Background: The metabolite and antibiotic export system of cyanobacteria is largely unexplored.
Results: Uptake of ethidium bromid by Anabaena sp. depends on porin-type activity while its seceretion relies on HgdD.
Conclusion: The antibiotic export of cyanobacteria involves a proton gradient-driven TolC activity and MFS-type proteins.
Significance: TolC of Anabaena sp. is placed in the context of antibiotic uptake and export.
Keywords: Cyanobacteria, Membrane Biogenesis, Membrane Proteins, Secretion, Transport, Omp85-independent OMP Insertion, TolC, Antibiotics and Metabolite Export, Porin
Abstract
The role of TolC has largely been explored in proteobacteria, where it functions as a metabolite and protein exporter. In contrast, little research has been carried out on the function of cyanobacterial homologues, and as a consequence, not much is known about the mechanism of cyanobacterial antibiotic uptake and metabolite secretion in general. It has been suggested that the TolC-like homologue of the filamentous, heterocyst-forming cyanobacterium Anabaena sp. PCC 7120, termed heterocyst glycolipid deposition protein D (HgdD), is involved in both protein and lipid secretion. To describe its function in secondary metabolite secretion, we established a system to measure the uptake of antibiotics based on the fluorescent molecule ethidium bromide. We analyzed the rate of porin-dependent metabolite uptake and confirmed the functional relation between detoxification and the action of HgdD. Moreover, we identified two major facilitator superfamily proteins that are involved in this process. It appears that anaOmp85 (Alr2269) is not required for insertion or assembly of HgdD, because an alr2269 mutant does not exhibit a phenotype similar to the hgdD mutant. Thus, we could assign components of the metabolite efflux system and describe parameters of detoxification by Anabaena sp. PCC 7120.
Introduction
Detoxification by secretion systems is essential for all Gram-negative bacteria. It involves an energizing machine in the plasma membrane and a TolC (tolerance to colicins) protein in the outer membrane (1–4). The latter is involved in the secretion across the outer membrane of a variety of essential factors. For example, it is required for the assembly of the outer layer of the cell wall (e.g. see Ref. 5), for the uptake of iron (6–8), or for export of compounds that have entered the periplasm from the surrounding medium and would cause severe damage if not cleared (3).
The TolC-like proteins in general have a low sequence identity, but they appear to be highly conserved at the structural level (9). The biochemical and structural analysis revealed a homotrimer forming a single-channel tunnel with a membrane-inserted β-barrel and a periplasm-spanning α-helical barrel (e.g. see Ref. 10). To secrete its various substrates to the extracellular space, TolC proteins are known to be involved in three different secretion systems. First, TolC is part of the type I secretion system, where it interacts with different inner membrane ABC-transporters involved in the secretion of proteinaceous toxins (e.g. see Ref. 11). Second, it functions as an outer membrane factor for different superfamilies of multidrug efflux pumps, such as the resistance-nodulation-cell division type, AcrAB, or the major facilitator superfamily (MFS)2 type (Fig. 1) (see Refs. 1 and 12). Third, it has been described as part of cation efflux pumps for extrusion of toxic metal ions (13). Thus, the substrate specificity is determined by the periplasmic and the inner membrane components of the complexes that are engaged. The contact between TolC and the inner membrane permease is established in the periplasm with the help of adaptor proteins, also called membrane fusion proteins (14). The complexes are transient, and once substrate secretion has been completed, the complex disengages and reverts to the resting state (e.g. see Ref. 15).
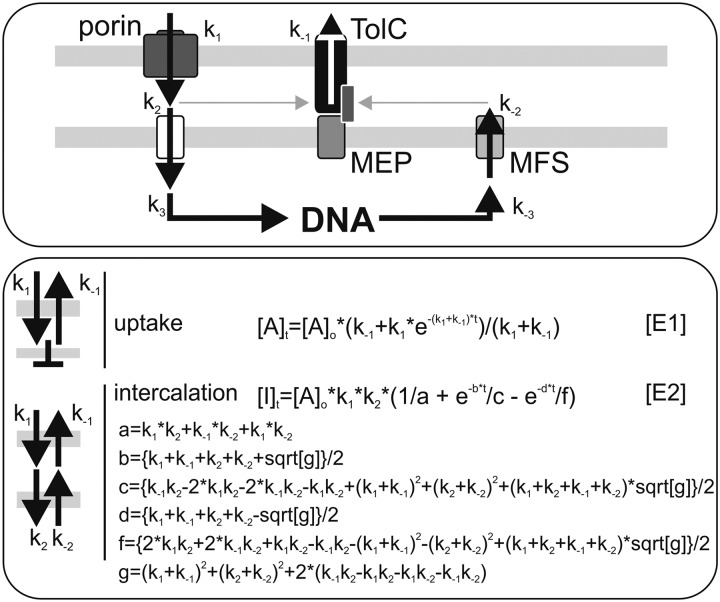
FIGURE 1.
Description of the uptake system used throughout the study. Shown is the scheme of the uptake and export cycle for metabolites (top) based on the current information for Gram-negative bacteria in general (1–4). Please note that parts thereof are only a model that has to be confirmed in the future by the identification of the components. The mathematical equations for the analysis of the transport across the outer membrane only or including the entire system as used to analyze results throughout this work are given (E1 and E2).
The best described model for TolC secretion complex assembly is the resistance-nodulation-cell division-type tripartite AcrAB-TolC system of Escherichia coli (e.g. see Ref. 3). The model that is currently favored suggests a transient recruitment of TolC by the transporter AcrB, which is enforced by the plasma membrane-anchored membrane fusion protein AcrA upon substrate interaction (16, 17). Substrate recognition can occur in the cytoplasm as well as in the periplasmic space, which requires the secretion of the substrate by MFS proteins. However, although the understanding of complex formation has improved in the last few years, it is still unclear how the recruitment of TolC by the right transporter in the right situation is regulated.
In contrast to proteobacteria, little is known about the function of TolC and Type I secretion systems in cyanobacteria. A TolC homologue of Anabaena sp. PCC 7120 (referred to hereafter as Anabaena sp.) has been identified by proteomic analysis (18, 19), and its relation to the TolC family was confirmed by homology modeling and analysis of the corresponding mutant (5, 7, 20). Anabaena sp. is a filamentous, photosynthetic cyanobacterium, which under nitrogen deficiency develops specialized nitrogen-fixing cells, the heterocysts (21–23). It was shown that the transcript of the TolC-like gene is enhanced upon nitrogen starvation. In line with this, the mutant resulting in a loss of function displays an impairment in heterocyst development due to the missing heterocyst-specific glycolipid layer and was therefore termed HgdD (heterocyst glycolipid depositioning D) (5). Preliminary secretome analysis demonstrated the correlation of this phenotype with an altered secretome, leading to the proposal that HgdD is involved in protein secretion (5). Subsequent studies demonstrated an impairment of the same mutant in siderophore and glycolipid secretion (7, 21). Although it has been proposed that three distinct substrate categories (proteins, lipids and siderophores) are secreted by HgdD, the molecular properties of HgdD and its involvement in secretion of secondary metabolites, such as antibiotics and toxins, in cyanobacteria has so far not been described. In this paper, we examine the secretion system in Anabaena sp. and demonstrate that the TolC-like protein HgdD is a major component of the metabolite export system of this cyanobacterium.
EXPERIMENTAL PROCEDURES
Growth of Anabaena sp., DNA Isolation, and PCR
Anabaena sp. wild type and mutants were grown photoautotrophically at 30 °C in liquid BG11 medium (24, 25) with and without (in the case of BG110) 17.6 mm NaNO3 (7). Mutant strains were grown in the presence of 3 μg/ml of streptomycin and 3 μg/ml of spectinomycin. Isolation of DNA from Anabaena sp. was performed as described (26). Techniques for manipulation of plasmid DNA (27), PCR using the Triple master PCR System (Eppendorf), and conjugal transfer of plasmids into Anabaena sp. (28) have been described.
Generation of Anabaena sp. Mutants
The present study was carried out with Anabaena sp. strain PCC 7120 and several mutant derivatives described in Table 1. For the generation of alr2215 and all5346 (hgdC) single-recombinant insertion mutants, an internal fragment of the corresponding coding region was amplified by PCR on Anabaena sp. genomic DNA (oligonucleotides in supplemental Table S1) introducing EcoRI/EcoRV restriction sites. PCR products were subsequently cloned into pCSEL24, replacing the nucA-nuiA region and producing plasmids pMS1 and pMS2 (Table 2). For the generation of mutant strains, DNA transfer from Escherichia coli Anabaena sp. was performed by conjugation as described previously (29).
TABLE 1.
Anabaena sp. strains used in this study
| Strain | Resistancea | Genotype | Properties | Reference/Source |
|---|---|---|---|---|
| Anabaena sp. PCC 7120 | Wild type | |||
| CSR10 | SpR/SmR | alr4167::SpRSmR | Gene interruption by plasmid pCSV3 | Ref. 70 |
| AFS-I-alr2887 | SpR/SmR | alr2887::SpRSmR | Gene interruption by plasmid pCSV3 | Ref. 5 |
| AFS-I-alr2215 | SpR/SmR | alr2215::pMS1 | Gene interruption by plasmid pMS1 | This work |
| CSCW2 | SpR/SmR | all4025::pCSW11 | Gene interruption by gene cassette C.S3 (with partial gene deletion) | Ref. 7 |
| AFS-I-HgdC | SpR/SmR | alr5346::pMS2 | Gene interruption by plasmid pMS2 | This work |
| AFS-I-alr2269 | SpR/SmR | alr2269::SpRSmR | Gene interruption by plasmid pCSV3 | Ref. 54 |
| DR181 | NmR | Δalr2887::C.K3 | Partial replacement of alr2887 by C.K3 resistance cassette | Ref. 5 |
a Sp, spectinomycin; Sm, streptomycin; Nm, neomycin.
TABLE 2.
