Background: Tn4 B cells from a patient with Tn antigen-positive leukocytes lack transcripts of Cosmc.
Results: The Cosmc promoter in Tn4 cells is methylated and 5-aza-2′-deoxycytidine treatment restores Cosmc transcription and normal O-glycans.
Conclusion: Methylation-dependent epigenetic silencing of Cosmc occurs in Tn4 cells and results in Tn antigen expression.
Significance: These findings provide a novel mechanism underlying aberrant expression of Tn antigen in human diseases.
Keywords: Chaperone Chaperonin, DNA Methylation, Epigenetics, Glycoprotein, Glycosylation, Glycosyltransferases, Cosmc, T-synthase, Tn Antigen
Abstract
Cosmc is the specific molecular chaperone in the endoplasmic reticulum for T-synthase, a Golgi β3-galactosyltransferase that generates the core 1 O-glycan, Galβ1–3GalNAcα-Ser/Thr, in glycoproteins. Dysfunctional Cosmc results in the formation of inactive T-synthase and consequent expression of the Tn antigen (GalNAcα1-Ser/Thr), which is associated with several human diseases. However, the molecular regulation of expression of Cosmc, which is encoded by a single gene on Xq24, is poorly understood. Here we show that epigenetic silencing of Cosmc through hypermethylation of its promoter leads to loss of Cosmc transcripts in Tn4 cells, an immortalized B cell line from a male patient with a Tn-syndrome-like phenotype. These cells lack T-synthase activity and express the Tn antigen. Treatment of cells with 5-aza-2′-deoxycytidine causes restoration of Cosmc transcripts, restores T-synthase activity, and reduces Tn antigen expression. Bisulfite sequencing shows that CG dinucleotides in the Cosmc core promoter are hypermethylated. Interestingly, several other X-linked genes associated with glycosylation are not silenced in Tn4 cells, and we observed no correlation of a particular DNA methyltransferase to aberrant methylation of Cosmc in these cells. Thus, hypermethylation of the Cosmc promoter in Tn4 cells is relatively specific. Epigenetic silencing of Cosmc provides another mechanism underlying the abnormal expression of the Tn antigen, which may be important in understanding aberrant Tn antigen expression in human diseases, including IgA nephropathy and cancer.
Introduction
O-Glycans are commonly found on Ser/Thr residues of animal cell glycoproteins and serve to regulate many aspects of glycoprotein function and recognition (1–7). The generation of the most common core 1 O-glycan, Galβ1–3GalNAcα-Ser/Thr (T antigen),2 from GalNAcα1-Ser/Thr (Tn antigen) requires action of a single enzyme, the core 1 β3-galactosyltransferase (T-synthase) (8). Interestingly, biosynthesis of active T-synthase requires an endoplasmic reticulum-localized molecular chaperone, Cosmc (core 1 β3-galatosyltransferase-specific molecular chaperone), that prevents the aggregation and subsequent proteasomal degradation of T-synthase (9–12). Human T-synthase is encoded by a multiexon functional gene on 7p14-p13 (8), whereas human Cosmc is encoded by a single exon gene on Xq24 (9). Deletion of either T-synthase or Cosmc in mice causes embryonic lethality (4, 13), which is associated with bleeding due to defective angiogenesis. Both Cosmc and T-synthase are ubiquitously and coordinately expressed in all tissues of mice and humans (8, 10).
Although under normal physiological conditions, functional Cosmc and T-synthase ensure the conversion of Tn antigen to the core 1 structure (T antigen), aberrant expression of Tn antigen has been observed in several human diseases, including Tn syndrome (14), IgA nephropathy (IgAN) (15), and human tumors (16). In some cases, patients have acquired somatic mutations in the open reading frame (ORF) of Cosmc, as seen in hematopoietic stem cells in patients with Tn syndrome (17, 18) and in human and mouse tumor cell lines (19, 20). In other cases, such as human cervical cancer and melanoma cells, tumor cells have the gene deletion, or loss of heterozygosity, of Cosmc (19). Interestingly, Thurnher et al. (21) and Felner et al. (22) observed that the T-synthase activity was regained after treatment of the Tn-positive T cells from patients with Tn syndrome with 5-azacytidine or sodium n-butyrate, and they concluded that the suppression of T-synthase presumably by methylation was the mechanism for Tn syndrome. Because both human Cosmc and T-synthase were not cloned then, it was not possible for them to examine the methylation status of these critical genes. In addition, in IgAN, aberrant O-glycosylation of the hinge region of IgA1 (e.g. Tn and sialyl-Tn expression) is considered a hallmark for pathogenesis in this most common glomerulonephritis (15), but the molecular mechanism for undergalactosylation of IgA1 is not well understood.
In an immortalized B cell line termed Tn4, derived from a male blood donor with a Tn-positive phenotype, a C428T change in the Cosmc (C1GALT1C1) exon encoded an A143V substitution, and the transcripts of Cosmc were not detected. However, erythrocytes from the Tn4 donor were Tn-negative, suggesting that the C428T conservative substitution is not the cause of Tn antigen expression in donor Tn4 (17). Here we investigated in greater detail the lack of Cosmc transcripts and Tn expression in Tn4 cells. Our results show that Cosmc in Tn4 cells is completely silenced due to hypermethylation of the core promoter. Inhibition of DNA methylation by 5-aza-2′-deoxycytidine (5-Aza-dC) (23) causes the restoration of transcription of Cosmc, restores T-synthase activity, and corrects the O-glycans in Tn4 cells. These results identify a unique epigenetic mechanism for dysregulation of Cosmc that may shed light on human diseases associated with aberrant Tn antigen expression.
EXPERIMENTAL PROCEDURES
Reagents
All chemical reagents, including 5-Aza-dC were purchased from Sigma and Fisher. DNA oligonucleotides (primers) were from Integrated DNA Technologies Inc. (Coralville, IA).
Cell Lines and Culture
Tn4 cells are the Epstein-Barr virus (EBV)-transformed B lymphocytes from a male individual (Tn4) whose leukocytes express Tn antigen (17). EBV-transformed human B-lymphoblast DAKIKI cells were purchased from ATCC (TIB-206). C4 B cells are the EBV-transformed B lymphocytes from peripheral blood of a healthy male donor. All cells were grown in RPMI1640 (Invitrogen) containing 20% heat-inactivated fetal bovine serum at 37 °C, 5% CO2. For 5-Aza-dC treatment, Tn4 cells were seeded at a density of 1 × 106 cells/ml and 0.5–10 μm (final concentration). 5-Aza-dC was added into the growth medium as indicated in the figures.
Preparation of Cytosolic Fraction and Nuclear Extracts
Cell pellets containing 5–6 × 106 cells were processed to generate the cytosolic fractions and nuclear extracts using the Nuclear Extract Kit from Active Motif (Carlsbad, CA). Protein concentrations were measured with a bicinchoninic acid (BCA) kit (Pierce) with bovine serum albumin (BSA) as a standard.
Flow Cytometry
Cells (1 × 106) in 200 μl of Hanks' balanced buffer were stained with 1 μg/ml Alexa488-labeled mouse anti-Tn mAb (IgM, CA3638), or FITC-labeled antibodies, anti-IgA, IgA1, IgA2, CD19, and CD59, and isotype controls in Hanks' balanced buffer and analyzed on a flow cytometer (FACSCalibur, BD Biosciences) as described previously (19).
RT-PCR and PCR Analysis
Total RNA and genomic DNA from Tn4 and DAKIKI cells were extracted with the RNeasy minikit and FlexiGene DNA kit (Qiagen, Valencia, CA), respectively. RT-PCR was carried out with 500 ng of mRNA as template using the SuperScript first-strand synthesis system for RT-PCR (Invitrogen). The regular PCRs were done with the Advantage GC genomic PCR polymerase kit from Clontech. The primers are listed in Table 1, and annealing temperatures for each set of primers are listed in Table 2.
TABLE 1.
PCR primers
| Name | Sequence |
|---|---|
| Cosmc-F9 | 5′-TTCTCCATAGAGGAGTTGTTGC-3′ |
| Cosmc-R10 | 5′-TGTGGTTATACCAGTGCCACC-3′ |
| Cosmc-cDNA-F1 | 5′-CGTGAGAGGAAACCCGTG-3′ |
| Cosmc-cDNA-R1 | 5′-TGTGTGGTTATACCAGTGCC-3′ |
| Cosmc-cDNA-F3 | 5′-TTTAGCGAGACCAACGAG-3′ |
| Cosmc-cDNA-R3 | 5′-GAACTCTGCTTTGTCACAGTG-3′ |
| Cosmc-5′-F2 | 5′-AGCGGGCCCACTGTCAGAAAT-3′ |
| Cosmc-5′-R2 | 5′-TGGCTGCTGTCCTTAAGTGTTTTGA-3′ |
| Tsyn-cDNA-F1 | 5′-TCTTACAGAAATACACTTTCGG-3′ |
| Tsyn-cDNA-R1 | 5′-ATTTTAACACACTTCACAGCTC-3′ |
| Tsyn-cDNA-F3 | 5′-GGTTGACACCCAGCCTAATG-3′ |
| Tsyn-cDNA-R3 | 5′-TTTTCAGTCCCACAGCAGGG-3′ |
| GAPDH-F | 5′-TGGGGAAGGTGAAGGTCGG-3′ |
| GAPDH-R | 5′-GGGATCTCGCTCCTGGAAG-3′ |
| Cosmc-M-F | 5′-GTTTAGTCGTCGGTTTTTTTATTTC-3′ |
| Cosmc -M-R | 5′-AAACGATATTCTCTCGTTAATCTCG-3′ |
| Cosmc -U-F | 5′-TTTAGTTGTTGGTTTTTTTATTTTGT-3′ |
| Cosmc -U-R | 5′-AACAATATTCTCTCATTAATCTCACT-3′ |
| Tsyn-110F | 5′-TTTTTTTTTGGGGTTGGTG-3′ |
| Tsyn+138R | 5′-CCTTTACTTCTCTAAACAAC-3′ |
| Tsyn-89MF | 5′-CGGCGGAGACGTTAGCGTTAG-3′ |
| Tsyn+63MR | 5′-TCCCCGACAACGACAACACAA-3′ |
| Tsyn-89UF | 5′-TGGTGGAGATGTTAGTGTTAG-3′ |
| Tsyn+63UR | 5′-TCCCCAACAACAACAACACAA-3′ |
| hIgA-F | 5′-AATCCCAGCCAGGATGTGAC-3′ |
| hIgA-R | 5′-AGGCAGCAGTGCAAGTGAAG-3′ |
TABLE 2.
