Background: Phospholipase D2 harbors a newly described GEF activity.
Results: The domains and residues responsible for the GEF activity of PLD2 and their physiological relevance in vivo have been identified.
Conclusion: The catalytic site makeup provides a new mechanism of action.
Significance: Significance of the PX domain in Rac2GEF activity of PLD2 is shown.
Keywords: Cell Motility, Leukocyte, Lipase, Phagocytosis, Site-directed Mutagenesis, GEF, I-TASSER Modeling, Catalytic Site, Phospholipase, Structure-Function
Abstract
We have demonstrated that phospholipase D2 (PLD2) is a guanine nucleotide exchange factor (GEF) for Rac2 and determined the PLD2 domains and amino acid site(s) responsible for its GEF activity. Experiments using GST fusion proteins or GST-free counterparts, purified proteins revealed that the PX domain is sufficient to exert GEF activity similar to full-length PLD2. The PLD2-GEF catalytic site is formed by a hydrophobic pocket of residues Phe-107, Phe-129, Leu-166, and Leu-173, all of which are in the PX domain. A nearby Arg-172 is also important in the overall activity. PX mutants altering any of those five amino acids fail to have GEF activity but still bind to Rac2, while their lipase activity was mostly unaffected. In addition to the PX domain, a region in the pleckstrin homology domain (Ile-306–Ala-310) aids in the PX-mediated GEF activity by providing a docking site to hold Rac2 in place during catalysis. We conclude that PLD2 is a unique GEF, with the PX being the major catalytic domain for its GEF activity, whereas the pleckstrin homology domain assists in the PX-mediated activity. The physiological relevance of this novel GEF in cell biology is demonstrated here in chemotaxis and phagocytosis of leukocytes, as the specific PX and PH mutants abolished cell function. Thus, this study reveals for the first time the catalytic site that forms the basis for the mechanism behind the GEF activity of PLD2.
Introduction
Phospholipase D2 (PLD2),2 as the name suggests, is a lipase that catalyzes the breakdown of the phospholipid phosphatidylcholine to phosphatidic acid (PA) and choline (1). PA acts as a central lipid second messenger for many signaling pathways and, therefore, is implicated in a variety of physiological processes such as cell proliferation, migration, and cytoskeletal organization (2). PA also aids in the activation of Ras-GTPase by recruiting its exchange factor Sos (3) or Rac1 (4) to the plasma membrane, which mediates cell spreading (4). Also, PLD2-derived PA mediates the interaction between mTOR and raptor, which is critical for nutrient sensing in human cancer cells (5). On the other hand, it is increasingly evident that PLD2 itself in the absence of PA interacts with many proteins, like growth receptor-bound protein 2 (Grb2), epidermal growth factor receptor (EGFR), and Wiscott Aldrich Syndrome protein (WASp) (6–8). These protein-protein interactions play a significant role in key cellular processes.
PLD2 activity is regulated by tyrosine kinases, serine/threonine kinases, and also by small GTPases like those of the ADP-ribosylation factor (Arf) and Rho families (9, 10). Understanding the mechanism(s) that modulates GTPases is important, as many cancers, like colorectal and breast cancers, express PLD2 at high levels, suggesting a role for PLD2 in cell invasion/metastasis (11). Another small GTPase, Rac2, has received greater attention recently. PLD2 and the small GTPase Rac2 bind together in vivo, and their interaction has an effect on physiological functions such as phagocytosis and chemotaxis (12). Whereas overexpression of PLD2 along with Rac2 enhances phagocytosis, silencing of PLD2 significantly reduced Rac2-mediated phagocytosis as a result of the ability of PLD2 to regulate Rac2 (12–14).
Aside from its well defined lipase activity, PLD2 has recently been shown to bear an additional and novel catalytic function; that of a guanine nucleotide exchange factor (GEF) for the small GTPase Rac2 (13, 15) via turnover and release of the inactive GDP-bound GTPase to that of the active GTP-bound GTPase. The GEF activity occurs in the absence of PLD activity (i.e. the production of PA). PLD2 has also been found to have a GTP/GDP exchange activity toward RhoA that mediates stress fiber formation (16).
The newly defined PLD2-GEF activity makes it a potent GEF for the small GTPase Rac2, which is responsible for multiple effector functions inside the cell (13). Therefore, it is crucial to identify the catalytic site for its GEF activity. As PLD2 lacks the conventional DH domain in tandem with a PH domain, we hypothesized and proved that the PLD2 PX domain could serve in the role of and function as that of the DH domain in conventional GEFs. Following methodical mutational analysis, we successfully determined that the residues phenylalanines 107 and 129, arginine 172, and leucines 166 and 173 in the PLD2 PX domain are the key amino acid residues responsible for its guanine nucleotide exchange reaction. We also found that the newly defined amino acids in the catalytic GEF domains abrogate cell function in vivo. Ascertaining the exact amino acids that encompass the catalytic activity will be crucial for future rational design of pharmaceuticals to down-regulate leukocytes in situations where they can be harmful to human tissues.
EXPERIMENTAL PROCEDURES
GEF Activity; GDP Dissociation from Rac2
To examine the effect of PX, PH, or PXPH on [3H]GDP dissociation from Rac2, 19 pmol of Rac2 was preloaded with 2 μm [3H]GDP. Simultaneously, GST-PX, GST-PH, or GST-PXPH was incubated for 10 min in 20 mm Tris-HCl, pH 7.5, 0.1 mm DTT, 80 mm NaCl, 0.5 mm MgCl2, 0.8 mm AMP-PNP, and 1 mm GTP (80 μl volume). PLD2-WT (19 pmol) or GST alone was used as positive and negative controls, respectively. Preloaded [3H]GDP-bound Rac2 samples were mixed with buffer containing GST-PX, GST-PH, GST-PXPH, PLD2-WT, or GST alone from the previous step (100-μl volume). Aliquots were taken at different times to measure the amount of radiolabeled [3H]GDP bound to Rac2, spotted on Millipore BA85, air-dried, and washed 3× for 5 min with ice-cold 20 mm Tris-HCl, pH 7.5, 100 mm NaCl, and 10 mm MgCl2. The amount of [3H]GDP-bound to Rac2 was measured by scintillation spectrometry. The assay was performed similarly with immunoprecipitated PLD2 or full-length or mutant PXPH (purified).
GEF Activity; GTP Binding to Rac2
To examine the effect of PX, PH, or PXPH on GTP binding, [35S]GTPγS bound to Rac2 in the presence of GST-PX, GST-PH, or GST-PXPH was measured. PLD2-WT (19 pmol) or GST alone was used as the positive and negative control, respectively. 19 pmol of Rac2 were incubated with 8 μm GDP, 6 mm MgCl2 (20-μl volume) for 10 min at room temperature. GDP-bound Rac2 was added to 25 μl of 100 μm AMP-PNP, 1 mm MgCl2, and 1 μm [35S]GTPγS in the absence or presence of GST-PX, GST-PH, or GST-PXPH (75-μl volume). The relative amount of [35S]GTPγS-bound to Rac2 was measured by scintillation spectrometry. GTP binding assays were performed similarly with the immunoprecipitated PLD2 or full-length or mutant PXPH (purified). As a positive control we used a well characterized Rac2 GEF, Vav-1, purchased from OriGene (Rockville, MD).
Immunoprecipitation, SDS-PAGE, and Western Blot Analyses
To confirm the presence of overexpressed protein, we performed SDS-PAGE and Western blot analyses of myc- or HA-tagged mutant proteins that were overexpressed in COS-7 cells. Two days post-transfection, mutant proteins were immunoprecipitated using anti-myc or anti-HA antibodies. Samples were analyzed by SDS-PAGE and Western blot analysis to confirm the presence of mutant PLD2 proteins in the cell lysates. Immunoprecipitated mutants were also used to measure relative GDP dissociation from Rac2.
Rac2 PBD Pulldown Assays
Rac2 PBD pulldown assays were also conducted with purified, recombinant (Rac2 and PLD2 fusion) proteins to measure the activation of Rac2. Five μl of PAK-1-PBD-agarose was added to each sample and incubated at 4 °C for 30 min in the presence of magnesium lysis buffer (25 mm HEPES, 150 mm NaCl, 1% Igepal CA-630, 10 mm MgCl2, 1 mm EDTA, and 2% glycerol). Samples were loaded onto gels, transferred to blotting membranes, and probed with α-HA antibodies to detect recombinant, GTP-bound Rac2. HRP-conjugated secondary antibodies were incubated with PVDFs, and products were visualized using ECL reagents.
Generation of Recombinant PLD2 Mutants Using a Bacterial Expression System
PLD2 PX and PH domain point mutants were created from pcDNA-3.1-mycPLD2-WT, which was used as the template (17), as performed by Mutagenex (Hillsborough, NJ). The molecular identity of all mutants was confirmed by direct sequence analysis per the manufacturer. These PLD2 mutant plasmids were then transfected into COS-7 cells using Lipofectamine and Plus reagents and used for subsequent immunoprecipitation experiments.
