Background: Telomerase is essential for cancer cell growth.
Results: MiR-498 is a novel 1,25(OH)2D3 target gene that decreases telomerase, induces cell death, and suppresses tumor growth.
Conclusion: MiR-498 is an important mediator of the anti-tumor activity of 1,25(OH)2D3.
Significance: The studies define a new mechanism of telomerase regulation by small non-coding RNAs in response to 1,25(OH)2D3.
Keywords: Apoptosis, Cancer Chemoprevention, Microarray, Nuclear Receptors, Steroid Hormone, VDR, Cancer, MicroRNA, Telomerase, Vitamin D
Abstract
Telomerase is an essential enzyme that counteracts the telomere attrition accompanying DNA replication during cell division. Regulation of the promoter activity of the gene encoding its catalytic subunit, the telomerase reverse transcriptase, is established as the dominant mechanism conferring the high telomerase activity in proliferating cells, such as embryonic stem and cancer cells. This study reveals a new mechanism of telomerase regulation through non-coding small RNA by showing that microRNA-498 (miR-498) induced by 1,25-dihydroxyvitamin D3 (1,25(OH)2D3) decreases the mRNA expression of the human telomerase reverse transcriptase. MiR-498 was first identified in a microarray analysis as the most induced microRNA by 1,25(OH)2D3 in ovarian cancer cells and subsequently validated by quantitative polymerase chain reaction assays in multiple human cancer types. A functional vitamin D response element was defined in the 5-prime regulatory region of the miR-498 genome, which is occupied by the vitamin D receptor and its coactivators. Further studies showed that miR-498 targeted the 3-prime untranslated region of human telomerase reverse transcriptase mRNA and decreased its expression. The levels of miR-498 expression were decreased in malignant human ovarian tumors as well as human ovarian cancer cell lines. The ability of 1,25(OH)2D3 to decrease human telomerase reverse transcriptase mRNA and to suppress ovarian cancer growth was compromised when miR-498 was depleted using the sponges in cell lines and mouse tumor models. Taken together, our studies define a novel mechanism of telomerase regulation by small non-coding RNAs and identify miR-498 as an important mediator for the anti-tumor activity of 1,25(OH)2D3.
Introduction
Telomeres maintain genomic stability by protecting the linear chromosome ends from being recognized as DNA breaks that would need repair. They undergo frequent remodeling during important biological events such as development, proliferation, and neoplastic transformation, etc. (1). The telomeric TTAGGG repeats are replenished by telomerase, a ribonucleoprotein complex that consists of a catalytic reverse transcriptase protein subunit (e.g. TERT) (2), a template RNA (e.g. TERC) (3), dyskerin (4), and other accessory proteins (5). Telomerase activity is very low in adult somatic cells, and the inability of DNA replication machinery to reproduce telomeres is the reason why cells show progressive telomere shortening with ongoing cell division until some telomeres reach a critically shortened length and induce a DNA damage checkpoint that causes replicative senescence. Proliferative cells such as embryonic stem and cancer cells contain high telomerase activity regulated primarily at the level of hTERT transcription (6). Consistently, hTERT is overexpressed in almost 90% of human cancer cells by up to 100-fold over their normal counterpart, which confers a strong selective advantage for continued growth of malignant cells (7). Besides promoter regulation of the hTERT gene, little is known about other mechanisms that control telomerase activity, although post-translational modification of TERT has been reported (8).
MicroRNAs (miRNAs)3 are noncoding, single-stranded RNA molecules of 20–24 nucleotides that typically repress gene expression (9). Different from small interference RNAs (siRNAs), which are often from exogenous sources, miRNAs are encoded by genes transcribed by RNA polymerase II (10, 11). Once transcribed, primary miRNA transcripts are cleaved in the nucleus by the Drosha complex or spliced into precursor miRNAs of about 70 nucleotides and then cleaved in the cytoplasm by the Dicer complex into mature miRNA duplexes of about 22 nucleotides. One of the mature miRNA duplex strands is incorporated into the argonaute-containing RNA-induced silencing complex, whereas the other strand is released and degraded (12). The miRNA strand in the RNA-induced silencing complex interacts with target messenger RNA (mRNA) molecules by either perfect or near perfect matches in the 3′-untranslated region (3′-UTR) whereby they induce mRNA degradation or translational inhibition (9, 13). In humans, about 700 miRNAs have been identified, and more than one-third of all human genes have been predicted to be miRNA targets (9, 14). These small RNA molecules are involved in essentially every cellular process investigated to date (15, 16).
The active metabolite of vitamin D, calcitriol (1,25-dihydroxyvitamin D3 (1,25(OH)2D3)) is not only known for its crucial role in maintaining mineral homeostasis and skeletal functions (17, 18) but is also widely recognized as a natural nutraceutical compound with great potential for tumor prevention and treatment (19). 1,25(OH)2D3 exhibits antiproliferative, proapoptotic, and differentiation-inducing properties as well as immunomodulatory effects in a variety of human cancers (20). The biological effects of 1,25(OH)2D3 are mediated through the vitamin D receptor (VDR), which belongs to the superfamily of steroid/thyroid nuclear hormone receptors (21, 22). After ligand binding, the VDR forms a heterodimer with retinoid X receptor (RXR) and binds to vitamin D-responsive elements (VDREs) in the regulatory region of target genes to activate or repress their transcription (23). Previous studies have shown that the growth of multiple human ovarian cancers was suppressed by 1,25(OH)2D3 (24, 25) and that 1,25(OH)2D3-induced apoptosis involved down-regulation of telomerase activity (26). This study identifies miR-498 as a primary target gene for 1,25(OH)2D3, which binds to the 3′-UTR of hTERT and decreases its mRNA expression to induce cell death. The studies reveal a new mechanism of telomerase regulation by vitamin D and define a role for miRNAs in mediating the tumor suppressive activity of the nutraceutical compound.
EXPERIMENTAL PROCEDURES
Materials, Cell Lines, and Human Tissues
1,25(OH)2D3 was from Calbiochem. Baculovirus-expressed human VDR protein and human RXRβ protein were from Affinity BioReagents Inc. (Golden, CO). Anti-VDR antibody was from Chemicon International (Temecula, CA). All oligonucleotides were synthesized by Integrated DNA Technologies (San Diego, CA). OVCAR3, A2780, A2780-CP, and C13 were cultured in RPMI 1640 medium supplemented with 2 mm l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 5% FCS. OV2008, CAOV3, MCF-7, Ishikawa, and HeLa cells were maintained in DMEM containing 2 mm l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 5% FCS. BG1 cells were cultured in DMEM/F-12 medium supplemented with 2 mm l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 5% FCS. All cells are maintained at 37 °C in a humidified incubator with 5% CO2. Frozen human borderline and malignant ovarian tumor tissues were obtained from the Tissue Procurement Facility at H. Lee Moffitt Cancer Center, and their usage for the studies was approved by the institutional review board at the University of South Florida.
miRNA Microarray
For miRNA microarray analyses, OVCAR3 cells of about 70% confluence were treated with vehicle control (ethanol (EtOH)) or 10−7 m 1,25(OH)2D3 for 6 days. Small RNAs were isolated from treated cells using the mirVanaTM miRNA isolation kit (Ambion, Austin, TX) according to the manufacturer's instructions. The miRNA microarray analysis was performed by LC Sciences (Houston, TX). Ten micrograms of total RNA were labeled with Cy3 (EtOH) or Cy5 (VD) fluorescent dyes, which produces a green and red color, respectively. Dye switching was performed to eliminate dye bias. Pairs of labeled samples were hybridized to dual-channel microarrays. Microarray assays were performed on a μParaFlo microfluidics chip with each of the detection probes containing a nucleotide sequence of coding segment complementary to a specific miRNA sequence. The arrays were designed to detect miRNA transcripts corresponding to 327 miRNAs contained in the Sanger miRBase Release 8.0. The maximum signal level of threshold and background probes was 190. Data were analyzed by subtracting the background followed by signal normalization with a LOWESS (locally weighted regression) filter. The ratio of the 2 sets of detected signals (log2 transformed and balanced) and the p values of the t test were calculated. Differentially detected signals were those with p values less than 0.01.
RT-PCR and Quantitative RT-PCR
Total RNA was extracted using TRIzol (Invitrogen) according to the manufacturer's protocol. cDNA was reverse-transcribed from 1 μg of total cellular RNA with random hexamer primers and thermostable reverse transcriptase (Invitrogen). Analyses of hTERT and GAPDH expression by RT-PCR were performed as described previously (26). To quantify hTERT and GAPDH expression, PCR products were subjected to electrophoresis in a 15% polyacrylamide gel. PCR products were hybridized with a gene-specific probe, and signals were detected using a CSPD (Chloro-5-substituted adamantyl-1,2-dioxetane phosphate) chemiluminescence-based Southern blot analysis system according to the manufacturer's instructions.
Small RNAs were isolated from tissues and cells using the mirVanaTM miRNA isolation kit (Ambion) according to the manufacturer's instructions. Drosha, miR-498, and RNU6 (U6) expressions were measured by RT-PCR with following primers. Drosha primers were 5′-CACCTGTTCTAGCAGCTCAGAC-3′ and 5′-CTCCTCCCACTGAAGCATATTG-3′, miR-498 primers were 5′-TTTCAAGCCAGGGGGCGTTTTTC-3′ and 5′-GCTTCAAGCTCTGGAGGTGCTTTTC-3′, and U6 primers were 5′-AACGCTTCACGAATTTGCGT-3′ and 5′-CTCGCTTCGGCAGCACA-3′. The amplification of target genes was carried out using 1 μl of reverse transcription product in a total reaction volume of 25 μl. The PCR reaction included a 5-min denaturation at 94 °C followed by 32 cycles of denaturing at 94 °C for 30 s, annealing at 60 °C for 45 s, and extension at 72 °C for 15 s with a final extension phase of 10 min. The PCR products were separated on a 1.5% agarose gel and visualized by staining with ethidium bromide.
