Background: Ataxia telangiectasia is a genetic disease caused by biallelic mutations in ATM gene.
Results: Dexamethasone induces a noncanonical splicing that leads to translation of a shortened ATM variant retaining kinase activity.
Conclusion: ATM may be restored by a new molecular mechanism that overcomes most of mutations so far described in ATM gene.
Significance: Drug-induced noncanonical splicing may provide new approaches for genetic diseases.
Keywords: Alternative Splicing, Ataxia, Genetic Diseases, Glucocorticoids, Translation, SDR-mediated Splicing
Abstract
Ataxia telangiectasia (AT) is a rare genetic disease, still incurable, resulting from biallelic mutations in the ataxia telangiectasia-mutated (ATM) gene. Recently, short term treatment with glucocorticoid analogues improved neurological symptoms characteristic of this syndrome. Nevertheless, the molecular mechanism involved in glucocorticoid action in AT patients is not yet known. Here we describe, for the first time in mammalian cells, a short direct repeat-mediated noncanonical splicing event induced by dexamethasone, which leads to the skipping of mutations upstream of nucleotide residue 8450 of ATM coding sequence. The resulting transcript provides an alternative ORF translated in a new ATM variant with the complete kinase domain. This miniATM variant was also highlighted in lymphoblastoid cell lines from AT patients and was shown to be likely active. In conclusion, dexamethasone treatment may partly restore ATM activity in ataxia telangiectasia cells by a new molecular mechanism that overcomes most of the mutations so far described within this gene.
Introduction
Ataxia telangiectasia (AT)4 is a rare autosomal recessive disorder resulting from biallelic mutations in the ATM (ataxia telangiectasia-mutated) gene, which codifies for a protein kinase belonging to the PI 3-kinase family (1). AT patients show a complex phenotype characterized primarily by an early-onset progressive neurodegeneration with cerebellar ataxia, accompanied by oculocutaneous telangiectasias, immunodeficiency, and high incidence of infections and tumors (2–5). Unfortunately, up to now, no therapy is available for treating this condition. Recently, the short term treatment of AT patients with glucocorticoid analogues was shown to improve their neurological symptoms and, in some cases, reverse their cerebellar atrophy (6–10). Thus, ataxic patients are looking with great interest to treatments with glucocorticoid. However, the molecular mechanism involved in glucocorticoid action in AT is still unknown (11).
ATM gene is composed of 66 exons spreading over a genomic region of about 150 kb (12, 13) whose transcription produces 27 different mRNAs, 20 alternatively spliced variants, and 7 unspliced forms (www.ncbi.nlm.nih.gov/IEB/Research/Acembly). Alternative splicing is a frequently used method to increase transcriptome complexity in higher eukaryotes, but recently emerging evidences suggest that noncanonical splice events could notably amplify that complexity (14–16). Distinct from the classic splice-site boundary sequences, short direct repeated (SDR) sequences have been recently found to mediate unusual post-transcriptional processes in plant genes (17, 18).
Given the rapid turnaround of the neurodegenerative process provided by glucocorticoid therapy, in this study, we hypothesized that dexamethasone (dexa) could somehow restore a residual ATM activity, possibly promoting alternative splicing in the ATM gene. These events skip mutations so as to directly replace the deficiency that causes the disorder.
EXPERIMENTAL PROCEDURES
Cell Lines
Lymphoblastoid cell lines (LCLs), established from AT patients (ATM−/− (Table 1)) and healthy donors (ATM+/+), were maintained in RPMI 1640 medium supplemented with 2 mmol/liter l-glutamine, 100 μg/ml streptomycin, 100 units/ml penicillin, and 15% fetal calf serum in 5% CO2 at 37 °C. Cells were treated with 100 nm dexa for 24, 48, or 72 h, respectively for RNA extraction, protein extraction, and ATM-dependent phosphorylation evaluations.
TABLE 1.
Genotypes of AT lymphoblastoid cell lines
Lymphoblastoid cell lines were established from both AT patients (ATM−/−) and healthy volunteers (ATM+/+). In this table, the genotypes of AT cell lines are reported; as shown, all the investigated mutations lead to truncated protein with no detectable ATM levels. Hmz, homozygotes; cpd Htz, compound heterozygotes.
| Genotype | Allele 1 |
Allele 2 |
|||
|---|---|---|---|---|---|
| Mutation | Consequence | Mutation | Consequence | ||
| AT28RM | cpd Htz | 5692C→T | Nonsense/truncation | IVS37 + 2T→C | Splicing/truncation |
| AT50RM | cpd Htz | 7792C→T | Nonsense/truncation | 8283delTC | Deletion/truncation |
| AT129RM | Hmz | 8283delTC | Deletion/truncation | 8283delTC | Deletion/truncation |
RT-PCR
Total RNA was extracted from LCLs treated or not with dexa using the RNeasy Plus mini kit (Qiagen), and cDNA was obtained by PowerScript reverse transcriptase (Clontech). Long distance PCRs were performed by the Advantage 2 PCR kit (Clontech) with specific primers to amplify the CDS of the ATM gene, whereas all other routine PCRs were carried out using the Hot-Rescue DNA polymerase kit (Diatheva).
S1 Nuclease Protection Assay
To perform the S1 nuclease protection assay, a specific DNA probe complementary to ATMdexa1 junction was designed. Labeling of the DNA probe was performed by [α-32P]dCTP (PerkinElmer Life Sciences) incorporation using Klenow fragment, and probe was purified by gel permeation. Total RNA extracted from treated AT cell lines was hybridized with the radiolabeled probe for 18 h at 68 °C after which the hybridization mixture was subjected to S1 nuclease (Promega) digestion. Digested products were finally analyzed on 4% urea-PAGE, and dried gel exposed over Bio-Rad imaging screen cassette was subsequently analyzed using the Bio-Rad GS 250 molecular imager system.
Quantitative Real Time PCR for ATMdexa1 Quantification
One-microgram aliquots of total RNA were reverse-transcribed using random hexamers as primers and the SMARTScribe reverse transcriptase from Clontech according to the manufacturer's instructions. The synthesized cDNAs were then diluted 1/5 and used as templates in SYBR Green quantitative real time PCR assays, performed with the Hot-Rescue real time PCR kit (Diatheva). PCR reactions were set up in a volume of 25 μl containing 0.3 μm of gene-specific primers, 3.5 mm MgCl2, 0.625 units of Hot-Rescue DNA polymerase, and 1 μl of diluted cDNA. DNA amplifications were carried out in the ABI PRISM 7500 sequence detection system platform (Applied Biosystems). Each sample was analyzed in triplicate, and multiple blanks were included in each analysis. Quantitative real time PCR primers were obtained from Sigma-Genosys. HPRT1 gene from Homo sapiens was used as internal standard (HPRT1 forward TATGCTGAGGATTTGGAAAGGG and reverse AGAGGGCTACAATGTGATGG). Primer sequences designed to specifically recognize the ATMdexa1 variant were: forward ATCTAGATCGGCATTCAGATTCCA and reverse GTTTAGTAATTGGCTGGTCTGC, whereas primer sequences designed to amplify the native ATM were: forward AAATTCTAGTGCCAGTCAGAGC and reverse TTGCTTTAATCACATGCGATGG. Cycle conditions were 95 °C for 10 min followed by 40 two-step cycles of 20 s at 94 °C and 40 s at 68 °C for HPRT1 and native ATM, whereas for ATMdexa1 amplification, we set up 40 three-step cycles (15 s at 94 °C, 20 s at 65 °C, and 30 s at 72 °C). Amplification plots were analyzed using the 7500 System SDS software (Applied Biosystems), and relative expression data were calculated with the 2−ΔΔCt method as described by Winer et al. (19). The specificity of the amplification products was confirmed by examining thermal denaturation plots, by sample separation in a 2% DNA-agarose gel, and by amplicon sequencing.