Information on PCR with different sets of primers
| Primer sets | Annealing temperature | Size of product |
|---|---|---|
| °C | bp | |
| Cosmc-F9/-R10 | 59 | 1333 |
| Cosmc-cDNA-F1/-R1 | 62 | 1156 |
| Cosmc-cDNA-F3/-R3 | 53 | 437 |
| Cosmc-5′-F2/-R2 | 63 | 5092 |
| T-syn-cDNA-F1/-R1 | 62 | 1299 |
| T-syn-cDNA-F3/-R3 | 54 | 305 |
| GAPDH-F/-R | 62 | 245 |
| Cosmc-M-F/-M-R | 58 | 165 |
| Cosmc -U-F/-U-R | 58 | 165 |
| Tsyn-110F/+138R | 48 | 248 |
| Tsyn-89MF/+63MR | 55 | 152 |
| Tsyn-89UF/+63UR | 50 | 152 |
Enzyme Activity Assays
The activity of T-synthase was fluorescently assayed using GalNAc-α-(4-MU) as acceptor substrate and UDP-Gal as the donor as described previously (24). The neutral α-mannosidase activity, including α-mannosidase II, was assayed using mannose-α-(4-MU) (Sigma) as the substrate. Briefly, the 50-μl reaction system contained 1 mm mannose-α-(4-MU) in 100 mm Tris-HCl (pH 7.5) and 10 μl of cell extracts. The blank reaction was set up the same except for 10 μl of cell extraction boiled prior to addition to the reaction. The rest of the procedure was the same as the T-synthase activity assay (24).
Analysis of DNA Methylation by Bisulfite Sequencing
Genomic DNA was treated by sodium bisulfite with EZ DNA Methylation-Gold kit (Zymo Research, Irvine, CA). PCR primers are listed in Table 1, and PCR conditions used are shown in Table 2. The PCR products were analyzed on 2% agarose gel, and the bands were excised and purified using a Qiagen gel extraction kit (Qiagen) and subjected to sequencing.
Western Blot
Protein samples (20–30 μg) after denaturing under reducing conditions were separated on a 4–12% precast gel (SDS-PAGE) from Bio-Rad and transferred to nitrocellulose membranes. The membranes were blotted with anti-human DNMT-1, -3a, or -3b from IMGENEX (San Diego, CA) and anti-human DNMT-2 and -3L and anti-human OGT, SP1 (specificity protein 1), SP3, lamin B, and β-actin from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) as described previously (10).
RESULTS
Characterization of Tn4 Cells
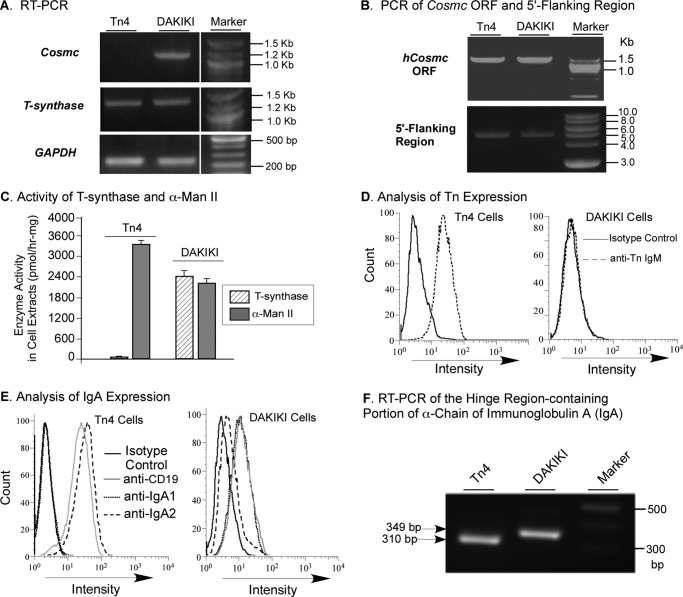
Tn4 cells are an EBV-immortalized B cell line originally derived from a patient with Tn-positive blood lymphocytes (17). We characterized Tn4 cells and showed that these cells lacked transcripts of Cosmc but contained normal transcripts for T-synthase (Fig. 1A), whereas DAKIKI cells, a human B cell line from a patient with myeloma, had transcripts for both Cosmc and T-synthase. To investigate the cause of the lack of Cosmc transcripts, we examined the integrity of Cosmc, which is a single exon gene on human Xq24. We found that the ORF and 5′-flanking region, including the promoter, of Cosmc in Tn4 cells were intact and comparable with those in DAKIKI cells, as shown by PCR amplification of genomic DNA from both cell lines (Fig. 1B). Lack of Cosmc transcript would indicate that no Cosmc protein is being made, which will result in an inactive T-synthase. Indeed, Tn4 cells lacked T-synthase activity (Fig. 1C). To compare with another Golgi enzyme as a control, the N-glycan processing glycosidase α-mannosidase II was chosen because its activity can be assayed at neutral pH using mannose-α-(4-MU) as the substrate, which is similar to the T-synthase activity assay using GalNAc-α-(4-MU) as the acceptor substrate. As shown in Fig. 1C, Tn4 cells had robust activity of α-mannosidase II, whereas DAKIKI cells had both T-synthase and α-mannosidase II activity. Inactive T-synthase in cells leads to expression of truncated O-glycans, including the Tn antigen, on the cell surface. Accordingly, Tn4 cells, but not DAKIKI cells, expressed Tn antigen on their surfaces (Fig. 1D). Flow cytometry analysis with anti-CD19 staining confirmed that both Tn4 and DAKIKI cells were B cells (Fig. 1E). Initially, we expected to isolate a subclone from Tn4 cells that secreted IgA1 with Tn antigen exclusively expressed in its hinge region, assuming that these cells were a mixed population of B cells. Such B cells could be very useful for IgAN study because aberrant O-glycosylation, such as Tn and STn antigens in the hinge region of human IgA1, is considered the major pathogenesis for IgAN. Therefore, we stained the cells with FITC-labeled anti-human IgA1 and IgA2 antibodies. Surprisingly, staining revealed that Tn4 cells uniformly expressed IgA2 but not IgA1, whereas DAKIKI cells produced IgA1 but not IgA2 (Fig. 1E). To further confirm this interesting result, RT-PCR using primers covering the hinge regions for both human IgA1 and IgA2 showed that the product from Tn4 cells was 310 bp (Fig. 1F), smaller than the size of the product with 349 bp from DAKIKI cells, an IgA1-producing cell line. Sequencing of RT-PCR products confirmed that Tn4 cells expressed IgA2, whereas DAKIKI cells produced IgA1. Thus, Tn4 cells were characterized as clonal, IgA2-producing B cells with a dysfunctional Cosmc. These characteristics of Tn4 cells differentiate them completely from the nature of Tn syndrome, in which some blood cells of all lineages, including erythrocytes and leukocytes, are affected. Although the clinical information for this blood donor is not available, it is possible that the Tn4 donor has an IgA2-producing myeloma.
FIGURE 1.
Characterization of Tn4 Cells. A, RT-PCR analysis of Cosmc and T-synthase expression. The mRNA from Tn4 and DAKIKI cells was isolated, and the cDNA was synthesized using universal primers. Using GAPDH as an internal control, PCRs for Cosmc (Cosmc-cDNA-F1/-R1) and T-synthase (Tsyn-cDNA-F1/-R1) were carried out, and products were analyzed on 1.2% agarose gel. The lanes were run on the same gel but were noncontiguous. B, PCR analysis of the ORF and 5′-flanking region of Cosmc. The genomic DNA from Tn4 and DAKIKI cells was extracted and analyzed for the ORF and 5′-flanking region, including the cis-elements of Cosmc, using PCR. C, enzyme activity assay. The extracts from Tn4 and DAKIKI cells were made and assayed for T-synthase activity and for α-mannosidase II activity (mean ± S.D. (error bars)) in duplicates using GalNAc-α-(4-MU) and mannose-α-(4-MU), respectively. The data represent two independent experiments. D, analysis of Tn expression. Tn4 and DAKIKI cell lines were stained with Alexa488-labeled mouse anti-Tn and isotype control mouse IgM and analyzed by flow cytometry. E, analyses of immunoglobulin A and CD19 expression. Tn4 and DAKIKI cells were stained with FITC-labeled mouse anti-human CD19, IgA1, and IgA2 as well as isotype control and analyzed by flow cytometry. F, RT-PCR analysis of IgA expression. The cDNA made from mRNA of Tn4 and DAKIKI cells in Fig. 1A was amplified by the primers covering human IgA hinge region using PCR. The products were analyzed on 1.2% agarose gel.
