Background: The lipid and protein contributions to the mechanical properties of the influenza viral envelope are unknown.
Results: The influenza viral envelope is 10 times softer than a viral protein-capsid coat but stiffer than a liposome.
Conclusion: Membrane-associated proteins contribute to the mechanical stiffness of the viral envelope.
Significance: The mechanical properties of the envelope are critical for the viral pH-regulated life cycle.
Keywords: Atomic Force Microscopy, Influenza Virus, Liposomes, Membrane Biophysics, Virus
Abstract
The envelope of the influenza virus undergoes extensive structural change during the viral life cycle. However, it is unknown how lipid and protein components of the viral envelope contribute to its mechanical properties. Using atomic force microscopy, here we show that the lipid envelope of spherical influenza virions is ∼10 times softer (∼0.05 nanonewton nm−1) than a viral protein-capsid coat and sustains deformations of one-third of the virion's diameter. Compared with phosphatidylcholine liposomes, it is twice as stiff, due to membrane-attached protein components. We found that virus indentation resulted in a biphasic force-indentation response. We propose that the first phase, including a stepwise reduction in stiffness at ∼10-nm indentation and ∼100 piconewtons of force, is due to mobilization of membrane proteins by the indenting atomic force microscope tip, consistent with the glycoprotein ectodomains protruding ∼13 nm from the bilayer surface. This phase was obliterated for bromelain-treated virions with the ectodomains removed. Following pH 5 treatment, virions were as soft as pure liposomes, consistent with reinforcing proteins detaching from the lipid bilayer. We propose that the soft, pH-dependent mechanical properties of the envelope are critical for the pH-regulated life cycle and support the persistence of the virus inside and outside the host.
Introduction
The influenza virus causes yearly epidemics of respiratory infections in millions of human beings, and the threat of a pandemic caused by new strains of this virus stimulates investigation of its pathogenic properties (1). The influenza virus belongs to the orthomyxoviridae family of enveloped viruses (2). Influenza virions are made out of a ribonucleoprotein (RNP)5 core composed of eight RNA-protein complexes (3) associated with viral polymerase (4). The virions obtain a lipid envelope containing two types of spike glycoproteins, hemagglutinin (HA) (5, 6) and neuraminidase (NA) (7) by budding through the host cell membrane. The matrix protein M1 forms a layer associated with the inside of the membrane and connects RNPs to the envelope (8, 9). The architecture of the influenza virus has been widely studied using electron microscopy (EM) (8–12), but critical details of its structural arrangements and pH-dependent rearrangements remain unknown. Specifically, very little is known about the relationship between the structure and the mechanical properties of the viral envelope and associated proteins. This, however, is critical for the membrane dynamics during viral assembly and budding, viral entry, and membrane fusion as well as for viral environmental persistence (13, 14). In order to develop more detailed molecular models of these membrane dynamics, it is crucial to resolve the mechanical contribution of the lipid and protein components of the viral structure.
The viral envelope is enriched in sphingolipids and cholesterol that associate with plasma membrane proteins to form dynamic rafts at the nanoscale; these are thought to merge by cross-linking to form larger platforms (13, 15, 16). The exact composition and physical state of the lipid component is still controversial, but they have important implications for the mechanical properties (17). Although virions are thought to bud from liquid-ordered state raft domains of the infected cells (18), recent data suggest that most lipids in the envelope are in a liquid-disordered state at physiological temperature (19, 20). HA and NA consist of a prominent ectodomain (or “spike”) that protrudes ∼13–14 nm from the surface of the lipid bilayer, a transmembrane, and a small cytoplasmic tail domain (6, 18, 21). Whereas HA mediates viral attachment to and entry into the host cell in a pH-dependent manner, NA cleaves the host receptor and enables release of progeny virions. The rigidity and dense packing of the spike glycoproteins in the viral envelope seen in EM suggest a possible functional role for the mechanical stiffness of the virus particle (8, 9, 22). The matrix protein M1 might also contribute to the stiffness because it oligomerizes and seems to form a helical network adjacent to the membrane, at least in filamentous virions (8, 9, 22).
The aim of our study here was to investigate the mechanical stiffness of single native influenza virions. In order to address the contribution of the lipid and protein components of the viral envelope we used simplified model systems, such as pure liposomes, as well as reconstituted virosomes made of defined lipids and glycoproteins. We also studied the effect of low pH as well as the effect of removing the ectodomains of the glycoproteins on the stiffness of the viral envelope. Our data revealed that influenza virions are about 10 times softer than virus particles enclosed by a protein capsid shell, such as DNA-bacteriophage and cowpea chlorotic mottle virus (23, 24), or enveloped viruses with a submembranous capsid, such as herpes simplex, HIV, or murine leukemia virus particles (25–27).
EXPERIMENTAL PROCEDURES
Liposome and Virosome Preparation
Small unilamellar vesicles were prepared by dissolving phosphatidylcholine from egg yolk (egg PC; Avanti Polar Lipids) in chloroform followed by drying under vacuum for 40 min as described previously (28). The lipids were resuspended in phosphate-buffered saline (PBS) to 1 mm and sonicated for 10 min in a bath sonicator. PBS contained 145 mm NaCl, 10 mm sodium phosphate, pH 7.3. After five freeze-thaw cycles, 10 extrusions were performed under N2 pressure on stacks of four 100- or 200-nm pore filters. This procedure yields ≥90% unilamellar vesicles (28). Size and unilamellarity of the vesicles were checked using dynamic light scattering and electron microscopy. For mixed lipid liposomes, 30% cholesterol and 70% egg PC were mixed in chloroform before being dried and resuspended in PBS as described above. Virosomes were prepared as described (29). In brief, purified Aichi/H3N2 X-31 (A/X31) influenza hemagglutinin dispersed in PBS plus 1% octyl glucoside (Melford) was added to extruded egg PC vesicles. The molar ratio of HA to lipid was 1:100 (mol/mol). In this condition, the detergent was incorporated into the liposomes at saturating concentration without the liposomes becoming dispersed into lipid/detergent micelles. This was checked using light scattering. After mixing HA, detergent, and liposomes, the sample was incubated for 3 h at room temperature. Then the detergent was removed by dialysis against PBS and Bio-Beads (Bio-Rad). The virosomes were finally separated from liposomes and unincorporated HA on a sucrose gradient.
Virus Preparation
A/X-31 viruses were grown and purified as described previously (30). Briefly, viruses were grown in the allantoic cavity of 10-day-old embryonated hen eggs. Allantoic fluid was extracted and then clarified by centrifugation, and virus particles were finally purified on 10–30% sucrose gradients. Viruses were resuspended in PBS to 10 mg ml−1. To prepare “bald” viruses, we removed the ectodomain of the spike glycoproteins by bromelain digestion (31). Both untreated and bromelain-treated viruses were inspected by electron microscopy. In order to study the effect of low pH treatment on the mechanical properties of virus particles, viruses were incubated for 5 min in a PBS buffer adjusted to pH 4.9 by the addition of 200 mm sodium citrate before the solution was readjusted to pH 7.3 by the addition of 1 m Tris solution, as described previously (32). Bromelain-treated A/X-31 virus samples were analyzed by 12.5% SDS-PAGE in non-reducing conditions. Gels were stained with Coomassie Blue. Molecular weight markers Seeblue Plus2 were obtained from Invitrogen.
Electron Microscopy
For cryo-electron microscopy (cryo-EM), the virus preparation was applied to glow-discharged Quantifoil grids, blotted manually, and plunged into liquid ethane. Micrographs were taken with Eastman Kodak Co. SO163 film in low dose conditions on a Philips Tecnai T12 electron microscope at 120 kV and a magnification of ×42,000. The micrographs were digitized with a step size of 7 μm on a KZA scanner (Medical Research Council, Cambridge, UK), corresponding to 3.33 Å on the object scale. For negative stain electron microscopy, the virus or liposome samples were diluted in PBS, pH 7.3. The samples were applied to UV-treated, continuous carbon-coated copper grids and negatively stained with 1% sodium phosphotungstate, pH 7.0. Images were taken in low dose conditions at ×50,000 magnification.
Dynamic Light Scattering
Dynamic light scattering was used to measure the hydrodynamic radius of the particles. The samples (0.1 mm lipid or less) were analyzed in a quartz cuvette using a Visocotek Omnisize apparatus.