Taqman microRNA assays (Applied Biosystems) were used to quantify mature miRNA expression as previously described (27). U6 was used as endogenous control for miRNA expression studies. PCR reactions were performed in a 20-μl reaction mixture containing 1.33 μl of reverse transcription product, 1.0 μl of 5× TaqMan micro-assay mix, and 10 μl TaqMan 2× universal PCR Master Mix. Reactions were run on the ABI Prism 7900 Fast Real-Time PCR system in triplicate. The PCR reaction was conducted by incubation at 95 °C for 10 min followed by 40 cycles of 15 s of denaturing at 95 °C and 1 min of annealing at 60 °C. The miRNA expression level at each time point was normalized with cognate U6 level by subtracting the Cycle threshold (Ct) value for U6 from Ct for miR498 to produce a ΔCt. The -fold of induction over vehicle control was calculated based on the formula 2−(ΔCt(1,25(OH)2D3) − ΔCt(vehicle)).
Plasmids, Constructions, and Site-directed Mutagenesis
p91023B-VDR (28) has been described previously. To construct miR-498 target validation vector, the 3′-UTR fragment of hTERT containing the miR-498 binding site was amplified by PCR (forward, 5′-GATCACTAGTTGGCCACCCGCCCACA-3′; reverse, 5′-TCGAAAGCTTCCCCCATCCAGGTGCAGCTGGGT-3′) to generate SpeI and HindIII sites at the 5′ and 3′ ends, respectively. To generate the control vector, 3′-UTR regions lacking the target site were amplified by PCR (forward, 5′-GATCACTAGTCCCGGGACGACGCT-3′; reverse, 5′-TCGAAAGCTTGGGTCCTTCTCAGGGT-3′). The amplified fragments were cloned into the SpeI and HindIII sites of pMIR-Report luciferase vector (Ambion).
To construct the miR-498 expression vector, pre-miR-498 double-stranded oligos were synthesized and inserted into the EcoRI site of pBS/U6 vector. The sequence of the pre-miR-498 sense strand from 5′ end to 3′ end is: CCGAATTCAACCCTCCTTGGGAAGTGAAGCTCAGGCTGTGATTTCAAGCCAGGGGGCGTTTTTCTATAACTGGATGAAAAGCACCTCCAGAGCTTGAAGCTCACAGTTTGAGAGCAATCGTCTAAGGAAGTTGAATTCCGG.
To generate pGL3-VDRE1 luciferase, the VDRE1 (585 bp) fragment was generated by PCR using primers 5′-CGGCTAGCGGAATTACCCTCCCCGCTCGAGTTCATCAGGTG-3′ (forward) and 5′-CCCCCCGGGTTAAAACCCAGACTCTGAGACCA-3′ (reverse) and cloned into the NheI and SmaI sites of pGL3-promoter vector. pGL3-VDRE2 luciferase was constructed similarly by generating a NheI site at the 5′-end and a SmaI at the 3′ end of the VDRE2 fragment (407 bp). The sequence of VDRE2 primers is 5′-CGGCTAGCTATCTGTCGACGCGTTTGCT-3′ (forward) and 5′-CCCCCCGGGAGGCCCTCATTCTGTTACTTCTT-3′ (reverse).
To mutate VDRE2 luciferase reporter, CA dinucleotides in the second half-site were replaced with TT using PCR-based site-directed mutagenesis (QuikChange, Stratagene, La Jolla, CA). The forward (5′-GCTTAATACCTAGCTCAATAGGTTTTGCAAACCACCACGG-3′) and reverse (5′-CCGTGGTGGTTTGCAAAACCTATTGACCTAGGTATTAAGC-3′) primers were designed according to manufacturer's instructions. The PCR reactions were started with incubation at 95 °C for 2 min followed by 18 cycles of 20 s at 95 °C, 10 s at 60 °C, and 30 s at 68 °C and a final 5-min extension at 68 °C. The PCR products were digested with 10 units of DpnI for 1 h at 37 °C before being transformed into the XL10-Gold ultracompetent cells. After transformed colonies were screened by PCR, the mutant plasmids produced from the clones were isolated, purified, and sequenced.
Vectors expressing control and miR-498 “sponges” were constructed based on a method described in previous reports (16, 29, 30). Oligos with seven repetitive miR-498 complementary sequences were synthesized by Integrated DNA Technologies (Coralville, IA). Each segment was designed with no spacers for miR-498 sponge (miR-498-SP) or with a four-nucleotide spacer at positions 9–12 for bulged sponge (miR-498-bSP). The entire cassette was cloned into the pBabe-puro vector digested with BamHI and SalI. The control sponge was designed based on a non-existent miRNA targeting CXCR4 as described previously (29).
siRNA, Transfections, and Reporter Assays
Drosha-specific siRNA was synthesized by Thermo Fischer Scientific (Waltham, MA). The sense and antisense sequences of Drosha siRNA oligos were 5′-AAGGACCAAGUAUUCAGCAAGdTdT-3′ and 5′-CUUGCUGAAUACUUGGUCCUUdTdT-3′, respectively. Nonspecific scrambled siRNA oligos were used as controls. 2′-O-Methyl (2′-O-me) siRNA oligonucleotides (antisense inhibition of miR-498) were synthesized by Ambion. The sequence of 2′-O-me-anti-miR-498 (2′-O-me-miR-498) is 5′-UUUCAAGCCAGGGGGCGUUUUUC-3′. 2′-O-me-scrambled miR (2′-O-me-control) (5′-AAAACCUUUUGACCGAGCGUGUU-3′) was used as a control.
For transfections, cells were plated at 1.5 × 105 cells/well in 6-well plates. One day after plating, transfections were performed with Lipofectamine following the protocol from Invitrogen. Forty-eight hours later, cells were treated with either vehicle (EtOH)) or 10−7 m 1,25(OH)2D3. For longer treatments, cells were placed in fresh medium with new compounds every 2 days. Luciferase and β-galactosidase activities were determined as described previously (24).
For the establishment of stable clones, OVCAR3 or OV2008 cells were transfected with 2 μg of pBABE vector expressing control or MiR-498 sponges and the pGL3 control plasmid (Promega, Madison, WI). The stable clones were obtained through selection with 2 μg/ml puromycin for about 5 weeks.
Telomerase Assays
Telomerase activity was measured by the telomeric repeat amplification protocol assay using the telomerase PCR-enzyme-linked immunosorbent assay kit (Roche Applied Science) as described (26).
Ribonuclease Protection Assays (RPAs)
The expression levels of mature miRNAs in tissue samples and cells were also measured by RPAs with a mirVana miRNA detection kit (Ambion). Small RNA from different tissues and cells were isolated for RT-PCR. Probes for miRNA detection were synthesized by Integrated DNA Technologies (Coralville, IA) and end-labeled with 32P using the mirVana probe and marker kit (Ambion). Briefly, 1 μg total RNA was incubated with 32P-labeled probes, which were prepared by [γ-32P]ATP using a T4 polynucleotide kinase labeling system. After digestion to remove the probe that was not bound by target miRNA, the radiolabeled products were separated by denaturing polyacrylamide gel electrophoresis in a 15% gel. The signals were visualized and quantified with a phosphorimaging system (Bio-Rad).
Electrophoretic Mobility Shift Assay (EMSA)
EMSA was performed as described previously (31) with modifications. Briefly, double-stranded oligonucleotides were end-labeled with 32P using a T4 polynucleotide kinase labeling system (Invitrogen). 1 μl of radiolabeled probe (about 50,000 cpm) was mixed with 19 μl of DNA binding reaction mixture that contained 250 ng of VDR and RXR in 10 mm Tris-HCl (pH 7.9), 100 mm KCl, 0.1 mm EDTA, 15% glycerol, 100 μg/ml poly(dI-dC), 0.1 μg/μl bovine serum albumin, 1 mm dithiothreitol, and 10−7 m 1,25(OH)2D3. The mixture was incubated at room temperature for 30 min. For competition and supershift experiments, VDR and RXR proteins were preincubated with a 100-fold excess of cold VDRE1 or VDRE2 probes and 2 μg of VDR antibody, respectively, on ice for 20 min before EMSA reactions. The reaction mixture was resolved in a 5% non-denaturing polyacrylamide gel, and protein-oligo complexes were revealed by autoradiography. The sequence in sense orientation of oligonucleotides used for producing complementary double-strand oligos is: VDRE1, 5′-CATCAGGTGAGGAAGTTGAGACC-3′; VDRE2, 5′-CCTAGGTCAATAGGTTCAGCAA-3′; hOC, 5′-ACTCACCGGGATGAACGGGGGCATT-3′.
Chromatin Immunoprecipitation (ChIP) Assay
For ChIP assays, OVCAR3 cells were treated with EtOH or 10−7 m 1,25(OH)2D3 for 60 min and cross-linked with 1% formaldehyde. Then the cells were lysed in buffer containing 5 mm PIPES (pH 8.0), 85 mm KCl, 0.5% Nonidet P-40, and protease inhibitor mixture. Cell nuclei were pelleted and resuspended in buffer containing 50 mm Tris-Cl (pH 8.1), 10 mm EDTA, 1% SDS, and protease inhibitor mixture. Soluble chromatin was prepared by sonication and diluted in buffer containing 16.7 mm Tris-HCl (pH 8.1), 0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 167 mm NaCl, and protease inhibitor mixture. Immunoprecipitates were prepared with rat anti-VDR antibody or rat IgG (Sigma). 24-Hydroxylase (CYP24) was used as a positive control. DNA was extracted from the immunocomplexes using a QIAquick spin kit. 2 μl (of 30 μl) of the DNA extracts were used for PCR with primers: VDRE1, forward, 5′- ATGATTCCTAGCTAGCGAGATATAGATGATATCAGA-3′; reverse, 5′- AACCATAGTCTTCCCCCGGGTGGGTGTGAATTCCCAGT-3′; VDRE2, forward, 5′-ACTGGAACCTTGCCTGGCCA-3′; reverse, 5′-GCCAGGCCCTCATTCTGTTA-3′; CYP24, forward, 5′-CAGACGCGGCAGCTTTTCTG-3′; reverse, 5′-CGTTTCCTCCTGTCCCTCTC-3′. PCR products were resolved on a 2% agarose gel and stained with ethidium bromide.