The GC-rich DNA-binding Proteins, SP1/3 Transcriptional Factors Essential for Transcription of Human Cosmc, Are Expressed in Tn4 Cells
In our previous study (19), we found that human melanoma LOX cells lack transcripts of Cosmc due to gene deletion. By contrast, Tn4 cells had no transcripts of Cosmc, while having an apparent intact gene. This information indicates that the mechanism underlying the lack of Cosmc transcripts in Tn4 cells is different from that in LOX cells. In studying transcription factors that might be important for Cosmc expression, our preliminary studies suggested that the GC-rich DNA-binding proteins, including the SP/KLF (specificity protein/Krüppel-like factor) family, were transcriptional factors needed for transcription of human Cosmc. Thus, we reasoned that the loss of transcripts of Cosmc could result from a lack of or decreased expression of the SP/KLF transcription factors, such as SP1 and SP3, in Tn4 cells. However, both Tn4 and DAKIKI cells were shown to have SP1/3 proteins (Fig. 2), and these proteins were also expressed in colorectal carcinoma LSB cells and breast cancer MCF-7 cells. Tn4 cells had an even higher expression level of SP1/3 than DAKIKI cells, suggesting that a mechanism independent of expression of these transcription factors underlies the loss of Cosmc transcripts in Tn4 cells.
FIGURE 2.
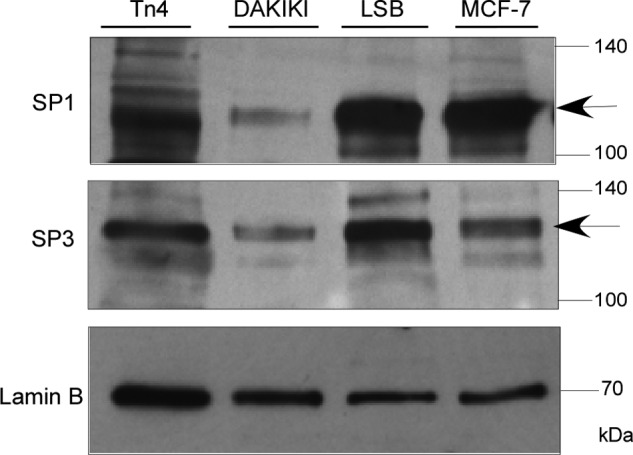
Expression of SP1/3 in Tn4 and DAKIKI Cells. The nuclear extracts made from Tn4, DAKIKI, LSB, and MCF-7 cells were analyzed by Western blot with mouse anti-human SP1 and SP3 monoclonal antibodies (IgG). The loading of SDS-PAGE was normalized with lamin B.
The DNA Methylation Inhibitor 5-Aza-dC Restores Transcription of Cosmc and T-synthase Activity in Tn4 Cells and Diminishes Tn Antigen Expression
In an attempt to understand the molecular mechanism underlying the absence of transcripts of Cosmc in Tn4 cells, we hypothesized that epigenetic dysregulation (e.g. hypermethylation of the Cosmc promoter) might be responsible for silencing Cosmc. We tested this hypothesis by treating Tn4 cells for several days with 5-Aza-dC, a DNA methylation inhibitor, and probed for Cosmc transcription by simply monitoring the T-synthase activity. Incubation with 5-Aza-dC at a concentration of 5 or 10 μm for 9 days did not significantly alter the gross morphology and viability of Tn4 cells. Within 2 days of treatment with 5-Aza-dC, however, the T-synthase activity in Tn4 cells could be detected, and it reached the highest level after 6 days (Fig. 3A); activity remained at a similar level at 9 days postinduction, whereas untreated Tn4 cells lacked any activity. We confirmed that Cosmc transcripts were recovered by the 5-Aza-dC treatment by RT-PCR. Although the untreated Tn4 cells had no product, Cosmc transcripts were detected by day 2 of 5-Aza-dC-treated cells and reached the highest level between days 6 and 9 (Fig. 3B), which was consistent with the expression of T-synthase activity (Fig. 3A). T-synthase transcripts were detected and were not affected by 5-Aza-dC, in both treated and untreated samples (Fig. 3B). Thus, 5-Aza-dC treatment results in restoration of Cosmc transcripts, allowing synthesis of Cosmc and formation of functional T-synthase enzyme. These results also indicate that Cosmc is completely silenced through a methylation-dependent process in Tn4 cells.
FIGURE 3.
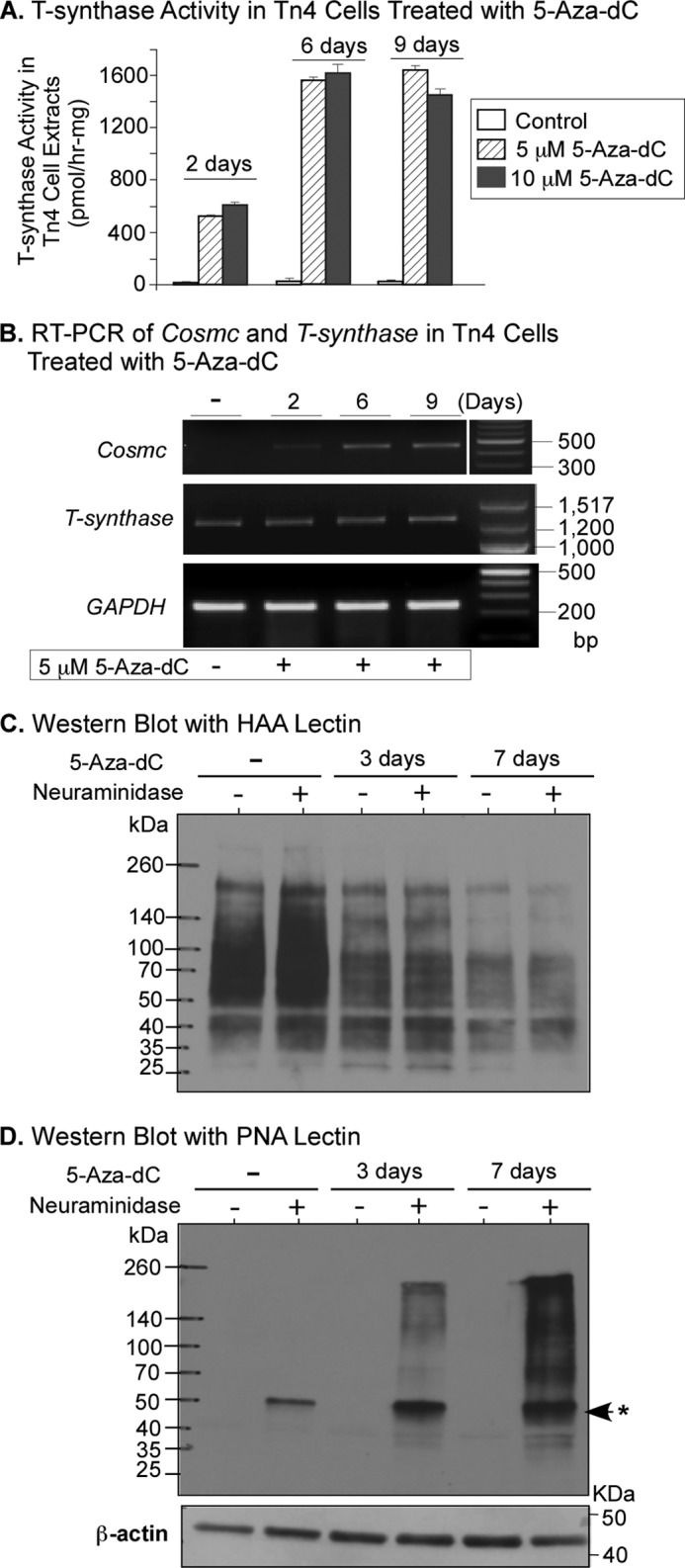
5-Aza-dC treatment of Tn4 cells resumes transcription of Cosmc, restores T-synthase activity, and consequently elongates O-glycans. A, T-synthase activity assay. Tn4 cells were treated with different concentrations of 5-Aza-dC as indicated and collected at 2, 6, and 9 days after treatment. Cell extracts were assayed for T-synthase activity (mean ± S.D. (error bars)) in duplicates. The data represent three independent experiments. B, RT-PCR analysis of Cosmc and T-synthase expression. Tn4 cells treated with 5 μm 5-Aza-dC for 2, 6, and 9 days were harvested and analyzed for expression of Cosmc (Cosmc-cDNA-F3/-R3) and T-synthase (Tsyn-cDNA-F1/-R1) using RT-PCR. The amount of transcripts was normalized by GAPDH. In the analysis of RT-PCR of Cosmc, the DNA marker was run at the opposite side on the same gel but was noncontiguous. C and D, lectin blotting. Cell extracts from Tn4 cells treated with 5 μm 5-Aza-dC for 3 and 7 days were incubated with or without neuraminidase and analyzed by Western blotting with lectins HAA (C) and PNA (D). *, neuraminidase band.