Generation of Purified PLD2 from a Baculoviral Expression System
To generate a purified, recombinant, and full-length PLD2 for use as a positive control in certain enzymatic reactions, a baculovirus expression system was used for the overexpression and subsequent purification of PLD2. Briefly, the PLD2-WT gene and the Bsu36I-digested BacPAK5 viral DNA were co-transfected into Sf21 insect cells in separate reactions, which rescued the very large viral DNA and effectively transferred the PLD2 gene into the AcMNPV genome. PLD2 baculoviral stock was selected that overexpressed PLD2-WT and was used to infect Sf21 insect cells for further overexpression of PLD2 and subsequent purifications of PLD2 using the TALON matrix, as previously described in Ref. 12.
Generation of GST-tagged Fusion Proteins Using a Bacterial Expression System
The PX, PH, or PXPH domain of PLD2 was subcloned into the pGEX-4T-1 vector. When single point mutations were required, mutations were generated by site-directed mutagenesis using pGEX-PXPH and were then verified by sequencing the constructs. Individual constructs were transformed into BL21 cells. BL21 culture was induced with 800 μm (GST-PX) or 100 μm isopropyl 1-thio-β-d-galactopyranoside (GST-PH and GST-PXPH) overnight at 16 °C (GST-PX or PH) or 25 °C (GST-PXPH). Cells were harvested, and cell lysates were prepared by resuspending the pellets in lysis buffer (5 mm HEPES, 100 μm sodium orthovanadate, and 0.4% Triton X-100 supplemented with protease inhibitor mixture just before use). Samples were sonicated to obtain crude lysates, which were then centrifuged at 5000 × g for 5 min to obtain the clarified lysates.
GST-tagged proteins in the clarified lysates were pulled down using glutathione-Sepharose beads overnight at 4 °C, after which samples were centrifuged to remove the unbound protein, and the beads were washed 3 times with 1 × PBS. GST-proteins bound to glutathione-Sepharose beads were analyzed via Western blot analysis using anti-GST antibody. GST-PX or GST-PH or GST-PXPH bound to glutathione-Sepharose beads were used to perform GDP dissociation and GTP binding assays. GST alone was used as a negative control.
Generation of GST-free Fusion Proteins
For large scale purification of GST-free recombinant proteins, an in-column digestion with thrombin protease was performed using a glutathione-SepharoseTM 4 fast flow column (GE Healthcare). Briefly, cell lysates were prepared by homogenization and centrifuged at 13,000 rpm for 30 min. The clarified lysates obtained were passed through the column at a flow rate of 0.2 ml/min, and the column was washed overnight with the lysis buffer. For the GST tag removal, thrombin protease (1 unit/100 μg) was injected into the column and incubated for 6–8 h at room temperature. GST-free proteins were eluted with 1× PBS, and the presence of protein in the eluates was analyzed by SDS gel and Coomassie Blue staining.
Multiple Sequence Alignment and Comparison of Three-dimensional Folding between Dbs, Tiam1, and PLD2
PLD2 was compared with two other known Rho family GEFs, Dbs and Tiam1, which activate GTPases Cdc42 and Rac1, respectively. They are the most similar of the GTPases in this protein family that share maximum sequence similarity with the Rac2 GTPase. The sequences of PX-PLD2 and the DH domains of Dbs and Tiam1 or PH domains of PLD2, Dbs, and Tiam1 were aligned using ClustalW. α-Helices and β-sheets of PLD2 were predicted by the PSPIRED server (University College of London, Department of Computer Science).
Three-dimensional folding of the PX-PH domains of PLD2 was derived using the I-TASSER server, as no crystal structure of human PLD2 is available as of yet. The Threading ASSEmbly Refinement algorithm through the server I-TASSER (Center for Computational Medicine and Bioinformatics University of Michigan) employs a hierarchical protein structure-modeling approach based on secondary structure-enhanced profile-profile threading alignment combined with ab initio and Monte Carlo simulation to arrive at low resolution structural predictions of a query amino acid sequence.
Independently of this, PH domains of GEFs, Dbs (PDB ID 1KZ7) and Tiam1 (PDB ID 1FOE) were fetched from model structure files protein data bank and were viewed using the structure viewer Chimera. The predicted PH-PLD2 structure was compared with the crystal structures of Dbs and Tiam1 to examine the secondary structural arrangement. The PDB identifications that were used to visualize Dbs and Tiam1 are 1KZ7 and 1FOE, respectively.
ELISA Assay for Binding Measurements
ELISA assays were performed to determine the level of binding and interaction between PXPH and Rac2. First, we determined the levels of saturation binding of recombinant, purified PXPH and the relevant mutants to the ELISA plate alone. Similar concentrations of protein were bound to the ELISA plate to yield similar binding affinities and were saturated with ∼2.3 pmol of each protein. Next, Rac2 at different concentrations was used to determine binding to either PXPH or the various mutant PXPHs that were seeded in separate ELISA plates at ∼2.3 pmol. After blocking with 2% BSA in Tris-buffered saline, 0.2% Tween 20, the PXPH-coated plate was then incubated with buffer only or with increasing amounts of Rac2 protein ranging from 0.043 to 96.15 pmol.
Bound Rac2 was detected immunologically by incubating the ELISA plate with an anti-HA monoclonal mouse antibody specific for the HA tag on the recombinant Rac2. PXPH/Rac2 binding was quantified spectroscopically at 620 nm after treatment with a secondary, anti-mouse IgG HRP conjugate and adding tobacco mosaic virus.
Fluorescence Resonance Emission Transfer (FRET) Analysis
In these experiments the FRET acceptor was YFP-PLD2, which was prepared by subcloning PLD2 into a mCitrine/YFP vector (pEYFP-C1). The FRET donor was prepared by subcloning Rac2 into a mCerulean/CFP vector (pCerulean-C1). Donor and acceptor plasmids were characterized by direct sequencing. Catalytic activities of the corresponding protein products were verified by PLD assay (for YFP-PLD2) and their ability to bind GTP (for CFP-Rac2). Images were collected and analyzed for FRET stoichiometry as described in Ref. 18 with appropriate excitation and emission filter sets (Chroma Corp.) for CFP, YFP, and FRET (CFP excitation and YFP emission).
Physiological Assays; Phagocytosis
Phagocytosis was measured in the LR5/RAW lines by using fluorescent-labeled (GFP) zymosan A (Saccharomyces cerevisiae) (Invitrogen). The particles were opsonized at 37 °C for 1 h using the zymosan A Bioparticles opsonizing reagent (derived from highly purified rabbit polyclonal IgG antibodies; Invitrogen) and applied to ∼1 × 107 cells such that the input was a ratio of 20 zymosan particles per each cell. After application of the zymosan, the 6-well plates containing the cells were sedimented at 800 × g for 5 min and then incubated at 37 °C for 15 min. Fluorescent-labeled zymosan particles ingested by the cells were counted manually in three different fields. Results are expressed as a percent phagocytosis by counting cells that have phagocytosed between 4–6 zymosan beads on average.
Physiological Assays; Chemotaxis
To ascertain the effect of the mutants on cell functionality, we measured cell migrations (chemotaxis) of macrophages. RAW264.7 cells overexpressing either WT or mutant PLD2 were resuspended at the density of 5 × 105 cells/ml in chemotaxis buffer (Dulbecco's modified Eagle's medium + 0.5% BSA). 200 μl were placed in the upper chambers (or “inserts”) of Transwell inserts that are separated from the lower wells by a 6.5-mm diameter, 8-μm pore polycarbonate membrane. For the study of chemotaxis, mouse macrophage colony stimulating factor was prepared fresh the day of the experiment in 1 × PBS + 0.5% BSA, pH 7.2, at a stock concentration of 1 mm.
When ready for chemotaxis, mouse macrophage colony stimulating factor was diluted to a 3 nm working concentration in 500 μl of chemotaxis buffer and placed into the lower wells of 24-well plates. Cell migration inserts were incubated for 1 h at 37 °C under a 5% CO2 atmosphere. The number of cells that migrated to the lower wells was calculated by placing 10-μl aliquots on a hemocytometer and counting 4 fields in duplicate.
Lipase Assay
COS-7 cells were overexpressed with PLD2-WT or mutants that were immunoprecipitated with anti-myc antibody 2 days post-transfection. Immunoprecipitated PLD2 was processed for PLD2 activity in PC8 liposomes and n-[3H]butanol beginning with the addition of the following reagents (final concentrations): 3.5 mm PC8 phospholipid, 45 mm HEPES, pH 7.8, and 1.0 μCi of n-[3H]butanol in a liposome form as indicated in Liscovitch et al. (19). Samples were incubated for 20 min at 30 °C with continuous shaking. The addition of 0.3 ml of ice-cold chloroform/methanol (1:2) stopped the reactions. Lipids were then isolated and resolved by thin layer chromatography. The amount of [3H]phosphatidylbutanol ([3H]PBut) that co-migrated with PBut standards was measured by scintillation spectrometry.
Statistical Analysis
Data are presented as the mean ± S.E. The difference between means was assessed by the single factor analysis of variance (ANOVA) test. Probability of p < 0.05 indicated a significant difference.
