Background: Ras signaling is known to be critical for tumor progression.
Results: Ras-PI3K regulates H3K56 acetylation (H3K56ac) via the MDM2-dependent degradation of CBP/p300. H3K56ac is revealed to be associated with the transcription, proliferation, and migration of tumor cells.
Conclusion: H3K56 acetylation is a critical component of the oncogenic Ras-PI3K pathway.
Significance: The Ras-PI3K-AKT-H3K56ac pathway is a potential target for cancer therapy.
Keywords: CBP, Epigenetics, Histone Modification, p300, Phosphatidylinositol 3-Kinase, Ras, Signal Transduction, MDM2, Histone Acetylation
Abstract
It is well established that the small GTPase Ras promotes tumor initiation by activating at least three different mediators: Raf, PI3K, and Ras-like (Ral) guanine nucleotide exchange factors. However, the exact mechanisms that underlie these different Ras signaling pathways, which are involved in tumor progression, remain to be elucidated. In this study, we report that the Ras-PI3K pathway, but not Raf or the Ral guanine nucleotide exchange factors, specifically targets the acetylation of H3 at lysine 56 (H3K56ac), thereby regulating tumor cell activity. We demonstrate that the Ras-PI3K-induced reduction in H3K56ac is associated with the proliferation and migration of tumor cells by targeting the transcription of tumor-associated genes. The depletion of the histone deacetyltransferases Sirt1 and Sirt2 rescues the Ras-PI3K-induced decrease in H3K56ac, gene transcription, tumor cell proliferation, and tumor cell migration. Furthermore, we demonstrate that the Ras-PI3K-AKT pathway regulates H3K56ac via the MDM2-dependent degradation of CREB-binding protein/p300. Taken together, the results of this study demonstrate that the Ras-PI3K signaling pathway targets specific epigenetic modifications in tumor cells.
Introduction
Ras mutations, often glycine 12 to valine (G12V), are known to be critical in both the formation and maintenance of human cancers. The constitutive expression of the active version of Ras promotes tumor initiation by activating different effectors, including Raf, PI3K, and Ras-like (Ral)2 guanine nucleotide exchange factors (RalGEFs). These pathways further activate/repress the activity of transcription factors (1–3) and participate in specific cellular functions. Raf is a serine/threonine kinase, and activated Raf proteins initiate a MAPK cascade that leads to transformed morphologies, anchorage-independent growth, and angiogenesis (4, 5). PI3K activation results in phosphoinositide phosphorylation, changes in cell morphology, increased angiogenesis, and cell survival (6). The RalGEFs are a family of guanine exchange factors that are activated by recruitment to the plasma membrane by GTP-Ras. It is at the membrane level that RalGEFs activate RalA and RalB (7, 8). Although Raf, PI3K and RalGEF are all required to initiate tumor growth in human cells, Ras exerts its oncogenic effects through different sets of effectors during different stages of tumorigenesis (9). Ras-PI3K activation appears to be more critical during tumor maintenance and may be particularly important for the treatment of Ras-driven human cancers (9).
Epigenetic modifications, such as histone acetylation, methylation, or DNA methylation, are associated with transcriptional activities and cellular functions (10). Aberrant changes in specific histone modifications, such as H3K27 methylation, as well as H3K9 and H3K56 acetylation, have been reported in a variety of human cancers (10–13). These modifications often affect chromatin structure and the subsequent transcriptional activity of genes (10), including that of oncogenes or tumor suppressor genes. A recent study suggested that certain epigenetic modifications can be targeted via specific signaling pathways (14). This finding raised the possibility that different cellular functions that are induced by specific signaling transduction events may be executed by distinct epigenetic machinery.
Here, we investigated the relationship between the Ras signaling pathways and histone modifications. We demonstrate that the Ras-PI3K signaling specifically down-regulates H3K56ac, a modification that is known to be involved in the response to multiple genotoxic agents (15) and that is considered to be a signal for the completion of the repair of double-strand breaks (16). We further demonstrate that the Ras-PI3K-induced reduction in H3K56ac is associated with the proliferation and migration of tumor cells by targeting the transcription of tumor-associated genes. The Ras-PI3K-induced decrease in H3K56ac and the concomitant alterations in transcription, proliferation, and tumor cell migration are partially rescued by the knockdown of the histone deacetylases Sirt1/Sirt2. Finally, we demonstrate that the Ras-PI3K-AKT pathway specifically targets H3K56ac via the MDM2-dependent degradation of CBP/p300. This work establishes a link between the Ras-PI3K signaling pathway and specific epigenetic machinery.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
The rabbit polyclonal anti-AKT antibody (antibody 9272), rabbit monoclonal anti-phospho-Akt (Ser-473) (antibody 4058) antibody and rabbit polyclonal anti-H3 (antibody 9715) antibody were purchased from Cell Signaling Technology. The rabbit polyclonal anti-H3K56ac antibody for ChIP (antibody 16908001, 9 μl/ChIP) was purchased from Active Motif. The rabbit polyclonal anti-H4K16ac (antibody 07-329) antibody was purchased from Millipore. The rabbit polyclonal anti-H4 (ab10158), anti-H3K18ac (ab1191), anti-H3K9ac (ab10812), and anti-H4K8ac (ab15823) antibodies were purchased from Abcam. Also purchased from Abcam were the rabbit monoclonal anti-H3K56ac (ab76307), anti-H3K14ac (ab52946), anti-H4K5ac (ab51997), anti-HDAC1 (ab31263), anti-HDAC2 (ab51832), anti-CBP (ab2832), and anti-p300 (ab14984) antibodies. The mouse monoclonal anti-MDM2 (antibody OP115) antibody was purchased from Calbiochem. The mouse monoclonal anti-β actin (antibody TA09), anti-HA tag (antibody TA04), and anti-GFP (antibody TA06) antibodies, as well as the rabbit polyclonal anti-His tag (antibody TA02) antibody, were purchased from the Zhongshan Golden Bridge Biotechnology Company. The rabbit monoclonal anti-human Sirt1 (antibody 1054-1) and Sirt2 (antibody 1999-1) antibodies were purchased from Epitomics. The MG132 (antibody C2211) and MTT (antibody M2128) were purchased from Sigma. H2O2 (antibody 386790) was purchased from Calbiochem. The Cy3 AffiniPure F(ab′)2 fragment goat anti-rabbit IgG (H+L) (antibody 111–166-003) was purchased from Jackson ImmunoResearch.
Plasmid Construction and siRNA
The coding regions of human H3, Asf1A, Gcn5, Sirt1 isoforms 1 and 2, Sirt2 isoforms 1 and 2, MDM2, and wild-type H-Ras were amplified from HeLa cDNA using PCR. The PCR products were subcloned into HA tag or His tag vectors and were sequenced. HA tag-CBP and HA tag-p300 were gifts from Professor Y. Eugene Chin. The MDM2C464A expression plasmid was provided by Dr. Martin R. Bennett. The pEGFP-H-RasG12V construct was mutated using site-directed mutagenesis. pEGFP-H-RasG12V/E37G, -H-RasG12V/Y40C, and -H-RasG12V/T35S were amplified from pBabe-H-RasG12V/E37G, -H-RasG12V/Y40C, and H-RasG12V/T35S, respectively, which were purchased from Addgene (catalogue number 12274, 12275, and 12276, respectively). These constructs were then subcloned into pEGFP-N1. siRNAs that were specific for MDM2, Sirt1, and Sirt2 were purchased from Shanghai GenePharma. The pEGFP-H3K56Q/A construct was constructed using the TaKaRa MutanBEST Kit (catalogue number D401), as recommend by the manufacturer.
