Background: Rad9, a cell cycle checkpoint and DNA repair protein, is functionally related to human prostate tumorigenesis.
Results: Rad9 deletion in prostate cancer cells impairs migration and invasion, sensitizes to anoikis, and down-regulates integrin β1.
Conclusion: Rad9 controls ITGB1 expression, migration, invasion, and anoikis resistance of prostate cancer cells.
Significance: This study reveals the significance of Rad9 in prostate tumor migration and invasion.
Keywords: Cell Invasion, Cell Migration, Gene Silencing, Prostate Cancer, Signal Transduction, Rad9, Anoikis, Cancer, Migration, Prostate
Abstract
Rad9 as part of the Rad9-Hus1-Rad1 complex is known to participate in cell cycle checkpoint activation and DNA repair. However, Rad9 can act as a sequence-specific transcription factor, modulating expression of a number of genes. Importantly, Rad9 is up-regulated in prostate cancer cell lines and clinical specimens. Its expression correlates positively with advanced stage tumors and its down-regulation reduces tumor burden in mice. We show here that transient down-regulation of Rad9 by RNA interference reduces DU145 and PC3 prostate cancer cell proliferation and survival in vitro. In addition, transient or stable down-regulation of Rad9 impairs migration and invasion of the cells. Moreover, stable reduction of Rad9 renders DU145 cell growth anchorage-dependent. It also decreases expression of integrin β1 protein and sensitizes DU145 and LNCaP cells to anoikis and impairs Akt activation. On the other hand, stable expression of Mrad9, the mouse homolog, in DU145/shRNA Rad9 cells restores migration, invasion, anchorage-independent growth, integrin β1 expression, and anoikis resistance with a concomitant elevation of Akt activation. We thus demonstrate for the first time that Rad9 contributes to prostate tumorigenesis by increasing not only tumor proliferation and survival but also tumor migration and invasion, anoikis resistance, and anchorage-independent growth.
Introduction
Prostate cancer is the most prevalent non-cutaneous type of cancer in men (1). Despite successes in treating localized primary prostate tumors, metastatic prostate cancer poses a real challenge and remains essentially incurable. As in most types of solid cancers, metastasis is the major morbidity and mortality factor for patients with prostate cancer. During progression to metastasis, tumor cells acquire multiple traits, including detachment from the extracellular matrix, anoikis resistance, migration and invasion through the basement membrane, intravasation to blood and lymphatic vessels, extravasation from the circulation to distant sites, the ability to stimulate angiogenesis, and, finally, formation of macroscopic metastases (2).
An integral part of the metastatic process is the integrins. These molecules are heterodimeric αβ transmembrane receptors that connect the extracellular matrix to the cytoskeleton (3) and play important roles in migration, invasion, and anoikis resistance (4). In particular, β1 integrin (ITGB1) is known to confer higher survival and metastatic capacity to a number of cancer cells, including those of prostate origin (5, 6). Integrin β1 is crucial for insulin-like growth factor I receptor-mediated prostate cancer cell proliferation and anchorage-independent growth (7).
The serine/threonine protein kinase Akt is a downstream effector of PI3K and an important regulator of various cellular functions including cell metabolism, transcription, survival, and proliferation (8). Up-regulation of the Akt kinase signaling pathway is frequently observed in prostate cancer. Akt is activated by phosphorylation at Thr-308 and Ser-473 in response to mitogenic stimulation as well as by integrin attachment to the extracellular matrix (9). Cancer cells rely heavily on active Akt to survive from a number of insults such as genotoxic stress, growth factor depletion, and anoikis, a form of apoptotic cell death when cells lose contact with the extracellular matrix (10).
Rad9 is a protein with an established role in the DNA damage response and DNA repair. As part of the Rad9-Hus1-Rad1 complex, it acts as a sensor of DNA damage that enables ATR kinase, independently recruited to the site of damage, to phosphorylate and activate its downstream effector Chk1 (11). However, Rad9 can interact with several other proteins outside the context of the 9-1-1 complex and checkpoint functions (12). Interestingly, Rad9 can act independently of its partners Hus1 and Rad1 to transactivate a number of genes including p21waf1/cip1 (13). Aberrant Rad9 expression has been associated with prostate, breast, lung, skin, thyroid, and gastric cancers (12). We have shown previously that Rad9 is overexpressed in human prostate cancer specimens as well as prostate cancer cell lines (14). Experiments designed to assess the contribution of Rad9 to prostate tumor growth revealed that down-regulation of Rad9 in PC3 and DU145 human tumor cell line xenografts impairs growth in nude mice. Importantly, ectopic expression of Rad9 in an immortalized, nontumorigenic, prostate cell line (PWR-1E) conferred upon them the ability to form aberrant growths in mice (14). Furthermore, immunohistochemical analysis of normal and tumor prostate specimens showed that Rad9 expression increased along with cancer progression stages, suggesting a role for Rad9 in prostate malignant progression (14).
The objective of this study was two-fold: to assess the effect of Rad9 down-regulation on prostate cancer cell proliferation and survival and to investigate the impact of Rad9 down-regulation on three cellular biological traits related to metastasis (migration, invasion, and anoikis sensitivity). In this report, we propose that Rad9 may influence cell migration and invasion by controlling the abundance of integrin β1, anoikis resistance, and activation of Akt.
EXPERIMENTAL PROCEDURES
Cell Culture
Prostate cancer cell lines DU145, PC-3, and LNCaP were grown at 37 °C, 5% CO2 in RPMI 1640 (Invitrogen), supplemented with 10% FBS (Atlanta Biologicals), 100 units/ml penicillin, and 100 μg/ml streptomycin (all from Cellgro). NIH3T3 cells were grown in DMEM supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin.
Plasmid Construction
ITGB1 cDNA was generated by mRNA extracted from DU145 cells and reverse-transcribed with a SuperScript III kit (Invitrogen). cDNA was amplified by PCR human ITGB1 gene (X07979) primers: forward, 5′-GGTACTGGATCCATGAATTTACAACCAATTTTCTGGA-3′ (BamHI underlined) and reverse, 5′-GTAGACTCGAGTCATTTTCCCTCATACTTCGGATTGA-3′ (XhoI underlined). PCR conditions were 94 °C for 2 min, 30 cycles of denaturation at 94 °C for 30 s, annealing and extension at 68 °C for 4 min, and a final extension at 72 °C for 7 min. The PCR-amplified product was ligated into pcDNA3 expression vector (Invitrogen).
The mouse Rad9 construct was made by inserting the mouse Rad9 ORF into the HindIII/NotI restriction sites of the pZeoSV2(+) vector (Invitrogen). The ORF was amplified using the following PCR primers: forward, 5′-TCGGTGAAGCTTACAATGAAGTGCCTGATCAC-3′ (HindIII underlined) and reverse, 5′-TTTAGAGCGGCCGCTCACCCTTCACCATCACTGTCTT-3′ (NotI underlined). For stable clones, the plasmid was transfected into DU145/shRad9 clone #2 using Lipofectamine 2000. Cells were incubated in the presence of 100 μg/ml zeocin, and stable Mrad9 clones were selected 2 weeks later. Cells were routinely cultured in 100 μg/ml zeocin and 1 μg/ml puromycin.
RNA Interference
The pSUPER.retro.puro Rad9 shRNA expression vector (Oligoengine, Inc.) and viral production have been described previously (14). Down-regulation of Rad9 protein was assessed by Western blotting with Rad9 antibodies. The clones with the greatest reduction in Rad9 expression were used for further studies.