Assessment of ATMdexa1 Translation in a Mammalian Cell Model
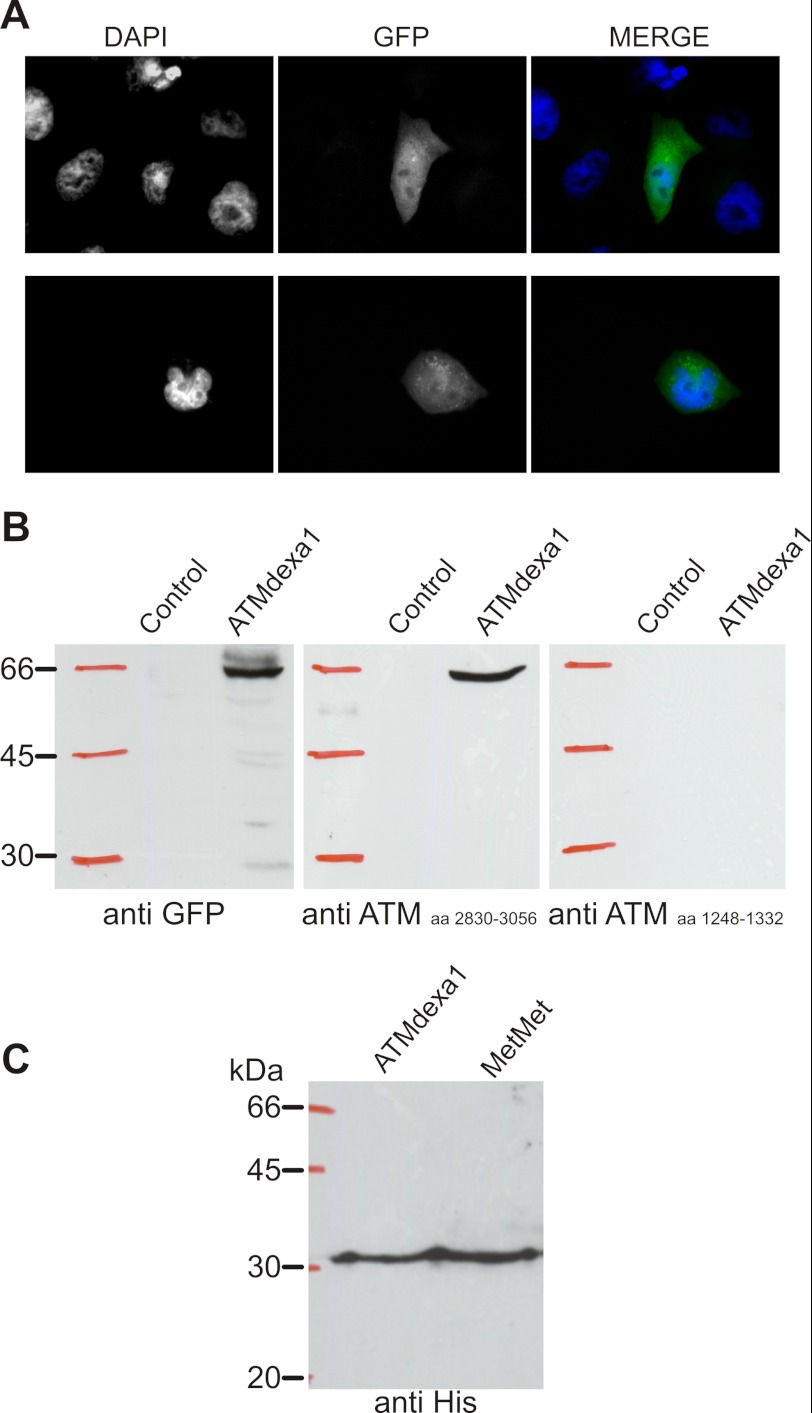
HeLa cells were transfected with the noncanonical spliced ATMdexa1 transcript to assess the mRNA translation. The mammalian expression vector p3×Flag-CMVTM-14 (Sigma) was used for HeLa transfection by ESCORTTM IV transfection reagent (Sigma). In detail, the cDNA of interest was cloned into p3×Flag vector in-frame with GFP so that any translation of ATMdexa1, harboring the PI3K domain in the correct frame, would determine the subsequent translation of the GFP. Transfected cells were subjected to fluorescent microscopy observation and Western blotting for the detection of the translated fusion protein. Microscopic observations were performed by Leica DMLB fluorescent microscope equipped with a DC300F CCD camera.
Start Codon Identification on ATMdexa1 mRNA
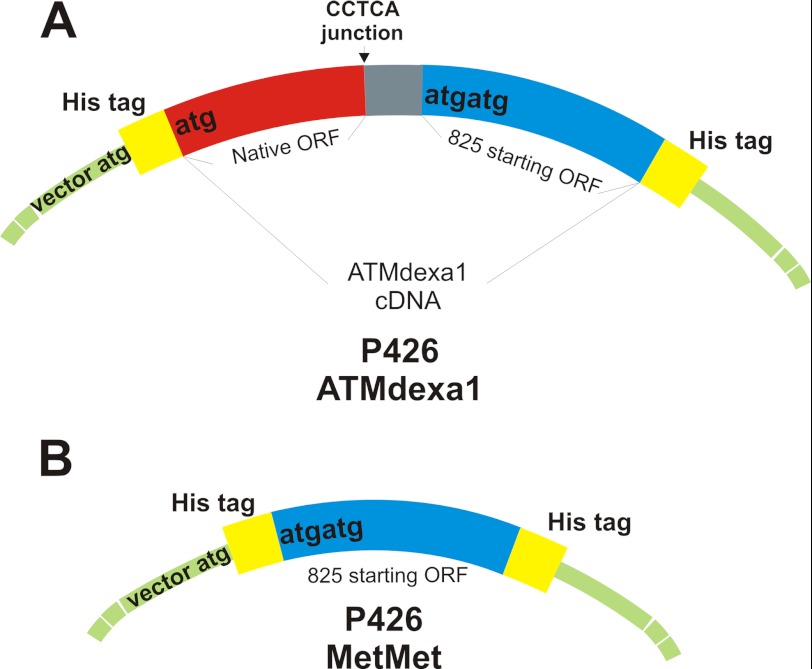
To confirm ATMdexa1 translation in yeasts and to identify the start codon used on ATMdexa1 mRNA, p426-based vectors were employed to transform INVsc1 yeast strain (Invitrogen). Briefly, full ATMdexa1 and the second half of the same cDNA, immediately downstream of the putative start codon (Met-825), were cloned in p426 expression vector to produce recombinant His-tagged proteins (Fig. 1). Recombinant proteins were purified through nickel-Sepharose high performance (Amersham Biosciences) affinity chromatography, from INVsc1 transformed with p426His-ATMdexa1-His and p426His-MetMet-His constructs, respectively, and analyzed by Western blotting.
FIGURE 1.
p426-ATMdexa1 and p426-MetMet His-tagged constructs. A, the entire ATMdexa1 cDNA was cloned into p426 expression vector carrying N-terminal and C-terminal His tags. Translation of the transcribed mRNA can potentially generate two recombinant proteins: an N terminus His-tagged protein starting from vector ATG codon (first 90 amino acids of the native protein, colored in red) and a C terminus His-tagged protein from a putative start codon following position 825 (maximum 252 amino acids, colored in blue). B, the second half of ATMdexa1 cDNA (825–1580 residues) was cloned into the p426 expression vector, generating a double-tagged recombinant protein (N terminus and C terminus His tags) of 252 amino acids.
Yeast Complementation Assay
Yeast strains lacking ATM homologue were used in the complementation assay to investigate the functionality of the translated ATMdexa1 protein. In particular, KSC1368 (single mutant for the ATM yeast homolog, tel1Δ) and KSC1402 (double mutant for homologues of ATM and ATR, mec1-81Δtel1Δ) yeast strains were kindly supplied by Prof. Katsunori Sugimoto (20). The ATMdexa1 complementation assay was performed using pYES2 (Invitrogen) or pYES2-ATMdexa1 to transform KSC yeasts. Recombinants were grown on solid or liquid selective medium. Serial dilutions of transformed cells were seeded on solid selective medium containing 5 μg/ml phleomycin at pH 8.0. Transfected yeasts were also inoculated in liquid selective medium supplemented with phleomycin, and growth was monitored for 18 h.
In Vitro Kinase Activity of Recombinant miniATM
Recombinant GST-miniATM was produced in Escherichia coli and purified, under native conditions, by GSH-agarose beads (Sigma Aldrich), to be employed in an in vitro kinase assay essentially as described by Sarkaria et al. (21). Recombinant ATM was diluted in kinase buffer containing 10 mm Hepes, pH 7.4, 50 mm NaCl, 10 mm MgCl2, 10 mm MnCl2, 1 mm DTT, 10 μm ATP, 20 μCi of [γ-32P]ATP (specific activity 6000 Ci/mmol), and 25 μg/μl PHAS-I (Sigma), with 1× protease inhibitor mixture (Roche Applied Science), 10 mm NaF, and 10 mm NaOV3, in a total volume reaction of 40 μl. Kinase reactions were conducted for 30 min at 30 °C. Different conditions were tested including two ATM concentrations both in the absence and in the presence of the specific PI 3-kinase inhibitor wortmannin. In the appropriate conditions, proteins were preincubated with 400 nm wortmannin for 30 min at room temperature before running the kinase reaction. Reactions were stopped with 4× Laemmli buffer and electrophoresed on 14% SDS-PAGE, and the radioactivity was revealed by a Bio-Rad GS 250 molecular imager.
Identification of miniATM in AT Cell Lines and Subcellular Localization
Human lymphoblastoid cell lines established from AT patients were treated with 100 nm dexa. After 48 h, total proteins were extracted from treated and untreated cells, 20 μg/sample was electrophoresed on 10% SDS-PAGE and blotted onto 0.2-μm nitrocellulose, and membrane was probed with anti-ATM antibodies. To determine the subcellular localization of miniATM, AT LCLs were transfected with a p3×Flag-based vector to express the identified miniATM protein fused to its C terminus with GFP or FLAG peptide. Transient transfection was performed through electroporation essentially as reported by Ref. 22; 1 × 108 cells were added with 30 μg of plasmid, subjected to electric shock, and grown for 48 h in complete RPMI. Protein expression was then assessed by fluorescence microscopy, and fluorescent emission was used to determine the subcellular localization of miniATM in expressing cells.
Localization was also performed by indirect immunofluorescence. Cells were layered on a polylysine-treated glass coverslip and subsequently fixed by 4% formaldehyde in PBS. After treatment in cold methanol and permeabilization with 0.2% Triton X-100 in PBS, surfaces were blocked by 3% BSA and 5% goat serum solution in PBS. Anti-FLAG antibody (Sigma-Aldrich) was used 1/80 in 1% BSA/PBS solution, whereas the secondary anti-mouse TRITC-conjugated antibody (Sigma-Aldrich) was utilized 1/150 in 1% BSA/PBS for 1 h at room temperature. After washing procedures, DNA was stained with 4,6-diamino-2-phenylindole (DAPI) at a final concentration of 0.2 μg/ml. Washed slides were mounted and embedded with ProLong antifade reagent (Invitrogen).
