Background: Plasmodium falciparum P2 (PfP2) protein plays nonribosomal roles through SDS- and DTT-resistant oligomerization.
Results: For SDS- and DTT-sensitive oligomerization, the 53rd cysteine of PfP2 plays an important role.
Conclusion: DTT- and SDS-resistant oligomerization of PfP2 was propagated by differentially expressed parasite proteins.
Significance: Analysis of regulation of PfP2 oligomerization in parasite-infected erythrocytes may help in understanding the export of P2 to erythrocyte surface.
Keywords: Aggregation, Cell Biology, Malaria, Parasite, Parasitology, Protein Export, Biochemistry, Biophysics, Molecular Biology, Protein Structure and Folding
Abstract
The eukaryotic 60 S-ribosomal stalk consists of P0, P1, and P2 proteins, which associate in a pentameric structure (P12-P0-P22). The Plasmodium falciparum protein P2 (PfP2) appears to play nonribosomal roles. It gets exported to the infected erythrocyte (IE) surface at 30 h post-merozoite invasion (PMI), concomitant with extensive oligomerization. Here we present certain biophysical properties of PfP2. Recombinant P2 (rPfP2) protein showed SDS-resistant oligomerization, which could be significantly abolished under reducing conditions. However, the protein continued to oligomerize even when both cysteine residues were mutated, and with up to 40 amino acids (aa) deleted from the C-terminal end. CD analysis of P2 showed largely α-helical and random coil domains. The SDS- and DTT-resistant oligomerization was studied further as it occurred in a development-specific manner in Plasmodium. In a synchronized erythrocytic culture of P. falciparum, the PfP2 protein was detected as part of the ribosomal complex (∼96 kDa) at 18 and 30 h PMI, and was SDS sensitive. However, at 30 h, large amounts of SDS-sensitive aggregates of >600 kDa were also seen. At 30 h PMI, each of the parasites, IE cytosol and IE ghost contained 60–80-kDa PfP2 complexes, which resolved to a single 65-kDa species on SDS-PAGE. Tetramethylrhodamine-labeled rPfP2 protein exhibited DTT- and SDS-resistant oligomerization when treated with P. falciparum parasite extracts only from 24 to 36 h PMI, and multiple proteins appeared to be required for this oligomerization. Understanding the regulation of oligomerization of PfP2 may help in the elucidation of the novel structure-function relationship in the export of PfP2 to the red cell surface.
Introduction
The ribosomal stalk consists of certain proteins, functionally conserved in all organisms (1–3). In eukaryotes, the stalk is composed of three types of P-proteins, P0, P1, and P2. The P0 protein, equivalent to the L10 protein in prokaryotes, forms the base of the stalk and directly interacts with the 28 S rRNA, and constitutes the binding site for the two protein dimers, (P1-P2)2 (4–6). In eukaryotes the dimers are made of two independent polypeptides, P1 and P2, which is different from that in prokaryotes, which contain two to three homodimers of L12 protein (4, 6). In lower eukaryotes, such as yeast and Trypanosoma, two additional subgroups are distinguished, comprising the P1α, P1β, P2α, and P2β proteins (7, 8), whereas an additional P3 protein has been recognized in plants (9).
It is believed that the P protein-pentameric complex binds to eukaryotic 28 S rRNA and plays an important role in the GTPase-associated center of eukaryotic ribosomes (5, 10–11). When added to prokaryotic 23 S rRNA, this P-protein complex changes the specificity of the ribosome to the eukaryotic elongation factors (12). This strong dependence on the P0-P1-P2 complex for the factors accessibility suggests a direct interaction between the protein complex and elongation factors. It has also been suggested that the pentameric P-complex modulates the structures of the sarcin/ricin domain of 23 S/28 S rRNA and makes them accessible to eukaryotic elongation factors (13). It has been reported that binding of P1 and P2 to P0 protein induces the binding activity of P0 to rRNA (10). Through several deletion constructs, it was shown that the C-terminal half of the P0 protein contains two neighboring sites for P1-P2 heterodimers (14). In yeast, each of the P1 or P2-null mutants is viable in rich medium and no significant effects are seen in the rates of peptide bond formation, although protein synthesis and growth rates are reduced (15). Heterogeneity of P-proteins in ribosomal composition has been observed, and ribosomes from stationary phase-deficient P1/P2 proteins have been reported (15). It is of interest that the pattern of protein expression in the absence of P1 and P2 proteins is distinct from that in the presence of these acidic proteins (16). It was shown that such a differential expression pattern was not due to translation error or termination suppression, but was postulated to be due to differential translation modulation, and/or due to extraribosomal properties of these acidic proteins (16).
Ribosomal proteins are known to play varied roles besides protein synthesis (17). We have earlier demonstrated that P0 protein plays a protective role at the merozoite surface (18–20). In Neisseria gonorrhoeae, the functional orthologue of P2 (L12), is shown to be surface exposed, and has been implicated in cell invasion (21). There have been associations of ribosomal protein expression with cancer, but those have been subscribed to altered cellular protein synthesis (22). In Plasmodium, neither P1 nor P2 protein is likely to be vital for the ribosomal functions, because in a complementation study in Saccharomyces cerevisiae, we have demonstrated that ribosomes containing just PfP0 (without any P1/P2 proteins) were capable of synthesizing proteins (23). We have not been able to knock-out Plasmodium P2 protein,2 and therefore surmise that P2 protein may possess important extraribosomal functions.
A nonribosomal role of Plasmodium P2 protein certainly appears to exist, because a translocation of the P2 protein, but not P0 or P1 proteins, to the infected erythrocyte (IE)3 surface during early cell division has been observed recently (24). Moreover, an unusual cell cycle arrest of Plasmodium occurs when IEs are treated with a panel of anti-PfP2-specific monoclonal antibodies (24). The IE surface-exposed PfP2 protein occurs during a short window of erythrocytic development and the exported P2 protein appears to exist exclusively as an SDS-resistant P2-homotetramer (24). This indicates a developmental regulation of oligomerization of the Plasmodium P2 protein.
The structure of the N-terminal dimerization domain of human P2 protein was determined recently by NMR (25). By homology modeling, a structural model of P1/P2 dimerization domain was proposed and this model predicted that helix-3 of P1 is not involved in P1/P2 dimerization, but plays an important role in formation of the P-complex (25). The hetero-oligomerization of P1/P2 proteins have been reported earlier, and these appear to be precursors to the pentameric ribosomal P-protein complex (26). However, P1 and P2 proteins have been found in the cytoplasmic pool, which exchange with those on the ribosome, and also form homo-oligomers (26). For P. falciparum P2 protein, so far the NMR structure has been possible only under denaturing conditions, because the recombinant PfP2 protein undergoes extensive oligomerization at concentrations of about 1 mm (27).
Here we report the biophysical properties of the ribosomal P2 protein of P. falciparum, with special emphasis on oligomerization. We demonstrate that the 53rd cysteine residue of PfP2 is vital for SDS-resistant DTT-sensitive oligomerization, and that the P2 protein shows anomalous folding due to the acidic C-terminal domain. We also show that apart from the SDS-sensitive pentameric complex, the P2 protein forms developmentally regulated DTT- and SDS-resistant oligomers, and that certain differentially expressed protein components of the parasite appear to play important roles in such oligomer formation.
EXPERIMENTAL PROCEDURES
Ethics Statement
Tata Institute of Fundamental Research (TIFR) Animal House is registered under CPCSEA (Committee for the Purpose of Control and Supervision of Experiments on Animals), Ministry of environment and forest, Government of India (registration number 56/1999/CPCSEA) for breeding and experiments on animals. This study was carried out under strict accordance with the guidelines of CPCSEA, India, for the care and use of laboratory animals. The study was approved by the institutional animal ethics committee, TIFR, Mumbai (Project number TIFR/IAEC/2008-1) formulated by CPCSEA.
Human blood was collected from volunteers after obtaining their written consents, for the in vitro cultures of Plasmodium falciparum. The procedure for such collection, details of informed consent, and the frequency of samples to be collected were in accordance with a detailed proposal approved by the Institutional Human Ethics Committee of TIFR. The Institutional Human Ethics Committee of TIFR is constituted as per the guidelines of Indian Council of Medical Research, Government of India.
Cloning and Expression of PfP2 (PFC0400w) Gene and Its Mutants in the pProExHTa Vector
The PfP2 gene was PCR amplified from P. falciparum (3D7) genomic DNA using the following primers containing EcoRI (New England BioLabs) at the 5′ end and XhoI (New England BioLabs) at the 3′ end restriction overhang, respectively, forward primer: 5′-CCCCGAATTCATGGCTATGAAATACGTTGCTG-3′; reverse primer: 5′-GGGGCTCGAGTTAACCAAATAAGGAAAATCCTAAGTC-3′. Both the PCR amplified PfP2 gene fragment and the pProExHTa vector (Lablife) DNA were digested using EcoRI and XhoI restriction enzymes at 37 °C, purified, and ligated at 16 °C for 16 h using T4 DNA ligase (Roche Applied Science, catalog number 10481220001). DH5α-competent cells were transformed by heat shock, and positive clones were identified through plasmid purification and restriction digestion. PfP2CΔ20 and PfP2CΔ40 were also cloned in the same vector following the same methodology, except that the amplifications were carried out using the following reverse primers: PfP2CΔ20, 5′-GGGCTCGAGTTATTCTTTCTTATCTTCTTTCTTAG-3′; PfP2CΔ40, 5′-GGGGCTCGAGTTAACCACCTCCAATATTTTG-3′, and keeping the forward primer as mentioned above.
The GST-PfP1 construct was made by cloning the PfP1 gene between EcoRI and XhoI sites in the pGEX-4T-3 vector (GE Healthcare). The PfP1 gene, with one postulated intron, was amplified from the P. falciparum (3D7) cDNA library using the following primers: forward: 5′-CCCCGAATTCATGGCATCAATTCCAGCATC-3′ and reverse: 5′-GGGGCTCGAGACCAAATAAGGAGAAACC-3′. The DNA (ORF) sequences of all the clones were confirmed by DNA sequencing. For generating mutants of the PfP2 gene, custom based site-directed mutagenesis was carried out and five different mutant clones of the PfP2 gene were generated, M1 (C12A), M2 (C53Y), M3 (C12A,C53Y), M4 (C53A), and M5 (C12A,C53A). The mutant genes were cloned in pProExHTa vector and the nucleotide sequence was confirmed by DNA sequencing through Bangalore Genei, India.
Recombinant Protein Expression and Purification
All constructs (P2, M1(C12A), M2 (C53Y), M3 (C12A,C53Y), M4 (C53A), M5 (C12A,C53A), P2CΔ20, P2CΔ40), were transformed in Escherichia coli BL21(DE3) strain and protein expression was induced by 0.5 mm IPTG (Sigma, catalog number I6758). However, M5 did not get expressed under similar conditions. Recombinant PfP2 (rPfP2), M1, M2, M3, M4, rPfP2CΔ20, and rPfP2CΔ40 proteins were purified using nickel-nitrilotriacetic acid beads (Qiagen, catalog number 30230). All recombinant proteins, rPfP2, M1, M2, M3, M4, rPfP2CΔ20, and rPfP2CΔ40, were fusion proteins containing an additional 30 amino acids (aa) at the N terminus, including 6-histidine, totaling 142 (for P2, M1, M2, M3, M4), 122 (for rPfP2CΔ20), and 102 aa (rPfP2CΔ40), respectively. PfP2 was cloned in pQE and pET vectors to obtain a nonfusion or a cleavable P2-recombinant protein. However, no stable expression of PfP2 protein could be achieved without these additional 30 amino acids. PfP1 protein was expressed in BL21 cells as a GST fusion protein and purified using GST beads. 10 mm reduced glutathione was used to elute PfP1-GST protein from the beads.
Parasite Culture and Synchronization
P. falciparum 3D7 strain parasites were maintained in culture as described earlier (28). Human blood, from healthy adults with B+ blood group, was collected in K2 EDTA vacutainers (BD Biosciences) as an anticoagulant. After removing the leukocytes, the erythrocytes were washed and resuspended in complete RPMI (cRPMI; RPMI with 0.5% Albumax). Asexual stages of the P. falciparum 3D7 strain were cultured in vitro and maintained at 5% hematocrit in complete RPMI at 37 °C in a humidified chamber containing 5% CO2. Synchronization of parasite was carried out using 5% sorbitol.