Cell Culture and Treatments
The HeLa cells were obtained from the Cell Culture Center of Peking Union Medical College and were maintained in DMEM that was supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin (complete DMEM). All of the cultures were incubated in a humidified atmosphere of 95% air and 5% CO2 at 37 °C.
Transfection
The HeLa cells (5 × 105/well) were seeded in 6-well plates and incubated overnight. The plasmids or siRNA were transfected alone or co-transfected using Lipofectamine 2000 (Invitrogen) as recommended by the manufacturer. Forty-eight hours post-transfection, the cells were collected and were analyzed using Western blotting or RT-PCR.
Cell Viability
The cells (5 × 103/well) were seeded in complete DMEM (100 μl/well) in 96-well plates and were incubated for 24 h. The plasmids were transiently transfected using Lipofectamine 2000. Untreated cultures were used as negative controls. After 48 h, 20 μl of MTT (5 mg/ml) was added to each well, and the plates were incubated at 37 °C for 4 h. Following this incubation, 100 μl of dimethyl sulfoxide was added to each well to lyse the cells. The absorbance was measured at 570 nm using a multiwell spectrophotometer (Emax; Molecular Devices, Sunnyvale, CA).
RNA Extraction and RT-PCR
The HeLa cells were lysed directly in tissue culture plates using TRIzol (Invitrogen) reagent, and the RNA was isolated as recommended by the manufacturer. DNase-I-treated total RNA was used for first strand cDNA synthesis using Moloney murine leukemia virus reverse transcriptase (Fermentas) and oligo(dT) primers (Invitrogen), as recommended by the manufacturers. The primer sequences are provided in the supplementary materials.
Real Time PCR
The real time PCR was performed using a QuantiTect SYBR Green PCR kit (Qiagen) as recommended by the manufacturer. Human GAPDH mRNA was used as a control, and all of the samples were analyzed in triplicate. The primer sequences are provided in the supplementary materials.
Soft Agar Colony Formation Assay
For the soft agar assay, the HeLa cells were suspended in DMEM that contained 0.35% low melting agarose. The cells were then plated onto solidified 0.6% agarose in DMEM in 6-well culture plates at a density of 1 × 103 cells/well. The number of the colonies were observed microscopically (40×) 3 weeks after seeding.
Transwell Migration Assay
HeLa cell migration was assayed using a Transwell system with 8-μm pore filters (Costar, Boston, MA). After filling the lower chamber with complete medium, 1 × 104 HeLa cells in 0.5 ml of serum-free medium were loaded into the upper chamber. Following 12 h of incubation at 37 °C, the cells that migrated to the bottom surface of the membrane were fixed with methanol, stained with 0.5% crystal violet, and microscopically inspected. The cells on the top surface of the membrane were removed with a cotton swab. The A570 of the cells that had been washed with acetic acid was measured.
Western Blot Analysis
The HeLa cells were collected 48 h following transfection and washed once with PBS. The whole cell lysates were prepared using lysis buffer (50 mm Tris-Cl, 150 mm NaCl, 0.02% NaN3, 1% Nonidet P-40, 0.1% SDS, 0.5% sodium deoxycholate, 5 mg/ml leupeptin, and 1 mg/ml aprotinin). The protein content of the cell lysates was quantified using a BCA protein assay kit (Novagen). The proteins were separated using SDS-PAGE and transferred to nitrocellulose membranes (Schleicher & Schuell). The membranes were blocked at room temperature (24–26 °C) for 1 h in 130 mm NaCl, 2.5 mm KCl, 10 mm Na2HPO4, 1.5 mm KH2PO4, 0.1% Tween 20, and 5% BSA (pH 7.4). The membranes were then incubated with the appropriate primary antibody overnight at 4 °C and were subsequently incubated with an HRP-conjugated anti-rabbit or anti-mouse secondary antibody for 1 h at room temperature. The blots were developed using the Western blotting Luminol Reagent (SC-2048; Santa Cruz Biotechnology, Santa Cruz, CA) as recommended by the manufacturer. The density of target bands was quantified using a computer-aided one-dimensional gel analysis system and Gel-Pro Analyzer 4.0 software. The antibodies were diluted to 1:1000 for the Western blot analysis.
Microarray Analysis
All of the microarray analyses were performed by CapitalBio Corporation.
Immunofluorescence
The HeLa, MCF-7, SMMC-7721, HL-7702, and PNAC-1 cells were maintained as described above. The cells were grown on coverslips to 70% confluence, transfected with pEGFP-H-RasG12V/Y40C, cultured for 48 h, and washed three times with PBS. The cells were then fixed in chilled 4% formaldehyde for 10 min, washed three times with PBS, and blocked for 30 min in a blocking solution (3% BSA in PBS). Next, the cells were incubated with the primary antibody (anti-H3K56ac or anti-H3K18ac diluted to 1:100 in blocking solution) for 1 h at room temperature. Following two PBS washes, the cells were incubated with the Cy3-AffiniPure F(ab′)2 fragment goat anti-rabbit secondary antibody for 1 h and washed three times with PBS. The cells were stained with DAPI for 5 min and washed twice with PBS. The images were captured using a confocal laser scanning microscope (Leica).
Flow Cytometric Analysis of Cell Cycle Distribution
The HeLa cells were harvested, washed twice in ice-cold PBS, and fixed in 1% (v/v) paraformaldehyde for 1 h at room temperature. The cells were then washed twice in ice-cold PBS and fixed in 70% ethanol for 2 h at 4 °C. The cells were rehydrated in PBS and then incubated for 30 min at room temperature in a propidium iodide staining solution (0.2 mg/ml propidium iodide, 0.2 mg/ml DNase-free RNase A (Roche Applied Science), and 0.1% Triton X-100 in PBS). A total of 4000 events were acquired using an Epics XL flow cytometer (Beckman Coulter, Fullerton, CA) for each sample. The percentage of cells in the G0/G1, S, and G2/M phases of the cell cycle was determined based on propidium-DNA fluorescence using the system software (Beckman Coulter).