T-synthase converts the Tn antigen to the T antigen, which is then further modified or extended to form complex O-glycans. To define whether restoration of T-synthase activity in Tn4 cells by 5-Aza-dC caused loss of Tn antigen and restoration of normal core 1 O-glycans, we performed lectin blotting with Helix aspersa agglutinin (HAA), which binds to terminal α-GalNAc on O-glycans, such as Tn antigen (GalNAcα-Ser/Thr), and with peanut agglutinin (PNA) from Arachis hypogaea, which recognizes the core 1 structure (Galβ1–3GalNAcα-Ser/Thr) and that disaccharide within core 2 O-glycans. HAA heavily stained the untreated cell extracts but showed little staining after 7-day treatment of Tn4 cells with 5-Aza-dC (Fig. 3C). By contrast, cell extracts of Tn4 cells treated with 5-Aza-dC for 3 days were recognized by PNA and were heavily stained by PNA after the 7-day induction with 5-Aza-dC. The PNA staining was only detected after desialylation by neuraminidase, indicating that sialyl core 1 or sialyl core 1-based O-glycans were present in these treated cells (Fig. 3D). These data demonstrate that inhibition of methylation by 5-Aza-dC in Tn4 cells resumes the transcription of Cosmc, restores T-synthase activity, and consequently allows expression of extended O-glycans.
The Restoration of Transcription of Cosmc by 5-Aza-dC Is Temporary and Reversible
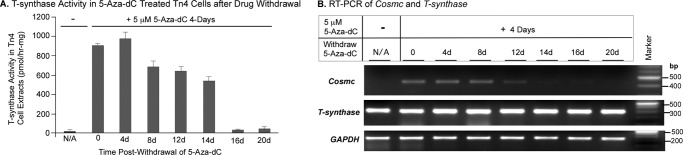
To address whether 5-Aza-dC-mediated restoration of Cosmc transcription was reversible and to establish the length of time for which Cosmc was fully silenced after withdrawal of 5-Aza-dC, we treated Tn4 cells with 5-Aza-dC for 4 days and then withdrew 5-Aza-dC and monitored the T-synthase activity and presence of Cosmc transcripts for 20 days. T-synthase activity decreased upon the removal of 5-Aza-dC and finally disappeared after 16 days (Fig. 4A), whereas the Cosmc transcripts were still decreasing by 8 days (Fig. 4B) and continued declining to an undetectable level after 16 days. Thus, activation of Cosmc resulting from the inhibition of DNA methylation in Tn4 cells was dependent on the presence of 5-Aza-dC and was fully reversible. These data further indicate that the DNA methylation is the cause of Cosmc silencing in Tn4 cells.
FIGURE 4.
Activation of Cosmc in Tn4 cells by treatment with 5-Aza-dC is reversible. A, T-synthase activity in Tn4 cells at different time periods after withdrawal of 5-Aza-dC. Untreated Tn4 cells and Tn4 cells treated for 4 days with 5-Aza-dC as well as treated cells at different periods of time after withdrawal of 5-Aza-dC were collected. Cell extracts were prepared, the concentration of protein in the extracts was measured by the BCA method, and the activity of T-synthase in extracts was assayed in duplicates (mean ± S.D. (error bars)) using a fluorescent substrate, GalNAc-α-(4-MU). The representative result shown is from two independent experiments. B, RT-PCR of Cosmc and T-synthase in Tn4 cells at different times after withdrawal of 5-Aza-dC. The mRNA of the Tn4 cells from A was isolated, and the cDNAs were synthesized using universal primers. After normalizing with GAPDH, PCR for Cosmc (Cosmc-cDNA-F3/-R3) and T-synthase (Tsyn-cDNA-F3/-R3) was carried out, and the products were analyzed on 1.2% agarose gel.
The Core Promoter of Cosmc in Tn4 Cells Is Hypermethylated
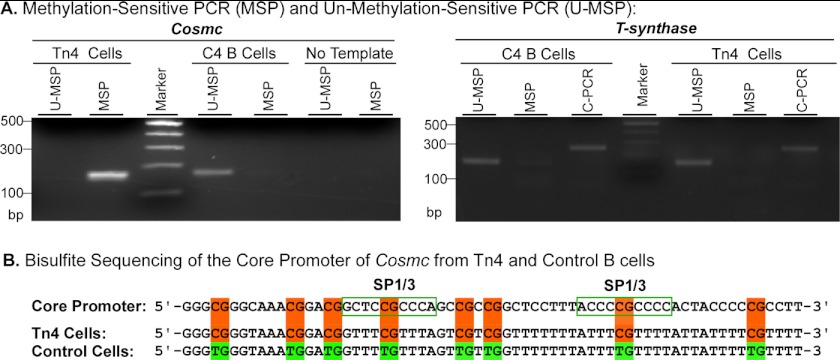
To define the status of methylation of Cosmc, we performed bisulfite sequencing. Sodium bisulfite converts the unmodified cytosine residue in genomic DNA to uracil, whereas the 5-methylated cytosine is nonreactive and remains intact. As a consequence, the unmodified cytosine is changed to thymine in the product of PCR during amplification, and the methylated cytosine remains unchanged after PCR amplification. To amplify the core promoter of Cosmc using the bisulfite-treated genomic DNA as the template, methylation-sensitive PCR (MSP) was carried out using methylation-sensitive primers, which are specific for methylated DNA, and unmethylation-sensitive PCR (U-MSP) was carried out using unmethylated primers specific for unmethylated DNA. As shown in Fig. 5A, the amplification product from Tn4 cells was only found in MSP, whereas no product was detected in the U-MSP. These results indicate that the core promoter of Cosmc is completely methylated in Tn4 cells. In a parallel control study, we examined the core promoter in a control B cell line (C4) immortalized from the peripheral B lymphocytes of a healthy male individual, where the opposite result was observed (i.e. the PCR product was only obtained in U-MSP and not in MSP), indicating that the core promoter of Cosmc in C4 B cells was not methylated. Also in control studies, the T-synthase promoter was only amplified by its U-MSP and not amplified in the MSP in both Tn4 and C4 B cells. It was amplified with a common primer (C-PCR) set with sequences that do not contain the CG dinucleotide (Fig. 5A), because only the cytosine in the CG dinucleotide could be methylated; with the C-PCR, the DNA can be amplified regardless of methylation status in the core region. Thus, the core promoter of T-synthase was not methylated in either Tn4 or C4 B cells, which was consistent with earlier findings that T-synthase itself was not silenced in Tn4 cells (Fig. 5A).
FIGURE 5.
The promoter of Cosmc in Tn4 cells is hypermethylated. A, methylation-sensitive PCR. The genomic DNA from Tn4 cells and C4 B cells was treated with sodium bisulfite. MSP and U-MSP covering the core promoters of human Cosmc and T-synthase were carried out. C-PCR for T-synthase was also performed. The PCR products were analyzed on 2.0% agarose gel. B, bisulfite sequencing of the Cosmc promoter. The MSP product from Tn4 cells and U-MSP product from C4 B cells were purified and subjected to sequencing. The portion of the core promoter sequence of Cosmc is shown. The CG dinucleotides are highlighted, and the two SP1/3 binding sites are boxed.
To further confirm that the promoter of Cosmc was hypermethylated, we sequenced the PCR products amplified by MSP from Tn4 cells and PCR product amplified with U-MSP from C4 B cells. All CG dinucleotides in the Cosmc core promoter, including two critical SP1/SP3 binding sites from Tn4 cells, remained unchanged, demonstrating that they were uniformly methylated (Fig. 5B), whereas other C residues were not methylated, and were efficiently converted to U (T in the PCR product) by the bisulfite treatment, indicating the efficient bisulfite conversion of the genomic DNA. By contrast, the C residues, including the one in CG dinucleotides in the core promoter of Cosmc from control C4 B cells, were completely converted to T (Fig. 5B). These results demonstrate that the core promoter of Cosmc from C4 B cells is not methylated. Thus, the core promoter of Cosmc, but not T-synthase, in Tn4 cells is hypermethylated. Our ongoing studies further suggest, as might be expected from these results, that the GC-rich DNA sequences in the core promoter of Cosmc are essential to maintain its promoter activity. This is the first example of a complete loss of Cosmc expression arising from epigenetic silencing through hypermethylation of the gene.
Other Glycosylation-relevant Genes on X Chromosome, OGT and PIG-A, Are Functional in Tn4 Cells
To test whether hypermethylation silencing of Cosmc is relatively specific to that locus, we examined the methylation status of two other “glycogenes” that are also on the X chromosome: OGT (Xq13), which encodes the O-linked β-O-acetylglucosaminyltransferase that adds O-GlcNAc to cytoplasmic and nuclear proteins (25), and PIG-A (Xp22), which encodes the phosphatidylinositol N-acetylglucosaminyltransferase subunit A that initiates glycosylphosphatidylinositol anchor synthesis and is often somatically mutated in the hematopoietic cells in patients with paroxysmal nocturnal hemoglobinuria (PNH) disease (26). Western blotting showed that OGT protein was expressed in Tn4 cells, indicating that OGT was active, which was also observed for several other cell lines tested (Fig. 6A). These results are consistent with the earlier observation that expression of OGT is essential for cell survival (25). CD59, a glycosylphosphatidylinositol-anchored glycoprotein was expressed normally on the Tn4 cell surface, as shown by flow cytometry after staining with FITC-labeled anti-human CD59 antibody (Fig. 6B). As a control, the PNH-N B cell line from a patient with PNH disease, which contains a defective PIG-A gene, lacked expression of CD59 (Fig. 6B), thus demonstrating that PIG-A is expressed and fully functional in Tn4 cells. Taken together, these results demonstrate that in contrast to Cosmc, these two X-linked glycogenes, OGT and PIG-A, are not silenced in Tn4 cells. Thus, the observed dysregulated epigenetic hypermethylation in Tn4 cells is relatively specific to the Cosmc locus.
