Background: Regulation of FYVE domain proteins by phosphoinositides other than PtdIns(3)P is not known.
Results: PtdIns(4,5)P2, PtdIns(3,4)P2, and PtdIns(3,4,5)P3 bind the FYVE domain of protrudin.
Conclusion: PtdIns(4,5)P2, PtdIns(3,4)P2, and PtdIns(3,4,5)P3 differentially regulate cellular protrudin function.
Significance: This study provides new insight into how phosphoinositides modulate neurite formation.
Keywords: Lipid-binding Protein, Neurite Outgrowth, Neurological Diseases, Phosphoinositides, PI 3-Kinase (PI3K), FYVE Domain
Abstract
Protrudin is a FYVE (Fab 1, YOTB, Vac 1, and EEA1) domain-containing protein involved in transport of neuronal cargoes and implicated in the onset of hereditary spastic paraplegia. Our image-based screening of the lipid binding domain library revealed novel plasma membrane localization of the FYVE domain of protrudin unlike canonical FYVE domains that are localized to early endosomes. The membrane binding study by surface plasmon resonance analysis showed that this FYVE domain preferentially binds phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2), phosphatidylinositol 3,4-bisphosphate (PtdIns(3,4)P2), and phosphatidylinositol 3,4,5-trisphosphate (PtdIns(3,4,5)P3) unlike canonical FYVE domains that specifically bind phosphatidylinositol 3-phosphate (PtdIns(3)P). Furthermore, we found that these phosphoinositides (PtdInsP) differentially regulate shuttling of protrudin between endosomes and plasma membrane via its FYVE domain. Protrudin mutants with reduced PtdInsP-binding affinity failed to promote neurite outgrowth in primary cultured hippocampal neurons. These results suggest that novel PtdInsP selectivity of the protrudin-FYVE domain is critical for its cellular localization and its role in neurite outgrowth.
Introduction
Hundreds of signaling proteins recognize specific phospholipids via lipid binding domains (LBD)3 with diverse structures and lipid specificity (1–3). Phospholipid-LBD interaction is critical in regulating the appropriate subcellular localization of proteins involved in diverse cellular processes, including cell signaling, vesicle trafficking, organization of the cytoskeleton, autophagy, and neurite outgrowth (1–3). Defects of proteins regulating these lipid-mediated processes cause various human diseases exact causative mechanisms of which have not been fully understood (4).
Patients with hereditary spastic paraplegia (HSP), an inherited neurological disease, suffer from progressive dysfunction of the nerves (5). The large group of HSP-related proteins appears to be involved in membrane trafficking (5). Previous reports showed that HSP-related proteins, spastin and kinesin superfamily protein 5 (KIF5), interact with protrudin (6). Protrudin is a FYVE domain-containing protein that also harbors a Rab11-binding domain (RBD11), two hydrophobic domains (HP-1 and HP-2), an FFAT motif, and a coiled-coil domain (7, 8). Protrudin is reported to play a role in membrane recycling (8) and in induction of neurite outgrowth through the delivery of neuronal cargoes (6, 7). It has also been suggested that protrudin is implicated in an autosomal dominant form of HSP (AD-HSP) (6, 8, 9). However, it is not known how the cellular function of protrudin is regulated and how protrudin is involved in neurite formation and the onset of HSP.
The FYVE domain is a zinc-containing module of 60–80 amino acids that specifically bind PtdIns(3)P (10–12). As expected from the endosomal localization of PtdIns(3)P and its role in vesicle trafficking, a large number of FYVE domain-containing proteins, including EEA1, Hrs, and FENS-1, are involved in endocytic vesicle trafficking (13–15). Some FYVE domain-containing proteins also function in cytoskeletal regulation (FGD1) (16) and growth factor signaling (SARA (17) and endofin (18)). High-resolution structures of different FYVE domains showed that they achieve high PtdIns(3)P specificity via the lipid binding pocket composed of three conserved motifs, the N-terminal WXXD, the central R + HHC + XCG, and the C-terminal RVC motifs, where + represents a basic residue and X any residue (19–22). To date, the specificity of FYVE domains for other PtdInsPs has not been reported. In this study, we found that the FYVE domain of protrudin has unique subcellular localization and novel selectivity for PtdIns(4,5)P2, PtdIns(3,4)P2, and PtdIns(3,4,5)P3 by means of imaging-based cell screening (23–25) and SPR analysis. Our study suggests that binding of PtdIns(4,5)P2, PtdIns(3,4)P2, and PtdIns(3,4,5)P3 to the FYVE domain may differentially regulate the shuttling of protrudin between the endosome and PM, which is critical for the role of protrudin in mediating neurite outgrowth.
EXPERIMENTAL PROCEDURES
DNA Constructs
LBDs were tagged with enhanced yellow fluorescence protein (EYFP) by a combination of pENTR-207 with pDSA03 (pEYFP-XB) using LR recombinase (Invitrogen). LBDs generally contain additional flanking sequences on both sides of the domain ends. Detailed information on constructed LBDs is listed in supplemental Table S1. Protrudin was a kind gift of Keiichi I. Nakayama (8) and transferred into pCitrine-C1 vector. To make FYVE domain-deleted protrudin, the FYVE domain was removed from positions 348 to 409 in full-length protrudin by using HindIII and KpnI. EYFP-Cb5 (26) and EYFP-Btk-PH domains were kindly provided from Takanari Inoue. PLCδ-PH domain and Akt-PH domain constructs were created in pEYFP-C1 vector (Clontech). 2× FYVE domain of FENS-1 was constructed by using compatible ends in the EYFP-C1 vector (Clontech). ARF6(Q67L)-EYFP and ARF6(T27N)-EYFP were generated in the pEYFP-N3 vector using BglII and KpnI sites, and EYFP-Rab5B(Q79L) was inserted in pEYFP-C1 vector (Clontech) using EcoRI and BamHI sites. Lyn was constructed using EcoRI and BamHI sites into the pEYFP-N1 vector (Clontech) and then EYFP of Lyn-EYFP was replaced with mCherry. The FYVE domain (318–409 amino acids) of protrudin was constructed in pRSET-B vector (Invitrogen) for protein purification and surface plasmon resonance (SPR) analysis.
Cell Culture and Transfection
NIH3T3 was obtained from American Type Culture Collection (ATCC, Manassas, VA) and maintained in Dulbecco's modified Eagle's medium (DMEM, PAA Laboratories GmbH) supplemented with 10% fetal bovine serum (FBS, Invitrogen) at 37 °C in an atmosphere of 10% CO2 and 95% humidity. Rat hippocampus was obtained from embryonic rat brain at embryonic day 18 (KOATECH). Primary rat hippocampal neurons were plated in Neurobasal medium containing B27 supplement (Invitrogen) and GlutaMAX (Invitrogen). Transfection was performed by using Lipofectamine 2000 (Invitrogen), Neon® Transfection System 10 μl (Invitrogen) or the CalPhosTM Mammalian Transfection kit (Clontech) (27) according to the manufacturer's instructions.
Live Cell Imaging and Image Acquisition
NIH3T3 cells expressing LBDs were observed using a ×60 lens (oil immersion objective, NA 1.40; Nikon) and primary cultured hippocampal neuron expressing protrudin was imaged using ×20 lens (air objective, NA 0.75; Nikon) of an A1R confocal microscope (Nikon) at room temperature. Images were acquired at a resolution of 512 × 512. Time-lapse images were captured with 30-s intervals.