Plasmids used in this study
| Plasmid | Markera | Properties | Reference/Source |
|---|---|---|---|
| pCSV3 | SpR/SmR | pRL500 with substituted ApR gene | Refs. 28 and 71 |
| pCSEL24 | ApRSpR/SmR | pBR322 containing Anabaena sp. 2-kb nucA-nuiA fragment and C.S3 cassette | Ref. 72 |
| pMS1 | SpR/SmR | pCSEL24 where nucA-nuiA was replaced by an internal fragment of alr2215 | This work |
| pMS2 | SpR/SmR | pCSEL24 where nucA-nuiA was replaced by an internal fragment of hgdC (alr5346) | This work |
a Ap, ampicilin; Sp, spectinomycin; Sm, streptomycin.
mRNA Isolation and RT-PCR
mRNA isolation from Anabaena sp. has been described (e.g. see Refs. 5 and 7). In brief, RNA was isolated from 15 ml of cells at A600 = 1 by centrifugation (5 min/3200 × g). The pellet was suspended in 200 ml of PBS, mixed with 350 ml of 4 m guanidine thiocyanate, 0.1 mm DTT, 20 mm sodium acetate, pH 5.2, 0.5% N-laurylsarcosin, 10 mg/ml β-mercaptoethanol, and two small spoons of glass beads. 300 ml of phenol (65 °C) was added, and the mixture was incubated at 65 °C for 10 min with vortexing, followed by centrifuging (5 min/20,800 × g/4 °C). The aqueous layer was mixed with an equal volume of phenol/chloroform/isoamylalcohol (25:24:1) and centrifuged (20,800 × g/5 min/4 °C), and the aqueous layer was again mixed with an equal volume of chloroform. After centrifugation (5 min/20 800 × g/4 °C), the aqueous layer was supplemented with 250 ml of 70% ethanol. Further RNA isolation was performed with the NucleoSpin RNA II isolation kit (Machery & Nagel), including DNase treatment according to the instructions of the manufacturer. Digestion of residual DNA was performed for 1 h at 37 °C using 10 units of RNase-free recombinant DNase I (Roche Applied Science). cDNA was derived from 1 μg of total RNA through reverse transcription (RT), which was performed using the SuperScriptTM III First-Strand Synthesis System for RT-PCR (Invitrogen) for 1 h at 42 °C.
Counting and Surface Estimation of Anabaena sp. Cells
For the estimation of Anabaena sp. cell count, cells were grown in BG11 to OD750 = 1 and subsequently fragmented by sonification to an average length of 2–10 cells/filament. Cells were counted using a Fuchs-Rosenthal counting chamber (Blaubrang).
To calculate the surface of an Anabaena sp. cell, the approximation for an ellipsoid surface (S) and the equation for the volume (V) of an ellipsoid were used.
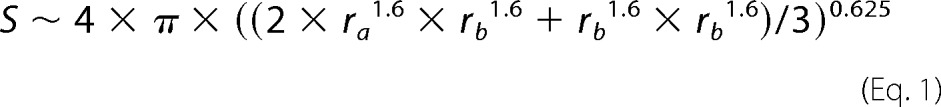 |
Microscopy
For microscopic analysis, a Zeiss Axiophot microscope (Zeiss) with a ×63/1.4 numerical aperture Plan-APOCHROMAT object lens was used, combined with a Color View XS photo system (Soft Imaging System). Transmission light and autofluorescence microscopy images were taken 30 min after ethidium bromide (EB) incubation.
Homology Search for MFS-type Proteins
Uniprot IDs assigned to the MFS were downloaded from PFAM (PF07690, PF13347, and PF05977; version 26.0). By a Python script using Biopython, the respective sequences and taxonomy information were downloaded from UniProt, and only bacterial sequences were selected for further processing. Redundant sequences were removed with cd-hit (30), and the remaining ∼55,000 MFS sequences were clustered with CLANS (31) for 100,000 iterations. Sequences from Anabaena sp. were assigned a putative function if they clustered closely with a known and functionally characterized MFS protein: AmpG (P0AE16), Bcr (P28246), EmrB (P0AEJ0), EmrD (P31442), EntS (P24077), SetA (P31675), ShiA (P76350), MelB (P02921), or MdfA (P0AEY8) of E. coli.
Ethidium Uptake Measured by Quantification in Extracellular Medium
Anabaena sp. cells were grown in BG11 medium to late logarithmic phase based on the growth curve analysis (OD750 = 1.5). To determine uptake, the cells were adjusted to OD750 = 1, concentrated 10-fold by centrifugation, and resuspended in BG11 medium containing 20 mm HEPES, pH 7. After the addition of EB, cells were transferred into mini filter spin columns (Sigma-Aldrich) containing a PET membrane with a pore size of 0.45 μm. After 2.5, 5, 15, and 30 min, the growth medium was separated from cells by centrifugation (30 s/4000 × g).
For the measurement of ethidium bromide secretion, cells were incubated for 30 min in different concentrations of EB and washed twice with BG11 medium using mini filter spin columns. EB concentration in the flow-through was determined by fluorescence measurement (excitation, 316 nm; emission, 620 nm) after the addition of 100 μg of herring sperm DNA with an Infinite® 200 PRO multimode reader (Tecan). The S.D. value was calculated on the basis of at least three independent experiments. For the competition of EB with erythromycin, both substances were mixed in a 10:1, 1:1, and 1:10 molar ratio before addition to the concentrated Anabaena sp. cell suspension.
Ethidium Uptake Measured by DNA Intercalation
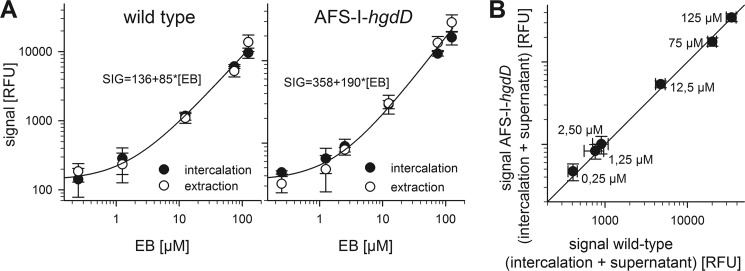
190 μl of concentrated Anabaena sp. cell suspension was loaded into a 96-well microplate (Corning Glass). After the addition of EB, the increase of fluorescence due to the intercalation of ethidium into endogenous nucleic acids after uptake was monitored every minute for 30 min (excitation, 316 nm; emission, 620 nm). The procedure was controlled by analysis of EB concentration by an independent method as outlined in Fig. 2. For that, the extraction was performed as follows. Cells were incubated in different EB concentrations and applied to filter spin columns. After 15 min, cells were separated from incubation medium by centrifugation at (30 s/4000 × g). Cells were recovered from the column in an equal volume of EB-free incubation medium and mechanically opened by using glass beads and a cutting mill (Retsch MM 300). After centrifugation (2 min/20,000 × g), supernatant was cleared from photosynthetic proteins by adding a small amount of DEAE-Sepharose CL-6B beads (Sigma-Aldrich). EB concentration was determined as described earlier.
FIGURE 2.
Justification of the reliability of the analysis by intercalation. A, the uptake of EB by wild type (left) or AFS-I-hgdD (right) incubated at the indicated EB concentration was measured by intercalation (black) and extraction of ethidium (white). The line represents the least square fit analysis of both data sets in combination by a linear function. B, the ethidium remaining in the solution (supernatant) and intercalated was determined for wild type and AFS-I-hgdD. The line represents the x = y. For A and B, the average of at least three independent results are shown Error bars, standard deviation. RFU, relative fluorescence units.
Measurement of Ethidium Uptake after Carbonyl Cyanide m-Chlorophenylhydrazone (CCCP) or Spermidine/Spermine Treatment
10 min prior to a measurement, 10 mm CCCP in DMSO was added to a final concentration of 200 μm to a concentrated Anabaena sp. cell suspension and incubated at room temperature. A control sample was treated in parallel with an equal amount of DMSO. The same procedure was applied for incubation with spermidine/spermine at the concentrations indicated in the Fig. 3 legend.
FIGURE 3.
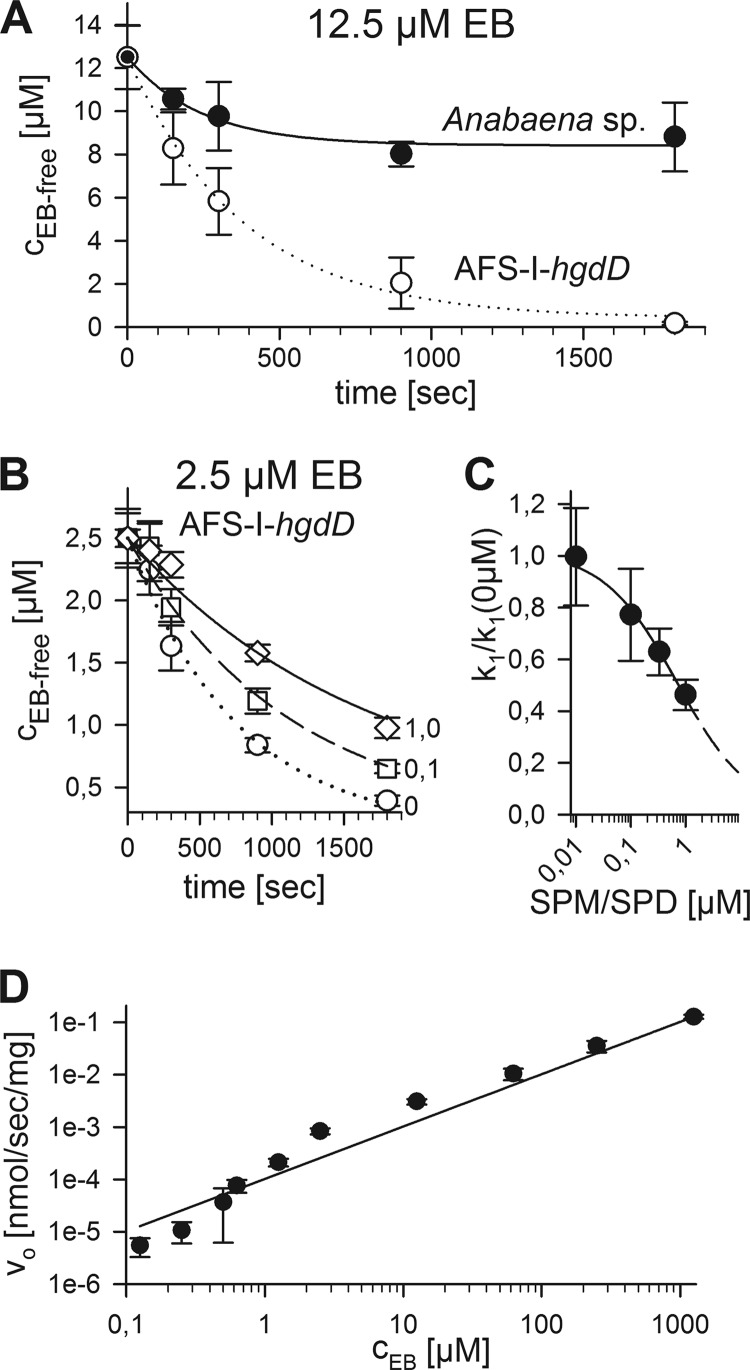
Ethidium uptake by Anabaena sp. and the hgdD mutant by analysis of the extracellular ethidium. A, the EB remaining in solution after incubation of wild type (black) or AFS-I-hgdD (white) with 12.5 μm EB was quantified after the indicated time periods. Lines indicate the analysis according to E1 (Fig. 1). B, the EB remaining in solution after incubation of AFS-I-hgdD with 2.5 μm EB in the presence of 0 (circles), 0.1 (squares), or 1.0 μm spermine/spermidine (diamonds) was quantified after the indicated time periods. Lines indicate the analysis according to E1 (Fig. 1). C, k1 determined in the presence of the indicated concentrations of spermine/spermidine that were set in relation to k1 in the absence of the polyamines. The line represents the least square fit analysis to a four-parameter logistic equation to determine the EC50 value. The dashed line represents the extrapolated result according to the parameters determined. D, the initial uptake rate (v0) was determined from measurements as in A for wild type and plotted against the EB concentration. The line represents the least square fit analysis to the equation described (34). Error bars, standard deviation.