For transient transfection experiments, the following siRNA were used: 50 nm siRad9#1, 5′-AGGCCCGCCAUCUUCACCA-3′ (sense); 25 nm siRad9#3, 5′- GUCUUUCCUGUCUGUCUUC-3′ (sense) and a commercial siRad9#2 (Ambion) at 12.5 nm; 50 nm of siLuc (luciferase control) 5′-CUUACGCUGAGUACUUCGA-3′ (sense); and 30 nm ITGB1 siRNA 5′-AAUGUAACCAACCGUAGCA-3′ (sense). Cells at ∼30% confluence were transfected with the indicated siRNA and Lipofectamine 2000 (Invitrogen), and protein abundance was tested 48–72 h later.
RNA Extraction and RT-PCR
Expression levels of ITGB1 mRNA were quantified relative to control GAPDH mRNA by quantitative RT-PCR. Cells were grown to ∼70% confluence, washed once with PBS, and then total cellular RNA was isolated using TRIzol reagent (Invitrogen) according to the instructions of the manufacturer. Total RNA (2 μg) was reverse-transcribed to cDNA with the Superscript III first-strand synthesis system (Invitrogen) using oligo(dT) primer following the protocol of the manufacturer. Subsequently, samples were subjected to quantitative PCR in an ABI 7300 real-time PCR system (Applied Biosystems) using Power SYBR Green PCR MasterMix, a cDNA template, and the following primers: for ITGB1, 5′-CCTACTTCTGCACGATGTGATG-3′ (forward) and 5′-CCTTTGCTACGGTTGGTTACATT-3′ (reverse) and GAPDH, 5′-CATCTCTGCCCCCTCTGCTG-3′ (forward) and 5′-CCCTCCGACGCCTGCTTCAC-3′ (reverse). The PCR products were analyzed on a 3% agarose gel stained with ethidium bromide.
Protein Stability Assays
ITGB1 protein stability was assessed by incubating cells with either 50 μg/ml or 10 μg/ml cycloheximide (Sigma) for the indicated times. The proteasomal inhibitor MG132 (Sigma) was added to the cells at 10 μm and incubated for 4 h. Cells were lysed at the end of the incubation in radioimmune precipitation assay buffer, and protein levels were measured by Western blotting.
Cell Cycle Analysis
Cells were harvested by trypsinization, washed in PBS, and fixed in 70% ethanol overnight at −20 °C. Fixed cells were subsequently washed in PBS once and incubated with propidium iodide/RNase staining buffer (BD Biosciences) for 30 min at 37 °C. Flow cytometry was performed by a FACScalibur with CellQuest software (BD Biosciences).
Cell Proliferation and Apoptosis
To determine proliferation, cells were seeded on 6-well plates and counted at the indicated intervals with a hemocytometer. To assess clonogenic survival, cells were transiently transfected with 100 nm siRNA against Rad9 or luciferase (control), and 72 h after transfection they were trypsinized, counted, and added in triplicate to 10-cm culture dishes containing complete medium. After 14 days in culture, cells were fixed in 100% methanol for 15 min and stained with Giemsa for 30 min. Colonies containing at least 50 cells were scored.
Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining was performed using an in situ cell death detection kit (Roche) according to the protocol of the manufacturer. Images were acquired with an Olympus BH-2 microscope and Olympus MicroSuite FIVE imaging software.
Cell Adhesion
Cells were seeded in 6-well plates (4 × 104 cells/well) coated with fibronectin (10 μg/ml, BD Biosciences). After a 30-min period, attached cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. The stain was recovered by extracting with 10% acetic acid, and optical density was measured at 595 nm.
Cell Migration and Invasion
Migration and invasion assays were performed using 6.5-mm diameter, 8-μm-pore-size, transwell membrane filter inserts in a 24-well tissue culture plate. For the migration assay, 40,000 cells in 150 μl of RPMI + 0.1% BSA were added to each top chamber, and the bottom chamber was filled with 500 μl of complete medium containing 10% FBS as a chemoattractant. The migration assay was performed at 37 °C for 6 h. Nonmigratory cells at the upper membrane surface were removed with several cotton swabs, and the migratory cells at the bottom of the membrane were stained with crystal violet (0.1% crystal violet in 0.1 m borate buffer (pH 9) and 20% ethanol) for 20 min at room temperature. After rinsing extensively with deionized water, the stain was extracted with 10% acetic acid, and optical density was measured at 595 nm. Haptotactic migration toward fibronectin was assayed by coating the lower side of the Boyden chamber with 10 μg/ml human plasma fibronectin. Cells were resuspended in serum-free RPMI containing 0.1% BSA, and 40,000 cells were added to each insert. Serum-free RPMI containing 0.1% BSA was added to the bottom wells. After 6 h of incubation at 37 °C, the cells were treated as described above.
An invasion assay was performed under the same conditions as the migration assay, with the exception that 200 μl of growth factor-reduced Matrigel (BD Biosciences) diluted 1:3 with RPMI + 0.1% BSA, supplemented with 10 μg/ml fibronectin, was added to the top chamber. Cells were allowed to invade for 40 h.
For the wound-healing migration assay, cell populations grown to confluency in 6-well plates were scratched with a sterile P200 pipette tip. For some experiments, the plates were precoated with either 2.5 μg/ml or 10 μg/ml fibronectin overnight at 4 °C. Detached cells were removed with Dulbecco's PBS. Attached cells were allowed to migrate in serum-free medium (RPMI/0.1% BSA). Photographs of the wounds were taken at 0 h, 4 h, 9 h, 24 h, 32 h, and 48 h. The wound gap was quantified by National Institutes of Health ImageJ software (http://rsbweb.nih.gov/ij/).
Anchorage-independent Growth Assay
Cells were suspended in 500 μl of RPMI/10% FBS medium containing 0.33% agarose and plated in duplicate on top of 1 ml of solidified 0.67% agarose/RPMI/10% FBS in 12-well plates (1000 cells/well). Cells were placed in a 37 °C and 5% CO2 incubator, and colonies were allowed to form for 2 weeks. Colonies were stained with 0.005% crystal violet for 2 h, and photographs of five fields/well were taken under the microscope. Colonies with a diameter greater than 20 μm were scored.
Anoikis
6-well plates were coated overnight with either 10 μg/ml fibronectin (at 4 °C) or the nonadhesive poly(2-hydroxyethyl methacrylate) (poly-HEMA) (20 mg/ml at room temperature). Cells were added to the plates and incubated for the indicated time periods. Cell death was detected by trypan blue exclusion assay. In some experiments, 50 μm of the PI3K inhibitor LY294002 (Invitrogen) was added to the cells.
Western Blot Analysis
Cells were lysed in radioimmune precipitation assay buffer (100 mm HEPES (pH 7.4), 10% glycerol, 150 mm NaCl, 1% Nonidet P-40, 0.1% SDS, 0.5% sodium deoxycholate, 30 mm NaF, 25 mm β-glycerophosphate, 1 mm Na3VO4, 2 mm EDTA, and complete protease inhibitors (Roche)) or in 1% Triton X-100 lysis buffer (100 mm HEPES (pH 7.4), 10% glycerol, 150 mm NaCl, 1% Triton X-100, 30 mm NaF, 25 mm β-glycerophosphate, 1 mm Na3VO4, 2 mm EDTA, and complete protease inhibitors (Roche)) for 30 min on ice. Lysates were cleared by centrifugation at 16,000 × g for 20 min. Protein was subjected to either 8% Tris-glycine or 4–12% Bis/Tris (Invitrogen) SDS-PAGE. Quantitation of band intensity was performed with ImageJ software.
To assay secreted fibronectin, cells were grown in serum-free RPMI/0.1% BSA for 24 h. Conditioned medium was collected, centrifuged to remove any floating cells, and 2 ml were concentrated with an Amicon Ultracell (Millipore) and immunoblotted for fibronectin. Attached cells were also collected, lysed, and immunoblotted with actin to ensure that conditioned medium had been collected from the same number of cells.