Lipid Modulation Systems
PLCδ-PH, Btk-PH, and Akt-PH domains were used as PtdIns(4,5)P2, PtdIns(3,4,5)P3, and PtdIns (3,4)P2/PtdIns(3,4,5)P3 biosensors, respectively (28–30). 2× FYVE domain of FENS-1 served as a biosensor for PtdIns(3)P (31). ARF6(Q67L)-EYFP and ARF6(T27N)-EYFP were applied to induce and block PIP5K activation followed by formation of PtdIns(4,5)P2-containing endocytic vesicles (32). EYFP-Rab5B(Q79L) was used to accumulate PtdIns(3)P in the endocytic vesicles (33). 50 μm LY29 (Sigma) or 5 nm PDGF (PreproTech) was used to inhibit and induce phosphoinositide 3-kinase (PI3K) activation at the PM, respectively (23, 34). LY30 (Sigma) served as a structural inactive analog for LY29 (23).
Image Analysis
Image analyses were performed using the NIS-element AR 64 bit version 3.1 and MetaMorph offline version 7.6.0.0 from MDS Analytical Technologies. Neurite length of primary culture hippocampal neurons was measured using NeuronJ 1.4.2 (National Institutes of Health). The relative abundance of the fluorescence protein-labeled protein at the PM in each cell was assessed by calculating the ratio of fluorescence intensity at the PM to that at the cytosol (=FPM/Fcytosol) as described (23). The fold-change of fluorescence intensity of a protein at the PM caused by each treatment was quantified as (F*PM/F*cytosol)/(FPM/Fcytosol) in which FPM/Fcytosol and F*PM/F*cytosol indicate the fluorescence ratio before and after each treatment, respectively. Fluorescence fold-change in cytosol (F*cytosol/Fcytosol) indicates the degree of translocation of each domain from the PM to cytosol upon PDGF treatment: i.e. a lower value means more PM translocation.
Protein Expression and Purification
The protrudin FYVE domain (residue number 318–409) was expressed as an N-terminal His6 fusion protein in Escherichia coli BL21(DE3) pLysS (Novagen). Cells were inoculated at 37 °C in 500 ml of Luria broth containing 50 μg/ml of kanamycin until the A600 reached 0.6–0.8. The overexpression of recombinant FYVE domains was induced by adding isopropyl β-d-thiogalactopyranoside to a final concentration of 1 mm. Then the cells were grown for 16 h at room temperature (16 °C) and harvested by centrifugation at 4 °C. The recombinant proteins were purified with affinity chromatography using nickel-nitrilotriacetic acid-agarose beads (Qiagen). Briefly, the cell pellets were resuspended in 20 ml of lysis buffer (50 mm Tris, 300 mm NaCl, 10 mm immidazole, 10% (v/v) glycerol, 50 μm phenylmethylsulfonyl fluoride, 5 mm 2-mercaptoethanol, pH 7.9), and the solutions were sonicated on an ice bath for 5 min (15 s of sonication followed by 15 s of cooling on ice). The cell lysates were centrifuged at 61,000 × g for 30 min at 4 °C, the supernatants were collected and mixed with 1 ml of 50% (v/v) nickel-nitrilotriacetic acid-agarose beads and incubated for 1 h at 4 °C with mild shaking at 60 rpm. The protein-bound beads were then loaded to a column and washed with an excess volume of washing buffer 1 (50 mm Tris, 300 mm NaCl, 20 mm immidazole, pH 7.9) followed by washing buffer 2 (20 mm Tris, 160 mm NaCl, 20 mm immidazole, pH 7.9). The FYVE domain proteins were then eluted with an elution buffer (20 mm Tris, 160 mm NaCl, 300 mm immidazole, pH 7.9). Finally, the buffer solution for the FYVE domains was exchanged to the SPR elution buffer (20 mm Tris-HCl, pH 7.4, 160 mm NaCl) using a gel filtration column, PD-10 (GE Healthcare), equilibrated with the same buffer. Protein purities were checked on a 20% polyacrylamide gel, and the protein concentrations were determined by a Bradford assay (Bio-Rad).
Lipid Vesicles Preparation and SPR Analysis
Large unilamellar vesicles were prepared using a Liposofast (Avestin) microextruder with a 100-nm polycarbonate filter. All SPR measurements were performed at 23 °C in 20 mm Tris-HCl, pH 7.4, containing 0.16 m NaCl using a lipid-coated L1 chip in the BIACORE T100 system. 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC)/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (POPS)/PtdInsP (77:20:3) vesicles and POPC vesicles were coated onto the active surface and the control surface, respectively. Vesicles were coated onto the corresponding sensor chip surfaces to yield the identical resonance units, ensuring the equal concentration of the coated lipids. Equilibrium SPR measurements were done at the flow rate of 5 μl/min to allow sufficient time for the response in the association phase to reach near-equilibrium values (Req) (35). A minimum of 5 different protein concentrations were injected to collect a set of Req values that were plotted against the protein concentrations (Po). An apparent dissociation constant (Kd) was then determined by nonlinear least squares analysis of the binding isotherm using the following equation, Req = Rmax/(1 + Kd/Po), where Rmax indicates the maximal Req value (36). Because the concentration of lipids coated on the sensor chip cannot be accurately determined, an apparent dissociation constant Kd is defined as the protein concentration yielding half-maximal binding with a given lipid concentration. The measurement was repeated at least three times to determine average and S.D. values.
Molecular Modeling
The molecular modeling was performed using the program CCP4 molecular graphics (37) by superposing the protrudin FYVE structure (PDB code 1X4U) onto the structure of 1,3-bisphosphate bound human EEA1-FYVE domain (PDB code 1JOC). The root mean square deviation score was calculated using the same program. For modeling the protrudin-FYVE domain structure in a PtdIns(4,5)P2-bound form, inositol 1,4,5-triphosphate was incorporated to the lipid binding pocket of the apo-form structure in a similar manner with 1,3-bisphosphate bound to the human EEA1-FYVE domain.
RESULTS
Identification of PM-localized LBDs
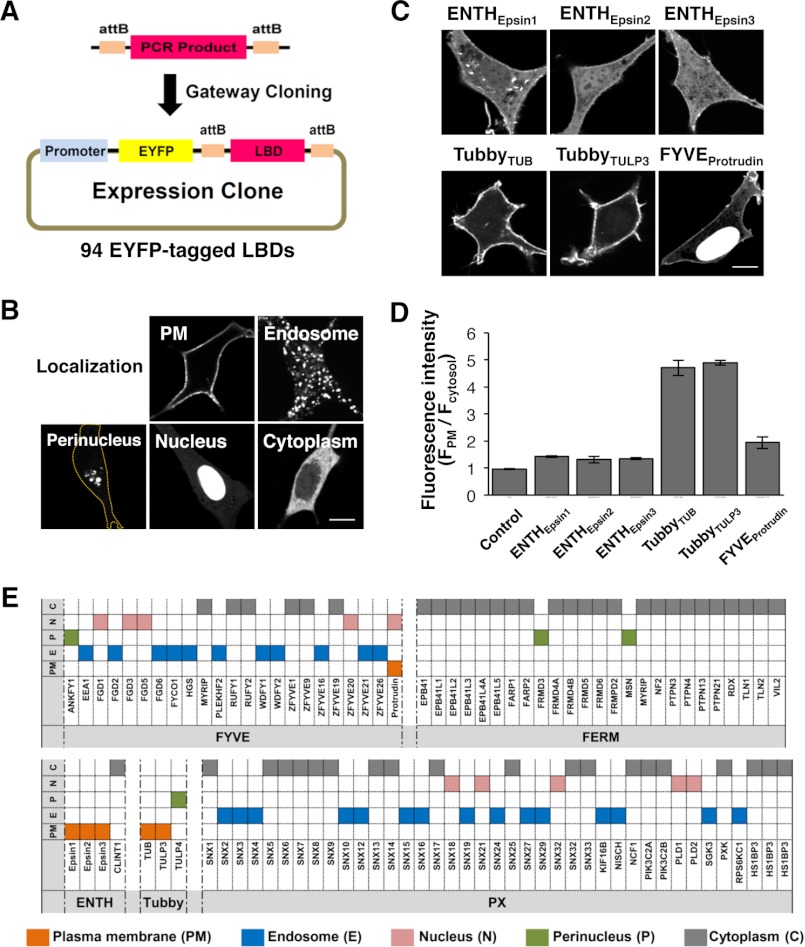
We generated a library of 94 LBDs tagged with EYFP (Fig. 1A) and then imaged NIH3T3 cells expressing each EYFP-LBD. 94 LBDs were divided into five groups based on the subcellular localization pattern (Fig. 1B). Our results revealed that all endosome-localized LBDs were FYVE and PX domains, consistent with previous studies (31, 38–41) (Fig. 1E). They include the FYVE domains of HGS, FYCO1, WDFY1 (also known as FENS-1), and EEA1, and the PX domain of KIF6B. Among six LBDs showing PM localization (Fig. 1, C and D), five of them belonged to the ENTH and Tubby domains (Fig. 1, C and E), which are known to interact with PtdIns(4,5)P2 (42–44). Intriguingly, the other PM-localized LBD was the FYVE domain of protrudin (Fig. 1, C and E), which was unexpected in light of the other FYVE domains that display endosomal localizations (45). Therefore, we further examined lipid binding of the FYVE domain of protrudin. It should be noted that a significant degree of nuclear localization of the protrudin-FYVE domain was seen in our study. Similar nuclear localization has been reported for many other isolated LBDs, including PH domains of Gab1, Gab2, and Grp1 (30, 46–48), which has been attributed to smaller sizes of isolated LBDs, potential exposure of a cryptic nuclear localization sequence, or nonspecific binding to nuclear lipids or nucleic acids.
