Background: Jak3 is a tyrosine kinase, and the mechanism of Jak3 interactions with cytoskeletal proteins is not known.
Results: Tyrosine autophosphorylation of SH2 domain of Jak3 facilitated the interactions between the Jak3-FERM domain and cytoskeletal proteins.
Conclusion: Results demonstrate the molecular mechanism of interactions between Jak3 and cytoskeletal proteins.
Significance: Understanding of Jak3 functions has important implications in transplant biology, epithelial wound repair, cancer metastasis, and immune cell migration.
Keywords: Actin, Jak Kinase, Protein-Protein Interactions, SH2 Domains, Wound Healing, FERM Domain, Gelsolin, In Vitro Kinase, Intramolecular Interactions, Villin-Jak3 Interactions
Abstract
Janus kinase 3 (Jak3) is a nonreceptor tyrosine kinase expressed in both hematopoietic and nonhematopoietic cells. Although mutations that abrogate Jak3 functions cause different immunological disorders, its constitutive activation leads to various types of cancer. Previously, we demonstrated that Jak3 interacted with actin-binding protein villin, thereby facilitating cytoskeletal remodeling and wound repair. In this study, we characterize the structural determinants that regulate the interactions between Jak3 and cytoskeletal proteins of the villin/gelsolin family. Functional reconstitution of kinase activity by recombinant full-length (wt) Jak3 using Jak3-wt or villin/gelsolin-wt as substrate showed that Jak3 autophosphorylation was the rate-limiting step during interactions between Jak3 and cytoskeletal proteins. Determination of kinetic parameters showed that phosphorylated (P) Jak3-wt binds to P-villin-wt with a dissociation constant (Kd) of 23 nm and a Hill's coefficient of 3.7. Pairwise binding between Jak3 mutants and P-villin-wt showed that the FERM domain of Jak3 was sufficient for binding to P-villin-wt with a Kd of 40.0 nm. However, the SH2 domain of Jak3 prevented P-villin-wt from binding to the FERM domain of nonphosphorylated protein. We demonstrate that the intramolecular interaction between the FERM and SH2 domains of nonphosphorylated Jak3 prevented Jak3 from binding to villin and that tyrosine autophosphorylation of Jak3 at the SH2 domain decreased these intramolecular interactions and facilitated binding of the FERM domain to villin. Thus we demonstrate the molecular mechanism of interactions between Jak3 and cytoskeletal proteins where tyrosine phosphorylation of the SH2 domain acted as an intramolecular switch for the interactions between Jak3 and cytoskeletal proteins.
Introduction
Janus kinases (Jaks) are a family of nonreceptor tyrosine kinase with four members: Jak1, Jak2, Jak3, and Tyk2. Like other members, Jak3 mediates signals initiated by cytokine through interactions with receptors for IL-2, IL-5, IL-7, IL-9, and IL-15 via the common γ chain of these receptors (1). Jak3 protein contains seven Jak homology (JH)2 domains common with other Jak proteins. The characteristic feature of the Jaks is the presence of a fully functional tyrosine kinase domain (JH1) and a catalytically inactive pseudokinase domain (JH2) (3). Apart from these two, Jaks also contain five other conserved regions. The recently described JH3-JH4 regions have homology with SH2 domains (5). Although the presence of the SH2 domain indicates interactions with other signaling molecule, the specific signaling partner(s) has not been identified. On the other hand, the JH6-JH7 domains have homologies with the FERM domain found in molecules such as Band 4.1, ezrin, radixin, and moesin. Although the FERM domains mediate intermolecular interactions with cytokine receptor (6), they are also involved in intramolecular binding to JH1 kinase domain, thereby enhancing the kinase activity (7). The crystal structure of the JH1 kinase domain has been solved (PDB ID:1 YVJ) but there is no report on the functions and/or crystal structure of other domains of Jak3. Previously, we reported that intestinal epithelial cells (IECs) express functional specific Jak3 (8, 9), whose biological functions had been presumed to be largely limited to lymphocyte and macrophage populations (1), and proposed mechanisms through which activated Jak3 regulated mucosal wound repair through its interactions with cytoskeletal protein villin (8) and mucosal homeostasis (9).