Human Cell Extract-based ATM Assay
To confirm a regained kinase activity in AT LCLs by dexa, likely through miniATM induction, we developed an in vitro assay along the lines of Shiotani and Zou (23). This assay provides an “ex vivo ” kinase assay thanks to native protein extraction and the addition of DNA double strand breaks. We optimized the assay to our needs using total protein extracts from control and treated LCLs, adding DNA double strand breaks with no overhangs to selectively induce ATM activity despite the activity of ATR (23) and investigating the phosphorylation of H2A.X substrate (24).
In particular, total protein extracts were obtained through mild sonication in nondenaturing lysis buffer (20 mm HEPES, 0.15 m NaCl, 1.5 mm MgCl2, and 1 mm EGTA, pH 7.4, containing 1 mm DTT, 1× protease inhibitor mixture, and 0.2% Tween 20). Equivalent amounts of protein (20 μg) were diluted in reaction buffer and ATP buffer, as reported (23), and incubated for 30 min at 37 °C in the presence of 160 ng of blunt-ended plasmid DNA in a total 40-μl volume reaction. Reaction was stopped by the addition of 4× Laemmli buffer and then subjected to SDS-PAGE and Western blotting. Nitrocellulose membranes were finally incubated with primary antibody against phospho-H2A.X in serine 139. Concurrently, the same assay using control protein extract from untreated AT LCLs supplemented with recombinant GST-miniATM was performed.
Cycloheximide Treatment
For ATM half-life determination, all LCLs were treated with 25 μm cycloheximide (Sigma; stock solution 25 mg/ml in ethanol, stored at −20 °C) 48 h after dimethyl sulfoxide (DMSO)/dexa addition. Samples were harvested just before cycloheximide treatment, for the time point 0, whereas the others were collected after 2, 4, and 8 h after the addition of the translation inhibitor. Cell extracts were prepared for Western immunoblot analysis.
Western Blotting
Antibodies used in Western blotting analyses were: anti-GFP, anti-ATM H-248, anti-ATM 2C1, and anti-B-glucuronidase (Santa Cruz Biotechnology); anti-ATM core protein (Sigma-Aldrich); anti-histidine tag (Serotec); anti-AKT and phospho-specific antibodies against AKT(Ser-473), c-RAF(Ser-338), ERK1/2(Thr-202/Tyr-204), Chk1(Ser-345) (purchased from Cell Signaling Technology); anti-H2A.XSer139 (Millipore); and anti-ATM 1B10 (Abnova).
RESULTS
Identification and Quantification of an Alternative ATM Transcript Induced by Dexa
With the initial aim of finding possible exon-skipping events, we performed long distance PCR (from start to stop codon of ATM CDS) on cDNA obtained from lymphoblastoid cell lines established from AT patients, treated or not with dexa. Although in control samples the ∼9000-bp amplicon representing the whole CDS of native ATM mRNA was detected, in tester samples a smaller PCR product was observed (Fig. 2A). The sequence revealed an unusual splicing event using nucleotides 255 and 7843 as alternative 5′ and 3′ excision sites (Fig. 2B). This causes the complete deletion of the exons from 4 to 51 and the joining of exon fragments 3 and 52. Unexpectedly, an SDR sequence of CCTCA at the 3′ end was identical to that of the immediate upstream 5′ end of the deleted sequence; one copy of the SDR was excised, and the other copy was retained in the resultant transcript. This new ATM transcript has been identified in all tested LCLs treated with dexa, including both wild type and ATM-mutated lines (see Fig. 3).
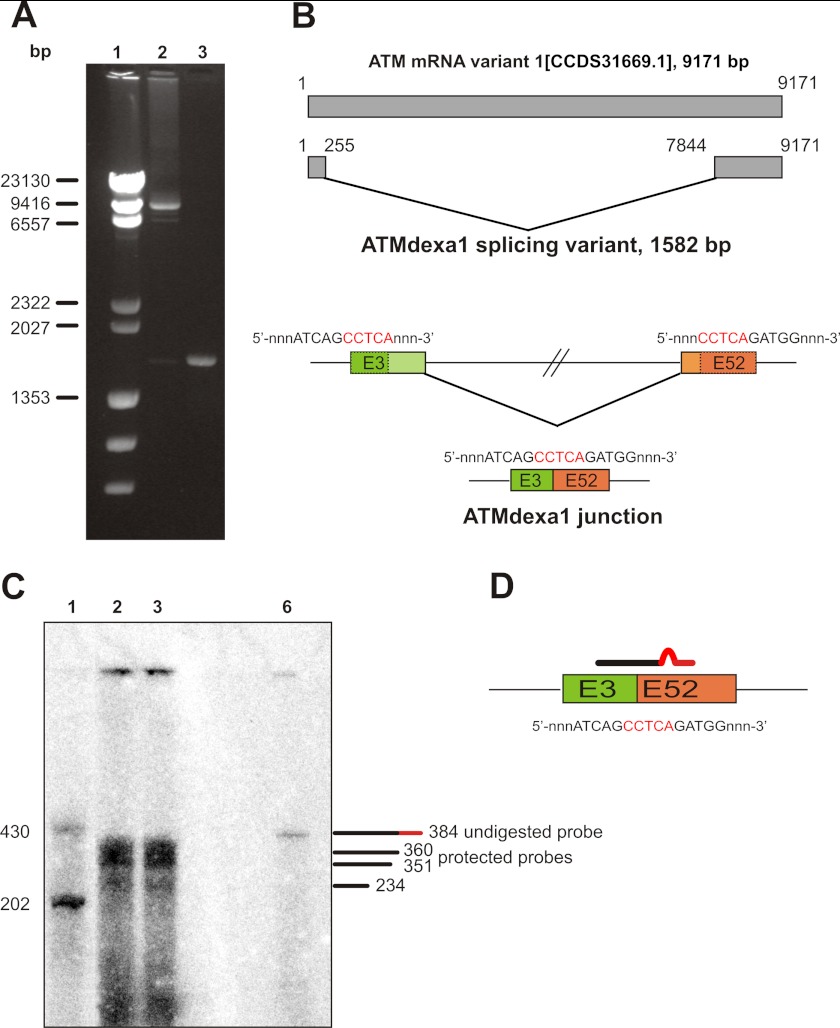
FIGURE 2.
Identification of an alternative ATM transcript induced by dexa. A, RT-PCR on AT lymphoblastoid cell lines. The entire CDS of ATM gene was amplified by RT-PCR on total cDNA templates obtained from AT lymphoblastoid cell lines treated or not with dexa. The full-length CDS was amplified in control cells, whereas a shorter band, lacking the central portion of ATM CDS, was identified in treated cells. Lane 1, λ HindIII DNA ladder; lane 2, untreated AT129RM; lane 3, treated AT129RM. B, scheme representing the alternative ATM splicing variant induced by dexa. Sequencing of the deleted band identified in AT LCLs reveals a noncanonical splicing event in the ATM gene, mediated by a short direct repeat (in red), which leads to the joining of exon 3 with exon 52. The resulting cDNA carries the first three exons and the last 11 exons of native CDS and lacks internal ones. C, S1 nuclease protection assay on AT lymphoblastoid cell lines. To prove the real attendance of SDR-directed spliced ATM mRNA (ATMdexa1), a S1 nuclease protection assay was performed. Total RNA from AT lymphoblastoid cell lines receiving 100 nm dexa was probed with a specific probe complementary to the identified junction and hybridization mixture subjected to S1 digestion. Protection of the probe demonstrates the presence of ATMdexa1 mRNA in both AT lines. Lane 1, ssDNA ladder; lane 2, AT28RM; lane 3, AT129RM; lane 6, undigested probe. D, exemplification of the probe designed for S1 protection assay overlapping the ATMdexa1 junction.
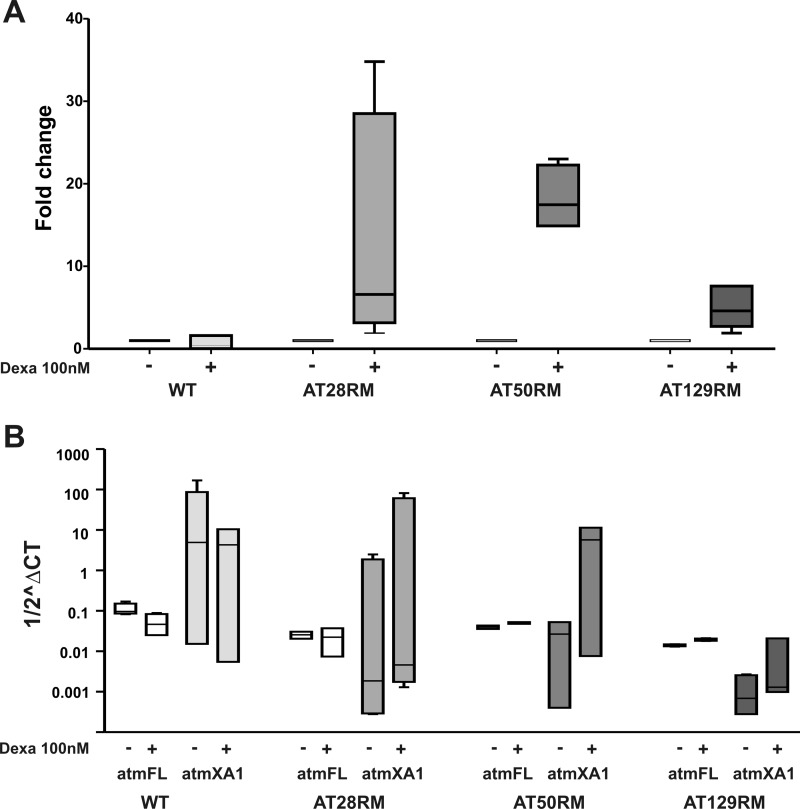
FIGURE 3.