Chromatin Immunoprecipitation
Approximately 3 × 106 cells/sample were cross-linked for 10 min in 1% formaldehyde at room temperature. The cells were washed twice with cold PBS and lysed with SDS lysis buffer (Upstate, catalogue number 20-163). The lysates were sonicated in an ultrasonic bath (Bioruptor, Diagenode) to sheer the DNA to an average length of 200–800 bp. Following the sonication, the samples were centrifuged at 15,000 × g. The supernatants were diluted with ChIP dilution buffer (Upstate, catalogue number 20-153) and immunoprecipitated overnight with rabbit anti-H3K56ac (2 μg) at 4 °C. The control immunoprecipitations were performed using 2 μg of normal anti-IgG antibody. The beads were washed sequentially for 5 min each in low salt (Upstate, catalogue number 20-154: 20 mm Tris-HCl, pH 8, 150 mm NaCl, 2 mm EDTA, 1% Triton X-100, and 0.1% SDS), high salt (Upstate, catalogue number 20-155: 20 mm Tris-HCl, pH 8, 500 mm NaCl, 2 mm EDTA, 1% Triton X-100, and 0.1% SDS), and LiCl buffers (Upstate, catalogue number 20-156: 10 mm Tris, pH 8.0, 1 mm EDTA, 250 mm LiCl, 1% Nonidet P-40, and 1% deoxycholate) at 4 °C. Lastly, the beads were washed twice in 1× TE (Upstate, catalogue number 20-157) for 2 min at room temperature. The DNA was eluted from the beads in 1% SDS, 100 mm NaHCO3 for 15 min at room temperature. NaCl was added to a final concentration of 200 mm, and the cross-links were reversed for 7 h at 65 °C. The eluted DNA was precipitated with ethanol overnight at −20 °C, treated with 20 μg of proteinase K, and purified using QIAquick PCR purification columns (Qiagen) as recommended by the manufacturer. The immunoprecipitated DNA (1.5 μl) and serial dilutions of the 10% input DNA (1:4, 1:20, 1:100, and 1:500) were analyzed using real time quantitative PCR. The primer sequences are provided in the supplementary materials.
Co-immunoprecipitation Analysis
The HeLa cells were transfected with plasmids expressing His-tagged Sirt1 or Sirt2, GFP-tagged Ras WT, or RasG12V/Y40C using Lipofectamine 2000. After 48 h, the cells were harvested, washed with ice-cold PBS, resuspended in BC300 buffer (20 mm Tris-Cl, pH 8.0, 300 mm NaCl, 0.2 mm EDTA, 10% glycerol, 0.2 mm PMSF, and 0.2% Tween 20), and pulse-sonicated (Branson digital sonifier) 10 times on ice (3 s with 30% efficiency). The cell lysates were incubated with normal mouse IgG (Santa Cruz Biotechnology, as a negative control) or anti-GFP antibodies (Santa Cruz Biotechnology) overnight at 4 °C. Fifty microliters of protein A/G-agarose beads was then added, and the lysates were incubated for another 3 h. The beads were washed five times with BC300 buffer and collected via centrifugation. The precipitated proteins were released by boiling in BC300 buffer, resolved by SDS-PAGE (15%), and probed for Sirt1 or Sirt2 using Western blotting, as indicated. The co-immunoprecipitations of MDM2 and CBP or p300 were performed in an analogous manner.
Statistical Analysis
The statistical analyses were performed using the Student's t test. A p value of < 0.05 was considered significant (*, p < 0.05; **, p < 0.01).
RESULTS
To investigate the relationship between the activation of the Ras pathway and epigenetic modifications, we produced cell lines that overexpressed H-RasG12V, which is known to constitutively activate the Ras protein (17). This overexpression resulted in an ∼60% reduction in H3K56ac (Fig. 1A).
FIGURE 1.
H3K56 acetylation is specifically regulated by the Ras-PI3K pathway but not by the MAPK or RalGEF pathways. A, HeLa cells were transfected with empty-pEGFP-N1 (vector), pEGFP-H-RasWT, pEGFP-H-RasG12V, pEGFP-H-RasG12V/E37G, pEGFP-H-RasG12V/Y40C, or pEGFP-H-RasG12V/T35S plasmids. The H3K56ac levels were then measured using Western blotting and densitometric analyses. B, the Ras-PI3K pathway activation level was measured based on the level of phosphorylated AKT (P - AKT). C, the H3K56ac level following the activation of the Ras-PI3K pathway was detected using immunofluorescence. Activated Ras-induced changes in the H3K56ac level in different cell lines (HeLa, SMMC-7721, and HL-7702 cells) that were transfected with pEGFP-H-RasG12V/Y40C or empty-pEGFP-N1 are shown. The percentage of cells that exhibited reduced H3K56ac levels is indicated in the right panel of C.
There are three major Ras effector pathways, namely, the Raf, PI3K, and RalGEF pathways (1–3). To test whether the impact of H-RasG12V on H3K56ac was specific, we next investigated whether H3K56ac was modulated by the activation of the three effector pathways. We expressed H-RasG12V/E37G, H-RasG12V/Y40C, or H-RasG12V/T35S, which are known to preferentially activate the RalGEF, PI3K, and MAPK pathways, respectively (Ref. 17 and supplemental Fig. S1A). A consistent reduction in H3K56ac was observed when the Ras-PI3K pathway was activated (Fig. 1A). However, no changes in H3K56ac were detected following the activation of either of the other two pathways. Using Western blot analyses, we also examined a number of other histone modifications, including H3K18ac, H3K9ac, H3K14ac, H4K16ac, H4K5ac, and H4K8ac; however, no obvious effects on these epigenetic modifications were observed following the expression of H-RasG12V/Y40C (supplemental Fig. S1B). The effect of H-RasG12V/Y40C on H3K56ac was also confirmed in several other cell lines using immunofluorescence analyses (Fig. 1C and supplemental Fig. S1, C and D). The results of these experiments indicate that the effect in H3K56ac is similar in the different tested cell lines. PI3K pathway activation in the transfected cells was confirmed by detecting the levels of phosphorylated AKT (Fig. 1B).
The specific impact of the Ras-PI3K-AKT pathway on H3K56ac may account for why this pathway seems to be more critical for cell proliferation, survival, and migration than are the other two pathways (9, 17, 18, 19). To demonstrate that the change in H3K56ac was functionally involved in Ras-PI3K-AKT-induced tumorigenesis, we next expressed an H3 mutant protein that mimics the constitutive acetylation of H3K56. This was achieved by replacing the lysine at 56 with a glutamine (H3K56Q). The presence of H3K56Q reduced RasG12V/Y40C-induced tumor cell proliferation by 65%, colony formation by 76%, and cell migration by 61% (Fig. 2, A–C). However, a mutation in histone H3 at lysine 18 (H3K18Q) had no obvious effect on these cells. These data suggest that H3K56ac is involved in Ras-PI3K-AKT pathway-induced tumorigenesis. We also verified these effects in HL-7702 (a normal liver cell line) and SMMC-7721 (a hepatoma carcinoma cell line) cells, for which we obtained the same results (supplemental Fig. S1E).
FIGURE 2.
