of breast cancer suppressor BRCA1 has been reported in sporadic breast cancers.
Results: SCFFBXO44 E3 ligase controls BRCA1 stability and is overexpressed in many sporadic breast cancers with low BRCA1 level.
Conclusion: SCFFBXO44-mediated BRCA1 stability may contribute to sporadic breast cancer development.
Significance: Regulation of BRCA1 stability provides new insights and targets on BRCA1-mediated sporadic breast cancer.
Keywords: BRCA1, Breast Cancer, E3 Ubiquitin Ligase, Protein Degradation, Ubiquitination
Abstract
BRCA1 mutations account for a significant proportion of familial breast and ovarian cancers. In addition, reduced BRCA1 protein is associated with sporadic cancer cases in these tissues. At the cellular level, BRCA1 plays a critical role in multiple cellular functions such as DNA repair and cell cycle checkpoint control. Its protein level is regulated in a cell cycle-dependent manner. However, regulation of BRCA1 protein stability is not fully understood. Our earlier study showed that the amino terminus of BRCA1 harbors a degron sequence that is sufficient and necessary for conferring BRCA1 degradation. In the current study, we used mass spectrometry to identify Skp1 that regulates BRCA1 protein stability. Small interfering RNA screening that targets all human F-box proteins uncovered FBXO44 as an important protein that influences BRCA1 protein level. The Skp1-Cul1-F-box-protein44 (SCFFBXO44) complex ubiquitinates full-length BRCA1 in vitro. Furthermore, the N terminus of BRCA1 mediates the interaction between BRCA1 and FBXO44. Overexpression of SCFFBXO44 reduces BRCA1 protein level. Taken together, our work strongly suggests that SCFFBXO44 is an E3 ubiquitin ligase responsible for BRCA1 degradation. In addition, FBXO44 expression pattern in breast carcinomas suggests that SCFFBXO44-mediated BRCA1 degradation might contribute to sporadic breast tumor development.
Introduction
Germ-line mutations of BRCA1 predispose women to breast and ovarian cancers. Studies in the past eighteen years have implicated BRCA1 in several important cellular functions including DNA repair, cell cycle checkpoint control, apoptosis, and transcriptional regulation (1, 2). The BRCA1 protein level is regulated in multiple physiological contexts (3–5). In particular, it has been shown that BRCA1 protein is degraded via a proteasome-dependent mechanism in G1 phase of the cell cycle (3), but its level arises in S/G2 phase, which is most likely coupled with its role in S phase checkpoint and DNA repair. In addition, we previously showed that BRCA1 is ubiquitinated and degraded during steroidogenesis in ovarian granulosa cells, which results in relief of BRCA1-mediated repression on steroidogenic gene expression (4). Reduced BRCA1 protein level has also been reported in a large number of sporadic breast and ovarian cancer cases. While this is likely contributed by promoter hypermethylation and thus reduced transcription of the BRCA1 gene (6–8), dysregulation at post-transcriptional levels including protein stability may also underlie the clinical observation (9–12).
BRCA1 encodes a protein of 1863 amino acids with two recognizable features: the N-terminal RING finger and the C-terminal BRCT domains (13). Missense cancer mutations of BRCA1 predominantly cluster in these two domains, highlighting their importance in the tumor suppressor function of the protein. The N-terminal RING domain interacts with another BRCT-containing protein, BARD1; and the BRCA1/BARD1 heterodimer possesses intrinsic ubiquitin E3 ligase activity that can auto-ubiquitinate BRCA1 (14–16) and also ubiquitinate other substrates (17). However, BRCA1 auto-ubiquitination, predominantly involving lysine 6 (K6) of ubiquitin for polyubiquitination, may not be directly responsible for its proteasome-sensitive degradation (3, 4, 18, 19). It has been suggested that the degradation-related ubiquitination of BRCA1 is catalyzed by other cellular E3 ligases (3, 4). We and others have shown that BRCA1 and BARD1 are both ubiquitinated under various cellular contexts (4, 14–16). Furthermore, our earlier study demonstrates that the first 324 amino acids (aa)3 of BRCA1 contains a degron that is both necessary and sufficient for BRCA1 degradation (4). Mutations within the BRCA1 degron sequence that abolish the BRCA1/BARD1 E3 ligase activity did not impair degradation of BRCA1 (4, 20), further supporting the notion that degradation-dependent ubiquitination of BRCA1 is catalized by ubiquitin E3 ligases other than BRCA1/BARD1. It was recently reported that HERC2 is a ubiquitin E3 ligase that mediates degradation of a subpopulation of BRCA1 that is not bound by BARD1 (20). As the bulk of cellular BRCA1 and BARD1 exist as a heterodimer and degradation of the two proteins can be triggered by the same signaling (4), it remains possible that ubiquitination/degradation of BRCA1 could be regulated by additional E3 ligases.
The Skp1-Cul1-F-box-protein (SCF) ubiquitin ligase is one of the most characterized mammalian E3 ligases. Extensive SCF studies reveal a well-conserved architecture for the multi-subunits of SCF complexes, with the divergent F-box proteins dictating substrate specificity (21, 22). The F-box proteins are further classified into three subfamilies: FBWs that contain WD-40 repeats and recognize specific Ser/Thr phosphorylation consensus sequences; FBLs contain leucine-rich repeats (LRRs) and also seem to prefer phosphorylated substrates characterized so far; and the rest of the F-box protein family (FBXs) that lack either WD-40 repeats or LRRs (21, 22). Several human FBXs (FBXO2, FBXO6, FBXO17, FBXO27, and FBXO44) share a conserved G domain (also known as FBA domain), which is responsible for recognizing N-glycan moieties by most FBXs in this group except FBXO44. FBXO44 does not bind to glycan and its physiological substrates remain to be identified (23–25).
In the present study, we combined mass spectrometry and screening of an F-box protein-targeted siRNA library to identify novel E3 ligase(s) that is responsible for degradation-related ubiquitination of BRCA1. Our work led to the identification of SCF complex as an important E3 ligase that regulates BRCA1 stability. Furthermore, we uncovered FBXO44 as an important F-box protein that mediates substrate recognition for BRCA1 ubiquitination both in vivo and in vitro.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
HEK293T and T47D cells were purchased from American Type Culture Collection (Manassas, VA). Transient plasmid DNA transfection was performed with Lipofectamine 2000 (Invitrogen).
Plasmids
pCR3-BRCA1, pCR3-Flag-BRCA1, pCR3-Flag-ΔN-BRCA1(303–1863aa), the untagged BRCA1-#1(1–324aa), and BRCA1-#1N(1–167aa) have been described (4). pCS2+Myc-Roc1 has been described (26). CBP-SBP-BRCA1#1 was generated by inserting cDNA corresponding to the N-terminal 1–324aa of BRCA1 to pNTAP-A, resulting in in-frame CBP tag followed by a SBP tag (Stratagene, #240103 InterPlay N-terminal Mammalian TAP System). pIRESpuro2-Flag-BRCA1, pcDNA3-Flag2AB-Skp1, Myc-hCul-1, and Myc-Skp1, pcDNA-Flag2AB-FBXO44–1 (NM_033182 from OriGene) and pcDNA-Flag2AB-FBXO44–2 (NM_183412 from Open Biosystems) cloning strategy will be provided upon request.