Force Spectroscopy
Force spectroscopy was carried out with a JPK Instruments (Berlin, Germany) atomic force microscope (AFM). In order to image the virions using the AFM, the virions were immobilized on coverslips. Because the virions did not attach to clean glass or mica, we functionalized the coverslips with an aminosilane to introduce a positive surface charge. The improved surface attachment of the viral particles was probably due to the prominent spike glycoprotein hemagglutinin interacting with positive charges above pH 7 (33). Microscope coverslips were cleaned using KOH and given a positive surface charge by coating them with 3-(2,2-aminoethylamino))-ethylaminopropyltrimethoxysilane (Sigma-Aldrich) as described (34). A single drop (20 μl) of 10 μg ml−1 virus or liposome in PBS (pH 7.3) was deposited on the silanized glass coverslip and left for 30 min to allow the particles to adhere to the surface. Cantilevers (Olympus BL-RC150) were prewetted with another 20 μl of PBS buffer. The temperature of the samples during the experiments was 27 ± 2 °C. Imaging was performed in dynamic mode, oscillating the cantilever at ∼9 kHz with an amplitude of ∼2 nm.
In order to determine the stiffness of all studied species, we generated force maps of individual particles. Each force map was obtained by generating force versus distance (FZ) curves at 2 μm s−1 (sampled at 12 kHz) in an array of 18 × 18 points, covering a square region of 250 × 250 nm encompassing the virus particle. FZ curves were obtained by extending the z-piezo until the cantilever tip established mechanical contact and indented the virus particle with forces up to 0.5 nN. Height images were reconstructed by plotting the z-piezo height of the contact point for each FZ curve. An automated routine selected the center of the particle and averaged curves obtained in a central circle with a 20-nm radius on top of each particle (typically five curves from the center region for each particle) because we found that the stiffness within this area was constant.
To convert the force-distance curves into force-indentation curves, we considered the cantilever and the particle as two springs in series. The spring constant of the cantilever was subtracted from the total spring constant to get the spring constant of the particle. We calibrated the spring constant κcant of each of the 60 cantilevers in our experiments by using the equipartition theorem (35), with κcant = 0.038 ± 0.07 nN nm−1 (mean ± S.D.) and subtracted the obtained value from the measured curves. In order to quantify the viral and liposome elasticity for indentation forces above 100 pN, we performed a linear fit between 100 and 200 pN. The height of the particles was determined by calculating the difference between the z-position of the contact point between the tip and the object (i.e. the onset of the indentation in the FZ curves) and the background FZ curves.
Modeling
Deformation of a spherical shell supported by a flat surface and indented by a hyperbolic AFM tip was calculated using the expected Young's modulus for a pure lipid bilayer and a protein shell, respectively. We used finite element methods (COMSOL Multiphysics 3.5, Stockholm, Sweden) to create a spherical model made of a homogeneous elastic material. The mechanical elements were described as thin shells where in-plane buckling and out-of-plane compression of the shell were ignored. This approach can be used for objects with a relatively thin shell and allows much faster computation (20, 36). We further reduced the model to a quarter-sphere of ∼2000 elements by making use of the symmetry. The sphere was supported by a flat surface and indented from the top with a parabolic tip with f(x) = x2/2p; the focal parameter p = 25 nm. We included these non-linear boundary conditions by using a contact-penalty stiffness method. The force resulting from indentation was obtained by integrating the vertical components of the load over the total contact area, and the horizontal components of the load cancelled out due to symmetry. Models were solved using a parametric nonlinear solver, where the solved parameter was the stepwise lowering of the tip onto the sphere.
The elastic properties of the shell were defined by its Young's modulus E and the shell thickness h. These two parameters are related to the bending rigidity Kb and the area compressibility modulus Ka, which are generally used to describe the elastic properties of lipid bilayers.
 |
 |
In agreement with the literature, we set the Poisson ratio ν at 0.5 and α = 24 (37). Because the ratio Kb/Ka was found to be almost constant at 3–5 × 10−19 m2 (38), we chose Kb/Ka = 4 × 10−19 m2 = h2/24. This gave an effective shell thickness of h = 3.1 nm. In this simplified model, the lipid bilayer, which consists of two coupled leaflets, was replaced by a single sheet. The single sheet had a reduced thickness compared with the real thickness of the lipid bilayer but had the same bending rigidity and an area compressibility modulus like the bilayer (20). E was varied to fit the experimental values we measured with the pure egg PC liposomes. A best match was obtained with E = 30 megapascals, corresponding to a Kb of 0.5 × 10−19 Nm.
RESULTS
Cryo-EM and Atomic Force Microscopy of Influenza Virions
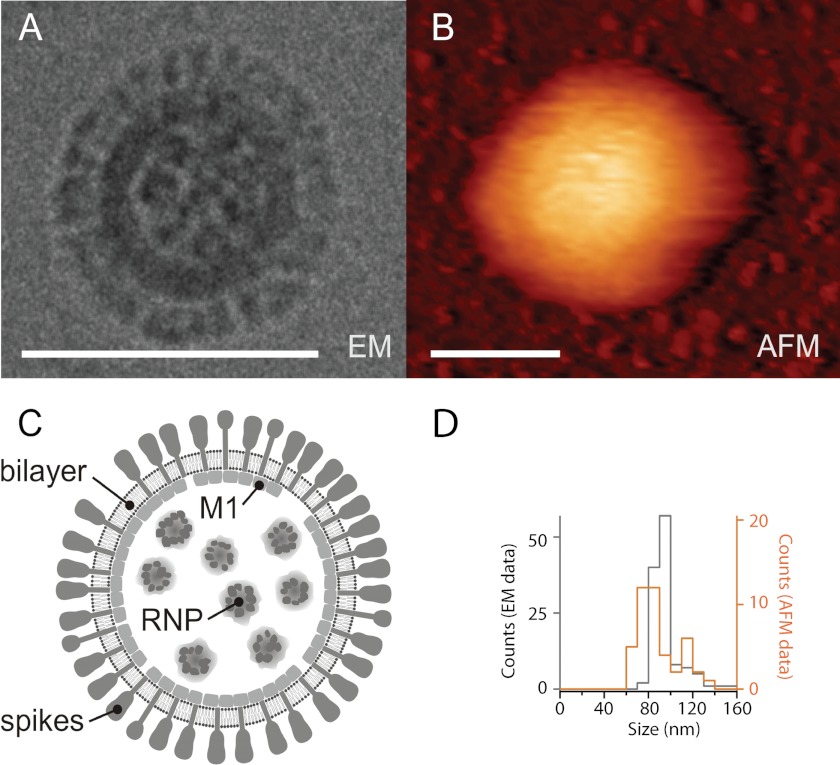
To ensure that we did not irreversibly deform or damage our samples during imaging and subsequent force mapping using atomic force spectroscopy, we compared the AFM-determined height of our virus preparation with the diameter of the same preparations measured using cryo-EM (Fig. 1). For the AFM studies, virions and liposomes were immobilized on coverslips functionalized with an aminosilane (see “Experimental Procedures”). In these conditions, the particles could still be displaced relatively easily, by pushing them with the AFM tip. This suggested relatively weak interactions between the particles and the surface, unlikely to affect the mechanical properties measured in our force mapping experiments. In order to ensure that all particles included in the study were roughly spherical in shape, we first determined their dimensions by AFM in the x/y plane (Fig. 1B). This image was distorted by the shape of the AFM tip but enabled us to obtain a rough estimate of the particle's aspect ratio. By probing them in the z-direction, we then determined the height of the particles with nanometer accuracy. This measurement was not affected by the shape of the AFM tip. The height of 43 virions used in the force mapping experiments and of 123 cryo-EM images of virions was analyzed. Both methods yielded comparable size distributions (Fig. 1D), indicating that AFM scanning neither destroyed nor extensively altered influenza virions.
FIGURE 1.
Imaging using AFM does not deform the virus particles. Shown are a cryo-electron micrograph (A) and AFM scan (dynamic mode) (B) of a native A/X-31 influenza virus. Scale bars, 100 nm. Whereas spike glycoproteins, HA, and NA could be identified by cryo-electron microscopy, the particles imaged by AFM appeared smooth. The lateral dimensions of the AFM scans are exaggerated due to tip-sample dilation. C, schematic representation of the influenza virus structure. The viral ribonucleoprotein core (RNP) inside the virus particle is surrounded by a lipid bilayer with associated matrix protein M1 interacting with the cytoplasmic tails of spike glycoproteins HA and NA. D, histograms of the diameter of single virus particles imaged by cryo-EM and measured by atomic force microscopy (height measurement, not affected by tip-sample dilation).