Apoptosis and Methylthiazole Tetrazolium Assays
To determine the apoptotic index, cells were treated with EtOH or 10−7 m 1,25(OH)2D3 for 6 days, harvested by trypsin digestion, and washed with phosphate-buffered saline. Cell suspensions were incubated with phycoerythrin (PE) and 7-amino-actinomycin according to manufacturer's protocol (PE Annexin V kit, BD Biosciences). Flow cytometry was performed in a FACScan (BD Biosciences). For each data point, duplicate samples were analyzed, and the experiment was reproduced three times.
To analyze cell growth, methylthiazole tetrazolium assays were performed as described previously (32). Cells were plated at 10 × 104 in 96-well plates and treated with EtOH or 10−7 m 1,25(OH)2D3 for the indicated times. Absorptions at 595 nm were measured with a MRX microplate reader (DYNEX Technologies, Chantilly, VA).
Xenograft Model
OV2008-Luciferase cells stably expressing control (control-bSP) or miR-498 (miR-498-bSP) sponges were harvested and resuspended in DMEM medium containing 5% FCS with Matrigel (1:1) at a concentration of 5 × 106 cells per 100 μl, which was delivered by intraperitoneal injections into the peritoneal cavity of 6-week-old female athymic nu/nu mice (Harlan Sprague-Dawley, Indianapolis, IN). The mice were fed with a vitamin D-deficient diet supplemented with 0.47% calcium (Harland Teklad, Madison, WI) for the duration of the study. Mice were randomized and treated every other day by gavage with either EtOH or EB1089 at 0.5 μg/kg body weight diluted in sesame oil in a volume of 50 μl. The treatments started on the day following cell inoculation. Mice body weights were monitored every 5 days over the course of treatment. Tumor growth and invasion were monitored with IVIS-200 Live imaging system at 15 and 30 days. After 30 days of treatment, mice were sacrificed, and tumor weight, number, and extent of overt metastases were quantified. Metastases were defined as visible tumor nodules >1 mm in diameter. Tumors were fixed in 10% neutral-buffered formalin for histological analysis. Serum calcium levels were measured as described previously (33). Mice studies were carried out according to the procedures approved by the Institutional Animal Care and Use Committee at University of South Florida.
Bioluminescence Imaging with IVIS
Mice were anesthetized with isoflurane inhalation and subsequently injected (intraperitoneal) with 100 μl of 15 mg/ml D-luciferin (Xenogen). Bioluminescence imaging with a CCD camera (IVIS-200 series, Xenogen) was initiated 5 min after luciferin injection with imaging times ranging from 1 to 2 min, depending on the amount of luciferase activity. Bioluminescence from the region of interest was defined manually, and the data were expressed as photon/s/cm2/steradian. Background photon counts were defined as the region of interest from a mouse that was not given an intraperitoneal injection of D-luciferin. All bioluminescent data were collected and analyzed using Living Imaging software 3.2.
Immunohistochemical Analyses
The proliferative and apoptotic indices of OV2008 xenograft tumors were determined as previously described (26). Briefly, tumors were fixed in formalin, embedded in paraffin, and sectioned at 5 μm. Tumor proliferation was assessed by Ki-67 immunostaining. Apoptotic cells in tumor sections were detected by in situ terminal deoxynucleotide transferase-mediated dUTP nick end labeling (TUNEL) using the Apoptag peroxidase in situ apoptosis detection kit (Serologicals). Apoptotic and proliferative indices were calculated by dividing the total number of cells with the number of TUNEL and Ki-67-positive cells, respectively, in 10 randomly selected microscopic (40× objective) fields.
RESULTS
Identification of MiR-498 as a Primary Target Gene for 1,25(OH)2D3
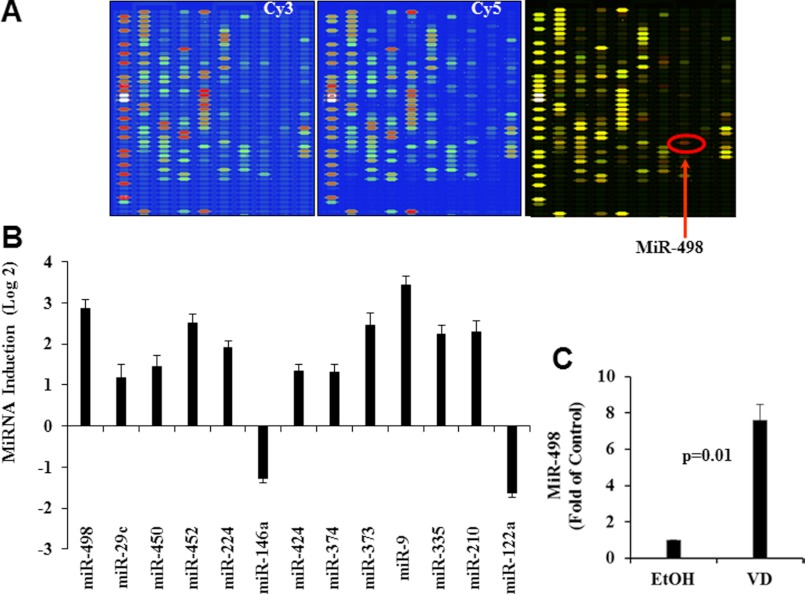
Our published studies showed that 1,25(OH)2D3 decreased hTERT mRNA stability (26), opening the possibility for miRNA involvement in hTERT regulation. To identify miRNA target genes for 1,25(OH)2D3, miRNA microarray analyses were performed in OVCAR3 cells treated with ethanol (EtOH) or 1,25(OH)2D3 for 6 days. Of 327 miRNA elements represented on the array, 190 miRNAs were expressed above background levels. Multiple miRNAs were increased (red color in the overlay image) by 1,25(OH)2D3 treatment, including miR-498 (red arrow) (Fig. 1, A and B). Quantification of the signal intensity of the miR-498 after normalization with LOWESS filter (Fig. 1C) revealed that miR-498 was significantly (n = 3, p = 0.01) increased (up to ∼7-fold) by 1,25(OH)2D3. The microarray analyses identified miR-498 as a potential target gene for the hormone.
FIGURE 1.
Microarray analyses identify a group of miRNA target genes for 1,25(OH)2D3. Total RNAs were isolated from OVCAR3 cells treated with EtOH or 10−7 m 1,25(OH)2D3 (VD) for 6 days labeled with Cy3 and Cy5, respectively, and subjected to independent miRNA microarray analyses. A, ChIP images from a representative region with pseudo-colored fluorescent signals are shown. The overlay image of Cy3/Cy5 is presented, and genes unchanged, increased, and decreased by VD were shown in yellow, red, and green colors, respectively. MiR-498 is indicated by a red arrow. B, VD miRNA target genes with Log2 values greater than one (relative to vehicle control) are presented as bar graphs. C, miR-498 signal intensity was quantified, and values are expressed as -fold change (mean ± S.D., n = 3). Statistical analyses were performed with Student's t test.
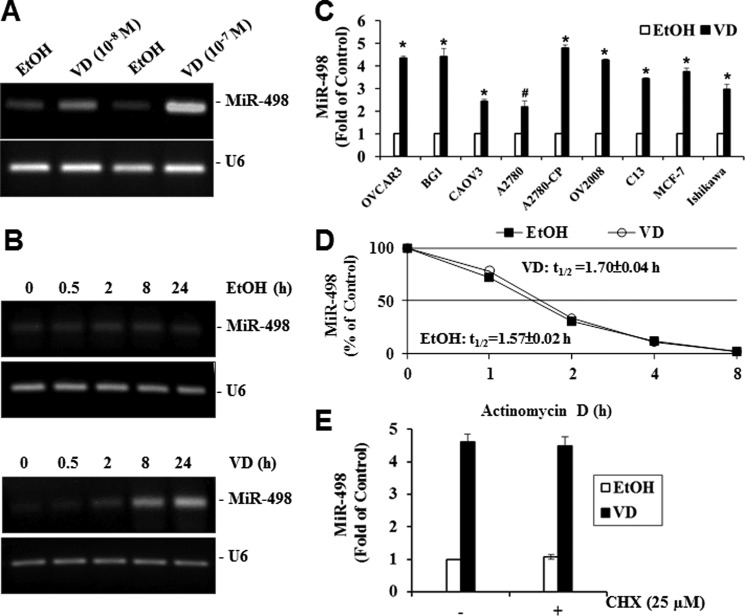
To validate the array data, OVCAR3 cells were treated with different concentrations of 1,25(OH)2D3 for 24 h, and then miR-498 expression levels were determined by RT-PCR. As shown in Fig. 2A, miR-498 was increased in a dosage-dependent manner by 1,25(OH)2D3. A subsequent time course study (Fig. 2B) showed that miR-498 was induced within 30 min by 10−7 m 1,25(OH)2D3, and the level of induction appeared maximized around 24 h, suggesting that miR-498 may be an immediate early response gene. In parallel analyses, the vehicle did not change MiR-498 levels, and U6 expression was not altered by 1,25(OH)2D3, showing that the induction is specific to miR-498. Quantitative RT-PCR analyses detected miRNA-498 induction by 1,25(OH)2D3 in multiple vitamin D-sensitive human OCa as well as breast (MCF-7) and endometrial (Ishikawa) cancer cells (Fig. 2C), showing that the regulation occurs in many human cancer types.
FIGURE 2.