Analysis of Uptake Results and Mathematical Concepts
The results were processed by Sigma Plot and, if not stated otherwise, analyzed by a least square fit analysis to the equations shown in Fig. 1. For the second equation in Fig. 1 (E2), the abbreviations used are listed in Fig. 1 as well. This equation is taken from Ref. 32.
RESULTS
Putative Model for Analysis of Metabolite Uptake
To describe the uptake and secretion of metabolites by Anabaena sp., we utilized the fluorescent dye ethidium bromide as a model substrate. We first defined a model for the analysis (Fig. 1, top), which is based on the established models for secretion by Gram-negative bacteria (1–4). We considered five independent reactions: the uptake by porins (k1), the transfer into the cytoplasm (k2), association and dissociation with and from DNA (k3, k−3), the export into the periplasm by MFS proteins (k−2), and the export by TolC (k−1) (33). In general, the initial process across the outer membrane is defined by passive diffusion through porins (e.g. see Refs. 34 and 35). However, for simplicity, we treated the reaction with a simple kinetic constant, which can be represented by k1. In a first step, we determined uptake by analyzing the ethidium remaining in the growth medium. In this case, we only considered the k1 and k−1, which can be analyzed by the first equation in Fig. 1 (E1). Using this method, we could determine the properties of the porins in the outer membrane and the rate of export catalyzed by the TolC-containing machinery because we do not discriminate between ethidium in the periplasm or cytosol.
In another step, we analyzed the uptake by measuring the intercalation of ethidium into intracellular DNA, which requires a two-state model. Given the dissociation constant of ethidium for DNA, about 4 μm (e.g. see Ref. 36), and the high concentration of DNA and RNA in the cell, it is possible to combine k2 and k3 as well as k−2 and k−3. With this simplification, the reaction can be described analytically (Fig. 1, E2) (32). In this way, we were able to describe differences of the components involved in the export cycle between the wild type and mutants. However, the combination of k2 and k3 must be taken into account for the interpretation of the rate constants.
Uptake of Ethidium by Anabaena sp. Involves a Porin-type Activity
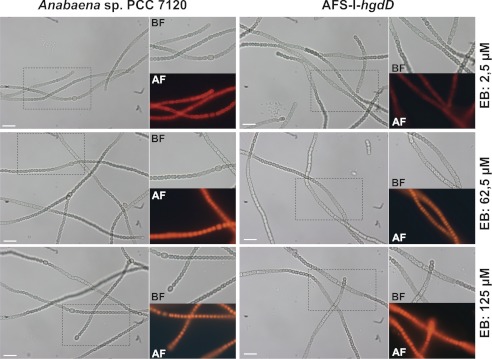
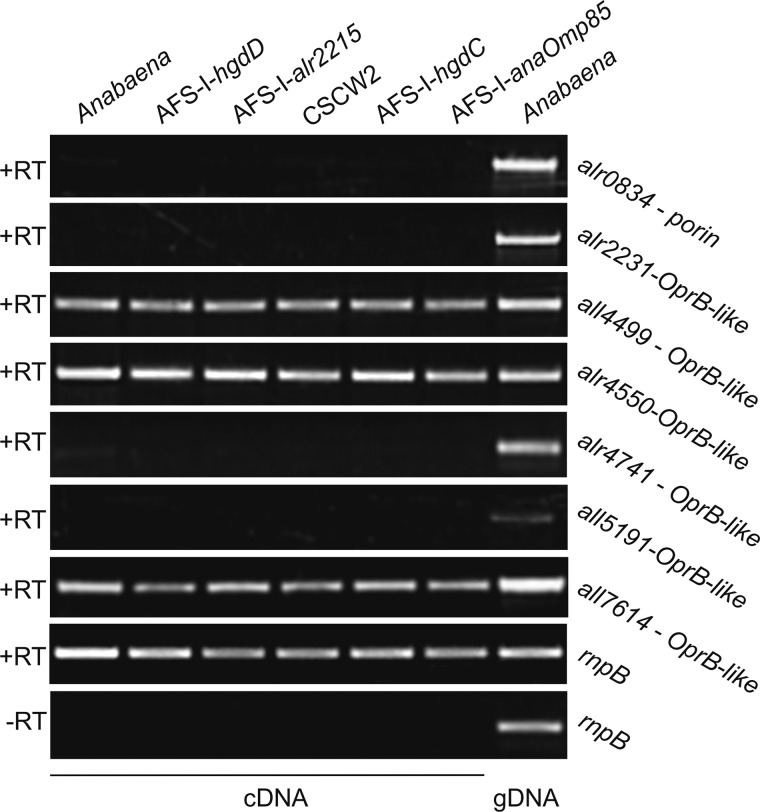
We analyzed the uptake of ethidium by Anabaena sp. and AFS-I-hgdD (5, 7) (Fig. 1, bottom, uptake). We incubated Anabaena sp. with various concentrations of ethidium bromide and analyzed the kinetics of uptake by least square fit analysis to Fig. 1 (E1) (Fig. 3A). The used ethidium bromide concentration did not alter the morphology of the cells and the filament of wild type Anabaena sp. or AFS-I-hgdD, respectively (Fig. 4) In the wild type, the equilibrium between uptake and export was reached after 500 s, whereas almost no export was detectable from the AFS-I-hgdD strain under the conditions used (Fig. 3A). The same result was observed for an independently generated hgdD deletion mutant, strain DR181 (5) (supplemental Fig. S1). This supports the notion that the reduced export is the result of the loss of HgdD function. Consistent with this notion, none of the Anabaena sp. genes coding for putative porins (18, 19, 22) were altered in their expression in the hgdD mutant used in this study (Fig. 5).
FIGURE 4.
Cellular morphology after ethidium bromide incubation. Shown are representative images of filaments of Anabaena wild type (left) of AFS-I-hgdD (right) incubated for 30 min with the indicated concentration of ethidium bromide. No obvious alteration of filament structure or length and no obvious alteration of the cellular integrity were realized. BF, bright field; AF, autofluorescence; scale bar, 10 μm.
FIGURE 5.
Expression of genes coding for putative porin-like proteins of Anabaena sp. RNA from Anabaena sp., AFS-I-hgdD, AFS-I-all2215, CSCW2, AFS-I-hgdC, and AFS-I-anaOmp85 grown in BG11 as well as genomic DNA from Anabaena sp. grown in BG11 were isolated and analyzed. RT-PCR/PCR was performed using oligonucleotides for the indicated genes listed in supplemental Table S1. The bottom panel shows the result in the absence of the reverse transcriptase. The genes for analysis were selected according to Refs. 18, 19, 22, and 24.
The likely functional importance of a porin-type activity was further probed by analysis of ethidium uptake by AFS-I-hgdD in the presence of known porin inhibitors (37, 38). Prominent and often-used inhibitors are the polyamines spermine and spermidine (37). For experimental reasons, the highest concentration that could be applied was 1 μm. However, even at this concentration, a significant decrease of ethidium uptake was observed (Fig. 3B). Analyzing the uptake kinetics by E1 (Fig. 1) revealed a decrease of k1 in relation to the spermine/spermidine concentration, which led to half the maximal inhibition at a concentration of about 0.7 μm (Fig. 3C). This is in the same range as the spermine concentration at which the half-maximum of the spermine-dependent change of closing frequency of OmpC or OmpF from E. coli was observed (38).
To further describe the properties of the porins, we subsequently determined the initial rate, V0. The uptake was analyzed by the equation, Vin = P × A × (Co − Cp), describing the relation for a passive pore (e.g. see Ref. 34), where Vin is the rate of uptake, P is the permeability coefficient (cm/s), A is the surface area of a cell (cm2/mg), and Co and Cp are the concentrations in the extracellular space and the periplasm, respectively. To apply this equation, we estimated the cell surface present in our assay as described below.
By counting cells, we observed an average of 2.8 × 107 cells/ml at OD750 nm = 1. A cell has an average length of 2.5 μm and an average width of 2.2 μm, as determined by electron microscopy (e.g. see Ref. 5). This leads to a cell volume of about 6.3 μm3 and a cell surface of ∼17 μm2. The cell-cell contact is ∼1.5 μm in diameter (leading to an area of ∼2 μm2). Thus, the contact sites reduce the surface of a single cell by about 15%, which is in the range of the error for the determination of cell dimensions and thus can be neglected. Accordingly, we estimated a cell volume of 1.8 mm3 and a surface of 48 cm2/ml cell culture at OD750 nm = 1. In addition, the cell density of OD750 nm = 1 reflects a dry mass of about 0.7 mg/ml, which at least in the case of the filamentous cyanobacterium Gloeotrichia echinulata was shown to be linearly related to the OD measured at this wavelength (e.g. Ref. 39). Thus, we had an estimated surface of 69 cm2/mg (dry weight) in our experiments.
Using the initial rate, the equation can be simplified to Vo = P × A × Co, reflecting a simple linear behavior (Fig. 2D). Vo was normalized to the dry weight of the cells used. As a result, we obtained for P × A (1.03 ± 0.08) × 10−4 cm3/s/mg and thus a permeability coefficient of 1.5 × 10−6 cm/s (Table 3). This value is comparable with the one obtained for porin-dependent transport of other, similarly sized compounds in proteobacterial systems (e.g. see Refs. 40 and 41).
TABLE 3.