H3K56ac is functionally involved in the oncogenic Ras signaling pathway. As detected using cell viability, soft agar, and Transwell assays, the presence of H3K56ac, which was mimicked by H3K56Q expression, reduced Ras-PI3K induced cell viability (A), colony formation (B), and cell migration (C). HeLa cells were transfected with pEGFP-N1, pEGFP-H3, pEGFP-H-RasG12V/Y40C, pEGFP-H3K56Q, or pEGFP-H3K18Q (indicated as GFP, H3, Ras, H3K56Q, and H3K18Q, respectively) plasmids. A, the H3, H3K18Q, or increasing amounts of the H3K56Q (0.5, 1, and 2 μg) expression plasmids were co-transfected with 0.6 μg of pEGFP-H-RasG12V/Y40C plasmid. pEGFP-H3K18Q was used as a control (2 μg). The numbers of colonies and the optical density (OD) values of the migrating cells are shown.
H3K56ac has been demonstrated to be critical for genomic stability and other cellular functions (15, 16, 20–27). Whether H3K56ac is involved in tumorigenesis via the regulation of gene expression is unclear. We performed microarray analyses using total RNA that was isolated from cells that co-expressed histone H3 and EGFP, histone H3 and RasG12V/Y40C, or RasG12V/Y40C and H3K56Q (Fig. 3, A and B). The results indicate that the activation of the Ras pathway resulted in the altered transcription of more than 400 genes. Among the affected genes, nearly two-thirds were up-regulated. Notably, the presence of H3K56Q resulted in the down-regulation of more than 83% of the genes that were up-regulated by Ras activation. Furthermore, one-third of the genes that were down-regulated by Ras activation were rescued to the same basal level as the controls in the presence of H3K56Q. Thus, this result supports a repressive role of H3K56ac in the transcriptional regulation of Ras-induced genes. The expression of an H3 mutant protein that mimics the constitutive loss of H3K56ac by replacing the lysine at 56 with an alanine (H3K56A) consistently resulted in the up-regulation of two-thirds of the affected genes (Fig. 3A).
FIGURE 3.
The transcription of Ras-PI3K-AKT-targeted genes can be partially rescued by increased H3K56ac. A, the genes that are differentially expressed following Ras-PI3K (Ras) activation alone and in combination with the expression of the acetylation mimic H3K56Q were determined using microarray analysis. The effect of the H3K56A mutation, which mimics the nonacetylation state of H3K56 via a lysine-to-alanine substitution, was measured by another set of microarray analyses. B, the number of genes that were differentially expressed is shown. C, the transcription of several genes was confirmed using real time PCR. The values are presented as the means ± S.E. percentage relative to GAPDH expression. D, as detected using ChIP, reduced levels of H3K56ac are present on the differentially expressed genes. ERK1 was used as a negative control. The values are presented as the means ± S.E. from three independent experiments.
There are certain genes that are up-regulated by Ras activation that are known to be associated with tumor progression. CYR61 was demonstrated to promote cell proliferation by inhibiting carboplatin-induced apoptosis (28), and CDC14A was observed to stabilize the complex of ErK3 and cyclin-D3, which is implicated in cellular proliferation (29). CDC14A also promotes cellular proliferation by dephosphorylating Sirt2 (30, 31). The expression of certain other genes, including CD36 and Arhgap5, has also been implicated in tumorigenesis via the negative regulation of cell-extracellular matrix adhesion and/or antiangiogenic effects (32, 33). The altered transcription of these genes that was observed following overexpression of constitutively active Ras was also confirmed using real time PCR (Fig. 3C). Similar to what was observed for the other genes, the transcription of these genes was rescued by ∼50% in the presence of H3K56Q.
To further confirm that H3K56ac was directly associated with the transcription of these genes, we next performed ChIP experiments (Fig. 3D). Consistent with the observed global reduction in H3K56ac that was detected by Western blotting, we observed a reduced level of H3K56ac at the promoters of these up-regulated genes following the activation of the Ras-PI3K-AKT pathway. This result supports the hypothesis that H3K56ac is largely a marker for silenced genes but that it also marks a significant fraction of activated genes. This finding is consistent with a previous study reporting that H3K56ac marks both transcriptionally active and inactive genes (34).
A recent study suggested that Sirt1 and Sirt2 deacetylate H3K56 (35). We therefore examined whether the Ras-induced reduction in H3K56ac and promotion of cell proliferation and migration could be rescued by depleting Sirt1 and Sirt2. HeLa cells were therefore co-transfected with pEGFP-H-RasG12V/Y40C and siRNAs against Sirt1 or Sirt2. As expected, the depletion of either Sirt1 or Sirt2 rescued the observed Ras-induced decrease in H3K56ac (Fig. 4A and supplemental Fig. S3, A and B). Consistent with this result, Sirt1 or Sirt2 depletion also attenuated the observed Ras-induced changes in the cell cycle, as well as the alterations that were observed in tumor cell migration and proliferation (Fig. 4, B–D). HDAC1 and HDAC2 have also been reported to deacetylate H3K56ac, and we confirmed that the depletion of HDAC1 and HDAC2 caused H3K56 hyperacetylation (Ref. 9 and supplemental Fig. S3E). We also observed a modest increase in H3K56ac under the background activation of Ras-PI3K-AKT (supplemental Fig. S3F); however, this increase was not as obvious as was observed following Sirt1/2 depletion. The association of Sirt1/Sirt2 and HDAC1/HDAC2 with the affected genes was analyzed using ChIP (supplemental Fig. S4, A and B).
FIGURE 4.
Sirt1 or Sirt2 depletion prevents Ras-PI3K activation-induced decreases in H3K56ac. A, the efficiency of siRNA-mediated Sirt1 or Sirt2 knockdown was determined (left panel). The depletion of Sirt1 (middle panel) or Sirt2 (right panel) prevented the Ras-PI3K activation-induced decrease in the H3K56ac. HeLa cells were co-transfected as indicated. Whole cell lysates were assayed using Western blotting. B–D, HeLa cells were co-transfected with pEGFP-H-RasG12V/Y40C or pEGFP-N1 plasmids and a Sirt1- or Sirt2-specific or control siRNA as indicated (Ras, GFP, si-Sirt1, si-Sirt2, or si-con, respectively). As detected using Transwell and cell viability assays, Sirt1 or Sirt2 depletion resulted in reduced cell migration and cell viability (B and C). Cell cycle progression was measured using flow cytometry (D). E, HeLa cells were transfected with siRNA to deplete Sirt1 and Sirt2. The transcription of genes was examined using real time PCR. The values are presented as the means ± S.E. percentage relative to GAPDH expression.
We performed real time PCR to ascertain whether the transcription of CYR61, CDC14A, CD36, and Arhgap5 was affected following the rescue of H3K56ac by Sirt1 and Sirt2 depletion. Indeed, Sirt1 or Sirt2 depletion partially rescued the transcription of these genes (Fig. 4E), supporting a role for Sirt1 and Sirt2 in Ras-PI3K-AKT signaling-induced gene expression. HDAC1 and HDAC2 appear to play a transcriptional role different from that of Sirt1 and Sirt2 in the regulated genes (Fig. 4E and supplemental Fig. S4A). Several Ras-induced genes were observed to be bound by HDAC1/2 (supplemental Fig. S4B), supporting a relationship between Ras-induced genes and these enzymes; however, the exact mechanisms of these regulators in the transcription of Ras-induced genes remain unclear.