Preparation of P. falciparum Parasite, Infected RBC Cytosol, and Ghost Proteins
P. falciparum 3D7-infected RBCs at about 7–8% parasitemia were pelleted at 500 × g for 5 min, and washed with complete RPMI once. RBC pellet was resuspended in 0.1% saponin and a protease inhibitor mixture (Sigma, catalog number P8340) and 1 mm PMSF in phosphate-buffered saline (PBS), pH 7.4, for 15 min at 37 °C. The sample was then centrifuged for 10 min at 10,000 × g at 4 °C to remove the parasite pellet, which was sonicated in lysis buffer (PBS, pH 7.4, with 0.1% Triton X-100 and protease inhibitors) to obtain the parasite protein and stored at −80 °C. About 60–70% of the opaque supernatant (RBC ghost and cytosol fraction) was gently separated to avoid contamination from the parasite pellet. This supernatant fraction was pelleted at 20,000 × g for 2 h at 4 °C, washed twice with PBS, pH 7.4, and stored at −80 °C for use as IE ghost. After ghost precipitation, the supernatant (IE cytosol) was stored at −80 °C for subsequent analysis. All buffers contained protease inhibitor mixture (Sigma, catalog number P8340).
Immunoblots of Recombinant and Parasite Proteins
The recombinant or parasite proteins were used for the immunoblots after protein estimation using Bradford reagent (Sigma). Before loading, the protein was mixed with gel loading buffer (50 mm Tris-Cl, pH 6.8, 10 mm DTT, 20 mm β-mercaptoethanol, 2% SDS, 0.1% bromphenol blue, 10% glycerol) and boiled for 10 min. Samples were resolved on 12% SDS-PAGE and proteins were transferred to methanol-activated polyvinylidene fluoride (PVDF) membrane (Millipore) using anode buffer (25 mm Tris-Cl, pH 10.4, 10% methanol) and the TransBlot Semi-Dry Transfer system (Bio-Rad). Membranes were blocked with 5% nonfat skim milk powder in 1× PBS overnight and probed with specific antibodies. Primary antibody dilution was made in 1× PBS with 0.2% Tween (PBST) and incubated with the membrane for 3 h at room temperature (RT) on a rocker. Primary antibody binding was detected by appropriate secondary antibodies conjugated to HRP (GE Healthcare, catalog number NXA931). Dilution of secondary antibody was made in PBST. After every incubation, the membrane was washed with PBST at least 6–7 times with 5-min intervals. The immunoblots were developed using the ECL PlusTM (Amersham Biosciences) and also the Super Signal West Pico chemiluminescent substrate (Thermo Scientific). The antibodies used were anti-PfP2 monoclonal antibody E2G12 (1:100) (24) and anti-β-actin antibody (1:1000) (Sigma, catalog number A1978).
Mass Spectrometry of Recombinant PfP2 Protein Using MALDI-TOF
Purified recombinant PfP2 protein was subjected to mass spectrometry to determine the molecular weight. For determination of mass, MALDI was used from Bruker Corporation (model number 201344). The protein was dissolved in Tris-Cl, pH 7.4, with 50 mm NaCl and reconstituted using 50% acetonitile and 0.1% TFA (500 μl of 100% acetonitrile + 499 μl of distilled water + 1 μl of TFA). The reconstituted protein was loaded on a MALDI plate using saturated solution of α-cyanohydroxyl cinnamic acid (Bruker 201344) (∼20 mg/ml) in 50% acetonitrile and 0.1% TFA. The proteins were ionized and the molecular weight was determined. In the y axis the intensity of the protein in atomic units (AU) was plotted and in the x axis the m/z was plotted.
Gel Filtration of Recombinant and Parasite Protein
For gel filtration, the AKTA device from GE Healthcare was used, which was coupled to a UV spectrometer. The gel filtration profiles of all recombinant rPfP2, mutants, PfP2CΔ20, and PfP2CΔ40 proteins were determined with a Superdex-75 column, using 1–2 mg of protein. For parasite crude protein fractionation, 3–4 mg of parasite protein extract was injected in to a Superdex-200 column. For each set of runs about 90 fractions were collected. The parameters of a gel filtration device were: pressure, 0.12 MPa; column bed volume, 120 ml; flow rate, 1 ml/min; fraction collection rate, 1 ml/min; empty loop volume, 40 ml. For runs under the reduced conditions, 10 mm DTT was added. Fractions were concentrated to 20% volume and 10-μl concentrated samples from each fraction were resolved on 12% SDS-PAGE followed by immunoblot using the anti-PfP2 monoclonal antibody E2G12 (24).
Interaction of P2 with Gel Filtration Beads
Gel beads from different column materials (Superdex, Sephadex G50, and Bio-Gel P60) were equilibrated in PBS and used as a 50% bead slurry for further binding experiments. 100 μg of P2 (1 μg/μl) in PBS was incubated with 50 μl of a 50% bead slurry of Superdex, Sephadex, and Bio-Gel beads at room temperature with constant mixing for 1 h. Additionally, some samples were incubated in PBS containing 500 mm NaCl. The beads were subsequently pelleted down by centrifugation at 1000 × g for 5 min and the supernatant was collected. The beads were then washed for the indicated number of times by resuspension in 20 bed volumes of PBS followed by centrifugation. The beads and post-bead supernatant were boiled with gel loading dye for 10 min before being resolved on a 12% SDS-PAGE followed by staining with Coomassie Brilliant Blue R-250.
Concanavalin A was used as a positive control while incubating P2 with Superdex 75 gel filtration beads. 100 μg of concanavalin A was incubated with gel filtration beads. Post-bead treatment and after 15 washes, beads and the post-bead supernatant were resolved in 12% SDS-PAGE followed by Coomassie staining.
Circular Dichroism (CD)
Far-UV circular dichorism (CD) spectra of the protein at 25 °C were recorded on a JASCO-J810 spectropolarimeter (Jasco, Hachioji, Japan) using 0.1-cm cell. Spectra for P2 and point mutants as well as deletion constructs were recorded. The protein concentration was 30 μm. The CD machine was pre-calibrated with iolar nitrogen for 1 h before starting. The samples at appropriate conditions were equilibrated at least 10–12 h before CD measurements. Each spectrum was an average of three scans (slit width of 2 nm). The data were plotted as millidegree versus the UV wavelength (from 190 to 250 nm) in the two-dimensional graph and compared with standards to estimate the extent of α helicity, β-sheet, and random coil. Near-UV CD spectra were recorded using a 1-cm cell, with protein concentrations at 1.5–2 mg/ml. Each spectrum was an average of 10 scans (slit width of 2 nm) and the data were plotted as the CD in millidegrees versus the UV wavelength (from 250 to 320 nm).
Fluorescence Labeling of Recombinant PfP2 and PfP1-GST Proteins with Tetramethylrhodamine Isothiocyanate(TRITC)
About 2 mg of recombinant PfP2 protein was buffer exchanged with 1 ml of 0.1 m sodium bicarbonate buffer, pH 8.5. While labeling the protein, the molar ratio of the protein versus the dye was 1:10 (10-fold more dye was used). 5 mg of tetramethylrhodamine (Invitrogen) was dissolved in 0.5 ml of dimethyl sulfoxide. The labeling of protein was done according to the manufacturer's protocol (Invitrogen). The final concentration of protein was 0.5 mg/ml. While stirring the protein solution slowly, 50–100 μl of dye solution was added. Protein and dye solution were incubated at room temperature for 1 h. 1.5 m hydroxylamine, pH 8.5, was added into the protein dye solution to terminate the reaction. A 3-kDa membrane filtration cut off device was used to remove the unbound dye and purify the labeled protein. Recombinant GST-PfP1 protein was also conjugated with tetramethylrhodamine using the same protocol.
Treatment of Tetramethylrhodamine-labeled rPfP2 Protein with Parasite Lysate and FPLC Fractions
Synchronized P. falciparum cultures were harvested at different time points post-merozoite invasion (PMI), and parasite lysates were prepared. Parasite lysate was centrifuged at 12,000 × g for 1 h at 4 °C, and the supernatant was collected. Supernatant protein was quantified using Bradford or BCA assays. About 5 μg each of parasite crude extract from different time points (PMI) was incubated with 1 ng of tetramethylrhodamine-labeled rPfP2 at 37 °C for 3 h in a water bath with intermittent mixing of the solution. Postincubation, the mixture was boiled with SDS-loading buffer containing β-mercaptoethanol and resolved on a 12% SDS-PAGE. The gel was scanned using Typhoon TRIO (GE Healthcare) to check the presence of SDS-resistant fluorescent protein oligomer bands. The PMT gain of the scanner was set at 250 volts. The λex of tetramethylrhodamine was 532 nm and λem was 580 nm. For the FPLC fractions, 25 μl of the pooled fractions were used for incubation of 1 ng of tetramethylrhodamine-labeled rPfP2 or GST-PfP1 protein at 37 °C for 3 h, and monitored for the SDS-resistant fluorescent protein bands on SDS-PAGE as described above.
RESULTS
SDS-resistant Oligomerization of rPfP2 and Its Mutants
The Plasmodium ribosomal protein P2 displays a novel nonribosomal role, as well as exhibits oligomerization at the trophozoite stages in the erythrocytic development cycle (24). The recombinant PfP2 (rPfP2) protein also exhibits oligomerization (24, 27). To evaluate the propensity of SDS-resistant and -sensitive oligomer formation, the recombinant protein rPfP2 and certain recombinant PfP2 mutant and deletion proteins (supplemental Fig. S1) were run on SDS-PAGE and through gel-filtration columns (Figs. 1 and 2).
FIGURE 1.