Kinetic parameters determined in this study
| Strain | Porina, P | Secretionb |
E1c |
E2d |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Km | Vmax | k1 | k−1 | k1 | k−1 | k2 | k−2 | ||
| cm s−1 | s−1 | s−1 | s−1 | s−1 | s−1 | s−1 | |||
| Wild type | 1.5 × 10−6 | 170 ± 10 μm | 420 ± 20 nm s−1 | (12 ± 1) × 10−4 | (20 ± 5) × 10−4 | (15 ± 2) × 10−4 | (9.7 ± 0.6) × 10−4 | (18 ± 3) × 10−4 | (34 ± 3) × 10−4 |
| AFS-I-hgdD | NDe | 40 ± 8 m | 25 ± 5 mm s−1 | (14 ± 4) × 10−4 | (0.8 ± 0.2) × 10−4 | (19 ± 2) × 10−4 | (6 ± 1) × 10−6f | (15 ± 5) × 10−4 | (27 ± 5) × 10−4 |
| AFS-I-hgdC | ND | ND | ND | ND | ND | (18 ± 2) × 10−4 | (6.9 ± 0.9) × 10−4 | (13 ± 2) × 10−4 | (35 ± 3) × 10−4 |
| CSCW2 | ND | ND | ND | (9 ± 2) × 10−4 | (31 ± 9) × 10−4 | (18 ± 4) × 10−4 | (15 ± 6) × 10−4g | (18 ± 8) × 10−4 | (5 ± 1) × 10−4f |
| AFS-I-all2215 | ND | ND | ND | (13 ± 2) × 10−4 | (24 ± 7) × 10−4d | (19 ± 5) × 10−4 | (14 ± 5) × 10−4g | (11 ± 4) × 10−4 | (2 ± 1) × 10−4f |
| AFS-I-anaOmp85 | ND | ND | ND | ND | ND | (3.7 ± 0.6) × 10−4f | (7 ± 2) × 10−4 | (19 ± 6) × 10−4 | (25 ± 7) × 10−4 |
Ethidium Export in Anabaena sp. Is HgdD-dependent
Next, we aimed to confirm that HgdD is involved in metabolite export. We incubated Anabaena sp. and the hgdD mutant with ethidium bromide, calculated the intracellular concentration of ethidium for each strain, and analyzed the export by its reoccurrence in the medium. We determined the initial rate (v0) as suggested for TolC-dependent efflux (34), which was analyzed with respect to the preincorporated concentration (Fig. 6). However, this method is error-prone at multiple levels. The starting concentration calculated was normalized to the reaction volume and not to the intracellular volume. Furthermore, we could not quantify to which extent ethidium was distributed between the periplasmic space and the cytoplasm. Thus, the values determined are only for comparison between the two genotypes and do not reflect the exact kinetic parameters of the TolC system.
FIGURE 6.
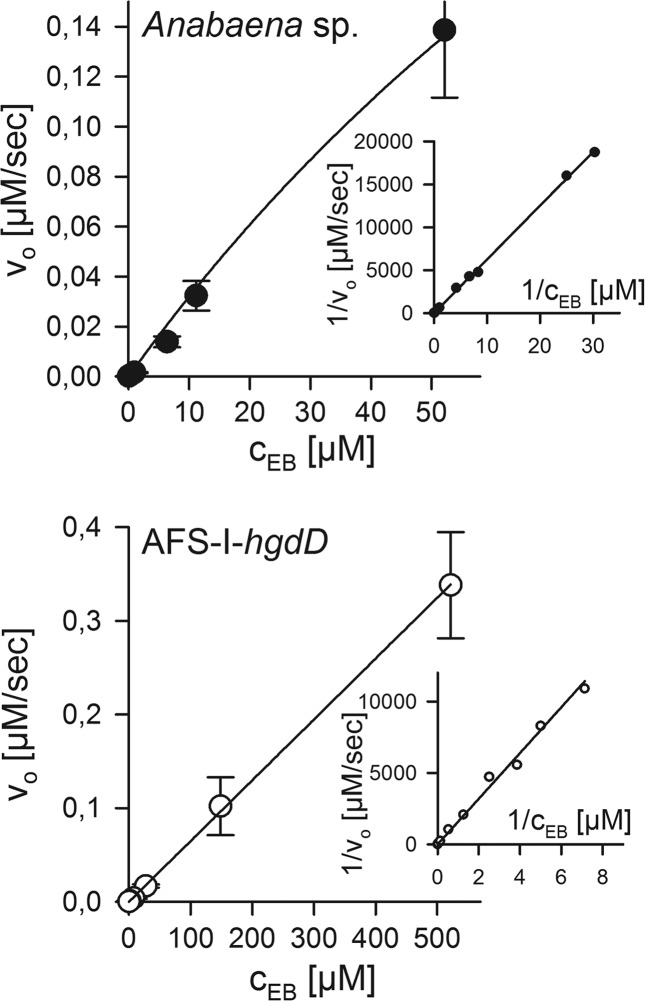
Ethidium release by Anabaena sp. and the hgdD mutant. The ethidium released from preloaded wild type (top) or AFS-I-hgdD cells (bottom) was analyzed as described under “Experimental Procedures.” Michaelis-Menten constants were determined by least square fit analysis and confirmed by Lineweaver-Burk analysis (inset). Values are listed in Table 3.
Comparing the Michaelis-Menten constant determined for the wild type and for AFS-I-hgdD, we observed a difference of 5 orders of magnitude. Because the concentration of ethidium in the Anabaena sp. cells was underestimated while considering the concentration of the entire reaction volume for the calculation, we could conclude that the TolC-independent export has a Michaelis-Menten constant of more than 40 M. The maximal rate is higher in the mutant than in the wild type as well (Fig. 6 and Table 3). This suggests that TolC-independent export from the mutant strain occurs by a process, most likely passive diffusion.
HgdD Is Involved in General Antibiotic Export in Anabaena sp.
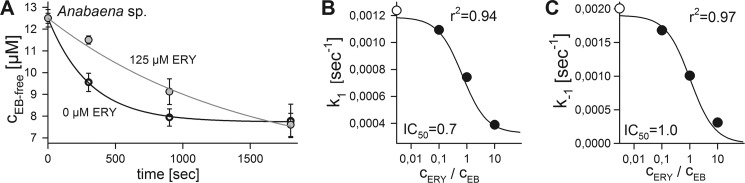
Based on the observations described above, one may assume that HgdD is involved in a general antibiotic and toxin export pathway. To test this hypothesis, we used erythromycin to compete for both uptake and export of ethidium. Increasing concentrations of erythromycin were added to the measurement of the uptake kinetics of ethidium (Fig. 7A). The analysis by E1 (Fig. 1) revealed the import and export rates in the absence, in the presence, and at a 10-fold excess of erythromycin compared with 12.5 μm ethidium. We analyzed the erythromycin concentration-dependent rates using a hyperbolic equation (Fig. 7B) and determined an IC50 value of 8.6 and 12.9 μm for k1 and k−1. This reflects an ethidium/erythromycin ratio of 0.7 for uptake and 1.0 for secretion for the applied concentration of ethidium. Considering the IC50 values that were obtained, we can conclude that the porin system and the TolC system can transport both ethidium (394 Da) and erythromycin (734 Da). However, the ratio between the uptake rates of ethidium and erythromycin might be concentration-dependent because the two compounds are structurally distinct. Nevertheless, by this observation, we can generalize the conclusion that HgdD is involved in export of secondary metabolites, such as antibiotics.
FIGURE 7.
Competition of ethidium uptake by Anabaena sp. and the hgdD mutant by analysis of the extracellular ethidium. A, comparison of ethidium uptake in the absence (white) and presence (gray) of 125 μm erythromycin (ERY) as measured in Fig. 2. Lines represent least square fit analysis by E1 (Fig. 1). The k1 (B) and k−1 rate (C) for different ratios between erythromycin and EB is plotted, and lines represent the least square fit to determine the IC50 value indicated. Error bars, standard deviation.
Intercalation of Ethidium by Anabaena sp. and AFS-I-hgdD
Next, we analyzed the uptake of 12.5 μm ethidium by Anabaena sp. and AFS-I-hgdD using the intercalation assay (Fig. 1). We observed a significant intercalation of ethidium by AFS-I-hgdD, but not by the wild type (Fig. 8). This confirms the observation made while measuring the uptake by the two strains (Figs. 3–7). Analyzing the intercalation by E2 (Fig. 1) (Fig. 8A, lines), we observed a 100-fold lower rate constant k−1 for AFS-I-hgdD than for wild type, which reflects that HgdD is indeed involved in export (Table 3). All other rate constants were in the same range.
FIGURE 8.
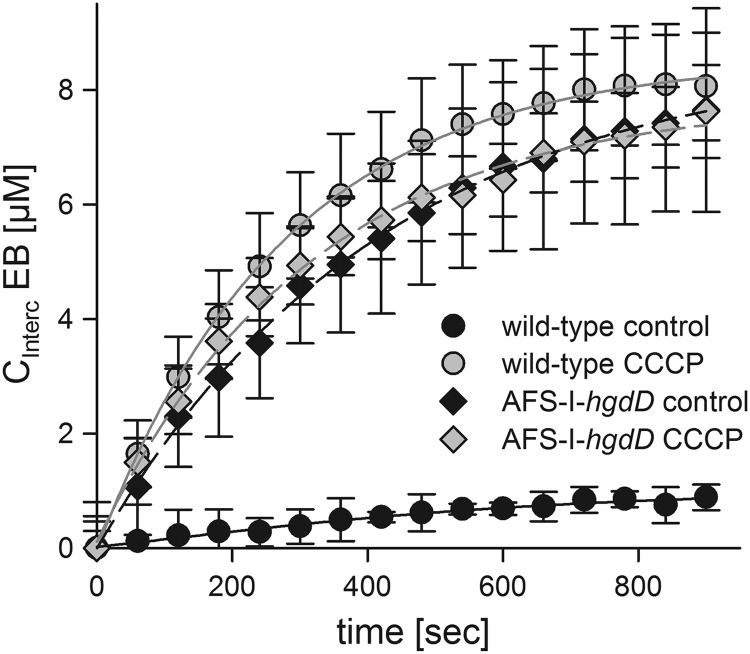
Analysis of ethidium uptake by Anabaena sp. and the hgdD mutant by intercalation. A, ethidium uptake (12.5 μm) in the absence (black) and presence (gray) of CCCP by wild type (circles) or AFS-I-hgdD cells (diamonds) was measured as described under “Experimental Procedures.” Lines represent least square fit analysis by E2 (Fig. 1). Error bars, standard deviation.
Because proteins of the TolC family usually represent the outer membrane factor of a tripartite secretion system, they have to interact with a plasma membrane-localized permease. Hence, we analyzed whether such putative HgdD-charging permease is proton gradient-dependent. For this, we abolished the proton gradient across membranes in the cyanobacterium by adding the protonophore CCCP. Measuring the uptake of ethidium by the wild type and AFS-I-hgdD mutant in the presence of CCCP, we observed a similar intercalation by both strains (Fig. 8). Furthermore, the intercalation in the presence of CCCP was comparable with the one observed for AFS-I-hgdD in the absence of the protonophore. Thus, the HgdD-dependent transport indeed requires the proton gradient, which is most likely produced by a proton pump-type permease (42, 43).