We next investigated how H3K56ac is regulated by the Ras-PI3K-AKT pathway. We first examined the transcription of the genes that encode the enzymes that regulate the acetylation and deacetylation of H3K56ac. These genes included the histone acetyltransferases Gcn5, CBP, and p300; the histone deacetylases Sirt1 and Sirt2 (20, 35), HDAC1, and HDAC2 (36); and the histone chaperone ASF1A, which is known to be required for H3K56ac (35). The transcriptional level of these genes was not affected when the Ras pathway was activated (supplemental Fig. S2A). Notably, a decrease in the protein levels of tagged p300 and CBP was observed. No such decreases were observed in association with the other factors (Fig. 5A and supplemental Fig. S2B). The histone acetyltransferases p300 and CBP are known to be involved in malignancy (37) and to regulate H3K56ac (35). To ensure that the observed reduction in the levels of tagged p300 and CBP were not the result of the artificial transgenic system we employed, we further analyzed endogenous p300 and CBP expression. We observed that, similar to the tagged version of these proteins, endogenous p300 and CBP were also consistently down-regulated following Ras-PI3K pathway activation (Fig. 5A). To further confirm whether p300 and CBP physically associated with the affected genes, we next performed a ChIP analysis. The results of these experiments demonstrate that ∼70% of the tested genes that exhibited reduced H3K56ac following Ras-PI3K activation also exhibited a reduction in p300 or CBP binding (Fig. 5B). These results support the hypothesis that p300 and CBP are responsible for the Ras-PI3K-induced reduction in H3K56ac.
FIGURE 5.
Ras-PI3K-AKT pathway activation is associated with CBP/p300 degradation. A, HeLa cells were transfected with 0.6 μg of pEGFP-H-RasG12V/Y40C, 0.6 μg of pEGFP-N1, and 3 μg of CBP-HA or p300-HA plasmids. The cell lysates were analyzed using Western blotting. B, the recruitment of CBP and p300 to the genes that exhibited changes in H3K56ac was analyzed using ChIP analysis, with IL-8 as a positive control and Mef2c, GATA4, and Nkx2.5 as negative controls. C, whole cell extracts from HeLa cells that were treated with MG132 were prepared, and the CBP and p300 protein levels were analyzed using Western blotting with antibodies against HA and GFP. Whole cell extracts from HeLa cells that were untreated (the samples were collected at 24, 48, 51, 54, and 60 h following transfection; upper panel) or treated with MG132 (the MG132 was added at 48 h following transfection, and the samples were collected at 24, 48, 51, 54, and 60 h following transfection; lower panel) were analyzed using Western blotting.
Because the expression of p300 and CBP was not affected at the transcriptional level, these proteins are likely regulated post-transcriptionally. To confirm this possibility, the transfected cells were treated with MG132, a known proteasome inhibitor. Our Western blotting analysis indicated that the presence of MG132 indeed reversed the observed reduction in p300 and CBP levels following the activation of the Ras-PI3K-AKT pathway (Fig. 5C, upper panels). Using the same experimental approach, we also confirmed that the addition of MG132 reversed the reduction in H3K56ac (Fig. 5C, lower panels). These results therefore suggest that the activation of the Ras-PI3K-AKT pathway likely decreases H3K56ac via the proteasome-mediated degradation of CBP/p300.
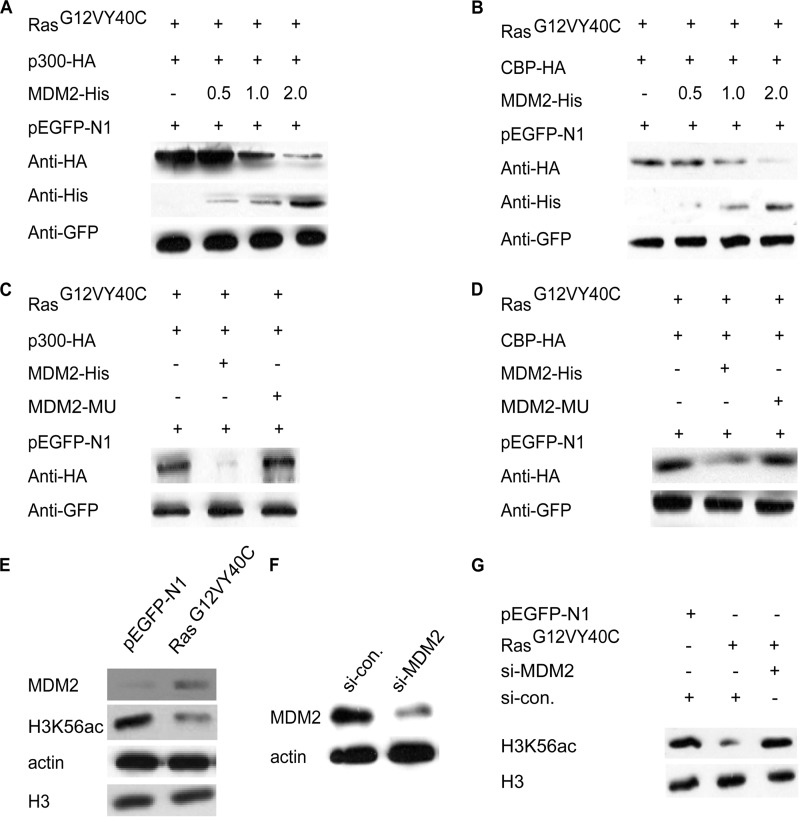
It is well established that the E3 ubiquitin ligase MDM2 is both the major substrate of AKT and is responsible for the degradation of several different histone acetyltransferases (38, 39). Therefore, we examined whether MDM2 is involved in the degradation of CBP/p300 following the activation of the Ras-PI3K-AKT pathway. HeLa cells were co-transfected with RasG12V/Y40C and HA-tagged p300 or CBP expression plasmids with or without an MDM2-His expression plasmid. Whole cell extracts were analyzed for p300 and CBP expression (Fig. 6, A and B). The results of these experiments indicated that the co-expression of H-RasG12V/Y40C and MDM2 indeed caused a significant reduction in HA-p300 and HA-CBP expression. However, when we performed the assay using an MDM2C464A mutant, which is mutated in the RING-finger domain and lacks ubiquitin ligase activity, no such changes in p300 and CBP protein levels were observed (Fig. 6, C and D). These results support the finding that MDM2 activity is required for the degradation of p300/CBP.
FIGURE 6.
The Ras-PI3K-AKT signaling pathway-induced degradation of p300 and CBP is mediated by MDM2. A and B, MDM2 induces the degradation of p300 (A) and CBP (B). HeLa cells were co-transfected with 1 μg of p300-HA or CBP-HA, 0.6 μg of pEGFP-H-RasG12V/Y40C, 0.1 μg of pEGFP-N1 plasmids and increasing amounts of MDM2-His plasmid (0.5, 1, and 2 μg). C and D, a mutation in MDM2 (MDM2-MU) that abolishes its ubiquitin ligase activity prevents p300 and CBP degradation. The HeLa cells were co-transfected as indicated. E, whole cell extracts from HeLa cells that were transfected with pEGFP-N1 or pEGFP-H-RasG12V/Y40C were analyzed using Western blotting. F, the efficiency of the siRNA-mediated MDM2 knockdown. G, The co-transfection of pEGFP-H-RasG12V/Y40C and MDM2-specific siRNA restores H3K56ac to normal levels.