The following antibodies were used in this study: Rad9, integrin β1, integrin β3, integrin α2, integrin αv, fibronectin monoclonal antibodies (BD Biosciences), β-actin, and α-tubulin monoclonal antibodies (Sigma), phospho-Akt (Ser-473), Akt, phospho-ERK1/2 (Thr-202/Tyr-204), phospho-FOXO3a (Ser-253), FOXO3a, phospho-GSK3β (Ser-9), and GSK3α/β polyclonal antibodies (Cell Signaling Technology).
RESULTS
Down-regulation of Rad9 Decreases Cell Proliferation
In a previous report, we assessed the contribution of Rad9 to prostate tumor growth. This investigation revealed that down-regulation of Rad9 impaired growth of PC3 and DU145 xenografts injected into nude mice (14). To assess the role of Rad9 in the proliferation of these two human prostate cancer cell lines, we transiently knocked down Rad9 expression by short interfering RNA (siRad9), or used control siRNA2 (luciferase, siLuc) and examined the impact on cell proliferation over a period of 4 days. Transfection with 100 nm siRad9 resulted in more than 90% down-regulation of Rad9 protein levels compared with Lipofectamine-only (Lip.) or siLuc-transfected cells (Fig. 1A, upper panel). Cell proliferation was reduced in siRad9-expressing cells compared with control cells (Fig. 1A, lower panel). The reduction of cell proliferation for siRad9-expressing cells became apparent after 3 days in culture, and it was ∼2.5-fold lower compared with the siLuc-expressing cells by the end of the experiment at day 4. Similar results were obtained with another prostate cancer cell line, PC3 (supplemental Fig. S1A). Likewise, we measured cell proliferation of DU145 cells stably transfected with short hairpin RNA against Rad9 (shRad9) or insertless vector (pSUPER-retro-puro vector; shControl), or untransfected parental cells. Those stably expressing shRad9 had reduced Rad9 protein levels by ∼90% compared with the levels in either parental or shControl cells (Fig. 1B, upper panel). After day 6 in culture, the DU145/shRad9 cell number was ∼2-fold lower than the DU145/shVector or DU145/parental cell populations (Fig. 1B, lower panel). Thus, the in vitro cell proliferation assay recapitulated the in vivo xenograph studies using the same cell lines. Cell proliferation results were also verified by colony formation for both PC3 and DU145 cells (supplemental Fig. S1B). In the case of DU145 cells, treatment with siRNA sustained suppression of endogenous Rad9 levels for at least 6 days (supplemental Fig. S1C). Results showed that transient reduction of Rad9 in these cells was sufficient to permanently impair proliferation by as much as 80%.
FIGURE 1.
Cell proliferation assay. A, upper panel, Western blot analysis of Rad9 expression levels in cells transiently transfected with Lipofectamine-only (Lip.), control siRNA (luciferase, siLuc), or Rad9 siRNA (siRad9). Lower panel, proliferation of DU145 cells transiently transfected with Lip., siLuc, or siRad9 was analyzed by counting with a hemocytometer at the indicated time intervals after plating an equal number of cells. B, upper panel, Western blot analysis of Rad9 expression levels in parental DU145 cells or cells stably expressing Rad9 shRNA (shRad9) or pSUPER-retro insertless vector (shControl). Lower panel, proliferation of parental DU145 cells or cells stably expressing shRad9 or shControl were analyzed as in A. C, down-regulation of Rad9 induces apoptosis. Apoptosis of DU145 cells transiently transfected with either siRad9 or siLuc was measured 4 days posttransfection by TUNEL assay. Shown are representative images of TUNEL-positive nuclei (green) counterstained with DAPI (blue). D, TUNEL assay results were quantified as a percentage of TUNEL-positive nuclei divided by the total number of nuclei scored. All data represent the mean ± S.D. from three independent experiments. Rad9 levels were normalized against α-tubulin and shown as percent expression relative to Rad9 levels in control (Lip.) cells (100%).
Rad9 Knock-down Reduces Cell Survival
To account for the decreased cell proliferation of DU145 and PC3 cells with knocked down Rad9 expression, we examined cell viability using the terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay. Immunofluorescence analysis showed that transient down-regulation of Rad9 in DU145 cells increased the number of TUNEL-positive cells (Fig. 1C). Quantitation of apoptotic cells 4 days post-transfection revealed an increase from 5% in control (transfected with siLuc) cells expressing normal levels Rad9 to 35% in the siRad9 cells (Fig. 1D). Likewise, Rad9 silencing in PC3 cells resulted in a 4-fold increase in apoptotic cells as compared with control (Luc) siRNA (supplemental Fig. S1C).
Suppression of Migration and Invasion Are Mediated by Rad9 Silencing
We showed previously that Rad9 expression levels correlate with metastatic progression in prostate cancer (14). Likewise, increased Rad9 expression has been correlated with tumor growth and local recurrence in breast cancer (16). We thus examined the impact of Rad9 down-regulation on DU145 and PC3 cell migration and invasion. To investigate DU145 cells for the role of Rad9 in these activities we used two different stable clones that show ∼60–95% Rad9 down-regulation (Fig. 3A) and compared their ability to migrate toward 10% fetal bovine serum or 10 μg/ml fibronectin (Fig. 2A) or invade a Matrigel matrix supplemented with 10 μg/ml fibronectin (B). Rad9 silencing resulted in a 3-fold less migration compared with DU145 parental cells or DU145 cells stably bearing an insertless pSUPER-puro vector (shControl) as well as a 2.5-fold lower invasion capacity. A well filled with bovine serum albumin served as a negative control (no chemoattractant) and defined basal invasion. Furthermore, DU145 cell migration was generally twice as efficient toward fibronectin as compared with 10% FBS (Fig. 2A).
FIGURE 3.
Expression of Mrad9 restores migration and invasion capability of DU145 cells with reduced endogenous levels of Rad9. A, Western blot analysis of expression of Rad9 in the DU145 parental cell line and cells that stably contain shControl, shRad9 (clone A), and shRad9 (clone 2) that also stably expresses either pSV2zeo or mouse Mrad9. The percent expression of Rad9 in various clones compared with Rad9 protein levels in parental DU145 is indicated. B, wound healing assay. Wound closure of DU145 parental cells, shControl, shRad9 (clones 2 and A), and shRad9 (clone 2) cells containing insertless vector (pSV2zeo) or with Mrad9 was monitored after 0, 24, and 48 h. Shown are photographs from a representative experiment. C, quantitation of wound migration assay. Three independent experiments ± S.D. were performed. At time 0 h, values were set at 100%, and wound closure was expressed as a percentage relative to the 0 h. D, ectopic expression of Mrad9 in DU145 cells with reduced levels of Rad9 restores the invasive capacity of these cells. Data are mean ± S.D. (n = 3).
FIGURE 2.