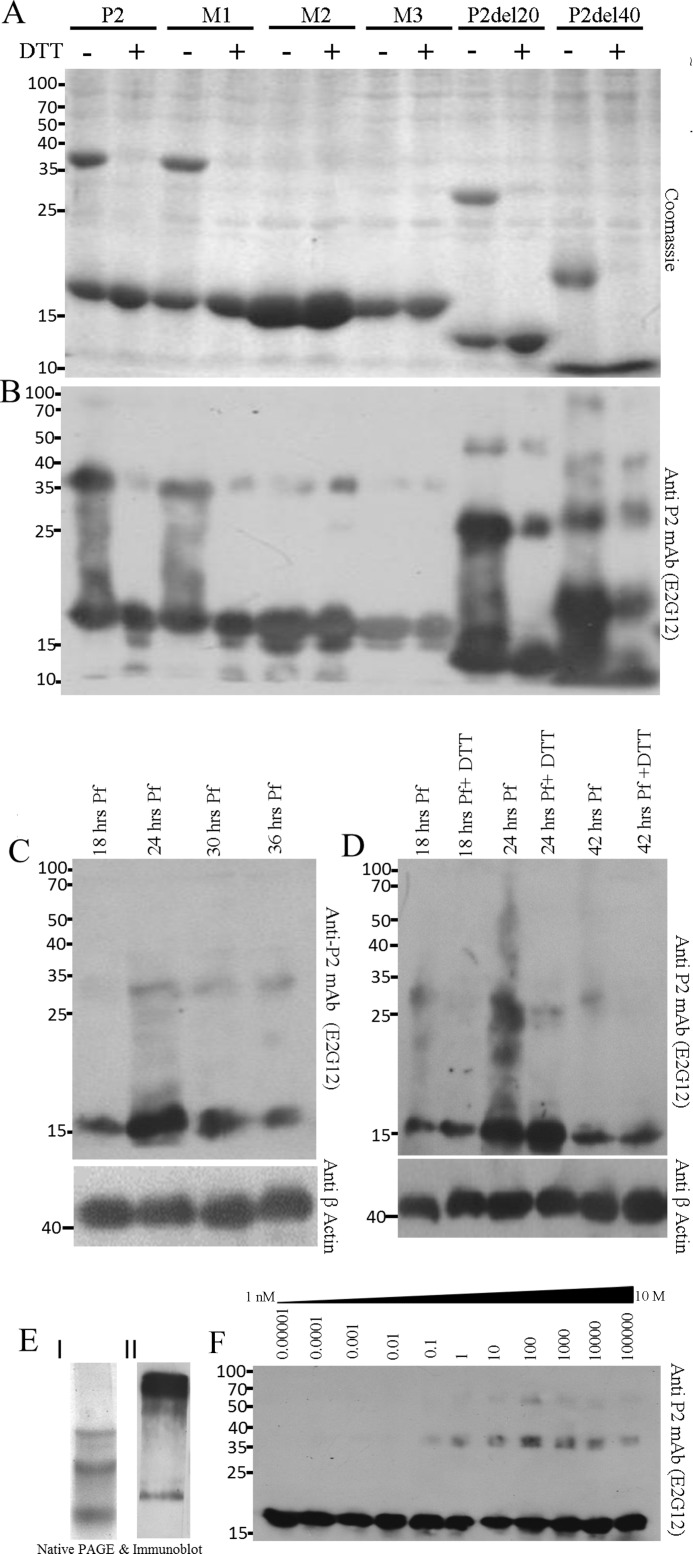
P2 oligomerization of rPfP2 as well as in the Plasmodium parasite. A, Coomassie-stained gel of 2 μg of protein each of recombinant purified PfP2, PfP2 mutants M1 (C12A), M2 (C53Y), M3 (C12A,C53Y), PfP2 deletions P2del20, and P2del40 (supplemental Fig. S1) proteins were resolved on 12% SDS-PAGE in the absence and presence of reducing agent (10 mm DTT). B, the same gel in A was immunoblotted and probed with anti-PfP2 specific monoclonal antibody (E2G12) (24). C, about 4 μg each of parasite protein extract obtained from synchronized P. falciparum cultures harvested at 18, 24, 30, and 36 h post-merozoite invasion, were separated on 12% SDS-PAGE in the presence of reducing agent (10 mm DTT), and the immunoblot was probed using anti-PfP2 monoclonal antibody E2G12 (24). D, 4 μg of crude P. falciparum parasite protein harvested at 18, 24, and 42 h post-merozoite invasion were separated on 12% SDS-PAGE without and with reducing agent, 10 mm DTT, and the immunoblot was probed using anti-PfP2 monoclonal antibody E2G12 (24). β-Actin was used as a loading control. E, I, Coomassie stain of rPfP2 and II, immunoblot of 10 μg of asynchronous P. falciparum parasite crude protein extract, run on 10% native PAGE, and probed using anti-PfP2 monoclonal antibody (E2G12) (24). F, to estimate the concentration-dependent oligomerization, various concentrations of rPfP2 solution in PBS, pH 7.4, ranging from 0.01 ng/ml to 100 mg/ml were incubated at 4 °C for 48 h, and 1 ng of rPfP2 from each solution, diluted in SDS-PAGE loading buffer, was resolved on 12% SDS-PAGE with reducing agent (10 mm DTT) followed by immunoblotting and probing with E2G12 (24).
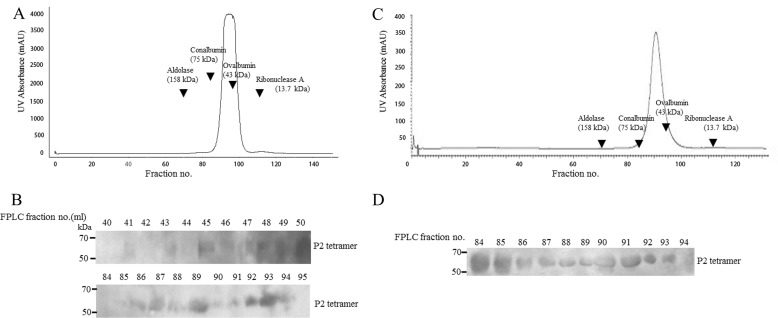
FIGURE 2.
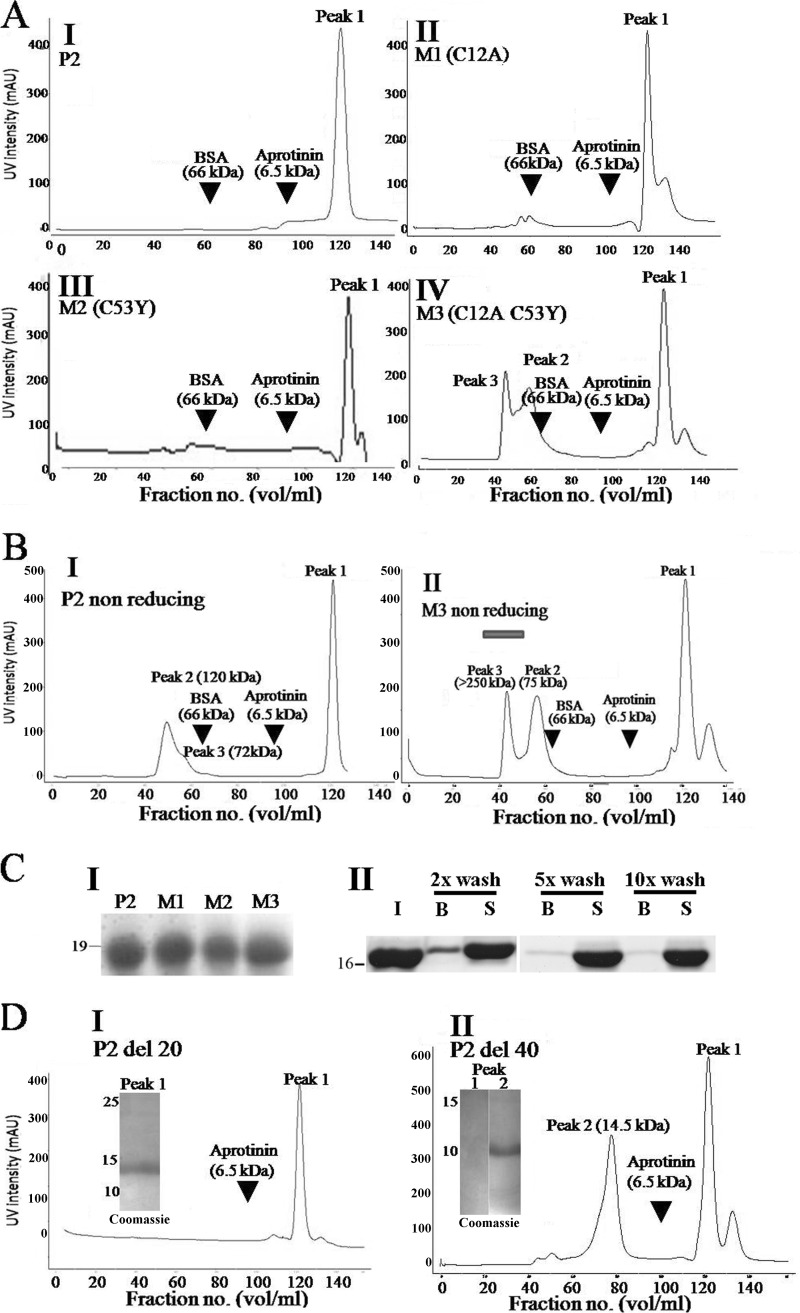
Gel filtration profile of rPfP2, rPfP2 mutants, and deletion proteins. A, FPLC profile of 1 mg each of purified recombinant proteins (P2, M1, M2, and M3). Arrowheads show the elution positions of marker proteins, BSA (66 kDa) and aprotinin (6.5 kDa). The y axis represents UV absorption of protein in arbitrary units (AU) and the x axis shows the fraction number. All gel filtration runs were performed under reducing conditions using buffer containing 10 mm DTT. B, I and II, gel filtration profiles for rPfP2 and M3 (C12A and C53Y) proteins under nonreducing conditions. About 1 mg each of rPfP2 and M3 proteins were injected in the Superdex-75 gel filtration column and fractions were eluted. Bar in B, II, indicates void volume (>300 kDa). C, I, Coomassie stain of 12% SDS-PAGE of peak 1 eluted from A I-IV FPLC runs, showing intact PfP2, M1, M2, and M3 proteins. II, rPfP2 was incubated with agarose-dextran cross-linked Superdex-75 beads and subjected to different wash stringencies (×2, 5, and 10 washes with PBS) to detect for weak binding interactions with the chromatography column material. I, input; B, beads; S, post-bead supernatant. D, I and II, gel filtration profiles of 1 mg each of recombinant P2del20 and P2del40 proteins, respectively, run under reducing conditions with 10 mm DTT. Insets show Coomassie stained SDS-PAGE profiles of peaks 1 and 2. Each run was repeated at least three times, and a representative plot is shown.
Coomassie-stained SDS-PAGE of the rPfP2 protein showed the monomer along with large amounts of SDS-resistant dimers (Fig. 1A, first lane), which were verified as PfP2 dimers (Fig. 1B, first lane) using PfP2-specific mAb E2G12 (24). To check the role of disulfide bonding in such dimer formation, rPfP2 was run in the presence of the reducing agent DTT. The dimers were found to be abolished in the presence of reducing agent (Fig. 1, A and B, second lane). In P. falciparum, two cysteine residues are found in the P2 protein at the 12th and 53rd positions for the 3D7 strain (supplemental Figs. S2–S4). These were confirmed by sequencing the rPfP2 clone, prepared from genomic DNA of the 3D7 strain. The sequence was identical in six different Indian isolates, as also in different laboratory strains of P. falciparum.4 Interestingly, although the 12th cysteine is conserved in all Plasmodium species, the 53rd cysteine is not present in the rodent species of Plasmodium, namely Plasmodium berghei, Plasmodium yoelii, and Plasmodium chabaudi (supplemental Fig. S4). Conversely, P. falciparum does not possess a tyrosine residue conserved at the 53rd position in the rodent and simian parasites. However, the tyrosine residue is also absent in the malaria parasite species of aves, Plasmodium gallinaceum, and of the chimpanzee, Plasmodium reichenowi (supplemental Fig. S4).
To test the contributions of the two cysteines of P. falciparum P2 protein toward dimerization, several different recombinant clones, M1 (C12A), M2 (C53Y), and M3 (C12A,C53Y) were generated (supplemental Fig. S1). The C53Y mutation for the M2 protein was guided by the observation that a tyrosine residue was found in certain species of Plasmodium P2 proteins at the 53rd position (supplemental Figs. S3 and S4). The recombinant P2 protein from the M1 clone showed that replacement of the Cys-12 residue had no significant effect on the DTT-sensitive rPfP2 dimer amounts, and behaved in a fashion similar to rPfP2 protein (Fig. 1, A and B). M2 protein, with a C53Y mutation, showed a large decline in dimerization. M3 protein, with the double mutations, behaved similar to the M2 mutant (Fig. 1, A and B). To rule out an influence of the tyrosine residue at the 53rd position, constructs M4 (C53A) and M5 (C12A,C53A) were also generated, shown schematically in supplemental Fig. S1. M4 behaved the same way as M2, and exhibited no detectable dimer on Coomassie-stained SDS-PAGE (supplemental Fig. S5IA). Mutant M5 did not show adequate protein expression (supplemental Fig. S5B), and therefore it was not possible to assess how the double mutant (C12A,C53A) of PfP2 would behave. However, from the results using M4, it could be deduced that the Cys-53 residue played a dominant role in the formation of SDS-resistant, DTT-sensitive dimers. However, the continued presence of small amounts of SDS- and DTT-resistant oligomers in rPfP2 indicated that although rPfP2 oligomerization was influenced by disulfide bonding, other interactions were also operative.