FIGURE 1.
Identification of PM-localized LBDs. EYFP-tagged (A) 94 different LBDs were imaged and grouped into five localization patterns (B). C, six distinct LBDs showed PM localization. The outline of the cell is marked as an orange line. Bars, 10 μm. D, quantitative analysis of the proportion of PM-localized LBDs in C. E, summary of image-based screening results.
Quantitative Lipid Binding Analysis of Protrudin-FYVE Domain
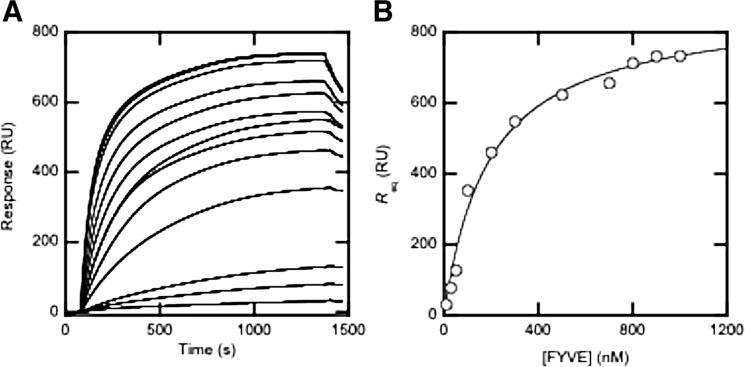
To validate our cell imaging data, we quantitatively measured lipid binding of the protrudin-FYVE domain by equilibrium SPR analysis. Specifically, we measured the PtdInsP selectivity of the protrudin-FYVE domain by determining its Kd values for POPC/POPS/PtdInsP (77:20:3) vesicles (see Fig. 2 for example). 30% PS was included to simulate the PS content of the inner layer of the PM of mammalian cells. As summarized in Table 1, the protrudin-FYVE domain has high affinity for vesicles containing PtdIns(3,4)P2, PtdIns(4,5)P2, and PtdIns(3,4,5)P3, respectively. However, it shows very low affinity for vesicles containing PtdIns(3)P, PtdIns(4)P, or PtdIns(5)P. The Kd values for binding of the protrudin-FYVE domain to vesicles containing PtdIns(3,4)P2, PtdIns(4,5)P2, and PtdIns(3,4,5)P3 range from 100 to 200 nm, which are comparable with that for PtdIns(3,4,5)P3-specific PH domains determined under similar conditions (30). These values also compare well with the Kd value for binding of other prototypical FYVE domains to vesicles containing PtdIns(3)P (49). Collectively, our SPR analysis shows that the protrudin-FYVE domain has high affinity for PtdIns(3,4)P2, PtdIns(4,5)P2, and PtdIns(3,4,5)P3.
FIGURE 2.
Determination of Kd for binding of the protrudin-FYVE domain to POPC/POPS/PtdIns(3,4,5)P3 (77:20:3) vesicles. A, the protrudin-FYVE domain was injected at 5 ml/min at varying concentrations (10, 30, 50, 100, 200, 300, 500, 700, 800, 900, and 1000 nm from bottom to top) over the POPC/POPS/PtdIns(3,4,5)P3 (77:20:3) surface and Req values were measured. B, a binding isotherm was generated from the Req (average of triplicate measurements) versus the concentration of FYVE plot. A solid line represents a theoretical curve constructed from Rmax (=875 ± 35) and Kd (=190 ± 30 nm) values determined by nonlinear least-squares analysis of the isotherm using the equation: Req = Rmax/(1 + Kd/Po), where Rmax indicates the maximal Req value. All measurements were performed at 23 °C in 20 mm Tris-HCl buffer, pH 7.4, with 0.16 m KCl.
TABLE 1.
PtdInsP selectivity of protrudin-FYVE determined by equilibrium SPR measurements
| Proteins |
Kda |
|||||
|---|---|---|---|---|---|---|
| PtdIns(3,4,5)P3 | PtdIns(4,5)P2 | PtdIns(3,4)P2 | PtdIns(3)P | PtdIns(4)P | PtdIns(5)P | |
| nm | ||||||
| WT | 190 ± 30 | 180 ± 20 | 125 ± 20 | >2000b | >2000 | >2000 |
| 4A mutant | >2000 | >2000 | >2000 | NMc | NM | NM |
| K367A/R369A | >2000 | >2000 | >2000 | NM | NM | NM |
a POPC/POPS/PtdInsP (77:20:3 in mol %) vesicles were used for Kd determination.
b Showing a binding signal that does not, however, approach saturation with the protein concentration up to 2 μm.
c Not measured.
Identification of Protrudin-FYVE Domain Residues Involved in the PtdInsP Selectivity
Comparison of the amino acid sequence of the protrudin-FYVE domain with that of the prototypical EEA1-FYVE domain reveals unique features of the protrudin-FYVE domain (Fig. 3A). To identify which residues of the FYVE domain of protrudin are involved in lipid selectivity, three-dimensional structures of this domain and the inositol 1,3-bisphosphate-bound human EEA1-FYVE domain were superposed with a root mean square deviation of 0.68 Å over 34 Cα atoms. Based on the structural superposition, we modeled the lipid binding pocket of the protrudin-FYVE domain, which is larger than that of the EEA1-FYVE domain, accommodating an inositol 1,4,5-triphosphate molecule (a PtdIns(4,5)P2 headgroup) (Fig. 3B). The model suggests that PtdIns(4,5)P2 associates with the predominantly positive surface of the FYVE domain and that Lys-367, Arg-369, Arg-381, and Lys-386 might be the residues that directly interact with the phosphate groups of PtdIns(4,5)P2.
FIGURE 3.