Tandem Affinity Purification and Mass Spectrometry
Ten 100-mm dishes of HEK293T cells were each transfected with 24 mg of pNTAP-BRCA1-#1 using Lipofectamine2000 (Invitrogen). 18 h after transfection, proteasome inhibitor MG132 (EMD, 474790) was added to a final concentration of 5 μm. 24 h after transfection, cells were harvested, and resuspended in lysis buffer (Stratagene). The TAP protocol was then performed according to the manufacturer's instructions (InterPlay TAP purification kit #240107). Eluted proteins were resolved on 10% SDS-polyacrylamide gels. The presence of BRCA1#1 was confirmed by immunoblotting with a BRCA1 antibody (EMD, OP92). Mass spectrometry was performed according to a previously reported protocol (27). SDS-PAGE slices were excised and digested by trypsin. The resulting peptides were extracted and analyzed by nanoscale LC-MS/MS on an LTQ-Orbitrap mass spectrometer (Thermo Finnigan). MS/MS spectra were searched against a protein target-decoy database for the identification of peptides/proteins. The search results were filtered to reduce protein false discovery rate to less than 0.1%.
Immunoblotting
The following antibodies were used: BRCA1 Ab1 (OP92; Calbiochem), BARD1 H-300 (sc-11438; Santa Cruz Biotechnology), α-tubulin (CD06; Calbiochem), α-Flag (F1804; Sigma), α-c-Myc (sc-789; Santa Cruz Biotechnology), α-Skp1 (sc-7163; Santa Cruz Biotechnology), α-Cullin-1 (sc-11384; Santa Cruz Biotechnology), α-β-Catenin (#9562, Cell Signaling Technology), α-β-TRCP (sc-15354; Santa Cruz Biotechnology), and α-FBXO44 (HPA003363; Sigma-Aldrich). Peroxidase-conjugated anti-mouse IgG and anti-rabbit IgG antibody (31430, 31460; Pierce) were used as the secondary antibodies for Western blotting. Blots were visualized with the enhanced chemiluminescence (Pierce).
Immunoprecipitation
18 h after transfection, MG132 was added (5 μm). 24 h after transfection, cells were harvested and lysed with 1 ml/60-mm dish of high salt lysis buffer (HSL: 50 mm Tris-HCl, pH 8.0, 1% Nonidet P-40, 500 mm NaCl) or RIPA Buffer (150 mm NaCl, 50 mm Tris pH8.0, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS), with freshly added phosphatase inhibitors and protease inhibitors mixture (20 mm NaF, 1 mm Na4P2O7, 1 mm Na3VO4, 1 mm phenylmethylsulfonyl fluoride, a.k.a. PMSF, 1 μg/ml pepstatin, 1 μg/ml leupeptin, 1 μg/ml aprotinin). The supernatants (1 ml) were mixed with 30 μl of anti-FLAG M2 beads at 4 °C overnight. The proteins bound to the beads were washed three times with either HSL or RIPA buffer. The sample was resolved by SDS-PAGE followed by immunoblotting.
In Vitro Protein Binding Assay
GST and GST fusion proteins were purified from Escherichia coli using glutathione-agarose beads and maintained as 50% slurry according to manufacturer's instruction (Pharmacia). FBXO44 was translated in vitro in the presence of [35S]methionine using TNT quick-coupled transcription/translation system (Promega, Inc.). Binding reaction in 500 ml buffer (150 mm NaCl, 50 mm Tris pH8, 1% Nonidet P-40, with the phosphatase and protease inhibitors mixture) were carried out at 4 °C for overnight, and washed four times with the same buffer and subjected to SDS-PAGE.
siRNA Knockdown
Control siRNA(d-001810), siRNA for Skp1(M-003323), Cullin-1(D-004086), FBXO44, Skp2, and FBXO5 SMARTpool siRNA were purchased from Thermo Scientific/Dharmacon. Human siGENOME siRNA library-Ubiquitin Conjugation Subset 2 was purchased from Thermo Scientific. Lipofectamine RNAiMAX (Invitrogen 13778-150) was used for siRNA transfection.
RNA Isolation and Real-time PCR
Total RNA was isolated using TRIzol Reagent (Invitrogen). RNA was reverse-transcribed using the random primers of the ImPrompII kit (Promega). The SYBR Green-based real-time PCR was conducted in an AB 7900HT system (Applied Biosystems). GAPDH was used for normalization. The sequences of PCR primers will be provided upon request.
Cycloheximide Chase Assay
BRCA1 stability was measured in the presence of cycloheximide after FBXO44 or Skp1 knockdown. 24 h after siRNA transfection, cells were split into multiple dishes. 72 h after siRNA transfection, cycloheximide was added (100 μg/ml), and cells were harvested at the indicated times.
In Vivo Ubiquitination Assays
HEK293T cells transfected with siRNA oligos were split to two 60-mm dishes 24 h after transfection. Cells were transfected again 24 h later with either pcDNA3-BRCA1-#1-N and empty vector or pcDNA3-BRCA1-#1-N and Topo-His-HA-Ub. 18 h after second DNA transfection, MG132 was added (5 μm). Cells were harvested 72 h after siRNA transfection and lysed in a phosphate/guanidine buffer (6 m guanidine-HCl; 10 mm Tris-HCl, pH 8.0; 50 mm Na2HPO4/NaH2PO4, pH 6.4; 100 mm NaCl; 10 mm imidazole; with freshly added 10 mm β-mercaptoethanol and 10 mm N-ethylmaleimide) with mild sonication. The ubiquitinated proteins were precipitated with Ni-NTA-agarose (QIA-GEN, Chatsworth, CA), followed by four washes with buffer (8 m urea; 50 mm Na2HPO4/NaH2PO4, pH 8.0; 100 mm NaCl). The precipitated proteins were eluted with NTA buffer (150 mm Tris-HCl, pH 6.8; 200 mm imidazole; 5% SDS; 30% glycerol; 0.72 m β-mercaptoethanol; 0.004% bromphenol blue), resolved by 17% SDS-PAGE, and analyzed by immunoblotting with α-BRCA1 antibody.