The deduced molecular mass of the parasite PfP2 protein is 12 kDa. The recombinant rPfP2 protein contains an additional 30 amino acids (from the vector pProExHTa) and the monomer and dimers correctly showed the expected mass of 15.884 and 31.336 kDa on MALDI mass spectrometry (supplemental Fig. S6). However, on SDS-PAGE, the monomeric and dimeric rPfP2, as also the PfP2 protein from parasite extracts, migrated at higher molecular masses of 18 and 36 kDa and 16 and 32 kDa, respectively (Fig. 1, A–D). Because the C-terminal region is negatively charged, it is possible that this repels the negative charge of SDS and hinders SDS binding. When SDS binding is less, mobility would be less, giving an anomalously high molecular weight. In silico prediction (DisEMBL 1.5) shows that amino acids from 72 to 112 of PfP2 protein are disordered in nature, and therefore this abnormality could also be due to this disordered acidic C-terminal region not conforming to globular structure, resulting in abnormal migration on SDS-PAGE. To test the contribution of the acidic C-terminal domain on oligomerization and mobility on SDS-PAGE, two deletion constructs, rPfP2CΔ20 and rPfP2CΔ40 proteins, with C-terminal 20 and 40 aa deletions respectively, were expressed, and run on SDS-PAGE (Fig. 1, A and B). Although rPfP2CΔ20 (deduced molecular mass 12 kDa) was still migrating abnormally, rPfP2CΔ40 (deduced molecular mass 10.6 kDa) behaved normally and migrated at the expected size of 11 kDa on SDS-PAGE (Fig. 1, A and B). Once again, the dimers of these deletion proteins were largely DTT-sensitive. However, significant amounts of SDS- and DTT- resistant oligomerization continued to occur in the rPfP2CΔ20 and rPfP2CΔ40 proteins (Fig. 1B). Indeed, the extent of oligomerization seemed to be slightly enhanced in the rPfP2CΔ40 protein (Fig. 1B). Thus the acidic C-terminal region hindered normal migration of the PfP2 protein on SDS-PAGE and the DTT-sensitive dimerization was crucially dependent on the Cys-53 residue. As expected, DTT-resistant oligomerization was independent of both cysteine residues.
The concentration of P2 protein in Plasmodium cells is around 20–50 nm, as determined through ELISA using rPfP2 as a standard. In the test tube, rPfP2 does not form SDS-resistant oligomers at that concentration, but were detected in parasite extracts even at these low concentrations (Fig. 1, C and D). An analysis of different developmental stages of the P. falciparum extract showed that SDS-resistant, DTT-sensitive oligomers (mainly dimers) were detected throughout the erythrocytic stages, but at certain specific stages, 24 to 36 h postmerozoite invasion (PMI), SDS- and DTT-resistant oligomers were produced (Fig. 1, C and D). In summary, PfP2 protein readily formed dimers that are SDS-resistant but largely DTT-sensitive. However, a fraction of the SDS-resistant oligomers are DTT-resistant, and these are developmentally regulated.
Multiple bands were seen in the native PAGE for both the recombinant and parasite PfP2 proteins (Fig. 1E), as seen through Coomassie staining and upon probing with anti-PfP2 mAb E2G12 (24). We were unable to obtain an NMR solution structure of rPfP2 because this protein formed aggregates at the concentration required for a solution NMR structure (>1 mm) (27). The formation of DTT- and SDS-resistant rPfP2 dimers and higher oligomers was concentration-dependent and in vitro the oligomers were observed at >0.1 μg/μl (about 10 μm) concentration (Fig. 1F).
Thus, both rPfP2 in vitro and PfP2 in vivo exhibited SDS-resistant oligomers. Although a large part of the DTT-sensitive forms were dimers, it was the DTT-resistant oligomers that exhibited developmentally regulated formation in the parasite.
Gel Filtration Profile of rPfP2, M1, M2, M3, rPfP2CΔ20, and rPfP2CΔ40 Proteins
All of the above data pertained to SDS-resistant oligomerization properties of PfP2 protein. To assess the nature of oligomeric protein complexes present in rPfP2 solution, 1 mg of purified rPfP2 and mutant proteins were run on a 120-ml Superdex-75 gel filtration column under reducing and nonreducing conditions (Fig. 2, A and B). Several molecular markers were run to calibrate the column. In the presence of DTT, each of the wild type rPfP2, M1, and M2 proteins eluted as a single peak at a 120-ml fraction volume, projecting at a globular size far smaller than aprotinin (6.5 kDa), which eluted at a 90-ml fraction volume (Fig. 2A). However, the M3 protein behaved differently and some amount of recombinant protein was eluted at the 60-ml fraction volume (75 kDa), whereas a larger proportion eluted at the same 120-ml fraction volume (Fig. 2A). In the absence of DTT, rPfP2 showed a peak that mapped to 120 kDa, and a minor peak at 75 kDa, whereas the M3 protein showed predominantly 75-kDa and large aggregates that eluted in the void volume (> 250 kDa) (Fig. 2B, I and II). Thus both the rPfP2 protein and the double mutant aggregated to various molecular sizes under nonreducing conditions. Peak 1 continued to occur in both rPfP2 and in the double cysteine mutant M3 under nonreducing conditions as well (Fig. 2B).
The gel filtration column separates proteins by their hydrodynamic volumes and for sets of proteins with similar conformation; the hydrodynamic volume is proportional to the molecular mass. The columns were calibrated with mainly globular proteins. If the rPfP2 is not a globular protein, its chromatographic behavior will change. Moreover, an interaction of rPfP2 with the Superdex column material may also retard movement of the protein through the column. The proteins may be degraded so that they migrate abnormally. Coomassie staining showed the presence of intact 18-kDa rPfP2/mutant proteins in the peak fractions, with no evidence of degradation (Fig. 2C, I). The other fractions were also run on SDS-PAGE and only the peak fractions at 120 ml showed the presence of recombinant PfP2 proteins. To test the possibility of an interaction of rPfP2 with the gel filtration column material, rPfP2 was incubated with Superdex-75 beads, and the beads were assessed for bound rPfP2 protein (Fig. 2C, II). It was observed that the rPfP2 protein does bind weakly to Superdex-75 gel filtration materials. The binding is weak and with multiple washes, the bound protein amount is reduced (Fig. 2C, II). Similar behavior was seen with Sephadex G-50 and Bio-Gel beads (supplemental Fig. S7, I). The presence of NaCl in the binding buffers did not make much difference to this weak binding (supplemental Fig. S7, II). With extensive washings the rPfP2 protein showed no signature of binding, although the sugar-binding protein concanavalin-A did remain bound to the beads as expected (supplemental Fig. S7, III). Therefore abnormal migration of the rPfP2 and mutants could be partly due to weak nonspecific interactions with the column matrix.
To check the effect of the disordered acidic C-terminal region on abnormal migration of rPfP2 on gel filtration, the two deletion constructs, rPfP2CΔ20 and rPfP2CΔ40 proteins, were run on FPLC (Fig. 2D). It was observed that the deletions did influence the abnormal migration of rPfP2, and the major amount of protein was eluted at the expected monomeric size of 11.0 kDa for the rPfP2CΔ40 protein (Fig. 2D). A peak was still observed at the 120-ml fraction volume, but no intact rPfP2CΔ40 protein was observed on SDS-PAGE (Fig. 2D), indicating a possible lack of stability in the rPfP2CΔ40 protein and collection of very small degradation products. Thus, the anomalous gel filtration behavior appears to be mainly due to shape aberrations and nonspecific interactions caused by the highly negatively charged C-terminal region. Eliminating this region resulted in an expected elution profile for a globular protein.
Circular Dichroism (CD) Measurements of rPfP2 and the Various Mutant/Deletion Proteins
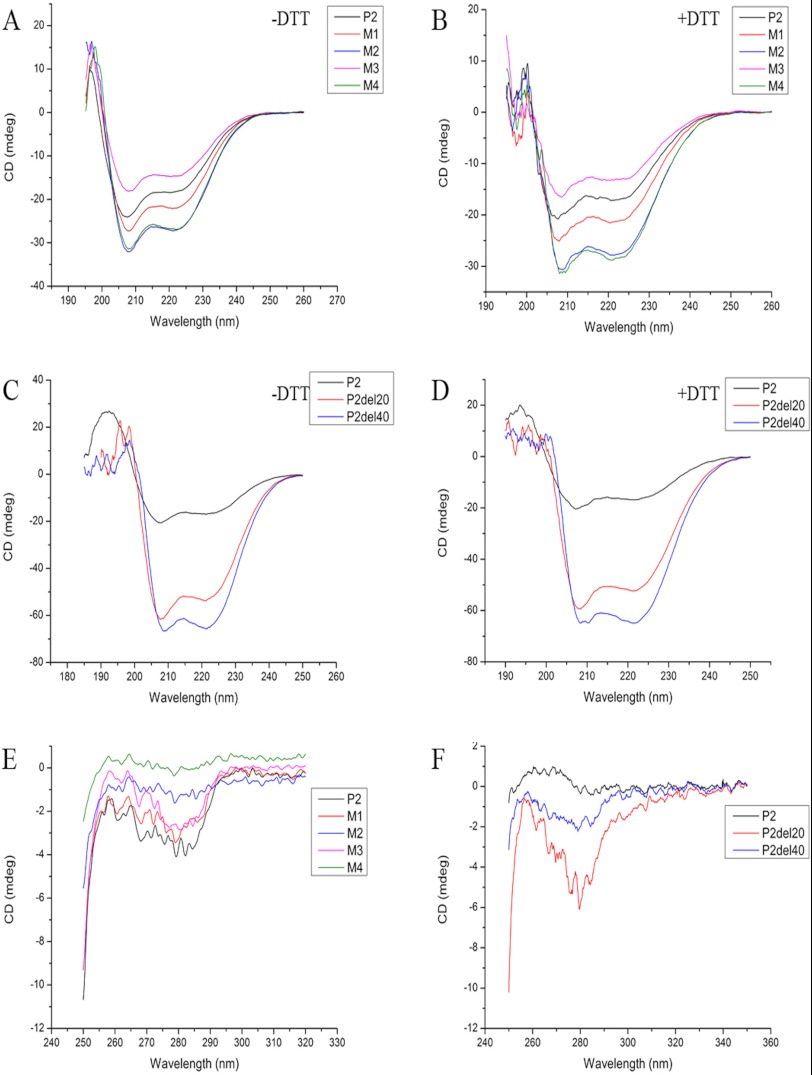
We have reported CD data of rPfP2 protein and rPfP2CΔ40 earlier (27). A CD study of the rPfP2 protein at the far-UV region shows features of largely α-helical structures, both in the presence and absence of reducing agent (Fig. 3, A and B). Comparison of the CD spectra of the P2 proteins under reducing and nonreducing conditions, as well as among different mutants, did not show any significant differences. As shown earlier (27), the CD spectra predicted about 30% α helicity. CD spectra of both deletion constructs, rPfP2CΔ20 and rPfP2CΔ40, also showed the canonical depths at 220 and 208 nm (Fig. 3, C and D), indicating the α-helical nature of both deletion proteins. The extent of the random coil was reduced considerably in the deletion proteins, which indicated that the carboxyl-terminal amino acids did contribute to the disordered random coil structures in the PfP2 protein. In both rPfP2CΔ20 and rPfP2CΔ40, the negative millidegree increased, which indicated that with deletion of the acidic C terminus, the α helicity of the proteins increased.
FIGURE 3.
P2 is largely an α-helical protein and helicity does not change upon cysteine replacements. Far-UV circular dichroism (CD) spectra of the protein samples were recorded under native condition as also in the presence of reducing agent DTT (5 mm) on a JASCO-J810 spectropolarimeter (Jasco, Hachioji, Japan) at 30 °C, using a 0.1-cm cell and a slit width of 2 nm. 30 μm of each recombinant protein in PBS, pH 7.4, were subjected for secondary CD. Each spectrum is the average of 3 wavelength scans. Near-UV CD spectra for all constructs were acquired under nonreducing conditions. Each spectrum is the average of 10 wavelength scans acquired using a 1-cm cell and a slit width of 2 nm. Samples were equilibrated for at least 10–12 h before CD measurements. A and B, overlay of far-UV CD spectra of P2 and point mutants under nonreducing (A) and reducing (B) conditions. C and D, overlay of far-UV CD spectra of P2 and its deletion constructs under nonreducing (C) and reducing (D) conditions. E and F, overlay of near-UV CD spectra of P2 and its point mutants (E) as well as deletion constructs (F).