FIGURE 6.
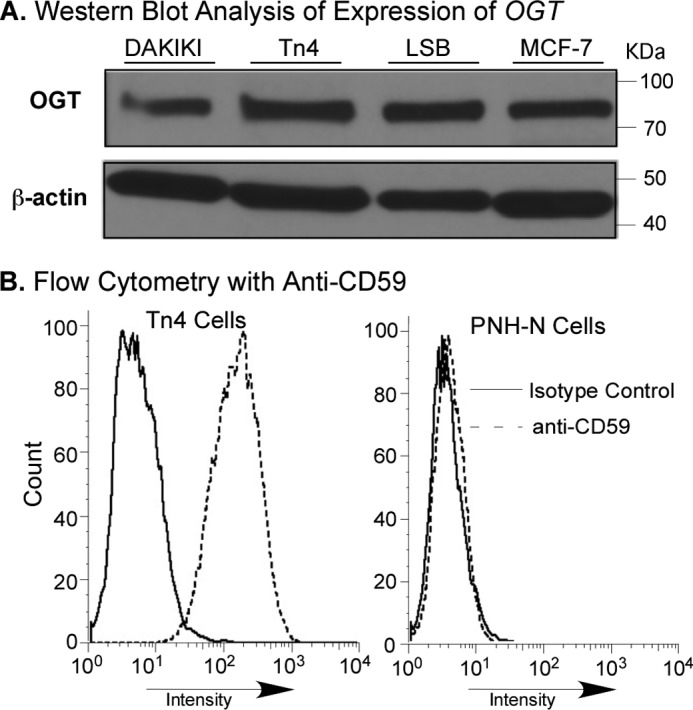
OGT and PIG-A are present or functional in Tn4 Cells. A, analysis of OGT on Western blot. The cytosolic fractions from Tn4, DAKIKI, LSB, and MCF-7 cells were analyzed by Western blot with rabbit anti-human OGT antibodies (IgG). B, flow cytometric analysis of CD59 for function of PIG-A. Tn4 and PNH-N B cells were stained with FITC-labeled anti-human CD59 and isotype control and analyzed by flow cytometry.
DNA Methyltransferases Are Not Significantly Up-regulated in Tn4 Cells
Thus far, all of the results demonstrate that hypermethylation is the mechanism underlying the Cosmc silencing in Tn4 cells. DNA methylation requires the de novo methylation activity of DNA methyltransferase-3a and -3b (DNMT-3a and DNMT-3b) as well as the regulator, DNMT-3L, whereas the maintenance of DNA methylation requires DNA methyltransferase-1 (DNMT-1). DNMT-2 lacks significant DNA methylation activity but may methylate RNA (27). To test whether hypermethylation in Tn4 cells was correlated with up-regulation in expression of these DNMTs or a particular DNMT, the nuclear extracts from Tn4 cells and control cell lines were analyzed by Western blot. DNMT-1, -2, -3a, -3b, and -3L were all expressed in Tn4 cells, whereas only DNMT-1, -3b, and -3L and a low level of DNMT-3a were expressed in DAKIKI cells (Fig. 7A). To investigate further whether expression of DNMT-3a, which is expressed at a high level in Tn4 cells and much less in DAKIKI cells, and DNMT-2, which is expressed in Tn4 cells but not in DAKIKI cells, were responsible for the aberrant methylation of Cosmc, we surveyed other cell lines (human colorectal carcinoma LSB and breast cancer cell MCF-7 lines), which have normal expression of Cosmc. Whereas LSB cells had an expression of DNMTs similar to that of DAKIKI cells, MCF-7 cells expressed all of the DNMTs in a pattern similar to that in Tn4 cells (Fig. 7A). More importantly, in contrast to Tn4 cells but similar to DAKIKI and LSB cells, MCF-7 cells had a robust T-synthase activity (Fig. 7B). These results suggest that solo expression of DNMTs does not correlate with the hypermethylation of Cosmc in Tn4 cells. We noticed that the expression of DNMT-3b in Tn4 cells was slightly higher than in the other cell lines, but we are unsure if this is important because expression levels of DNMT-3b in other cell lines do not correlate with altered Cosmc expression. Together, these results show that there is no apparent correlation of expression of a particular DNMT with the hypermethylation of Cosmc in Tn4 cells.
FIGURE 7.
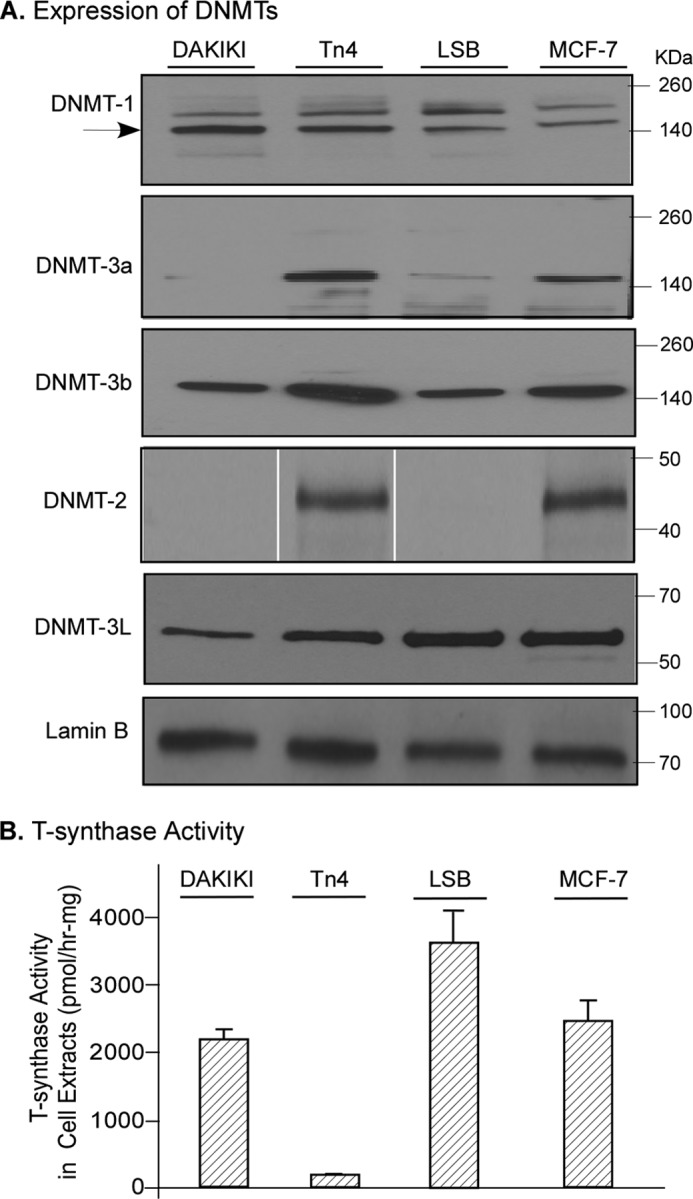
Expression of DNMTs and T-synthase activity in Tn4 and other cell lines. A, Western blot of DNMTs in different cell lines. The nuclear extracts prepared from Tn4, DAKIKI, LSB, and MCF-7 cells were analyzed by Western blot with mouse anti-human DNMT-1, -2, -3a, -3b, and -3L antibodies (IgG). The lanes of Western blot (WB) for DNMT-2 were run on the same gel but were noncontiguous. The loading of SDS-PAGE was normalized with lamin B. B. T-synthase activity. The cytosolic fractions from Tn4, DAKIKI, LSB, and MCF-7 cells were measured for protein concentration and for T-synthase activity in triplicates using a fluorescent substrate, GalNAc-α-(4-MU). The results are from two independent experiments (mean ± S.D. (error bars)).