In Vitro Ubiquitination Assay
Recombinant SCFControl and SCFFBXO44 complexes were isolated from transfected 293T cells, as previously described (28) with minor modifications. Components of the SCF complex (Myc-Skp1, Myc-Cul1, Myc-Roc1, and Flag-FBXO44) were expressed in HEK293T cells and immunopurified with anti-FLAG M2 beads (50% slurry) and used in an in vitro Ub ligation assay. HEK293T cells that stably expressed Flag-BRCA1 (≈1 × 107 cells) were lysed by a brief sonication in 1 ml of Flag lysis buffer (50 mm Tris-HCl, pH 7.9, 137 mm NaCl, 1% Triton X-100, 0.2% Sarkosyl, 10% glycerol) supplemented with 1 mm dithiothreitol, protease, and phosphatase inhibitors mixture, and deubiquitinase inhibitors (10 mm N-ethylmaleimide, 10 nm ubiquitin aldehyde) and immunoprecipitated with anti-FLAG M2 beads. The beads were washed three times with FLAG lysis buffer and once with Nonidet P-40 buffer III. Percipitated SCFControl or SCFFBXO44 was mixed with the Flag-BRCA1 immobilized on Flag-beads. Beads were washed once with ubiquitination reaction buffer (50 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 2 mm NaF). 20 μl of ubiquitination reaction mix was added to each tube, which contained 50 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 2 mm NaF, 10 nm okadaic acid, 2 mm ATP, 0.6 mm DTT, 2 μg of ubiquitin (Boston Biochem, U-100), 100 ng E1 (Boston Biochem, E-304), 400 ng E2-UbcH5b, (Boston Biochem, E2-622). Reactions were incubated for 2 h at 30 °C with agitation at 425 rpm. To terminate reaction, 8 ml 4× LDS (Invitrogen, NP0007) with DTT was added to the reaction mixture and inclubated 10min at 70 °C. Samples were separated on 3–8% gradient Tris acetate gels (Invitrogen), and blotted with BRCA1 antibody (OP92; Calbiochem).
Immunohistochemistry
Breast cancer samples and adjacent noncancerous tissues were obtained from 307 Hospital of Academy of Military Medical Sciences with the informed consent of patients and with IRB approval. The expression of FBXO44 and BRCA1 in the clinical samples was detected by immunohistochemistry as described previously (29). Briefly, the antigens were retrieved by autoclaving the sections in citric acids buffer and incubated with rabbit anti-FBXO44 antibody (Sigma) or mouse anti-BRCA1 antibody (Calbiochem) at a dilution of 1/100 (anti-FBXO44) or 1/50 (anti-BRCA1) for 1h at room temperature. Bound antibodies were detected by the addition of biotinylated anti-rabbit or anti-mouse secondary antibody and streptavidin-horseradish peroxidase (Zymed Laboratories Inc.. The color was developed with 3,3′-diaminobenzidine. The sections were counterstained using hematoxylin, dehydrated, cleared, and sealed. For negative controls, normal rabbit or mouse IgG (Santa Cruz Biotechnology) or PBS was substituted for the primary antibody. The expression of FBXO44 and BRCA1 was classified into two categories, depending on the percentage of cells stained and/or the intensity of staining: −, 0–10% positive tumor cells; +, >10% positive tumor cells.
Statistical Analysis
Differences between variables in immunohistochemistry were assessed by χ2 analysis. Statistical calculations were performed using SPSS 13.0. p values of less than 0.05 were considered statistically significant.
RESULTS
Skp1 Regulates BRCA1 Stability
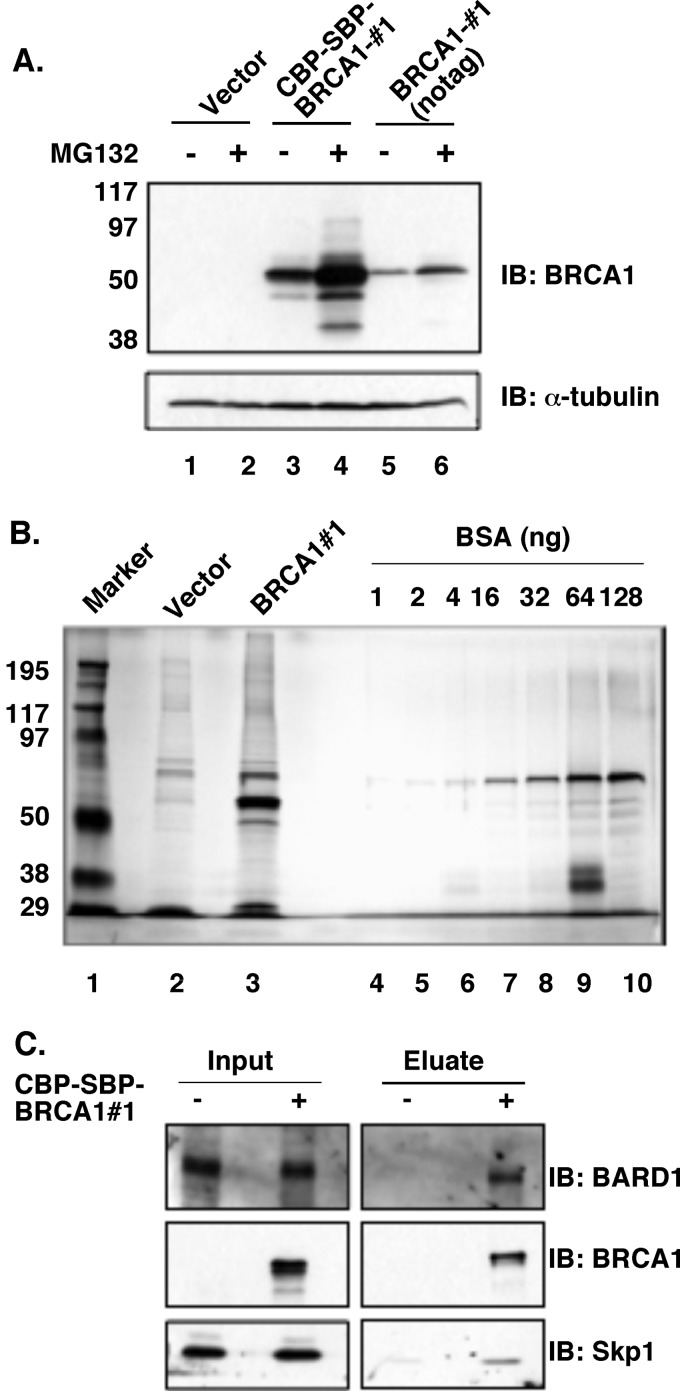
Our earlier studies identified a degron sequence within the BRCA1 N terminus that mediates BRCA1 degradation. To identify potential ubiquitin ligases that recognize the BRCA1 degron and subsequently target it for degradation, we employed tandem affinity purification (TAP) of BRCA1 N-terminal 324aa fragment (BRCA1#1) overexpressed in HEK293T cells, followed by mass spectrometry analysis of BRCA1#1-associated proteins. The BRCA1#1 fragment was tagged in tandem by sequences derived from calmodulin-binding protein (CBP) and streptavidin-binding protein (SBP), resulting in a fusion of CBP-SBP-BRCA1#1. Similar to the untagged BRCA1#1, CBP-SBP-BRCA1#1 was also stabilized by the presence of proteasome inhibitor MG132 (Fig. 1A), suggesting that the proteasome-mediated degradation of the BRCA1 degron was not affected by the addition of CBP and SBP tags. BRCA1#1 was the predominant species after affinity purification (Fig. 1B). Mass spectrometry of BRCA1-associated proteins indicates the presence of BARD1 (data not shown). In addition, Skp1, a key component in SCF ubiquitin E3 ligase, was also among the proteins associated with BRCA1#1. To confirm the mass spectrometry result, TAP purification of BRCA1#1 was repeated, and Skp1 was indeed enriched in BRCA1#1 eluate, as independently verified by Western blotting (Fig. 1C).