1,25(OH)2D3 increases miR-498 expression in OVCAR3 cells. A, shown is dose-dependent induction of miR-498 expression by VD. OVCAR3 cells were treated with EtOH or VD at the indicated concentrations for 24 h. Total RNA was extracted and subjected to RT-PCR analyses. B, time-dependent induction of miR-498 expression by VD is shown. Total RNA was extracted from OVCAR3 cells treated with EtOH or 10−7 m VD for the indicated times. RT-PCR was performed as in panel A. C, VD induces miR-498 in multiple cancer cell lines. Cells were treated with EtOH or 10−7 m VD for 6 days. Total RNAs were extracted and subjected to quantitative RT-PCR. The levels of miR-498 were normalized with corresponding U6 levels and expressed as -fold of the EtOH controls (*, p ≤ 0.0001; #, p = 0.0004). D, lack of a VD effect on miR-498 stability is shown. OVCAR3 cells were treated with EtOH or 10−7 m VD for 24 h followed by subsequent treatment with actinomycin D (5 μg/ml) for the indicated times. The levels of miR-498 and U6 were determined by quantitative RT-PCR. The miR-498 levels were normalized with corresponding U6 levels and presented as percentage of the miR-498 level of time 0. E, shown is the effect of cycloheximide (CHX) on the induction of miR-498 by VD. OVCAR3 cells were treated with EtOH or 10−7 m VD for 24 h in the presence or absence of 25 μm of cycloheximide. Quantitative RT-PCR was performed as in panel C, and miR-498 levels were normalized with U6 and expressed as -fold of the vehicle control. Values are presented as the mean ± S.D. (n = 3 in panels C–E). Statistical analyses were performed with Student's t test.
To test whether the miR-498 induction is due to RNA stabilization, we measured the half-life (t½) of miR-498 in the presence of RNA synthesis inhibitor, actinomycin D, by quantitative RT-PCR. As shown in Fig. 2D, the half-life of miR-498 was essentially the same in cells treated with 1,25(OH)2D3 or vehicle. Comparison of MiR-498 induction by 1,25(OH)2D3 in OVCAR3 cells treated with or without protein synthesis inhibitor, cycloheximide, demonstrated that the induction did not require new protein synthesis (Fig. 2E), identifying miR-498 as a primary target gene for the hormone.
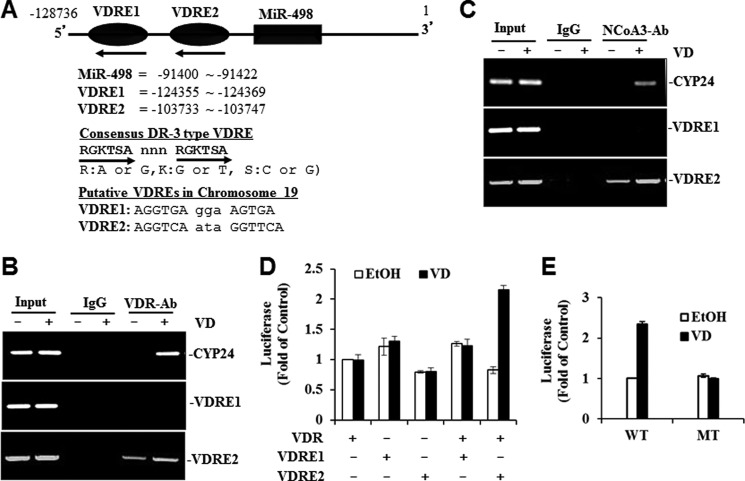
Sequence analyses of the miR-498 locus of human chromosome 19 clone CTC-360P9 (GenBankTM accession number AC011456) revealed the presence of two putative DR3-type VDREs, namely VDRE1 and VDRE2 (Fig. 3A), which matched the consensus VDRE (34) and the VDRE sequence of known target genes such as osteopontin (35), osteocalcin (36), and GADD45 (24). In EMSA assays, the recombinant receptor proteins and the VDRE2, but not VDRE1, probe formed a complex (data not shown). The complex formation was specific for the VDR-RXR heterodimer as neither VDR nor RXR alone formed a detectable complex. This specific DNA oligo-protein complex was reduced by a 100-fold excess of cold VDRE2 but not VDRE1 probe and up-shifted by a VDR-specific antibody (data not shown). Overall, the analyses suggest a specific interaction between the VDR and VDRE2 in vitro.
FIGURE 3.
A functional VDRE is identified in the upstream region of miR-498 genome. A, a schematic diagram of the human miR-498 locus in chromosome 19 shows the positions of miR-498 and the putative VDREs. The consensus DR3 sequences and the two putative miR-498 VDREs are listed. Hexameric VDRE half-sites are shown in capital letters, and the 3-bp spacer is shown in small letters. The numbering is based on Homo sapiens chromosome 19 clone CTC-360P9 (GenBankTM accession number AC011456). It should be noted that the miR-498 gene is on the strand complementary to the sequence of CTC-360P9. B and C, shown is the in vivo interaction of VDR (panel B) and co-activator (NCoA3) (panel C) with miR-498 VDRE2. OVCAR3 cells were treated with EtOH or 10−7 m VD for 60 min, and ChIP assays were performed with control (rat IgG), anti-VDR, or anti-NCoA3 antibody. PCR was performed with primers spanning CYP24 VDRE or the putative VDREs of miR-498. D, VD induces VDRE2 reporter activity. VDR-deficient HeLa cells were transfected with VDRE1 or VDRE2-Luc reporters and pCMVGal with or without p91023B-VDR. Transfected cells were treated with either 10−7 m VD or EtOH for 36 h. Luciferase activity was determined, normalized with cognate β-gal activity, and plotted as -fold of control. Values are presented as the mean ± S.D. (n = 3). E, mutational analyses of the VDRE2 reporter is shown. The second half-site of the VDRE2 was mutated as described under “Experimental Procedures.” The wild type (WT) and mutant (MT) VDRE2 reporters were transiently transfected into OVCAR3 cells. The cells were treated, and reporter activities were measured as described in panel D and presented as the mean ± S.D. (n = 3).
To find out whether the VDRE2 interacts with the receptors in vivo, ChIP assays were conducted in OVCAR3 cells. As shown in Fig. 3, B and C, antibodies against the VDR and its co-activator NCoA3 precipitated genomic DNA fragments containing the CYP24 VDRE and miR-498 VDRE2, but not VDRE1, in cells treated with 1,25(OH)2D3. Reduced amounts of DNA fragments were precipitated by the VDR and NCoA3 antibodies in vehicle-treated cells, and no DNA was detected in IgG precipitates, showing that the VDR and its coactivators were recruited to the miR-498 genomic DNA fragments containing VDRE2 and the recruitments induced by 1,25(OH)2D3.
To further validate the functionality of the VDREs of miR-498, the ability of VDRE1 and VDRE2 to mediate the activation of luciferase reporters was tested in HeLa cells that contain little endogenous VDR. In the reporter assays, VDRE2 reporter activity was increased by the hormone in a VDR-dependent manner, whereas VDRE1 reporter activity did not respond to 1,25(OH)2D3 treatment or VDR expression (Fig. 3D). More importantly, the mutation of the VDRE2 at two key nucleotides of the second half-site eliminated the induction by 1,25(OH)2D3 (Fig. 3E), showing that the VDRE2 in the miR-498 genome is functional.
Establishment of MiR-498 as the Mediator for hTERT Down-regulation by 1,25(OH)2D3
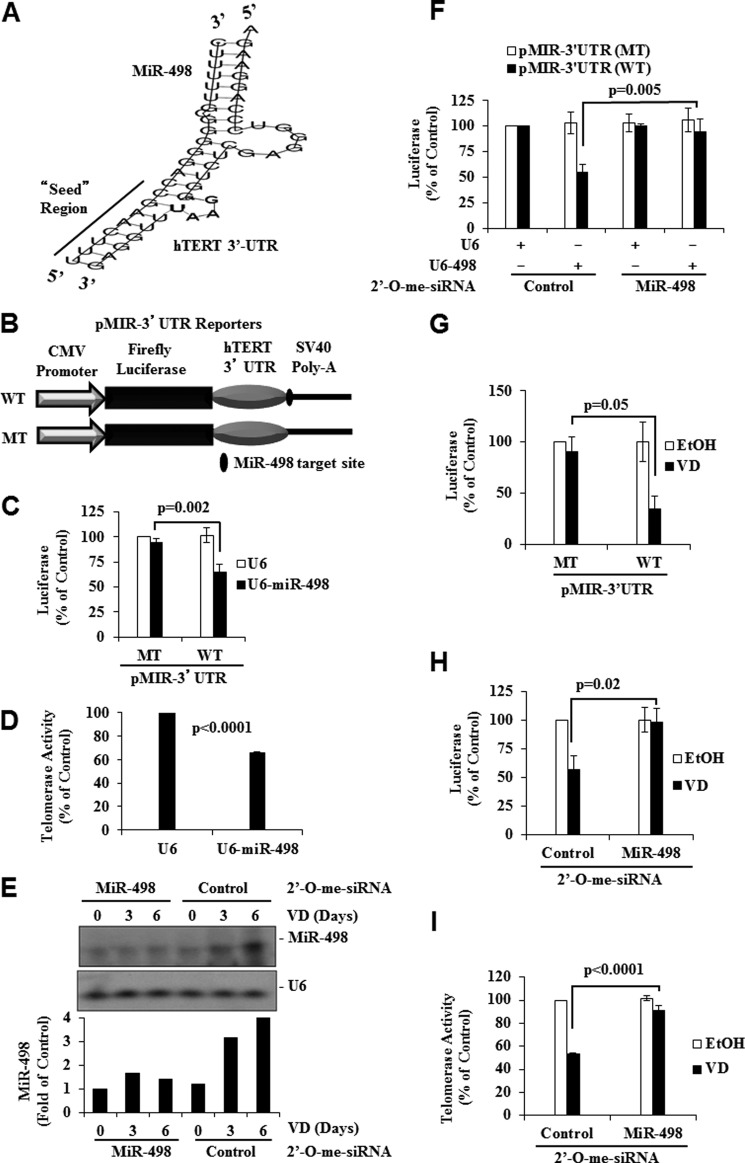
Sequence analyses revealed the presence of a potential miR-498 target site in the 3′-UTR of hTERT (nucleotides 21,800–21,831 of clone AF128894). Sequence alignment showed a complementary pairing in the “seed” region of miR-498 (Fig. 4A) and the 3′ “compensatory” site, which has the ability to compensate for mismatches in the seed region (37). The sequence analyses indicate that hTERT mRNA may be a direct target of miR-498. To test this idea, we constructed luciferase reporter genes (Fig. 4B) in which the hTERT 3′-UTR region with (pMIR-3′-UTR-WT) or without (pMIR-3′-UTR-MT) the putative miR-498 target site was inserted between luciferase cDNA and the poly(A) tail of the pMIR vector and tested its down-regulation by ectopic miR-498 expression in HeLa cells. As shown in Fig. 4C, the activity of the wild type reporter, not the mutant reporter devoid of the target site, was decreased by ectopic miR-498 expression. Consistent with the reporter data, ectopic miR-498 expression in OVCAR3 cells decreased the telomerase activity in telomeric repeat amplification protocol assays (Fig. 4D).