Amino acids responsible for lipid-binding specificity of the FYVE domain. A, amino acid sequence of the protrudin-FYVE domain was compared with that of the EEA1-FYVE domain. Lipid-binding pocket (purple) of the EEA1-FYVE domain is composed of a WXXD motif, R + HHC + XCG, and RVC regions. Corresponding sites in the protrudin-FYVE domain are colored pink. The CXXC motif or the predicted amino acids in the protrudin-FYVE domain were distinctly marked as green and red. B, a model of PtdIns(4,5)P2 binding to the protrudin-FYVE domain. The PtdIns(4,5)P2 molecule is shown in stick, and the FYVE domain in an electrostatic surface model. The positively charged residues (blue) presumed to interact with PtdIns(4,5)P2 are labeled. The structure is shown using PyMOL software (DeLano Scientific LLC). C, images of cells expressing the wild type FYVE domain, K367A/R369A, R386A, and 4A. Bars, 10 μm. D, quantitative analysis of the expression level of the FYVE domain on the PM (WT, n = 10; K367A, n = 28; K367A/R369A, n = 11; R381A, n = 20; R386A, n = 19; 2A, n = 12; 4A, n = 10). Error bars, mean ± S.E. E, images of cells expressing the full-length of protrudin (WT), protrudin containing K367A/R369A and 4A (K367A/R369A/R381A/K386A) mutations, and the FYVE domain-deleted protrudin (ΔFYVE), respectively. Bars, 10 μm.
To determine whether these four cationic residues of the protrudin-FYVE domain control its PtdInsP selectivity, we mutated them to alanine, individually or in combination, expressed them in NIH3T3 cells, and monitored their subcellular localization. The levels of protrudin-FYVE domain at the PM were quantified by calculating the ratio of fluorescence intensity at the PM to that in the cytoplasm. In cells expressing the FYVE domain with single alanine mutations such as K367A, R381A, and K386A, the relative fluorescent intensity at the PM decreased 77.8, 57.2, and 45.4%, respectively, when compared with that of the wild type (WT) (Fig. 3, C and D). PM localization of a double-site mutant K367A/R369A was completely abolished, as was K367A/R369A/R381A/K386A (referred to as 4A hereafter). The R381A/K386A mutant remained at the PM to a similar extent as K367A (Fig. 3D). When measured by SPR analysis, K367A/R369A and 4A mutants showed dramatically lower affinity than WT for vesicles containing PtdIns(4,5)P2, PtdIns(3,4)P2, or PtdIns(3,4,5)P3 (Table 1). These results indicate that two cationic residues, Lys-367 and Arg-369, are critically involved in PtdInsP selectivity of the protrudin-FYVE domain for PtdIns(4,5)P2, PtdIns(3,4)P2, and PtdIns(3,4,5)P3.
Differential Role of PtdIns(4,5)P2 and PtdIns(3,4)P2/PtdIns(3,4,5)P3 in the Endosome to PM Trafficking of Protrudin
To test the effect of the novel lipid-binding specificity of the FYVE domain on subcellular localization of protrudin, the above mutations were introduced to the full-length protrudin and the mutants were expressed in NIH3T3 cells. Although protrudin WT was localized weakly to the PM, vesicles, and the endoplasmic reticulum (ER), three mutants, i.e. K367A/R369A, 4A, and FYVE domain-deleted protrudin (ΔFYVE), with low to no affinity for PtdIns(4,5)P2, PtdIns(3,4)P2, and PtdIns(3,4,5)P3, were seen only in the ER (Fig. 3E).
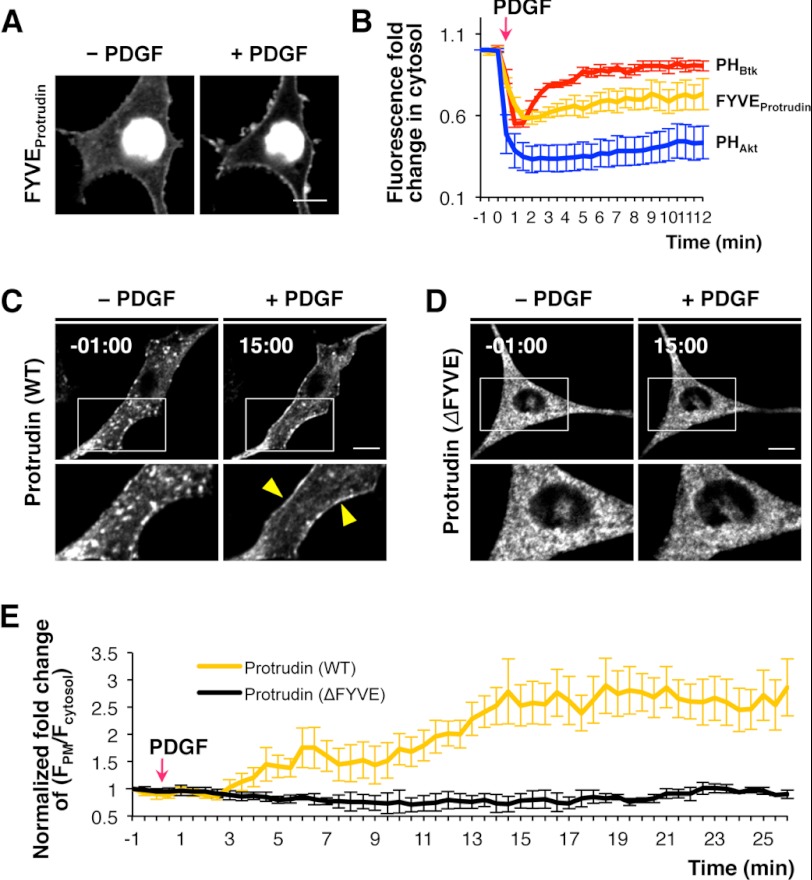
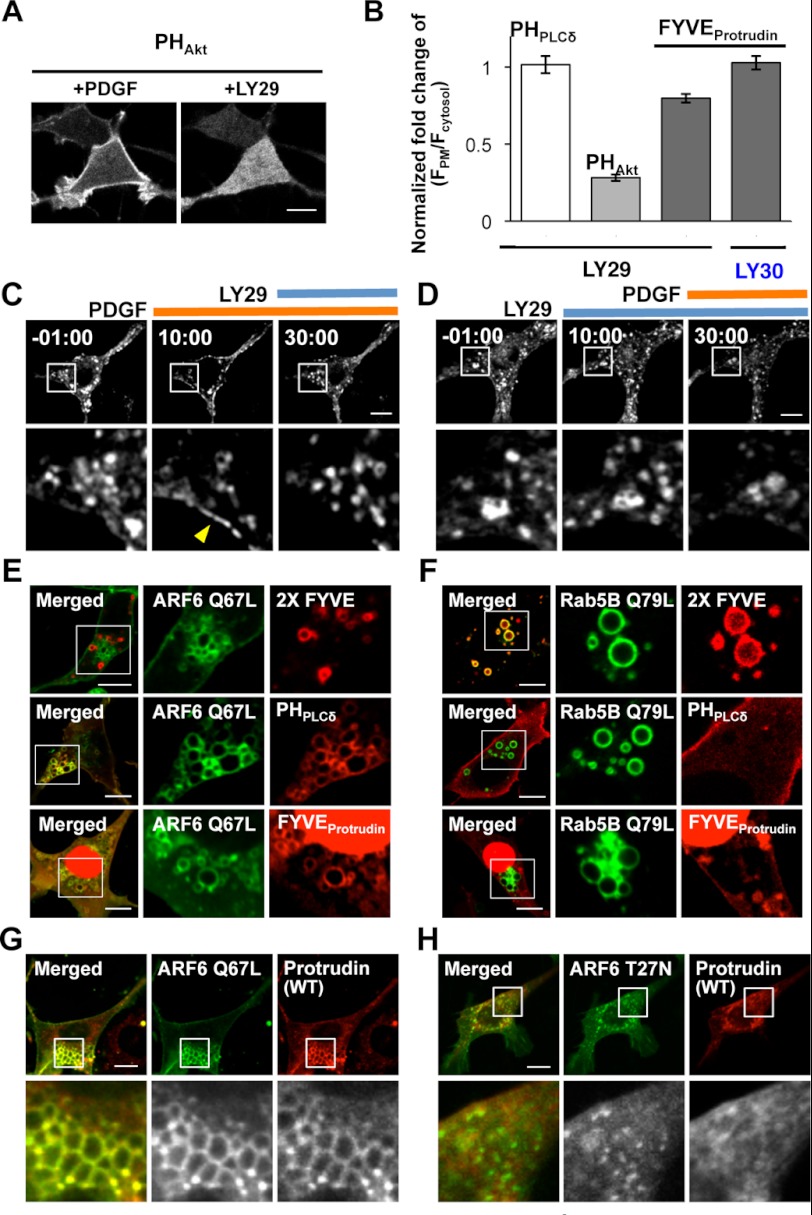
We then modulated the level of PtdIns(3,4)P2 and PtdIns(3,4,5)P3 at the PM by PDGF and LY29 treatments, which activates and inhibits PI3K, respectively. Levels of the protrudin-FYVE domain at the PM were increased when accumulation of PtdIns(3,4,5)P3 and PtdIns(3,4)P2 was induced by PDGF treatment (Fig. 4A) (34). Interestingly, the protrudin-FYVE domain exhibited sustained translocation from the cytosol to PM (Fig. 4B). It was previously reported that the Akt-PH domain interacting with both PtdIns(3,4)P2 and PtdIns(3,4,5)P3 showed similar sustained PM localization, whereas the PtdIns(3,4,5)P3-specific Btk-PH domain exhibited transient translocation to the PM (Fig. 4B) (30). Thus, the PM recruitment pattern of the protrudin-FYVE domain seems to be consistent with its PtdInsP specificity. As was the case with the isolated FYVE domain, full-length protrudin was translocated to the PM upon PDGF-triggered PtdIns(3,4)P2 and PtdIns(3,4,5)P3 increases at the PM (Fig. 4, C and E, and supplemental Movie 1). Treatment of NIH3T3 cells with LY29 reduced the PtdIns(3,4)P2 and PtdIns(3,4,5)P3 levels at the PM, as evidenced by displacement of the Akt-PH domain from the PM (Fig. 5A) but had no effect on the PM PtdIns(4,5)P2 level as seen by retention of the PLCδ-PH domain (Fig. 5B). Fig. 5C shows that LY29 treatment caused protrudin to localize at endosomes (see also supplemental