Identification of Putative MFS-type Proteins
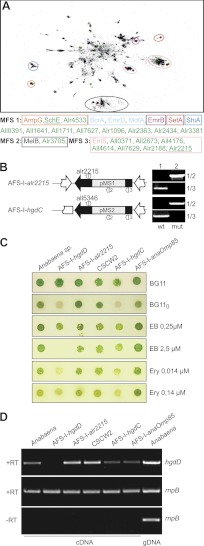
We have demonstrated that HgdD of Anabaena sp. is involved in detoxification. As depicted in Fig. 1, additional plasma membrane-inserted proteins of different families are involved in the process of uptake and export (33). To obtain further insights into putative factors involved in metabolite export, we used a bioinformatics approach to identify candidates for MFS proteins in the genome of Anabaena sp. Within the three MFS types annotated in the PFAM database (MFS 1, MFS 2, and MFS 3), we found in total 18 sequences from Anabaena sp. We performed a CLANS analysis (Fig. 9A) (e.g. see Ref. 31) to assign the identified sequences to the different MFS types.
FIGURE 9.
Putative MFSs involved in antibiotic resistance. A, a CLANS clustering of ∼55,000 non-redundant MFS proteins of the PFAM families MFS 1, MFS 2, and MFS 3 is shown. Clusters containing functionally characterized transporters (marked as red dots) are circled and are listed below the clustering. Sequences from Anabaena sp. are marked with green dots. B, AFS-I-alr2215 and AFS-I-hgdC were generated via insertion of pMS1 and pMS2, respectively, by single homologue recombination (primer combination 1/2). Both strains are fully segregated because no amplification of wild type genomic DNA could be observed under standard PCR conditions (primer combination 1/3). Wild type genomic DNA was used as a control (lane 1). C, Anabaena sp., AFS-I-hgdD, AFS-I-all2215, CSCW2, AFS-I-hgdC, and AFS-I-anaOmp85 were grown on the indicated media. Images were taken after 7 days. Ery, erythromycin. D, RNA from Anabaena sp., AFS-I-hgdD, AFS-I-all2215, CSCW2, AFS-I-hgdC, and AFS-I-anaOmp85 grown in BG11 and genomic DNA from Anabaena sp. grown in BG11 were isolated and analyzed. RT-PCR/PCR was performed using oligonucleotides for hgdD (top) or rnpB (middle and bottom). The bottom panel shows the result in the absence of the reverse transcriptase.
Two of these sequences are similar to the transporter for GlcNAc-1,6-anhydromuropeptides AmpG (MFS 1) (e.g. see Ref. 44), namely the schizokinon exporter SchE (All4025) (7) and Alr4533. Furthermore, a single sequence is related to sugar transporters described by the MFS 2-type (Alr3705), which is similar to, for example, MelB (45), UidB (46), YihP (47), and YihO (48).
Among sequences of the MFS 3-type, which contains transporters involved in enterobactin (49) or antibiotic efflux (50), seven proteins are from Anabaena sp. From the remaining eight proteins, which do not cluster with any known protein, only All0391 was previously identified as part of the putative schizokinen synthesis cluster (51).
To analyze whether such proteins are involved in the TolC-dependent export cycle, we analyzed the mutant strain CSCW2, which carries an insertion in schE, as an example of a MFS 1-type transporter. This mutant has been previously described as defective in secretion of the siderophore schizokinin (7). We also generated an insertion mutant of the MFS 3-type transporter gene all2215 (Fig. 9B; strain AFS-I-all2215). We excluded the MFS 2-type transporter from subsequent studies because they are known to be involved in sugar transport (45–48). A mutant of hgdC (AFS-I-hgdC), a plasma membrane ABC transporter involved in heterocyst development (all5346) (52), was used as a negative control. The strains generated were fully segregated, because no wild type gene was detectable (Fig. 9B, 1/3). In contrast to AFS-I-hgdD (5) and AFS-I-hgdC (52), AFS-I-all2215 was able to grow on the medium without a fixed nitrogen source (Fig. 9C). Testing the mutants with respect to their sensitivity to EB or erythromycin, we observed a growth reduction of AFS-I-hgdC, AFS-I-hgdD, and CSCW2, but not of AFS-I-alr2215 or AFS-I-anaOmp85. This observation supports the notion that the TolC-like protein HgdD performs multiple functions in concert with distinct plasma membrane permeases (7).
The expression of hgdD in different strains was probed by RT-PCR. Using identical amounts of mRNA, as controlled by amplification of RNase P RNA (rnpB) cDNA, we observed an enhanced expression of hgdD in AFS-I-alr2215 and CSCW2, whereas the transcript level did not change in AFS-I-hgdC or AFS-I-anaOmp85, suggesting a functional relation between HgdD and Alr2215 and SchE.
Intercalation of Ethidium by Different Mutant Strains
Having generated and confirmed the mutant strains, we analyzed their behavior based on the intercalation of ethidium. Additionally, we used AFS-I-anaOmp85, in which the outer membrane protein assembly was most likely affected (53, 54). However, it has to be mentioned that this mutant is not fully segregated, although a drastic phenotype can be observed (54).
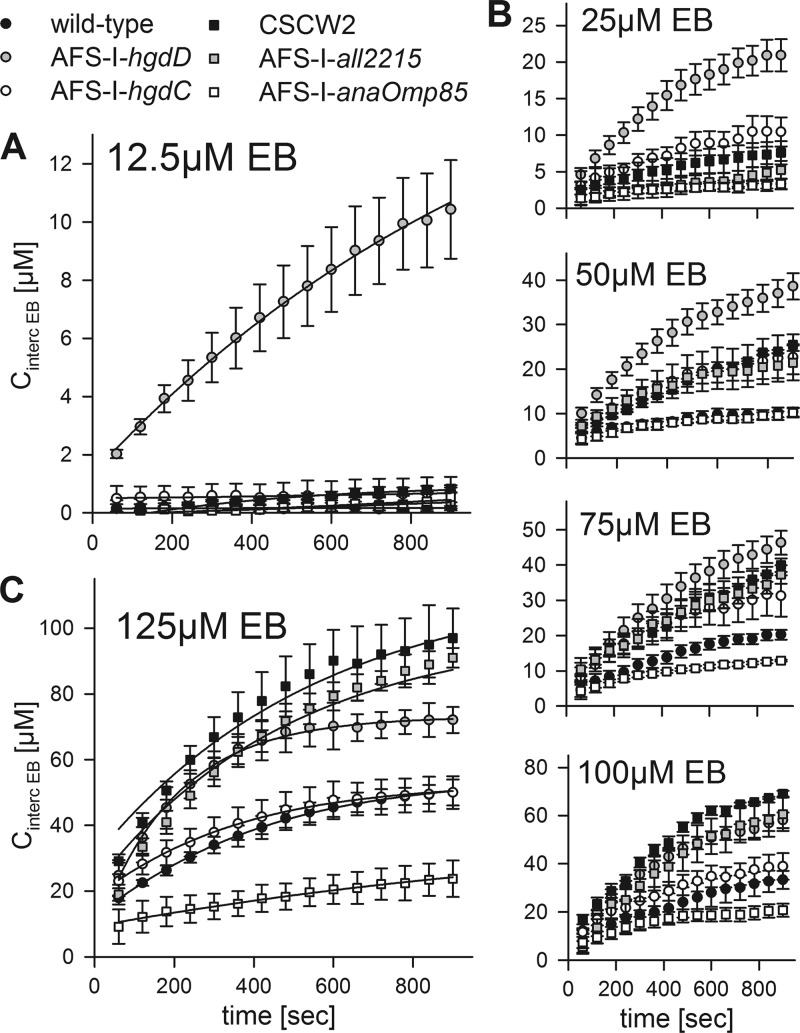
At a low concentration of ethidium bromide (12.5 μm; Fig. 10A), we did not observe a significant intercalation by the strains with the exception of AFS-I-hgdD. Notably, the result obtained for the strain AFS-I-anaOmp85 suggests that in this mutant, the proper targeting and assembly of HgdD is not affected.
FIGURE 10.
Analysis of ethidium uptake by wild type and different mutants of Anabaena sp. by intercalation. A, ethidium uptake at 12.5 μm EB by wild type (filled circle), AFS-I-hgdD (gray circle), AFS-I-hgdC (white circle), CSCW2 (filled square), AFS-I-all2215 (gray square), and AFS-I-anaOmp85 (white square) was measured as in Fig. 5. The lines represent least square fit analysis by E2 (Fig. 1). B, ethidium uptake in the presence of 25, 50, 75, and 100 μm EB was determined and presented as in A. C, ethidium uptake at 125 μm EB was determined and presented as in A. The determined rate constants are listed in Table 3. Error bars, standard deviation.
However, we observed a concentration-dependent increase in the intercalation of ethidium by the wild type (25–125 μm; Fig. 10, B and C, black circles). Consistent with this phenotype (Fig. 9B), intercalation by AFS-I-hgdC (Fig. 10, B and C, open circles) was largely similar to that in the wild type. In contrast, intercalation by AFS-I-all2215 (gray squares), CSCW2 (black squares), and AFS-I-hgdD (gray circles) exceeded the one of wild type, whereas intercalation by AFS-I-anaOmp85 was reduced (open squares).
We next described the data by least square fit analysis by E2 (Fig. 1) (lines in Fig. 10, A and C) and compared the determined rate constants. The rate constant reflecting the uptake across the outer membrane (k1) was only affected in AFS-I-anaOmp85, which is consistent with its function in the assembly of outer membrane proteins (Table 3). Similarly, the rate constant reflecting the export across the outer membrane (k−1) was strongly reduced in AFS-I-hgdD, whereas only slight alterations were observed for other mutants. Here, the slight increase of the rate observed for AFS-I-alr2215 or CSCW2 can be explained by the enhanced expression of hgdD in these two strains (Fig. 9).
The rate for uptake across the plasma membrane (k2) was not affected in any of these mutants, although slight variations existed (Table 3). The rate for export across the plasma membrane (k−2) was of the same order of magnitude as the rate of transport across the plasma membrane (k2). CSCW2 and AFS-I-all2215, but not AFS-I-hgdC (Table 3), showed a reduced rate for export across the plasma membrane, which was consistent with the enhanced sensitivity of these two strains against erythromycin (Fig. 9C). However, having two exporters explains the lower sensitivity against this antibiotic seen for each of the corresponding mutants than for AFS-I-hgdD and explains why only a reduction and not a complete loss of export across the plasma membrane was observed.