Villin is an actin-binding protein expressed in specific epithelial cells where it can nucleate, cap, sever, and bundle actin filaments (10). Villin knock-out mice showed compromised mucosal wound repair (11), and Jak3 mediated tyrosine phosphorylation of villin was necessary for its role in wound repair (8). However, the molecular mechanism of Jak3 interactions with villin is not known. In this study, we demonstrate a molecular switch on Jak3 that regulates its interactions with cytoskeletal proteins including epithelial-specific villin and ubiquitously expressed gelsolin.
EXPERIMENTAL PROCEDURES
Cell Culture, IL-2 Treatment, Wound Closure, and Cell Proliferation Assays
HT-29 Cl-19A is a human-derived differentiated IEC line. Methods for culture maintenance, IL-2 treatment, wound closure, and cell proliferations were reported before (8, 9).
Site-directed Mutagenesis
Mutations were created by introducing a stop codon (TGA) at different positions in full-length Jak3 cDNA as reported (12). Jak3-SH2 domain with N-terminal FLAG tag in p6X His-ET vector was synthesized by a commercial facility (Integrated DNA Technologies Inc.).
Expression and Purification of the Recombinant Proteins
Wild type or mutant Jak3 cDNAs cloned in pGEX-4T or p6X His-ET were expressed in Escherichia coli BL21 or TKX1 cells using protocols as reported before (12) and detailed in the supplemental Methods.
In Vitro Kinase Assay and Protein-Protein Interaction Study
In vitro kinase and pairwise binding assays were developed (available through the Texas A&M University System, Office of Technology Commercialization, Disclosure 3196HSC10). Kinetic parameters were determined as reported (12).
Stable Transfection
pCDNA-HA-Jak3-wt and pCDNA-HA-Jak3-V484* were stably transfected into the HT-29 CL19 A cells using methods as reported before (9).
Immunoprecipitations (IP), Immunoblotting (IB), and Immunofluorescence Microscopy (IM)
Standard methods for IP, IB, and IM were used as reported before (8), using villin, HA (Santa Cruz Biotechnology Cruz), pY20 (MP Biomedicals), GST (Millipore), His (GenScript), and FLAG (Sigma) antibody.
RESULTS
Recombinant Jak3 Autophosphorylates Itself and Transphosphorylates Cytoskeletal Proteins of Villin/Gelsolin Family
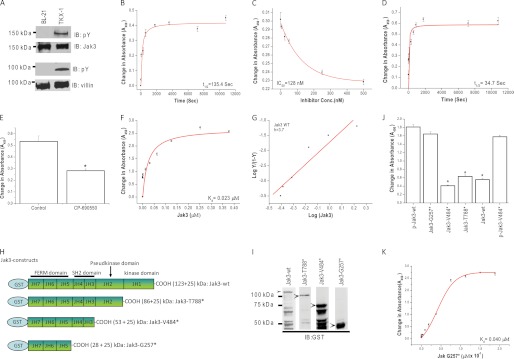
The molecular mechanism and the structural determinants that regulate Jak3 interactions with villin are not known. As a first step to determine these, we expressed and purified the phosphorylated (P) and nonphosphorylated forms of Jak3-wt and villin-wt using the TKX1 and BL21 expression systems, respectively (Fig. 1A). To determine whether the nonphosphorylated form of Jak3-wt was functionally active, we reconstituted its in vitro kinase activity. Since immunoprecipitated Jak3 autophosphorylates itself (15), we determined the autophosphorylation of recombinant Jak3-wt. As shown in Fig. 1B, Jak3-wt autophosphorylated itself in a time-dependent manner with a t½ (the time taken to reach half of the maximum phosphorylation) of autophosphorylation at 135 s. To further confirm these, we determined the dose effect of Jak3 inhibitor CP-690550. Using crystal structure studies, it was shown that CP-690550 directly bound to the kinase domain of nonphosphorylated Jak3 (14). Fig. 1C showed that CP-690550 inhibited Jak3 autophosphorylation in a dose-dependent manner with an inhibition constant (IC50) of 128 nm. Because autophosphorylation of Jak3 led to the activation of Jak3 (13), we determined whether the autophosphorylated Jak3-wt could transphosphorylate cytoskeletal proteins of the villin/gelsolin family. As shown in Fig. 1D, incubation of villin-wt with activated Jak3-wt (from Fig. 1B) led to