Although the virions were not deformed and the resolution of AFM imaging was sufficient for single protein detection (39), we did not resolve details on the surface of the native particles, which generally looked like smooth balls (Fig. 1B). This is probably due to the softness of our samples and to the flexibility of the spike glycoproteins anchored in the lipid envelope. Consistent with this, in a recent study in which influenza virions were also imaged using AFM, plaques and honeycomb structures indicating glycoprotein spikes could only be resolved on the surface of virions fixed with glutharaldehyde, which increases the sample rigidity (33). We could, however, clearly resolve individual spike glycoproteins in the cryo-electron micrographs of native influenza virions (Fig. 1, A and C), indicating that our virus samples were intact.
Stiffness of the Viral Envelope Measured Using Atomic Force Spectroscopy
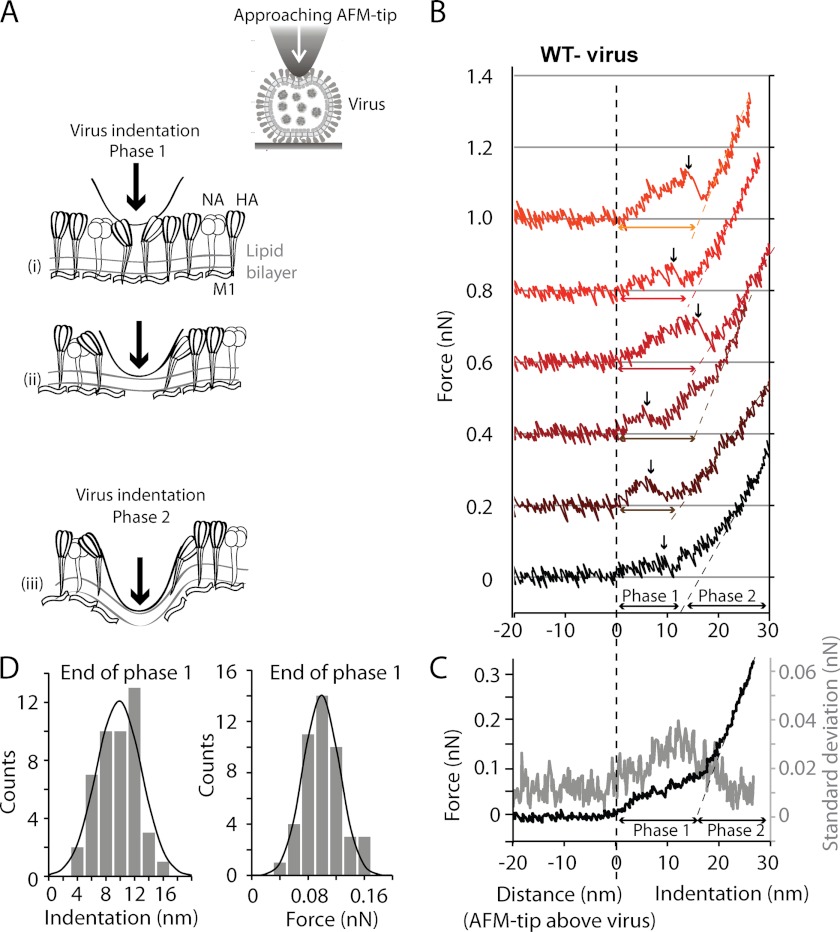
We used the AFM probe to indent the virus particles in order to measure their elastic response (Fig. 2A). Because the influenza virus lacks the geometrically well defined oligomeric structural subunits of a viral protein capsid, we expected a softer mechanical response and a stronger variability between individual particles. To minimize the error of the stiffness measurements, we obtained multiple force-indentation curves from each particle. The stiffness was mapped over a central square of 250 × 250 nm on top of each particle, using an automated data collection routine (see “Experimental Procedures”). In short, force-indentation maps were obtained from FZ curves generated by moving the z-piezo at 2 μm s−1 in an array of 18 × 18 points. Fig. 2B shows that the particle response to indentation was biphasic. The first phase was non-linear, more noisy, and observed at forces of ≤100 pN. This phase ended at a relatively large indentation of about 10 nm and mostly included a stepwise change in indentation (Fig. 2B, black arrow). The second phase, starting at forces of >100 pN, was approximately linear. In this regime, the force-indentation curves were fully reproducible and could be repeated tens of times, indicating an elastic behavior. We acquired force-indentation curves at forces up to 500 pN, which corresponds to a ∼30-nm indentation of the virion. Because forces exceeding 300 pN often led to the displacement of virus particles, we limited the forces to this value during force mapping. In Fig. 2C, the six force-indentation curves measured for a single virion in Fig. 2B were ensemble-averaged. The averaged curve (in black) confirmed the biphasic response, with the transition from phase 1 to phase 2 at around 100 pN of force and about 10-nm indentation. By extrapolating the linear part of the force-indentation curve in phase 2 to zero force, we determined the average indentation and average force at the end phase 1 for a larger data set. Fig. 2D shows the distribution of these forces (0.1 ± 0.02 nN; mean ± S.D.) and indentations (10 ± 3 nm; mean ± S.D.) from 46 force-indentation curves obtained from nine virus particles. The S.D. value of the ensemble average (Fig. 2C, gray curve) was increased for indentations up to about 20 nm. This reflects the stepwise change in stiffness at variable indentations in phase 1 of the individual force-indentation curves.
FIGURE 2.
Successive force-indentation curves obtained with an individual A/X-31 virion (diameter ∼116 nm) consistently show a biphasic response. A, schematic of the force spectroscopy experiment. During phase 1 of virus indentation, HA and NA, connected to the matrix protein M1 at the luminal side of the lipid bilayer, are pushed apart (i and ii). During phase 2 (iii), the lipid bilayer and associated proteins are indented by the AFM tip. B, six successive force-indentation curves performed on top of a single virion. The initial indentation up to about 10 nm (phase 1) contains steps (black arrows) and shows considerable variability between the curves. At increasing deformation, the curves appear more similar. Force-indentation curves were aligned using the part of the curve between 100 and 200 pN in phase 2. C, the averaged force-indentation curve (black) of the data shown in B shows the biphasic response, with a transition from soft to stiff at about 100 pN of force. The S.D. of the ensemble of the six curves is shown in gray. D, a linear fit was applied to each force-indentation curve for forces of ≥100 pN in phase 2. Extrapolation of each linear fit to zero force was used to determine the indentation in nanometers reached at the end of phase 1. The histograms show the distribution of indentation and of the forces reached at the end of phase 1 (46 force-indentation curves measured on nine virions).
Spike Glycoproteins Are Contributing to the Stiffness of the Viral Envelope
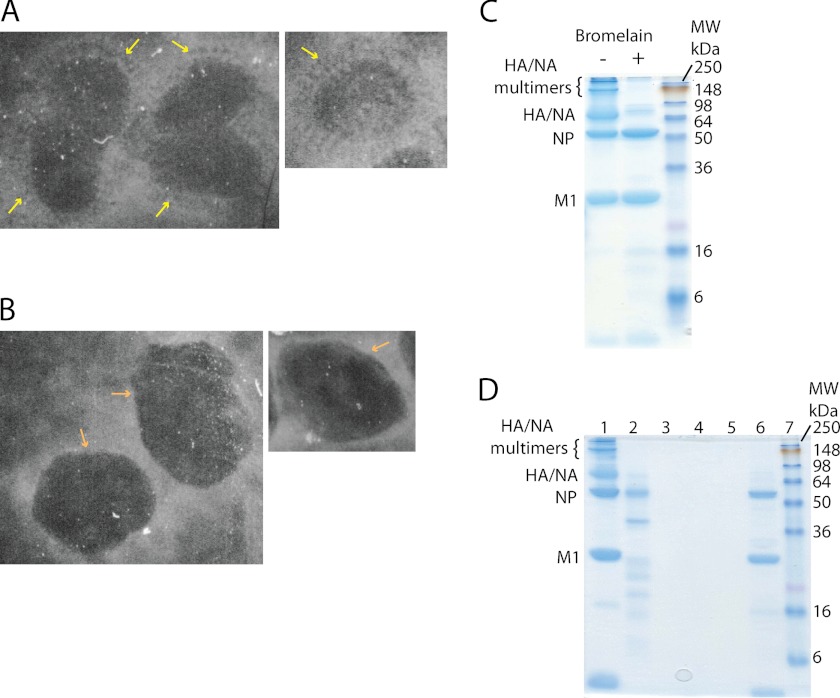
In order to test whether phase 1 of the force-indentation curves involved interactions of the AFM tip with the spike glycoproteins protruding from the viral membrane, we repeated our measurements on “bald” virions. To obtain bald virions, the ectodomain of the glycoproteins was enzymatically removed by bromelain (30). Fig. 3A shows negatively stained electron micrographs of native influenza virions with the typical dense cover of spike glycoproteins (yellow arrows) protruding from the membrane surface of the virus. In contrast, following treatment with bromelain, the surface of the viral particles appeared smooth, and protruding spikes were no longer visible (Fig. 3B). Otherwise, the virions retained their shape. Biochemical analysis of the bald virus preparations was carried out using SDS-PAGE (Fig. 3C). The gels confirmed that HA trimers and NA tetramers (molecular mass about 210 and 240 kDa, respectively) as well as full-length HA and NA monomers (molecular mass about 70 kDa) were present before bromelain treatment but were strongly reduced following enzymatic digestion with bromelain. The intraviral protein M1 was not affected and NP was only little affected by the treatment with bromelain. A small amount of NP was lost following the enzymatic treatment (Fig. 3D, lane 2), but no further release from the virus was observed during the three following wash steps (Fig. 3D, lanes 3–5).