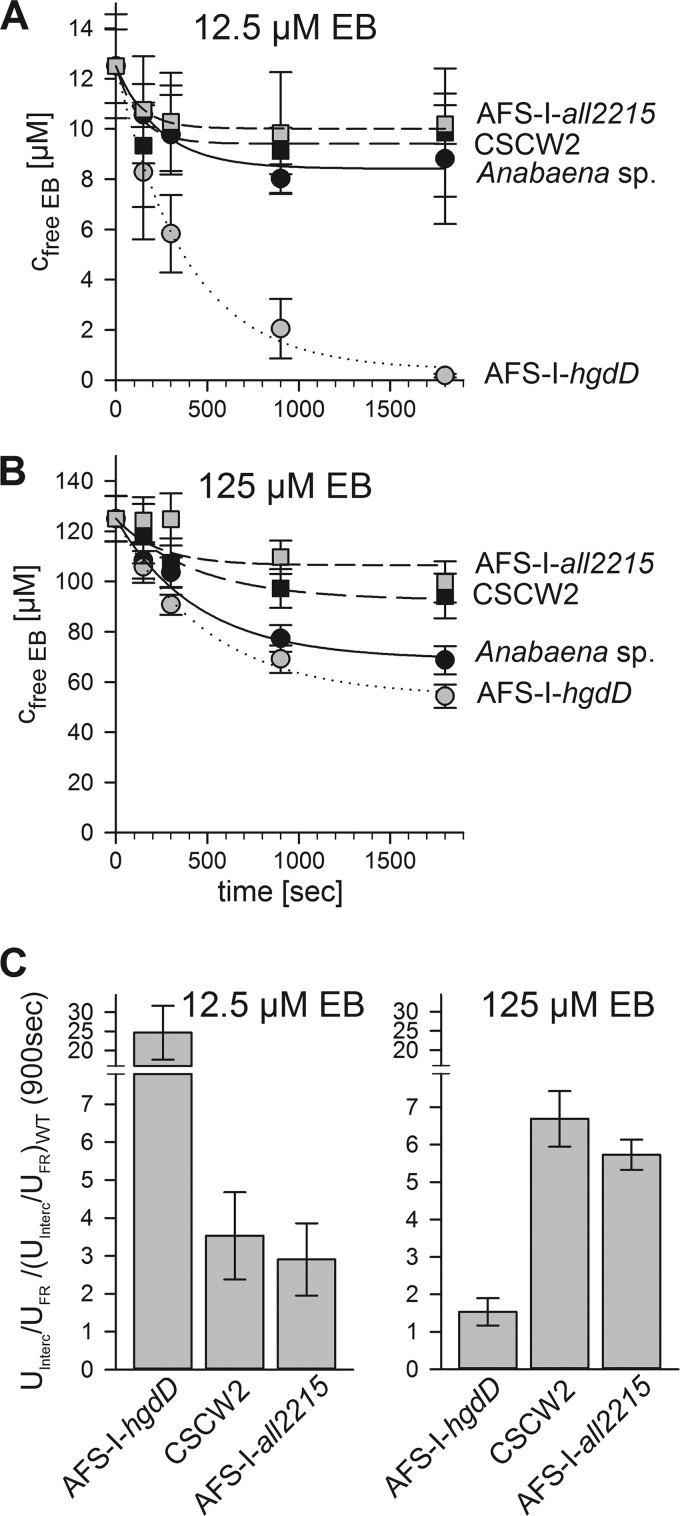
Uptake of Ethidium by CSCW2 and AFS-I-all2215
The observed enhanced intercalation by CSCW2 and AFS-I-all2215 at high ethidium bromide concentrations prompted us to analyze the uptake behavior by measuring the ethidium bromide remaining in the solution. At 12.5 μm, no significant difference in uptake by CSCW2 and AFS-I-all2215 when compared with the wild type was observed (Fig. 11A). However, the uptake of ethidium at 125 μm was reduced in both strains (Fig. 11B). This is noteworthy, considering the higher intercalation in comparison with the wild type at 125 μm EB (Fig. 10C). Analysis by E1 (Fig. 1) confirmed the slight increase of the TolC-dependent export rate (k−1) when compared with the wild type (Table 3). Thus, we suggest that the reduced uptake of both MFS mutants is a consequence of an enhanced export activity across the outer membrane (Fig. 9D). At the same time, the reduction in export across the plasma membrane (k−2) led to an accumulation of ethidium bromide in the cytosol, which was therefore no longer accessible for secretion by HgdD.
FIGURE 11.
Ethidium uptake by Anabaena sp. and mutants by analysis of the extracellular EB. The free ethidium bromide was quantified as described after incubation of wild type (filled circles), AFS-I-hgdD (gray circles), CSCW2 (filled squares), and AFS-I-all2215 (filled squares) with 12.5 μm EB (A) or 125 μm EB (B) for the indicated time periods. Lines indicate the analysis according to E1 (Fig. 1). C, the ratio between uptake (U) determined by intercalation (IN) and determined by measuring the EB remaining in solution (FR) was calculated and placed in relation to the value observed for the wild type strain (WT). Uptake values after 900 s were used. Error bars, standard deviation.
We next compared the ratio of intercalation and total uptake of ethidium for hgdD and both MFS mutants with the one obtained for the wild type. It became obvious that at low ethidium concentrations, the accumulation in the cytosol of AFS-I-hgdD was 25-fold higher when compared with the wild type, whereas at high concentration, the ratios became comparable (Fig. 11C). In contrast, for CSCW2 and AFS-I-all2215, the ethidium retention in the cytosol was about 6-fold higher compared with the wild type and at high ethidium concentration even compared with ASF-I-hgdD (Fig. 11C). These observations show that at low concentrations, TolC is sufficient to export ethidium from the periplasm, whereas at high concentrations, the process of secretion is rate-limiting by the export efficiency of ethidium across the plasma membrane.
DISCUSSION
An anaOmp85-independent Insertion of HgdD into the Outer Membrane?
Omp85 is thought to be a general factor for the insertion of outer membrane proteins in Gram-negative bacteria, mitochondria, and chloroplasts (e.g. see Refs. 53 and 55). Consistent with its global function, Omp85 (BamA) of E. coli is essential for the assembly and subsequently for the function of TolC (56). Interestingly, in Anabaena sp., Omp85 encoded by alr2269 influences the uptake activity (k1; Fig. 10 and Table 3), but not the secretion efficiency (k−1; Table 3). This suggests that in AFS-I-Omp85, the assembly of HgdD is not disturbed. Furthermore, the mutants of the other two Omp85-like genes (alr0075 and alr4893) (54, 57, 58) do not show an overlapping phenotype with the hgdD mutant (59) with respect to heterocyst development (5) or the sensitivity of AFS-I-hgdD to 30 mm lysozyme (59). A function of Alr0075 in HgdD insertion and assembly is further unlikely because this Omp85-like protein only exists in vegetative cells (54), whereas the expression of hgdD is enhanced in heterocysts (5). This could suggest that in Anabaena sp., the function of the Omp85 is not required for the assembly of HgdD, which, however, has to be further investigated.
Porin-mediated Permeability of the Outer Membrane of Anabaena sp. PCC 7120
The existence of classical porins in cyanobacteria is largely under debate. On the one hand, the porins found to be encoded in the genomes of different cyanobacteria or by proteomic approaches are generally larger than those found in proteobacteria (e.g. see Refs. 18, 19, and 60). The extension is thought to serve as a link of the proteins to the peptidoglycan layer (61), which has a defined distance to the outer membrane (e.g. see Ref. 62). On the other hand, the only experimentally approached porin-like proteins, SomA and SomB from Synechococcus (63–65), have a conductance of about 0.5 nanosiemens (66), which parallels the observed conductance of cell envelope fractions of Anabaena variabilis (67) but is about 10-fold smaller than that found for typical proteobacterial porins (e.g. see Ref. 68).
The reduced size could be interpreted as a remnant of the pore activity, which is no longer required as cyanobacterial “porins” if the hypothesis is made that they are involved in anchoring to the peptidoglycan rather than in transporting molecules. Alternatively, if a function in transport is considered, the pore activity could be regulated in a manner different from that found for proteobacterial porins. One could envision that such regulation includes a periplasmic section functioning as a “plug.” Both ideas would be consistent with the hypothesis that the periplasm of Anabaena sp. is involved in long distance transport of solutes in the filament (69).
However, our results put the previous notion into question that porins of cyanobacteria are smaller because of “the photoautotrophic lifestyle” of cyanobacteria, which requires porins that are only large enough to facilitate the uptake of small solutes, such as ions, from their environment, whereas biopolymers are synthesized by the bacteria themselves (61). We observed uptake of ethidium (394 Da) and erythromycin (734 Da; Figs. 3–11), and a permeability coefficient for Anabaena sp. (Fig. 3 and Table 3) comparable with the one found for proteobacteria (e.g. see Refs. 40 and 41). Further, uptake of ethidium is reduced in the Omp85 mutant (Table 3), the latter being involved in the insertion and assembly of porins (e.g. see Ref. 53) and by adding spermine/spermidine, well established inhibitors of porin activity (e.g. see Refs. 37 and 38). The effective concentration of the polyamines is in the same range as found to alter the closing probability of OmpC or OmpF from E. coli (Fig. 3) (38), which is consistent with the interpretation that a porin-like activity accounts for the uptake of ethidium. Thus, we provide evidence that a porin-like permeability exists in Anabaena sp., and future work will be needed to dissect which of the proposed β-barrel proteins (22) might serve this function.
The Antibiotic Resistance System
HgdD is the only TolC-like protein encoded in the Anabaena sp. genome (5). Including our results, four distinct functions have been assigned to it: protein secretion (5) and glycolipid (20), siderophore (7), and antibiotic export (Fig. 7). The Km for secretion of ethidium by the wild type is more than 5 orders of magnitude lower than in the strain with abolished HgdD function (Fig. 6). Interestingly, the metabolite export by HgdD described here is a proton gradient-dependent process (Fig. 8). The latter suggests that metabolite efflux is probably catalyzed by a proton gradient-dependent efflux pump protein, similar to the AcrAB system in E. coli (e.g. see Refs. 3 and 16). Thus, we conclude that HgdD engages different energizing complexes to perform the different functions observed. This notion is consistent with the phenotype (Fig. 9B) and the wild type-like ethidium uptake behavior of AFS-I-hgdC (Fig. 10, B and C, open circles). Thus, HgdC is important for glycolipid export but is not relevant for the drug efflux cycle.