phosphorylation of villin-wt in a time-dependent manner. The t½ for transphosphorylation of villin-wt by Jak3-wt was ∼35 s. Because in a reaction, the slowest step is considered as rate-limiting, these results showed that autophosphorylation of Jak3 was the rate-limiting step during tyrosine phosphorylation of villin by Jak3. To further confirm that Jak3-wt transphosphorylated villin-wt, the kinase reaction was carried out in the presence of CP-690550. As shown in Fig. 1E, CP-690550 inhibited the phosphorylation of villin-wt by Jak3-wt. To determine whether the phosphorylation by Jak3-wt was villin/gelsolin family-specific, the kinase reaction was also carried out using gelsolin-wt as substrate. Jak3-wt also phosphorylated recombinant gelsolin-wt in a time-dependent manner (supplemental Fig. S1A).
FIGURE 1.
Recombinant full-length Jak3 autophosphorylates itself and transphosphorylates villin. A, the expression and tyrosine phosphorylation of recombinant proteins were detected through Western analysis using Jak3, villin, and pY20 antibody. B, changes in tyrosine autophosphorylation of Jak3-wt were detected in the presence or absence (control) of ATP using a 96-well Multiplate coated with GST-Jak3-wt using pY20 antibody, horseradish peroxidase (HRP)-conjugated secondary antibody, and 3,3′,5,5′-tetramethylbenzidine substrate kit (Thermo Scientific). Absorbance was measured at 450 nm in an ELISA plate reader. C, similar experiments were performed as in B except in the presence or absence of different concentrations of Jak3 inhibitor CP-690505. D, similar experiments were performed as in B except the 96-well microtiter plates were precoated with villin-wt proteins and the phosphorylation was induced by the addition of P-Jak3-wt where P-Jak3-wt alone and villin-wt alone were taken as controls. E, similar experiments were performed as in D except in the presence of Jak3 inhibitor CP-690505 and a fixed reaction time of 5 min. F and G, P-Jak3 interacts with P-villin. F, Jak3 interactions with villin were determined through a pairwise binding assay using a 96-well Multiplate precoated with P-villin-wt. These plates were incubated with increasing concentrations of P-GST-Jak3-wt proteins, and the bound P-GST-Jak3-wt was detected using anti-GST antibody as mentioned for B. GST alone was used as a control. Curve fitting was done as reported before (11) using the Hyperbol-fit program in MicroCal Origin to calculate t½ (B and D), Kd (F), and IC50 (C). G, the Hill equation is rearranged as reported before (11) and plotted to show the relation between the log Y/(1 − Y) and log (Jak3), where Y is the fractional saturation of absorbance. The Hill coefficient (h) was derived from the slope. H, schematic representation of GST-Jak3-wt and mutants. I, these wt and mutant proteins were expressed and purified as in A, and Western analyses of the expressed proteins were done using anti-GST antibody. Lanes shown were from the same blot, but unrelated lanes were removed for clarity. Arrows indicate recombinant proteins. J, direct interactions between P-villin-wt and Jak3-wt (control) or its mutants or P-Jak3-wt or P-Jak3-V484* (from Fig. 2A). Interactions were determined using pairwise binding assay as described under “Experimental Procedures.” GST alone was taken as control. K, binding kinetics of GST-Jak3-G257* with P-villin-wt. The Kd for Jak3-G257* binding to P-villin-wt was calculated as in F. * indicates statistically significant differences from p-Jak3-wt p < 0.05, n = 3 experiments. All blots shown are representative from n = 3 experiments. B–G and J–K, values are mean ± S.E.* indicates statistically significant differences from control p < 0.05, n = 3 experiments.