FIGURE 1.
Tandem affinity purification (TAP) of BRCA1 N-terminal fragment 1(BRCA1#1) for mass spectrometry. A, CBP-SBP-BRCA1#1 and native BRCA1#1 can both be stabilized in HEK293T cells in the presence of 5 μm proteasome inhibitor MG132. B, TAP-purified CBP-SBP-BRCA1#1 fragment shown in a silver-stained polyacrylamide gel. The asterisk indicates the position of BRCA1#1 fragment. Different amounts of BSA are loaded on the right for estimation of BRCA1#1 yield. C, Skp1, as well as BARD1, is enriched in the eluate of BRCA1#1 TAP.
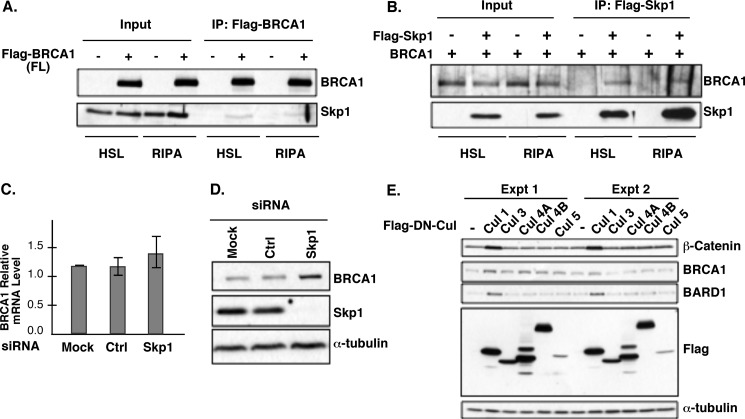
The interaction between BRCA1 and Skp1 was further validated by coimmunoprecipitation (Co-IP). Flag-tagged full-length BRCA1 was transiently transfected into HEK293T cells and immunoprecipitated by an anti-Flag antibody, the endogenous Skp1 was coimmunoprecipitated in either high salt lysis buffer (HSL) or RIPA buffer (Fig. 2A). Conversely, when Flag-Skp1 was immunoprecipitated by an anti-Flag antibody, ectopically expressed full-length BRCA1 (untagged) was also specifically coimmunoprecipitated (Fig. 2B). To determine whether steady-state level of the endogenous BRCA1 could be regulated by Skp1, small interfering RNA (siRNA) was used to knock down Skp1. Reduction of Skp1 protein led to accumulation of BRCA1 protein (Fig. 2D) without affecting the BRCA1 mRNA level (Fig. 2C), consistent with the notion that Skp1 might regulate BRCA1 protein stability. Since Skp1 functions as part of the SCF ubiquitin ligase complexes, we used a dominant negative mutant of Cul1, another key component of SCF, to further dissect the relationship between BRCA1 protein level and SCF ligases. Consistent with published data (30), the dominant negative mutant of Cul1 stabilized a well-established SCF substrate, β-catenin (Fig. 2E). In addition, the same mutant also resulted in increased BRCA1 and BARD1 protein level. The effect appears to be specific to Cul1, as dominant negative mutants of several other Cul proteins (Cul3, 4A, 4B) did not have the same impact on BRCA1 and BARD1 steady-state level (Fig. 2E). The impact of the dominant negative mutant of Cul5 remains inconclusive due to its low expression level. Taken together, our data indicate that the SCF complex plays a role in modulating the BRCA1 protein level.
FIGURE 2.
Skp1 interacts with and stabilizes BRCA1 protein. A, endogenous Skp1 is coimmunoprecipitated with Flag-BRCA1 in high salt lysis buffer (HSL) or RIPA buffer. B, overexpressed BRCA1 is coimmunoprecipitated with Flag-Skp1. C, Skp1 siRNA knockdown does not affect BRCA1 mRNA level, compared with mock treatment or control siRNA oligo. D, knockdown of Skp1 increases steady-state level of BRCA1. E, expression of a dominant negative mutant of Cul1, but not dominant negative mutants of other Cul proteins, increases steady-state level of BRCA1and BARD1. Shown here is an experiment with two independent (duplicate) transfection.
FBXO44 Mediates BRCA1 Degradation
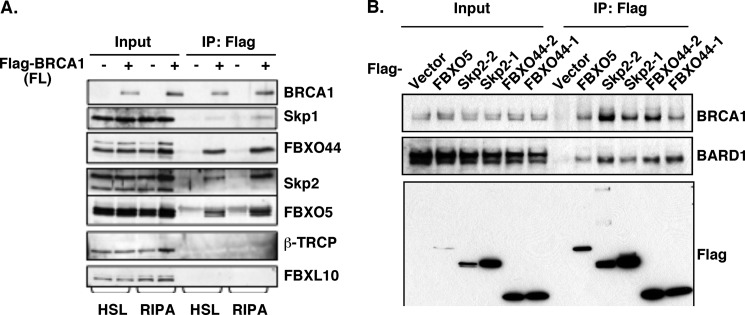
SCF complex is a family of multi-subunit ubiquitin E3 ligase, in which the F-box proteins determine the substrate specificity (21, 22). To identify potential F-box proteins that specifically recognize BRCA1 as a substrate, we conducted a screening using a siRNA library targeting all known human F-box proteins (supplemental Fig. S1, A and B). Among several potential candidates obtained from the initial screening, FBXO44, Skp2, and FBXO5 (No. 91, 56, and 33 in the screening, respectively) knockdown reproducibly resulted in accumulation of BRCA1 protein in multiple cell lines including HEK293T (supplemental Fig. S1, A and B), breast cancer cell lines T47D and MCF7 (data not shown). To determine the physical interaction between these three F-box proteins and BRCA1, we first carried out co-IP in HEK293T cells. Endogenous FBXO44, Skp2, and FBXO5 were specifically coimmunoprecipitated with Flag-BRCA1, while other F-box proteins, β-TRCP and FBXL10, were not (Fig. 3A). Conversely, when Flag-FBXO44/Skp2/FBXO5 was immunoprecipitated, the endogenous BRCA1, as well as the endogenous BARD1 was coimmunoprecipitated (Fig. 3B). The interaction between BRCA1 and these three F-box proteins appears to be stronger than that between BRCA1 and Skp1, as endogenous BRCA1 could be readily detected in Flag-FBXO44/Skp2/FBXO5 immunoprecipitates (Fig. 3B), while only overexpressed BRCA1 could be detected in the Flag-Skp1 immunoprecipitates (Fig. 2B). This is consistent with the notion that F-box proteins directly interact with their substrates while Skp1 interacts with substrates indirectly through other components of the SCF complexes. In addition, both Skp1 and Cul1 are present in the Flag-FBXO44/Skp2/FBXO5 immunoprecipitates (data not shown), consistent with the notion that each of these F-box proteins is an integral part of the SCF complex.