FIGURE 4.
hTERT 3′-UTR contains a target site for MiR-498 and is targeted by VD. A, shown is sequence alignment of human miR-498 with the 3′-UTR region of hTERT showing the complementary match between the seed sequence at the 5′ end of miR-498 and hTERT 3′-UTR. B, a schematic presentation of the reporters constructed with the hTERT 3′-UTR (WT) or the 3′-UTR lacking miR-498 target site (MT) in the pMIR backbone (pMIR-3′-UTR Reporters). C, shown is the effect of ectopic miR-498 on pMIR-3′-UTR reporters. HeLa cells were transfected with control (U6) or miR-498 expression vector (U6-miR-498) together with pMIR-hTERT 3′-UTR reporters and pCMVGal. Forty-eight hours later, luciferase activity was determined, normalized with cognate β-gal activity, and plotted as percentage of U6 control. D, OVCAR3 cells were transfected with control (U6) or the miR-498 expression vector (U6-MiR-498). Forty-eight hours later, cells were extracted and equal amounts of proteins subjected to PCR-telomeric repeat amplification protocol assays. E, knockdown of miR-498 by 2′-O-me-siRNA is shown. OVCAR3 cells were transfected with 2′-O-me-siRNA oligonucleotides targeting miR-498 (2′-O-me-MiR-498) or 2′-O-me-scrambled miRNA (2′-O-me-control). Transfected cells were treated with EtOH or 10−7 m VD. RPAs were performed, and MiR-498 was quantified by phosphorimaging and normalized with U6. F, shown is the effect of 2′-O-me-siRNA against miR-498 on the ability of ectopic miR-498 to decrease hTERT UTR reporters. HeLa cells were transfected as in C but with 2′-O-me-miR-498 or 2′-O-me-control. Luciferase activities were determined, normalized with cognate β-gal activity, and plotted as percentage of controls. G, shown is the effect of VD on pMIR-3′-UTR reporters. OVCAR3 cells were transfected with pMIR-hTERT 3′-UTR reporters and pCMVGal. Transfected cells were treated with EtOH or 10−7 m VD for 3 days. Luciferase activity was determined, normalized with cognate β-gal activity, and plotted as percentage of vehicle control. H, shown is the effect of 2′-O-me-siRNA against miR-498 on the ability of VD to decrease hTERT UTR reporters. OVCAR3 cells were transfected as in G but with 2′-O-me-miR-498 or 2′-O-me-control. Transfected cells were treated with EtOH or 10−7 m VD for 3 days. Luciferase activity was determined, normalized with cognate β-gal activity, and plotted as percentage of vehicle control. I, shown is the effect of 2′-O-me-siRNA against miR-498 on the ability of VD to decrease telomerase activity. OVCAR3 cells were transfected and treated as H. Cellular extracts with equal amounts of protein were subjected to PCR- telomeric repeat amplification protocol assays. Values are presented as the mean ± S.D. Statistical analyses were performed with Student's t test (n = 3 in panels C, D, and F–I).
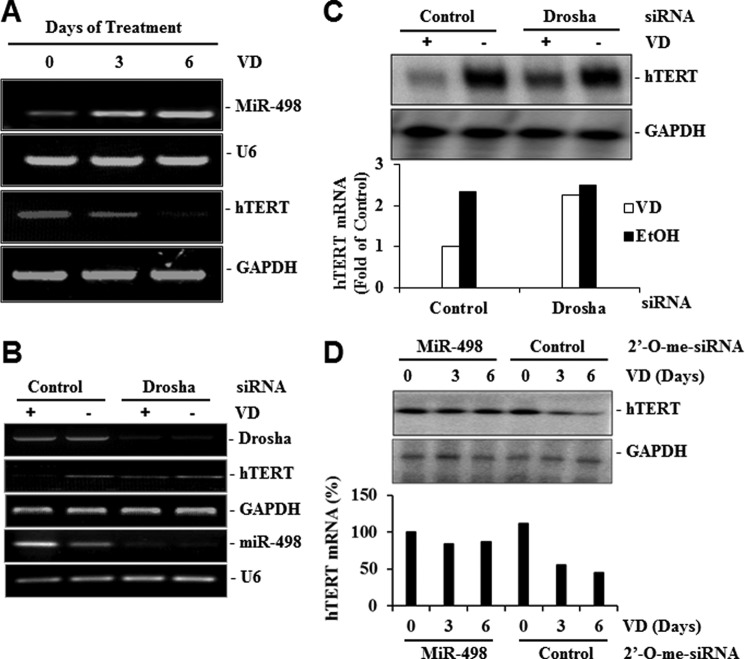
To test whether miR-498 indeed targets hTERT mRNA, 2′-O-methylated siRNA oligonucleotides against miR-498 were synthesized, and their effect on miR-498 expression was tested in RPAs developed for quantitative miR-498 measurement. The probe used in the RPAs was four nucleotides longer than miR-498, so the product of the RPAs was easily distinguishable from the undigested probe. The assay detected the mature miR-498 induced by 1,25(OH)2D3, which was diminished by the synthetic miR-498 siRNA oligos without an effect on U6 expression (Fig. 4E). The scrambled control siRNA did not alter the expression of either miR-498 or U6. Subsequent reporter assays showed that the miR-498 siRNA, but not the scrambled control oligos, diminished miR-498-induced decrease in the activity of the luciferase reporter gene containing wild type hTERT 3′-UTR but not the mutant reporter lacking the putative miR-498 target site (Fig. 4F). Consistently, 1,25(OH)2D3 decreased the activity of the wild type but not the mutant reporter (Fig. 4G), and the ability of 1,25(OH)2D3 to decrease the wild type hTERT reporter (Fig. 4H) and the telomerase activity (Fig. 4I) was compromised by the miR-498 siRNA but not the scrambled control oligos.
Consistent with a role for miR-498 in mediating the effect of 1,25(OH)2D3 on hTERT mRNA, RT-PCR analyses revealed an inverse correlation between hTERT mRNA and miR-498 in response to 1,25(OH)2D3 treatments (Fig. 5A) and Drosha siRNA diminished both miR-498 induction and hTERT down-regulation by 1,25(OH)2D3 (Fig. 5, B and C). The MiR-498 siRNA, but not the scrambled control, blunted a 1,25(OH)2D3-induced decrease in hTERT mRNA expression (Fig. 5D). These analyses establish miR-498 as the primary target gene that mediates hTERT down-regulation by 1,25(OH)2D3.
FIGURE 5.
MiR-498 mediates the hTERT down-regulation by VD. A, inverse correlation between hTERT and miR-498 regulation by VD is shown. OVCAR3 cells were treated with EtOH or 10−7 m VD for the indicated days, and miR-498, U6, and hTERT mRNA levels were detected by RT-PCR. B, shown is the effect of Drosha knockdown on the regulation of miR-498 and hTERT by VD. OVCAR3 cells were transfected with control or Drosha siRNA (20 μm). Forty-eight hours later, cells were treated with EtOH or 10−7 m VD for 3 days. Total RNAs were extracted, and RT-PCR analyses were performed. C, RT-PCR products of hTERT and GAPDH from panel B were blotted to nitrocellulose membrane, detected by Southern blot analyses, and quantified by phosphorimaging. The hTERT signals were normalized with cognate GAPDH signals and presented as -fold of the control. D, MiR-498 knockdown diminishes hTERT mRNA down-regulation by VD. OVCAR3 cells were transfected with 2′-O-me-MiR-498 or 2′-O-me-control and treated with 10−7 m VD or EtOH for the indicated times. The hTERT mRNA levels were determined as in panel C and presented in bar graphs as percentage of the time zero. GAPDH was included as a loading control.
In Vitro and in Vivo Role of MiR-498 in Ovarian Tumor Suppression by 1,25(OH)2D3 and Its Synthetic Analog EB1089
Our published studies have shown that 1,25(OH)2D3 suppressed the growth of OCa cells through multiple mechanisms and that the growth suppression is partly mediated through hTERT down-regulation (26). To assess the contribution of miR-498 to overall growth suppression by 1,25(OH)2D3, control and miR-498 sponges (Fig. 6A) were designed and stably expressed in OVCAR3 cells. As shown in Fig. 6, the miR-498 sponges, either standard or bulged, diminished the apoptosis induced by 1,25(OH)2D3 (Fig. 6B) and partially relieved the overall growth suppression (Fig. 6C). This shows that miR-498 is an important mediator of 1,25(OH)2D3 in suppressing OCa growth.
FIGURE 6.