Movie 1). Also, when cells were preincubated with LY29, protrudin did not show PM localization even after PDGF treatment (Fig. 5D). Treatment with LY303511 (LY30), an inactive structural analog of LY29, had no effect in NIH3T3 cells expressing the protrudin-FYVE domain (Fig. 5B). The LY29 treatment, which inhibits production of PtdIns(3)P as well as PtdIns(3,4)P2 and PtdIns(3,4,5)P3, did not interfere with formation of endocytic vesicles carrying protrudin (Fig. 5, C and D), suggesting that these vesicles contain primarily PtdIns(4,5)P2 instead of PtdIns(3)P. Furthermore, protrudin (ΔFYVE) remained in the ER and did not show any response to PDGF treatment (Fig. 4, D and E). Collectively, these results suggest that PtdIns(4,5)P2 and PtdIns(3,4)P2/PtdIns(3,4,5)P3 play differential roles in controlling protrudin trafficking through interaction with the FYVE domain: i.e. PtdIns(4,5)P2 is important for vesicular transport of protrudin, and PtdIns(3,4)P2 and PtdIns(3,4,5)P3 play a more direct role in the PM recruitment of protrudin.
FIGURE 4.
The role of PtdIns(3,4)P2 and PtdIns(3,4,5)P3 in protrudin localization through the FYVE domain. A, NIH3T3 cell expressing the protrudin-FYVE domain was treated with 5 nm PDGF. Bars, 10 μm. B, quantitative analysis for translocation of the Btk-PH domain (red, n = 5), Akt-PH domain (blue, n = 7), and protrudin-FYVE domain (orange, n = 12) to the PM under PDGF treatment. Fluorescence fold-change in cytosol (F*cytosol/Fcytosol) indicates the degree of translocation of each domain from PM to cytosol upon PDGF treatment: i.e. a lower value means more PM translocation. Error bars, mean ± S.E. PDGF-triggered translocation of protrudin (WT) (C) and ΔFYVE (D). E, quantitative analysis of the level of protrudin (WT, orange, n = 6) and ΔFYVE (black, n = 4) at the PM after PDGF treatment. The fold-change of fluorescence intensity of a protein at the PM caused by each treatment was quantified as (F*PM/F*cytosol)/(FPM/Fcytosol) in which FPM/Fcytosol and F*PM/F*cytosol, indicate the fluorescence ratio before and after each treatment, respectively. Error bars represent S.E.
FIGURE 5.
The roles of PtdIns(4,5)P2 in protrudin localization through the FYVE domain. A, Akt-PH domain interacting PtdIns(3,4)P2 and PtdIns(3,4,5)P3 stimulated by PDGF was completely removed from the PM under LY29. Bars, 10 μm. B, quantitative analysis of the level of controls, PLCδ-PH domain and Akt-PH domain, and protrudin-FYVE domain at the PM after LY29 or LY30 treatments. The fold-change of fluorescence intensity of each domain at the PM caused by chemical treatment was quantified as (F*PM/F*cytosol)/(FPM/Fcytosol), in which FPM/Fcytosol and F*PM/F*cytosol indicate the fluorescence ratio before and after each treatment, respectively. Error bars represent S.E. Cells expressing protrudin (WT) were sequentially treated with PDGF and LY29 (C) or vice versa (D). Yellow arrowhead indicates the PM. Bars, 10 μm. ARF6(Q67L)-EYFP (green) (E) or EYFP-Rab5B (Q79L) (green) (F) were cotransfected with the 2× FYVE domain (PtdIns(3)P biosensor, red), PLCδ-PH domain (PtdIns(4,5)P2 biosensor, red), and the protrudin-FYVE domain (red), respectively. ARF6(Q67L)-EYFP (green) (G) and ARF6(T27N)-EYFP (green) (H) were cotransfected with protrudin (WT, red).
ARF6 activates PtdIns(4)P 5-kinase (PIP5K) inducing local synthesis of PtdIns(4,5)P2 and regulates membrane trafficking between PM and endosomes (32, 50, 51). Rab5B induces PtdIns(3)P formation and vesicle trafficking through recruitment of class III PI3K (33). Thus, one would expect that overexpression of a constitutively active form of ARF6 (i.e. Q67L) and Rab5B (i.e. Q79L) will increase the local concentration of PtdIns(4,5)P2 and PtdIns(3)P, respectively, which can be monitored by a PtdIns(4,5)P2 probe (i.e. PLCδ-PH domain) or a PtdIns(3)P probe (i.e. tandem FENS1-FYVE domain (2× FYVE)) (Fig. 5, E and F). We found that, in cells expressing either ARF6(Q67L) or Rab5B(Q79L), the protrudin-FYVE domain was localized in PtdIns(4,5)P2-enriched vesicles (Fig. 5E), but not in PtdIns(3)P-accumulated vesicles (Fig. 5F), which is consistent with its PtdInsP specificity. As was the case of the isolated FYVE domain, vesicle-bound protrudin colocalized with PtdIns(4,5)P2-enriched endocytic vesicles resulting from PIP(5)K activation (Fig. 5G) and these protrudin-containing vesicles disappeared in the presence of a dominant-negative mutant of ARF6 (i.e. T27N) (Fig. 5H). These results suggest that vesicular localization of protrudin is dependent on the PtdIns(4,5)P2 binding activity of the FYVE domain and that PtdIns(4,5)P2-mediated endocytosis regulates endosomal trafficking of protrudin.