FIGURE 3.
FBXO44/Skp2/FBXO5 interacts with BRCA1. A, endogenous FBXO44, Skp2 and FBXO5, but not β-TRCP and FBXL10, are coimmunoprecipitated with Flag-BRCA1. B, both endogenous BRCA1 and BARD1 are coimmunoprecipitated with Flag-FBXO44/Skp2/FBXO5. Each of two isoforms of FBXO44 and Skp2 (designated 1 and 2) is transfected into HEK293T cells.
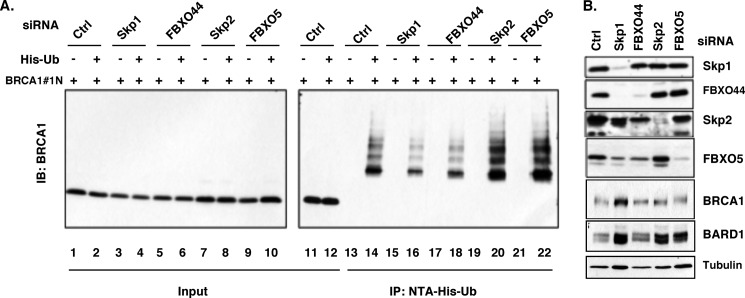
To further determine whether these F-box proteins affect the ubiquitination status of BRCA1 degron, we examined ubiquitination of the BRCA1 N-terminal fragment (1–167aa, BRCA1#1N) in the presence and absence of Skp1 or F-box proteins. BRCA1#1N and histidine-tagged ubiquitin (His-Ub) were coexpressed in HEK293T cells, where Skp1, FBXO44, Skp2 or FBXO5 was knocked down (Fig. 4B). Ubiquitinated BRCA1#1N was precipitated with Ni-NTA-agarose, and the amount of Ub-BRCA1#1N was probed with an anti-BRCA1 antibody. Consistent with our previous study, a high-molecular ladder representing the ubiquitinated BRCA1 fragment was readily detected (Fig. 4A, lane 14). Skp1 or FBXO44 knockdown significantly reduced the level of ubiquitination of BRCA1#1N (Fig. 4A, lanes 16 and 18), while Skp2 and FBXO5 knockdown did not (Fig. 4A, lanes 20 and 22). These data suggest that the FBXO44-containing SCF complex is involved in the ubiquitination of the BRCA1 N-terminal domain. In addition, the ubiquitination of BRCA1 degron by SCFFBXO44 could occur in a BARD1-independent manner, as mutations in BRCA1#1, C61G and I26A, that abolished either BARD1 interaction or its E3 activity can serve as SCFFBXO44 substrates, just like wild type BRCA1#1 (supplemental Fig. S2). Thus, findings from both in vivo and in vitro studies clearly demonstrate the ability of SCFFBXO44 E3 activity to directly ubiquitinate BRCA1 degron.
FIGURE 4.
Impact of SCFFBXO44 on BRCA1 ubiquitination. A, in vivo ubiquitination of BRCA1#1N is reduced when Skp1 or FBXO44 is knocked down. B, verification of Skp1 and FBXO44/Skp2/FBXO5 knockdown by Western blot analysis.
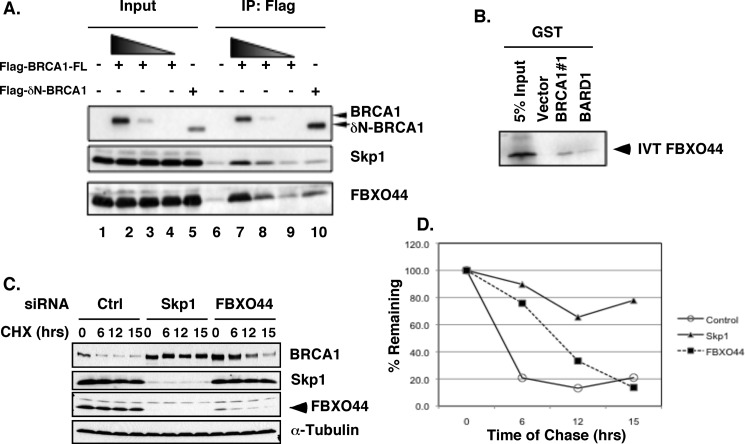
To determine whether the BRCA1 N terminus is essential for its interaction with the SCFFBXO44 complex, Flag-tagged full-length BRCA1 (FL) and N-terminal deletion of BRCA1 (ΔN, deleted first 302aa) were compared in a co-IP experiment. When a comparable amount of BRCA1-FL and BRCA1-ΔN was immunoprecipitated, significantly more Skp1 and FBXO44 were associated with full-length BRCA1 than the truncated protein (Fig. 5A, lanes 7 and 10), suggesting that the N-terminal domain of BRCA1 contributes significantly to the interaction between BRCA1 and SCFFBXO44. Furthermore, in vitro translated FBXO44 could interact with purified GST-BRCA1#1 in vitro (Fig. 5B).
FIGURE 5.
FBXO44 interacts with BRCA1 N terminus and regulates BRCA1 protein level. A, N-terminal deletion of BRCA1 significantly decreases its interaction with endogenous Skp1 and FBXO44. B, in vitro translated 35S-FBXO44 binds to GST-BRCA1#1 in vitro. C, knockdown of Skp1 or FBXO44 in HEK293T stabilizes BRCA1 protein in the presence of 100 μg/ml cycloheximide. The arrowhead indicates the position of FBXO44 (lower band of the doublet). The upper band of the doublet appears to be nonspecific. D, quantification of results from C.
To directly examine the effect of SCFFBXO44 on BRCA1 protein stability, we assessed the BRCA1 protein level in the presence of cycloheximide, an inhibitor of protein translation, after siRNA knockdown of Skp1 or FBXO44 (Fig. 5, C and D). The BRCA1 level in the control cells was significantly decreased six hours after cycloheximide treatment. In contrast, the BRCA1 level in Skp1 or FBXO44 knockdown cells was significantly higher than the control during the same period. Unlike Skp2, which expresses predominantly at S/G2 phases of cell cycle, the protein level of Skp1 and FBXO44 is constant throughout cell cycle (Fig. S3, A and B). Knockdown of Skp1 slightly increased while FBXO44 knockdown decreased S/G2/M cell population (supplemental Fig. S3C), suggesting that elevated BRCA1 protein level in FBXO44 knockdown cells was unlikely due to enriched S/G2 cells. Of note, we repeatedly observed that Skp1 knockdown led to reduction of FBXO44 protein level as well (Fig. 5C, lanes 5–8). The reason for the “co-knockdown” is not fully understood, but it suggests that FBXO44 may not be stable unless it is in a SCF complex. Taken together, these results strongly suggest that the FBXO44-containing SCF complex is responsible for BRCA1 protein degradation likely by targeting the N terminus of the protein.