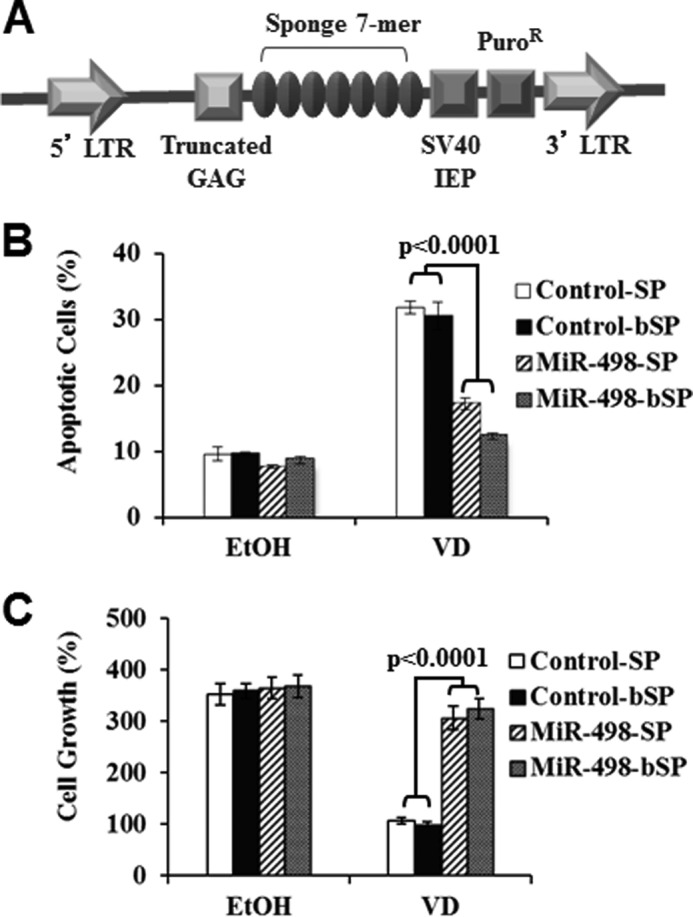
MiR-498 contributes to apoptosis induction by VD. A, a schematic presentation of pBABE plasmid expressing control or miR-498 sponges is shown. LTR, long term repeat; IEP, immediate early promoter. B, OVCAR3 cells were stably transfected with control or miR-498 sponges, treated with 10−7 m VD or EtOH for 6 days, and assayed for apoptosis by phycoerythrin annexin V method. The data were reproduced three times. SP, sponges. C, OVCAR3 cells stably transfected with control or miR-498 sponges were treated with 10−7 m VD or EtOH for 6 days. Cell growth was assessed by methylthiazole tetrazolium assays. For each data point, triplicate samples were analyzed, and the experiment was reproduced three times.
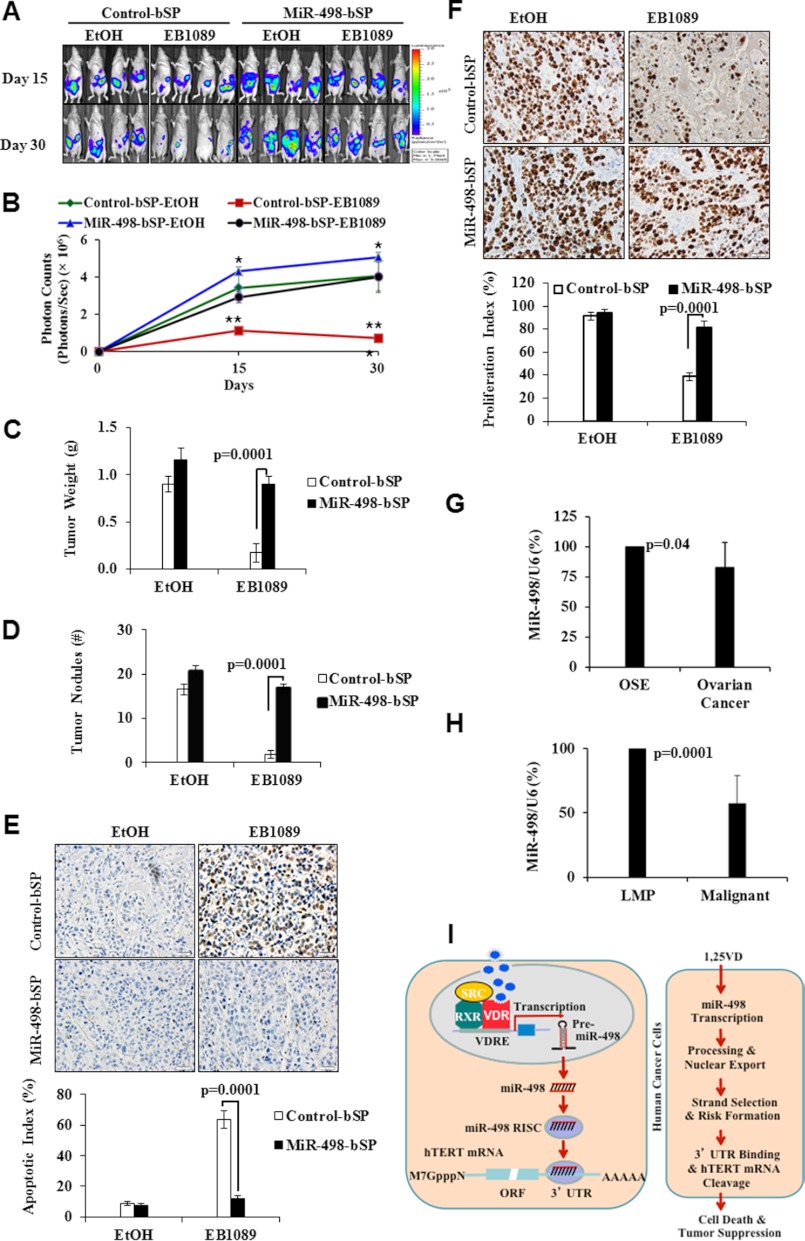
To investigate the role of miR-498 in the antitumor activity of 1,25(OH)2D3 compounds in vivo, OV2008 cells stably transfected with the firefly luciferase gene together with either the control or miR-498 sponges were injected into the peritoneal cavity of nu/nu mice and treated with a synthetic 1,25(OH)2D3 analog EB1089. As shown in Fig. 7, A and B, the growth of tumors expressing control sponges was significantly reduced by EB1089. The expression of miR498 sponges enhanced the basal growth of tumors treated with the vehicle, indicating a role of basal miR-498 expression in suppressing ovarian tumor growth. More importantly, miR-498 sponges significantly diminished the suppressive effect of EB1089, causing an increase in both tumor volumes (Fig. 7C) and nodules of tumors invading the abdominal wall (Fig. 7D).
FIGURE 7.
MiR-498 contributes to in vivo ovarian tumor suppression by a synthetic VD analog EB1089. A–F, EB1089 inhibits invasive ovarian tumor growth in vivo through miR-498. OV2008-Luc cells (5 × 106) expressing control or miR-498 sponges were mixed with Matrigel and injected intraperitoneally into female athymic nu/nu mice. Mice bearing OV2008 tumor xenografts were treated every other day with EB1089 (0.5 μg/kg) or vehicle (EtOH) in sesame oil. After 15 and 30 days of treatments, tumor growth was monitored by bioluminescent imaging (panel A). The bioluminescence intensity (photons/sec/cm2/steradian), a measurement of viable tumor volume, was presented as the mean ± S.D. (n = 8) (panel B). Statistical analyses were done with Student's t test. *. p = 0.05 (control-bSP-EtOH versus miR-498-bSP-EtOH); **, p = 0.0001; ***, p = 0.0002 (control-bSP-EB1089 versus miR-498-bSP-EB1089). Tumor weight (cumulative weight of tumor tissues) (panel C) and the number of tumor nodules (>1 mm in diameter) (panel D) from mice treated with either EtOH or EB1089 for 30 days are presented as the mean ± S.D. Apoptotic (panel E) and proliferation (panel F) indices of OV20008 tumors treated with either EtOH or EB1089 for 30 days were determined by tissue TUNEL assays and Ki-67 staining, respectively. G and H, small RNAs were extracted from immortalized human OSE (n = 4) and OCa (n = 8) cells (panel G) or tissues of frankly malignant human ovarian tumors (n = 9) and tumors of low malignant potential (LMP) (n = 5) (panel H). miR-498 and U6 expression levels were determined by RPA analyses and quantified by phosphorimaging. The averages of quantified miR-498 levels after being normalized with cognate U6 are shown as bar graphs. Statistical analyses were done with Student's t test. I, a model shows how VD induces tumor suppression and apoptosis through miRNA-498 induction and hTERT down-regulation. See the discussion section for details.
Tumor TUNEL assays (Fig. 7E) and Ki-67 immunostaining (Fig. 7F) showed that the percentage of apoptotic cells in tumors expressing control sponges was significantly increased, whereas the percentage of Ki-67-positive cells decreased by EB1089. In tumors expressing miR-498 sponges, the effects of EB1089 on both cell apoptosis and proliferation were diminished. The data suggest that the mechanistic information obtained in cell line studies is translatable into tumor suppression in vivo. The body weight of mice was not affected by the treatment with EB1089, and serum calcium levels remained in the normal range during EB1089 treatment (data not shown). These analyses show that EB1089 inhibits invasive ovarian tumor growth and that miR-498 is an important mediator of its antitumor activity.
Consistent with its role in the suppression of ovarian tumor growth in vitro and in vivo, miR-498 was expressed at reduced levels in a panel of human OCa cells when compared with non-tumorigenic OSE cells (Fig. 7G). Analyses of human tumor samples with the same assay revealed a significantly decreased miR-498 expression in frankly malignant human ovarian tumors in comparison to tumors of low malignant potential (Fig. 7H). Together with the fact that hTERT expression and telomerase activity are known to be higher in malignant tumor cells, the analyses support the existence of an overall inverse relationship between miR-498 and hTERT expression. The data also implicate miR-498 as a potential marker to monitor ovarian tumor progression.
DISCUSSION
These studies are the first to define a role for a miRNA in regulating hTERT mRNA stability in response to a natural compound. Overall, the studies support a model of 1,25(OH)2D3 actions involving a miRNA in cancer cells as depicted in Fig. 7I. 1,25(OH)2D3 induces the activation of VDR-RXR heterodimer and its binding to the functional VDRE located in the 5′ regulatory region of miR-498 gene to increase its production through recruitment of coactivators. After processing, nuclear export, strand selection, and assembling into RNA-induced silencing complex, the mature miR-498 binds to the target site in the 3′-UTR of hTERT mRNA, resulting in its degradation. The miRNA-mediated decrease in hTERT expression contributes to cell death and the growth suppression of human tumor cells by 1,25(OH)2D3. Although the majority of the data were collected in ovarian tumor cells, the induction was also detected in other human tumor cells such as cells of human breast and endometrial cancers. Consistently, the down-regulation of hTERT by 1,25(OH)2D3 was also observed in cancer cells other than OCa (Fig. 2C). The analyses indicate that the 1,25(OH)2D3 action through miR-498 to control telomerase is not limited to OCa and can be generalized to other vitamin D-sensitive human cancers. Given the important role of telomerase in tumor cell growth and survival, targeted delivery of miR-498 or its induction by a nutraceutical may represent a new strategy for targeted telomerase inhibition to treat various human cancers.