Increase of T-synthase Activity in Tn4 Cells Responding to 5-Aza-dC Is in a Dose- and Time-dependent Manner but Does Not Correlate with Inhibition of a Particular DNMT
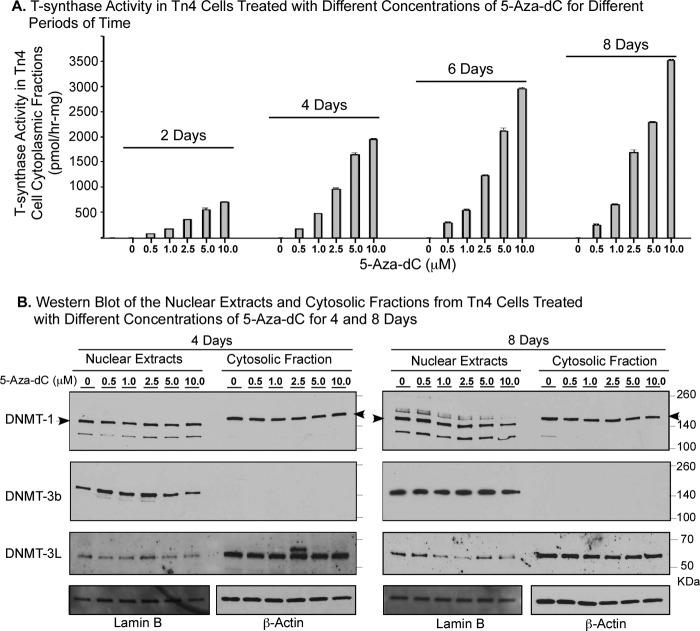
In regard to the mechanism for 5-Aza-dC inhibition of DNA methylation, studies have suggested that 5-Aza-dC could selectively target the DNMT-1 to degradation by 26S proteasome (28) or trap the DNMTs on the DNA after its incorporation into the replicated DNA (23, 29). To investigate the insight that 5-Aza-dC inhibited DNA methylation, leading to the restoration of the transcription of Cosmc and T-synthase activity in Tn4 cells, we treated Tn4 cells with 0, 0.5, 1.0, 2.5, 5.0, and 10.0 μm 5-Aza-dC for 2, 4, 6, and 8 days, respectively, and then monitored the T-synthase activity in the cytosolic fractions (Fig. 8A) and examined the DNMT-1, -3b, and -3L in both nuclear extracts and cytosolic fractions in the treated Tn4 cells (Fig. 8B). In responding to 5-Aza-dC, the T-synthase activity steadily increased in a dose-dependent manner at each time point; the activity was also increased along with the length of time of treatment at the same dose of 5-Aza-dC, although the most significant changes were seen within 4–6 days. These results further confirm the conclusion that Cosmc in Tn4 cells is silenced through hypermethylation. Interestingly, as shown in Fig. 8B, the protein levels of DNMT-1, -3b, and -3L, the major DNA methyltransferases in both nucleus and cytosol of Tn4 cells, were not significantly changed at any concentration of 5-Aza-dC and at any time point. These data indicate that inhibition of methylation of Cosmc in Tn4 cells by 5-Aza-dC is not directly related either to promoting degradation or to trapping of DNMTs but rather to the mechanism by which direct incorporation of 5-Aza-dC into the newly replicated DNA leads to the blocking of methylation. We noticed that the molecular mass of the major DNMT-1 in the nuclear extracts migrated as a ∼160-kDa protein, whereas it appeared as a ∼200-kDa protein in the cytosolic fractions, suggesting that the cytosolic DNMT-1 carries significant modifications (30). Whereas DNMT-1 and -3b were mainly in nuclear extracts, the major fraction of DNMT-3L was seen in the cytosolic fractions. The nature of these differences is unclear. Taken together, the unchanged protein level of DNMTs in 5-Aza-dC-treated and control Tn4 cells further indicates that no particular DNMT in Tn4 cells is associated with the aberrant methylation of the Cosmc locus.
FIGURE 8.
Increase of T-synthase activity in Tn4 cells responding to 5-Aza-dC is in a dose- and time-dependent manner but does not correlate with inhibition of a particular DNMT. Tn4 cells were treated with 0, 0. 5, 1.0, 2.5, 5.0, and 10.0 μm 5-Aza-dC for 2, 4, 6, and 8 days, respectively, and the nuclear extracts and cytosolic fractions were prepared. A, the T-synthase activity in the cytosolic fractions of Tn4 cells was assayed in triplicates using a fluorescent substrate GalNAc-α-(4-MU). The results are from two independent experiments (mean ± S.D. (error bars)). B, the protein levels of DNMT-1, -3b, and -3L in both nuclear extracts and cytosolic fractions were analyzed using Western blot. Lamin B was used as the loading control for nuclear extracts, and β-actin was used as the loading control of cytosolic fractions.
DISCUSSION
Our studies show that Tn4 cells, which were originally derived from the leukocytes in blood of a male patient with Tn-positive lymphocytes, are IgA2-producing B cells that lack transcripts for Cosmc due to epigenetic silencing of Cosmc through hypermethylation of its promoter. The treatment of Tn4 cells by the DNA methylation inhibitor 5-Aza-dC causes restoration of Cosmc transcription and consequent restoration of T-synthase activity. Such results also confirm that both Cosmc and T-synthase are genetically intact in Tn4 cells. This is the first example of a ubiquitously expressed glycomics-relevant gene to be completely and reversibly silenced through an epigenetic dysregulation. This finding could shed light on our full understanding of abnormal expression of the Tn and sialyl Tn antigens in many human diseases through non-heritable, acquired mechanisms.
It is unusual that Cosmc, but not other X-linked glycomics-relevant genes or glycogenes tested, is silenced through hypermethylation in Tn4 cells. Despite numerous examples of hypermethylation-associated gene silencing events identified in human tumor cells and other pathological situations (31, 32), it is not known why particular CpG islands are susceptible to aberrant methylation. Epigenetic regulation has a variety of important functions in mammals, including control of gene expression, cellular differentiation and development, preservation of chromosomal integrity, parental imprinting, and X chromosome inactivation (33). DNA methylation is carried out by the DNMTs, including DNMT-1, which functions as a maintenance enzyme, and DNMT-2, which may have some activity toward RNA, whereas DNMT-3a, -3b, and -3L methylate DNA de novo (33, 34). It is well documented that dysregulation of epigenetic pathways, such as methylation, including hypo- and hypermethylation, is one of the important mechanisms for oncogene activation or tumor suppressor gene suppression in human tumors and perhaps as important as genetic mutations. Three models for aberrant methylation have been proposed. One model is up-regulation or overexpression of DNMT-1 or -3b. In our case, however, we observed no significant change in the expression of the major DNMTs nor apparent correlation between the expression of DNMTs and aberrant methylation of the Cosmc promoter in Tn4 cells. The second model is the loss of protection of promoter by the DNA-binding proteins or transcription factors, such as SP1 for the binding sequences. This is not the case for Cosmc hypermethylation in Tn4 cells, because SP1 and SP3 are expressed in Tn4 cells. Furthermore, Cosmc can be transcribed in the presence of 5-Aza-dC, further suggesting that these transcription factors are present and functional in Tn4 cells. The third model is the loss of protection by the hypermethylation of Alu elements surrounding the promoter. Cosmc has clusters of Alu elements surrounding its promoter and the coding region (ORF), and the methylation status of these Alu elements in genes from Tn4 cells is not yet known. Evidence that two other glycogenes, OGT (25, 35) and PIG-A (36), which are also X-linked, are not hypermethylated in Tn4 cells strongly suggests that the propensity for the CpG islands to become aberrantly methylated comes from the intrinsic characteristics of the promoter sequence and structure of Cosmc, such as the Alu clusters. Presumably, the methylated Cosmc promoter is recognized and bound by methyl CpG-binding domain-containing proteins, which then recruit the histone deacetylase-containing transcription repression complex to shut down transcription of Cosmc (23, 37). Interestingly, the DNA methylation inhibitor 5-Aza-dC can restore transcription of Cosmc and consequent expression of active T-synthase in Tn4 cells in a dose- and time-dependent manner but not through a specific inhibition of a particular DNMT (Fig. 8). Usually, 5-Aza-cytidine inhibits the methylation of DNA through targeting DNMT-1 selectively to proteasome degradation (28) and/or trapping or sequestering the DNMTs after being incorporated into the replicated DNA (23, 29). In our experiment, however, 5-Aza-dC treatment did not cause any significant change of any level of DNMT-1, -3b, and -3L, three major DNMTs expressed in Tn4 cells. This strongly suggests that the third mechanism of incorporation of 5-Aza-dC into the DNA leads to blocking of methylation, resulting in an inability to silence the Cosmc promoter in Tn4 cells, thus explaining the restoration of Cosmc transcription by 5-Aza-dC treatment of Tn4 cells. Interestingly, this restoration is reversible, strongly suggesting that the promoter of Cosmc is susceptible to de novo methylation, presumably by DNMT-3a or -3b regulated by DNMT-3L, which is then maintained by DNMT-1. This also suggests that there might be other factors protecting the Cosmc promoter from methylation by DNMTs, although DNMTs play a direct role in the hypermethylation. Identification of these factors in future studies should aid in understanding the regulation of epigenetic pathways in both biology and cancer.
Expression of glycosyltransferases is likely to be regulated through both transcription factors and epigenetic mechanisms. Emerging evidence has shown that aberrant methylation on glycosyltransferase genes is associated with some changes in the glycome in human tumors or tumor cell lines, as seen for the ST3Gal6 in HCT15 colorectal carcinoma cells (38) and the 3-OST-2 in human breast, colon, lung, and pancreatic cancers (39) and 3-O-sulfotransferases in H-EMC-SS chondrosarcoma cells (40). In addition, hypermethylation of several glycosyltransferases resulted in silencing of these enzymes in normal gastrointestinal mucosa and in gastric and colorectal cancer cells, leading to aberrant glycosylation and expression of tumor-associated carbohydrate antigens (41). Similar findings were reported for ABO blood group glycosyltransferase silencing in oral squamous cell carcinoma (42). However, our findings are novel in this regard because Cosmc and T-synthase are ubiquitous and coordinately expressed genes in all animal cells. Although X-linked Cosmc has one inactivated allele in female somatic cells mainly through methylation, all normal somatic cells have a single constitutively active gene. Thus, Tn4 cells represent a novel example of a ubiquitously expressed X-linked gene in a male cell that is completely silenced.