Physiological Roles of PtdIns(4,5)P2 and PtdIns(3,4)P2/PtdIns(3,4,5)P3 Binding of Protrudin
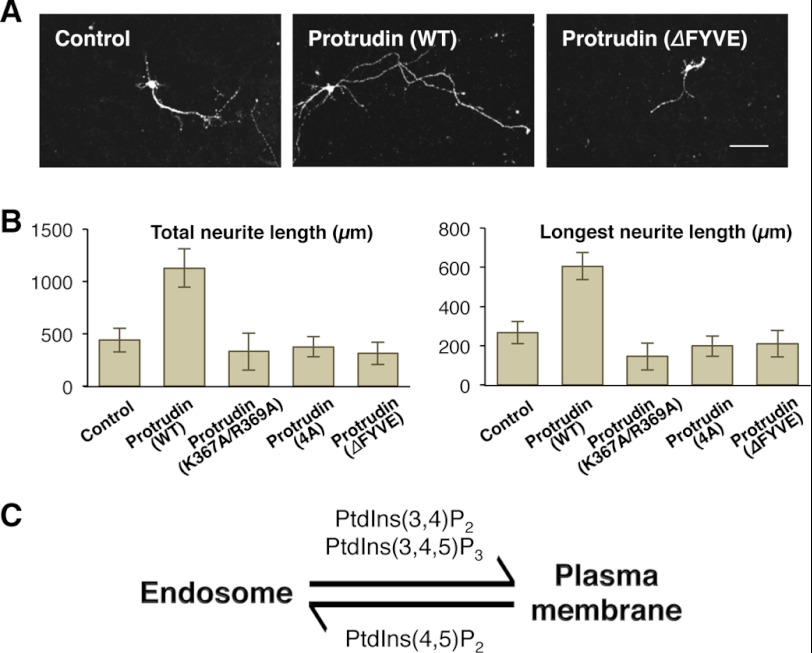
Previous studies suggest that spastin and KIF5, protrudin-interacting proteins, induce neurite formation through microtubule-related membrane trafficking (6, 52). To demonstrate the physiological significance of novel PtdInsP selectivity of the FYVE domain of protrudin, we measured the neurite length of primary cultured hippocampal neurons expressing either protrudin (WT) or various protrudin mutants 36 h post-transfection. As expected, protrudin (WT) promoted 2.56- and 2.26-fold longer neurite outgrowth than the control when the total and maximum length of neurite was measured, respectively (Fig. 6, A and B). However, expression of protrudin harboring K367A/R369A or 4A mutations, or the ΔFYVE mutant had no significant effect on promotion of neurite outgrowth (Fig. 6, A and B). Overall, these data suggest that interaction of the FYVE domain of protrudin with PtdIns(4,5)P2, PtdIns(3,4)P2, and PtdIns(3,4,5)P3 is essential for the function of protrudin in promoting neurite outgrowth.
FIGURE 6.
The effect of the FYVE domain in neurite outgrowth promoted by protrudin. A, images of primary cultured hippocampal neurons expressing Lyn-mCherry together with control (citrine only), citrine-protrudin (WT), or citrine-protrudin (without FYVE). Lyn-mCherry, a PM marker, was used to clearly show neurites. Bars, 10 μm. B, total and maximum neurite length was measured by NeuronJ (control, n = 16, WT, n = 13; K367A/R369A, n = 16; 4A, n = 28; without FYVE, n = 13). Error bars indicate S.E. C, a hypothetical model of the PM to endosome translocation regulated by different PtdInsPs.
DISCUSSION
Many signaling proteins are regulated by complex interactions between their modular domains and numerous factors. LBDs play a key role in precise spatiotemporal regulation of proteins through their interaction with various lipids. To study roles of key signaling lipids, PtdIns(4,5)P2, and PtdIns(3,4,5)P3 in the regulation of cellular proteins in a simple, comprehensive, and explicit manner, we generated a library of distinct LBDs obtained from diverse cellular proteins and determined their subcellular localization. Our study showed that most FYVE and PX domains are endosome-localized, whereas ENTH and Tubby domains are PM-associated, as expected from their reported lipid specificity (42–44). The study also revealed that the FYVE domain of protrudin is localized to PM and the nucleus instead of endosomes, unlike typical FYVE domains (45).
The FYVE domains are known to show endosomal localization and interact with PtdIns(3)P (10–12). Unlike typical FYVE domains, the FYVE domain of protrudin did not show endosomal localization but was instead found at the PM. It was recently reported that the protrudin-FYVE domain binds to PtdIns(3)P when measured by a lipid blot assay (52). Our quantitative SPR analysis demonstrate, however, that the protrudin-FYVE domain has high affinity for PtdIns(4,5)P2 and two main products of the class I PI3K (34), PtdIns(3,4)P2 and PtdIns(3,4,5)P3, but not for PtdIns(3)P. Comparison of the amino acid sequences of the protrudin-FYVE domain and the prototypical EEA1-FYVE domain reveals that the protrudin-FYVE domain has unique structural variations in lipid binding motifs. Furthermore, molecular modeling suggests that a larger lipid binding pocket of the protrudin-FYVE domain may allow interaction with bulkier multi-phosphorylated PtdInsPs. Our modeling as well as mutational analysis indicates that Lys-367 and Arg-369 are most critically involved in specific recognition of 3-, 4-, and/or 5-phosphate groups of PtdIns(3,4)P2, PtdIns(4,5)P2, and PtdIns(3,4,5)P3.
The novel PtdInsP selectivity of the protrudin-FYVE domain is consistent with the observed subcellular localization pattern of the domain and the intact protrudin. PDGF triggered PM localization of protrudin, implying that PI3K activation is involved in dynamic PM recruitment of protrudin. Also, reduction of the PtdIns(3,4)P2 and PtdIns(3,4,5)P3 levels at PM by LY29, which inhibits production of 3-phosphorylated PtdInsPs, caused protrudin to localize at endosomes. Thus, PtdIns(4,5)P2 binding activity may be more important for vesicular trafficking of protrudin than its PM recruitment. ARF6 regulates membrane trafficking between PM and endosomes by activating PIP5K that locally synthesizes PtdIns(4,5)P2 from PtdIns(4)P (51). We observed that the PtdIns(4,5)P2-rich endocytic vesicles formed by a constitutively active ARF6 were colocalized with protrudin-containing vesicles, and coexpression of a dominant-negative mutant of ARF6 blocked localization of protrudin to the vesicles. This suggests that PIP5K-mediated PtdIns(4,5)P2 synthesis and endocytosis carry protrudin to endosomes for recycling. The fact that the protrudin mutant lacking the FYVE domain is localized at the ER indicates that ER localization of protrudin is driven not by lipid binding but by protein binding. In our model, ER-resident or vesicle-bound protrudin is recruited to the PM in response to local synthesis of PtdIns(3,4)P2 and PtdIns(3,4,5)P3 through PI3K activation and recycled to endosomes via PtdIns(4,5)P2-enriched vesicles when PtdIns(3,4)P2 and PtdIns(3,4,5)P3 levels decrease. By this mechanism, PtdIns(4,5)P2 and PtdIns(3,4)P2/PtdIns(3,4,5)P3 differentially modulate the endosome to PM trafficking of protrudin (Fig. 6C).