Determination of Kinetic Parameters for Jak3 Interactions with Villin
Because Jak3-wt phosphorylated villin-wt, we determined the binding kinetics of P-Jak3-wt to P-villin-wt. Pairwise binding studies showed that P-Jak3-wt interacted with P-villin-wt in a dose-dependent manner with a Kd of 23 nm and a Hill's coefficient of 3.7 (Fig. 1, F–G). This showed that the binding between P-rJak3 and P-villin was cooperative.
FERM Domain of Jak3 Is Sufficient for the Interactions between Jak3 and Villin
We determined the structural determinants of Jak3 responsible for these interactions. Fig. 1H shows the schematic diagram for truncation mutants of Jak3. Jak3-wt and these mutants were expressed and purified using the BL21 expression system (Fig. 1I) and were allowed to interact with P-villin-wt using a pairwise binding assay. The binding between P-Jak3-wt and P-villin-wt was ∼4-fold higher as compared with binding between Jak3-wt and P-villin-wt (Fig. 1J), which indicated that tyrosine phosphorylation of Jak3 was important for the interactions between Jak3 and villin. Next, we determined whether truncation of Jak3 had an effect on the interactions between P-villin-wt and Jak3. Truncation of either kinase or kinase along with pseudokinase domain (Jak3-T788* and Jak3-V484*, respectively) resulted in decreased binding between Jak3 and P-villin-wt, which was comparable with Jak3-wt. However, truncation of the kinase, pseudokinase, and SH2 domains altogether (Jak3-G257*) resulted in substantial increase in binding between Jak3 and P-villin-wt, which was comparable with P-Jak3-wt. Fig. 1K shows that the FERM domain of Jak3 interacted with P-villin-wt in a dose-dependent manner with a Kd of 40 nm. Taken together, these data suggested that in nonphosphorylated Jak3 protein, the presence of the SH2 domain prevented the FERM domain from binding to P-villin-wt.
Tyrosine Phosphorylation of Jak3-SH2 Domain Facilitates the Interactions between Villin and Jak3
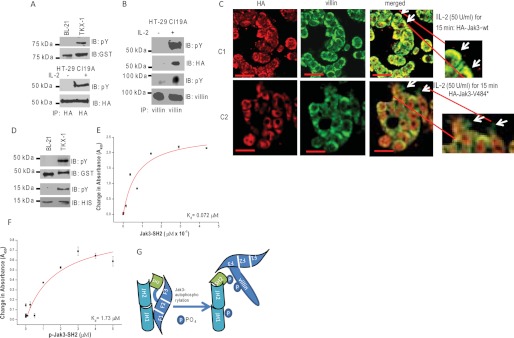
Because the interactions of Jak3-G257* (FERM) with P-villin-wt increased substantially as compared with Jak3-V484* (FERM+SH2), we investigated the possibility whether Jak3-V484* could be tyrosine-phosphorylated in vitro and in a mammalian cell culture model and whether these phosphorylations facilitated the interactions between Jak3 and villin. As shown in Fig. 2A, Jak3-V484* was tyrosine-phosphorylated both in vitro using the TKX1 expression system (first and second panels) and in stably transfected mammalian cells (HT-29Cl-19A) upon IL-2 activation (third and fourth panels). Previously, we showed that IL-2 treatment led to tyrosine phosphorylation of Jak3 in HT-29cl19A cells that facilitated its interactions with villin that enhanced IL-2-induced wound repair (8). Here we showed that IL-2 treatment also led to tyrosine phosphorylation of Jak3-V484* that co-immunoprecipitated with villin only in the presence of IL-2 (Fig. 2B). To further confirm these, we used a pairwise binding assay using tyrosine-phosphorylated Jak3-V484* and P-villin-wt generated using TKX1 cells. As shown in Fig. 1J (sixth bar from the left), tyrosine phosphorylation of the SH2 domain in Jak3-V484* was necessary for the interactions of the FERM domain with P-villin-wt. Determination of IL-2 functions in IEC showed that deletion of the pseudokinase and kinase domain resulted in increased deposition of villin-HA-Jak3V484* complex at the cell margin with defective cell periphery (Fig. 2C) and F-actin turnover (data not shown), which were absent in HA-Jak3-wt-expressing cells. However, all untreated control cells showed no difference (supplemental Fig. S1C). Moreover, IL-2-induced wound closure and cell proliferation were decreased in these mutant cells compared to their wt counterparts (data not shown).