Our results could provide more insight into the mechanisms that lead to dysfunctional Cosmc, which may aid us in fully understanding the Tn/STn antigen expression in human tumors. Tn and STn are recognized as the most common tumor antigens (6, 16, 43). One mechanism for Tn antigen expression was found to be genetic mutations of Cosmc (6, 19, 20), or loss-of-heterozygosity, in Cosmc (19). Our finding that Cosmc is epigenetically silenced in Tn4 cells may be related to studies of Tn antigen expression in human leukemic cells, such as myeloma (44), lymphoma (45), and chronic lymphocytic leukemia (46). Aller et al. (46) reported that the B cells from six of six patients with B-cell chronic lymphocytic leukemia were Tn-positive, whereas the blood cells from five normal individuals were negative for Tn expression. Yoo et al. (47) examined the ORF of Cosmc in human breast and colon cancer samples using PCR-SSCP (single-stranded conformation polymorphism) and did not detect mutations, but they did not determine whether there were mutations outside the Cosmc ORF or an entire gene deletion, and no information about the methylation status of the Cosmc promoter region was provided.
Our findings may also be related to early studies on Tn syndrome, which is a rare blood disorder characterized by the Tn antigen on a subpopulation of blood cells of all lineages (14). We reported that a major genetic basis for Tn syndrome is acquired genetic mutations in the ORF of X-linked Cosmc (17, 18). However, an earlier study on the mechanism of loss of activity of T-synthase in immortalized T cells from one male and one female patient with Tn syndrome reported that treatment of the cells with 5-aza-cytidine restored some T-synthase activity in these cells, with the hypothesis that the T-synthase might be suppressed by the methylation in those Tn syndrome patients (21, 22). Thus, it is very possible that those immortalized T cells from the male patient with Tn syndrome also contained the same silencing of the Cosmc gene as we observed in Tn4 cells. In the Tn(+) T-cells from the female patient, the mechanism underlying the dysfunctional Cosmc could be either a nonsense mutation in the ORF of the active allele of Cosmc or silencing of both alleles by methylation; in either case, inhibition of DNA methylation would reactivate the transcription of Cosmc to restore the T-synthase activity if the mutation on the active allele of Cosmc is not dominant negative.
We found that Tn4 cells express IgA2, but Tn4 cells could also be very useful in studies on O-glycosylation of human IgA1. Aberrant O-glycosylation (e.g. Tn and STn antigens on the hinge region of IgA1) is recognized as the major pathogenesis of IgAN (15, 48). Several studies suggested that the transcript levels of Cosmc and/or T-synthase are reduced in the B cells of patients with IgAN (48–54). However, the mechanism for how Cosmc and T-synthase are suppressed is unknown. Most recently, Qin et al. (55) reported that hypermethylation in peripheral blood B cells might play some role in the down-regulation of Cosmc in IgAN patients, but no direct evidence was provided in this regard, and the authors did not fully take into account the fact that Cosmc is X-linked and that individuals are functionally hemizygous at this locus. The Tn4 cells could be useful in future studies in this regard because the transcription of Cosmc in Tn4 cells can be turned on and off or even turned on at adjustable levels by treatment with DNA methylation inhibitor at different concentrations as shown in Fig. 8.
In summary, we have demonstrated that in Tn4 cells, which are immortalized B cells from a patient with Tn(+) leukocytes in his blood, Cosmc is silenced because of hypermethylation of the gene promoter. These observations, together with those of our previous studies, demonstrate that both genetic mutations and epigenetic silencing can generate a dysfunctional Cosmc and lead to an inactive T-synthase and consequent expression of the truncated O-glycans Tn and STn antigens in pathologic conditions. A fuller understanding of these regulatory pathways may shed light on expression of the Tn and STn antigens in human tumors and IgAN.
Acknowledgments
We thank Dr. David Araten at New York University School of Medicine for kindly providing the PNH-N B cell line. We thank Drs. Jamie Heimburg-Molinaro and David F. Smith for help in editing the manuscript. We thank Dr. Xiaodong Cheng for helpful suggestions regarding this work.
This work was supported, in whole or in part, by National Institutes of Health Grant R01DK80876 (to T. J.).
- T antigen
- Galβ1–3GalNAcα1-Ser/Thr
- Tn antigen
- GalNAcα1-Ser/Thr
- T-synthase
- core 1 β3-galatosyltransferase (UDP-Gal:N-acetylgalactosaminyl-α1-Ser/Thr β3-galactosyltransferase)
- 5-Aza-dC
- 5-Aza-2′-deoxycytidine
- 4-MU
- 4-methylumbelliferone
- DNMT
- DNA methyltransferase
- OGT
- O-linked N-acetylglucosaminyltransferase (UDP-N-acetylglucosamine:polypeptide N-acetylglucosaminyltransferase)
- PIG-A
- phosphatidylinositolglycan class A (phosphatidylinositol N-acetylglucosaminyltransferase subunit A)
- MSP
- methylation-sensitive PCR
- U-MSP
- unmethylation-sensitive PCR
- C-PCR
- PCR using the common primer set
- IgAN
- IgA nephropathy
- EBV
- Epstein-Barr virus
- HAA
- Helix aspersa agglutinin
- PNA
- peanut agglutinin
- PNH
- paroxysmal nocturnal hemoglobinuria.
REFERENCES
- 1. Homeister J. W., Thall A. D., Petryniak B., Malý P., Rogers C. E., Smith P. L., Kelly R. J., Gersten K. M., Askari S. W., Cheng G., Smithson G., Marks R. M., Misra A. K., Hindsgaul O., von Andrian U. H., Lowe J. B. (2001) The α(1,3)fucosyltransferases FucT-IV and FucT-VII exert collaborative control over selectin-dependent leukocyte recruitment and lymphocyte homing. Immunity 15, 115–126 [DOI] [PubMed] [Google Scholar]
- 2. Yeh J. C., Hiraoka N., Petryniak B., Nakayama J., Ellies L. G., Rabuka D., Hindsgaul O., Marth J. D., Lowe J. B., Fukuda M. (2001) Novel sulfated lymphocyte homing receptors and their control by a Core1 extension β1,3-N-acetylglucosaminyltransferase. Cell 105, 957–969 [DOI] [PubMed] [Google Scholar]
- 3. McEver R. P., Moore K. L., Cummings R. D. (1995) Leukocyte trafficking mediated by selectin-carbohydrate interactions. J. Biol. Chem. 270, 11025–11028 [DOI] [PubMed] [Google Scholar]
- 4. Xia L., Ju T., Westmuckett A., An G., Ivanciu L., McDaniel J. M., Lupu F., Cummings R. D., McEver R. P. (2004) Defective angiogenesis and fatal embryonic hemorrhage in mice lacking core 1-derived O-glycans. J. Cell Biol. 164, 451–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fu J., Gerhardt H., McDaniel J. M., Xia B., Liu X., Ivanciu L., Ny A., Hermans K., Silasi-Mansat R., McGee S., Nye E., Ju T., Ramirez M. I., Carmeliet P., Cummings R. D., Lupu F., Xia L. (2008) Endothelial cell O-glycan deficiency causes blood/lymphatic misconnections and consequent fatty liver disease in mice. J. Clin. Invest. 118, 3725–3737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ju T., Otto V. I., Cummings R. D. (2011) The Tn antigen-structural simplicity and biological complexity. Angew. Chem. Int. Ed. Engl. 50, 1770–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang Y., Jobe S. M., Ding X., Choo H., Archer D. R., Mi R., Ju T., Cummings R. D. (2012) Platelet biogenesis and functions require correct protein O-glycosylation. Proc. Natl. Acad. Sci. U.S.A. 109, 16143–16148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ju T., Brewer K., D'Souza A., Cummings R. D., Canfield W. M. (2002) Cloning and expression of human core 1 β1,3-galactosyltransferase. J. Biol. Chem. 277, 178–186 [DOI] [PubMed] [Google Scholar]
- 9. Ju T., Cummings R. D. (2002) A unique molecular chaperone Cosmc required for activity of the mammalian core 1 β3-galactosyltransferase. Proc. Natl. Acad. Sci. U.S.A. 99, 16613–16618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ju T., Aryal R. P., Stowell C. J., Cummings R. D. (2008) Regulation of protein O-glycosylation by the endoplasmic reticulum-localized molecular chaperone Cosmc. J. Cell Biol. 182, 531–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Aryal R. P., Ju T., Cummings R. D. (2010) The endoplasmic reticulum chaperone Cosmc directly promotes in vitro folding of T-synthase. J. Biol. Chem. 285, 2456–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aryal R. P., Ju T., Cummings R. D. (2012) Tight complex formation between Cosmc chaperone and its specific client non-native T-synthase leads to enzyme activity and client-driven dissociation. J. Biol. Chem. 287, 15317–15329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang Y., Ju T., Ding X., Xia B., Wang W., Xia L., He M., Cummings R. D. (2010) Cosmc is an essential chaperone for correct protein O-glycosylation. Proc. Natl. Acad. Sci. U.S.A. 107, 9228–9233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Berger E. G. (1999) Tn-syndrome. Biochim. Biophys. Acta 1455, 255–268 [DOI] [PubMed] [Google Scholar]