Quantification of ATMdexa1 and full-length ATM versus ATMdexa1, by quantitative real time PCR. A, ATMdexa1 transcript was quantified in wild type and AT lymphoblastoid cells receiving 100 nm dexa with respect to untreated LCLs. In AT samples, the drug treatment leads to ATMdexa1 overexpression, whereas WT resulted in minimal or negative affect by the drug. AT cells are more sensitive to dexa exposure, ranging from 4- to 35-fold change overexpression. (p < 0.05 Wilcoxon matched pairs test n = 6). B, raw data from full-length ATM and ATMdexa1 mRNA quantification were compared by box and whiskers plot. Target amounts are reported in Log10 ^ΔCT (HPRT1 as housekeeping gene). Although the amount of ATMdexa1 resulted in marked increase only in treated AT LCLs (p < 0.05 Wilcoxon matched pairs test), the full-length mRNA was unaffected by dexa (p < 0.05 Wilcoxon matched pairs test n = 6). Based on these results, ATMdexa1 formation is independent of full-length ATM mRNA synthesis, suggesting that the alternative spliced ATMdexa1 variant is directly prompted by dexa. atmFL, full-Length ATM; atmXA1, ATMdexa1. Error bars indicate mean ± S.D.
Because experimental evidences prove that some of the noncanonical splicing events described in the literature could be in vitro artifacts generated by reverse transcriptase (25), we performed an S1 nuclease protection assay to directly prove the new ATM variant authenticity (Fig. 2C). We designed a DNA probe complementary to the mRNA region surrounding the noncanonical exons 3–52 junction of ATM (Fig. 2D). The probe contained 6 mismatching nucleotides 20 residues before its 5′ terminus (total length 384 nucleotides) to distinguish the protected probe from the undigested one. As reported in Fig. 2C, we were able to detect the protected probe (360 residues long), proving the actual existence of the alternative spliced ATM variant induced by dexa (ATMdexa1). Surprisingly, an additional band just below the expected one was identified. By in silico analysis, we found an alternative hybridization event that leads to a larger mismatch producing a shorter protected probe (351 nucleotides in length). Moreover, the hybridization of the probe on the precursor of the ATM transcript generates two truncated bands because of the binding upstream and downstream of the junction (bands of 131 bp, out of gel resolution range, and of 234 bp, respectively).
The extent of ATMdexa1 transcript expression, in the presence and in the absence of dexa stimulus, was evaluated by quantitative real time PCR. Suitable primers, surrounding the noncanonical spliced ATMdexa1 junction, were designed to selectively amplify the newly identified transcript at the expense of the full-length one. The average Ct values for ATMdexa1 were normalized against average Ct values for the HPRT1 transcript used as housekeeping gene. The expression differences in three AT and one WT LCLs were extrapolated. In the presence of dexa stimulus, ATMdexa1 expression resulted in marked increase (up to 35-fold change increase) in all tested ATM−/− LCLs (Fig. 3A). On the contrary, ATMdexa1 expression in wild type LCL was minimally or negatively affected by the drug.
To prove that the action of dexamethasone relies on the noncanonical splicing induction and exclude a generic effect on the overall ATM expression, the expression of the native full-length messenger was also estimated. Only in two LCLs (AT50RM and AT129RM), a very slightly induction, statistically not significant, of the transcript was observed (with a magnitude up to 100-fold lower when compared with ATMdexa1), whereas in WT and in AT28RM, a down-expression was evidenced (Fig. 3B).
Translation of the ATMdexa1 Transcript
The sequence analysis of the ATMdexa1 transcript revealed that the canonical ATM start codon leads to a truncated protein due to a reading frameshift occurring after the described exon junction (Fig. 4). At any rate, various start codons following native ATG could theoretically prime the translation of several ORFs. The translation of these ORFs would escape the mutations contained in the analyzed cDNA maintaining the complete PI3K domain of the native protein. To verify the actual recognition and translation of one of the ORFs, the whole isolated cDNA of ATMdexa1 was cloned in a mammalian expression vector, fused in its 3′ terminus to a GFP, and subsequently transfected into HeLa cells. As reported in Fig. 5A, the GFP fluorescence was detectable in transfected cells, proving that alternative start codons of ATMdexa1 could be effectively recognized and translated in vivo. Protein extracts from transfected and untransfected cells were also tested by Western blotting using anti-GFP and two different anti-ATM antibodies recognizing the core (amino acids 1248–1332) and the C terminus (H-248, amino acids 2830–3056) of the WT protein, respectively. Immunoreactivity with anti-GFP and ATM H-248 revealed, in transfected cells, the same band with an electrophoretic mobility of about 60 kDa corresponding to the fusion protein ATMdexa1-GFP (predicted molecular mass of the longest ORF fused with GFP ∼56 kDa) (Fig. 5B). No bands were detected with both antibodies in untransfected cells. As further evidence, the antibody raised against the amino acids 1248–1332 was not able to recognize the fused protein (last panel) because this region is deleted in the putative ATM variant protein.
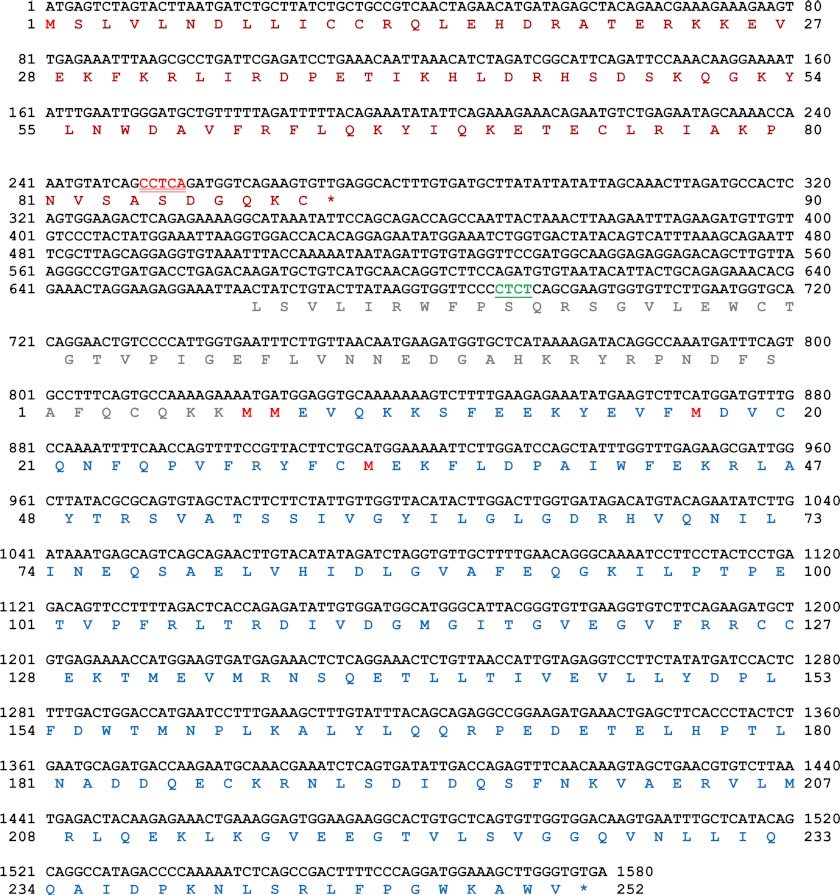
FIGURE 4.
Sequence of ATMdexa1 cDNA and computational analysis of ORF prediction. The nucleotide sequence of the identified noncanonical-spliced ATMdexa1 transcript is depicted in black font; the SDR sequence that mediates this splicing event is indicated in the nucleotide sequence by underlined red font, whereas the region surrounding the mutation 8283ΔTC is in underlined green font. The ORF prediction is shown under the nucleotide sequence. The computational analysis revealed at least two ORFs, the first one starting from nucleotide 1 (native ATG) up to nucleotide 271 (red amino acid sequence) and the second one starting from nucleotide 825 up to nucleotide 1577 (native STOP codon, blue amino acid sequence).
FIGURE 5.
Translation of the ATMdexa1 transcript. A, fluorescence imaging of HeLa cells transfected with p3×Flag-ATMdexa1-GFP fusion protein. GFP translation has been placed in subordination to ATMdexa1 PI3K domain translation in the p3×Flag expression vector and used to transfect HeLa cells. B, Western blots performed on protein extracts from control and transfected HeLa cells revealed a fusion protein recognized both by anti-GFP and by anti-ATM antibodies. In particular, ATM antibody H-248 raised against the C terminus of ATM protein cross-reacted with ATMdexa1-GFP protein, whereas ATM antibody raised against amino acids (aa) in the central portion of the native protein did not. C, identification of the start codon on ATMdexa1 was performed by cloning both the full-length cDNA of ATMdexa1 and the second half of the same cDNA downstream of the putative ATG revealed by in silico ORF prediction (named MetMet, see also Fig. 4). Both constructs were transformed in yeast INVsc strain, and recombinant His-tagged proteins were purified; immunoblot revealed the same band from both recombinants, proving that translation is prompted by the putative ATG at position 825 or downstream giving a protein with the entire PI3K domain (miniATM).