FIGURE 2.
Tyrosine phosphorylation of SH2 domain regulates the interactions between FERM domain of Jak3 and villin. A, tyrosine phosphorylation of Jak3-V484* in vitro and in human IEC. Western analysis of recombinant GST-Jak3-V484* (expressed and purified as in Fig. 1A) was done using pY20 (the top panel) and anti-GST (second from the top) antibodies. Tyrosine phosphorylation of Jak3-V484* in human IEC was determined by stable transfection of pCDNA-HA-Jak3-V484* into HT-29 Cl-19A cells treated with or without 50 units/ml of IL-2 for 15 min as reported before (8). Equal amounts of proteins from the cell lysates were subjected to IP using anti-HA antibody followed by IB using either pY20 (third from the top) or anti-HA (fourth from the top) antibody. B, tyrosine-phosphorylated Jak3-V484* interacts with villin in a human IEC. Similar experiments were done as in A, and the cell lysates were subjected to IP using anti-villin antibody followed by IB using pY20 (first and third from the top), anti-HA (second from the top), and anti-villin (the bottom panel) antibody. C–F, Jak3-V484* induces defective IL-2 functions in IEC. C, localization of HA-tagged Jak3-wt or Jak3-V484* and villin was examined in stably transfected HT-29 Cl-19A cells treated with IL-2 using IM as described under “Experimental Procedures.” Yellow color in the merged panels shows co-localization of HA-tagged protein and villin. Arrows indicate the defective periphery in cells expressing HA-Jak3-V484* (Scale bar-14 μm). D, GST-Jak3-G257* and His-Jak3-SH2 are tyrosine-phosphorylated. Similar experiments were done as in Fig. 1A to generate phosphorylated and nonphosphorylated proteins of GST-Jak3-G257* and His-Jak3-SH2. The expression and tyrosine phosphorylation of these proteins were detected through Western analysis using pY20 antibody (first and third from the top for JAk3-G257* and Jak3-SH2, respectively), anti-GST antibody (second from the top for GST-Jak3-G257*), and anti-His antibody (the bottom panel for Jak3-SH2). E and F, determination of Kd for His-Jak3-SH2 (E) or P-His-Jak3SH2 (F) binding to GST-Jak3-G257*. Similar experiments were done as in Fig. 1K to determine the Kd. G, proposed model for Jak3 interactions with villin. Tyrosine phosphorylation of the SH2 domain of Jak3 disrupts the interactions between the Jak3-FERM domain and Jak3-SH2 domain, making the F3 subdomain of the FERM domain available to interact with phosphorylated villin. A, B, and D, blots shown are representative of n = 3 with similar results. E and F, values are mean ± S.E. * indicates statistically significant differences from control (p < 0.05, n = 6).