The above result raised the question of whether the reduction in p300 and CBP levels following RAS-PI3K-AKT activation is a consequence of an up-regulation of MDM2. We therefore used an anti-MDM2 antibody to detect changes in the levels of endogenous MDM2 protein following the activation of Ras signaling. The result of this experiment indicated that the MDM2 protein level was indeed increased severalfold (Fig. 6E). This result is in agreement with a previous study that demonstrated that MDM2 protein levels increased in RasG12V-overexpressing NIH3T3 cells and induced CBP/p300 degradation (40). However, the transcription of the MDM2 gene appeared to be unaffected following the activation of the Ras pathway (supplemental Fig. S2E).
To confirm that the up-regulation of MDM2 caused the observed degradation of p300/CBP, we next knocked down MDM2 expression in H-RasG12V/Y40C-expressing cells using MDM2 siRNA. The results of this experiment indicated that the decrease in H3K56ac was indeed prevented (Fig. 6, F and G), suggesting that Ras-PI3K-AKT regulates H3K56ac via MDM2-mediated CBP/p300 degradation. We also confirmed the connection between CBP/p300, MDM2, and H3K56ac in HL-7702 cells and SMMC-7721 cells following the activation of the Ras-PI3K pathway (supplemental Figs. S5 and S6).
DISCUSSION
It is well established that Ras promotes the initiation of several types of human tumors. However, among the three Ras effector pathways, the PI3K/AKT pathway appears to be more important than the others in terms of tumor maintenance (9, 17). It is therefore of interest to determine the mechanism by which the Ras-PI3K pathway participates in tumorigenesis. In this study, we demonstrated that the Ras-PI3K effector pathway, but not the Ras-MAPK or RalGEF pathways, specifically regulates H3K56ac. This result supports the hypothesis that the Ras pathways have different functions, possibly as a result of specific epigenetic modifications.
Histone H3K56 acetylation was demonstrated to be critical for genome stability (15, 16, 20) and to be associated with multiple types of cancer (35). In yeast, H3K56ac regulates a number of cellular events, including gene activity (41–43), histone replacement (44), chromatin assembly (15), DNA replication, DNA repair, and gene silencing (46, 47). H3K56 acetylation is believed to exert an effect on the nucleosome and chromatin via the recruitment of an SWI/SNF-like nucleosome remodeling complex (41, 46, 48). Recent studies have demonstrated that H3K56ac is present on active and inactive genes (34, 49, 50). The results of the present study support both a negative and positive role of H3K56ac in gene transcription. However, given that the genes that were observed to be up-regulated by Ras-PI3K pathway activation are associated with a reduction in H3K56ac in their promoter regions, H3K56ac appears to negatively regulate transcription in this context. This negative regulation is also supported by the fact that the presence of H3K56Q (the acetylation mimic) also induced reductions in transcription (Fig. 3). Furthermore, the loss of H3K56ac, which was achieved by the expression of the H3K56A mutant, resulted in increased transcription of nearly two-thirds of the affected genes (Fig. 3A).
The exact role of H3K56ac in the transcriptional repression of Ras-induced genes is unclear. Given that H3K56ac also participates in DNA damage repair and other chromatin activities, one cannot exclude the possibility that the observed changes in gene transcription may be an indirect effect of reduced H3K56ac on the chromatin. Our ChIP analysis of H3K56ac regulators confirms that the reduction in H3K56ac correlates well with reduced levels of p300 and CBP. These proteins are both histone acetyltransferases and are known to be associated with transcriptional activity. Sirt1/2 also appear to be involved in Ras-induced cellular activity. However, we did not detect Sirt1/2 in the ChIP analysis of genes that were affected by Ras up-regulation. It is unclear whether the observed effects of Sirt1/2 on Ras-induced cellular and gene activity were indirect or were due to the specificity/sensitivity of the antibody used, which failed to detect the presence of or any change in the levels of these regulators among the tested genes. Overall, our results are consistent with a report that H3K56ac marks both transcriptionally active and inactive genes (34).
How is H3K56ac specifically regulated following Ras-PI3K pathway activation? We demonstrated that the decrease in H3K56ac is mediated by CBP/p300 (Figs. 5 and 6). However, other modifications, such as H3K18ac, are also known to be induced by CBP/p300 (51). Surprisingly, no clear changes in H3K18ac were observed following Ras pathway activation. Similarly, the acetylation of several other lysine residues, including H3K9, H3K14, H4K16, H4K5, and H4K8, which is regulated by the same enzymes as H3K56, was unaffected (supplemental Fig. S1B). This result implies that the precise regulation of the specific modification is more complicated than has been believed. In a previous study, it was also shown that depletion of CBP or p300 only affected specific histone acetylation (51). Notably, the viral protein E1A and the regulatory protein Twist bind to p300 and inhibit its activity (52–54), and E1A has a stimulatory effect on the histone acetyltransferase activity of CBP (55). The recruitment of CBP/p300 onto a specific gene promoter can be regulated via specific protein-protein interaction (56, 57) and the finding that CBP/p300 is a cell type-specific modulator of transcription (58) all support that the histone acetyltransferase activity of CBP/p300 is regulated by a number of other factors. Therefore, the specific effect of CBP/p300 on H3K56 may be due to the requirement for specific partners or the formation/function of CBP/p300 complexes, either of which would allow for specific recognition of the H3K56 acetylation following Ras signaling activation. Alternatively, Ras-induced changes in other histone acetyltransferases may compensate for the observed reduction in these modifications (45). Future studies are required to explore these possibilities.
We provide evidence that the reduction in CBP/p300 levels was due to the MDM2-mediated protein degradation. This conclusion is supported both by the degradation of tagged p300/CBP in the presence of MDM2 and increased endogenous MDM2 protein levels following Ras activation. The transcriptional level of the MDM2 gene did not appear to be affected (supplemental Fig. S2E), suggesting that MDM2 is post-transcriptionally regulated. The precise mechanism of MDM2 up-regulation following RAS-PI3K activation is unclear. Our results suggest that MDM2 is involved in the down-regulation of CBP/p300 and H3K56ac given that the depletion of MDM2, under the condition of Ras activation, rescued both the degradation of both CBP and p300 and the reduction in H3K56ac (Fig. 6).
Taken together, our findings demonstrate that H3K56ac is specifically targeted by the oncogenic Ras-PI3K pathway. We also demonstrate a regulatory mechanism that links the Ras-PI3K signaling pathway to specific regulators of H3K56 acetylation in tumorigenesis; this mechanism may serve as a novel therapeutic target for tumor prevention.

This article contains supplemental Table S1 and Figs. S1–S6.
- Ral
- Ras-like
- RalGEF
- Ral guanine nucleotide exchange factor
- CREB
- cAMP-responsive element-binding protein
- CBP
- CREB-binding protein
- H3K56ac
- H3K56 acetylation
- HDAC
- histone deacetylase.