ATMdexa1 start codon identification was performed by cloning both the entire ATMdexa1 cDNA and the second half of the transcript downstream of position 825 (Fig. 4, blue sequence) in a double-His-tagged yeast expression vector (tags in the N and C terminus). Recognition of the start codon at position 825 or downstream would lead to a hypothetical protein of 221–252 amino acids with a predicted molecular mass of between 25 and 29 kDa. The recombinant proteins produced by differently transfected yeasts were then purified. From both transformants, we were able to purify the same translated His-tagged protein (Fig. 5C) with an electrophoretic mobility of about 30 kDa. These results lead to the conclusion that the vector start codon near the 5′ end of ATMdexa1 RNA is somehow disadvantaged and that the translation actually starts only from nucleotide 825 or, less probably, from downstream ATG, in any case producing a protein with the entire PI3K domain of ATM (referred to as miniATM).
Functionality of the miniATM
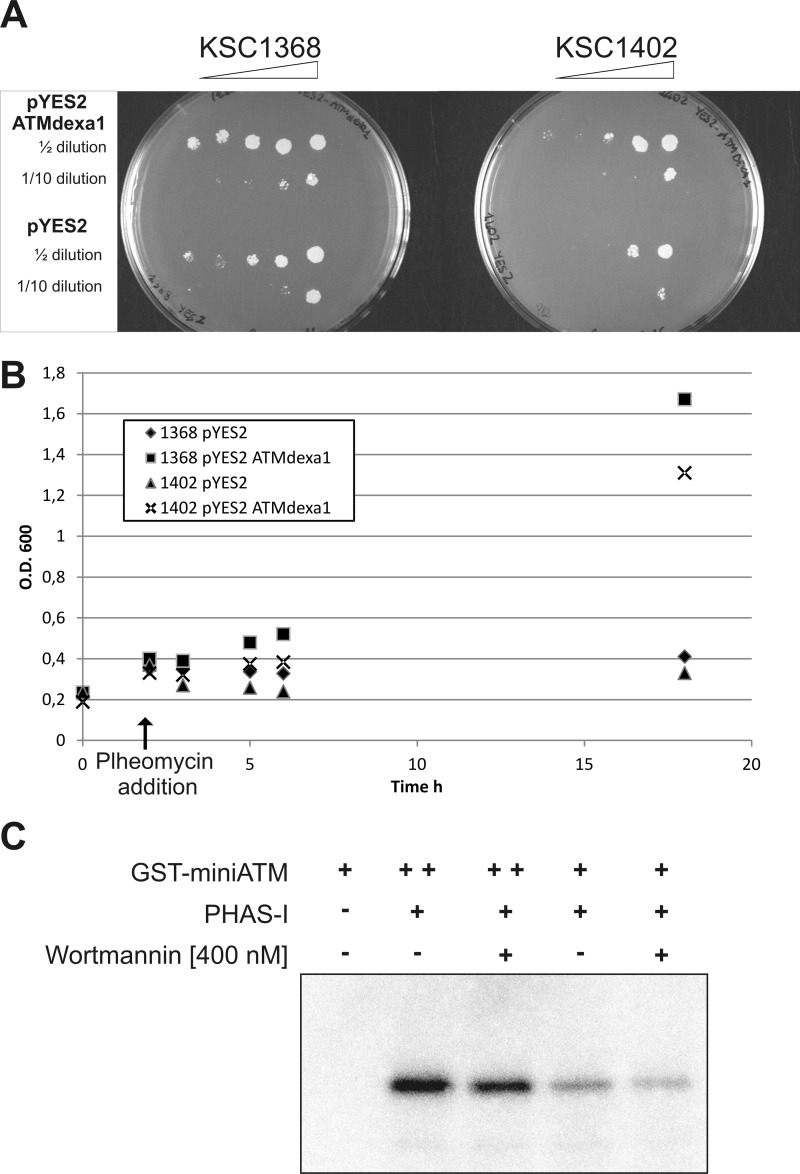
The activity of the produced protein was tested in yeast complementation assays with KSC1402 and KSC1368 yeast strains. The KSC1368 strain is lacking the endogenous ATM homologous gene (TEL1), whereas the strain KSC1402 is a double mutant also lacking the ATR homologous gene (MEC1). Once more, the full sequence of ATMdexa1 was directionally cloned in pYES2 plasmid. Recombinant colonies carrying ATMdexa1 or the empty vector were tested in a growth assay by solid and liquid selective medium containing the chemical DSB inducer phleomycin as described previously (20). As reported in Fig. 6, A and B, the ATMdexa1 recombinant colonies showed an improved growth rate when compared with the controls transfected with the empty vector, suggesting that the produced protein was able to cooperate with other cellular components to complement Tel1 and Mec1 deficiencies and counteract the effect of phleomycin.
FIGURE 6.
Functionality of miniATM. A, yeast complementation assay was performed with tel1Δ (KSC1368) and mec1-81tel1Δ (KSC1402) yeast mutants transfected with pYES2-ATMdexa1 or empty pYES2 constructs. Recombinants were grown on solid selective medium or liquid selective medium supplemented with the DSB inducer phleomycin (B) to assess the ability of ATMdexa1 to complement TEL1 and TEL1-MEC1 deficiencies. O.D., optical density. C, miniATM kinase assay. Recombinant GST-miniATM was tested for in vitro phosphorylation of PHAS-I substrate in the presence of [γ-32P]ATP. Two kinase concentrations were tested, with or without wortmannin inhibition.
To further demonstrate the retained catalytic activity of the identified ATM variant, we produced a recombinant GST-tagged miniATM to be employed in a kinase assay using the PHAS-I substrate. As shown in Fig. 6C, radioactivity was transferred by ATP to PHAS-I, confirming the ability of miniATM to catalyze kinase reactions, using a classic substrate of the native protein, at least in vitro. Phosphorylation assays were also performed after inhibition by wortmannin; in these cases, a decreased phosphorylation rate was demonstrated.
Identification and Subcellular Localization of miniATM in AT LCLs
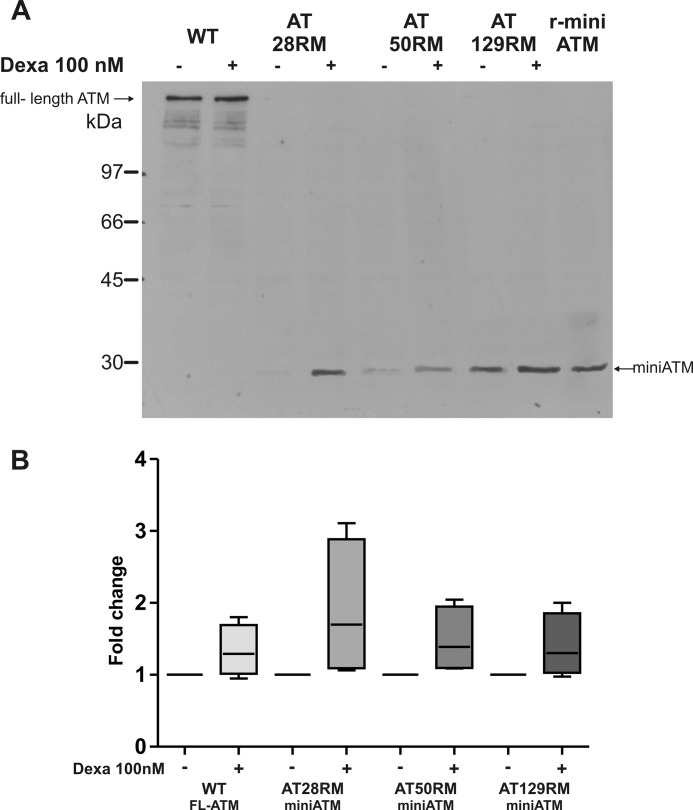
After demonstrating that ATMdexa1 is translated into a functional protein, which carries the C-terminal domain of full-length protein, we looked for this miniATM in AT cell lines treated with dexa. Immunoblots using anti-ATM 2C1 or 1B10, raised against the C-terminal portion of the protein, revealed a band with an electrophoretic mobility nearby 29 kDa (in agreement with the predicted molecular mass, Fig. 7A) that co-migrate with the recombinant miniATM deprived of GST tag. This band was remarkable also in AT-untreated lines but was more represented in treated cells, suggesting that the identified new transcript of ATM induced by dexa was effectively translated into a miniATM protein, which carries the C-terminal catalytically active portion of native protein. The increase of miniATM ranged from 40 to 100% (Fig. 7B) in all the tested AT cell lines, whereas it was not detectable in the WT cell line. From the same immunoblot, the full-length ATM was noticeable in WT cells. Curiously, dexa also induced the full-length protein production of about 40%. As expected, the full-length ATM was not detectable in AT cell lines.
FIGURE 7.
ATM identification and protein quantification in lymphoblastoid cell lines. A, total protein extracts from wild type and AT LCLs, treated or not with dexa, were run on 10% SDS-PAGE, transferred to nitrocellulose membrane, and probed with anti-ATM 1B10, which recognizes the C terminus of the native protein. As expected, full-length protein was found only in wild type LCL, whereas the band of miniATM (29 kDa) was detected in all tested AT LCLs and overexpressed by dexa. The full-length ATM was slightly induced by dexa in WT as well. Recombinant “tag-free” miniATM (r-miniATM), reported in the last lane, showed the same electrophoretic mobility of AT miniATM. B, box and whiskers plot obtained by densitometric quantification of Western blot protein bands of full-length ATM (WT LCL) and miniATM (AT LCLs) by using 1C2 or 1B10 anti-ATM antibodies, after normalization to B-glucuronidase. -Fold change values are reported for each treated cell line. Both full-length ATM and miniATM quantities resulted in increase after dexa addition (p < 0.05 Wilcoxon matched pairs test n = 7). FL-ATM, full-Length ATM. Error bars indicate mean ± S.D.