RESULTS
PLD2 Not Only Interacts with Rac2 in Vivo but Affects Rac2-mediated Phagocytosis
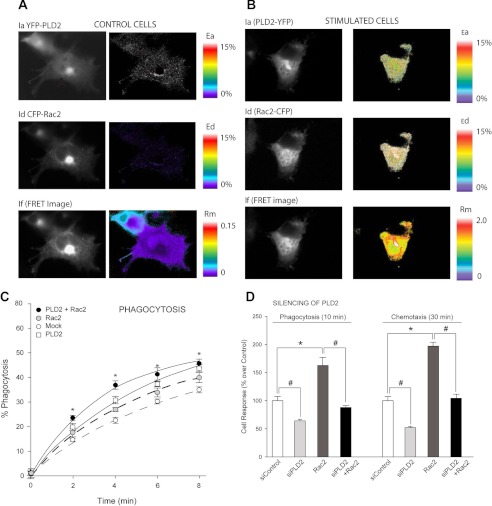
We have detected PLD2-Rac2 association in living mouse RAW264.7 macrophages using FRET. Control, unstimulated cells showed no FRET signal in either acceptor (Ea) or donor (Ed) channels with their ratio (Rm) being almost negligible (Fig. 1A). However, in stimulated cells (Fig. 1B), a high FRET was present in both channels (Ea and Ed) and a robust ratio of Rm ∼ 2.0. Thus, FRET microscopy indicates a Rac2/PLD2 association meaning that these two molecules are in close spatial proximity (at less than 70 nm) and physically interact in living cells.
FIGURE 1.
Demonstration of a molecular association in vivo between Rac2 and PLD2 by FRET and the associated physiological relevance. Shown is FRET between YFP-PLD2 and CFP-Rac2 in control (unstimulated) cells (A) and in stimulated RAW264.7 mouse macrophages (EGF, 3 nm, 10 min) (B). The color bars to the far right indicate the magnitude as either % FRET for the acceptor (Ea), YFP-PLD2 and the donor (Ed), CFP-Rac2, respectively, or the ratio of acceptor to donor molecules expressed in the cell (Rm). C, RAW 264.7 cells were transiently overexpressed with PLD2 and/or Rac2. Results from three independent experiments performed in duplicate are shown as the mean ± S.E. and are expressed in terms of percentage phagocytosis, which is calculated by dividing the number of labeled zymosan-containing cells by the total number of cells. D, phagocytosis or chemotaxis in Transwells of RAW 264.7 cells after transfection or silencing is shown. Data are the mean ± S.E. and are expressed as percent of total cells migrated.
Based on the above evidence, we reasoned that such an interaction must have some physiological significance. Among possible physiological consequences, we chose to perform phagocytosis to determine the effect of PLD2 on Rac2 function. Transient overexpression of PLD2 alone increased phagocytosis (Fig. 1C, open squares). Moreover, overexpression of PLD2 along with Rac2 significantly enhanced phagocytosis of macrophages toward opsonized zymosan (Fig. 1C, filled circles). All this suggests that PLD2 and Rac2 not only interact but that the interaction modulates the function of Rac2. We present physiologically relevant data in Fig. 1D that show when silencing PLD2, the Rac2 effect on both phagocytosis and chemotaxis is more significantly decreased.
PLD2 GEF Activity Resides in Its PX-PH Protein Domains
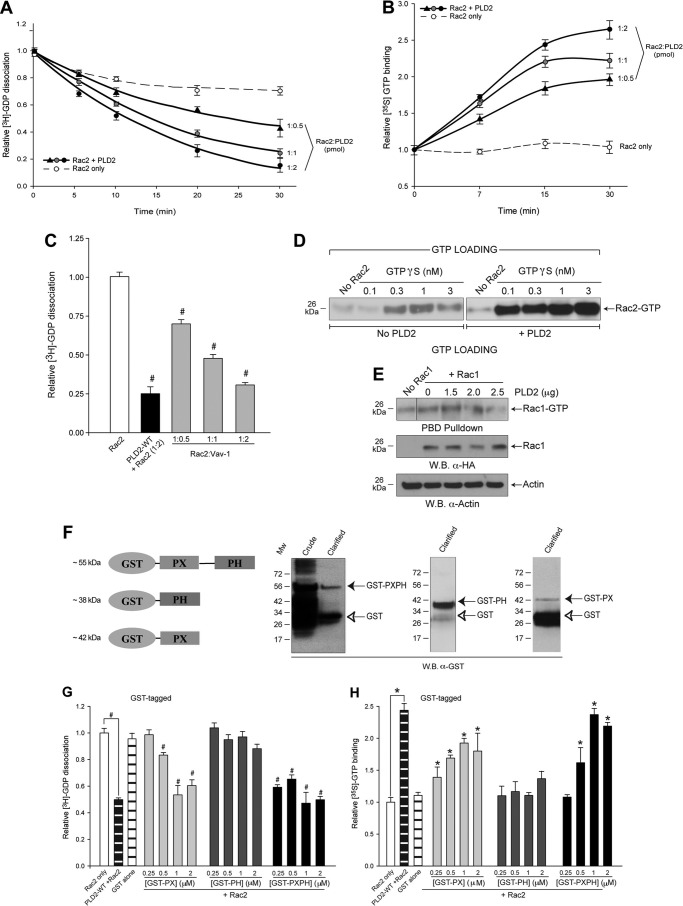
PLD2 intrinsic GEF activity is demonstrated with recombinant PLD2 and Rac2 proteins (Fig. 2, A, B, and D). The GEF function is defined using two different assays: GDP dissociation (Fig. 2A) and GTP binding (Fig. 2B). This GEF activity is time-dependent with a noticeable effect as early as 5 min. Furthermore, at constant concentrations of Rac2, the extent of the turnover varies with the concentration of PLD2. Fig. 2C shows that PLD2 as a GEF was comparable with that of Vav-1, a well known Rac2 GEF (20).
FIGURE 2.
Recombinant PLD2 has a GEF activity (GTP/GDP exchange) on Rac2. The effect of purified, baculoviral PLD2 on [3H]GDP dissociation (A) or [35S]GTP binding of purified (B), baculoviral Rac2 alone (empty circles) or in the presence of 1:0.5 PLD2:Rac2 (triangles), 1:1 PLD2-Rac2 (gray circles) or 1:2 PLD2:Rac2 (black circles) as a function of time was measured. Triplicate results are the mean ± S.E. and are expressed in terms of relative [35S]GTP binding or [3H]GDP dissociation. C, effect of Vav1 on [3H]GDP dissociation of Rac2 alone or in the presence of increasing Vav1. Vav1, a known Rac2 GEF, was used as a positive GEF control. # denotes differences between means that were statistically significant (p < 0.05) by ANOVA and below control (Rac2 alone) as more GEF/GDP dissociation activity (the lower the bars, the higher the activity). D and E, PBD pulldown assays are shown. Purified recombinant, baculoviral proteins were used for the PBD pulldown assay to ascertain the effect of PLD2 on GTP binding to Rac2 (D) or Rac1 (E). D, the left panel is the negative control with no exogenous PLD2, whereas the right panel is a positive control with exogenous, recombinant PLD2. Shown is a representative experiment among three performed with similar results. Equal concentrations of Rac1/2 (19 pmol) were used for the PBD pulldown assays. E, the left lane is the negative control in the absence of both Rac1 and PLD2. The second lane from the left is the Rac1 control in the absence of PLD2. W.B., Western blot. F, PLD2 fusion proteins GST-PX, -PH or -PXPH have GEF activity, as do purified, recombinant PX, PH or PXPH free of GST tags. GST-tagged PX, PH and PXPH proteins are shown in the schematic. All proteins were produced using pGEX-4T-1-PXPH-overexpressing BL21 cells. Expression of respective proteins is shown in Western blots. Empty vector producing GST only is used as a negative control. Each panel shows the expression of each protein in the uninduced or induced (Crude and Clarified) supernatants of bacterial lysates. G and H, GEF activity is shown. Dose response of GST fusion proteins on GDP dissociation (G) and GTP binding (H) is shown. Concentrations of glutathione-Sepharose-bound GST-tagged proteins that were used for the assay are 0.25, 0.5, 1 and 2 μm. # denote differences between means that were statistically significant (p < 0.05) by ANOVA and below control (Rac2 alone) as more GEF/GDP dissociation activity (the lower the bars, the higher the activity) is found with PLD2 and fusion proteins. * denotes differences between means that were statistically significant (p < 0.05) by ANOVA and above control (Rac2 alone) as more GEF/GTP binding activity (the higher the bars, the higher the activity) is found with PLD2 and fusion proteins.
Rac2 activation by PLD2 was also measured by PBD pulldown assay, which specifically detects GTP-bound Rac2. Fig. 2D shows that purified, recombinant PLD2 enhanced GTP binding to Rac2 over samples devoid of PLD2. At the highest concentrations of GTPγS, the density of Rac2-GTP that is pulled down in the presence of PLD2 is almost 8-fold more than that of the negative control without PLD2. We also tested the effect of PLD2 on Rac1, a close homolog of Rac2. Fig. 2E shows that PLD2 did not affect the levels of Rac1 activation when compared with that of Rac2. In conclusion, all data in Fig. 2 show that PLD2 significantly and specifically increased guanine nucleotide exchange on the Rac2 GTPase and, thus, functions as a potent GEF for Rac2.
The next logical experiments were to locate the catalytic site of this activity. Therefore, we examined the PLD2 N-terminal domains, PX and PH. GST alone, GST-PX, GST-PH or GST-PXPH fusion proteins were generated. SDS-PAGE and Western blot analysis of the GST proteins confirmed the presence of the truncated PLD2 domains (Fig. 2F). We have used equal amounts of protein as shown in the gels. Fig. 2G indicates that there is an increasing trend in GDP dissociation measured at 15 min with increasing concentrations of the fusion proteins (0.25, 0.5, 1, and 2 μm). Similar is the case with GTP binding (Fig. 2H). GST alone, which did not reveal any effect, served as a negative control. Thus, these results indicate that the PX domain exerts GEF activity on Rac2 GTPase and recapitulate the GEF activity of the full-length PLD2 protein.