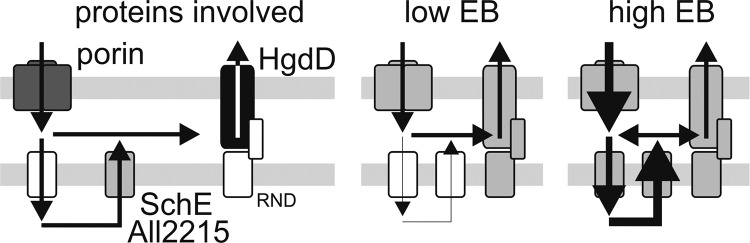
We observed that metabolite export in Anabaena sp. occurs in at least two separate stages, which become apparent at different toxin concentrations (Fig. 12). At low concentrations of toxins, the export from the periplasm by the action of HgdD, charged by a proton pump, is sufficient for secretion because equilibrium between uptake and secretion is observed after a few min (Figs. 3 and 11), whereas intercalation of ethidium into cellular DNA is very low (Fig. 8). Under these conditions, the function of HgdD seems to be essential, whereas the two MFS-like proteins (SchE and All2215) that were analyzed seem to play only a minor role, as reflected by the very low intercalation of ethidium in the corresponding mutants (Figs. 3, 8, 10, and 11). Therefore, under these conditions, the periplasm serves as a threshold reservoir from which toxins and antibiotics are efficiently exported before crossing the plasma membrane (Fig. 12, center).
FIGURE 12.
Model of secondary metabolite export. On the left, the characterized components in the outer (top) and plasma membrane (bottom) are depicted. In the middle, the components involved in export at low toxin/antibiotic concentrations are shown; on the right, the components involved in export at high concentrations are shown in gray, and the path of metabolites/antibiotics is given by arrows.
At high extracellular concentrations, the “buffer capacity” of the periplasm is exceeded, which is reflected by an enhanced uptake (Fig. 11) and intercalation (Fig. 10) by the wild type. At this stage, the function of the MFS proteins becomes relevant (Fig. 12, right). At high ethidium bromide concentrations, we observed an enhanced intercalation for the mutants, the MFS genes alr2215 and schE, which even exceeds the intercalation of HgdD. The ratio between intercalation and uptake in the MFS mutants is at least 5-fold higher than in the wild type and the AFS-I-hgdD mutant. This suggests that ethidium is effectively retained in the cytoplasm, most likely by the impaired transport to the periplasm in these mutants (Fig. 11C). However, the ethidium uptake (Fig. 12) is still lower compared with wild type activity, which might be explained by the slightly enhanced export activity across the outer membrane as reflected by k−1. This parallels the enhanced expression of hgdD in both MFS mutant strains (Fig. 9 and Table 3), which most likely is a response to the impaired MFS system. This up-regulation shifts the buffer capacity of the periplasm to higher concentrations and prevents toxins from reaching the cytosol. Interestingly, the two MFS proteins act in parallel and cannot complement each other, because both mutants show an impaired but not abolished export activity (Table 3).
Thus, we have documented that HgdD is involved in the export of toxins in a proton gradient-dependent manner. This underlines the importance of this protein previously found to modulate the protein secretion pattern (5) and the lipid export activity (20) as well.
Acknowledgments
We thank Enrique Flores (Sevilla) and Martin Pos (Frankfurt) for constant support and helpful discussions and Nassia von Mende and Tihana Bionda (Frankfurt) for critical reading and discussion of the manuscript.
This work was supported by the Center of Membrane Proteomics (Goethe University Frankfurt) (to A. H.), the Cluster of Excellence Frankfurt “Macromolecular Complexes” (to E. S.), and Deutsche Forschungsgemeinschaft Grant SCHL 585/2 (to E. S.).

This article contains supplemental Table S1 and Fig. S1.
- MFS
- major facilitator superfamily
- EB
- ethidium bromide
- CCCP
- carbonyl cyanide m-chlorophenylhydrazone.
REFERENCES
- 1. Nikaido H. (1996) Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 178, 5853–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Piddock L. J. (2006) Multidrug-resistance efflux pumps. Not just for resistance. Nat. Rev. Microbiol. 4, 629–636 [DOI] [PubMed] [Google Scholar]
- 3. Pos K. M. (2009) Drug transport mechanism of the AcrB efflux pump. Biochim. Biophys. Acta 1794, 782–793 [DOI] [PubMed] [Google Scholar]
- 4. Mirus O., Hahn A., Schleiff E. (2010) Outer membrane proteins. in Prokaryotic Cell Wall Compounds: Structure and Biochemistry (König H., Claus H., Varma A., eds) pp. 175–230, Springer-Verlag, Berlin/ Heidelberg [Google Scholar]
- 5. Moslavac S., Nicolaisen K., Mirus O., Al Dehni F., Pernil R., Flores E., Maldener I., Schleiff E. (2007) A TolC-like protein is required for heterocyst development in Anabaena sp. strain PCC 7120. J. Bacteriol. 189, 7887–7895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bleuel C., Grosse C., Taudte N., Scherer J., Wesenberg D., Krauss G. J., Nies D. H., Grass G. (2005) TolC is involved in enterobactin efflux across the outer membrane of Escherichia coli. J. Bacteriol. 187, 6701–6707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nicolaisen K., Hahn A., Valdebenito M., Moslavac S., Samborski A., Maldener I., Wilken C., Valladares A., Flores E., Hantke K., Schleiff E. (2010) The interplay between siderophore secretion and coupled iron and copper transport in the heterocyst-forming cyanobacterium Anabaena sp. PCC 7120. Biochim. Biophys. Acta 1798, 2131–2140 [DOI] [PubMed] [Google Scholar]
- 8. Newton S. M., Trinh V., Pi H., Klebba P. E. (2010) Direct measurements of the outer membrane stage of ferric enterobactin transport. Postuptake binding. J. Biol. Chem. 285, 17488–17497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Koronakis V., Eswaran J., Hughes C. (2004) Structure and function of TolC. The bacterial exit duct for proteins and drugs. Annu. Rev. Biochem. 73, 467–489 [DOI] [PubMed] [Google Scholar]
- 10. Koronakis V., Sharff A., Koronakis E., Luisi B., Hughes C. (2000) Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405, 914–919 [DOI] [PubMed] [Google Scholar]
- 11. Awram P., Smit J. (1998) The Caulobacter crescentus paracrystalline S-layer protein is secreted by an ABC transporter (type I) secretion apparatus. J. Bacteriol. 180, 3062–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bolhuis H., van Veen H. W., Poolman B., Driessen A. J., Konings W. N. (1997) Mechanisms of multidrug transporters. FEMS Microbiol. Rev. 21, 55–84 [DOI] [PubMed] [Google Scholar]
- 13. Goldberg M., Pribyl T., Juhnke S., Nies D. H. (1999) Energetics and topology of CzcA, a cation/proton antiporter of the resistance-nodulation-cell division protein family. J. Biol. Chem. 274, 26065–26070 [DOI] [PubMed] [Google Scholar]
- 14. Delepelaire P. (2004) Type I secretion in Gram-negative bacteria. Biochim. Biophys. Acta. 1694, 149–161 [DOI] [PubMed] [Google Scholar]
- 15. Thanabalu T., Koronakis E., Hughes C., Koronakis V. (1998) Substrate-induced assembly of a contiguous channel for protein export from E. coli. Reversible bridging of an inner membrane translocase to an outer membrane exit pore. EMBO J. 17, 6487–6496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Blair J. M., Piddock L. J. (2009) Structure, function and inhibition of RND efflux pumps in Gram-negative bacteria. An update. Curr. Opin. Microbiol. 12, 512–519 [DOI] [PubMed] [Google Scholar]
- 17. Nikaido H., Takatsuka Y. (2009) Mechanisms of RND multidrug efflux pumps. Biochim. Biophys. Acta 1794, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moslavac S., Bredemeier R., Mirus O., Granvogl B., Eichacker L. A., Schleiff E. (2005) Proteomic analysis of the outer membrane of Anabaena sp. strain PCC 7120. J. Proteome Res. 4, 1330–1338 [DOI] [PubMed] [Google Scholar]
- 19. Moslavac S., Reisinger V., Berg M., Mirus O., Vosyka O., Plöscher M., Flores E., Eichacker L. A., Schleiff E. (2007) The proteome of the heterocyst cell wall in Anabaena sp. PCC 7120. Biol. Chem. 388, 823–839 [DOI] [PubMed] [Google Scholar]
- 20. Staron P., Forchhammer K., Maldener I. (2011) Novel ATP-driven pathway of glycolipid export involving TolC protein. J. Biol. Chem. 286, 38202–38210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang C. C., Laurent S., Sakr S., Peng L., Bédu S. (2006) Heterocyst differentiation and pattern formation in cyanobacteria. A chorus of signals. Mol. Microbiol. 59, 367–375 [DOI] [PubMed] [Google Scholar]
- 22. Nicolaisen K., Hahn A., Schleiff E. (2009) The cell wall in heterocyst formation by Anabaena sp. PCC 7120. J. Basic Microbiol. 49, 5–24 [DOI] [PubMed] [Google Scholar]
- 23. Flores E., Herrero A. (2010) Compartmentalized function through cell differentiation in filamentous cyanobacteria. Nat. Rev. Microbiol. 8, 39–50 [DOI] [PubMed] [Google Scholar]
- 24. Ladig R., Sommer M. S., Hahn A., Leisegang M. S., Papasotiriou D. G., Ibrahim M., Elkehal R., Karas M., Zickermann V., Gutensohn M., Brandt U., Klösgen R. B., Schleiff E. (2011) A high-definition native polyacrylamide gel electrophoresis system for the analysis of membrane complexes. Plant J. 67, 181–194 [DOI] [PubMed] [Google Scholar]
- 25. Rippka R., Deruelies J., Waterbury J. B., Herdman M., Stanier R. Y. (1979) Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111, 1–61 [Google Scholar]
- 26. Cai Y. P., Wolk C. P. (1990) Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J. Bacteriol. 172, 3138–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, chapter 1.21–1.85, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 28. Elhai J., Wolk C. P. (1988) Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 167, 747–754 [DOI] [PubMed] [Google Scholar]
- 29. Elhai J., Vepritskiy A., Muro-Pastor A. M., Flores E., Wolk C. P. (1997) Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J. Bacteriol. 179, 1998–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li W., Godzik A. (2006) Cd-hit. A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 [DOI] [PubMed] [Google Scholar]