It was originally thought that miRNAs mainly work through translational inhibition due to imperfect pairing between miRNAs and their target sites. Because the pairing between miR-498 and the complementary region in hTERT 3′-UTR is imperfect, it might thus appear more reasonable to expect a translational inhibition of hTERT by miR-498 instead of mRNA degradation. It becomes increasingly clear now that miRNAs can work through translational inhibition, mRNA degradation, or both (38, 39). There are numerous examples for miRNA-induced mRNA degradations in different species. As a matter of fact, the realization that miRNA can promote mRNA degradation has allowed the identification of miRNA targets using mRNA microarrays (40). Contradictory to early belief that perfect pairing is important for small RNA-mediated mRNA cleavage and degradation, the present consensus is that, except the match in the seed region, the extent of miRNA pairing to complementary region of the target mRNA often does not necessarily play an essential role in determining the ability of miRNA to decrease target mRNA molecules (9). Nevertheless, it is important to point out that the present studies do not exclude the involvement of translational inhibition in hTERT down-regulation by miR-498. It remains possible that miR-498 works though both mechanisms to exert its negative effect or that the mRNA degradation revealed in the current studies is a consequence of blocked translation. Although degradation can be uncoupled from translation, it is often the case that the increased mRNA degradation by miRNAs is because of deadenylation, decapping, and exonucleolytic digestion of the mRNA (41) instead of argonaute-catalyzed mRNA cleavage as for siRNA.
OCa is the fifth leading cause of cancer death among women in the Western world and has the highest mortality rate of all gynecological cancers (42), primarily due to the fact that patients relapse quickly from cisplatin-based chemotherapy. Published studies have shown that 1,25(OH)2D3 and its synthetic analogs suppress OCa cell growth (24, 25) and induce apoptosis (26, 33), raising the potential of using synthetic vitamin D analogs as an alternative or complementary method for OCa intervention (43). Published studies have identified EGF receptor (44) and GADD45 (24) as primary target genes for 1,25(OH)2D3 to induce cell cycle arrests in OCa cells. The present studies define a functional VDRE in the miR-498 loci and identified miR-498 as a primary and immediate early response gene to mediate the cell death induction by the hormone. EMSA assays show that miR-498 regulation is mediated through a VDR-RXR heterodimer. ChIP assays demonstrate that in vivo binding of VDR to the VDRE is ligand-induced, which also occurred in CYP24 VDRE as well as the functional VDREs in GADD45(24) and osteopontin promoter (45). To the best of our knowledge, miR-498 is the first miRNA primary target gene identified to mediate the effects of 1,25(OH)2D3 on OCa cells. The studies lay a foundation for continued studies to fully define the mechanisms of vitamin D action in OCa cells and to test 1,25(OH)2D3 and its analogs either alone or in combination with other therapeutic modalities for OCa intervention.
miRNA molecules can act as either a tumor suppressor or an oncogene (46, 47) and are frequently deregulated in cancers (48) through different mechanisms, including transcriptional deregulation, epigenetic alterations, dysfunction of key proteins in the miRNA biogenesis pathway, mutation, and DNA copy number abnormalities. So far, literature information for miR-498 in cancers is very limited except that its expression is reported to correlate with the probability of recurrence-free survival by multivariate analysis in stage II colon cancers (49) and that it is one of many miRNAs identified in profiling as being differentially expressed in cancer cells and tissues (50). The present studies suggest that miR-498 is down-regulated in malignant ovarian tumors. Together with its negative effect on telomerase and cell survival, miR-498 appears to function as a tumor suppressor. The concept is consistent with the fact that the miR-498 gene is located in chromosome 19q13.42, a region in which loss of heterozygosity has been reported in human ovarian carcinoma (51). In addition, chromosome 19 is reported to be most frequently deleted in OCa (52), and high hTERT expression or telomerase activity is associated with poor prognosis in most human malignancies (53) including OCa (54). A recent study of exome capture and sequencing analyses of DNA isolated from more than 300 serous OCa samples revealed hTERT as one of the new tightly localized amplification peaks (55). Overall, the information suggests that the imbalance between miR-498 and telomerase may play an important role in ovarian tumorigenesis.
Besides cancer cells, high telomerase activity is also essential for the functionality of embryonic stem cells. Because the VDR is known to be expressed in embryonic stem cells (56), the induction of miR-498 by 1,25(OH)2D3 in embryonic stems cells appears possible, which may have profound implications in aging and lifespan regulation. Interestingly, miR-498 belongs to a cluster of primate-specific miRNA molecules absent in mice and rats (57), and the miR-498 target sequence in hTERT also appears missing in the 3′-UTR of mouse TERT. Although the selective pressure and evolution advantages are at present unclear, the gain of miR-498 expression in primates is likely to contribute to their complex telomerase biology and regulation. It is established that, in many aspects, telomere biology is different between mice and humans, with the former having longer telomere length and higher telomerase activity detected in somatic cells. The establishment of a role of a primate-specific miRNA in telomerase expression and its regulation by vitamin D advances the current understanding about species-specific telomerase regulation, which is an important issue to be considered when using rodents in modeling the human aging process and related diseases.
Acknowledgments
We thank Dr. Kenneth S. Korach for BG1 and Dr. Jeanne Becker for Ishikawa cells. The IVIS mouse imaging was carried in the Small Animal Imaging Lab (SAIL), and the immunohistochemistry was performed by the Tissue Core facility at H. Lee Moffitt Cancer Center.
This work was supported, in whole or in part, by National Institutes of Health Grants CA111334 and CA93666 (Public Health Service). This work was also supported by an idea grant from Breast Cancer Research Program of the United States Department of Defense Grant BC085205 and a program development grant from Ovarian Cancer Research Fund.
- miRNA
- microRNA
- hTERT
- human telomerase reverse transcriptase
- 1,25(OH)2D3
- 1,25-dihydroxyvitamin D3
- RXR
- retinoid X receptor
- VDR
- vitamin D (VD) receptor
- VDRE
- VD-responsive element
- MiR-498
- microRNA-498
- 2′-O-me
- 2′-O-methyl
- RPA
- ribonuclease protection assay
- U6
- RNU6
- bSP
- bulged sponge.
REFERENCES
- 1. Blackburn E. H. (2001) Switching and signaling at the telomere. Cell 106, 661–673 [DOI] [PubMed] [Google Scholar]
- 2. Harrington L., McPhail T., Mar V., Zhou W., Oulton R., Bass M. B., Arruda I., Robinson M. O. (1997) A mammalian telomerase-associated protein. Science 275, 973–977 [DOI] [PubMed] [Google Scholar]
- 3. Blasco M. A., Funk W., Villeponteau B., Greider C. W. (1995) Functional characterization and developmental regulation of mouse telomerase RNA. Science 269, 1267–1270 [DOI] [PubMed] [Google Scholar]
- 4. Cohen S. B., Graham M. E., Lovrecz G. O., Bache N., Robinson P. J., Reddel R. R. (2007) Protein composition of catalytically active human telomerase from immortal cells. Science 315, 1850–1853 [DOI] [PubMed] [Google Scholar]
- 5. Venteicher A. S., Abreu E. B., Meng Z., McCann K. E., Terns R. M., Veenstra T. D., Terns M. P., Artandi S. E. (2009) A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 323, 644–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang S., Zhu J. (2003) Evidence for a relief of repression mechanism for activation of the human telomerase reverse transcriptase promoter. J. Biol. Chem. 278, 18842–18850 [DOI] [PubMed] [Google Scholar]
- 7. Smith L. L., Coller H. A., Roberts J. M. (2003) Telomerase modulates expression of growth-controlling genes and enhances cell proliferation. Nat. Cell Biol. 5, 474–479 [DOI] [PubMed] [Google Scholar]
- 8. Liu K., Hodes R. J., Weng N. (2001) Cutting edge. Telomerase activation in human T lymphocytes does not require increase in telomerase reverse transcriptase (hTERT) protein but is associated with hTERT phosphorylation and nuclear translocation. J. Immunol. 166, 4826–4830 [DOI] [PubMed] [Google Scholar]
- 9. Bartel D. P. (2009) MicroRNAs. Target recognition and regulatory functions. Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee Y., Kim M., Han J., Yeom K. H., Lee S., Baek S. H., Kim V. N. (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23, 4051–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rodriguez A., Griffiths-Jones S., Ashurst J. L., Bradley A. (2004) Identification of mammalian microRNA host genes and transcription units. Genome Res. 14, 1902–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maniataki E., Mourelatos Z. (2005) A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 19, 2979–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. He L., Hannon G. J. (2004) MicroRNAs. Small RNAs with a big role in gene regulation. Nat. Rev. Genet 5, 522–531 [DOI] [PubMed] [Google Scholar]