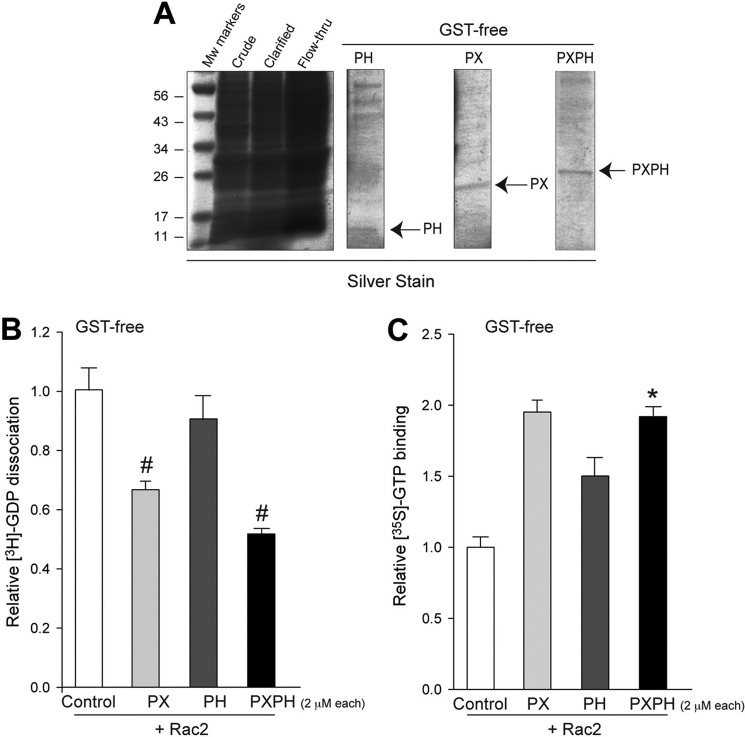
Nevertheless, as the possibility exists that the GST tag itself might interfere with the GEF activity of the PX/PH fusion proteins, we removed the GST tag and performed GEF activity assays in vitro. Fig. 3A shows the purified PX, PH or PXPH proteins. These recombinant proteins still had GEF activity toward Rac2, as measured by GDP dissociation (Fig. 3B) or GTP binding (Fig. 3C). As with the case of GST-tagged proteins described before, GST-free PX showed a greater effect in catalyzing guanine nucleotide exchange when compared with the GST-free PH domain alone. However, the effect is maximal with the GST-free PXPH domain. This implicates that the PH domain assists in PX-mediated guanine nucleotide exchange.
FIGURE 3.
Removal of the GST tag with thrombin protease did not alter the ability of PX/PH domains to catalyze the exchange reaction. A, shown is SDS-PAGE of the purified proteins stained with Gel-code Blue showing GST-free PX, PH, and PXPH proteins at the expected molecular weights. Relative GDP dissociation (B) and GTP binding (C) of Rac2 in the presence of recombinant GST-free PX, PH, or PXPH (2 μm each) is shown. Rac2 alone (white) was used as a control.
Sequence Similarities between the PX Domains of PLD2 and Catalytic DH Domains of Known GEF Proteins
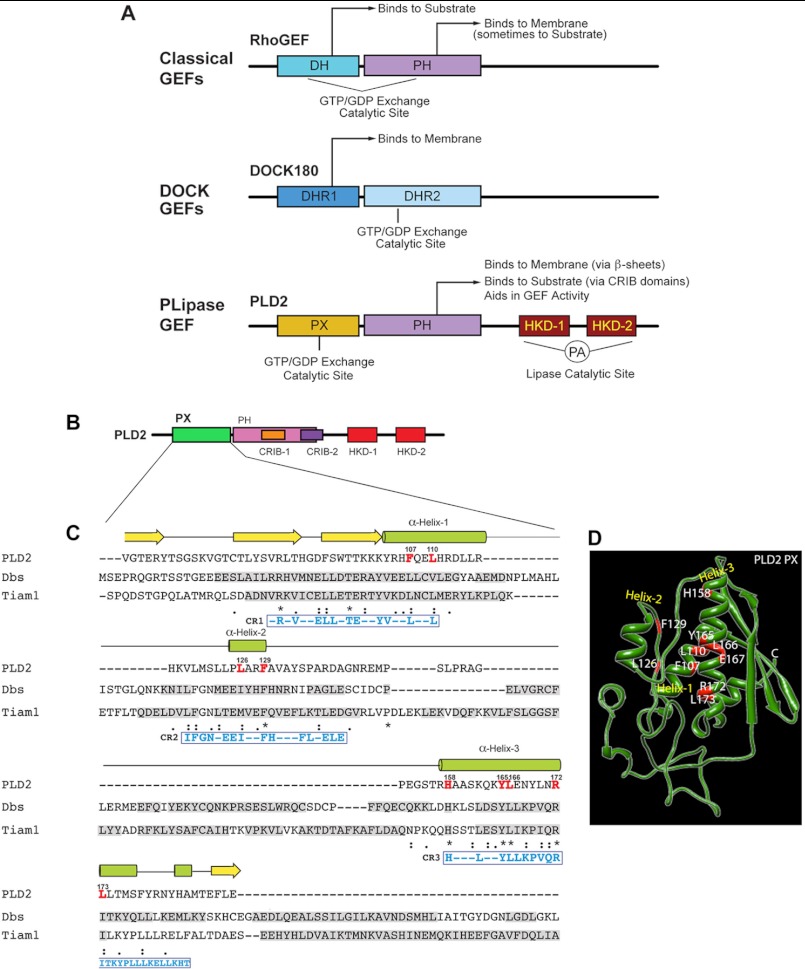
As presented in Figs. 2 and 3, the PX and PH domains of PLD2 have GEF activity (particularly the former). The next question we asked is, What are the specific amino acid residues within the PX and PH domains that are essential for GEF activity of PLD2? An appropriate approach for this was to create mutations to find the essential amino acids involved in the function. However, we had at our disposal very few leads, as PLD2 is a non-conventional GEF. Classical Rho GEFs have a DH-PH tandem, whereas Dedicator of cytokinesis (DOCK) GEFs have DHR domains (Fig. 4A). Because PLD2 has no DH domain (but it has PH) (Fig. 4B), we hypothesized that PX can take the place of DH particularly as Fig. 3 shows that the PX fusion protein does have GEF activity. We decided to perform multiple sequence alignment between PLD2 PX domain and two of the known Rho family GEFs, Dbs and Tiam1, using ClustalW to see if candidate amino acids emerged. Dbs and Tiam1 were chosen for comparison because Rac2 is homologous to both Cdc42 and Rac1 GTPases and because these Rho family GTPases are substrates of Dbs and Tiam1, respectively (12).
FIGURE 4.
Sequence alignments of PX in PLD2 with DH in Dbs and Tiam1. A, shown are schemes depicting the major classes of Rho GEFs with their catalytic GEF domains, as well as the newly-defined PLD2-GEF in this study. B, shown is a schematic representation of PLD2 with the PX domain highlighted in green. C, sequence alignments of PLD2 PX, Dbs DH and Tiam1 DH domains were performed using ClustalW (29). α-Helices and β-sheets of PLD2 are shown on the top of the sequence in green and yellow, respectively. Sequences of α-helices and β-sheets of Dbs and Tiam1 are shown shaded in gray within their respective sequences. Residues conserved within the DH domains of Dbs and Tiam1 and within the helices of the PLD2 PX domain are highlighted in red letters. Identical amino acids, similar amino acids and nearly similar amino acids are shown underneath the sequences as stars, as a single dot, or as double dots respectively. D, shown is a predicted three-dimensional structure schematic of PLD2 PX generated by I-TASSER. Also shown are the mutations considered in this study.
The multiple sequence alignments between the PX domain of PLD2 and the DH domains of Dbs and Tiam1 are shown in Fig. 4C. The secondary structures of PLD2 are presented above the PLD2 sequence. From this alignment, we noticed three conserved regions among PX-PLD2, DH-Dbs, and DH-Tiam1, respectively, that we named CR1, CR2, and CR3. These conserved regions are similar to a key region where the GEF Dbs binds to its substrate GTPase (21).
Within these conserved regions 1–3 we decided to concentrate on several amino acids within the α-helices (shown in red and labeled as α-Helix-1, α-Helix-2 and α-Helix-3) that we hypothesized are implicated in the GEF activity of PLD2. This is because the PX domain of PLD2 is mainly comprised of α-helices (Fig. 4D) like that of the DH domains in the other two GEFs. The amino acids outlined in red in Fig. 4, C and D were targeted for site-directed mutagenesis in subsequent experiments.