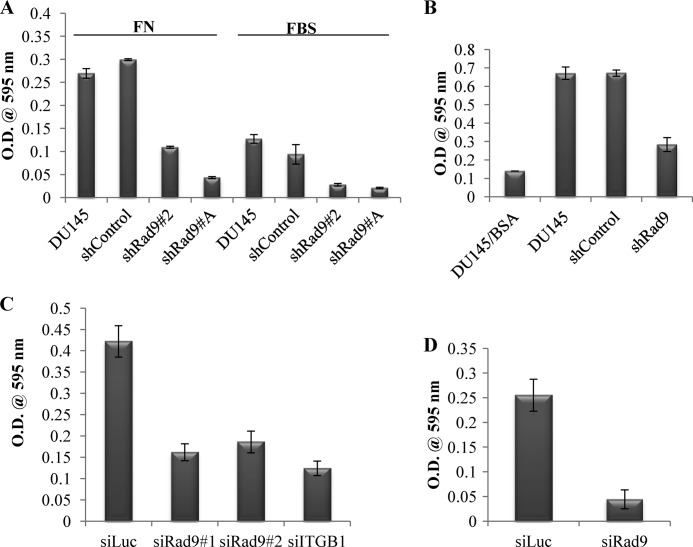
Rad9 regulation of DU145 cell migration and invasion. A, cell migration toward either FN or 10% FBS was performed with parental DU145 or those cells stably expressing pSUPER-puro insertless vector (shControl) or two clones with shRad9 (2, A) for 6 h using 24-well Transwell inserts. B, cell invasion was performed with Matrigel containing 10 μg/ml fibronectin. Cell invasion with parental (DU145), shRad9, or shControl were allowed to invade for 40 h using FBS (10%) as chemoattractant or 0.1% bovine serum albumin as negative control (DU145/BSA). C, DU145 cells were transiently transfected with control siRNA (siLuc), two Rad9 siRNAs (siRad9#1 and siRad9#2), or siRNA against integrin β1 (siITGB1) and assayed for migration toward fibronectin for 6 h. D, invasion of DU145 cells transiently down-regulating Rad9 (siRad9) or control (siLuc) was carried out as in C. All data are mean ± S.D. of a representative experiment of three independent experiments performed in triplicate.
We also assessed migration and invasion by transiently down-regulating Rad9 using two non-overlapping siRNAs that were able to reduce Rad9 protein expression by ∼90% (Fig. 5A). We completed these assays with 50 nm siRad9 (siRad9#1) or 12.5 nm siRad9 (siRad9#2) within 72 h post-transfection, when cell proliferation is similar in siRad9 and control cells (Fig. 1A). As a negative control, we used siRNA against luciferase (siLuc, 50 nm), whereas an additional siRNA against integrin β1 (siITGB1, 30 nm) was used as a positive control. Integrin β1 (ITGB1), when complexed with integrin α5, is a major fibronectin receptor in these cells (17). Rad9 silencing in DU145 cells caused impaired capacity to migrate toward fibronectin (Fig. 2C) or invade through Matrigel/fibronectin (Fig. 2D). Quantitation analysis revealed a 2.5-fold decreased migration and 5-fold reduced invasion in cells with diminished levels of Rad9 compared with control cells (siLuc) expressing normal levels of Rad9.
FIGURE 5.
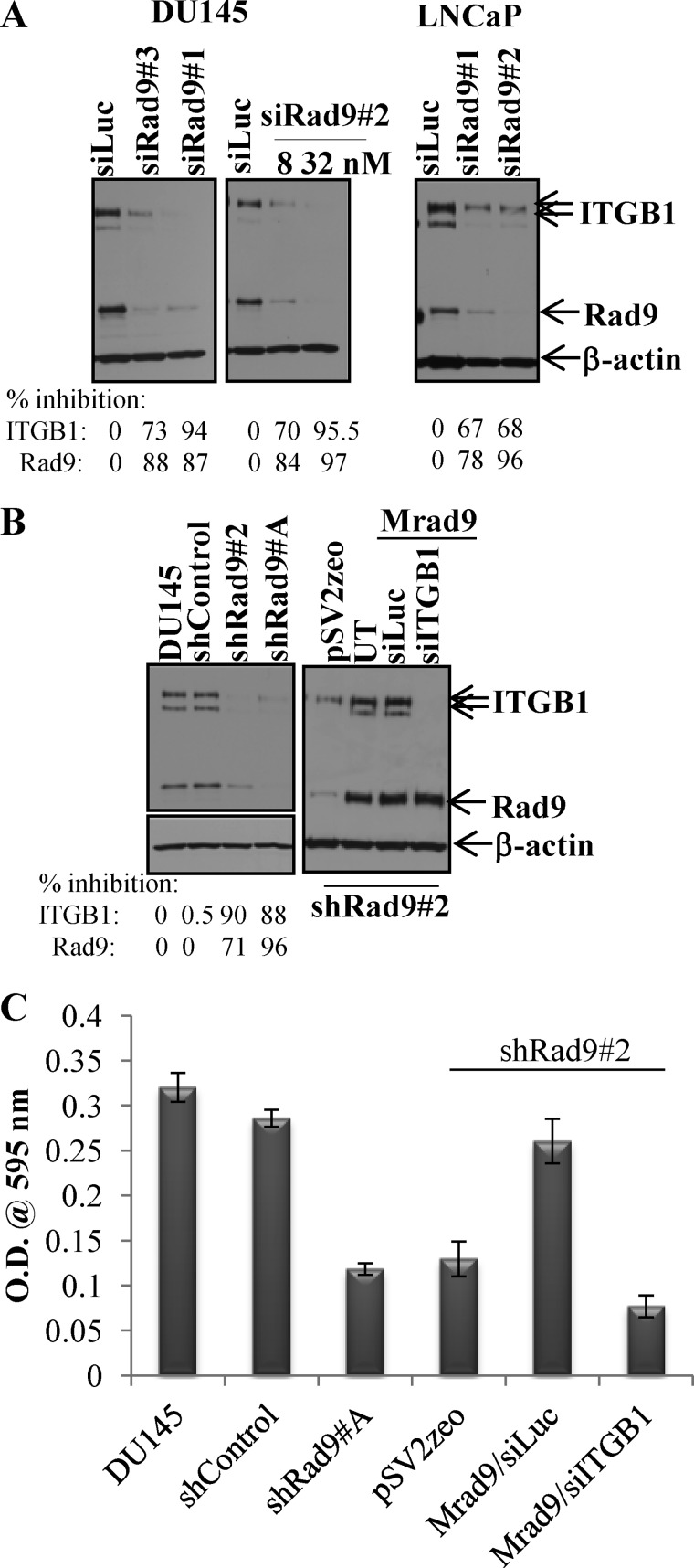
Rad9 expression modulates ITGB1 protein levels. A, transient silencing of Rad9 in either DU145 or LNCaP cells using three different siRad9 (#1, #2, #3) suppresses ITGB1 expression. siRad9#2 was used at two different concentrations. The percent inhibitions of ITGB1 and Rad9 compared with siLuc are indicated. B, stable down-regulation of Rad9 in DU145 (clones 2 and A) results in diminished levels of ITGB1 protein compared with parental DU145 or shControl cells, whereas stable expression of Mrad9 in the DU145/shRad9#2 clone restores ITGB1 levels. The percent inhibitions of ITGB1 and Rad9 compared with parental DU145 are indicated. UT, untransfected. C, DU145/shRad9 invasive capability is restored by expressing Mrad9 only when ITGB1 levels are maintained. Data are mean ± S.D. (n = 3).
In addition to DU145 cells, Rad9 down-regulation, by transiently transfecting with a Rad9-specific siRNA, affected migration and invasion of PC3 cells when comparing migration toward 10% fetal bovine serum (chemoattractant) and invasion to Matrigel for these cells as well as siLuc-transfected controls. Rad9 down-regulation diminished migration (supplemental Fig. S2A) and invasion (supplemental Fig. S2B) of PC3 cells 2-fold compared with the control siLuc. Therefore, we conclude that Rad9 mediates tumor cell migration and invasion in vitro.