Intramolecular Interactions between FERM and SH2 Domains of Jak3 Prevent Jak3 Interactions with Villin
Because nonphosphorylated Jak3-FERM (Jak3-G257*) domain interacted with P-villin-wt but only the phosphorylated form of the Jak3-FERM plus SH2 (Jak3-V484*) domains interacted with P-villin-wt, we determined how the SH2 domain regulated the interactions between Jak3 and villin. To do that, first we determined whether the Jak3-FERM domain interacted with the Jak3-SH2 domain and whether these interactions were affected by tyrosine phosphorylation of either of these domains. Phosphorylated and nonphosphorylated forms of GST-tagged Jak3-FERM and His-tagged Jak3-SH2 domain proteins were expressed and purified using the BL21 and TKX1 expression systems, respectively (Fig. 2D). Pairwise binding kinetics showed that the nonphosphorylated Jak3-FERM domain interacted with the Jak3-SH2 domain with a Kd of 72 nm (Fig. 2E); however, tyrosine phosphorylation of the Jak3-SH2 domain resulted in decreased affinity between the SH2 and FERM domain, which was reflected by an increase in Kd from 72 nm to 1.73 μm (Fig. 2F). On the other hand, tyrosine phosphorylation of the Jak3-FERM domain increased its affinity for the SH2 domain, and the affinity was highest when the FERM domain was phosphorylated and the SH2 domain was not phosphorylated (supplemental Fig. S2, A and B, and supplemental Table ST1). Taken together these results showed that there were intramolecular interactions between the FERM and SH2 domains of Jak3 that prevented its interactions with phosphorylated villin. Moreover, tyrosine phosphorylation of Jak3 specifically at the SH2 domain was necessary to decrease the interactions between the SH2 and FERM domains, making the FERM domain available to interact with phosphorylated villin (Fig. 2G).
DISCUSSION
Jak3 is involved in regulation of different signal transduction pathways mainly through its association and activation of the common γ chain of diverse cytokine receptor including IL-2 (1). Inactivating mutation of Jak3 leads to childhood immunodeficiency (3, 15), and its abnormal activation is associated with hematologic and epithelial malignancies (3, 4), indicating that regulation of its activity is essential for normal hematopoietic and epithelial functions. Although nonhematopoietic expression of this protein has been reported, the understandings of Jak3 functions in these cells were very limited (1, 2). Recently, we showed that Jak3 plays an essential role during mucosal wound repair and homeostasis (8, 9). Interestingly, although Jak3 interactions with cytoskeletal protein villin were essential for its wound repair function (8), the mechanism of Jak3 interactions with cytoskeletal proteins were not known. To achieve this, we characterized the kinetics parameters of Jak3 autophosphorylation and its transphosphorylation of villin. Consistent with our previous study where we showed that Jak3 kinase domain phosphorylated villin (8), the current study showed that full-length Jak3 also phosphorylated villin and gelsolin where the t½ of villin/gelsolin transphosphorylation was lower than that of Jak3 autophosphorylation (Fig. 1). This indicated that Jak3 autophosphorylation was rate-limiting during Jak3-cytoskeletal protein interactions. We further confirmed these interactions by inhibition studies where CP-690550 inhibited both Jak3 autophosphorylation and villin transphosphorylation by Jak3. Although CP-690550 had been reported to bind directly to the kinase domain of truncated Jak3 with IC50 of 35 nm (14), we showed that full-length Jak3 also bound to this inhibitor, albeit with a higher IC50 (128 nm). These indicated that the presence of other domains in Jak3-wt decreased the binding affinity of inhibitor to its kinase domain.