Effect of PX Mutations on Full-length PLD2 GEF Activity
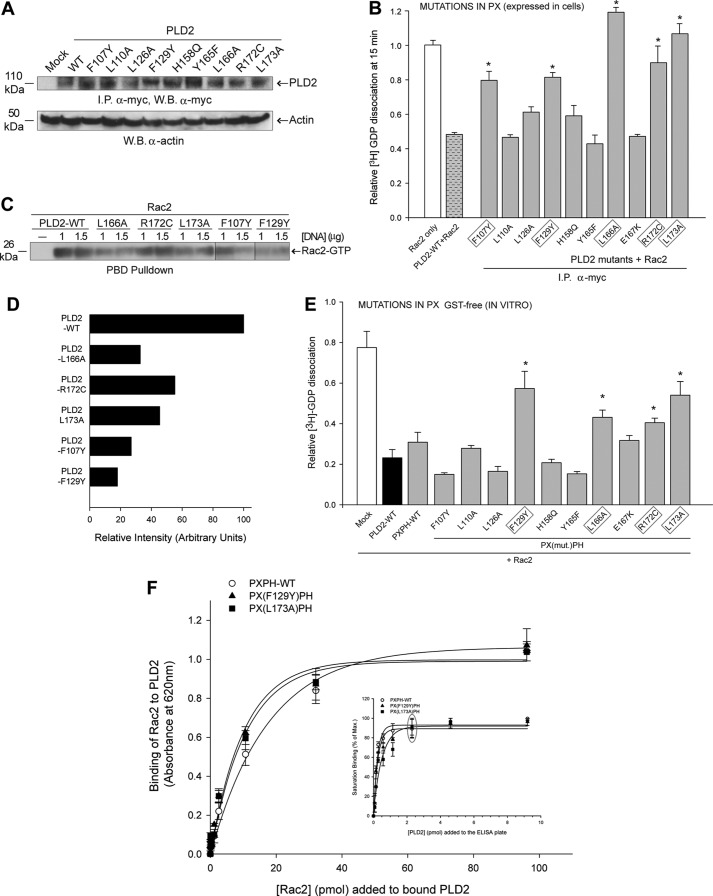
Substitution mutations in PLD2 that were made for the selected amino acid residues in the PX predicted catalytic sites in α-helix-1, α-helix-2 and α-helix-3 (as shown in Fig. 4D) were F107Y, L110A, L126A, F129Y, H158Q, Y165F, L166A, E167K, R172C and L173A. These mutants were generated in full-length pcDNA3.1-mycPLD2 to determine their ability to catalyze guanine nucleotide exchange in immunoprecipitates. Fig. 5A shows a similar level of protein expression with all the mutants compared with wild type. Fig. 5B shows GEF activity in whole cells as we overexpressed PLD2-WT or mutants in COS-7 cells and immunoprecipitated (I.P.) PLD2WT or mutants using recombinant Rac2 as the substrate. As indicated in Fig. 5B, 5 PX mutants (F107Y, F129Y, L166A, R172C and L173A) had a greater effect on abrogating the GEF activity, whereas L110A, L126A, H158Q, Y165F and E167K did not have any effect on GEF activity. These results indicate that the residues Phe-107, Phe-129, Leu-166, Arg-172 and Leu-173 are directly or indirectly involved in PLD2 GEF activity. In addition to GDP dissociation, we also measured GTP binding to Rac. Fig. 5C shows in vivo Rac2 activation (PBD pulldown as Rac2-GTP) of all mutants except R172C, which decreased the GTP-loading activity of PLD2-GEF and possibly could be due to an unknown compensatory factor in the cellular milieu.
FIGURE 5.
Effect of mutations in the PX domain on PLD2 GEF activity. The PX point mutations were made in pcDNA3.1-mycPLD2 (full-length). Panel A shows protein expression levels of the PLD2 GEF mutants generated for this study using Western blot (W.B.) analysis and immunoprecipitation (I.P.). B, the effect of single point mutations of the PLD2 PX domain on Rac2 GDP dissociation was determined by two approaches. PLD2 residues involved in GEF activity in whole cells are outlined in gray boxes. We called this GEF activity in whole cells because PLD2 was overexpressed and immunoprecipitated (I.P.) from COS-7 cells (first approach). Activity of I.P.-PLD2WT or mutants was determined using recombinant Rac2 as the substrate. C, in vivo Rac2 activation (PBD pulldown) using PLD2-WT and relevant PLD2 mutants overexpressed in cells. D, horizontal bar graph representing relative intensity of Rac2-GTP in panel C. E, Rac2 GDP dissociation in the presence of purified recombinant GST-free PXPH (second approach) with mutations in PX domain is shown. Error bars are S.E., and significant (p < 0.05) differences with controls are shown by asterisk (*). Residues found to affect GEF activity using purified, recombinant proteins are outlined in gray boxes. F, PLD2 Rac2 binding is shown. The inset indicates that the PXPH-WT or mutants were bound to ELISA plates until saturation (gray ellipse). Increasing amounts of baculoviral, purified HA-Rac2 (0.043–96.15 pmol) were laid on top of the PXPH-coated wells, and bound Rac2 was detected with specific antibody. Protein binding was measured by spectrometry at A620 nm. Results represent the mean ± S.E. for four independent experiments.
To further confirm the residues involved in GEF activity, we followed the mutants that showed diminished GEF activity in the immunoprecipitates through a series of GEF activity assays using recombinant proteins. As the PXPH fragment is comparable to PLD2 full-length in exerting GEF activity, we introduced the substitution mutations F107Y, F129Y, L110A, L166A, E167K, R172C and L173A into the pGEX-4T-1-PXPH construct one at a time. All these constructs were transformed and overexpressed, and GST-free proteins were prepared. Using these recombinant PXPH full-length or mutant proteins, in vitro GEF assays were performed (this approach was referred to as in vitro because recombinant proteins were used for GEF activity assays). The results shown in Fig. 5D indicate that the mutants F129Y, L166A, R172C and L173A have severely reduced GEF activity. Regarding F107Y, even though it affected GEF activity in the immunoprecipitates (Fig. 5B), it did not do so in the assays with recombinant proteins (Fig. 5E). This could be due to the loss of a putative cofactor during the process of recombinant protein purification necessary to exert full GEF function in whole cells.
Combining data from Fig. 5, B–E, we arrived at the conclusion that Phe-107, Phe-129, Leu-166, Arg-172 and Leu-173 in the PX domain are crucial for PLD2 GEF activity (with Leu-172 being the weakest of them all). These conclusions are based on the performance of the mutants in (a) cell-based GTP pulldown assays, (b) GEF activity of the immunoprecipitates and (c) GEF activity of the purified proteins. These results provide proof that these amino acids comprise the major catalytic GEF site on PLD2. To ascertain that differences seen with the PXPH mutants were not a result of inhibition of PLD2 binding to Rac2, we determined the amount of PLD2 required to saturate binding sites on ELISA plates using PXPH, PXPH-F129Y and PXPH-L173A (Fig. 5F). As shown in Fig. 5F inset, PXPH and both mutant PXPH proteins have essentially the same binding capacity for the ELISA plate, and saturation was achieved at 2.3 pmol each. Next, Rac2 was used to determine binding to PXPH, PXPH-F129Y or PXPH-L173A (Fig. 5F), and we found that the three proteins have similar binding patterns to Rac2. This indicates that the differences in GEF activities seen with the mutants in Fig. 5, B–E are not due to defects in binding to the substrate but occur as a result of authentic differences in its ability to act as a GEF.
There Is Extensive Three-dimensional Structural Similarity between the PH Domain of PLD2 and PH Domains of Known GEFs
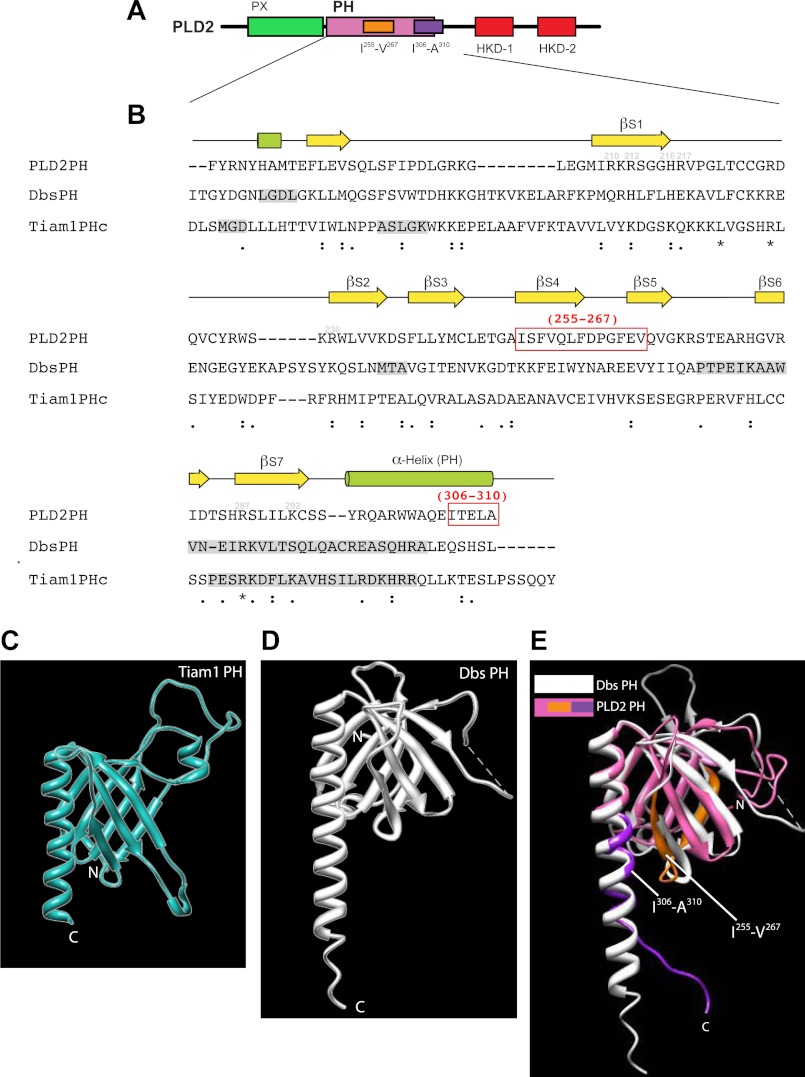
As indicated in Fig. 3, the PH domain aids in GEF activity; therefore, we studied it here further. PH sequence (Fig. 6, A and B) and structure (Fig. 6, C–E) similarity analyses with the PH domains of Dbs and Tiam1 were performed. Similar to other PH domains, the PLD2 PH domain (Fig. 6A) is primarily composed of β-strands and a C-terminal α-helix (Fig. 6B). The similarity of the PLD2-PH domain when compared with the PH domains of Dbs and Tiam1 is very extensive (Fig. 6, B–E). We noticed that in the sequence alignment there is a predominant α-helix that we named α-Helix-PH, and we hypothesized that this was a suitable site to further investigate. Additionally, three-dimensional modeling of PLD2-PH with the PH domains of Dbs and Tiam1 yielded extensive overlaps. Interestingly, we were able to visualize that the α-Helix-PH was close to β-sheet regions that we named βS4 and βS5. Thus, we prepared two deletion mutants in the PH domain, ΔIle-255–Val-267 (in βS4 and βS5) and ΔIle-306–Ala-310 (in α-helix-PH) for mutational analysis focusing on the amino acid regions that we just described (Fig. 6B, highlighted in red boxes).