Expression of Mrad9 Restores Migration and Invasion Potential and Anchorage-independent Growth of DU145 Cells with Reduced Human Rad9 Levels
To investigate whether ectopic Rad9 expression in DU145 clones with reduced endogenous Rad9 can reverse the observed phenotype of diminished migration and invasion, we stably expressed mouse Rad9 (Mrad9) in DU145/shRad9 cells. Mrad9 protein level was not affected by human shRad9 (supplemental Fig. S3A). We isolated a clone that expressed ∼3-fold more Mrad9 compared with Rad9 expression in parental DU145 (Fig. 3A). A clone with much higher levels of Mrad9 displayed signs of increased cell death. This is not surprising, as it has been reported that Rad9 can also act as a cell death inducer when overexpressed (18). Stable silencing of Rad9 did not increase cell death as judged by the trypan viability assay (supplemental Fig. S3B) and, in agreement with published reports (19, 20), neither did it affect cell cycle phase distribution (supplemental Fig. S3C). We measured cell migration with a wound healing assay (Fig. 3B). DU145 parental cells or various clones were grown to confluency and scratched with a pipette tip (time, 0 h). Twenty-four hours later, DU145, shControl, and Mrad9-expressing cells showed an ∼80% wound closure. In contrast, clones with reduced levels of Rad9 (shRad9#A, shRad9#2, shRad9#2/pSV2zeo) displayed only 20–30% wound closure (Fig. 3C). By 48 h, cells expressing high levels of Rad9 had closed the wound completely, whereas Rad9 knockdown cells showed 60–70% wound closure (Fig. 3C). The wound healing assay was also repeated with plates that had been coated with either 10 μg/ml or 2.5 μg/ml fibronectin (FN) or left uncoated. Although migration of shRad9 was not affected by the presence of exogenous fibronectin, migration of shControl and Mrad9 cells was somewhat impaired with increasing concentration of fibronectin (supplemental Fig. S4, A–C). Although the remaining wound gap after 48 h was ∼20% in shControl and Mrad9 cells in the uncoated plates, it was ∼40% in plates coated with 10 μg/ml FN in the same time. We analyzed fibronectin secretion in the different DU145 stable cell lines and found that shControl and Mrad9 stables secrete more FN than shRad9 clones (clones A and 2) (supplemental Fig. S4D). The speed of cell migration depends on substrate extracellular-matrix concentration, integrin concentration, and activity and follows a bell-shaped response curve (21). At a very high concentration of extracellular matrix proteins, the migration speed can even be reversed. An example is SHARPIN (SHANK-associated RH-domain interacting protein), an inhibitor of ITGB1. Down-regulation of SHARPIN by siRNA results in greater migration (measured by scratch assay) for PC3 cells in 0.1 μg/ml collagen-coated plates, whereas the difference is abrogated at 5 μg/ml and reversed (siRNA SHARPIN cells migrate faster) at 50 μg/ml of collagen (22).
In addition to the wound healing assay, Mrad9 expression was sufficient to restore the invasive capability of DU145 cells with diminished endogenous Rad9 expression through Matrigel/fibronectin (Fig. 3D).
In vitro anchorage-independent growth of cancer cells correlates with their ability to produce experimental metastasis in vivo (23). Therefore, we examined the effect of Rad9 down-regulation on anchorage-independent growth. The clones with reduced expression of Rad9 were not able to sustain anchorage-independent growth as assessed by colony formation in soft agarose (Fig. 4). However expression of Mrad9 restored this ability to the levels of parental and shControl DU145 cells (Fig. 4).
FIGURE 4.
Effect of Rad9 on anchorage-independent growth. A, an equal number of cells from different groups was assayed for colony formation in soft agarose. The numbers of colonies with a diameter greater than 20 μm were quantified 2 weeks later. Shown is one representative experiment of three independent experiments, each performed in duplicate. B, representative images of colonies in soft agarose.
Rad9 Silencing Suppresses Integrin β1 Expression
As DU145 cells migrated better toward fibronectin when Rad9 levels were high, we hypothesized that Rad9 may affect cell migration by controlling the abundance of integrin β1. Down-regulation of Rad9 by transiently transfecting DU145 cells with three non-overlapping siRNAs (#1, #2, #3) reduced levels of total integrin β1 protein from 73% to 95%, depending on the siRNA (Fig. 5A, left panel). Varying the degree of Rad9 silencing using 8 nm (84% Rad9 suppression) or 32 nm (97% inhibition) of siRad9#2 reduced expression of ITGB1 proportionally, that is 70% and 95.5% (Fig. 5A, center panel). Furthermore, suppression by ∼70% of ITGB1 protein via reducing levels of Rad9 was also observed with the LNCaP prostate cancer cell line (Fig. 5A, right panel). DU145 cells with stably reduced levels of Rad9 protein (clones shRad9#2 and shRad9#A) displayed reduced levels of ITGB1 protein (by ∼90%) when compared with DU145 cells or shControl cells (Fig. 5B, left panel). Rad9 knockdown specifically down-regulated ITGB1, as other integrins, ITGA2 and ITGAV, were not affected (supplemental Fig. S5A). In contrast, ITGB3 expression was increased in the cells in which ITGB1 was reduced (supplemental Fig. S5A). This is in agreement with published reports that ITGB1 exerts an inhibitory effect on ITGB3 by destabilizing ITGB3 mRNA (24).
Conversely, stable or transient expression of Mrad9 in DU145 cells that stably suppress expression of endogenous Rad9 showed a 3-fold increased expression of ITGB1 (Fig. 5B, right panel and supplemental Fig. S5B). Expression of Mrad9 in DU145/shRad9 cells is able to restore invasion in these cells (Fig. 3D). To assess whether ITGB1 can influence Mrad9-induced cell invasion, endogenous ITGB1 was silenced by a specific siRNA in DU145/shRad9/Mrad9 cells. As shown in Fig. 5C, silencing of ITGB1 reduced invasion of cells and negated the effect of expressed Mrad9 to levels comparable with those of cells with knockdown Rad9. These results suggest that Rad9-induced invasion may proceed through the regulation of ITGB1 expression. Re-expressing ITGB1 in DU145/shRad9 cells with reduced endogenous ITGB1 predictably should phenocopy the Mrad9 effect on invasion. However, ectopically expressing ITGB1 in these cells caused increased cell death, so invasion could not be assessed.
To investigate the mechanism by which Rad9 controls ITGB1 levels, we first asked whether Rad9 controls ITGB1 expression at the transcriptional level. We carried out quantitative RT-PCR experiments and compared ITGB1 mRNA levels from DU145/shControl, DU145/shRad9, and DU/shRad9/Mrad9 cells relative to GAPDH mRNA levels that served as an internal control. We found that ITGB1 mRNA did not change with the status of Rad9 in these cells, albeit a modest increase was observed in the cells expressing Mrad9 (supplemental Fig. S6A). We conclude that Rad9 does not control ITGB1 at the transcriptional level. Next, we examined ITGB1 protein stability in DU145 cells expressing either shRad9/Mrad9 or shRad9. We blocked protein synthesis for up to 6 h by treating cells with the protein synthesis inhibitor cycloheximide (50 μg/ml) and monitored ITGB1 protein levels by Western blotting. As expected, expression of ITGB1 in DU145/shRad9 cells was lower than in DU145/shRad9/Mrad9 cells. However, the precursor form of ITGB1 half-life in control cells was ∼6 h, and diminishing levels of ITGB1 coincided with the disappearance of Rad9. In contrast, the (precursor) ITGB1 half-life in DU145/shRad9 cells was much shorter (less than 2 h) (Fig. S6B). The mature form of ITGB1 showed the same reduction, but persisted longer that the newly synthesized form, apparently because of the fact that it is glycosylated and, thus, more stable than the nascent ITGB1 polypeptide (25). After 9 h of incubation with 10 μg/ml cycloheximide (a lower concentration of the drug was necessary to avoid cell death), the mature ITGB1 (upper band) started diminishing as well. Similar results were also recorded when shControl cells were treated with cycloheximide (supplemental Fig. S6B). When we treated DU145/shControl, DU145/shRad9/Mrad9, and DU145/shRad9 cells with the proteasomal inhibitor MG132 (10 μm) for 4 h, we found that the inhibitor had a minimal effect on ITGB1 protein stabilization (supplemental Fig. S6C), and, therefore, the shorter half-life of ITGB1 protein in shRad9 cells cannot be attributed to a proteasome-dependent degradation pathway. On the basis of these findings, we conclude that Rad9 affects ITGB1 protein stability, although the exact mechanism is presently not known.