- 31. Frickey T., Lupas A. (2004) CLANS. A Java application for visualizing protein families based on pairwise similarity. Bioinformatics 20, 3702–3704 [DOI] [PubMed] [Google Scholar]
- 32. Chrastil J. (1993) Determination of the first-order consecutive reversible reaction kinetics. Comp. Chem. 17, 103–106 [Google Scholar]
- 33. Tal N., Schuldiner S. (2009) A coordinated network of transporters with overlapping specificities provides a robust survival strategy. Proc. Natl. Acad. Sci. U.S.A. 106, 9051–9056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nagano K., Nikaido H. (2009) Kinetic behavior of the major multidrug efflux pump AcrB of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 106, 5854–5858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pagès J. M., James C. E., Winterhalter M. (2008) The porin and the permeating antibiotic. A selective diffusion barrier in Gram-negative bacteria. Nat. Rev. Microbiol. 6, 893–903 [DOI] [PubMed] [Google Scholar]
- 36. Baranovsky S. F., Bolotin P. A., Evstigneev M. P., Chernyshev D. N. (2009) Interaction of ethidium bromide and caffeine with DNA in aqueous solution. J. Appl. Spectrosc. 76, 132–139 [Google Scholar]
- 37. Iyer R., Wu Z., Woster P. M., Delcour A. H. (2000) Molecular basis for the polyamine-ompF porin interactions. Inhibitor and mutant studies. J. Mol. Biol. 297, 933–945 [DOI] [PubMed] [Google Scholar]
- 38. Iyer R., Delcour A. H. (1997) Complex inhibition of OmpF and OmpC bacterial porins by polyamines. J. Biol. Chem. 272, 18595–18601 [DOI] [PubMed] [Google Scholar]
- 39. Wyman M., Fay P. (1986) Interaction between light quality and nitrogen availability in the differentiation of akinetes in the planktonic cyanobacterium Gloeotrichia echinulata. Br. Phycol. J. 21, 147–153 [Google Scholar]
- 40. Sen K., Hellman J., Nikaido H. (1988) Porin channels in intact cells of Escherichia coli are not affected by Donnan potentials across the outer membrane. J. Biol. Chem. 263, 1182–1187 [PubMed] [Google Scholar]
- 41. Plésiat P., Nikaido H. (1992) Outer membranes of gram-negative bacteria are permeable to steroid probes. Mol. Microbiol. 6, 1323–1333 [DOI] [PubMed] [Google Scholar]
- 42. Apte S. K., Thomas J. (1986) Membrane electrogenesis and sodium transport in filamentous nitrogen-fixing cyanobacteria. Eur. J. Biochem. 154, 395–401 [DOI] [PubMed] [Google Scholar]
- 43. Nitschmann W. H., Peschek G. A. (1986) Oxidative phosphorylation and energy buffering in cyanobacteria. J. Bacteriol. 168, 1205–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Park J. T., Uehara T. (2008) How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan). Microbiol. Mol. Biol. Rev. 72, 211–227, table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pourcher T., Bassilana M., Sarkar H. K., Kaback H. R., Leblanc G. (1990) The melibiose/Na+ symporter of Escherichia coli. Kinetic and molecular properties. Philos. Trans. R. Soc. Lond. B Biol. Sci. 326, 411–423 [DOI] [PubMed] [Google Scholar]
- 46. Liang W. J., Wilson K. J., Xie H., Knol J., Suzuki S., Rutherford N. G., Henderson P. J., Jefferson R. A. (2005) The gusBC genes of Escherichia coli encode a glucuronide transport system. J. Bacteriol. 187, 2377–2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Herzberg M., Kaye I. K., Peti W., Wood T. K. (2006) YdgG (TqsA) controls biofilm formation in Escherichia coli K-12 through autoinducer 2 transport. J. Bacteriol. 188, 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Poolman B., Knol J., van der Does C., Henderson P. J., Liang W. J., Leblanc G., Pourcher T., Mus-Veteau I. (1996) Cation and sugar selectivity determinants in a novel family of transport proteins. Mol. Microbiol. 19, 911–922 [DOI] [PubMed] [Google Scholar]
- 49. Furrer J. L., Sanders D. N., Hook-Barnard I. G., McIntosh M. A. (2002) Export of the siderophore enterobactin in Escherichia coli. Involvement of a 43 kDa membrane exporter. Mol. Microbiol. 44, 1225–1234 [DOI] [PubMed] [Google Scholar]
- 50. Lebel S., Bouttier S., Lambert T. (2004) The cme gene of Clostridium difficile confers multidrug resistance in Enterococcus faecalis. FEMS Microbiol. Lett. 238, 93–100 [DOI] [PubMed] [Google Scholar]
- 51. Nicolaisen K., Moslavac S., Samborski A., Valdebenito M., Hantke K., Maldener I., Muro-Pastor A. M., Flores E., Schleiff E. (2008) Alr0397 is an outer membrane transporter for the siderophore schizokinen in Anabaena sp. strain PCC 7120. J. Bacteriol. 190, 7500–7507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fan Q., Huang G., Lechno-Yossef S., Wolk C. P., Kaneko T., Tabata S. (2005) Clustered genes required for synthesis and deposition of envelope glycolipids in Anabaena sp. strain PCC 7120. Mol. Microbiol. 58, 227–243 [DOI] [PubMed] [Google Scholar]
- 53. Knowles T. J., Scott-Tucker A., Overduin M., Henderson I. R. (2009) Membrane protein architects. The role of the BAM complex in outer membrane protein assembly. Nat. Rev. Microbiol. 7, 206–214 [DOI] [PubMed] [Google Scholar]
- 54. Nicolaisen K., Mariscal V., Bredemeier R., Pernil R., Moslavac S., López-Igual R., Maldener I., Herrero A., Schleiff E., Flores E. (2009) The outer membrane of a heterocyst-forming cyanobacterium is a permeability barrier for uptake of metabolites that are exchanged between cells. Mol. Microbiol. 74, 58–70 [DOI] [PubMed] [Google Scholar]
- 55. Schleiff E., Soll J. (2005) Membrane protein insertion. Mixing eukaryotic and prokaryotic concepts. EMBO Rep. 6, 1023–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Werner J., Misra R. (2005) YaeT (Omp85) affects the assembly of lipid-dependent and lipid-independent outer membrane proteins of Escherichia coli. Mol. Microbiol. 57, 1450–1459 [DOI] [PubMed] [Google Scholar]
- 57. Ertel F., Mirus O., Bredemeier R., Moslavac S., Becker T., Schleiff E. (2005) The evolutionarily related β-barrel polypeptide transporters from Pisum sativum and Nostoc PCC7120 contain two distinct functional domains. J. Biol. Chem. 280, 28281–28289 [DOI] [PubMed] [Google Scholar]
- 58. Bredemeier R., Schlegel T., Ertel F., Vojta A., Borissenko L., Bohnsack M. T., Groll M., von Haeseler A., Schleiff E. (2007) Functional and phylogenetic properties of the pore-forming β-barrel transporters of the Omp85 family. J. Biol. Chem. 282, 1882–1890 [DOI] [PubMed] [Google Scholar]
- 59. Tripp J., Hahn A., Koenig P., Flinner N., Bublak D., Brouwer E. M., Ertel F., Mirus O., Sinning I., Tews I., Schleiff E. (2012) Structure and conservation of the periplasmic targeting factor Tic22 from plants and cyanobacteria. J. Biol. Chem. 287, 24164–24173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Huang F., Hedman E., Funk C., Kieselbach T., Schröder W. P., Norling B. (2004) Isolation of outer membrane of Synechocystis sp. PCC 6803 and its proteomic characterization. Mol. Cell Proteomics 3, 586–595 [DOI] [PubMed] [Google Scholar]
- 61. Hoiczyk E., Hansel A. (2000) Cyanobacterial cell walls. News from an unusual prokaryotic envelope. J. Bacteriol. 182, 1191–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wilk L., Strauss M., Rudolf M., Nicolaisen K., Flores E., Kühlbrandt W., Schleiff E. (2011) Outer membrane continuity and septosome formation between vegetative cells in the filaments of Anabaena sp. PCC 7120. Cell. Microbiol. 13, 1744–1754 [DOI] [PubMed] [Google Scholar]
- 63. Hansel A., Schmid A., Tadros M. H., Jürgens U. J. (1994) Isolation and characterisation of porin from the outer membrane of Synechococcus PCC 6301. Arch. Microbiol. 161, 163–167 [DOI] [PubMed] [Google Scholar]
- 64. Hansel A., Pattus F., Jürgens U. J., Tadros M. H. (1998) Cloning and characterization of the genes coding for two porins in the unicellular cyanobacterium Synechococcus PCC 6301. Biochim. Biophys. Acta 1399, 31–39 [DOI] [PubMed] [Google Scholar]
- 65. Kashiwagi S., Kanamuru K., Mizuno T. (1995) A Synechococcus gene encoding a putative pore-forming intrinsic membrane protein. Biochim. Biophys. Acta 1237, 189–192 [DOI] [PubMed] [Google Scholar]
- 66. Hansel A., Tadros M. H. (1998) Characterization of two pore-forming proteins isolated from the outer membrane of Synechococcus PCC 6301. Curr. Microbiol. 36, 321–326 [DOI] [PubMed] [Google Scholar]
- 67. Benz R., Böhme H. (1985) Pore formation by an outer membrane protein of the cyanobacterium Anabaena variabilis. Biochim. Biophys. Acta 812, 286–292 [Google Scholar]
- 68. Nikaido H. (1992) Porins and specific channels of bacterial outer membranes. Mol. Microbiol. 6, 435–442 [DOI] [PubMed] [Google Scholar]
- 69. Flores E., Herrero A., Wolk C. P., Maldener I. (2006) Is the periplasm continuous in filamentous multicellular cyanobacteria? Trends Microbiol. 14, 439–443 [DOI] [PubMed] [Google Scholar]
- 70. Pernil R., Picossi S., Mariscal V., Herrero A., Flores E. (2008) ABC-type amino acid uptake transporters Bgt and N-II of Anabaena sp. strain PCC 7120 share an ATPase subunit and are expressed in vegetative cells and heterocysts. Mol. Microbiol. 67, 1067–1080 [DOI] [PubMed] [Google Scholar]
- 71. Valladares A., Rodríguez V., Camargo S., Martínez-Nöel G. M., Herrero A., Luque I. (2011) Specific role of the cyanobacterial PipX factor in the heterocysts of Anabaena sp. strain PCC 7120. J. Bacteriol. 193, 1172–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Olmedo-Verd E., Muro-Pastor A. M., Flores E., Herrero A. (2006) Localized induction of the ntcA regulatory gene in developing heterocysts of Anabaena sp. strain PCC 7120. J. Bacteriol. 188, 6694–6699 [DOI] [PMC free article] [PubMed] [Google Scholar]