SCFFBXO44 Ubiquitinates Full-length BRCA1
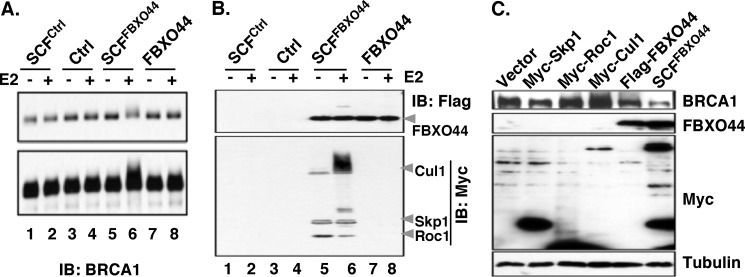
To demonstrate a direct enzyme-substrate relationship between SCFFBXO44 and BRCA1, we immunopurified the SCFFBXO44 E3 ligase and examined its ability to directly ubiquitinate immunopurified full-length BRCA1 in vitro. When ectopically coexpressed with Flag-FBXO44, all Myc-tagged subunits of SCF complex (myc-Cul1, myc-Roc1, and myc-Skp1) were present in FLAG-FBXO44 immunoprecipitates (Fig. 6B, lanes 5 and 6). When incubated with the SCFFBXO44 complex in vitro, immunoprecipitated full-length BRCA1 gave rise to a high-molecular smear above the original BRCA1 band (Fig. 6A, lane 6). The presence of BRCA1-related smear was dependent upon the presence of ubiquitin E2, strongly indicating that the high-molecular species represented polyubiquitinated BRCA1 (Fig. 6A, compare lanes 5 and 6). Immunoprecipitates from lysates that contained all ectopical SCF subunits except FBXO44 (SCFctrl) did not produce any detectable polyubiquitinated products (Fig. 6A, lane 2), nor did immunoprecipiated FBXO44 alone without co-expressing the other SCF subunits (Fig. 6A, lane 8). This strongly suggests that the entire SCFFBXO44 complex is required for polyubiquitination of BRCA1. As BRCA1 alone in the absence of SCF immunoprecipitates did not display any polyubiquitination either (Fig. 6A, lane 4), we interpret this to mean that the observed polyubiquitination was not due to BRCA1 autoubiquitination. Consistent with the notion that SCFFBXO44 mediates BRCA1 degradation, overexpression of SCFFBXO44 markedly reduced endogenous BRCA1 level (Fig. 6C).
FIGURE 6.
A, in vitro ubiquitination of full-length BRCA1 by immunoprecipitated SCFFBXO44. Lower panel shows a longer exposure of the same blot. B, immunoblotting of Flag-FBXO44, myc-Skp1, myc-Cul1, and myc-Roc1 in immunopurified SCFFBXO44. The smear and ladders above the designated molecule (lane 6) reflect autoubiquitination of these SCF subunits. BRCA1 ubiquitination shows a tail-like smear, due to high molecular weight of BRCA1. C, overexpression of SCFFBXO44 reduces endogenous BRCA1 protein level in HEK293T cells.
Expression of FBXO44 in Breast Cancer Cell Lines and Tumor Specimens
It is believed that BRCA1 functions as a tumor suppressor mainly by its role in DNA repair. To determine whether SCFFBXO44 could be involved in BRCA1-mediated DNA damage response, colocalization of BRCA1 and FBXO44 was examined by immunofluorescence in U2OS cells in response to DNA damage produced by laser microirradiation. Unlike BRCA1 and γH2AX, which are colocalized at laser stripes corresponding to damaged DNA sites, FBXO44 does not localize at stripes (supplemental Fig. S4), suggesting that SCFFBXO44 may not be directly involved in DNA repair function of BRCA1.
To determine whether SCFFBXO44 could contribute to breast cancer development, FBXO44 protein level was knocked down in breast cancer cell lines T47D and MCF7, and its impact on cell growth was examined 3–5 days after siRNA transfection. Compared with control siRNA, FBXO44-specific siRNA knockdown led to a moderate, but statistically significant growth inhibition (data not shown). We also examined the expression of FBXO44 by immunohistochemistry in 72 pairs of human breast tumors and matched non-tumor breast tissues. FBXO44 was distributed predominantly in nuclei (Fig. 7A). 80.6% (58/72) of cancers expressed FBXO44, while only 22.2% (16/72) of non-cancerous tissues stained positive for FBXO44 (p < 10−4). Focusing on paired tumor and normal tissues, none of normal tissues expressed higher levels of FBXO44 than cancers; in 58.3% (42/72) of patients, the expression levels of FBXO44 in tumors were higher than those in adjacent normal tissues; and in 41.7% (30/72) of patients, normal tissue and breast cancer had similar staining patterns. The specificity of the staining was confirmed by immunobloting with anti-FBXO44 (supplemental Fig. S5A) and immunohistochemistry using normal serum substituted for anti-FBXO44 antibody (supplemental Fig. S5B). Intriguingly, in 97 breast cancer patients examined, FBXO44 inversely correlated with BRCA1 expression (p = 0.001) (Fig. 7B). These data suggest that SCFFBXO44-mediated BRCA1 degradation may contribute to sporadic breast cancer development.
FIGURE 7.
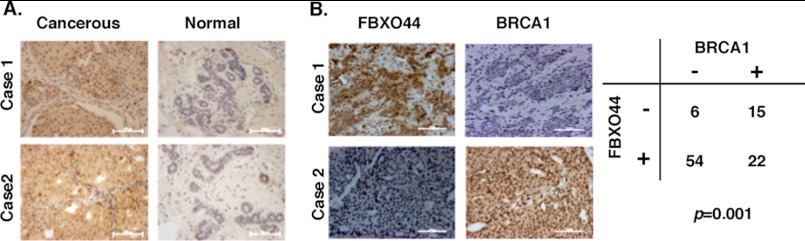
FBXO44 expression in human breast tumors and matched non-tumor breast tissues and its correlation with BRCA1 expression. A, representative cases of breast carcinomas showed positive immunohistochemical staining of cancerous cells compared with negative staining of adjacent non-cancerous cells using anti-FBXO44. Original magnification, ×20. Scale bar, 100 μm. B, inverse correlation between FBXO44 and BRCA1 expression in human breast cancer samples. Representative immunohistochemical staining of FBXO44 and BRCA1 proteins was shown in the upper panel. A summary of 97 samples is shown in the lower panel. The p value was generated by Chi-square test. Original magnification, ×20. Scale bar, 100 μm.