Information on the structure-function relationship of the Jak3-cytoskeletal protein complex showed that P-Jak3-wt interacted with P-villin-wt in a dose-dependent manner with a Kd of 23 nm and a Hill's coefficient of 3.7, indicating a high affinity and cooperative binding between Jak3 and villin (Fig. 1). Due to lack of crystallographic data, the structure-function relationship between Jaks and their interacting partners still remains largely elusive. However, most of the Jaks bind to their cytokine receptor through the N-terminal FERM domain, which encompasses from JH7 through a part of JH4 domain (16). In the case of Jak3, the FERM domain also interacted with the JH1 kinase domain, thereby activating it (14). Our data showed that tyrosine phosphorylation of Jak3 was necessary for its interactions with villin and that these interactions took place through direct contact between the FERM domain of Jak3 and villin (Fig. 1). Tyrosine phosphorylation of the FERM domain per se was not necessary for this interaction; however, the tyrosine phosphorylation of the SH2 domain was essential. This was because under nonphosphorylated conditions, there were intramolecular interactions between the FERM and SH2 domain of Jak3 that prevented villin from interacting with Jak3. Tyrosine phosphorylation of the SH2 domain decreased its affinity for the FERM domain (F1–F3), resulting in disruption of the interactions between these two domains, and this facilitated the interactions between the (now free) FERM domain of Jak3 and villin (Fig. 2G). Conversely and interestingly, our data also showed that tyrosine phosphorylation of the FERM domain increased its affinity for the SH2 domain. However, there was substantial decrease in affinity for the FERM domain when the SH2 domain was tyrosine-phosphorylated irrespective of the phosphorylation status of the FERM domain (supplemental Table ST1). Moreover, pairwise binding showed that nonphosphorylated Jak3-G257* directly bound to villin. These data, combined with data using the IEC culture model, which showed that IL-2 activation resulted in tyrosine phosphorylation-dependent interactions between Jak3-V484* and villin, indicated that tyrosine phosphorylation of the SH2 domain of Jak3 played a major role during interactions between Jak3 and villin. It was reported that the clover-shaped FERM domain is composed of three subdomains: F1 with a ubiquitin-like β-grasp fold, F2 with an acyl-CoA-binding protein-like fold, and F3 that shares the fold of the phosphotyrosine binding or pleckstrin homology domains (3). We predict that the F3 subdomain may be responsible for the intermolecular interactions between tyrosine-phosphorylated villin and the FERM domain (Fig. 2G). Previously, we reported that Jak3 regulated IL-2-induced wound closure and proliferation in IEC (8, 9). The present study showed that although Jak3-V484* interacted with villin, lack of kinase and pseudokinase domains compromised Jak3-mediated physiological functions. This could be due to defective F-actin turnover and failed signal integration leading to surface defects induced by Jak3 mutants (Fig. 2C) and loss of cobblestone morphology as seen in HA-Jak3-wt-expressing epithelial cells (Fig. S1B). Taken together, these results showed for the first time that the molecular mechanism of the interactions between Jak3 and cytoskeletal proteins of the villin/gelsolin family and the structural determinants of Jak3 are responsible for these interactions.
This work was supported, in whole or in part, by National Institutes of Health Grant DK081661 to N. K.). This work was also supported by grants from the Crohn's & Colitis Foundation of America (CCFA Reference Numbers 1351 and 2188).

This article contains supplemental Methods, Figs. S1 and S2, and Table ST1.
- JH
- Jak homology
- IEC
- intestinal epithelial cell
- IP
- immunoprecipitation(s)
- IB
- immunoblotting
- IM
- immunofluorescence microscopy
- P
- phosphorylated.
REFERENCES
- 1. Safford M. G., Levenstein M., Tsifrina E., Amin S., Hawkins A. L., Griffin C. A., Civin C. I., Small D. (1997) JAK3: expression and mapping to chromosome 19p12–13.1. Exp. Hematol. 25, 374–386 [PubMed] [Google Scholar]