FIGURE 6.
Sequence alignments of PH in PLD2 with PH in Dbs and Tiam1. A, shown is a schematic representation of PLD2 with the PH domain highlighted in magenta. B, shown is a comparative alignment between PLD2-PH, Dbs-PH and Tiam1-PH domains using ClustalW. Ile-255–Val-267 and Ile-306–Ala-310 that were chosen to make deletion mutations are highlighted in red boxes. Identical amino acids, similar amino acids and nearly similar amino acids are shown underneath the sequences as stars, as a single dot or as double dots, respectively. C–E, three-dimensional structure schematics of PLD2 PH and Dbs and Tiam1 PH are shown. The PH domains of Tiam1 (C), Dbs (D) and PLD2 PH are overlaid with Dbs-PH (E). Shown are the Ile-255- Val-267 and Ile-306-Ala-310 regions, as studied in this paper. The three-dimensional structures of PLD2 were obtained by I-TASSER predictions. The PDB IDs that were used to visualize the structures of Dbs and Tiam1 are 1KZ7 and 1FOE, respectively. All structures were viewed using the structure viewer Chimera.
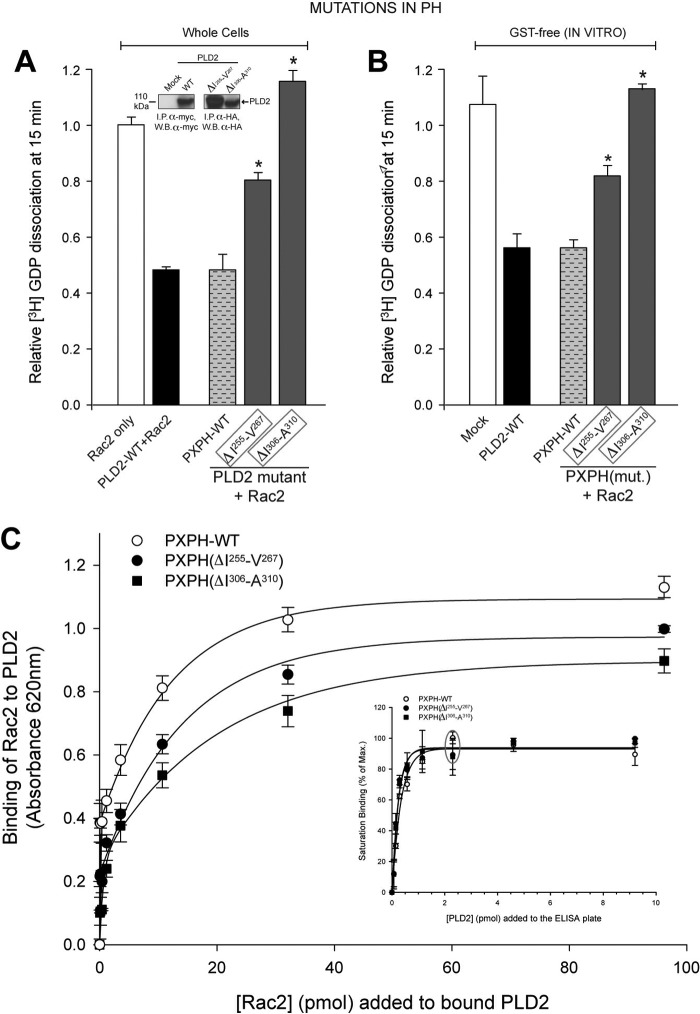
The GEF activity of the PH domain deletion mutants (Fig. 7A) in whole cells was found to be decreased (particularly ΔIle-306–Ala-310), suggesting that the PH domain should be intact to assist PX-mediated GEF activity. Similar results were obtained using the same GST-free purified proteins (Fig. 7B). Results from ELISA binding assays as shown in Fig. 7C indicate that contrary to that of the PX mutations, PH deletion decreases Rac2-PLD2 complex formation. Thus, the PH plays a crucial role in the catalysis that is primarily mediated by PX. Because Rac2-PLD2 binding is compromised when Ile-306–Ala-310 are deleted, we advanced the notion that this region is necessary to hold Rac2 in place, whereas the PX domain catalyzes the nucleotide exchange reaction to activate Rac2.
FIGURE 7.
Mutation analysis of PLD2 PH domain and the effect on its GEF activity. The PH point mutations were made in pcDNA3.1-mycPLD2 (full-length). Effects of mutation of PLD2 PH domain on Rac2 GDP dissociation from whole cells (first approach, immunoprecipitate (I.P.)-PLD2WT or mutants from COS-7 cells were analyzed for GEF activity using recombinant Rac2 as substrate) (A) or from purified, recombinant protein (second approach, purified recombinant PXPH with or without mutations in the PH domain along with recombinant Rac2 were used in the reaction mixture) (B). The residues targeted are Ile-255–Val-267 and Ile-306–Ala-310, as mentioned in Fig. 7. Mutants found to be involved in GEF activity in whole cells are outlined in gray boxes. The inset shows immunoprecipitation of representative myc-tagged mutants, verified by SDS-PAGE and Western blot analysis. C and inset, Figure 5. PLD2 Rac2 binding is shown. The inset indicates that PXPH-WT or mutants were bound to ELISA plates until saturation (gray ellipse). Increasing amounts of baculoviral, purified HA-Rac2 (0.043–96.15 pmol) were laid on top of the PXPH-coated wells and bound Rac2 was detected with specific antibody. Protein binding was measured by spectrometry at A620 nm. Results represent the mean ± S.E. for four independent experiments.
Effect of PLD2 GEF Inactive Mutants on Cell Function
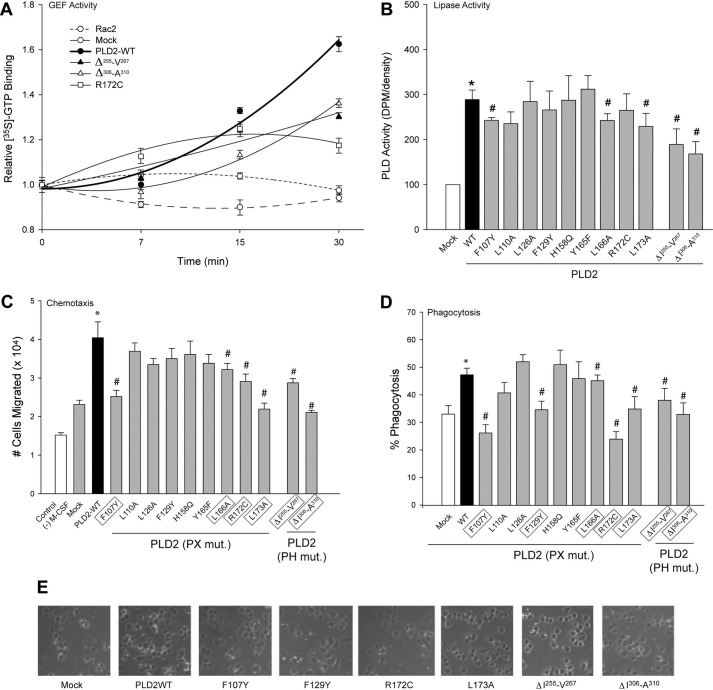
Mutants F107Y, F129Y, R172C and L173A in the PX domain and the two deletion mutants ΔIle-255–Val-267 and ΔIle-306–Ala-310 in the PH domain of PLD2 affect GEF activity as GDP dissociation from Rac2. We next analyzed GTP binding, for which three representative mutants R172C, ΔIle-255–Val-267 and ΔIle-306–Ala-310 were chosen. As indicated in Fig. 8A, these mutants are significantly lower than wild type in catalyzing GTP binding to Rac2 and further confirms that these residues need to be intact for PLD2 to be an efficient GEF.