DISCUSSION
BRCA1 is a multi-functional tumor suppressor. Although BRCA1 mutations mainly affect familial breast and ovarian cancers, reduced BRCA1 protein level has been associated with some sporadic breast and ovarian cancer cases (9–12). A wealth of information has been accumulated over the last decade regarding BRCA1 functions, but regulation of BRCA1 protein level and stability remains underinvestigated. In this report, we extended our previous finding on BRCA1 degron by identifying a SCF complex, SCFFBXO44, as one ubiquitin E3 ligase that targets BRCA1 for degradation. We also provide both in vitro and in vivo evidence for a direct substrate-enzyme relationship between SCFFBXO44 and BRCA1. It has been long suggested that other ubiquitin E3 ligases distinct from BRCA1/BARD1-mediated autoubiquitination are required for BRCA1 degradation. The identification of SCFFBXO44 as a ubiquitin E3 ligase for BRCA1 not only supports the long-held notion, but also offers new players for elucidating regulation of BRCA1 stability in physiological and pathological contexts. Down-regulation of BRCA1 in sporadic breast cancer patients has been reported in the literature. The mechanism underlying this observation was attributed to BRCA1 promoter methylation. We demonstrated for the first time another novel mechanism, i.e. BRCA1 could be degraded by FBXO44, which is overexpressed in most breast cancer patients.
Another ubiquitin E3 ligase, HERC2, has recently been reported to interact with the BRCA1 degron and target BRCA1 for degradation (20), although it remains to be demonstrated that HERC2 could ubiquitinate full-length BRCA1. Despite the similar strategy used in the published and current studies, Skp1 was the only potential E3 identified in our mass spectrometry and HERC2 was not one of the hits. This could be due to different purification approaches and/or sensitivity of mass spectrometry. It is conceivable that different E3 ligases may be involved in different physiological processes for BRCA1 degradation by targeting different pools of BRCA1 molecules. Consistent with this notion, SCFFBXO44 and HERC2 clearly display several differences in their shared role of BRCA1 degradation. For example, HERC2 targets BARD1-unbound BRCA1 for degradation. Knockdown of HERC2 stabilizes BRCA1 only when BARD1 is depleted (20). Indeed, when compared side by side with knockdown of SCFFBXO44, HERC2 depletion alone did not result in stabilization of BRCA1 protein.4 Furthermore, both BRCA1 and BARD1 were coimmunoprecipitated with Flag-FBXO44 and stability of both proteins were affected by knockdown of SCFFBXO44, suggesting that FBXO44 could recognize BRCA1/BARD1 complex.
BRCA1 plays an important role in DNA repair and G2-M checkpoint (31). Interestingly, the BRCA1 expression level peaks at G2-M while its ubiquitination and degradation peak at S phase (3). We observed that overexpression of SCFFBXO44 could reduce endogenous BRCA1 level. It remains to be determined how BRCA1 ubiquitination/degradation is integrated into its function in DNA repair and cell cycle checkpoint control.
FBXO44 belongs to a subfamily of the F-box protein (FBA family) that shares a conserved G domain for substrate binding. In addition to the sequence homology, all members of the FBA family except FBXO44 recognize glycosylated substrates (23–25). The current study uncovers the first substrate for SCFFBXO44. It will be interesting to determine whether the same SCFFBXO44 complex can target other proteins that share common recognition motif with BRCA1. It also remains to be tested whether SCFFBXO44-mediated ubiquitination of BRCA1 is preceded by phosphorylation events, a common theme in SCF-mediated protein degradation (21, 22). In the database, FBXO44 exists in two isoforms, 255aa and 224aa in length, with diverged C-terminal sequence resulting from alternative splicing. Available antibodies and siRNA oligos do not distinguish these two isoforms. However, only one single band could be detected in multiple cell lines (HeLa, HEK293, T47D, MCF7, and KGN). Because both isoforms interact with BRCA1 when expressed exogenously, the FBXO44–1 (255kD) was used in subsequent analyses involving overexpression of FBXO44. The potential functional difference of the two isoforms remains to be addressed in future studies.
Acknowledgments
We thank Drs. Wei Gu, David Livingston, and Zhengqiang Pan for plasmids used in this study, and Tao Kang for technical assistance. We thank Dr. Rong Li for critical reading of the manuscript.
This study was supported, in whole or in part, by National Institutes of Health Grant (to Y. H.) R01CA118578 and the cancer center support grant P30CA054174, a Research Scholar Grant RSG-09-181-01 from the American Cancer Society (to J. P.) and Major State Basic Research Development Program 2011CB504202 and National Natural Science Foundation 81072173 (to Q. Y.).

This article contains supplemental Figs. S1–S5.
Y. Lu, unpublished data.
- aa
- amino acids
- SCF
- Skp1-Cul1-F-box-protein
- TAP
- tandem affinity purification
- FBX
- F-box protein family
- CBP
- calmodulin-binding protein
- SBP
- streptavidin-binding protein.