To assess tertiary features of the P2 protein, near-UV CD data were recorded (Fig. 3, E and F). The data indicates a lack of significant tertiary structure in each of the rPfP2 proteins and mutants. However, as expected, the lack of disordered regions among the deletion proteins, rPfP2CΔ20 and rPfP2CΔ40, gave rise to some tertiary structure. It was interesting to note that deletion of the penultimate 20 aa (71 to 91 residues), containing a large number of alanine residues, reduced the extent of tertiary structure, as seen with CD data of rPfP2CΔ40 protein. Through NMR analysis, we have demonstrated earlier that both rPfP2 and rPfP2CΔ40 proteins form large aggregates (27). Despite such inter-molecular associations in solution, the near-UV CD data showed the absence of significant tertiary structural features in both rPfP2 and rPfP2CΔ40.
Gel Filtration Profile of P. falciparum Parasite Protein
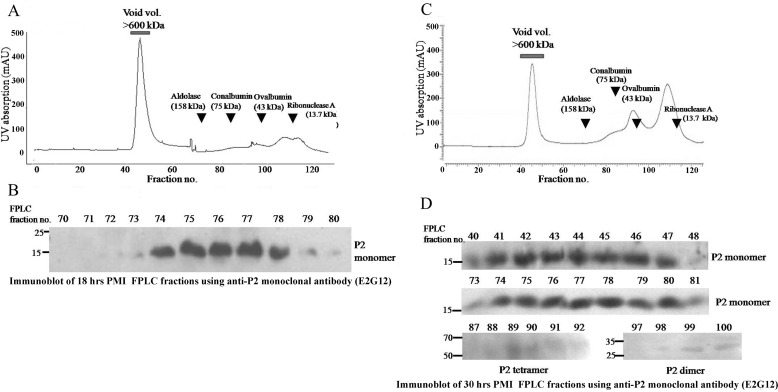
To observe the state of the PfP2 complexes in the parasite, total parasite lysates were run on FPLC and the fractions were tested for the presence of various oligomers of PfP2. Because we were aware that critical changes in SDS- and DTT-resistant oligomerization occur around 24 h PMI, elution profiles of parasite lysates at 18 and 30 h PMI were assessed on a Superdex 200 column in the presence of DTT under reduced conditions (Fig. 4, A and C). Fractions of 1 ml were collected, and a standard curve was generated using several molecular weight standards. The presence of PfP2 protein was determined by running every fraction on SDS-PAGE under reducing conditions and probing with the PfP2-specific monoclonal antibody E2G12 (24). No PfP2 molecules were detected at fractions corresponding to 16- and 32-kDa sizes, indicating that there was not a significant presence of free PfP2 monomers or dimers at either of these stages. A PfP2 protein peak was noted around fractions 76/77 (about 90–100 kDa) in both stages (Fig. 4). The ribosomal pentameric P0–2P1–2P2 complex calculates to 90 kDa theoretically, with molecular masses of P0 as 38 kDa, P1 as 13 kDa, and P2 as 12 kDa. However, all three P. falciparum proteins migrate at higher molecular masses in SDS-PAGE (24) and exhibit sizes of PfP0, 35–37 kDa; PfP1, 17–18 kDa; and PfP2, 16 kDa, respectively. This would estimate a size of about 90–105 kDa for the pentameric complex and the peak at fractions 76/77 would be consistent with this size. No other molecular species of PfP2 was detected at the 18-h PMI. The PfP2 present in this complex was resolved entirely as a monomeric 16-kDa PfP2 band on SDS-PAGE, showing the absence of detergent-resistant oligomers at 18 h PMI.
FIGURE 4.
Gel filtration profiles of PfP2 from total parasite protein extracts from a synchronized P. falciparum culture at 18 and 30 h postmerozoite invasion. A and C, gel filtration profiles of about 3–4 mg of total parasite protein extracts in PBS from 18- and 30-h PMI parasite cultures, respectively, was resolved on a Superdex-200 gel filtration column and fractionated. Arrows indicates the elution positions of marker proteins. B and D, fractions eluted from A and C respectively, were run on 12% SDS-PAGE under reducing conditions and immunoblotted using anti-PfP2 monoclonal antibody (E2G12) (24). All fractions were tested, and only those fractions that showed reactivity with anti-PfP2 antibody (E2G12) are shown.
In contrast, a dominant presence of additional large aggregates (>600 kDa) were noted during the 30-h stage (Fig. 4, C and D). These large aggregates also resolved into PfP2 monomers and did not show any other SDS-resistant species. However, small amounts of PfP2 complexes, observed at fractions 97–102, and 84–85 and 88–91 were detected in the 30-h PMI sample (Fig. 4D) mapping to average molecular sizes of 29 and 60–80 kDa, respectively (as determined through markers on the gel-filtration column). The 29-kDa fractions would map to the dimeric PfP2 (32 kDa) protein, and the 60–80-kDa fractions would correspond to a tetrameric 64-kDa species on SDS-PAGE. Thus at 30 h PMI under reducing conditions, the parasite appeared to contain no native free monomer of PfP2, small quantities of dimers and tetramers of PfP2, which were SDS-resistant, and predominantly the pentameric P-protein complex and large aggregates, which were SDS-sensitive (Fig. 4D).
A mild saponin treatment of infected RBCs (iRBCs) typically yields a parasite pellet, iRBC cytosol, and iRBC ghost membranes. The 30-h iRBC cytosol and ghost membrane fractions were resolved on FPLC and the fractions were probed for PfP2 protein (Fig. 5, A–D). Neither of these extracts exhibited the prominent 76/77 peak of PfP2, which presumably represents the pentameric ribosomal P-complex. Nor did either of the extracts exhibit any monomeric P2 protein on SDS-PAGE. This is consistent with our earlier observations of a lack of monomeric PfP2 proteins in immunoblots of iRBC ghost and cytosolic preparations (24). The iRBC cytosol and the iRBC membrane fractions from the 30-h PMI parasite preparation showed the presence of only a 64-kDa PfP2 band on the immunoblot. In the iRBC cytosol, this SDS-resistant tetrameric species occurred in gel fractions corresponding to molecular complexes ranging from 50 to 80 kDa, as also in the large aggregate complexes with a size >600 kDa that eluted with the void volume (Fig. 5B). However, the iRBC ghost membrane fractions did not show any large complexes but showed the PfP2 species exclusively at ranges of 50 to 70 kDa (Fig. 5D). Mass spectrometry analysis of this band shows predominantly peptides of the P2 protein and no other peptide with a significant score (24). No PfP2 bands could be detected in any fractions when the 18-h iRBC cytosol and ghost membrane samples were run through gel-filtration columns (data not shown).
FIGURE 5.
Gel filtration profile of PfP2 from infected RBC cytosol (A and B) and RBC ghost (C and D) prepared from a synchronized P. falciparum culture at 30 h postmerozoite invasion. A and C, gel filtration profiles of 3–4 mg of protein from 30-h PMI P. falciparum-infected RBC cytosol and ghost, respectively, using Superdex-200 gel filtration column. Arrows indicate the elution positions of marker proteins. B and D, fractions eluted from A and C were run on 12% SDS-PAGE under reducing conditions and immunoblotted using anti-PfP2 monoclonal antibody (E2G12) (24). All fractions were tested, and only those fractions that showed reactivity with anti-PfP2 antibody (E2G12) are shown.
These results showed that large aggregate complexes containing PfP2 occur in the parasite as also in the red cell cytoplasm at 30 h PMI. Only the SDS-resistant tetramers move out through the red cell cytoplasm as large complexes. At the iRBC membrane, PfP2 is detected exclusively as 60–80-kDa size species, possibly representing the tetrameric PfP2. On SDS-PAGE both the RBC cytosol and membranes were resolved to a uniform 64-kDa SDS-resistant species. All of the above studies were carried out under reducing conditions and therefore represent DTT-resistant PfP2 species.
Effect of Parasite Extracts on Oligomerization of rPfP2
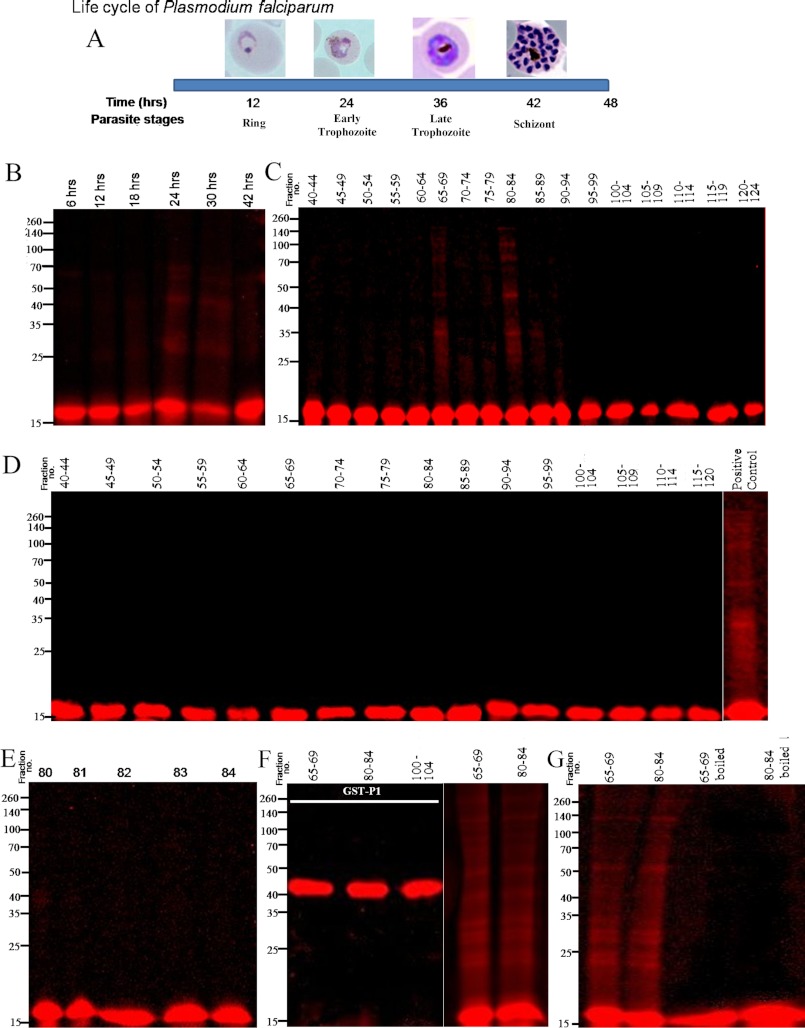
The erythrocytic stages of P. falciparum cycles is about 48 h, moving through ring, trophozoite, and schizont stages (Fig. 6A), within which SDS- and DTT-resistant oligomers of PfP2 were produced in the parasites beyond 24 h PMI (Figs. 1, 4, and 5). To test if certain protein components of the parasite were promoting such oligomerization, the rPfP2 protein was treated with total parasite lysates at different PMI stages and run on SDS-PAGE (Fig. 6B). The rPfP2 was labeled with tetramethylrhodamine, so that selectively the changes in rPfP2 protein could be tracked through the fluorescence. The reaction was carried out at 1 ng/μl concentration of labeled rPfP2, where no self-association of rPfP2 would be observed (Fig. 1F). It was observed that 24- and 30-h parasite lysates could indeed propel aggregation, whereas 6-, 12-, 18-, and 42-h extracts were not so effective in such oligomer formation (Fig. 6B). To dissect out the P. falciparum proteins responsible for such oligomerizations, pools of 5 FPLC fractions from 30- and 18-h PMI samples (Fig. 4) were used for the treatment of rhodamine-labeled rPfP2 and such incubated fractions were run on SDS-PAGE (Fig. 6, C and D). Although none of the 18-h samples exhibited significant oligomerization (Fig. 6D), certain pools of proteins from the 30-h PMI parasite sample caused distinct rPfP2 oligomerizations (Fig. 6C). The oligomerization was observed upon incubation with mainly fractions 65–69 (molecular size 230–183 kDa) and 80–84 (molecular size 85–70 kDa). However, when individual fractions from these pools were tested, no oligomerization was observed. Representative data of treatment with individual fraction numbers 80–84 are shown in Fig. 6E. The effect was specific to PfP2, because rhodamine-labeled PfP1-GST protein did not exhibit such oligomer formation (Fig. 6F). The pooled fractions 80–84 always showed oligomer formation and was used typically as a positive control (Fig. 6, D, F, and G). The properties of the effective pooled fractions were likely to be due to protein components, because heat treatment abolished such oligomerizing capability (Fig. 6G). Thus specific protein components of the 24–30-h PMI parasites appear to propel oligomerization of PfP2 as seen through our rhodamine-labeled rPfP2 assay.