FIGURE 8.
Effect of mutated PLD2 on physiological assays. A, shown is the effect of representative mutants (R172C, ΔIle-255–Val-267 and ΔIle-306–Ala-310) on GTP binding of Rac2 over time. B, shown is the effect of PX substitution mutants and PH deletion mutants on PLD2 lipase activity. Phagocytosis and chemotaxis activities of PLD2 mutants (mut) are shown in panels C and D, respectively. The PH point mutations were made in pcDNA3.1-mycPLD2 (full-length). Cells were transfected with a constant amount of PLD2 DNA (2 μg) and used for either phagocytosis of zymosan particles or chemotaxis using Transwell inserts. Data are the means ± S.E. from at least three independent experiments performed in duplicate. * or # represent differences between means, as determined by ANOVA, above or below the negative control levels. PLD2 residues involved in chemotaxis and phagocytosis in vivo are outlined in a gray box. E, shown are immunofluorescence photomicrographs of cells overexpressing PLD2-WT and mutants during phagocytosis of zymosan beads.
Next, we asked if any of the mutants generated in this study would affect the structure of PLD2 and inactivate its lipase activity. Fig. 8B indicates that the lipase activity of most of the PX mutants does not differ to a greater extent when compared with the wild type with only Phe-107, Leu-166 and Leu-173 losing a small (∼25%) amount of lipase activity. However, ΔIle-255–Val-267 and ΔIle-306–Ala-310 definitely showed a loss of lipase activity (of >40%) over the wild type, which probably might be due to deletion of more than one amino acid.
We previously demonstrated that the cellular processes of phagocytosis and chemotaxis rely on PLD2-mediated Rac2 activation (12, 13). Therefore, we chose these two assays using the mouse macrophage cell line RAW264.7/LR5 to test the effect of the mutants we generated for this study. Cells overexpressing PLD2 (WT or mutants) were tested for chemotaxis or phagocytosis. The results with chemotaxis (Fig. 8C) were very similar to that of phagocytosis as shown in Fig. 8D. Results in Fig. 8, C and D indicate that the mutants F107Y, F129Y, L166A, R172C, L173A (in the PX domain), ΔIle-255–Val-267 and ΔIle-306–Ala-310 (in the PH domain) affect the cell's ability to perform chemotaxis and phagocytosis. The strongest effects are seen with the Phe-129 and Leu-173 mutants. These results are in agreement with the GEF activity results, suggesting that the mutants affect the cell function by reducing PLD2 ability to activate the substrate GTPase.
DISCUSSION
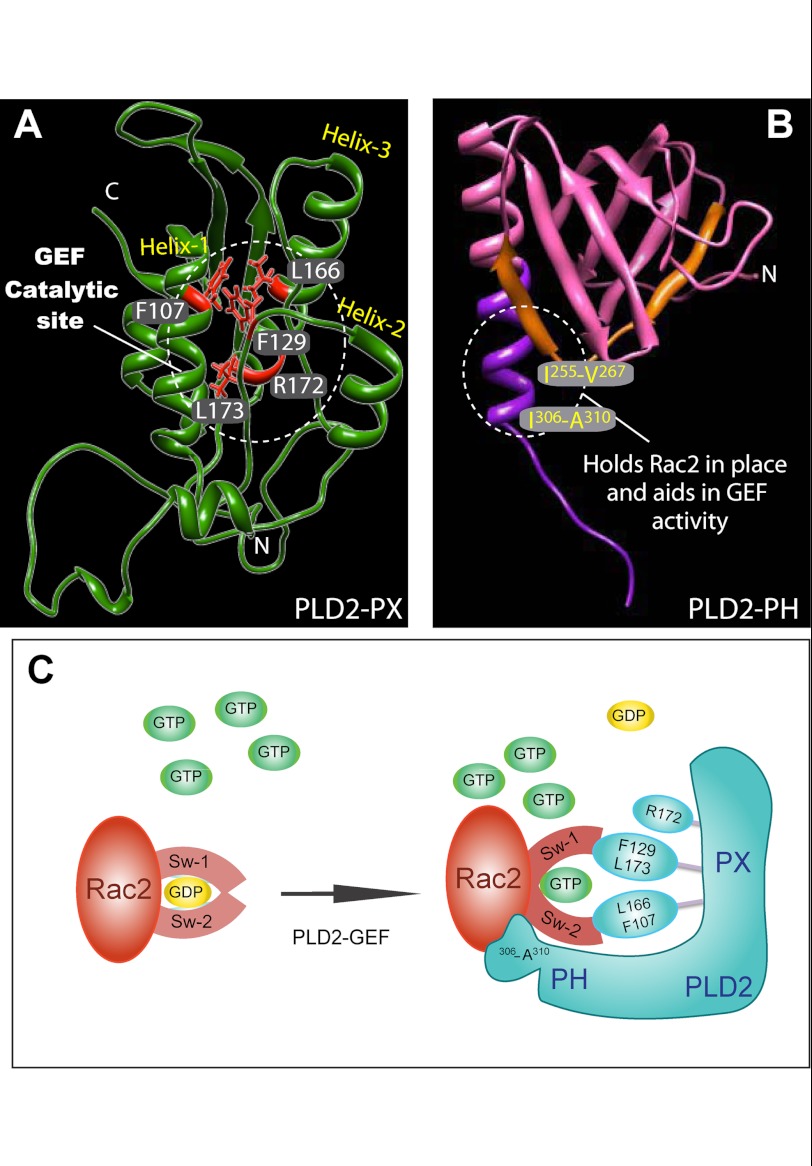
This study demonstrated that recombinant PLD2 itself is a novel class of GEF that lacks the DH-PH domain but possesses the a PX-PH domain. As the experiments indicate, recombinant PLD2 has intrinsic GEF activity, and the catalytic site resides in the PX domain, specifically at Phe-107, Phe-129, Leu-166 and Leu-173 (with contribution from Arg-172), whereas the PH domain at the region Ile-306–Ala-310 facilitates PX-mediated nucleotide exchange by maintaining Rac2 in place. All this evidence defines a new concept in the biology of GEF and is presented in Fig. 9A and B.
FIGURE 9.
Proposed GEF activity sites in PLD2 and the possible mechanism of action. A and B, shown are the proposed catalytic sites of PLD2 GEF activity according to this study. C, mechanism of PLD2-mediated nucleotide exchange is shown. Key residues on PLD2, phenylalanine 129, leucine 166, phenylalanine 107 and leucine 173 are shown as part of the catalytic site that forms a hydrophobic pocket. Phe-107 may need the interaction with an as-yet unknown cofactor in whole cells to function. In addition to this, arginine 172, the only charged residue, could aid in the GEF function. The model depicted in C indicates that Rac2 holds onto GDP with its switch 1 and 2 domains. PLD2 and Rac2 form a complex in such a way that switch 1/2 on Rac2 is perturbed, leading to GDP dissociation from Rac2. Followed by GTP binding to Rac2, switch 1/2 returns to the original conformation, leading to the separation of PLD2 from Rac2.
When the residues of the GEF catalytic site were presented in the three-dimensional model (Fig. 9A), it became very evident that four of the residues form a hydrophobic pocket with the side chains of leucines and phenylalanines pointing toward a common point that we speculate is where the Rac2 GDP-GTP exchange takes place. The α helices of the PX domain with these particular residues provide enough hydrophobicity so PLD2 functions properly as a GEF.
Apart from this, the mutant R172C was also found to yield lower GEF activity; this is not very extensive, meaning that electrostatic charges as those in arginine do not seem to play a major role in the GEF activity. This makes sense, as no covalent bonds are broken or created, only a switch of ligands (GDP → GDP).
The possible mechanism involving the previously mentioned amino acids might form important contacts with Rac2 and further aid in changing the confirmations of switch I/II of Rac2, thereby causing GDP dissociation and aiding in GTP binding, as depicted in Fig. 9C. We hypothesized that when PLD2 is complexed with Rac2, PLD2 makes contact with the Rac2 nucleotide binding pocket, specifically with Phe-107, Phe-129, Leu-166, Arg-172 and Leu-173, whereas the PH (Ile-306–Ala-310) holds Rac2 in place. By this mechanism, PLD2 perturbs Rac2 and reduces the nucleotide binding capacity. Thus, PLD2 acts as unique guanine nucleotide exchange factor for the small GTPase Rac2 and increases Rac2 activation.
The results indicate that the PLD2-GEF differs from the known Rho-GEF families, as they utilize either the DH domain or the Dedicator of cytokinesis (DOCK) homology region 2 (DHR2) domain for catalytic function (22–26). PLD2 can neither be defined as a Dbl family GEF, as it lacks the DH domain, nor can it be defined as a Dedicator of cytokinesis (DOCK) family GEF, because it lacks the DHR1/2 domains, which defines PLD2 as a novel class of GEF possessing a PX domain in tandem with a PH domain, as shown in the schematic representation in Fig. 4A. Certain non-redundant Dbl family GEFs, like Dbs, possess a PH domain that interacts with the substrate GTPase and also aids in guanine nucleotide exchange (21). PLD2 certainly falls into this category where the PX domain is certainly involved in catalyzing the exchange activity, but it is even more enhanced if the PX domain is in tandem with the PH domain.