REFERENCES
- 1. King M., Marks J., Mandell J., and Group, New York Breast Cancer Study Group (2003) Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science 302, 643–646 [DOI] [PubMed] [Google Scholar]
- 2. Narod S. A., Foulkes W. D. (2004) BRCA1 and BRCA2: 1994 and beyond. Nat. Rev. Cancer 4, 665–676 [DOI] [PubMed] [Google Scholar]
- 3. Choudhury A. D., Xu H., Baer R. (2004) Ubiquitination and proteasomal degradation of the BRCA1 tumor suppressor is regulated during cell cycle progression. J. Biol. Chem. 279, 33909–33918 [DOI] [PubMed] [Google Scholar]
- 4. Lu Y., Amleh A., Sun J., Jin X., McCullough S. D., Baer R., Ren D., Li R., Hu Y. (2007) Ubiquitination and proteasome-mediated degradation of BRCA1 and BARD1 during steroidogenesis in human ovarian granulosa cells. Mol. Endocrinol. 21, 651–663 [DOI] [PubMed] [Google Scholar]
- 5. Marquis S. T., Rajan J. V., Wynshaw-Boris A., Xu J., Yin G.-Y., Abel K. J., Weber B. L., Chodosh L. A. (1995) The developmental pattern of Brca1 expression implies a role in differentiation of the breast and other tissues. Nat. Genet. 11, 17–26 [DOI] [PubMed] [Google Scholar]
- 6. Catteau A., Harris W. H., Xu C. F., Solomon E. (1999) Methylation of the BRCA1 promoter region in sporadic breast and ovarian cancer: correlation with disease characteristics. Oncogene 18, 1957–1965 [DOI] [PubMed] [Google Scholar]
- 7. Chan K. Y., Ozçelik H., Cheung A. N., Ngan H. Y., Khoo U. S. (2002) Epigenetic factors controlling the BRCA1 and BRCA2 genes in sporadic ovarian cancer. Cancer Res. 62, 4151–4156 [PubMed] [Google Scholar]
- 8. Esteller M., Silva J. M., Dominguez G., Bonilla F., Matias-Guiu X., Lerma E., Bussaglia E., Prat J., Harkes I. C., Repasky E. A., (2000) Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. Journal of the National Cancer Institute 5, 564–569 [DOI] [PubMed] [Google Scholar]
- 9. Lambie H., Miremadi A., Pinder S., Bell J., Wencyk P., Paish E., Macmillan R., Ellis I. (2003) Prognostic significance of BRCA1 expression in sporadic breast carcinomas. J. Pathol. 200, 207–213 [DOI] [PubMed] [Google Scholar]
- 10. Russell P., Pharoah P., Foy K. D., Ramus S., Symmonds I., Wilson A., Scott I., Ponder B., Gayther S. (2000) Frequent loss of BRCA1 mRNA and protein expression in sporadic ovarian cancers. Int. J Cancer 87, 317–321 [DOI] [PubMed] [Google Scholar]
- 11. Yoshikawa K., Honda K., Inamoto T., Shinohara H., Yamauchi A., Suga K., Okuyama T., Shimada T., Kodama H., Noguchi S., Gazdar A., Yamaoka Y., Takahashi R. (1999) Reduction of BRCA1 protein expression in Japanese sporadic breast carcinomas and its frequent loss in BRCA1-associated cases. Clin. Cancer Res. 5, 1249–1261 [PubMed] [Google Scholar]
- 12. Yoshikawa K., Ogawa T., Baer R., Hemmi H., Honda K., Yamauchi A., Inamoto T., Ko K., Yazumi S., Motoda H., Kodama H., Noguchi S., Gazdar A., Yamaoka Y., Takahashi R. (2000) Abnormal expression of BRCA1 and BRCA1-interactive DNA-repair proteins in breast carcinomas. Int. J Cancer 88, 28–36 [PubMed] [Google Scholar]
- 13. Miki Y., Swensen J., Shattuck-Eidens D., Futreal P. A., Harshman K., Tavtigian S., Liu Q., Cochran C., Bennett L. M., Ding W. (1994) A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266, 66–71 [DOI] [PubMed] [Google Scholar]
- 14. Baer R., Ludwig T. (2002) The BRCA1/BARD1 heterodimer, a tumor suppressor complex with ubiquitin E3 ligase activity. Curr. Opin. Genet. Dev. 12, 86–91 [DOI] [PubMed] [Google Scholar]
- 15. Ohta T., Fukuda M. (2004) Ubiquitin and breast cancer. Oncogene 23, 2079–2088 [DOI] [PubMed] [Google Scholar]
- 16. Wu L. C., Wang Z. W., Tsan J. T., Spillman M. A., Phung A., Xu X. L., Yang M. C., Hwang L. Y., Bowcock A. M., Baer R. (1996) Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat. Genet. 14, 430–440 [DOI] [PubMed] [Google Scholar]
- 17. Horwitz A. A., Affar el B., Heine G. F., Shi Y., Parvin J. D. (2007) A mechanism for transcriptional repression dependent on the BRCA1 E3 ubiquitin ligase. Proc. Natl. Acad. Sci. U.S.A. 104, 6614–6619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nishikawa H., Ooka S., Sato K., Arima K., Okamoto J., Klevit R. E., Fukuda M., Ohta T. (2004) Mass spectrometric and mutational analyses reveal Lys-6-linked polyubiquitin chains catalyzed by BRCA1-BARD1 ubiquitin ligase. J. Biol. Chem. 279, 3916–3924 [DOI] [PubMed] [Google Scholar]
- 19. Wu-Baer F., Lagrazon K., Yuan W., Baer R. (2003) The BRCA1/BARD1 heterodimer assembles polyubiquitin chains through an unconventional linkage involving lysine residue K6 of ubiquitin. J. Biol. Chem. 278, 34743–34746 [DOI] [PubMed] [Google Scholar]
- 20. Wu W., Sato K., Koike A., Nishikawa H., Koizumi H., Venkitaraman A. R., Ohta T. (2010) HERC2 is an E3 ligase that targets BRCA1 for degradation. Cancer Res. 70, 6384–6392 [DOI] [PubMed] [Google Scholar]
- 21. Cardozo T., Pagano M. (2004) The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5, 739–751 [DOI] [PubMed] [Google Scholar]
- 22. Petroski M. D., Deshaies R. J. (2005) Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 [DOI] [PubMed] [Google Scholar]
- 23. Glenn K. A., Nelson R. F., Wen H. M., Mallinger A. J., Paulson H. L. (2008) Diversity in tissue expression, substrate binding, and SCF complex formation for a lectin family of ubiquitin ligases. J. Biol. Chem. 283, 12717–12729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ilyin G. P., Sérandour A.-L., Pigeon C., Rialland M., Glaise D., Guguen-Guillouzo C. (2002) A new subfamily of structurally related human F-box proteins. Gene 296, 11–20 [DOI] [PubMed] [Google Scholar]
- 25. Winston J. T., Koepp D. M., Zhu C., Elledge S. J., Harper J. W. (1999) A family of mammalian F-box proteins. Curr. Biol. 9, 1180–1182 [DOI] [PubMed] [Google Scholar]
- 26. Lin H.-R., Chuang L.-C., Boix-Perales H., Philpott A., Yew P. R. (2006) Ubiquitination of cyclin-dependent kinase inhibitor, Xic1, is mediated by the Xenopus F-box protein xSkp2. Cell Cycle 5, 304–314 [DOI] [PubMed] [Google Scholar]
- 27. Xu P., Duong D. M., Seyfried N. T., Cheng D., Xie Y., Robert J., Rush J., Hochstrasser M., Finley D., Peng J. (2009) Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 137, 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Strohmaier H., Spruck C. H., Kaiser P., Won K.-A., Sangfelt O., Reed S. I. (2001) Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature 413, 316–322 [DOI] [PubMed] [Google Scholar]
- 29. Zhang H., Xie X., Zhu X., Zhu J., Hao C., Lu Q., Ding L., Liu Y., Zhou L., Liu Y., Huang C. C., Wen C., Ye Q. (2005) Stimulatory cross-talk between NFAT3 and estrogen receptor in breast cancer cells. J. Biol. Chem. 280, 43188–43197 [DOI] [PubMed] [Google Scholar]
- 30. Jin J., Ang X. L., Shirogane T., Wade Harper J. (2005) Identification of substrates for F-box proteins. Methods Enzymol. 399, 287–309 [DOI] [PubMed] [Google Scholar]
- 31. Huen M. S. Y., Shirley M. H. S., Chen J. (2010) BRCA1 and its toolbox for the maintenance of genome integrity. Nat. Rev. Mol. Cell Biol. 11, 139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]