FIGURE 3.
A, negative stain electron micrographs of WT influenza virions. The yellow arrows mark the ectodomains of the glycoproteins protruding from the particle surface. B, negative stain electron micrographs of WT virions after bromelain treatment. The ectodomains of the glycoproteins are no longer visible, and the surface of the particles (orange arrows) appears smooth. C, bromelain-treated A/X31 virus samples were analyzed by 12.5% SDS-PAGE in non-reducing conditions. Gels were stained with Coomassie Blue. D, 12.5% SDS-PAGE in non-reducing conditions. Lane 1, untreated virus; lane 2, supernatant of bromelain-treated sample after ultracentrifugation; lanes 3–5, three pellet washes following bromelain treatment and ultracentrifugation; lane 6, pellet of bromelain-treated virus after ultracentrifugation.
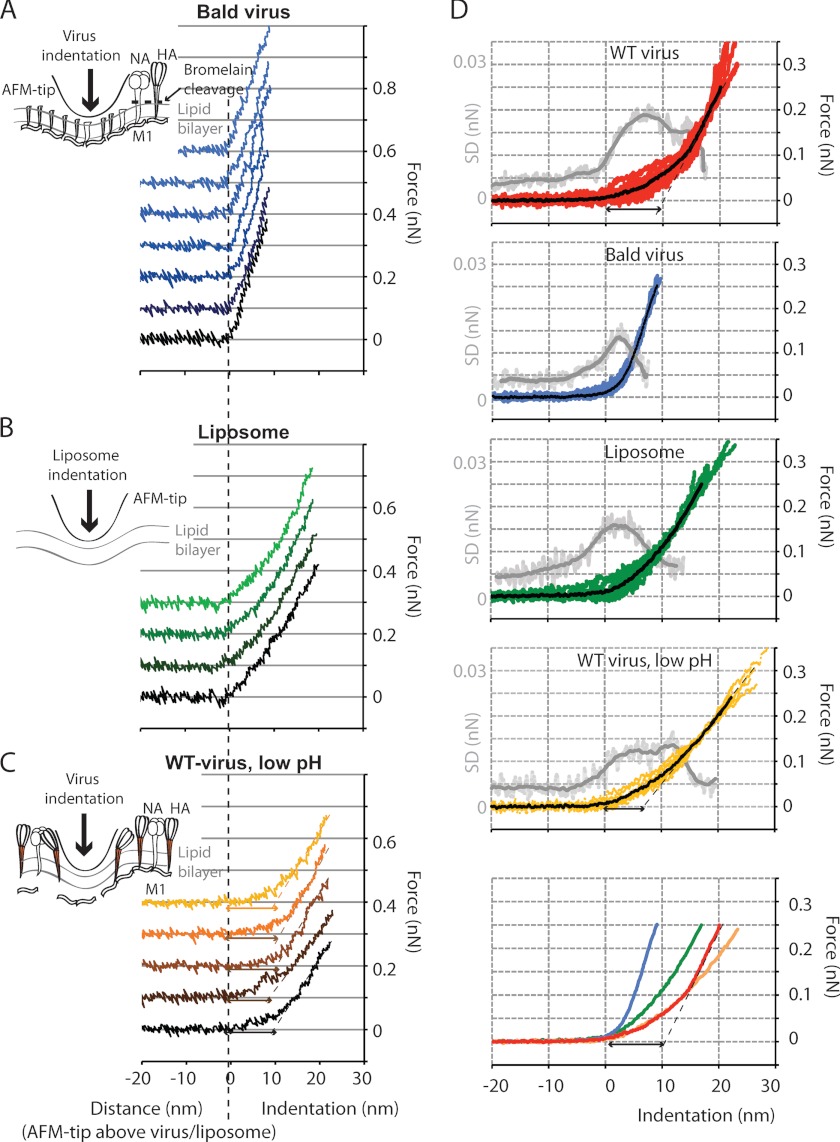
The stiffness measurements showed that notably the soft, non-linear phase 1 of the force-indentation curves we had observed for untreated virions was no longer seen after bromelain treatment. At all forces, the response of bald virions was elastic and quasilinear (Fig. 4), comparable with phase 2 for the untreated virus. To compare the stiffness of the different particle species in this study, we plotted ensembles of force-indentation curves for each species (Fig. 4D). Intriguingly, the bald virions had a higher stiffness over the full range of indentations compared with the untreated virus. Similar to what had been done for the untreated virus, we applied a linear fit to the averaged indentation curves of the bald virions for forces between 100 and 200 pN. Even at forces of >100 pN, the bald virions were nearly twice as stiff (0.05 nN nm−1) compared with the untreated virus (0.03 nN nm−1 for phase 2; particles with a diameter of 75–100 nm; Table 1).
FIGURE 4.
Comparison of force-indentation curves obtained with untreated (WT), bromelain-treated (bald), pH 5-treated virus, and liposomes of similar size (viruses 85–95 nm in diameter, liposomes 75–95 nm in diameter). A, seven successive force-indentation curves were measured on a bald virion. The soft initial response (phase 1) and stepwise indentation at ∼100 pN observed with untreated virus are absent. The stiffness was higher than that of the untreated virus or liposomes. B, the response of liposomes was similar to that of bald viruses, but the stiffness was lower. C, the pH 5-treated virus shows a response similar to untreated virus. The stiffness in phase 2 was slightly lower and more similar to that of the liposome. D, for nine untreated virions, the average curves (red) were calculated from 3–8 individual indentation curves/virus. The average of these is shown in black. The light gray curve shows the S.D. of the ensemble, and the mean of the S.D. is shown in dark gray and was calculated over a running window of 60 data points. For nine bald virus particles (eight liposomes and seven pH 5-treated virions, respectively), the averaged curves were calculated from 4–16 individual force-indentation curves/particle (3–5 individual force-indentation curves/particle for the liposomes and the pH 5-treated virions, respectively). The bottom panel shows all averaged force-indentation curves in one plot.
TABLE 1.
Averaged stiffness values determined for viruses, liposomes, and virosomes
Shown is a summary of the averaged stiffness values determined for bromelain-treated, untreated, and pH 5-treated viruses (phase 2), liposomes with or without cholesterol, and virosomes for a particle size of 75–100 nm in diameter. The data for all individual particles analyzed are shown in Fig. 6. The S.D. value was obtained from the difference between each data point and the fitted value.
| WT virus | Bald virus | Liposome | WT virus, low pH | Liposome + cholesterol | Virosome | |
|---|---|---|---|---|---|---|
| No. of particles (d = 75–100 nm) | 24 | 25 | 9 | 12 | ||
| No. of FZ curves analyzed | 121 | 185 | 39 | 47 | ||
| Stiffness (nN/nm) (mean ± S.D.) | 0.03 ± 0.01 | 0.05 ± 0.02 | 0.02 ± 0.01 | 0.02 ± 0.01 | ||
| Mean height (nm) (mean ± S.D.) | 85 ± 7 | 85 ± 5 | 82 ± 7 | 91 ± 7 | ||
| No. of particles (all) | 43 | 48 | 43 | 31 | 13 | 6 |
| Global fit stiffness k ± S.E. (nN/nm) for d = 100 nm | 0.023 ± 0.002 | 0.037 ± 0.002 | 0.014 ± 0.001 | 0.017 ± 0.001 | 0.015 ± 0.002 | 0.010 ± 0.002 |
Stiffness of the Influenza Viral Envelope at Low pH
In order to investigate how changes in the viral envelope structure at low pH encountered in the endosome affect the mechanical properties of the virus, we performed stiffness measurements on virions that had been incubated for 5 min at pH 4.9 at 37 °C before the pH of the solution was readjusted to pH 7.3 (Fig. 4C). Overall, the response to indentation was similar to what was observed with untreated virions. The response was biphasic with the stiffness of the second phase but only two-thirds of that of the untreated virus. Furthermore, the stepwise indentation during phase 1 was still observed but more rarely compared with the untreated virions.