- 2. Takahashi T., Shirasawa T. (1994) Molecular cloning of rat JAK3, a novel member of the JAK family of protein tyrosine kinases. FEBS Lett. 342, 124–128 [DOI] [PubMed] [Google Scholar]
- 3. Cornejo M. G., Boggon T. J., Mercher T. (2009) JAK3: a two-faced player in hematological disorders. Int. J. Biochem. Cell Biol. 41, 2376–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lin Q., Lai R., Chirieac L. R., Li C., Thomazy V. A., Grammatikakis I., Rassidakis G. Z., Zhang W., Fujio Y., Kunisada K., Hamilton S. R., Amin H. M. (2005) Constitutive activation of JAK3/STAT3 in colon carcinoma tumors and cell lines: inhibition of JAK3/STAT3 signaling induces apoptosis and cell cycle arrest of colon carcinoma cells. Am. J. Pathol. 167, 969–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kampa D., Burnside J. (2000) Computational and functional analysis of the putative SH2 domain in Janus Kinases. Biochem. Biophys. Res. Commun. 278, 175–182 [DOI] [PubMed] [Google Scholar]
- 6. Huang L. J., Constantinescu S. N., Lodish H. F. (2001) The N-terminal domain of Janus kinase 2 is required for Golgi processing and cell surface expression of erythropoietin receptor. Mol. Cell 8, 1327–1338 [DOI] [PubMed] [Google Scholar]
- 7. Zhou Y. J., Chen M., Cusack N. A., Kimmel L. H., Magnuson K. S., Boyd J. G., Lin W., Roberts J. L., Lengi A., Buckley R. H., Geahlen R. L., Candotti F., Gadina M., Changelian P. S., O'Shea J. J. (2001) Unexpected effects of FERM domain mutations on catalytic activity of Jak3: structural implication for Janus kinases. Mol. Cell 8, 959–969 [DOI] [PubMed] [Google Scholar]
- 8. Kumar N., Mishra J., Narang V. S., Waters C. M. (2007) Janus kinase 3 regulates interleukin 2-induced mucosal wound repair through tyrosine phosphorylation of villin. J. Biol. Chem. 282, 30341–30345 [DOI] [PubMed] [Google Scholar]
- 9. Mishra J., Waters C. M., Kumar N. (2012) Molecular mechanism of interleukin-2-induced mucosal homeostasis. Am. J. Physiol. Cell Physiol. 302, C735–C747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Friederich E., Pringault E., Arpin M., Louvard D. (1990) From the structure to the function of villin, an actin-binding protein of the brush border. Bioessays 12, 403–408 [DOI] [PubMed] [Google Scholar]
- 11. Ferrary E., Cohen-Tannoudji M., Pehau-Arnaudet G., Lapillonne A., Athman R., Ruiz T., Boulouha L., El Marjou F., Doye A., Fontaine J. J., Antony C., Babinet C., Louvard D., Jaisser F., Robine S. (1999) In vivo, villin is required for Ca2+-dependent F-actin disruption in intestinal brush borders. J. Cell Biol. 146, 819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kumar N., Zhao P., Tomar A., Galea C. A., Khurana S. (2004) Association of villin with phosphatidylinositol 4,5-bisphosphate regulates the actin cytoskeleton. J. Biol. Chem. 279, 3096–3110 [DOI] [PubMed] [Google Scholar]
- 13. Zhou Y. J., Hanson E. P., Chen Y. Q., Magnuson K., Chen M., Swann P. G., Wange R. L., Changelian P. S., O'Shea J. J. (1997) Distinct tyrosine phosphorylation sites in JAK3 kinase domain positively and negatively regulate its enzymatic activity. Proc. Natl. Acad. Sci. U.S.A. 94, 13850–13855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chrencik J. E., Patny A., Leung I. K., Korniski B., Emmons T. L., Hall T., Weinberg R. A., Gormley J. A., Williams J. M., Day J. E., Hirsch J. L., Kiefer J. R., Leone J. W., Fischer H. D., Sommers C. D., Huang H. C., Jacobsen E. J., Tenbrink R. E., Tomasselli A. G., Benson T. E. (2010) Structural and thermodynamic characterization of the TYK2 and JAK3 kinase domains in complex with CP-690550 and CMP-6. J. Mol. Biol. 400, 413–433 [DOI] [PubMed] [Google Scholar]
- 15. Macchi P., Villa A., Giliani S., Sacco M. G., Frattini A., Porta F., Ugazio A. G., Johnston J. A., Candotti F., O'Shea J. J., et al. (1995) Mutations of Jak-3 gene in patients with autosomal severe combined immune deficiency (SCID). Nature 377, 65–68 [DOI] [PubMed] [Google Scholar]
- 16. Haan C., Kreis S., Margue C., Behrmann I. (2006) Jaks and cytokine receptors–an intimate relationship. Biochem. Pharmacol. 72, 1538–1546 [DOI] [PubMed] [Google Scholar]