FIGURE 6.
Probing in vitro oligomerization of tetramethylrhodamine-labeled rPfP2 protein using parasite extracts. A, schematic depiction of the time scale of P. falciparum parasite development in erythrocyte. In a 48-h life cycle, the parasite changes its morphology from ring, trophozoite to multinucleated schizont stages. All subsequent panels depict reactions where 1 ng of tetramethylrhodamine-conjugated rPfP2 protein was incubated in 40 μl of various parasite protein preparations at 37 °C for 3 h. The mixture was treated with SDS loading dye and resolved on a 12% SDS-PAGE under reducing conditions. B, labeled rPfP2 was incubated with 4 μg of parasite protein extracts from each of 6-, 12-, 18-, 24-, 30-, and 42-h synchronized P. falciparum cultures. C, labeled rPfP2 was incubated with 25 μl of pooled gel filtration fractions obtained from 30-h parasite cultures Fig. 4C. D, labeled rPfP2 was incubated with 25 μl of pooled gel filtration fractions obtained from 18-h parasite cultures (Fig. 4A). E, labeled rPfP2 was incubated with 5 μl of individual gel filtration fractions (number 80–84) from 30-h parasite culture Fig. 4C. F, rhodamine-labeled recombinant GST-PfP1 (24) protein was incubated with 25 μl of pooled gel filtration fractions (65–69 and 80–84) obtained from 30-h PMI parasite cultures Fig. 4C. Labeled rPfP2 was used a positive control. G, about 25 μl of the two pooled gel filtration fractions (65–69 and 80–84) were boiled for 10 min. The supernatant was incubated with rhodamine-labeled rPfP2. Rhodamine-labeled rPfP2 incubated with the respective fractions without boiling was used as a control.
DISCUSSION
In this paper we have analyzed the biophysical properties of the P. falciparum acidic ribosomal protein PfP2. The secondary structure of rPfP2 is largely α-helical. The extensive oligomerization states of the recombinant and parasite PfP2 proteins have been detailed. The PfP2 protein shows SDS-resistant oligomers that are sensitive to reducing conditions. The 53rd cysteine residue, but not the 12th cysteine residue, appears to be mainly responsible for such DTT-sensitive, SDS-resistant dimerization. The acidic disordered C-terminal 40 aa residues contributed toward abnormal migration of the recombinant protein in gel filtration columns. The PfP2 protein in the parasite lysate was largely associated with the pentameric ribosomal P-protein complex, at more or less the expected size and did not exhibit the retarded behavior shown by the recombinant protein on gel filtration column. A lack of free C-terminal acidic ends of the PfP2 protein within the parasite extract would explain this difference in migration of the rPfP2 and the parasite PfP2 protein on gel filtration chromatography. Apart from the ribosomal P-protein complex, larger complexes containing the PfP2 protein were observed in 30-h PMI samples, and these complexes were resolved to DTT- and SDS-resistant tetrameric structures in SDS-PAGE. Rhodamine-labeled rPfP2 protein was used for monitoring oligomer formation on SDS-PAGE. Using this simple visual technique, it was demonstrated that multiple parasite protein components are apparently involved in the stage-specific formation of PfP2 oligomers.
Progressively it is becoming clear that several proteins function as complexes in the cell. Among these, the proportion that form self-complexes (or with very similar peptide chains) are considerable (29). An estimate of such homo-oligomeric proteins have indicated that there are a significantly larger number of self-interacting proteins than would be expected randomly (30, 31). Apart from general folding and stability issues, such a form of generating new protein surfaces using a single peptide chain would provide the cell with new functions and regulation without additional burden on the gene pool. Facilitating such oligomerization with development-specific protein(s) could regulate new activities, trafficking, and pathways.
Disulfide Bond and PfP2 Protein Oligomerization
The role of disulfide bonding in protein oligomerization is well established. The importance of cysteine residues in such a process, with consequences to protein functions, has been demonstrated in numerous experiments using chemical modification techniques and cysteine scanning mutagenesis (32, 33). Several malaria vaccine candidates' surface antigens, such as MSP1, AMA1, and EBA175, possess cysteine-rich motifs, which are engaged in disulfide bonding. Critical conformation domains belonging to these proteins are sensitive to reducing conditions (34–36). It is curious that the P2 protein of most organisms, other than a few protozoans, do not possess any cysteine residues (supplemental Fig. S3). The Apicomplexan species closest to Plasmodium, Toxoplasma and Babesia, possess no cysteine residues (supplemental Fig. S3). Thus it would be expected that disulfide bonding or reducing conditions would play no roles in the structural versatility of these P2 proteins. Several of the protozoan P2 proteins such as those from Entamoeba histolytica, Theileria annulata, and Leishmania mexicana contain a single cysteine residue within 12 to 28 aa positions (supplemental Fig. S3). Rodent malarial parasites P. yoelii, P. berghei, and P. chabaudi also possess only one cysteine residue at the 12th position (supplemental Fig. S4). This Cys-12 is conserved in all Plasmodium P2 proteins, but did not appear to play a role in the SDS-resistant DTT-sensitive oligomerization. The cysteine residue around the 53rd position is seen in all Plasmodium species, other than the rodent species (supplemental Fig. S4). It occurs at the 52nd position in the related simian/human malaria Plasmodium vivax and Plasmodium knowlesi, at the 54th position in avian P. gallinaceum and at the 53rd position in P. falciparum and P. reichenowi. The chimpanzee malaria parasite P. reichenowi is phylogenetically closest to the human parasite P. falciparum (37), and hence the similarity in the groups is understandable. Interestingly the apparently conserved tyrosine residue at the 53rd position in the rodent and simian parasites is not observed in the other species (supplemental Fig. S3 and S4).
From the results presented in this report it is apparent that the conserved cysteine at the 12th position does not seem to play a role in DTT-sensitive oligomerization in Plasmodium. On the other hand, the 53rd cysteine residue in P. falciparum is important for such DTT-sensitive oligomers (largely dimers). Neither of the functions of DTT-sensitive dimers nor DTT-resistant oligomers of PfP2 in the parasite is clear at present. The DTT-sensitive dimers exist in P. falciparum throughout all developmental stages, but it is the DTT-resistant oligomers that seem to occur in a developmentally dependent fashion. Not only are these oligomers generated at certain stages of the parasite (24–30 h PMI), some of these oligomers get exported through the red cell cytosol to the erythrocyte membrane (24). The erythrocyte membrane at 30 h PMI showed the presence of the 60–80-kDa species that eluted from the gel-filtration column, but these resolved to a single 65-kDa SDS- and DTT-resistant PfP2 on SDS-PAGE (Fig. 5). In contrast, the predominant species of the PfP2 protein in the parasite protein preparation consisted of large aggregates (>600 kDa) and the pentameric complex, which yielded monomeric PfP2 on SDS-PAGE (Fig. 4). Small quantities of SDS- and DTT-resistant dimer and tetramers were also detected in the parasite lysate. The infected RBC ghost membrane showed no large aggregates but only the 60–80-kDa species that was resolved exclusively to the SDS- and DTT-resistant tetrameric component and no monomer. However, it was interesting to note that the infected RBC cytosol contained several complexes, one set around >600 kDa, and another set ranging from 60 to 80 kDa. All of these complexes resolved to 65-kDa SDS- and DTT-resistant PfP2 on SDS-PAGE, and no monomer was detected (Fig. 5). As expected, the pentameric complex was limited to the parasite, whereas the species transported to the RBC cytosol and membrane belonged to different complexes. This is consistent with the ribosomal role of the pentameric complex. It is also apparent that translocation through the RBC cytosol was occurring through a range of molecular complexes, with mainly the smaller 60–80-kDa complex localized to the RBC membrane. As far as the SDS- and DTT-resistant components are concerned, within the parasite largely monomeric species existed (with small quantities of dimeric and tetrameric species), whereas the IE cytosol and ghost showed the exclusive presence of a SDS- and DTT-resistant 65-kDa tetrameric component.
We have earlier shown that the SDS- and DTT-resistant 65-kDa band consisted exclusively of the PfP2 peptides, analyzed through immunoprecipitation and mass spectrometry analysis (24). It has also been shown that PfP2 is exposed on the infected red cell surface and that blocking of PfP2 with specific monoclonal antibodies arrested the parasite at the onset of cell division (24). With the demonstration of various PfP2 oligomeric complexes in the IE cytosol (the present study), an analysis of the composition of such PfP2 complexes in the IE cytosol may help to elucidate the mechanism of translocation of the PfP2 protein to the red cell surface.
Regulation of SDS-resistant Oligomer Formation
Oligomerization and aggregation of proteins occur extensively in nature (29). The pathological state of various neurological disorders is associated with the accumulation of insoluble amyloid fibrils. In Alzheimer disease these amyloid fibrils are formed by multimerization of the 39–42-aa residue amyloid peptide (Aβ) (38, 39). Many types of oligomeric amyloid-β assemblies have been described (40). It has been reported that Aβ42 forms a noncovalent, SDS-resistant species with apparent molecular masses of dimer, trimer, and/or tetramer (41). A growing body of evidence indicates that prefibrillar oligomeric forms (Aβ) may represent the primary pathological species and not the mature amyloid fibrils that accumulate in plaque deposits; and the monomer to oligomer transition perhaps converts a benign protein to a neurotoxic one (41–44). Diverse experimental systems have been used to monitor amyloid-β self-association (45–48). It has been surmised that amyloid-β possesses an intrinsic potential to move through different assembly pathways, providing uncertainty about amyloid-β oligomeric states and their relevance to the final amyloid formation. Similarly Parkinson disease is associated with abnormal aggregation of the protein α-synuclein (49). In the presence of dopamine, recombinant human α-synuclein produces nonfibrillar, SDS-resistant oligomers, whereas β-sheet-rich fibril formation is inhibited. Pharmacologic elevation of the cytoplasmic dopamine level increased the formation of SDS-resistant oligomers in DA-producing neuronal cells (49). Thus, in this case, neurotransmitter signaling appears to regulate peptide oligomerization and secretion of α-synuclein oligomers.
Apart from disease-associated amyloids, there are several instances of functional amyloids that utilize the amyloid-fold to fulfill important physiological functions (50, 51). Yeast prions are possibly the first proven example of the heritable traits controlled by transmissible amyloids, and it is observed that fungal prions may be pathogenic, neutral, or beneficial (52, 53). Functional amyloids provide the cell with a stable protein structure that can be utilized in many different ways. Such functional amyloids are involved in the generation of important proteinaceous components in various organisms, such as the production of bacterial biofilms, insect chorion, and human Pmel17 amyloid templates in the formation of melanin (50, 54–57). Each of these display oligomeric intermediates in the amyloid-folding pathway, and because amyloid fibers are cytotoxic, it is postulated that highly controlled pathways are used to assemble such functional amyloids. The occurrence of higher oligomers at a particular erythrocytic stage in Plasmodium is also reflective of a very specific onset and reversal of P2 oligomerization by the parasite during its development.
The PfP2 protein has mainly α-helical structures, whereas the amyloid-folding proteins are characterized by the β-sheet structure of the peptides. However, this applies mainly to the peptide regions of amyloid proteins that are directly involved in the formation of amyloid axis. For β-sheet structures it has been postulated that hydrophobic and aromatic residues influence peptide self-assembly (58, 59). Apart from intrinsic properties, self-associative structures are influenced by modulators such as metal ions (60, 61) and drugs such as curcumin or chloroquine (47, 62). Interestingly, the disordered N-terminal octapeptide region of mammalian prion protein PrP, which is not essential for prion infectivity, plays a major role in the self-association of the prion protein (64, 65). Although the PfP2 protein has a strong disordered region at the C-terminal end, in the del40 recombinant PfP2, DTT- and SDS-resistant oligomerization is seen to persist, suggesting that the acidic C-terminal disordered region is not an absolute necessity for oligomerization. Recent examinations of the structural properties of amyloid fibrils formed spontaneously by recombinant mammalian PrP indicated a major refolding of the C-terminal domain from the α-helix to β-structure (66, 67). Such a possibility of a DTT- and SDS-resistant oligomeric structure arising from such a conversion of α-helix-rich PfP2 to a β-structure, therefore, may exist. Indeed, an NMR assessment of the PfP2 protein in the denatured state indicates a β-structure propensity on the basis of secondary chemical shift analysis (27). Thus, although the PfP2 protein primary sequence appears to be quite distinct from the amyloid proteins, the possibility remains that PfP2 may represent an amyloid species. Determination of the structure of the PfP2 oligomers will help to resolve such a possibility.