Stiffness of Pure Liposomes
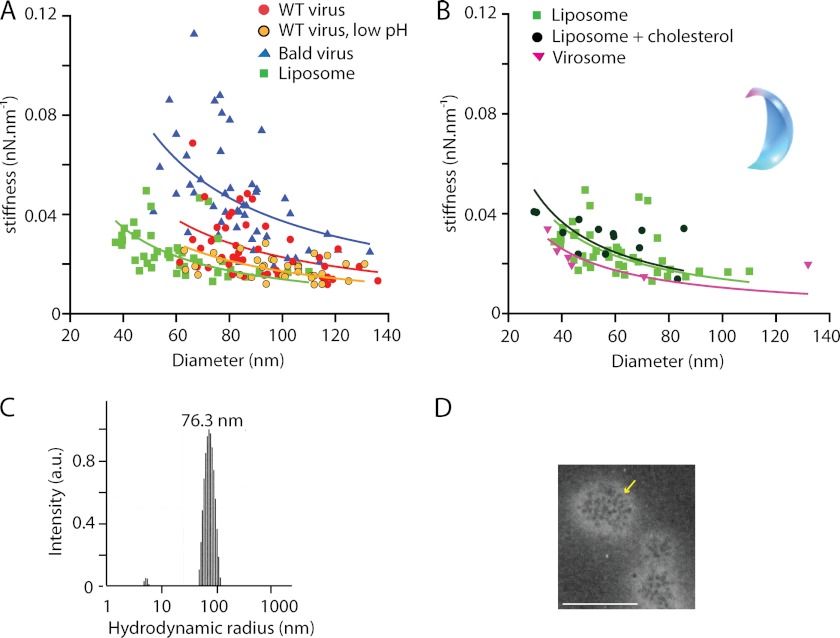
The fact that the influenza virions were an order of magnitude softer than protein capsid-coated or enveloped virus particles with submembranous protein-capsids (0.2–0.6 nN nm−1) (25, 26, 40) suggested that the mechanical properties of the influenza virus were critically dependent on the lipid component of the envelope. We tested this idea by studying the mechanics of small unilamellar vesicles made out of pure egg PC. Similar to what was observed for bald virions, the mechanical response of liposomes upon indentation was almost linear and elastic (Fig. 4B). There was no biphasic behavior. The average stiffness of liposomes with a diameter of 75–100 nm was 0.02 ± 0.01 nN nm−1 (Table 1), comparable with pH 5-treated virus. Thus, the liposomes were 2–3 times softer than the bald virions of similar size, revealing the contribution of viral protein components such as M1 and the viral genome to the viral envelope's mechanical response. Because the influenza viral envelope has a high cholesterol content (∼44% of the total viral lipid), which was shown to be critical for viral budding and host cell entry (41, 42), we also studied the stiffness of mixed liposomes containing 70% egg PC and 30% cholesterol. Furthermore, we reconstituted influenza virosomes by mixing egg PC liposomes with purified HA protein at a 100:1 molar ratio. This enabled us to study the effect of one of the glycoproteins in the absence of their native compact arrangement, as observed in viruses. Negative stain electron micrographs (Fig. 6D) of the virosomes showed that HA integrated into the lipid membrane but at a lower density compared with native virus. For both the cholesterol-enriched liposomes and virosomes, the measured stiffness was similar to that of pure egg PC liposomes (Tables 1 and 2).
FIGURE 6.
The stiffness of phase 2 for the untreated virions, the pH 5-treated virions, the bald virions, and the liposomes scales with the reciprocal of the particle size. All stiffness measurements were quantified by performing a linear fit in the force range of 100–200 pN to obtain a spring constant for each particle. The solid lines are 1/R fits to each data set. A, for all species, the reciprocal of the spring constant scaled with the diameter of the particle. The bald virions (blue) were stiffer than the untreated virions (red), and the liposomes (green) and pH 5-treated virions (yellow) were softest. B, comparison of the stiffness of pure egg PC liposomes (green), egg PC plus 30% cholesterol liposomes (black), and reconstituted virosomes (pink). We calculated the response for a liposome from a finite element model (inset). The model shows the deformed quarter of a sphere after indentation by an AFM tip (see “Experimental Procedures”). C, size distribution of unilamellar liposomes from egg PC extruded with 200-nm pore filters and determined in bulk by dynamic light scattering. With 100-nm pore filters, the distribution of the hydrodynamic radius was centered at 51.5 nm, with radii ranging from 30 to 80 nm. D, negative stain electron micrograph of reconstituted virosomes made of egg PC and hemagglutinin. The yellow arrow marks HA ectodomains protruding from the virosome surface. Scale bar, 100 nm.
TABLE 2.
Statistical analysis of the stiffness measurements
Statistical analysis of the stiffness measurements is shown. The probabilities were calculated from the fitted mean stiffness for particle diameter 75–100 nm, the S.E., and the error function.
| All particles, error function (probability) | |
|---|---|
| Bald virus versus WT virus | 7e−4 |
| Bald virus versus liposome | 2e−9 |
| Bald virus versus pH 5-treated WT virus | 1e−8 |
| Bald virus versus liposome + cholesterol | 4e−6 |
| Bald virus versus virosome | 7e−7 |
| WT virus versus liposome | 0.003 |
| WT virus versus WT virus low pH | 0.03 |
| WT virus versus virosome | 0.002 |
| WT virus versus liposome + cholesterola | 0.05 |
| WT virus low pH versus virosomea | 0.16 |
| WT virus low pH versus liposome + cholesterola | 0.45 |
| Liposome versus WT virus low pHa | 0.05 |
| Liposome versus liposome + cholesterola | 0.83 |
| Liposome versus virosomea | 0.35 |
| Liposome + cholesterol versus virosomea | 0.36 |
a For these compared species, no statistically significant difference was found (3% level).
S.D. Value of the Ensemble of Force-Indentation Curves
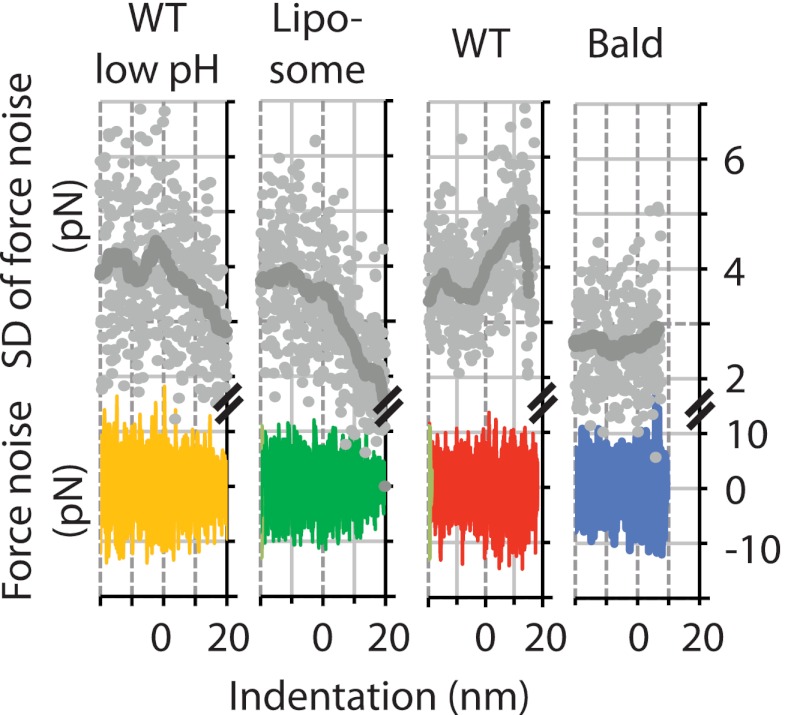
We also compared the increase in S.D. value during phase 1 of the ensemble of force-indentation curves for the different particle species (Fig. 4D, gray curves). We found that for the bald virions and the liposomes, the S.D. of the ensemble was increased over the range of −10 to +10-nm indentation. This can be explained by the alignment of the curves. In contrast, for the samples with glycoprotein spikes protruding from the membrane surface (i.e. untreated virus and pH 5-treated virus), the S.D. of the ensembles was increased over a much broader range (i.e. −10 to +20-nm indentation). To eliminate the effect of curve alignment on the S.D. of the ensemble, we subtracted the increase in force with increasing indentation for each force-indentation curve included in the ensemble. We then plotted the ensemble of the remaining force noise for each species (Fig. 5). This noise analysis confirmed that only the untreated virus showed a clear increase in force noise for indentations up to 20 nm. For the pH 5-treated virions, only a very small increase for indentations up to 10 nm was observed. In contrast, for liposomes and bald virions, no increase in force noise with increasing indentation was found.