Involvement of protrudin in an AD-HSP has been suggested (6, 8, 9). The large group of HSP-related proteins is related to membrane trafficking. Previous studies on protrudin showed that protrudin plays a role in promoting neurite outgrowth through dynamic movement from the ER to the PM (7, 8). It is also involved in delivery of neuronal cargoes by interacting with KIF5, a molecular motor protein, suggesting that microtubules can be involved in protrudin-regulated vesicle trafficking. Our present study provides new insight into how its FYVE domain with novel PtdInsP selectivity contributes to the cellular function and regulation of protrudin. That is, differential regulation of endosome to PM trafficking of protrudin by binding of PtdIns(4,5)P2 and PtdIns(3,4)P2/PtdIns(3,4,5)P3 to its FYVE domain is crucial for its function in facilitating transport of neuronal cargo proteins. Undoubtedly, further studies are necessary to fully understand the mechanism underlying this complex process. Nevertheless, our discovery of the unique PtdInsP specificity of the FYVE domain of protrudin should help better understand how protrudin is linked to the onset of neurodegenerative disease such as AD-HSP via lipid signaling.
Acknowledgments
We thank Dr. Keiichi I. Nakayama and Dr. Takanari Inoue for kindly providing construct and localization marker for ER, respectively. We are also thankful to Min Jee Jang and Dr. Yoonkey Nam for helping isolate hippocampus from rat brain and to all of my lab members for helpful discussion.
This work was supported by Korea Healthcare Technology R&D Project, Ministry of Health & Welfare, Republic of Korea Grant A101204.

This article contains supplemental Table S1 and Movie S1.
- LBD
- lipid binding domain
- AD-HSP
- autosomal dominant form of hereditary spastic paraplegia
- Btk
- Bruton's tyrosine kinase
- EYFP
- enhanced yellow fluorescence protein
- HSP
- hereditary spastic paraplegia
- LY29
- LY294002
- PtdIns
- phosphatidylinositol
- PLCδ
- phospholipase Cδ
- PM
- plasma membrane
- POPC
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPS
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine
- PtdIns
- phosphatidylinositol
- PtdIns(3)P
- phosphatidylinositol 3-phosphate
- PtdIns(4)P
- phosphatidylinositol 4-phosphate
- PtdIns(5)P
- phosphatidylinositol 5-phosphate
- PtdIns(3,4)P2
- phosphatidylinositol 3,4-bisphosphate
- PtdIns(4,5)P2
- phosphatidylinositol 4,5-bisphosphate, PtdIns(3,4,5)P3, phosphatidylinositol 3,4,5-trisphosphate
- SPR
- surface plasmon resonance
- ER
- endoplasmic reticulum.
REFERENCES
- 1. DiNitto J. P., Cronin T. C., Lambright D. G. (2003) Membrane recognition and targeting by lipid-binding domains. Sci. STKE 2003, re16. [DOI] [PubMed] [Google Scholar]
- 2. Cho W., Stahelin R. V. (2005) Membrane-protein interactions in cell signaling and membrane trafficking. Annu. Rev. Biophys. Biomol. Struct. 34, 119–151 [DOI] [PubMed] [Google Scholar]
- 3. Lemmon M. A. (2008) Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9, 99–111 [DOI] [PubMed] [Google Scholar]
- 4. Wymann M. P., Schneiter R. (2008) Lipid signalling in disease. Nat. Rev. Mol. Cell Biol. 9, 162–176 [DOI] [PubMed] [Google Scholar]
- 5. Blackstone C., O'Kane C. J., Reid E. (2011) Hereditary spastic paraplegias. Membrane traffic and the motor pathway. Nat. Rev. Neurosci. 12, 31–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Matsuzaki F., Shirane M., Matsumoto M., Nakayama K. I. (2011) Protrudin serves as an adaptor molecule that connects KIF5 and its cargoes in vesicular transport during process formation. Mol. Biol. Cell 22, 4602–4620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Saita S., Shirane M., Natume T., Iemura S., Nakayama K. I. (2009) Promotion of neurite extension by protrudin requires its interaction with vesicle-associated membrane protein-associated protein. J. Biol. Chem. 284, 13766–13777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shirane M., Nakayama K. I. (2006) Protrudin induces neurite formation by directional membrane trafficking. Science 314, 818–821 [DOI] [PubMed] [Google Scholar]
- 9. Mannan A. U., Krawen P., Sauter S. M., Boehm J., Chronowska A., Paulus W., Neesen J., Engel W. (2006) ZFYVE27 (SPG33), a novel spastin-binding protein, is mutated in hereditary spastic paraplegia. Am. J. Hum. Genet. 79, 351–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gillooly D. J., Simonsen A., Stenmark H. (2001) Cellular functions of phosphatidylinositol 3-phosphate and FYVE domain proteins. Biochem. J. 355, 249–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kutateladze T. G. (2006) Phosphatidylinositol 3-phosphate recognition and membrane docking by the FYVE domain. Biochim. Biophys. Acta 1761, 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Corvera S., D'Arrigo A., Stenmark H. (1999) Phosphoinositides in membrane traffic. Curr. Opin. Cell Biol. 11, 460–465 [DOI] [PubMed] [Google Scholar]
- 13. Burd C. G., Emr S. D. (1998) Phosphatidylinositol 3-phosphate signaling mediated by specific binding to RING FYVE domains. Mol. Cell 2, 157–162 [DOI] [PubMed] [Google Scholar]
- 14. Patki V., Lawe D. C., Corvera S., Virbasius J. V., Chawla A. (1998) A functional PtdIns3P-binding motif. Nature 394, 433–434 [DOI] [PubMed] [Google Scholar]
- 15. Lawe D. C., Patki V., Heller-Harrison R., Lambright D., Corvera S. (2000) The FYVE domain of early endosome antigen 1 is required for both phosphatidylinositol 3-phosphate and Rab5 binding. Critical role of this dual interaction for endosomal localization. J. Biol. Chem. 275, 3699–3705 [DOI] [PubMed] [Google Scholar]
- 16. Estrada L., Caron E., Gorski J. L. (2001) Fgd1, the Cdc42 guanine nucleotide exchange factor responsible for faciogenital dysplasia, is localized to the subcortical actin cytoskeleton and Golgi membrane. Hum. Mol. Genet. 10, 485–495 [DOI] [PubMed] [Google Scholar]
- 17. Tsukazaki T., Chiang T. A., Davison A. F., Attisano L., Wrana J. L. (1998) SARA, a FYVE domain protein that recruits Smad2 to the TGFβ receptor. Cell 95, 779–791 [DOI] [PubMed] [Google Scholar]
- 18. Seet L. F., Hong W. (2001) Endofin, an endosomal FYVE domain protein. J. Biol. Chem. 276, 42445–42454 [DOI] [PubMed] [Google Scholar]
- 19. Misra S., Hurley J. H. (1999) Crystal structure of a phosphatidylinositol 3-phosphate-specific membrane-targeting motif, the FYVE domain of Vps27p. Cell 97, 657–666 [DOI] [PubMed] [Google Scholar]
- 20. Mao Y., Nickitenko A., Duan X., Lloyd T. E., Wu M. N., Bellen H., Quiocho F. A. (2000) Crystal structure of the VHS and FYVE tandem domains of Hrs, a protein involved in membrane trafficking and signal transduction. Cell 100, 447–456 [DOI] [PubMed] [Google Scholar]
- 21. Dumas J. J., Merithew E., Sudharshan E., Rajamani D., Hayes S., Lawe D., Corvera S., Lambright D. G. (2001) Multivalent endosome targeting by homodimeric EEA1. Mol. Cell 8, 947–958 [DOI] [PubMed] [Google Scholar]
- 22. Kutateladze T., Overduin M. (2001) Structural mechanism of endosome docking by the FYVE domain. Science 291, 1793–1796 [DOI] [PubMed] [Google Scholar]