Rad9 Knockdown Sensitizes Cells to Anoikis and Impairs Akt Activity
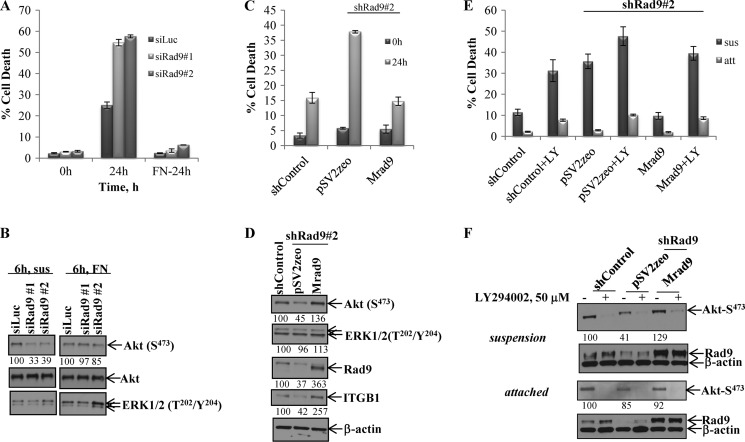
Resistance to anoikis, that is the induction of programmed cell death upon cell matrix detachment, is considered one of the in vitro markers of metastasis, and anoikis resistance is a prerequisite for tumor cells to remain viable when they lose attachment to the extracellular matrix (10, 26). The PI3K/Akt and Raf-1/ERK1/2 signaling pathways are intimately related to anoikis resistance (27). We sought to determine whether Rad9 expression affects anoikis resistance. Transient down-regulation of Rad9 by two non-overlapping siRad9 (#1, #2) caused increased anoikis compared with control DU145 cells (siLuc). After 24 h in suspension, cell populations with reduced levels of Rad9 showed 55% cell death in the population, whereas control cells displayed ∼25% cell death. In contrast, Rad9 silencing did not have an impact on cell survival of DU145 cells attached to fibronectin-coated plates (Fig. 6A). As Akt and ERK1/2 activation confer anoikis resistance, we examined phosphorylation levels at Ser-473 of Akt and Thr-202/Tyr-204 of ERK1/2. After 6 h in suspension, control cells maintained higher levels of phospho-Akt than cells with diminished Rad9 by ∼60% (Fig. 6B). In contrast, there was no difference in the phosphorylation of Akt between control and Rad9 knockdown cells plated onto fibronectin for the same time period (6 h). In agreement with these results, stable (supplemental Fig. S7A) or transient (supplemental Fig. S7B) down-regulation of Rad9 in DU145 cells did not affect cell adhesion to fibronectin, and neither was phosphorylation of Akt affected by Rad9 levels upon attachment to plastic or fibronectin (supplemental Fig. S7C). ERK1/2 phosphorylation was similar in all cells irrespective of Rad9 expression levels (Fig. 6B). These findings imply that Rad9 has a specific role in the protection of cancer cells from anoikis by maintaining elevated levels of Akt phosphorylation/activation. In support of this conclusion, stable expression of Mrad9 in DU145/shRad9 cells decreased cell death from ∼40% to 15% after 24 h in suspension (Fig. 6C). At the biochemical level, stable ectopic expression of Mrad9 in DU145/shRad9 cells resulted in an ∼3-fold increase in Akt phosphorylation at Ser-473 compared with DU145/shRad9 cells when in suspension for 6 h (Fig. 6D). Phospho-ERK1/2 levels remained unchanged. To establish the requirement of active Akt in anoikis resistance, we treated the cells with 50 μm LY294002 (a PI3K inhibitor) and assessed cell death after 24 h in suspension. As shown in Fig. 6E, inhibition of Akt resulted in ∼3-fold (for shControl) to 4-fold (Mrad9) more death. Interestingly, inhibition of Akt in shRad9, which express less active Akt (Fig. 6F) and have already a high cell death index, had a much lower impact on cell death. Treatment with the PI3K inhibitor impaired survival of attached cells to the same extent as Rad9 does not appear to have an effect on Akt activation in attached cells (Fig. 6F).
FIGURE 6.
Rad9 confers anoikis resistance and Akt activation. A, DU145 cells transiently transfected with siLuc or two different siRad9 (#1, #2) were incubated in suspension or plated onto fibronectin-coated plates (FN-24h) for 24 h, and cell viability was tested by trypan blue exclusion assay. Data represent mean ± S.D. (n = 3). B, Akt and ERK1/2 phosphorylation levels were assayed in siRNA-treated DU145 cells maintained either in suspension (sus) or after adhering to FN for 6 h. The percent of phospho-Akt levels, normalized against total Akt, in siRad9 cells compared with siLuc cells is indicated. C, expression of Mrad9 restores anoikis resistance to DU145 cells that stably express shRad9 (shRad9#2). Data represent mean ± S.D. (n = 3). D, activity of Akt and ERK1/2 in DU145 cells stably expressing shControl or shRad9#2 that express either a vector (pSV2zeo) or mouse Rad9 (Mrad9) maintained in suspension for 6 h. The percent expression of phospho-Akt, phospho-ERK, Rad9, and ITGB1 was normalized against expression of β-actin. E, Akt activation is required for anoikis resistance. Attached cells (att) or cells in suspension (sus) were treated with 50 μm LY294002, and 24 h later, cell viability was tested by trypan blue assay. Data represent mean ± S.D. (n = 3). F, Akt activation in attached and suspended cells treated with 50 μm LY294002 (+) or DMSO (-) was assessed by immunoblotting 2 h after treatment. The percent expression of phospho-Akt and phospho-ERK1/2 was normalized against β-actin expression and compared with shControl expression (100%).
The functional relationships among Rad9 protein levels, anoikis resistance, and Akt activation were also confirmed by using a second prostate cancer cell line. When LNCaP cells transiently down-regulating Rad9 independently by two different siRNA were incubated in suspension for 24 h, they showed an ∼2.5-fold increase in cell death, as measured by the trypan blue viability assay, compared with cells that were expressing a control siRNA (siLuc) (supplemental Fig. S7D). Rad9 was required for survival from anoikis, as LNCaP cells incubated in suspension for 3 h displayed an ∼65% inhibition in phosphorylation levels of Akt when Rad9 was suppressed by two non-overlapping specific Rad9 siRNA. In contrast to DU145, however, phospho-ERK1/2 was also reduced by 61–65% in the siRad9-expressing cells (supplemental Fig. S7E).
DISCUSSION
In this study, we demonstrate that Rad9 promotes prostate tumor cell growth and invasiveness. This is primarily on the basis of two findings: 1) Rad9 knockdown results in decreased cell proliferation and increased apoptosis; and 2) Rad9 down-regulation impairs in vitro migration and invasion of DU145 and PC3 cells and anchorage-independent growth of DU145 cells and renders DU145 and LNCaP cells more sensitive to anoikis, whereas restoration of Rad9 expression results in increased cell motility, invasion, anchorage-independent growth, and anoikis resistance.