FIGURE 5.
Noise analysis of the force-indentation curves. To analyze the noise in the force measurements with increasing indentation, we subtracted the average force value over a running window of 40 data points from each force-indentation curve. For low pH-treated WT virus, liposomes, WT virus, and bald virus, n = 8, 8, 16, and 16 force-indentation curves were included, respectively. Only for the untreated WT virus, an increase in the S.D. of the force noise for indentations up to 20 nm was observed.
Change in Stiffness with Particle Size
We finally investigated the relationship between the stiffness of the particles and particle size (Fig. 6, A and B). In addition to measuring the aspect ratio in the x/y plane and the height of the individual particles using atomic force microscopy, we also used dynamic light scattering as a bulk measurement to determine the size distribution of the liposome samples (Fig. 6C). The average size of the vesicles included in the mechanical measurements by AFM was 61 ± 18 nm (n = 43). The stiffnesses plotted in Fig. 6 had been determined by a linear fit to the force-indentation curves between 100 and 200 pN for each sample. We found that the stiffness k was related to the reciprocal of the diameter R of the particles, with k ≈ 1/R, as expected for spherical shells with radius R (24). Although we maximized the precision of our measurements by performing multiple measurements per particle, there was still a ∼40% spread (S.D.) around the 1/R fit. This spread can be attributed in part to the uncertainty in tip calibration (∼10%) (35) and to variations in the AFM tip size. Finite element modeling confirmed the measured stiffness to be affected by ±10% when the tip radius varies between 15 and 35 nm. The remaining variation in our data was attributed to heterogeneity of our samples. Comparing the stiffness of the different species over a diameter range of 40–140 nm showed no difference between liposomes made of pure egg PC or mixtures of egg PC and cholesterol, virosomes reconstituted from egg PC and hemagglutinin, or influenza virions following treatment at low pH. In contrast, WT and even more so bald virions were significantly stiffer over the entire range of particle diameter (Tables 1 and 2). This indicates that the stiffness of the influenza envelope is determined by a combination of the stiffness of the lipid component and the membrane-associated viral protein components.
DISCUSSION
In recent years, atomic force microscopy has emerged as a technique to measure the mechanical properties of nanometer-sized biological shells (43), such as protein-capsid-coated viruses (24, 40, 44), and yielded new insight into the biological role of their structural arrangement. It was shown that viral protein capsids form stiff shells that can withstand the high internal pressure resulting from genome packaging. In this study, we used atomic force microscopy to obtain the first measurements of the lipid and protein contribution to the mechanical stiffness of individual influenza virions. We found that the stiffness of virions was on the same order of magnitude as that of pure liposomes (20). We also found that the response of untreated, native influenza virions to indentation by the AFM tip was biphasic. The first phase was non-linear and more noisy, mostly included a stepwise change in stiffness, and was observed at forces of <100 pN. The second phase was approximately linear and highly reproducible.
A simple explanation for the increase in force noise during phase 1 over the initial 20-nm indentation of the native virus are variations in the interaction between the AFM tip and the ectodomains of spike glycoproteins. These variations were no longer observed for bald virions with the ectodomains enzymatically removed. Following this argument, the stepwise change in stiffness at about 10-nm indentation could be explained by the displacement of HA and NA with their ectodomains protruding about 13–14 nm from the lipid bilayer (6, 18, 21). The fact that the stepwise change in stiffness was observed at about 100 pN of force is consistent with fairly immobile glycoproteins in the native viral envelope, so that they have to be forced laterally by the indenting AFM tip. This is consistent with previous fluorescence studies that revealed a very small diffusion coefficient of D ≈ 0.03–0.1 μm2 s−1 for HA in the plasma membrane of COS7 cells or fibroblasts expressing the glycoprotein (45, 46). Such low mobility, possibly partly due the glycoproteins being concentrated in membrane microdomains (18), was observed even in the absence of the matrix protein M1. M1 has been suggested to cross-link the cytoplasmic tail domains of HA and NA in the native virus (8, 9, 13, 22, 47, 48). In our experiments, the estimated “clearance” of a viral membrane area of about 3 × 10−4 μm2 from glycoproteins in about 10 ms by our indenting AFM tip could be achieved by thermal motion of the glycoproteins with a mobility of at least 0.03 μm2 s−1. However, because this “clearance” was only observed at about 100 pN of indenting force, it rather suggests largely immobile membrane proteins in the viral envelope.
Interestingly, we did not always observe a clear stepwise indentation for repeated force-indentation curves on a single virion, as shown in Fig. 2B (black curve). This could be explained by regions on the envelope devoid of spike glycoproteins, as described in cryo-electron tomography analyses (9). In those studies, these bare regions with 20–50-nm diameter were co-localized with discontinuities in the M1 layer on the inside of the virions, which is consistent with our ∼40-nm diameter AFM tip interacting sometimes directly with the lipid surface of the virion, without interaction with the ectodomains of the spike glycoproteins.
The stiffness in phase 2 of the untreated virus was similar to the stiffness measured for liposomes or virosomes of similar size. However, there was still a statistically significant difference below the 5% level (Student's t test; Table 2). In contrast, we did not find a significant difference at that level between pure egg PC liposomes, egg PC liposomes with 30% cholesterol, or virosomes made of egg PC and HA glycoprotein. This suggests that the membrane-associated proteins contribute to the stiffness of the viral envelope but only if the proteins are natively organized in the membrane, in other words in a more compact fashion compared with the virosomes and possibly even in a symmetrical or periodical arrangement of the ectodomains of the glycoproteins.
Consistent with this, the stiffness of bald virions was 2–3 times higher than that of reconstituted liposomes of similar size. This difference may reflect a lower fluidity of the viral lipid bilayer and/or a reinforcing effect of the membrane-associated proteins. The bald virions had only the ectodomains of the glycoproteins removed but retained the structural arrangement of the transmembrane part of the glycoproteins and M1. Previous EM studies resolved M1 adjacent to the viral membrane, with regular interactions between M1 and membrane components (8, 9), probably HA and NA (47, 48). This would be consistent with our finding that only at about 100 pN of force, a stepwise change in stiffness was observed on untreated virions. Furthermore, this phenomenon was no longer seen with the bald virions, where the indenting AFM could no longer interact with the glycoprotein ectodomains. This suggests that the glycoproteins, immobilized in the lipid bilayer by interactions with lipids and the matrix layer M1, are contributing to the mechanical stability of the virions.
We found no statistically significant difference in stiffness between the influenza virions following treatment at low pH and the reconstituted virosomes or liposomes. In addition, the stepwise indentation during phase 1 was still observed, but more rarely compared with the untreated virions. Previous studies suggested that following treatment at low pH, the M1 layer detaches at least in part from the lipid membrane and the glycoproteins. This suggests that the latter become more mobile at low pH (8) and that mechanically important structural arrangements of the membrane-associated proteins at pH 7 are no longer sustained. Intriguingly, the stiffness of virions treated at low pH was about 2–3 times lower compared with that of bald virions kept at pH 7. Using the difference in stiffness between bald and pH 5-treated virions, we can estimate the contribution of the proteins to the overall stiffness of the influenza virions, with κbald − κvirus pH 5 ≈ 0.03 nN nm−1. Apart from the glycoproteins and M1, RNPs attached to the membrane via M1 might also contribute to this stiffness.
We found that the stiffness of egg PC liposomes (in liquid-disordered phase) was lower but still on the same order of magnitude as that of influenza virions. It has been suggested that influenza virions originate from liquid-ordered state lipid raft domains (13, 18), whereas other studies concluded that the disordered state fraction of influenza lipids increases drastically above 0 °C to reach a totally disordered state at physiological temperatures (19). Modeling the mechanical response of our cholesterol-rich bald influenza virions (Fig. 6) yielded a relatively low bending rigidity (Kb) of 2.5 × 10−19 Nm, more consistent with disordered lipid bilayers (49). For ordered bilayers, the bending rigidity would be at least 2-fold higher (50, 51).