REFERENCES
- 1. Cagnol S., Chambard J. C. (2010) ERK and cell death. Mechanisms of ERK-induced cell death. Apoptosis, autophagy and senescence. FEBS J. 277, 2–21 [DOI] [PubMed] [Google Scholar]
- 2. Yuan T. L., Cantley L. C. (2008) PI3K pathway alterations in cancer. Variations on a theme. Oncogene 27, 5497–5510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cascone I., Selimoglu R., Ozdemir C., Del Nery E., Yeaman C., White M., Camonis J. (2008) Distinct roles of RalA and RalB in the progression of cytokinesis are supported by distinct RalGEFs. EMBO J. 27, 2375–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Morrison D. K., Cutler R. E. (1997) The complexity of Raf-1 regulation. Curr. Opin. Cell Biol. 9, 174–179 [DOI] [PubMed] [Google Scholar]
- 5. Shields J. M., Pruitt K., McFall A., Shaub A., Der C. J. (2000) Understanding Ras. “It ain't over 'til it's over.” Trends. Cell Biol. 10, 147–154 [DOI] [PubMed] [Google Scholar]
- 6. Luo J., Manning B. D., Cantley L. C. (2003) Targeting the PI3K-Akt pathway in human cancer. Rationale and promise. Cancer Cell 4, 257–262 [DOI] [PubMed] [Google Scholar]
- 7. Feig L. A. (2003) Ral-GTPases. Approaching their 15 minutes of fame. Trends. Cell Biol. 13, 419–425 [DOI] [PubMed] [Google Scholar]
- 8. Lim K. H., Baines A. T., Fiordalisi J. J., Shipitsin M., Feig L. A., Cox A. D., Der C. J., Counter C. M. (2005) Activation of RalA is critical for Ras-induced tumorigenesis of human cells. Cancer Cell 7, 533–545 [DOI] [PubMed] [Google Scholar]
- 9. Carnero A. (2010) The PKB/AKT Pathway in Cancer. Curr. Pharm. Des. 16, 34–44 [DOI] [PubMed] [Google Scholar]
- 10. Kouzarides T. (2007) Chromatin modifications and their function. Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 11. Chari R., Thu K. L., Wilson I. M., Lockwood W. W., Lonergan K. M., Coe B. P., Malloff C. A., Gazdar A. F., Lam S., Garnis C., MacAulay C. E., Alvarez C. E., Lam W. L. (2010) Integrating the multiple dimensions of genomic and epigenomic landscapes of cancer. Cancer Metastasis Rev. 29, 73–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Seligson D. B., Horvath S., McBrian M. A., Mah V., Yu H., Tze S., Wang Q., Chia D., Goodglick L., Kurdistani S. K. (2009) Global levels of histone modifications predict prognosis in different cancers. Am. J. Pathol. 174, 1619–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fraga M. F., Ballestar E., Villar-Garea A., Boix-Chornet M., Espada J., Schotta G., Bonaldi T., Haydon C., Ropero S., Petrie K., Iyer N. G., Pérez-Rosado A., Calvo E., Lopez J. A., Cano A., Calasanz M. J., Colomer D., Piris M. A., Ahn N., Imhof A., Caldas C., Jenuwein T., Esteller M. (2005) Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 37, 391–400 [DOI] [PubMed] [Google Scholar]
- 14. Cha T. L., Zhou B. P., Xia W., Wu Y., Yang C. C., Chen C. T., Ping B., Otte A. P., Hung M. C. (2005) Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science 310, 306–310 [DOI] [PubMed] [Google Scholar]
- 15. Downs J. A. (2008) Histone H3 K56 acetylation, chromatin assembly, and the DNA damage checkpoint. DNA Repair 7, 2020–2024 [DOI] [PubMed] [Google Scholar]
- 16. Chen C. C., Carson J. J., Feser J., Tamburini B., Zabaronick S., Linger J., Tyler J. K. (2008) Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell 134, 231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lim K. H., Counter C. M. (2005) Reduction in the requirement of oncogenic Ras signaling to activation of PI3K/AKT pathway during tumor maintenance. Cancer Cell 8, 381–392 [DOI] [PubMed] [Google Scholar]
- 18. Martelli A. M., Chiarini F., Evangelisti C., Grimaldi C., Ognibene A., Manzoli L., Billi A. M., McCubrey J. A. (2010) The phosphatidylinositol 3-kinase/AKT/mammalian target of rapamycin signaling network and the control of normal myelopoiesis. Histol. Histopathol. 25, 669–680 [DOI] [PubMed] [Google Scholar]
- 19. Gibbons J. J., Abraham R. T., Yu K. (2009) Mammalian target of rapamycin. Discovery of rapamycin reveals a signaling pathway important for normal and cancer cell growth. Semin. Oncol. 36, (Suppl. 3) S3–S17 [DOI] [PubMed] [Google Scholar]
- 20. Tjeertes J. V., Miller K. M., Jackson S. P. (2009) Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J. 28, 1878–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaplan T., Liu C. L., Erkmann J. A., Holik J., Grunstein M., Kaufman P. D., Friedman N., Rando O. J. (2008) Cell cycle- and chaperone-mediated regulation of H3K56ac incorporation in yeast. PLoS Genet. 4, e1000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hiraga S., Botsios S., Donaldson A. D. (2008) Histone H3 lysine 56 acetylation by Rtt109 is crucial for chromosome positioning. J. Cell Biol. 183, 641–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thaminy S., Newcomb B., Kim J., Gatbonton T., Foss E., Simon J., Bedalov A. (2007) Hst3 is regulated by Mec1-dependent proteolysis and controls the S phase checkpoint and sister chromatid cohesion by deacetylating histone H3 at lysine 56. J. Biol. Chem. 282, 37805–37814 [DOI] [PubMed] [Google Scholar]
- 24. Maas N. L., Miller K. M., DeFazio L. G., Toczyski D. P. (2006) Cell cycle and checkpoint regulation of histone H3 K56 acetylation by Hst3 and Hst4. Mol. Cell 23,109–119 [DOI] [PubMed] [Google Scholar]
- 25. Erkmann J. A., Kaufman P. D. (2009) A negatively charged residue in place of histone H3K56 supports chromatin assembly factor association but not genotoxic stress resistance. DNA Repair 8, 1371–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vempati R. K., Jayani R. S., Notani D., Sengupta A., Galande S., Haldar D. (2010) p300 mediated acetylation of histone H3 lysine 56 functions in DNA damage response in mammals. J. Biol. Chem. 10, 1074–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hebert C., RoestCrollius H. (2010) Nucleosome rotational setting is associated with transcriptional regulation in promoters of tissue-specific human genes. Genome Biol. 11, R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee K. B., Byun H. J., Park S. H., Park C. Y., Lee S. H., Rho S. B. (2012) CYR61 controls p53 and NF-κB expression through PI3K/Akt/mTOR pathways in carboplatin-induced ovarian cancer cells. Cancer Lett. 315, 86–95 [DOI] [PubMed] [Google Scholar]
- 29. Hansen C. A., Bartek J., Jensen S. (2008) A functional link between the human cell cycle-regulatory phosphatase Cdc14A and the atypical mitogen-activated kinase Erk3. Cell Cycle 7, 325–334 [DOI] [PubMed] [Google Scholar]