We investigated mechanistic details of the observed phenotypes and discovered that silencing of Rad9 leads to a marked down-regulation of integrin β1. Conversely, ectopic expression of Mrad9 restores integrin β1 levels when endogenous Rad9 expression is knocked down in DU145 cells. Furthermore, reduction of ITGB1 protein levels by a specific siRNA negated the effect of Mrad9 on migration and invasion, suggesting that Rad9 affects these processes through the activity of integrin β1.
When cell adhesion to the extracellular matrix is lost, cells normally undergo anoikis. However, malignant cells have developed mechanisms to evade anoikis and either proliferate without matrix support or enter quiescence until a more suitable environment is presented (27, 28). Anoikis resistance is, therefore, a prerequisite of tumor metastasis and is considered a hallmark of cancer. Akt kinase plays a pivotal role in the resistance of malignant cells to anoikis (27). Our results show that Rad9 down-regulation impaired Akt phosphorylation when DU145 and LNCaP cells were maintained in suspension. Conversely, when Mrad9 was ectopically expressed in DU145 with reduced levels of endogenous Rad9, Akt phosphorylation was restored, and cells became more resistant to anoikis. ERK1/2 phosphorylation changes in response to Rad9 expression levels were variable and cell type-specific. In DU145 cells, ERK1/2 activation was not dependent on Rad9, as neither down-regulation of Rad9 by siRNA nor expression of Mrad9 had any effect on the phosphorylation of ERK1/2. In contrast, Rad9 down-regulation in LNCaP resulted in clear reduction of ERK1/2 phosphorylation. Finally, unlike anoikis, manipulation of Rad9 protein levels did not have an effect on the adhesion of DU145 cells to fibronectin. The downstream effectors of Akt activation are currently not known. Neither FOXO3a nor GSK3β seem to be involved, as their phosphorylation state is not affected by Akt in either DU145 cells (data not shown). It has been reported previously that Akt is not able to phosphorylate GSK-3β in LNCaP cells maintained in suspension (29).
Rad9 does not appear to directly control transcription of the ITGB1 gene because its knockdown reduces ITGB1 protein levels but not ITGB1 mRNA levels. In contrast, the absence of Rad9 increases the degradation rate of ITGB1 protein. When protein synthesis is inhibited by cycloheximide, ITGB1 protein half-life decreases from about 6 h in cells that express MRad9 to less than 2 h in shRad9 cells. Treatment with a proteasomal inhibitor cannot stabilize protein levels, and, therefore, the proteasome pathway is not involved. What, then, could be the mode of action of Rad9 on ITGB1 protein stability? ITGB1 protein levels are controlled by posttranslation modifications, most notably glycosylation. Interference with N-glycosylation in the Golgi renders the immature form of ITGB1 susceptible to degradation (23). Therefore, we can hypothesize that Rad9 is required for a maturation step of ITGB1 protein or that Rad9 acts as a transcription (co-) factor of a glycosyltransferase that, in turn, is required for ITGB1 glycosylation. Interestingly, many of the enzymes involved in glycoprotein metabolism are differentially expressed in cancer cells compared with normal cells and affect many aspects of cancer cell pathophysiology, including cell adhesion, migration, invasion, and, ultimately, metastasis (30–32).
In addition to Rad9, a DNA repair-independent function has been demonstrated previously for other components of DNA repair machinery. Ku80, a protein that is essential for non-homologous end joining, can also regulate cell adhesion to the extracellular matrix and promote cell invasion by physically interacting with and activating matrix metalloproteinase 9 (33). Plk1, a kinase important for mitosis, is also involved in acquisition of invasiveness in vimentin-expressing cells via regulating cell surface β1 integrin levels (34). Furthermore, strong evidence implicates BRCA1 in the regulation of spreading and motility of breast cancer cells through its localization to the plasma membrane as well as its interaction with ezrin, radixin, and moesin proteins (35). Flap endonuclease 1 (FEN1) is an enzyme with an established role in DNA repair and, more specifically, in base excision repair after oxidative stress (36). However, a recent report indicates that FEN1 can also regulate cell survival, MAPK p38 phosphorylation, and actin cytoskeleton reorganization through control of RhoA activation (37). The apurinic/apyrimidinic endonuclease 1/redox factor-1 (APE1), an essential gene in the base excision repair pathway, also functions as a transcriptional cofactor involved in cancer initiation and progression (38). Recently, it was shown that the transcriptional target LL-37 of APE-1 enhances keratinocyte migration on collagen I and fibronectin and induces Slug and Snail transcription factors, which are closely associated with keratinocyte migratory activity during wound healing (39). Interestingly, Rad9 interacts both physically and functionally with FEN1 and APE1, and enhances their enzymatic activities (40, 15). Therefore, it is possible that Rad9 impacts cell motility through its interactions with these two proteins.
In summary, our findings reveal that Rad9 is important not only for prostate cancer initiation but also for progression, as it controls cell motility and invasion as well as anoikis resistance. In the future, we will seek to elucidate the mechanism by which Rad9 affects expression of ITGB1 as well as other upstream events that culminate in Akt activation and anoikis resistance. The information gained will help define new molecular events in carcinogenesis and could potentially reveal targets for novel anti-cancer therapies.
This work was supported, in whole or in part, by National Institutes of Health Grants R01CA130536, R01GM079107, and P01CA49062 (to H. B. L).

This article contains supplemental Figs. S1–S7.
- siRNA
- short interfering RNA
- siLuc
- luciferase
- FN
- fibronectin.
REFERENCES
- 1. Shen M. M., Abate-Shen C. (2010) Molecular genetics of prostate cancer. New prospects for old challenges. Genes Dev. 24, 1967–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Valastyan S., Weinberg R. A. (2011) Tumor metastasis. Molecular insights and evolving paradigms. Cell 147, 275–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hynes R. O. (2002) Integrins. Bidirectional, allosteric signaling machines. Cell 110, 673–687 [DOI] [PubMed] [Google Scholar]
- 4. Margadant C., Monsuur H. N., Norman J. C., Sonnenberg A. (2011) Mechanisms of integrin activation and trafficking. Curr. Opin. Cell Biol. 23, 607–614 [DOI] [PubMed] [Google Scholar]
- 5. Hood J. D., Cheresh D. A. (2002) Role of integrins in cell invasion and migration. Nat. Rev. Cancer 2, 91–100 [DOI] [PubMed] [Google Scholar]
- 6. Zeng Z. Z., Jia Y., Hahn N. J., Markwart S. M., Rockwood K. F., Livant D. L. (2006) Role of focal adhesion kinase and phosphatidylinositol 3′-kinase in integrin fibronectin receptor-mediated, matrix metalloproteinase-1-dependent invasion by metastatic prostate cancer cells. Cancer Res. 66, 8091–8099 [DOI] [PubMed] [Google Scholar]
- 7. Goel H. L., Breen M., Zhang J., Das I., Aznavoorian-Cheshire S., Greenberg N. M., Elgavish A., Languino L. R. (2005) β1A integrin expression is required for type 1 insulin-like growth factor receptor mitogenic and transforming activities and localization to focal contacts. Cancer Res. 65, 6692–6700 [DOI] [PubMed] [Google Scholar]