In summary, our data suggest that the mechanical response of untreated (WT) virions to indentation results from two events: (i) during phase 1 displacement of the largely immobile spike glycoproteins and possibly a partial disconnection from the cross-linking matrix protein M1 by 100 pN of force imposed by the indenting AFM tip and (ii) during phase 2 elastic indentation of the viral envelope with its associated protein complexes (i.e. M1 layer and associated RNPs (8, 9)).
Finally, we used finite element modeling to test whether the stiffnesses, measured for the simplified model systems (liposomes and bald virions) could be explained by the known physical properties of a lipid bilayer or a compact protein shell, as found for capsid-coated virus species. The theoretical force-indentation curve matched the experimental data for a bending rigidity of the lipid bilayer Kb = 0.5 × 10−19 Nm. This is in good agreement with previously measured values of 0.4–2 × 10−19 Nm for egg PC bilayers (49). Despite the non-linear boundary conditions (increasing contact area during indentation), we found that the stiffness increased almost linearly with the reciprocal of the radius, consistent with our experimental data. Two different methods were used to model the bald virus mechanical response. (i) We calculated the response of an 80-nm diameter protein shell with a 1.6-nm thickness and 1-gigapascal Young's modulus (36, 40), which yielded a stiffness of 0.1 nN nm−1. This value is about twice as high as our experimental data for the bald virions and inconsistent with the calculated contributions of the viral core components, based on the difference between bald viruses and liposomes. However, this would be consistent with the matrix protein M1 forming a discontinuous shell underneath the viral envelope as recently reported (8, 9). (ii) Assuming that the mechanical properties of bald virions would be dominated by the properties of the lipid bilayer, Kb had to be increased to 2.5 × 10−19 Nm to match the measured stiffness. This increase in Kb could be accounted for by effects of the membrane-spanning domains of the spike glycoproteins and their cytoplasmic tails cross-linked by the M1 layer, and possibly also M1-attached RNPs, stiffening the membrane.
AFM has been used in the past to measure the stiffness of protein capsid-coated virions (24) and to discriminate submembranous protein capsids at different stages of maturation (26, 27, 40). Even at low forces, the stiffness of those particles was always between 0.2 and 0.6 nN nm−1. Compared with these virions that possess regular and symmetric protein-based capsids, we found the stiffness of influenza virions to be about an order of magnitude less. If influenza core proteins formed an 80-nm diameter protein shell with a 1.6-nm effective thickness and a 1-gigapascal Young's modulus (as described for capsids and other protein shells), it would contribute about 0.1 nN nm−1 to the stiffness of virions, even more if the capsid would be connected to the lipid envelope. Although the stiffness of bromelain-treated bald virions was higher than that of egg PC liposomes, it remained more than twice as soft as a theoretical protein capsid, and the inferred bending rigidity of the viral envelope remained in the known range for fluid bilayers (49). This indicates that the stiffness of the influenza viral envelope is dominated by the lipid component but reinforced by the membrane-associated viral proteins, such as the glycoproteins, M1, and RNPs.
It is tempting to speculate that the rigid design of protein-capsid-coated viruses has been selected due to its ability to withstand the high pressure introduced by the packaging of its genome and to persist outside of the host. Compared with the classical icosahedral viruses, including the picornaviruses, the environmental persistence of influenza virus is less but not negligible (52–54). In order to persist within the infected host cell, the influenza virus requires flexibility of its envelope to enable dynamics in membrane curvature during the viral life cycle, including host cell receptor binding, viral fusion with the host endosome, and budding during viral assembly. We propose that these functions can be achieved by a fluid yet stable lipid bilayer, reinforced in a regulated (pH-dependent) fashion by membrane-associated proteins, including spike glycoproteins, M1, and possibly RNPs. Although the influenza virions are remarkably soft at physiological temperatures, they withstand deformations at least as large as the ones applied in our AFM experiment (30%). This makes the flexible viral envelope a stable container for the virus genome both inside and outside the host.
Acknowledgments
Iwan Schaap and Frederic Eghiaian contributed equally to this work. We thank Steven Wharton, Rose Gonsalves, and David Stevens for technical assistance with liposome and virus production and purification, and we thank John Skehel, John McCauley, and Justin Molloy for scientific discussions.
This work was supported by the Medical Research Council, the Royal Society, Deutsche Forschungsgemeinschaft Grant SFB 863, and the Centre of Nanosciences, Ludwig-Maximilians Universität München, Germany.
- RNP
- ribonucleoprotein
- NA
- neuraminidase
- PC
- phosphatidylcholine
- A/X-31
- Aichi/H3N2 X-31
- AFM
- atomic force microscope
- N
- nN, and pN, newton(s), nanonewton(s), and piconewton(s), respectively.
REFERENCES
- 1. Ahmed R., Oldstone M. B., Palese P. (2007) Protective immunity and susceptibility to infectious diseases: lessons from the 1918 influenza pandemic. Nat. Immunol. 8, 1188–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Knipe D. M., Howley P. M., Griffin D. E., Lamb R. A., Martin M. A., Roizman B., Straus S. E. (2007) in Field's Virology, 5th Ed., Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 3. Coloma R., Valpuesta J. M., Arranz R., Carrascosa J. L., Ortín J., Martín-Benito J. (2009) The structure of a biologically active influenza virus ribonucleoprotein complex. PLoS Pathog. 5, e1000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Area E., Martín-Benito J., Gastaminza P., Torreira E., Valpuesta J. M., Carrascosa J. L., Ortín J. (2004) Three-dimensional structure of the influenza virus polymerase complex. Localization of subunit domains. Proc. Natl. Acad. Sci. U.S.A. 101, 308–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stevens J., Corper A. L., Basler C. F., Taubenberger J. K., Palese P., Wilson I. A. (2004) Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science 303, 1866–1870 [DOI] [PubMed] [Google Scholar]
- 6. Wilson I. A., Skehel J. J., Wiley D. C. (1981) Structure of the hemagglutinin membrane glycoproteon of influenza-virus at 3 Å resolution. Nature 289, 366–373 [DOI] [PubMed] [Google Scholar]
- 7. Varghese J. N., Laver W. G., Colman P. M. (1983) Structure of the influenza-virus glycoprotein antigen neuraminidase at 2.9 Å resolution. Nature 303, 35–40 [DOI] [PubMed] [Google Scholar]
- 8. Calder L. J., Wasilewski S., Berriman J. A., Rosenthal P. B. (2010) Structural organization of a filamentous influenza A virus. Proc. Natl. Acad. Sci. U.S.A. 107, 10685–10690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Harris A., Cardone G., Winkler D. C., Heymann J. B., Brecher M., White J. M., Steven A. C. (2006) Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc. Natl. Acad. Sci. U.S.A. 103, 19123–19127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Noda T., Sagara H., Yen A., Takada A., Kida H., Cheng R. H., Kawaoka Y. (2006) Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature 439, 490–492 [DOI] [PubMed] [Google Scholar]
- 11. Ruigrok R. W., Calder L. J., Wharton S. A. (1989) Electron microscopy of the influenza virus submembranal structure. Virology 173, 311–316 [DOI] [PubMed] [Google Scholar]
- 12. Yamaguchi M., Danev R., Nishiyama K., Sugawara K., Nagayama K. (2008) Zernike phase contrast electron microscopy of ice-embedded influenza A virus. J. Struct. Biol. 162, 271–276 [DOI] [PubMed] [Google Scholar]
- 13. Rossman J. S., Lamb R. A. (2011) Influenza virus assembly and budding. Virology 411, 229–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harrison S. C. (2008) Viral membrane fusion. Nat. Struct. Mol. Biol. 15, 690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Scheiffele P., Rietveld A., Wilk T., Simons K. (1999) Influenza viruses select ordered lipid domains during budding from the plasma membrane. J. Biol. Chem. 274, 2038–2044 [DOI] [PubMed] [Google Scholar]
- 16. Coskun U., Simons K. (2011) Cell membranes. The lipid perspective. Structure 19, 1543–1548 [DOI] [PubMed] [Google Scholar]