- 30. North B. J., Verdin E. (2007) Mitotic regulation of SIRT2 by cyclin-dependent kinase 1-dependent phosphorylation. J. Biol. Chem. 282, 19546–19555 [DOI] [PubMed] [Google Scholar]
- 31. Mailand N., Lukas C., Kaiser B. K., Jackson P. K., Bartek J., Lukas J. (2002) Deregulated human Cdc14A phosphatase disrupts centrosome separation and chromosome segregation. Nat. Cell Biol. 4, 317–322 [DOI] [PubMed] [Google Scholar]
- 32. Rosman D. S., Phukan S., Huang C. C., Pasche B. (2008) TGFBR1*6A enhances the migration and invasion of MCF-7 breast cancer cells through RhoA activation. Cancer Res. 68, 1319–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kaur B., Cork S. M., Sandberg E. M., Devi N. S., Zhang Z., Klenotic P. A., Febbraio M., Shim H., Mao H., Tucker-Burden C., Silverstein R. L., Brat D. J., Olson J. J., Van Meir E. G. (2009) Vasculostatin inhibits intracranial glioma growth and negatively regulates in vivo angiogenesis through a CD36-dependent mechanism. Cancer Res. 69, 1212–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xie W., Song C., Young N. L., Sperling A. S., Xu F., Sridharan R., Conway A. E., Garcia B. A., Plath K., Clark A. T., Grunstein M. (2009) Histone H3 lysine 56 acetylation is linked to the core transcriptional network in human embryonic stem cells. Mol. Cell 33, 417–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Das C., Lucia M. S., Hansen K. C., Tyler J. K. (2009) CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature 459, 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Miller K. M., Tjeertes J. V., Coates J., Legube G., Polo S. E., Britton S., Jackson S. P. (2010) Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat. Struct. Mol. Biol. 17, 1144–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Iyer N. G., Ozdag H., Caldas C. (2004) p300/CBP and cancer. Oncogene 23, 4225–4231 [DOI] [PubMed] [Google Scholar]
- 38. Legube G., Linares L. K., Lemercier C., Scheffner M., Khochbin S., Trouche D. (2002) Tip60 is targeted to proteasome-mediated degradation by Mdm2 and accumulates after UV irradiation. EMBO J. 21, 1704–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jin Y., Zeng S. X., Lee H., Lu H. (2004) MDM2 mediates p300/CREB-binding protein-associated factor ubiquitination and degradation. J. Biol. Chem. 279, 20035–20043 [DOI] [PubMed] [Google Scholar]
- 40. Sánchez-Molina S., Oliva J. L., García-Vargas S., Valls E., Rojas J. M., Martínez-Balbás M. A. (2006) The histone acetyltransferases CBP/p300 are degraded in NIH 3T3 cells by activation of Ras signalling pathway. Biochem. J. 398, 215–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xu F., Zhang K., Grunstein M. (2005) Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 121, 375–385 [DOI] [PubMed] [Google Scholar]
- 42. Schneider J., Bajwa P., Johnson F. C., Bhaumik S. R., Shilatifard A. (2006) Rtt109 is required for proper H3K56 acetylation. A chromatin mark associated with the elongating RNA polymerase II. J. Biol. Chem. 281, 37270–37274 [DOI] [PubMed] [Google Scholar]
- 43. Williams S. K., Truong D., Tyler J. K. (2008) Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. Proc. Natl. Acad. Sci. U.S.A. 105, 9000–9005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rufiange A., Jacques P. E., Bhat W., Robert F., Nourani A. (2007) Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol. Cell 27, 393–405 [DOI] [PubMed] [Google Scholar]
- 45. Allis C. D., Berger S. L., Cote J., Dent S., Jenuwien T., Kouzarides T., Pillus L., Reinberg D., Shi Y., Shiekhattar R., Shilatifard A., Workman J., Zhang Y. (2007) New nomenclature for chromatin-modifying enzymes. Cell 131, 633–636 [DOI] [PubMed] [Google Scholar]
- 46. Xu F., Zhang Q., Zhang K., Xie W., Grunstein M. (2007) Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Mol. Cell 27, 890–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Miller A., Yang B., Foster T., Kirchmaier A. L. (2008) Proliferating cell nuclear antigen and ASF1 modulate silent chromatin in Saccharomyces cerevisiae via lysine 56 on histone H3. Genetics 179, 793–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Masumoto H., Hawke D., Kobayashi R., Verreault A. (2005) A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436, 294–298 [DOI] [PubMed] [Google Scholar]
- 49. Martens J. A., Winston F. (2003) Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev. 13, 136–142 [DOI] [PubMed] [Google Scholar]
- 50. Kaeser M. D., Aslanian A., Dong M. Q., Yates J. R., 3rd, Emerson B. M. (2008) BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. J. Biol. Chem. 283, 32254–32263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Horwitz G. A., Zhang K., McBrian M. A., Grunstein M., Kurdistani S. K., Berk A. J. (2008) Adenovirus small E1A alters global patterns of histone modification. Science 321, 1084–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chakravarti D., Ogryzko V., Kao H. Y., Nash A., Chen H., Nakatani Y., Evans R. M. (1999) A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell 96, 393–403 [DOI] [PubMed] [Google Scholar]
- 53. Hamamori Y., Sartorelli V., Ogryzko V., Puri P. L., Wu H. Y., Wang J. Y., Nakatani Y., Kedes L. (1999) Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell 96, 405–413 [DOI] [PubMed] [Google Scholar]
- 54. Perissi V., Dasen J. S., Kurokawa R., Wang Z., Korzus E., Rose D. W., Glass C. K., Rosenfeld M. G. (1999) Factor-specific modulation of CREB-binding protein acetyltransferase activity. Proc. Natl. Acad. Sci. U.S.A. 96, 3652–3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ait-Si-Ali S., Ramirez S., Barre F. X., Dkhissi F., Magnaghi-Jaulin L., Girault J. A., Robin P., Knibiehler M., Pritchard L. L., Ducommun B., Trouche D., Harel-Bellan A. (1998) Hitone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature 396, 184–186 [DOI] [PubMed] [Google Scholar]
- 56. Ogryzko V. V., Schiltz R. L., Russanova V., Howard B. H., Nakatani Y. (1996) The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87, 953–959 [DOI] [PubMed] [Google Scholar]
- 57. Korzus E., Torchia J., Rose D. W., Xu L., Kurokawa R., McInerney E. M., Mullen T. M., Glass C. K., Rosenfeld M. G. (1998) Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science 279, 703–707 [DOI] [PubMed] [Google Scholar]
- 58. Hosoda H., Kato K., Asano H., Ito M., Kato H., Iwamoto T., Suzuki A., Masushige S., Kida S. (2009) CBP/p300 is a cell type-specific modulator of CLOCK/BMAL1-mediated transcription. Mol. Brain 2, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]