- 23. Heo W. D., Inoue T., Park W. S., Kim M. L., Park B. O., Wandless T. J., Meyer T. (2006) PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science 314, 1458–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heo W. D., Meyer T. (2003) Switch-of-function mutants based on morphology classification of Ras superfamily small GTPases. Cell 113, 315–328 [DOI] [PubMed] [Google Scholar]
- 25. Inoue T., Heo W. D., Grimley J. S., Wandless T. J., Meyer T. (2005) An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat. Methods 2, 415–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Komatsu T., Kukelyansky I., McCaffery J. M., Ueno T., Varela L. C., Inoue T. (2010) Organelle-specific, rapid induction of molecular activities and membrane tethering. Nat. Methods 7, 206–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jiang M., Chen G. (2006) High Ca2+-phosphate transfection efficiency in low-density neuronal cultures. Nat. Protoc 1, 695–700 [DOI] [PubMed] [Google Scholar]
- 28. Watton S. J., Downward J. (1999) Akt/PKB localization and 3′-phosphoinositide generation at sites of epithelial cell-matrix and cell-cell interaction. Curr. Biol. 9, 433–436 [DOI] [PubMed] [Google Scholar]
- 29. Stauffer T. P., Ahn S., Meyer T. (1998) Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr. Biol. 8, 343–346 [DOI] [PubMed] [Google Scholar]
- 30. Manna D., Albanese A., Park W. S., Cho W. (2007) Mechanistic basis of differential cellular responses of phosphatidylinositol 3,4-bisphosphate- and phosphatidylinositol 3,4,5-trisphosphate-binding pleckstrin homology domains. J. Biol. Chem. 282, 32093–32105 [DOI] [PubMed] [Google Scholar]
- 31. Ridley S. H., Ktistakis N., Davidson K., Anderson K. E., Manifava M., Ellson C. D., Lipp P., Bootman M., Coadwell J., Nazarian A., Erdjument-Bromage H., Tempst P., Cooper M. A., Thuring J. W., Lim Z. Y., Holmes A. B., Stephens L. R., Hawkins P. T. (2001) FENS-1 and DFCP1 are FYVE domain-containing proteins with distinct functions in the endosomal and Golgi compartments. J. Cell Sci. 114, 3991–4000 [DOI] [PubMed] [Google Scholar]
- 32. Brown F. D., Rozelle A. L., Yin H. L., Balla T., Donaldson J. G. (2001) Phosphatidylinositol 4,5-bisphosphate and Arf6-regulated membrane traffic. J. Cell Biol. 154, 1007–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Murray J. T., Panaretou C., Stenmark H., Miaczynska M., Backer J. M. (2002) Role of Rab5 in the recruitment of hVps34/p150 to the early endosome. Traffic 3, 416–427 [DOI] [PubMed] [Google Scholar]
- 34. Toker A., Bachelot C., Chen C. S., Falck J. R., Hartwig J. H., Cantley L. C., Kovacsovics T. J. (1995) Phosphorylation of the platelet p47 phosphoprotein is mediated by the lipid products of phosphoinositide 3-kinase. J. Biol. Chem. 270, 29525–29531 [DOI] [PubMed] [Google Scholar]
- 35. Ananthanarayanan B., Stahelin R. V., Digman M. A., Cho W. (2003) Activation mechanisms of conventional protein kinase C isoforms are determined by the ligand affinity and conformational flexibility of their C1 domains. J. Biol. Chem. 278, 46886–46894 [DOI] [PubMed] [Google Scholar]
- 36. Cho W., Bittova L., Stahelin R. V. (2001) Membrane binding assays for peripheral proteins. Anal. Biochem. 296, 153–161 [DOI] [PubMed] [Google Scholar]
- 37. Potterton E., McNicholas S., Krissinel E., Cowtan K., Noble M. (2002) The CCP4 molecular-graphics project. Acta Crystallogr. D Biol. Crystallogr. 58, 1955–1957 [DOI] [PubMed] [Google Scholar]
- 38. Pankiv S., Alemu E. A., Brech A., Bruun J. A., Lamark T., Overvatn A., Bjørkøy G., Johansen T. (2010) FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J. Cell Biol. 188, 253–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gaullier J. M., Ronning E., Gillooly D. J., Stenmark H. (2000) Interaction of the EEA1 FYVE finger with phosphatidylinositol 3-phosphate and early endosomes. Role of conserved residues. J. Biol. Chem. 275, 24595–24600 [DOI] [PubMed] [Google Scholar]
- 40. Blatner N. R., Wilson M. I., Lei C., Hong W., Murray D., Williams R. L., Cho W. (2007) The structural basis of novel endosome anchoring activity of KIF16B kinesin. EMBO J. 26, 3709–3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Miura S., Takeshita T., Asao H., Kimura Y., Murata K., Sasaki Y., Hanai J. I., Beppu H., Tsukazaki T., Wrana J. L., Miyazono K., Sugamura K. (2000) Hgs (Hrs), a FYVE domain protein, is involved in Smad signaling through cooperation with SARA. Mol. Cell. Biol. 20, 9346–9355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ford M. G., Mills I. G., Peter B. J., Vallis Y., Praefcke G. J., Evans P. R., McMahon H. T. (2002) Curvature of clathrin-coated pits driven by epsin. Nature 419, 361–366 [DOI] [PubMed] [Google Scholar]
- 43. Itoh T., Koshiba S., Kigawa T., Kikuchi A., Yokoyama S., Takenawa T. (2001) Role of the ENTH domain in phosphatidylinositol 4,5-bisphosphate binding and endocytosis. Science 291, 1047–1051 [DOI] [PubMed] [Google Scholar]
- 44. Quinn K. V., Behe P., Tinker A. (2008) Monitoring changes in membrane phosphatidylinositol 4,5-bisphosphate in living cells using a domain from the transcription factor Tubby. J. Physiol. 586, 2855–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Misra S., Miller G. J., Hurley J. H. (2001) Recognizing phosphatidylinositol 3-phosphate. Cell 107, 559–562 [DOI] [PubMed] [Google Scholar]
- 46. Yu M., Lowell C. A., Neel B. G., Gu H. (2006) Scaffolding adapter Grb2-associated binder 2 requires Syk to transmit signals from FcϵRI. J. Immunol. 176, 2421–2429 [DOI] [PubMed] [Google Scholar]
- 47. Maroun C. R., Naujokas M. A., Park M. (2003) Membrane targeting of Grb2-associated binder-1 (Gab1) scaffolding protein through Src myristoylation sequence substitutes for Gab1 pleckstrin homology domain and switches an epidermal growth factor response to an invasive morphogenic program. Mol. Biol. Cell 14, 1691–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Park W. S., Heo W. D., Whalen J. H., O'Rourke N. A., Bryan H. M., Meyer T., Teruel M. N. (2008) Comprehensive identification of PIP3-regulated PH domains from C. elegans to H. sapiens by model prediction and live imaging. Mol. Cell 30, 381–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Blatner N. R., Stahelin R. V., Diraviyam K., Hawkins P. T., Hong W., Murray D., Cho W. (2004) The molecular basis of the differential subcellular localization of FYVE domains. J. Biol. Chem. 279, 53818–53827 [DOI] [PubMed] [Google Scholar]
- 50. Martin T. F. (2001) PI(4,5)P(2) regulation of surface membrane traffic. Curr. Opin. Cell Biol. 13, 493–499 [DOI] [PubMed] [Google Scholar]
- 51. D'Souza-Schorey C., Chavrier P. (2006) ARF proteins. Roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 7, 347–358 [DOI] [PubMed] [Google Scholar]
- 52. Pantakani D. V., Czyzewska M. M., Sikorska A., Bodda C., Mannan A. U. (2011) Oligomerization of ZFYVE27 (Protrudin) is necessary to promote neurite extension. PLoS One 6, e29584. [DOI] [PMC free article] [PubMed] [Google Scholar]