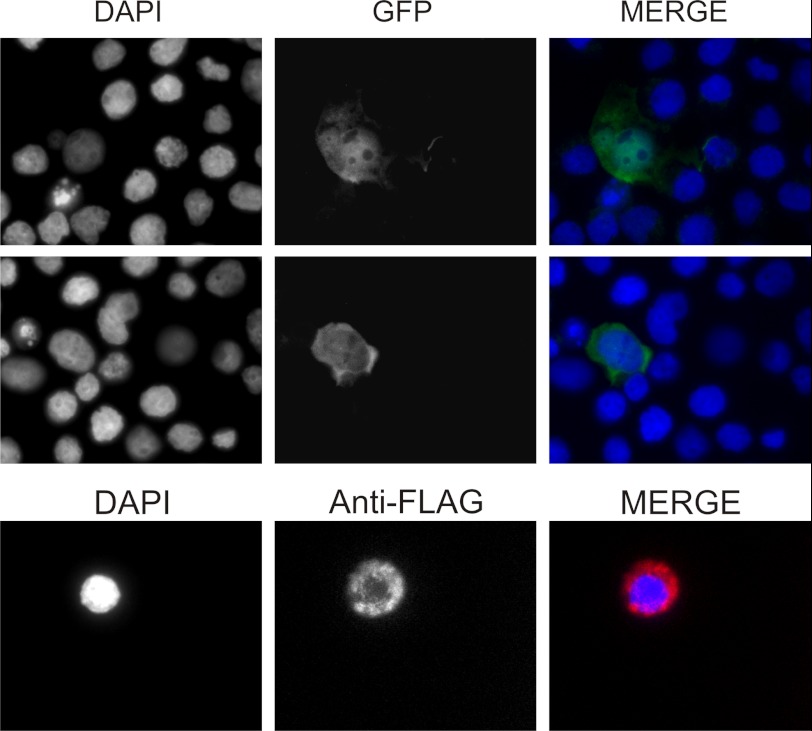
Subcellular localization of miniATM in expressing cells was determined through expression of recombinant protein fused to its C terminus with GFP or FLAG peptide, making indirect immunolocalization with anti-FLAG antibody possible. Fluorescence observations revealed that the protein localizes mainly in cytosol even if some cells also show nuclear accumulation (Fig. 8).
FIGURE 8.
Subcellular localization of miniATM in lymphoblastoid cell lines. Cell distribution of miniATM was observed by C-terminal fused GFP and by indirect immunofluorescence versus C-terminal fused FLAG peptide in AT-expressing cell lines. Cytosolic localization seems to be predominant, but nuclear accumulation is also present.
Dexa Differentially Affects ATM Stability
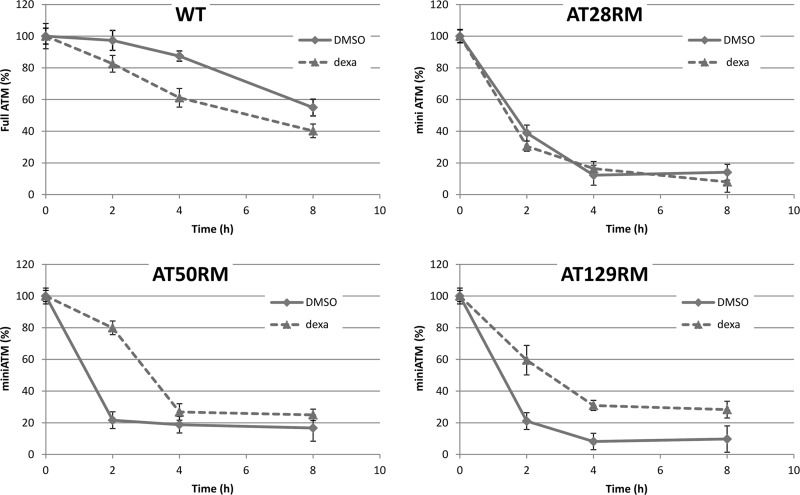
To determine whether alteration of miniATM stability may occur upon dexa exposure, thus contributing to protein cellular accumulation, degradation assay experiments were performed (Fig. 9). Although in the WT LCL, dexa slightly quickens the degradation of full-length ATM (half-life time shifted from about 8 to 6 h in control and treated samples, respectively), miniATM half-life time was increased from 1.5 to 3 h because of dexa stimulus in two of the tested AT LCLs. No differences were detected in the AT28RM sample. By comparing full-length protein with miniATM variant, the variant proved to be less stable in the absence of drug exposure, as demonstrated by the respective half-life time in control WT and AT cells.
FIGURE 9.
Cycloheximide chase assay. Wild type and AT LCLs were prestimulated for 48 h with 100 nm dexa and then treated with 25 μm cycloheximide. Cell lysates were prepared at 0, 2, 4, and 8 h following cycloheximide administration. The amount of full-length and miniATM was determined using Western blotting with 1B10 antibody. Data represent the percentages of ATM quantity derived from densitometric quantification of three independent cycloheximide chase assays (mean ± S.D.); all values are referred to the time 0 point, assumed as 100%. Dexa differentially affected ATM stability in the tested cell lines. DMSO, dimethyl sulfoxide.
ATM Kinase Activity in AT Cell Lines Treated with dexa
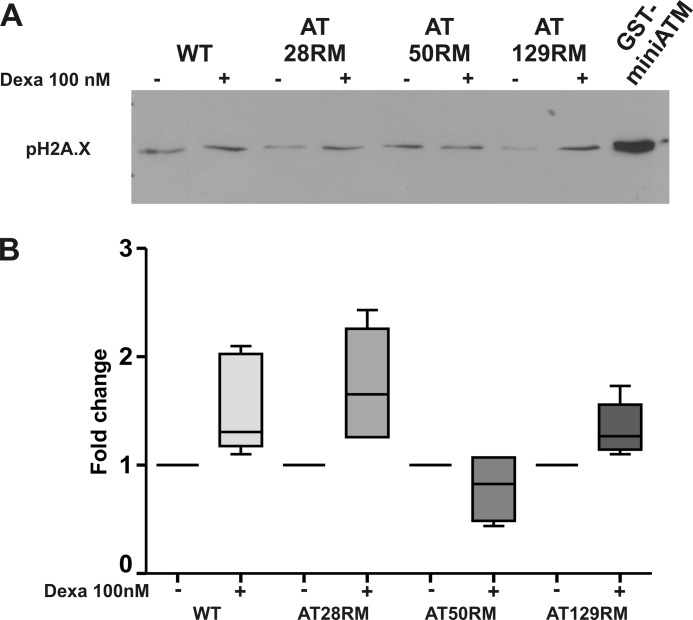
To demonstrate a regained kinase activity in AT LCLs by dexa, likely through miniATM induction, we set up an ex vivo assay using native protein extract from wild type and AT cell lines, as described under “Experimental Procedures.” In particular, we investigated the activation of the well known ATM substrate H2A.X as an early player in DNA damage response (24). In Fig. 10, A and B, we report the activation pattern obtained in the four studied lymphoblastoid cell lines exposed or not to dexamethasone treatment. Although in wild type LCL, γH2A.X activation was little affected by dexa, in AT28RM and AT128RM extracts, γH2A.X was improved by the drug, so that they regained sensitivity to DSB exposure to the same magnitude as the WT sample. The extract from AT50RM showed a nonsignificant reduction. To directly prove that the effect of dexamethasone could be mediated by miniATM induction, we performed the same assay using control protein extract from untreated AT LCLs added with recombinant GST-miniATM. As reported in Fig. 10A (last lane), the recombinant protein was able to exacerbate H2A.X phosphorylation in control extracts after DSB exposure, even in absence of the drug treatment.
FIGURE 10.
ATM kinase activity in AT cell lines treated with dexa. A, lymphoblastoid cell extract-based ATM assay was carried out by adding blunt-ended DNA to native protein extracts from wild type and AT cells. DNA damage response activation was assessed by H2A.X phosphorylation in Ser-139, which is a direct substrate of ATM. Activation was constantly present in wild type LCL and little affected by dexa, whereas AT28RM and AT129RM samples regained the same level of sensitivity to blunt end DNA exposure as the WT sample thanks to dexa treatment. In the last lane, recombinant miniATM strongly promoted H2A.X phosphorylation. B, box and whiskers plot from densitometric quantitation of γH2A.X ex vivo assay. -Fold change values are reported for each treated cell line. WT, AT28RM, and AT129RM showed an increase of γH2A.X quantity; AT samples restored the response to DSB (p < 0.05 Wilcoxon matched pairs test n = 4). The sample AT50RM result was unaffected by dexa treatment and showed a basal level of γH2A.X already comparable with the WT level. Error bars indicate mean ± S.D.