Distinct states of oligomerization of substrate proteins resulting in differential trafficking have also been documented. For instance, angiopoietin 2 modulates the endothelial receptor tyrosine kinase Tie2 and induces Tie2 translocation to the specific cell-matrix contact sites located at the distal end of focal adhesions. It is reported that the different oligomeric/multimeric forms of the angiopoietins cause induction of distinct patterns of Tie2 trafficking (68). Similarly phosphorylation and dimerization regulate nucleocytoplasmic shuttling of mammalian STE20-like kinase (69). Such post-translational modifications of proteins, resulting in multiple states of oligomerization that lead to differential functions/trafficking, have been extensively documented (29). The P2 protein has been shown to be phosphorylated in mammals and yeast at the serine residues at the carboxyl-terminal end (70). PfP2 does not possess these serine residues, but there is a conserved C-terminal serine residue at the 109th position (supplemental Fig. S4). However, the observation that deletion of the C-terminal 40 amino acids does not affect SDS- and DTT-resistant PfP2 oligomerization indicates that this serine may not be important for such an oligomerization. P. falciparum possesses other serine/threonine amino acids at positions 21/22 and 52, but these are not conserved in the rodent malarial parasite (supplemental Fig. S4). Because we see definite oligomerization in P. berghei and P. yoelii parasites, these amino acid residues are unlikely to be crucial. The two serine/threonine residues at the 47th and 58th positions, and a tyrosine at position 9 are conserved throughout Plasmodium, and these may be sites for possible phosphorylation. Our observations that rPfP2 oligomerization occurs without any added source of ATP (Fig. 6) indicates that such an in vitro process precludes active kinase reaction(s). It does not exclude possibilities such as nucleation of the complex with phosphorylated P2 protein present in those appropriate fractions, or the process of dephosphorylation of PfP2 through phosphatase(s). Mass spectrometry data on stage-specific parasite PfP2 protein will be informative regarding the post-translational modifications present at various positions in PfP2.
The most common method of analysis for assessment of SDS-resistant oligomerization is through immunoblotting using peptide-specific antibodies (47, 63), as was used in our studies of PfP2 as well. However, the labeling of rPfP2 with rhodamine allowed us an easy visual assay for the activation of formation of SDS-resistant oligomers (Fig. 6). Rhodamine labeling of peptides is an old and established technique, and such labeled proteins are typically assessed for their solution structures. However, in our case, we have used this simple method to search for parasite factors that may modulate SDS-resistant oligomerization. Because SDS-resistant oligomerization of peptides is a potential problem in various disease pathologies, the cellular factors responsible for such oligomerization may be accessed using this simple method.
Such a technique, using rhodamine-labeled rPfP2 incubated with various parasite extracts, confirmed that there were developmental stage-dependent factors in Plasmodium that propagated oligomerization of rPfP2 in vitro. Furthermore, using gel filtration-fractionated parasite proteins at the appropriate stage, we showed that a combination of multiple parasite protein components is required to promote oligomerization. Oligomerization of rPfP2 is a concentration-dependent phenomenon and does not seem to depend on any other extrinsic factors in vitro (Fig. 1F); indicating that the intrinsic structure of the PfP2 molecule has the propensity to form oligomers. However, at very low concentrations, the oligomerization of rPfP2 is propelled through the presence of a combination of certain parasite proteins belonging to specific developmental stages (Fig. 6). This happens without any added source of ATP, once again pointing to the noncovalent nature of aggregation. Analysis of the post-translational modifications on PfP2 and further dissection of parasite proteins to identify specific components that promote oligomerization will allow further understanding of the mechanism of developmental regulation of Plasmodium P2 protein oligomerization, and export of the P2 protein through the infected red cell to its surface.
Acknowledgments
We thank Shashidhar Dolas and Dr. Rukmini Govekar from Advanced Centre for Treatment, Research and Education in Cancer (ACTREC, Mumbai, India) for helping us with the MALDI. We also thank Reshma Korde for parasite cultures. We are grateful to A. S. R Koti for comments on the manuscript and Dr. K. Rajeshwari (Bioklone Pvt Ltd., India) for excellent monoclonal antibody services.
This work was supported by an intramural grant from Tata Institute of Fundamental Research, DAE, India. Part of this work was presented at the Gordon Research Conference on Malaria, 31st July-5th August, 2011, Pisa, Italy, as a poster (by S. Das).

This article contains supplemental Figs. S1–S7.
R. Tewari and S. Sharma, unpublished results.
H. Joshi and S. Sharma, unpublished data.
- IE
- infected erythrocyte
- TIFR
- Tata Institute of Fundamental Research
- TRITC
- tetramethylrhodamine isothiocyanate
- PMI
- postmerozoite invasion
- aa
- amino acid(s)
- iRBC
- infected RBC.
REFERENCES
- 1. Wahl M. C., Möller W. (2002) Molecular mechanism of P-glycoprotein assembly into cellular membranes. Curr. Protein Pept. Sci. 3, 485–486 [DOI] [PubMed] [Google Scholar]
- 2. Gonzalo P., Reboud J. P. (2003) The puzzling lateral flexible stalk of the ribosome. Biol. Cell. 95, 179–193 [DOI] [PubMed] [Google Scholar]
- 3. Tchórzewski M. (2002) The acidic ribosomal P proteins. Int. J. Biochem. Cell Biol. 34, 911–915 [DOI] [PubMed] [Google Scholar]
- 4. Wool I. G., Chan Y. L., Glück A., Suzuki K. (1991) The primary structure of rat ribosomal proteins P0, P1, and P2 and a proposal for a uniform nomenclature for mammalian and yeast ribosomal proteins. Biochimie 73, 861–870 [DOI] [PubMed] [Google Scholar]
- 5. Uchiumi T., Hori K., Nomura T., Hachimori A. (1999) Replacement of L7/L12.L10 protein complex in Escherichia coli ribosomes with the eukaryotic counterpart changes the specificity of elongation factor binding. J. Biol. Chem. 274, 27578–27582 [DOI] [PubMed] [Google Scholar]
- 6. Santos C., Ballesta J. P. (2005) Characterization of the 26 S rRNA-binding domain in Saccharomyces cerevisiae ribosomal stalk phosphoprotein P0. Mol. Microbiol. 58, 217–226 [DOI] [PubMed] [Google Scholar]
- 7. Ilag L. L., Videler H., McKay A. R., Sobott F., Fucini P., Nierhaus K. H., Robinson C. V. (2005) Heptameric (L12)6/L10 rather than canonical pentameric complexes are found by tandem MS of intact ribosomes from thermophilic bacteria. Proc. Natl. Acad. Sci. U.S.A. 102, 8192–8197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Skeiky Y. A., Benson D. R., Guderian J. A., Sleath P. R., Parsons M., Reed S. G. (1993) Trypanosoma cruzi acidic ribosomal P protein gene family. Novel P proteins encoding unusual cross-reactive epitopes. J. Immunol. 151, 5504–5515 [PubMed] [Google Scholar]
- 9. Szick K., Springer M., Bailey-Serres J. (1998) Evolutionary analyses of the 12-kDa acidic ribosomal P-proteins reveal a distinct protein of higher plant ribosomes. Proc. Natl. Acad. Sci. U.S.A. 95, 2378–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shimizu T., Nakagaki M., Nishi Y., Kobayashi Y., Hachimori A., Uchiumi T. (2002) Interaction among silkworm ribosomal proteins P1, P2, and P0 required for functional protein binding to the GTPase-associated domain of 28 S rRNA. Nucleic Acids Res. 30, 2620–2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hanson C. L., Videler H., Santos C., Ballesta J. P., Robinson C. V. (2004) Mass spectrometry of ribosomes from Saccharomyces cerevisiae. Implications for assembly of the stalk complex. J. Biol. Chem. 279, 42750–42757 [DOI] [PubMed] [Google Scholar]
- 12. Uchiumi T., Honma S., Nomura T., Dabbs E. R., Hachimori A. (2002) Translation elongation by a hybrid ribosome in which proteins at the GTPase center of the Escherichia coli ribosome are replaced with rat counterparts. J. Biol. Chem. 277, 3857–3862 [DOI] [PubMed] [Google Scholar]
- 13. Uchiumi T., Honma S., Endo Y., Hachimori A. (2002) Ribosomal proteins at the stalk region modulate functional rRNA structures in the GTPase center. J. Biol. Chem. 277, 41401–41409 [DOI] [PubMed] [Google Scholar]
- 14. Hagiya A., Naganuma T., Maki Y., Ohta J., Tohkairin Y., Shimizu T., Nomura T., Hachimori A., Uchiumi T. (2005) A mode of assembly of P0, P1, and P2 proteins at the GTPase-associated center in animal ribosome. In vitro analyses with P0 truncation mutants. J. Biol. Chem. 280, 39193–39199 [DOI] [PubMed] [Google Scholar]
- 15. Saenz-Robles M. T., Remacha M., Vilella M. D., Zinker S., Ballesta J. P. (1990) The acidic ribosomal proteins as regulators of the eukaryotic ribosomal activity. Biochim. Biophys. Acta. 1050, 51–55 [DOI] [PubMed] [Google Scholar]
- 16. Remacha M., Santos C., Bermejo B., Naranda T., Ballesta J. P. (1992) Stable binding of the eukaryotic acidic phosphoproteins to the ribosome is not an absolute requirement for in vivo protein synthesis. J. Biol. Chem. 267, 12061–12067 [PubMed] [Google Scholar]
- 17. Wool I. G. (1996) Extraribosomal functions of ribosomal proteins. Trends Biochem. Sci. 21, 164–165 [PubMed] [Google Scholar]
- 18. Lobo C. A., Kar S. K., Ravindran B., Kabilan L., Sharma S. (1994) Novel proteins of Plasmodium falciparum identified by differential immunoscreening using immune and patient sera. Infect. Immun. 62, 651–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chatterjee S., Singh S., Sohoni R., Kattige V., Deshpande C., Chiplunkar S., Kumar N., Sharma S. (2000) Characterization of domains of the phosphoriboprotein P0 of Plasmodium falciparum. Mol. Biochem. Parasitol. 107, 143–154 [DOI] [PubMed] [Google Scholar]
- 20. Singh S., Sehgal A., Waghmare S., Chakraborty T., Goswami A., Sharma S. (2002) Surface expression of the conserved ribosomal protein P0 on parasite and other cells. Mol. Biochem. Parasitol. 119, 121–124 [DOI] [PubMed] [Google Scholar]
- 21. Spence J. M., Clark V. L. (2000) Role of ribosomal protein L12 in gonococcal invasion of Hec1B cells. Infect. Immun. 68, 5002–5010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kasai H., Nadano D., Hidaka E., Higuchi K., Kawakubo M., Sato T. A., Nakayama J. (2003) Differential expression of ribosomal proteins in human normal and neoplastic colorectum. J. Histochem. Cytochem. 51, 567–574 [DOI] [PubMed] [Google Scholar]
- 23. Aruna K., Chakraborty T., Rao P. N., Santos C., Ballesta J. P., Sharma S. (2005) Functional complementation of yeast ribosomal P0 protein with Plasmodium falciparum P0. Gene 357, 9–17 [DOI] [PubMed] [Google Scholar]