- 8. Manning B. D., Cantley L. C. (2007) AKT/PKB signaling. Navigating downstream. Cell 129, 1261–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brazil D. P., Yang Z. Z., Hemmings B. A. (2004) Advances in protein kinase B signalling. AKTion on multiple fronts. Trends Biochem. Sci. 29, 233–242 [DOI] [PubMed] [Google Scholar]
- 10. 26. Sakamoto S., Kyprianou N. (2010) Targeting anoikis resistance in prostate cancer metastasis. Mol. Aspects Med. 31, 205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lieberman H. B., Bernstock J. D., Broustas C. G., Hopkins K. M., Leloup C., Zhu A. (2011) The role of RAD9 in tumorigenesis. J. Mol. Cell Biol. 3, 39–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Broustas C. G., Lieberman H. B. (2012) Contributions of RAD9 to tumorigenesis. J. Cell Biochem. 113, 742–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yin Y., Zhu A., Jin Y. J., Liu Y. X., Zhang X., Hopkins K. M., Lieberman H. B. (2004) Human RAD9 checkpoint control/proapoptotic protein can activate transcription of p21. Proc. Natl. Acad. Sci. U.S.A. 101, 8864–8869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhu A., Zhang C. X., Lieberman H. B. (2008) Rad9 has a functional role in human prostate carcinogenesis. Cancer Res. 68, 1267–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Balakrishnan L., Brandt P. D., Lindsey-Boltz L. A., Sancar A., Bambara R.A. (2009) Long patch base excision repair proceeds via coordinated stimulation of the multienzyme DNA repair complex. J. Biol. Chem. 284, 15158–15172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cheng C. K., Chow L. W., Loo W. T., Chan T. K., Chan V. (2005) The cell cycle checkpoint gene Rad9 is a novel oncogene activated by 11q13 amplification and DNA methylation in breast. Cancer Res. 65, 8646–8654 [DOI] [PubMed] [Google Scholar]
- 17. Witkowski C. M., Rabinovitz I., Nagle R. B., Affinito K. S., Cress A. E. (1993) Characterization of integrin subunits, cellular adhesion and tumorgenicity of four human prostate cell lines. J. Cancer Res. Clin. Oncol. 119, 637–644 [DOI] [PubMed] [Google Scholar]
- 18. Komatsu K., Miyashita T., Hang H., Hopkins K. M., Zheng W., Cuddeback S., Yamada M., Lieberman H. B., Wang H. G. (2000) Human homologue of S. pombe Rad9 interacts with BCL-2/BCL-xL and promotes apoptosis. Nat. Cell Biol. 2, 1–6 [DOI] [PubMed] [Google Scholar]
- 19. Hirai I., Wang H. G. (2002) A role of the C-terminal region of human Rad9 (hRad9) in nuclear transport of the hRad9 checkpoint complex. J. Biol. Chem. 277, 25722–25727 [DOI] [PubMed] [Google Scholar]
- 20. St Onge R. P., Besley B. D., Pelley J. L., Davey S. (2003) A role for the phosphorylation of hRad9 in checkpoint signaling. J. Biol. Chem. 278, 26620–26628 [DOI] [PubMed] [Google Scholar]
- 21. Palecek S. P., Loftus J. C., Ginsberg M. H., Lauffenburger D. A., Horwitz A. F. (1997) Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness. Nature 385, 537–540 [DOI] [PubMed] [Google Scholar]
- 22. Rantala J. K., Pouwels J., Pellinen T., Veltel S., Laasola P., Mattila E., Potter C. S., Duffy T., Sundberg J. P., Kallioniemi O., Askari J. A., Humphries M. J., Parsons M., Salmi M., Ivaska J. (2011) SHARPIN is an endogenous inhibitor of β1-integrin activation. Nat. Cell Biol. 13, 1315–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cifone M. A., Fidler I. J. (1980) Correlation of patterns of anchorage-independent growth with in vivo behavior of cells from a murine fibrosarcoma. Proc. Natl. Acad. Sci. U.S.A. 77, 1039–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Retta S. F., Cassarà G., D'Amato M., Alessandro R., Pellegrino M., Degani S., De Leo G., Silengo L., Tarone G. (2001) Cross talk between β(1) and α(V) integrins. β(1) affects β(3) mRNA stability. Mol. Biol. Cell 12, 3126–3138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang L., Liang Y., Li Z., Cai X., Zhang W., Wu G., Jin J., Fang Z., Yang Y., Zha X. (2007) Increase in β1–6 GlcNAc branching caused by N-acetylglucosaminyltransferase V directs integrin β1 stability in human hepatocellular carcinoma cell line SMMC-7721. J. Cell Biochem. 100, 230–241 [DOI] [PubMed] [Google Scholar]
- 26. Frisch S. M., Screaton R. A. (2001) Anoikis mechanisms. Curr. Opin. Cell Biol. 13, 555–562 [DOI] [PubMed] [Google Scholar]
- 27. Frisch S. M., Francis H. (1994) Disruption of epithelial cell-matrix interactions induces apoptosis. J. Cell Biol. 124, 619–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bretland A. J., Lawry J., Sharrard R. M. (2001) A study of death by anoikis in cultured epithelial cells. Cell Prolif. 34, 199–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yu J. T., Foster R. G., Dean D. C. (2001) Transcriptional repression by RB-E2F and regulation of anchorage-independent survival. Mol. Cell Biol. 21, 3325–3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ono M., Hakomori S. (2004) Glycosylation defining cancer cell motility and invasiveness. Glycoconj. J. 20, 71–78 [DOI] [PubMed] [Google Scholar]
- 31. Zhao Y., Sato Y., Isaji T., Fukuda T., Matsumoto A., Miyoshi E., Gu J., Taniguchi N. (2008) Branched N-glycans regulate the biological functions of integrins and cadherins. FEBS J. 275, 1939–1948 [DOI] [PubMed] [Google Scholar]
- 32. Janik M. E., Lityńska A., Vereecken P. (2010) Cell migration-the role of integrin glycosylation. Biochim. Biophys. Acta 1800, 545–555 [DOI] [PubMed] [Google Scholar]
- 33. Muller C., Paupert J., Monferran S., Salles B. (2005) The double life of the Ku protein. Facing the DNA breaks and the extracellular environment. Cell Cycle 4, 438–441 [DOI] [PubMed] [Google Scholar]
- 34. Rizki A., Mott J. D., Bissell M. J. (2007) Polo-like kinase 1 is involved in invasion through extracellular matrix. Cancer Res. 67, 11106–11110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Coene E. D., Gadelha C., White N., Malhas A., Thomas B., Shaw M., Vaux D. J. (2011) A novel role for BRCA1 in regulating breast cancer cell spreading and motility. J. Cell Biol. 192, 497–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zheng L., Jia J., Finger L. D., Guo Z., Zer C., Shen B. (2011) Functional regulation of FEN1 nuclease and its link to cancer. Nucleic Acids Res. 39, 781–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guerra L., Guidi R., Slot I., Callegari S., Sompallae R., Pickett C. L., Åström S., Eisele F., Wolf D., Sjögren C., Masucci M. G., Frisan T. (2011) Bacterial genotoxin triggers FEN1-dependent RhoA activation, cytoskeleton remodeling and cell survival. J. Cell Sci. 124, 2735–2742 [DOI] [PubMed] [Google Scholar]
- 38. Tell G., Fantini D., Quadrifoglio F. (2010) Understanding different functions of mammalian AP endonuclease (APE1) as a promising tool for cancer treatment. Cell Mol. Life Sci. 67, 3589–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Carretero M., Escámez M. J., García M., Duarte B., Holguín A., Retamosa L., Jorcano J. L., Río M. D., Larcher F. (2008) In vitro and in vivo wound healing-promoting activities of human cathelicidin LL-37. J. Invest. Dermatol. 128, 223–236 [DOI] [PubMed] [Google Scholar]
- 40. Wang W., Brandt P., Rossi M. L., Lindsey-Boltz L., Podust V., Fanning E., Sancar A., Bambara R. A. (2004) The human Rad9-Rad1-Hus1 checkpoint complex stimulates flap endonuclease 1. Proc. Natl. Acad. Sci. U.S.A. 101, 16762–16767 [DOI] [PMC free article] [PubMed] [Google Scholar]