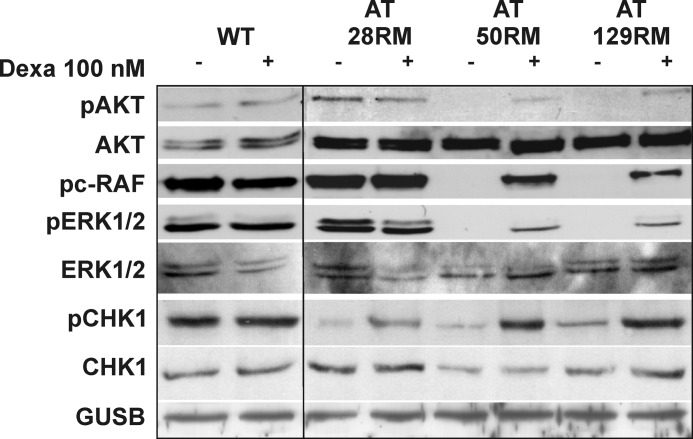
Finally, the effects of miniATM expression and activity on cellular signaling were evaluated by investigating a series of ATM-dependent phosphorylation events in AT and WT cell lines treated or not with dexa. After a 72-h treatment, two (AT50RM and AT129RM) out of three AT cell lines receiving the drug presented a remarkable activation of AKT, assessed by phosphorylation in Ser-473, which is totally absent in untreated cells (Fig. 11). WT sample was not affected by drug treatment. The same “on-off” activation pattern was highlighted for c-RAF and ERK1/2, suggesting the activation of the growth stimulus pathway. AT28RM showed no induced phosphorylation pattern for AKT and c-RAF and only a slight activation of p42 ERK. The direct ATM substrate Chk1 was also tested. Phosphorylation on serine 345 was assayed, and higher levels of activation in all treated AT cell lines were detected. Again, the WT sample was not responsive to drug treatment.
FIGURE 11.
Effects of miniATM induction by dexa were investigated on ATM-dependent cellular signaling. The phosphorylation pattern revealed the rescue of phospho-AKT (pAKT) in serine 473, phospho-c-RAF (pc-RAF), and phospho-Erk1/2 (pERK1/2) in two AT cell lines and the boost of phospho-Chk1 (pChk1) in serine 345 in all tested LCLs.
DISCUSSION
In this study, we describe a drug-induced SDR-mediated splicing event in the ATM precursor mRNA of lymphoblastoid cell lines. To our knowledge, this is the first description of such an event in mammalian cells, and this opens the possibility that SDR-mediated splicing, already described in plants (18), could also affect transcriptome complexity in mammals. The final consequence is that this event can, to some extent, act as exon-skipping therapy in genetic diseases. Clearly, the necessary condition to reach such a result is the translation of the obtained transcript into a functional protein. For this reason, the identification of ATMdexa1 transcript and miniATM protein in AT LCLs treated with dexa is of great clinical interest. In fact, if dexa is somehow able to promote the identified splicing event on ATM precursor mRNA, leading to the translation and overexpression of a shortened ATM variant, treatment with the drug could be an exceptional bailout for AT patients.
As hypothesized, we demonstrated that dexa effectively induces the ATMdexa1 isoform and that this occurs only in AT cells. Because we demonstrated that dexa is able to increase the ratio between ATMdexa1 and native ATM, the drug action is reasonably directed to the promotion of the SDR-mediated splicing event rather than to the overall increase of ATM transcription and precursor mRNA synthesis.
Subsequently, the translation of ATMdexa1 transcript has been validated first in a mammalian cell model (HeLa), then in yeast, and finally, in human lymphoblastoid cell lines. As reported, the noncanonical splicing and the choice of the alternative ORF could produce a C terminus ATM variant starting from codon at position 8450 of CDS overpassing all the mutations upstream to this nucleotide. How the new mRNA variant drives the start of translation to the internal ATG is odd. The transcript itself should contain an internal ribosome entry site, permitting the translation of the ATMdexa1 RNA into the miniATM protein. Furthermore, the ATMdexa1 RNA structure should in the same way hamper or abolish the ribosome entry in its 5′ terminus. In support of this hypothesis, in the yeast expression experiments, performed with the p426ATMdexa1 plasmid, the His-tagged protein encoded by the 5′ of the cloned cassette was not detectable. This hypothetical structure should be intrinsic to the new RNA molecule formed by SDR-mediated alternative splicing. Unlike ATMdexa1 mRNA, miniATM protein was found only in AT cells, suggesting that in wild type, the transcript does not give rise to protein translation. Accordingly, with mRNA induction, miniATM was promoted by the drug in AT samples, even if the extent of protein induction was decidedly lower than mRNA. This must be explained by poor ATMdexa1 translation due to the disadvantaged recognition of internal start codons by the ribosomal apparatus. In support of this statement, the production of miniATM protein was definitely lower in cells transfected with the full ATMdexa1 cDNA than in those transfected with the only miniATM coding sequence. The efficacy of translation may also be reduced by the presence of other ATGs upstream to the miniATM open reading frame, which might be recognized by the ribosome, giving rise to shorter translated products. Also, the strength of the hypothetical internal ribosome entry site might be weaker than the regular ones, thus leading to a poor level of miniATM synthesis. Moreover, considering that from cycloheximide experiments dexa seems to induce miniATM stabilization in the cell, the translation efficiency of ATMdexa1 should be even lower. All the above mentioned points could partially account for the discrepancy between ATMdexa1 RNA and miniATM protein outputs.
As previously implied, the absence of miniATM protein in wild type LCL, despite the presence of the ATMdexa1 mRNA, might be due to the lack of interacting partners and/or substrates that are engaged by native ATM, thus allowing the miniATM rapid removal. It is to be noted that in silico analysis revealed four potential PEST sequences in miniATM (data not shown). On the contrary, in AT cells, the absence of full-length ATM provides to miniATM the possibility of entry as a substitute of native ATM, therefore rescuing itself from degradation.
Obviously, miniATM cannot fulfill all the functions of the native protein, but it could, however, retain some residual kinase activity or acquire new functions. First of all, the newly identified miniATM seems to be constitutively active and can counteract the effect of phleomycin in yeast complementation assay, suggesting that it is able to induce DNA repair effectors. Moreover, recombinant miniATM was demonstrated to be active by the kinase assay test. Finally, ATM kinase activity was investigated in wild type and AT cells after dexa treatment. The drug was able to regain sensitivity to DSBs in two mutated LCLs as demonstrated by γH2A.X triggering. Clearly, the classic ATM kinase test cannot be performed because serine 1981 is lacking in miniATM. Regarding p53 activation, phosphorylation on serine 15 was slightly improved in the AT129RM cell line after dexa (data not shown). Intriguingly, the same sample showed higher basal miniATM levels. In the light of the above, miniATM seems to be active. It is noteworthy that a catalytically active truncated ATM protein was recently identified in ATM−/− mice (26).
The above mentioned results are in disagreement with the notion that the N terminus of the protein, which carries nuclear localization sequences and binding sites for chromatin and ATM-interacting protein, is essential for protein functions (27). Because the newly discovered ATM variant induced by dexa lacks the N terminus, it would be expected that it could not exert the full nuclear function. Young et al. (27) reported that mutagenized ATM proteins lacking the N terminus are unable to fully localize in the nucleus and activate the DNA repair effectors. However, the intricate role of ATM and its full functions in cellular compartments are still unclear.
Despite the well known nuclear role of ATM, a cytoplasmic role has been lately proposed (28–31), especially in neurons (32–34). Boehrs et al. (32) suggest that cytoplasmic ATM promotes neuron survival in an insulin-dependent manner. Viniegra et al. suggested that ATM is required for full activation of AKT, the key kinase of the insulin pathway (29). Finally, Kim et al. demonstrated that AKT and ERK1/2 are constitutively down-regulated in ATM−/− neurospheres (35). Consistent with direct replacement of ATM protein, we found that dexa rescues the switch-on of AKT, c-RAF, and ERK1/2, most likely in an ATM-dependent manner, and enhances the activation of Chk1, a direct substrate of ATM.
We are conscious that there is not a full correlation between the extent of miniATM induction and the effects on AT cells. Nevertheless, we should consider that the overall cellular response is the result of a dynamic equilibrium where miniATM activity interrelates with other overlapping proteins. Besides, it is difficult to make quantitative comparisons because the WT and mutant cells were derived from different individuals, with different genetic variability, drug sensitivity, and ATM genotypes. As a final point, we cannot exclude either the existence of other ATM variants, possibly induced by dexa, or the cumulative effects of dexa on further cellular components.
Putting all the pieces together, here we demonstrated that dexamethasone is able to promote a noncanonical splicing event in the ATM gene producing a new ATMdexa1 transcript variant; ATMdexa1 can be translated into a functional protein, which maintains the kinase domain of native ATM, providing to the cell a second chance to produce the protein, albeit with reduced functions. The accumulation into the cell of miniATM may rescue AT cells from the lack of a pivotal protein such as ATM.
In conclusion, the amazing induction of a truncated protein retaining kinase activity could partly justify the dramatic effect of dexamethasone on neurological deterioration of AT patients accumulated over the years and could also represent one of the mechanisms by which the drug acts in treated AT patients.
Acknowledgments
We thank Prof. Katsunori Sugimoto (International Center for Public Health, Newark, NJ) for supplying KSC1368 and KSC1420 yeast strains. We thank Prof. Luigia Rossi for critical discussions and comments. We thank Dr. Francesca Andreoni for DNA sequencing and Dr. Anna Maria Gioacchini for MS/MS sequencing.
This work was supported in part by grants from FanoAteneo and Associazione Nazionale AT “Davide De Marini.”
- AT
- ataxia telangiectasia
- ATM
- ataxia telangiectasia-mutated
- ATR
- ATM and Rad3-related
- SDR
- short direct repeat
- dexa
- dexamethasone
- LCL
- lymphoblastoid cell line
- SPA
- S1 nuclease protection assay
- CDS
- coding DNA sequence
- PI
- phosphatidylinositol
- DSB
- double strand break
- TRITC
- tetramethylrhodamine isothiocyanate.