- 24. Das S., Basu H., Korde R., Tewari R., Sharma S. (2012) Arrest of nuclear division in Plasmodium through blockage of erythrocyte surface exposed ribosomal protein P2. PLoS Pathog. 8, e1002858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee K. M., Yu C. W., Chan D. S., Chiu T. Y., Zhu G., Sze K. H., Shaw P. C., Wong K. B. (2010) Solution structure of the dimerization domain of ribosomal protein P2 provides insights for the structural organization of eukaryotic stalk. Nucleic Acids Res. 38, 5206–5216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tchórzewski M., Boguszewska A., Dukowski P., Grankowski N. (2000) Oligomerization properties of the acidic ribosomal P-proteins from Saccharomyces cerevisiae. Effect of P1A protein phosphorylation on the formation of the P1A-P2B hetero-complex. Biochim Biophys Acta 1499, 63–73 [DOI] [PubMed] [Google Scholar]
- 27. Mishra P., Das S., Panicker L., Hosur M. V., Sharma S., Hosur R. V. (2012) NMR insights into folding and self-association of Plasmodium falciparum P2. PLoS One 7, e36279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rajeshwari K., Patel K., Nambeesan S., Mehta M., Sehgal A., Chakraborty T., Sharma S. (2004) The P domain of the P0 protein of Plasmodium falciparum protects against challenge with malaria parasites. Infect. Immun. 72, 5515–5521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hashimoto K., Nishi H., Bryant S., Panchenko A. R. (2011) Caught in self-interaction. Evolutionary and functional mechanisms of protein homooligomerization. Phys Biol. 8, 035007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schomburg I., Chang A., Ebeling C., Gremse M., Heldt C., Huhn G., Schomburg D. (2004) BRENDA, the enzyme database. Updates and major new developments. Nucleic Acids Res. 32, (Database issue) D431–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ispolatov I., Yuryev A., Mazo I., Maslov S. (2005) Binding properties and evolution of homodimers in protein-protein interaction networks. Nucleic Acids Res. 33, 3629–3635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Norrgård M. A., Hellman U., Mannervik B. (2011) Cys-X scanning for expansion of active-site residues and modulation of catalytic functions in a glutathione transferase. J. Biol. Chem. 286, 16871–16878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lynch B. A., Koshland D. E., Jr. (1991) Disulfide cross-linking studies of the transmembrane regions of the aspartate sensory receptor of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 88, 10402–10406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kaslow D. C., Hui G., Kumar S. (1994) Expression and antigenicity of Plasmodium falciparum major merozoite surface protein (MSP1(19)) variants secreted from Saccharomyces cerevisiae. Mol. Biochem. Parasitol. 63, 283–289 [DOI] [PubMed] [Google Scholar]
- 35. Hodder A. N., Crewther P. E., Anders R. F. (2001) Specificity of the protective antibody response to apical membrane antigen 1. Infect. Immun. 69, 3286–3294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sim B. K., Chitnis C. E., Wasniowska K., Hadley T. J., Miller L. H. (1994) Receptor and ligand domains for invasion of erythrocytes by Plasmodium falciparum. Science 264, 1941–19444 [DOI] [PubMed] [Google Scholar]
- 37. Prugnolle F., Ayala F., Ollomo B., Arnathau C., Durand P., Renaud F. (2011) Plasmodium falciparum is not as lonely as previously considered. Virulence 2, 71–76 [DOI] [PubMed] [Google Scholar]
- 38. Glenner G. G., Wong C. W. (1984) Alzheimer disease. Initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem. Biophys. Res. Commun. 120, 885–890 [DOI] [PubMed] [Google Scholar]
- 39. Roychaudhuri R., Yang M., Hoshi M. M., Teplow D. B. (2009) Amyloid β-protein assembly and Alzheimer disease. J. Biol. Chem. 284, 4749–4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lazo N. D., Maji S. K., Fradinger E. A., Bitan G., Teplow D. B. (2005) in Amyloid Proteins: The Beta Sheet Conformation and Disease (Sipe J. D., ed) pp. 385–491, Wiley, New York [Google Scholar]
- 41. Soreghan B., Kosmoski J., Glabe C. (1994) Surfactant properties of Alzheimer Aβ peptides and the mechanism of amyloid aggregation. J. Biol. Chem. 269, 28551–28554 [PubMed] [Google Scholar]
- 42. McLean C. A., Cherny R. A., Fraser F. W., Fuller S. J., Smith M. J., Beyreuther K., Bush A. I., Masters C. L. (1999) Soluble pool of Aβ amyloid as a determinant of severity of neurodegeneration in Alzheimer's disease. Ann. Neurol. 46, 860–866 [DOI] [PubMed] [Google Scholar]
- 43. Lesné S., Koh M. T., Kotilinek L., Kayed R., Glabe C. G., Yang A., Gallagher M., Ashe K. H. (2006) A specific amyloid-β protein assembly in the brain impairs memory. Nature 440, 352–357 [DOI] [PubMed] [Google Scholar]
- 44. Shankar G. M., Li S., Mehta T. H., Garcia-Munoz A., Shepardson N. E., Smith I., Brett F. M., Farrell M. A., Rowan M. J., Lemere C. A., Regan C. M., Walsh D. M., Sabatini B. L., Selkoe D. J. (2008) Amyloid-β protein dimers isolated directly from Alzheimer brains impair synaptic plasticity and memory. Nat. Med. 14, 837–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bernstein S. L., Dupuis N. F., Lazo N. D., Wyttenbach T., Condron M. M., Bitan G., Teplow D. B., Shea J. E., Ruotolo B. T., Robinson C. V., Bowers M. T. (2009) Amyloid-β protein oligomerization and the importance of tetramers and dodecamers in the aetiology of Alzheimer disease. Nat. Chem. 1, 326–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen Y. R., Glabe C. G. (2006) Distinct early folding and aggregation properties of Alzheimer amyloid-β peptides Aβ40 and Aβ42. Stable trimer or tetramer formation by Aβ42. J. Biol. Chem. 281, 24414–24422 [DOI] [PubMed] [Google Scholar]
- 47. Bagriantsev S. N., Kushnirov V. V., Liebman S. W. (2006) Analysis of amyloid aggregates using agarose gel electrophoresis. Methods Enzymol. 412, 33–48 [DOI] [PubMed] [Google Scholar]
- 48. Nag S., Sarkar B., Bandyopadhyay A., Sahoo B., Sreenivasan V. K., Kombrabail M., Muralidharan C., Maiti S. (2011) Nature of the amyloid-β monomer and the monomer-oligomer equilibrium. J. Biol. Chem. 286, 13827–13833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lee H. J., Baek S. M., Ho D. H., Suk J. E., Cho E. D., Lee S. J. (2011) Dopamine promotes formation and secretion of nonfibrillar α-synuclein oligomers. Exp. Mol. Med. 43, 216–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fowler D. M., Koulov A. V. (2007) Functional amyloid–from bacteria to humans. Trends Biochem. Sci. 32, 217–224 [DOI] [PubMed] [Google Scholar]
- 51. Blanco L. P., Evans M. L., Smith D. R., Badtke M. P., Chapman M. R. (2012) Diversity, biogenesis, and function of microbial amyloids. Trends Microbiol. 20, 66–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liebman S. W., Chernoff Y. O. (2012) Prions in yeast. Genetics 191, 1041–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Uptain S. M., Lindquist S. (2002) Prions as protein-based genetic elements. Annu. Rev. Microbiol. 56, 703–741 [DOI] [PubMed] [Google Scholar]
- 54. Wang X., Smith D. R., Jones J. W., Chapman M. R. (2007) In vitro polymerization of a functional Escherichia coli amyloid protein. J. Biol. Chem. 282, 3713–3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dos Reis S., Coulary-Salin B., Forge V., Lascu I., Bégueret J., Saupe S. J. (2002) The HET-s prion protein of the filamentous fungus Podospora anserina aggregates in vitro into amyloid-like fibrils. J. Biol. Chem. 277, 5703–5706 [DOI] [PubMed] [Google Scholar]
- 56. Iconomidou V. A., Hamodrakas S. J. (2008) Natural protective amyloids. Curr. Protein Pept. Sci. 9, 291–309 [DOI] [PubMed] [Google Scholar]
- 57. Bowerman C. J., Liyanage W., Federation A. J., Nilsson B. L. (2011) Tuning β-sheet peptide self-assembly and hydrogelation behavior by modification of sequence hydrophobicity and aromaticity. Biomacromolecules. 12, 2735–2745 [DOI] [PubMed] [Google Scholar]
- 58. Gazit E. (2002) A possible role for πi-stacking in the self-assembly of amyloid fibrils. FASEB J. 16, 77–83 [DOI] [PubMed] [Google Scholar]
- 59. Arnesano F., Scintilla S., Calò V., Bonfrate E., Ingrosso C., Losacco M., Pellegrino T., Rizzarelli E., Natile G. (2009) Copper-triggered aggregation of ubiquitin. PLoS One 16, e7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kostka M., Högen T., Danzer K. M., Levin J., Habeck M., Wirth A., Wagner R., Glabe C. G., Finger S., Heinzelmann U., Garidel P., Duan W., Ross C. A., Kretzschmar H., Giese A. (2008) Single particle characterization of iron-induced pore-forming α-synuclein oligomers. J. Biol. Chem. 283, 10992–11003 [DOI] [PubMed] [Google Scholar]
- 61. Bernard K., Wang W., Narlawar R., Schmidt B., Kirk K. L. (2009) Curcumin cross-links cystic fibrosis transmembrane conductance regulator (CFTR) polypeptides and potentiates CFTR channel activity by distinct mechanisms. J. Biol. Chem. 284, 30754–30765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Neumeyer T., Schiffler B., Maier E., Lang A. E., Aktories K., Benz R. (2008) Clostridium botulinum C2 toxin. Identification of the binding site for chloroquine and related compounds and influence of the binding site on properties of the C2II channel. J. Biol. Chem. 283, 3904–3914 [DOI] [PubMed] [Google Scholar]
- 63. Urakov V. N., Vishnevskaya A. B., Alexandrov I. M., Kushnirov V. V., Smirnov V. N., Ter-Avanesyan M. D. (2010) Interdependence of amyloid formation in yeast. Implications for polyglutamine disorders and biological functions. Prion 4, 45–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Goldfarb L. G., Brown P., McCombie W. R., Goldgaber D., Swergold G. D., Wills P. R., Cervenakova L., Baron H., Gibbs C. J., Jr., Gajdusek D. C. (1991) Transmissible familial Creutzfeldt-Jakob disease associated with five, seven, and eight extra octapeptide coding repeats in the PRNP gene. Proc. Natl. Acad. Sci. U.S.A. 88, 10926–10930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hundt C., Gauczynski S., Leucht C., Riley M. L., Weiss S. (2003) Intra- and interspecies interactions between prion proteins and effects of mutations and polymorphisms. Biol. Chem. 384, 791–803 [DOI] [PubMed] [Google Scholar]
- 66. Cobb N. J., Sönnichsen F. D., McHaourab H., Surewicz W. K. (2007) Molecular architecture of human prion protein amyloid. A parallel, in-register β-structure. Proc. Natl. Acad. Sci. U.S.A. 104, 18946–18951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tycko R., Savtchenko R., Ostapchenko V. G., Makarava N., Baskakov I. V. (2010) The α-helical C-terminal domain of full-length recombinant PrP converts to an in-register parallel β-sheet structure in PrP fibrils. Evidence from solid state nuclear magnetic resonance. Biochemistry 49, 9488–9497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Pietilä R., Nätynki M., Tammela T., Kangas J., Pulkki K. H., Limaye N., Vikkula M., Koh G. Y., Saharinen P., Alitalo K., Eklund L. (2012) Ligand oligomerization state controls Tie2 receptor trafficking and angiopoietin-2-specific responses. J. Cell Sci. 125, 2212–2223 [DOI] [PubMed] [Google Scholar]
- 69. Lee K. K., Yonehara S. (2002) Phosphorylation and dimerization regulate nucleocytoplasmic shuttling of mammalian STE20-like kinase (MST). J. Biol. Chem. 277, 12351–12358 [DOI] [PubMed] [Google Scholar]
- 70. Hasler P., Brot N., Weissbach H., Parnassa A. P., Elkon K. B. (1991) Ribosomal proteins P0, P1, and P2 are phosphorylated by casein kinase II at their conserved carboxyl termini. J. Biol. Chem. 266, 13815–13820 [PubMed] [Google Scholar]