- 17. Kenworthy A. K., Nichols B. J., Remmert C. L., Hendrix G. M., Kumar M., Zimmerberg J., Lippincott-Schwartz J. (2004) Dynamics of putative raft-associated proteins at the cell surface. J. Cell Biol. 165, 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Takeda M., Leser G. P., Russell C. J., Lamb R. A. (2003) Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc. Natl. Acad. Sci. U.S.A. 100, 14610–14617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Polozov I. V., Bezrukov L., Gawrisch K., Zimmerberg J. (2008) Progressive ordering with decreasing temperature of the phospholipids of influenza virus. Nat. Chem. Biol. 4, 248–255 [DOI] [PubMed] [Google Scholar]
- 20. Li S., Eghiaian F., Sieben C., Herrmann A., Schaap I. A. (2011) Bending and puncturing the influenza lipid envelope. Biophys. J. 100, 637–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Collins P. J., Haire L. F., Lin Y. P., Liu J., Russell R. J., Walker P. A., Skehel J. J., Martin S. R., Hay A. J., Gamblin S. J. (2008) Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants. Nature 453, 1258–1261 [DOI] [PubMed] [Google Scholar]
- 22. Burleigh L. M., Calder L. J., Skehel J. J., Steinhauer D. A. (2005) Influenza A viruses with mutations in the M1 helix six domain display a wide variety of morphological phenotypes. J. Virol. 79, 1262–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Michel J. P., Ivanovska I. L., Gibbons M. M., Klug W. S., Knobler C. M., Wuite G. J., Schmidt C. F. (2006) Nanoindentation studies of full and empty viral capsids and the effects of capsid protein mutations on elasticity and strength. Proc. Natl. Acad. Sci. U.S.A. 103, 6184–6189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ivanovska I. L., de Pablo P. J., Ibarra B., Sgalari G., MacKintosh F. C., Carrascosa J. L., Schmidt C. F., Wuite G. J. (2004) Bacteriophage capsids. Tough nanoshells with complex elastic properties. Proc. Natl. Acad. Sci. U.S.A. 101, 7600–7605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kol N., Gladnikoff M., Barlam D., Shneck R. Z., Rein A., Rousso I. (2006) Mechanical properties of murine leukemia virus particles. Effect of maturation. Biophys. J. 91, 767–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kol N., Shi Y., Tsvitov M., Barlam D., Shneck R. Z., Kay M. S., Rousso I. (2007) A stiffness switch in human immunodeficiency virus. Biophys. J. 92, 1777–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Roos W. H., Radtke K., Kniesmeijer E., Geertsema H., Sodeik B., Wuite G. J. (2009) Scaffold expulsion and genome packaging trigger stabilization of herpes simplex virus capsids. Proc. Natl. Acad. Sci. U.S.A. 106, 9673–9678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mui B., Chow L., Hope M. J. (2003) Extrusion technique to generate liposomes of defined size. Methods Enzymol. 367, 3–14 [DOI] [PubMed] [Google Scholar]
- 29. Rigaud J. L., Lévy D. (2003) Reconstitution of membrane proteins into liposomes. Methods Enzymol. 372, 65–86 [DOI] [PubMed] [Google Scholar]
- 30. Skehel J. J., Schild G. C. (1971) The polypeptide composition of influenza A viruses. Virology 44, 396–408 [DOI] [PubMed] [Google Scholar]
- 31. Brand C. M., Skehel J. J. (1972) Crystalline antigen from the influenza virus envelope. Nat. New Biol. 238, 145–147 [DOI] [PubMed] [Google Scholar]
- 32. Ruigrok R. W., Wrigley N. G., Calder L. J., Cusack S., Wharton S. A., Brown E. B., Skehel J. J. (1986) Electron microscopy of the low pH structure of influenza virus hemagglutinin. EMBO J. 5, 41–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Giocondi M. C., Ronzon F., Nicolai M. C., Dosset P., Milhiet P. E., Chevalier M., Le Grimellec C. (2010) Organization of influenza A virus envelope at neutral and low pH. J. Gen. Virol. 91, 329–338 [DOI] [PubMed] [Google Scholar]
- 34. de Pablo P. J., Schaap I. A., MacKintosh F. C., Schmidt C. F. (2003) Deformation and collapse of microtubules on the nanometer scale. Phys. Rev. Lett. 91, 098101 [DOI] [PubMed] [Google Scholar]
- 35. Burnham N. A., Chen X., Hodge C. S., Matei G. A., Thoreson E. J., Roberts C. J., Davies M. C., Tendler S. J. B. (2003) Comparison of calibration methods for atomic-force microscopy cantilever. Nanotechnology 14, 1–6 [Google Scholar]
- 36. Schaap I. A., Carrasco C., de Pablo P. J., MacKintosh F. C., Schmidt C. F. (2006) Elastic response, buckling, and instability of microtubules under radial indentation. Biophys. J. 91, 1521–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rawicz W., Olbrich K. C., McIntosh T., Needham D., Evans E. (2000) Effect of chain length and unsaturation on elasticity of lipid bilayers. Biophys. J. 79, 328–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bloom M., Evans E., Mouritsen O. G. (1991) Physical properties of the fluid lipid bilayer component of cell membranes. A perspective. Q. Rev. Biophys. 24, 293–397 [DOI] [PubMed] [Google Scholar]
- 39. Schmitz S., Schaap I. A., Kleinjung J., Harder S., Grainger M., Calder L., Rosenthal P. B., Holder A. A., Veigel C. (2010) Malaria parasite actin polymerization and filament structure. J. Biol. Chem. 285, 36577–36585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Carrasco C., Carreira A., Schaap I. A., Serena P. A., Gómez-Herrero J., Mateu M. G., de Pablo P. J. (2006) DNA-mediated anisotropic mechanical reinforcement of a virus. Proc. Natl. Acad. Sci. U.S.A. 103, 13706–13711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Biswas S., Yin S. R., Blank P. S., Zimmerberg J. (2008) Cholesterol promotes hemifusion and pore widening in membrane fusion induced by influenza hemagglutinin. J. Gen. Physiol. 131, 503–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sun X., Whittaker G. R. (2003) Role for influenza virus envelope cholesterol in virus entry and infection. J. Virol. 77, 12543–12551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Laney D. E., Garcia R. A., Parsons S. M., Hansma H. G. (1997) Changes in the elastic properties of cholinergic synaptic vesicles as measured by atomic force microscopy. Biophys. J. 72, 806–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Roos W. H., Ivanovska I. L., Evilevitch A., Wuite G. J. (2007) Viral capsids. Mechanical characteristics, genome packaging and delivery mechanisms. Cell Mol. Life Sci. 64, 1484–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hess S. T., Gould T. J., Gudheti M. V., Maas S. A., Mills K. D., Zimmerberg J. (2007) Dynamic clustered distribution of hemagglutinin resolved at 40 nm in living cell membranes discriminates between raft theories. Proc. Natl. Acad. Sci. U.S.A. 104, 17370–17375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shvartsman D. E., Kotler M., Tall R. D., Roth M. G., Henis Y. I. (2003) Differently anchored influenza hemagglutinin mutants display distinct interaction dynamics with mutual rafts. J. Cell Biol. 163, 879–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen B. J., Leser G. P., Jackson D., Lamb R. A. (2008) The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J. Virol. 82, 10059–10070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen B. J., Leser G. P., Morita E., Lamb R. A. (2007) Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. J. Virol. 81, 7111–7123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lipowski R., Sackmann E. (2004) Structure and Dynamics of Membranes, Elsevier, Amsterdam, The Netherlands [Google Scholar]
- 50. Delorme N., Fery A. (2006) Direct method to study membrane rigidity of small vesicles based on atomic force microscope force spectroscopy. Phys. Rev. E 74, 030901 [DOI] [PubMed] [Google Scholar]
- 51. Dimova R., Pouligny B., Dietrich C. (2000) Pretransitional effects in dimyristoylphosphatidylcholine vesicle membranes. Optical dynamometry study. Biophys. J. 79, 340–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dublineau A., Batéjat C., Pinon A., Burguière A. M., Leclercq I., Manuguerra J. C. (2011) Persistence of the 2009 pandemic influenza A (H1N1) virus in water and on non-porous surface. PLoS One 6, e28043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Greatorex J. S., Digard P., Curran M. D., Moynihan R., Wensley H., Wreghitt T., Varsani H., Garcia F., Enstone J., Nguyen-Van-Tam J. S. (2011) Survival of influenza A (H1N1) on materials found in households. Implications for infection control. PLoS One 6, e27932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wood J. P., Choi Y. W., Chappie D. J., Rogers J. V., Kaye J. Z. (2010) Environmental persistence of a highly pathogenic avian influenza (H5N1) virus. Environ. Sci. Technol. 44, 7515–7520 [DOI] [PubMed] [Google Scholar]