REFERENCES
- 1. Abraham R. T. (2004) PI 3-kinase related kinases: 'big' players in stress-induced signaling pathways. DNA Repair 3, 883–887 [DOI] [PubMed] [Google Scholar]
- 2. Biton S., Barzilai A., Shiloh Y. (2008) The neurological phenotype of ataxia-telangiectasia: solving a persistent puzzle. DNA Repair 7, 1028–1038 [DOI] [PubMed] [Google Scholar]
- 3. Chun H. H., Gatti R. A. (2004) Ataxia-telangiectasia, an evolving phenotype. DNA Repair 3, 1187–1196 [DOI] [PubMed] [Google Scholar]
- 4. Gilad S., Chessa L., Khosravi R., Russell P., Galanty Y., Piane M., Gatti R. A., Jorgensen T. J., Shiloh Y., Bar-Shira A. (1998) Genotype-phenotype relationships in ataxia-telangiectasia and variants. Am. J. Hum. Genet. 62, 551–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lavin M. F. (2008) Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat. Rev. Mol. Cell Biol. 9, 759–769 [DOI] [PubMed] [Google Scholar]
- 6. Broccoletti T., Del Giudice E., Amorosi S., Russo I., Di Bonito M., Imperati F., Romano A., Pignata C. (2008) Steroid-induced improvement of neurological signs in ataxia-telangiectasia patients. Eur. J. Neurol. 15, 223–228 [DOI] [PubMed] [Google Scholar]
- 7. Broccoletti T., Del Giudice E., Cirillo E., Vigliano I., Giardino G., Ginocchio V. M., Bruscoli S., Riccardi C., Pignata C. (2011) Efficacy of very-low-dose betamethasone on neurological symptoms in ataxia-telangiectasia. Eur. J. Neurol. 18, 564–570 [DOI] [PubMed] [Google Scholar]
- 8. Buoni S., Zannolli R., Sorrentino L., Fois A. (2006) Betamethasone and improvement of neurological symptoms in ataxia-telangiectasia. Arch. Neurol. 63, 1479–1482 [DOI] [PubMed] [Google Scholar]
- 9. Russo I., Cosentino C., Del Giudice E., Broccoletti T., Amorosi S., Cirillo E., Aloj G., Fusco A., Costanzo V., Pignata C. (2009) In ataxia-telangiectasia betamethasone response is inversely correlated to cerebellar atrophy and directly to antioxidative capacity. Eur. J. Neurol. 16, 755–759 [DOI] [PubMed] [Google Scholar]
- 10. Zannolli R., Buoni S., Betti G., Salvucci S., Plebani A., Soresina A., Pietrogrande M. C., Martino S., Leuzzi V., Finocchi A., Micheli R., Rossi L. N., Brusco A., Misiani F., Fois A., Hayek J., Kelly C., Chessa L. (2012) A randomized trial of oral betamethasone to reduce ataxia symptoms in ataxia telangiectasia. Mov Disord. 27, 1312–1316 [DOI] [PubMed] [Google Scholar]
- 11. Gatti R. A., Perlman S. (2009) A proposed bailout for A-T patients? Eur. J. Neurol. 16, 653–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Platzer M., Rotman G., Bauer D., Uziel T., Savitsky K., Bar-Shira A., Gilad S., Shiloh Y., Rosenthal A. (1997) Ataxia-telangiectasia locus: sequence analysis of 184 kb of human genomic DNA containing the entire ATM gene. Genome Res. 7, 592–605 [DOI] [PubMed] [Google Scholar]
- 13. Savitsky K., Platzer M., Uziel T., Gilad S., Sartiel A., Rosenthal A., Elroy-Stein O., Shiloh Y., Rotman G. (1997) Ataxia-telangiectasia: structural diversity of untranslated sequences suggests complex post-transcriptional regulation of ATM gene expression. Nucleic Acids Res. 25, 1678–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li H., Wang J., Mor G., Sklar J. (2008) A neoplastic gene fusion mimics trans-splicing of RNAs in normal human cells. Science 321, 1357–1361 [DOI] [PubMed] [Google Scholar]
- 15. Shao X., Shepelev V., Fedorov A. (2006) Bioinformatic analysis of exon repetition, exon scrambling, and trans-splicing in humans. Bioinformatics. 22, 692–698 [DOI] [PubMed] [Google Scholar]
- 16. Horiuchi T., Aigaki T. (2006) Alternative trans-splicing: a novel mode of pre-mRNA processing. Biol. Cell 98, 135–140 [DOI] [PubMed] [Google Scholar]
- 17. Niu X., Luo D., Gao S., Ren G., Chang L., Zhou Y., Luo X., Li Y., Hou P., Tang W., Lu B. R., Liu Y. (2010) A conserved unusual posttranscriptional processing mediated by short, direct repeated (SDR) sequences in plants. J. Genet. Genomics 37, 85–99 [DOI] [PubMed] [Google Scholar]
- 18. Fan J., Niu X., Wang Y., Ren G., Zhuo T., Yang Y., Lu B. R., Liu Y. (2007) Short, direct repeats (SDRs)-mediated post-transcriptional processing of a transcription factor gene OsVP1 in rice (Oryza sativa). J. Exp. Bot. 58, 3811–3817 [DOI] [PubMed] [Google Scholar]
- 19. Winer J., Jung C. K., Shackel I., Williams P. M. (1999) Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal. Biochem. 270, 41–49 [DOI] [PubMed] [Google Scholar]
- 20. Nakada D., Shimomura T., Matsumoto K., Sugimoto K. (2003) The ATM-related Tel1 protein of Saccharomyces cerevisiae controls a checkpoint response following phleomycin treatment. Nucleic Acids Res. 31, 1715–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sarkaria J. N., Tibbetts R. S., Busby E. C., Kennedy A. P., Hill D. E., Abraham R. T. (1998) Inhibition of phosphoinositide 3-kinase related kinases by the radiosensitizing agent wortmannin. Cancer Res. 58, 4375–4382 [PubMed] [Google Scholar]
- 22. Xia F., Liber H. L. (1995) Electroporation of human lymphoblastoid cells. Methods Mol. Biol. 48, 151–160 [DOI] [PubMed] [Google Scholar]
- 23. Shiotani B., Zou L. (2011) A human cell extract-based assay for the activation of ATM and ATR checkpoint kinases. Methods Mol. Biol. 782, 181–191 [DOI] [PubMed] [Google Scholar]
- 24. Yuan J., Adamski R., Chen J. (2010) Focus on histone variant H2AX: to be or not to be. FEBS Lett. 584, 3717–3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Houseley J., Tollervey D. (2010) Apparent non-canonical trans-splicing is generated by reverse transcriptase in vitro. PLoS. One. 5, e12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li J., Chen J., Vinters H. V., Gatti R. A., Herrup K. (2011) Stable brain ATM message and residual kinase-active ATM protein in ataxia-telangiectasia. J. Neurosci. 31, 7568–7577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Young D. B., Jonnalagadda J., Gatei M., Jans D. A., Meyn S., Khanna K. K. (2005) Identification of domains of ataxia-telangiectasia mutated required for nuclear localization and chromatin association. J. Biol. Chem. 280, 27587–27594 [DOI] [PubMed] [Google Scholar]
- 28. Halaby M. J., Hibma J. C., He J., Yang D. Q. (2008) ATM protein kinase mediates full activation of Akt and regulates glucose transporter 4 translocation by insulin in muscle cells. Cell Signal. 20, 1555–1563 [DOI] [PubMed] [Google Scholar]
- 29. Viniegra J. G., Martínez N., Modirassari P., Hernández Losa J., Parada Cobo C., Sánchez-Arévalo Lobo V. J., Aceves Luquero C. I., Alvarez-Vallina L., Ramón y Cajal S., Rojas J. M., Sánchez-Prieto R. (2005) Full activation of PKB/Akt in response to insulin or ionizing radiation is mediated through ATM. J. Biol. Chem. 280, 4029–4036 [DOI] [PubMed] [Google Scholar]
- 30. Wu Z. H., Shi Y., Tibbetts R. S., Miyamoto S. (2006) Molecular linkage between the kinase ATM and NF-κB signaling in response to genotoxic stimuli. Science 311, 1141–1146 [DOI] [PubMed] [Google Scholar]
- 31. Zhang L., Tie Y., Tian C., Xing G., Song Y., Zhu Y., Sun Z., He F. (2006) CKIP-1 recruits nuclear ATM partially to the plasma membrane through interaction with ATM. Cell Signal. 18, 1386–1395 [DOI] [PubMed] [Google Scholar]
- 32. Boehrs J. K., He J., Halaby M. J., Yang D. Q. (2007) Constitutive expression and cytoplasmic compartmentalization of ATM protein in differentiated human neuron-like SH-SY5Y cells. J. Neurochem. 100, 337–345 [DOI] [PubMed] [Google Scholar]
- 33. Barlow C., Ribaut-Barassin C., Zwingman T. A., Pope A. J., Brown K. D., Owens J. W., Larson D., Harrington E. A., Haeberle A. M., Mariani J., Eckhaus M., Herrup K., Bailly Y., Wynshaw-Boris A. (2000) ATM is a cytoplasmic protein in mouse brain required to prevent lysosomal accumulation. Proc. Natl. Acad. Sci. U.S.A. 97, 871–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oka A., Takashima S. (1998) Expression of the ataxia-telangiectasia gene (ATM) product in human cerebellar neurons during development. Neurosci. Lett. 252, 195–198 [DOI] [PubMed] [Google Scholar]
- 35. Kim J., Wong P. K. (2009) Loss of ATM impairs proliferation of neural stem cells through oxidative stress-mediated p38 MAPK signaling. Stem Cells 27, 1987–1998 [DOI] [PubMed] [Google Scholar]