PLD2 and Rac2 are significantly implicated in leukocyte chemotaxis and phagocytosis due to their ability to regulate actin cytoskeleton (27, 28). In a previous study, we showed that these two proteins interact with each other (12). Activated Rac2 reorganizes the actin cytoskeleton and mediates membrane ruffle formation and cell migration, all key events in leukocyte physiology. The current study provides the molecular basis of the previously found GEF activity of PLD2.
In summary, the novel Rac2 GEF, PLD2, does not have a DH domain like other conventional Rho family GEFs, but its PX domain in tandem with its PH domain catalyzes the GEF reaction, which allow for PLD2 to be classified as a novel class of GEF. We were successful in identifying the key residues Phe-107, Phe-129, Leu-166, Arg-172 and Leu-173 in PLD2 (four of which are highly hydrophobic) as central for PLD2's ability to catalyze the exchange reaction, and modification of these residues will completely abolish its activity. Knowing the exact amino acids where the catalytic activity resides will be crucial for the rational design of drugs that can suppress this activity in chronic inflammation or cardiovascular disease, where leukocytes can be harmful to human tissues.
Acknowledgment
We thank Dr. Joel Swanson of University of Michigan for providing Rac2-CFP and FRET analysis and Dr. Dianne Cox (Albert Einstein) for providing the RAW/LR5 macrophages.
This work was supported, in whole or in part, by National Institutes of Health Grant HL056653 (to J. G.-C.). This work was also supported by Boonshoft School of Medicine Grant 229102 and State of Ohio Research Incentive Grant 668372 (to J. G.-C.).
- PLD2
- phospholipase D2
- PA
- phosphatidic acid
- PH
- pleckstrin homology
- GEF
- guanine nucleotide exchange factor
- AMP-PNP
- adenosine 5′-(β,γ-imino)triphosphate
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- ANOVA
- analysis of variance
- CFP
- cyan fluorescent protein
- PBD
- pak21-binding domain
- PC
- phosphatidylcholine
- PH
- phox homology
- YFP
- yellow fluorescent protein.
REFERENCES
- 1. Morris A. J., Hammond S. M., Colley C., Sung T. C., Jenco J. M., Sciorra V. A., Rudge S. A., Frohman M. A. (1997) Regulation and functions of phospholipase D. Biochem. Soc. Trans. 25, 1151–1157 [DOI] [PubMed] [Google Scholar]
- 2. Zhang Y., Du G. (2009) Phosphatidic acid signaling regulation of Ras superfamily of small guanosine triphosphatases. Biochim. Biophys. Acta 1791, 850–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhao C., Du G., Skowronek K., Frohman M. A., Bar-Sagi D. (2007) Phospholipase D2-generated phosphatidic acid couples EGFR stimulation to Ras activation by Sos. Nat. Cell Biol. 9, 706–712 [DOI] [PubMed] [Google Scholar]
- 4. Chae Y. C., Kim J. H., Kim K. L., Kim H. W., Lee H. Y., Heo W. D., Meyer T., Suh P. G., Ryu S. H. (2008) Phospholipase D activity regulates integrin-mediated cell spreading and migration by inducing GTP-Rac translocation to the plasma membrane. Mol. Biol. Cell 19, 3111–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xu L., Salloum D., Medlin P. S., Saqcena M., Yellen P., Perrella B., Foster D. A. (2011) Phospholipase D mediates nutrient input to mammalian target of rapamycin complex 1 (mTORC1). J. Biol. Chem. 286, 25477–25486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Di Fulvio M., Frondorf K., Henkels K. M., Lehman N., Gomez-Cambronero J. (2007) The Grb2/PLD2 interaction is essential for lipase activity, intracellular localization, and signaling in response to EGF. J. Mol. Biol. 367, 814–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Slaaby R., Jensen T., Hansen H. S., Frohman M. A., Seedorf K. (1998) PLD2 complexes with the EGF receptor and undergoes tyrosine phosphorylation at a single site upon agonist stimulation. J. Biol. Chem. 273, 33722–33727 [DOI] [PubMed] [Google Scholar]
- 8. Kantonen S., Hatton N., Mahankali M., Henkels K. M., Park H., Cox D., Gomez-Cambronero J. (2011) A novel phospholipase D2-Grb2-WASp heterotrimer regulates leukocyte phagocytosis in a two-step mechanism. Mol. Cell. Biol. 31, 4524–4537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Houle M. G., Bourgoin S. (1999) Regulation of phospholipase D by phosphorylation-dependent mechanisms. Biochim. Biophys. Acta 1439, 135–149 [DOI] [PubMed] [Google Scholar]
- 10. Exton J. H. (2002) Regulation of phospholipase D. FEBS Lett. 531, 58–61 [DOI] [PubMed] [Google Scholar]
- 11. Oshimoto H., Okamura S., Yoshida M., Mori M. (2003) Increased activity and expression of phospholipase D2 in human colorectal cancer. Oncol. Res. 14, 31–37 [DOI] [PubMed] [Google Scholar]
- 12. Peng H. J., Henkels K. M., Mahankali M., Dinauer M. C., Gomez-Cambronero J. (2011) Evidence for two CRIB domains in phospholipase D2 (PLD2) that the enzyme uses to specifically bind to the small GTPase Rac2. J. Biol. Chem. 286, 16308–16320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mahankali M., Peng H. J., Henkels K. M., Dinauer M. C., Gomez-Cambronero J. (2011) Phospholipase D2 (PLD2) is a guanine nucleotide exchange factor (GEF) for the GTPase Rac2. Proc. Natl. Acad. Sci. U.S.A. 108, 19617–19622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peng H. J., Henkels K. M., Mahankali M., Marchal C., Bubulya P., Dinauer M. C., Gomez-Cambronero J. (2011) The dual effect of Rac2 on phospholipase D2 regulation that explains both the onset and termination of chemotaxis. Mol. Cell. Biol. 31, 2227–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gomez-Cambronero J. (2011) The exquisite regulation of PLD2 by a wealth of interacting proteins: S6K, Grb2, Sos, WASp, and Rac2 (and a surprise discovery: PLD2 is a GEF). Cell. Signal. 23, 1885–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jeon H., Kwak D., Noh J., Lee M.N., Lee C.S., Suh P. G., Ryu S.H. (2011) Phospholipase D2 induces stress fiber formation through mediating nucleotide exchange for RhoA. Cell Signal 23, 1320–1326 [DOI] [PubMed] [Google Scholar]
- 17. Lu Z., Hornia A., Joseph T., Sukezane T., Frankel P., Zhong M., Bychenok S., Xu L., Feig L. A., Foster D. A. (2000) Phospholipase D and RalA cooperate with the epidermal growth factor receptor to transform 3Y1 rat fibroblasts. Mol. Cell. Biol. 20, 462–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hoppe A., Christensen K., Swanson J. A. (2002) Fluorescence resonance energy transfer-based stoichiometry in living cells. Biophys. J. 83, 3652–3664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liscovitch M., Czarny M., Fiucci G., Tang X. (2000) Phospholipase D. Molecular and cell biology of a novel gene family. Biochem. J. 345, 401–415 [PMC free article] [PubMed] [Google Scholar]
- 20. Ming W., Li S., Billadeau D. D., Quilliam L. A., Dinauer M. C. (2007) The Rac effector p67phox regulates phagocyte NADPH oxidase by stimulating Vav1 guanine nucleotide exchange activity. Mol. Cell. Biol. 27, 312–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rossman K. L., Worthylake D. K., Snyder J. T., Siderovski D. P., Campbell S. L., Sondek J. (2002) A crystallographic view of interactions between Dbs and Cdc42. PH domain-assisted guanine nucleotide exchange. EMBO J. 21, 1315–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rossman K. L., Der C. J., Sondek J. (2005) GEF means go. Turning on RHO GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol. 6, 167–180 [DOI] [PubMed] [Google Scholar]
- 23. García-Mata R., Burridge K. (2007) Catching a GEF by its tail. Trends Cell Biol. 17, 36–43 [DOI] [PubMed] [Google Scholar]
- 24. Côté J. F., Vuori K. (2002) Identification of an evolutionarily conserved superfamily of DOCK180-related proteins with guanine nucleotide exchange activity. J. Cell Sci. 115, 4901–4913 [DOI] [PubMed] [Google Scholar]
- 25. Meller N., Merlot S., Guda C. (2005) CZH proteins. A new family of Rho-GEFs. J. Cell Sci. 118, 4937–4946 [DOI] [PubMed] [Google Scholar]
- 26. Kwofie M. A., Skowronski J. (2008) Specific recognition of Rac2 and Cdc42 by DOCK2 and DOCK9 guanine nucleotide exchange factors. J. Biol. Chem. 283, 3088–3096 [DOI] [PubMed] [Google Scholar]
- 27. Knapek K., Frondorf K., Post J., Short S., Cox D., Gomez-Cambronero J. (2010) The molecular basis of phospholipase D2-induced chemotaxis. Elucidation of differential pathways in macrophages and fibroblasts. Mol. Cell. Biol. 30, 4492–4506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang H., Sun C., Glogauer M., Bokoch G. M. (2009) Human neutrophils coordinate chemotaxis by differential activation of Rac1 and Rac2. J. Immunol. 183, 2718–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Thompson J. D., Higgins D. G., Gibson T. J. (1994) ClustalW. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]