- 14. Lewis B. P., Burge C. B., Bartel D. P. (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 [DOI] [PubMed] [Google Scholar]
- 15. Kumar M. S., Lu J., Mercer K. L., Golub T. R., Jacks T. (2007) Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 39, 673–677 [DOI] [PubMed] [Google Scholar]
- 16. Valastyan S., Reinhardt F., Benaich N., Calogrias D., Szász A. M., Wang Z. C., Brock J. E., Richardson A. L., Weinberg R. A. (2009) A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell 137, 1032–1046 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17. Dusso A. S., Brown A. J., Slatopolsky E. (2005) Vitamin D. (2012) Am. J. Physiol. Renal Physiol. 289, F8–F28 [DOI] [PubMed] [Google Scholar]
- 18. Nagpal S., Na S., Rathnachalam R. (2005) Noncalcemic actions of vitamin D receptor ligands. Endocr. Rev. 26, 662–687 [DOI] [PubMed] [Google Scholar]
- 19. Holick M. F. (2007) Vitamin D deficiency. N. Engl. J. Med. 357, 266–281 [DOI] [PubMed] [Google Scholar]
- 20. Garland C. F., Garland F. C., Gorham E. D., Lipkin M., Newmark H., Mohr S. B., Holick M. F. (2006) The role of vitamin D in cancer prevention. Am. J. Public Health 96, 252–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Evans R. M. (1988) The steroid and thyroid hormone receptor superfamily. Science 240, 889–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tsai M. J., O'Malley B. W. (1994) Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu. Rev. Biochem. 63, 451–486 [DOI] [PubMed] [Google Scholar]
- 23. Zou A., Elgort M. G., Allegretto E. A. (1997) Retinoid X receptor (RXR) ligands activate the human 25-hydroxyvitamin D3–24-hydroxylase promoter via RXR heterodimer binding to two vitamin D-responsive elements and elicit additive effects with 1,25-dihydroxyvitamin D3. J. Biol. Chem. 272, 19027–19034 [DOI] [PubMed] [Google Scholar]
- 24. Jiang F., Li P., Fornace A. J., Jr., Nicosia S. V., Bai W. (2003) G2/M arrest by 1,25-dihydroxyvitamin D3 in ovarian cancer cells mediated through the induction of GADD45 via an exonic enhancer. J. Biol. Chem. 278, 48030–48040 [DOI] [PubMed] [Google Scholar]
- 25. Li P., Li C., Zhao X., Zhang X., Nicosia S. V., Bai W. (2004) p27(Kip1) stabilization and G(1) arrest by 1,25-dihydroxyvitamin D(3) in ovarian cancer cells mediated through down-regulation of cyclin E/cyclin-dependent kinase 2 and Skp1-Cullin-F-box protein/Skp2 ubiquitin ligase. J. Biol. Chem. 279, 25260–25267 [DOI] [PubMed] [Google Scholar]
- 26. Jiang F., Bao J., Li P., Nicosia S. V., Bai W. (2004) Induction of ovarian cancer cell apoptosis by 1,25-dihydroxyvitamin D3 through the down-regulation of telomerase. J. Biol. Chem. 279, 53213–53221 [DOI] [PubMed] [Google Scholar]
- 27. Chen C., Ridzon D. A., Broomer A. J., Zhou Z., Lee D. H., Nguyen J. T., Barbisin M., Xu N. L., Mahuvakar V. R., Andersen M. R., Lao K. Q., Livak K. J., Guegler K. J. (2005) Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33, e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Baker A. R., McDonnell D. P., Hughes M., Crisp T. M., Mangelsdorf D. J., Haussler M. R., Pike J. W., Shine J., O'Malley B. W. (1988) Cloning and expression of full-length cDNA encoding human vitamin D receptor. Proc. Natl. Acad. Sci. U.S.A. 85, 3294–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ebert M. S., Neilson J. R., Sharp P. A. (2007) MicroRNA sponges. Competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 4, 721–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Valastyan S., Weinberg R. A. (2009) Assaying microRNA loss-of-function phenotypes in mammalian cells. Emerging tools and their potential therapeutic utility. RNA Biol. 6, 541–545 [DOI] [PubMed] [Google Scholar]
- 31. Li P., Lee H., Guo S., Unterman T. G., Jenster G., Bai W. (2003) AKT-independent protection of prostate cancer cells from apoptosis mediated through complex formation between the androgen receptor and FKHR. Mol. Cell. Biol. 23, 104–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mosmann T. (1983) Rapid colorimetric assay for cellular growth and survival. Application to proliferation and cytotoxicity assays. J. Immunol. Methods 65, 55–63 [DOI] [PubMed] [Google Scholar]
- 33. Zhang X., Jiang F., Li P., Li C., Ma Q., Nicosia S. V., Bai W. (2005) Growth suppression of ovarian cancer xenografts in nude mice by vitamin D analogue EB1089. Clin. Cancer Res. 11, 323–328 [PubMed] [Google Scholar]
- 34. Toell A., Polly P., Carlberg C. (2000) All natural DR3-type vitamin D response elements show a similar functionality in vitro. Biochem. J. 352, 301–309 [PMC free article] [PubMed] [Google Scholar]
- 35. Noda M., Vogel R. L., Craig A. M., Prahl J., DeLuca H. F., Denhardt D. T. (1990) Identification of a DNA sequence responsible for binding of the 1,25-dihydroxyvitamin D3 receptor and 1,25-dihydroxyvitamin D3 enhancement of mouse secreted phosphoprotein 1 (SPP-1 or osteopontin) gene expression. Proc. Natl. Acad. Sci. U.S.A. 87, 9995–9999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morrison N. A., Shine J., Fragonas J. C., Verkest V., McMenemy M. L., Eisman J. A. (1989) 1,25-Dihydroxyvitamin D-responsive element and glucocorticoid repression in the osteocalcin gene. Science 246, 1158–1161 [DOI] [PubMed] [Google Scholar]
- 37. Vella M. C., Choi E. Y., Lin S. Y., Reinert K., Slack F. J. (2004) The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′-UTR. Genes Dev. 18, 132–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu L., Belasco J. G. (2008) Let me count the ways. Mechanisms of gene regulation by miRNAs and siRNAs. Mol. Cell 29, 1–7 [DOI] [PubMed] [Google Scholar]
- 39. Nilsen T. W. (2007) Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 23, 243–249 [DOI] [PubMed] [Google Scholar]
- 40. Guo H., Ingolia N. T., Weissman J. S., Bartel D. P. (2010) Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Carthew R. W., Sontheimer E. J. (2009) Origins and Mechanisms of miRNAs and siRNAs. Cell 136, 642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jemal A., Siegel R., Ward E., Hao Y., Xu J., Thun M. J. (2009) Cancer statistics, 2009. CA Cancer J. Clin. 59, 225–249 [DOI] [PubMed] [Google Scholar]
- 43. Zhang X., Nicosia S. V., Bai W. (2006) Vitamin D receptor is a novel drug target for ovarian cancer treatment. Curr. Cancer Drug Targets 6, 229–244 [DOI] [PubMed] [Google Scholar]
- 44. Shen Z., Zhang X., Tang J., Kasiappan R., Jinwal U., Li P., Hann S., Nicosia S. V., Wu J., Zhang X., Bai W. (2011) The coupling of epidermal growth factor receptor down-regulation by 1α,25-dihydroxyvitamin D3 to the hormone-induced cell cycle arrest at the G1-S checkpoint in ovarian cancer cells. Mol. Cell. Endocrinol. 338, 58–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yamamoto H., Shevde N. K., Warrier A., Plum L. A., DeLuca H. F., Pike J. W. (2003) 2-Methylene-19-nor-(20S)-1,25-dihydroxyvitamin D3 potently stimulates gene-specific DNA binding of the vitamin D receptor in osteoblasts. J. Biol. Chem. 278, 31756–31765 [DOI] [PubMed] [Google Scholar]
- 46. Ventura A., Jacks T. (2009) MicroRNAs and cancer. Short RNAs go a long way. Cell 136, 586–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hammond S. M. (2007) MicroRNAs as tumor suppressors. Nat. Genet. 39, 582–583 [DOI] [PubMed] [Google Scholar]
- 48. Deng S., Calin G. A., Croce C. M., Coukos G., Zhang L. (2008) Mechanisms of microRNA deregulation in human cancer. Cell Cycle 7, 2643–2646 [DOI] [PubMed] [Google Scholar]
- 49. Schepeler T., Reinert J. T., Ostenfeld M. S., Christensen L. L., Silahtaroglu A. N., Dyrskjøt L., Wiuf C., Sørensen F. J., Kruhøffer M., Laurberg S., Kauppinen S., Ørntoft T. F., Andersen C. L. (2008) Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 68, 6416–6424 [DOI] [PubMed] [Google Scholar]
- 50. Porkka K. P., Pfeiffer M. J., Waltering K. K., Vessella R. L., Tammela T. L., Visakorpi T. (2007) MicroRNA expression profiling in prostate cancer. Cancer Res. 67, 6130–6135 [DOI] [PubMed] [Google Scholar]
- 51. Bicher A., Ault K., Kimmelman A., Gershenson D., Reed E., Liang B. (1997) Loss of heterozygosity in human ovarian cancer on chromosome 19q. Gynecol. Oncol. 66, 36–40 [DOI] [PubMed] [Google Scholar]
- 52. Amfo K., Neyns B., Teugels E., Lissens W., Bourgain C., De Sutter P., Vandamme B., Vamos E., De Grève J. (1995) Frequent deletion of chromosome 19 and a rare rearrangement of 19p13.3 involving the insulin receptor gene in human ovarian cancer. Oncogene 11, 351–358 [PubMed] [Google Scholar]
- 53. Bièche I., Noguès C., Paradis V., Olivi M., Bedossa P., Lidereau R., Vidaud M. (2000) Quantitation of hTERT gene expression in sporadic breast tumors with a real-time reverse transcription-polymerase chain reaction assay. Clin. Cancer Res. 6, 452–459 [PubMed] [Google Scholar]
- 54. Sun P. M., Wei L. H., Luo M. Y., Liu G., Wang J. L., Mustea A., Könsgen D., Lichtenegger W., Sehouli J. (2007) The telomerase activity and expression of hTERT gene can serve as indicators in the anti-cancer treatment of human ovarian cancer. Eur. J. Obstet. Gynecol. Reprod. Biol. 130, 249–257 [DOI] [PubMed] [Google Scholar]
- 55. Cancer Genome Atlas Research Network (2011) Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Xie C. Q., Jeong Y., Fu M., Bookout A. L., Garcia-Barrio M. T., Sun T., Kim B. H., Xie Y., Root S., Zhang J., Xu R. H., Chen Y. E., Mangelsdorf D. J. (2009) Expression profiling of nuclear receptors in human and mouse embryonic stem cells. Mol. Endocrinol. 23, 724–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bentwich I., Avniel A., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Sharon E., Spector Y., Bentwich Z. (2005) Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 37, 766–770 [DOI] [PubMed] [Google Scholar]