- 15. Mestecky J., Tomana M., Moldoveanu Z., Julian B. A., Suzuki H., Matousovic K., Renfrow M. B., Novak L., Wyatt R. J., Novak J. (2008) Role of aberrant glycosylation of IgA1 molecules in the pathogenesis of IgA nephropathy. Kidney Blood Press. Res. 31, 29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Springer G. F. (1984) T and Tn, general carcinoma autoantigens. Science 224, 1198–1206 [DOI] [PubMed] [Google Scholar]
- 17. Crew V. K., Singleton B. K., Green C., Parsons S. F., Daniels G., Anstee D. J. (2008) New mutations in C1GALT1C1 in individuals with Tn positive phenotype. Br. J. Haematol. 142, 657–667 [DOI] [PubMed] [Google Scholar]
- 18. Ju T., Cummings R. D. (2005) Protein glycosylation. Chaperone mutation in Tn syndrome. Nature 437, 1252. [DOI] [PubMed] [Google Scholar]
- 19. Ju T., Lanneau G. S., Gautam T., Wang Y., Xia B., Stowell S. R., Willard M. T., Wang W., Xia J. Y., Zuna R. E., Laszik Z., Benbrook D. M., Hanigan M. H., Cummings R. D. (2008) Human tumor antigens Tn and sialyl Tn arise from mutations in Cosmc. Cancer Res. 68, 1636–1646 [DOI] [PubMed] [Google Scholar]
- 20. Schietinger A., Philip M., Yoshida B. A., Azadi P., Liu H., Meredith S. C., Schreiber H. (2006) A mutant chaperone converts a wild-type protein into a tumor-specific antigen. Science 314, 304–308 [DOI] [PubMed] [Google Scholar]
- 21. Thurnher M., Rusconi S., Berger E. G. (1993) Persistent repression of a functional allele can be responsible for galactosyltransferase deficiency in Tn syndrome. J. Clin. Invest. 91, 2103–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Felner K. M., Dinter A., Cartron J. P., Berger E. G. (1998) Repressed β-1,3-galactosyltransferase in the Tn syndrome. Biochim. Biophys. Acta 1406, 115–125 [DOI] [PubMed] [Google Scholar]
- 23. Patra S. K., Bettuzzi S. (2009) Epigenetic DNA-(cytosine-5-carbon) modifications. 5-Aza-2′-deoxycytidine and DNA-demethylation. Biochemistry 74, 613–619 [DOI] [PubMed] [Google Scholar]
- 24. Ju T., Xia B., Aryal R. P., Wang W., Wang Y., Ding X., Mi R., He M., Cummings R. D. (2011) A novel fluorescent assay for T-synthase activity. Glycobiology 21, 352–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shafi R., Iyer S. P., Ellies L. G., O'Donnell N., Marek K. W., Chui D., Hart G. W., Marth J. D. (2000) The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. U.S.A. 97, 5735–5739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brodsky R. A. (2008) Advances in the diagnosis and therapy of paroxysmal nocturnal hemoglobinuria. Blood Rev. 22, 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jin B., Li Y., Robertson K. D. (2011) DNA methylation. Superior or subordinate in the epigenetic hierarchy? Genes Cancer 2, 607–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ghoshal K., Datta J., Majumder S., Bai S., Kutay H., Motiwala T., Jacob S. T. (2005) 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol. Cell. Biol. 25, 4727–4741 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29. Christman J. K. (2002) 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation. Mechanistic studies and their implications for cancer therapy. Oncogene 21, 5483–5495 [DOI] [PubMed] [Google Scholar]
- 30. Bronner C. (2011) Control of DNMT1 abundance in epigenetic inheritance by acetylation, ubiquitylation, and the histone code. Sci. Signal. 4, pe3. [DOI] [PubMed] [Google Scholar]
- 31. Nai H. S., Lau Q. C. (2012) Advent of the cancer methylome. Comb. Chem. High Throughput Screen. 15, 216–220 [DOI] [PubMed] [Google Scholar]
- 32. Shenker N., Flanagan J. M. (2012) Intragenic DNA methylation. Implications of this epigenetic mechanism for cancer research. Br. J. Cancer 106, 248–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hermann A., Gowher H., Jeltsch A. (2004) Biochemistry and biology of mammalian DNA methyltransferases. Cell. Mol. Life Sci. 61, 2571–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jurkowska R. Z., Jurkowski T. P., Jeltsch A. (2011) Structure and function of mammalian DNA methyltransferases. Chembiochem 12, 206–222 [DOI] [PubMed] [Google Scholar]
- 35. O'Donnell N., Zachara N. E., Hart G. W., Marth J. D. (2004) Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol. Cell. Biol. 24, 1680–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Takeda J., Miyata T., Kawagoe K., Iida Y., Endo Y., Fujita T., Takahashi M., Kitani T., Kinoshita T. (1993) Deficiency of the GPI anchor caused by a somatic mutation of the PIG-A gene in paroxysmal nocturnal hemoglobinuria. Cell 73, 703–711 [DOI] [PubMed] [Google Scholar]
- 37. Bogdanović O., Veenstra G. J. (2009) DNA methylation and methyl-CpG binding proteins. Developmental requirements and function. Chromosoma 118, 549–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chachadi V. B., Cheng H., Klinkebiel D., Christman J. K., Cheng P. W. (2011) 5-Aza-2′-deoxycytidine increases sialyl Lewis X on MUC1 by stimulating β-galactoside:α2,3-sialyltransferase 6 gene. Int. J. Biochem. Cell Biol. 43, 586–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miyamoto K., Asada K., Fukutomi T., Okochi E., Yagi Y., Hasegawa T., Asahara T., Sugimura T., Ushijima T. (2003) Methylation-associated silencing of heparan sulfate d-glucosaminyl 3-O-sulfotransferase-2 (3-OST-2) in human breast, colon, lung and pancreatic cancers. Oncogene 22, 274–280 [DOI] [PubMed] [Google Scholar]
- 40. Bui C., Ouzzine M., Talhaoui I., Sharp S., Prydz K., Coughtrie M. W., Fournel-Gigleux S. (2010) Epigenetics. Methylation-associated repression of heparan sulfate 3-O-sulfotransferase gene expression contributes to the invasive phenotype of H-EMC-SS chondrosarcoma cells. FASEB J. 24, 436–450 [DOI] [PubMed] [Google Scholar]
- 41. Kawamura Y. I., Toyota M., Kawashima R., Hagiwara T., Suzuki H., Imai K., Shinomura Y., Tokino T., Kannagi R., Dohi T. (2008) DNA hypermethylation contributes to incomplete synthesis of carbohydrate determinants in gastrointestinal cancer. Gastroenterology 135, 142–151.e3 [DOI] [PubMed] [Google Scholar]
- 42. Gao S., Worm J., Guldberg P., Eiberg H., Krogdahl A., Liu C. J., Reibel J., Dabelsteen E. (2004) Genetic and epigenetic alterations of the blood group ABO gene in oral squamous cell carcinoma. Int. J. Cancer 109, 230–237 [DOI] [PubMed] [Google Scholar]
- 43. Desai P. R. (2000) Immunoreactive T and Tn antigens in malignancy. Role in carcinoma diagnosis, prognosis, and immunotherapy. Transfus. Med. Rev. 14, 312–325 [DOI] [PubMed] [Google Scholar]
- 44. Roxby D. J., Pfeiffer M. B., Morley A. A., Kirkland M. A. (1992) Expression of the Tn antigen in myelodysplasia, lymphoma, and leukemia. Transfusion 32, 834–838 [DOI] [PubMed] [Google Scholar]
- 45. Wallner M., Waldner R. (1985) Tn polyagglutinability occurring in a patient with B cell lymphoma. Blut 51, 355–360 [DOI] [PubMed] [Google Scholar]
- 46. Aller C. T., Kucuk O., Springer G. F., Gilman-Sachs A. (1996) Flow cytometric analysis of T and Tn epitopes on chronic lymphocytic leukemia cells. Am. J. Hematol. 52, 29–38 [DOI] [PubMed] [Google Scholar]
- 47. Yoo N. J., Kim M. S., Lee S. H. (2008) Absence of COSMC gene mutations in breast and colorectal carcinomas. APMIS 116, 154–155 [DOI] [PubMed] [Google Scholar]
- 48. Suzuki H., Moldoveanu Z., Hall S., Brown R., Vu H. L., Novak L., Julian B. A., Tomana M., Wyatt R. J., Edberg J. C., Alarcón G. S., Kimberly R. P., Tomino Y., Mestecky J., Novak J. (2008) IgA1-secreting cell lines from patients with IgA nephropathy produce aberrantly glycosylated IgA1. J. Clin. Invest. 118, 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Qin W., Zhong X., Fan J. M., Zhang Y. J., Liu X. R., Ma X. Y. (2008) External suppression causes the low expression of the Cosmc gene in IgA nephropathy. Nephrol. Dial. Transplant. 23, 1608–1614 [DOI] [PubMed] [Google Scholar]
- 50. Qin W., Zhou Q., Yang L. C., Li Z., Su B. H., Luo H., Fan J. M. (2005) Peripheral B lymphocyte β1,3-galactosyltransferase and chaperone expression in immunoglobulin A nephropathy. J. Intern. Med. 258, 467–477 [DOI] [PubMed] [Google Scholar]
- 51. Inoue T., Sugiyama H., Hiki Y., Takiue K., Morinaga H., Kitagawa M., Maeshima Y., Fukushima K., Nishizaki K., Akagi H., Narimatsu Y., Narimatsu H., Makino H. (2010) Differential expression of glycogenes in tonsillar B lymphocytes in association with proteinuria and renal dysfunction in IgA nephropathy. Clin. Immunol. 136, 447–455 [DOI] [PubMed] [Google Scholar]
- 52. Inoue T., Sugiyama H., Kitagawa M., Takiue K., Morinaga H., Kikumoto Y., Maeshima Y., Fukushima K., Nishizaki K., Akagi H., Hiki Y., Makino H. (2011) Abnormalities of glycogenes in tonsillar lymphocytes in IgA nephropathy. Adv. Otorhinolaryngol. 72, 71–74 [DOI] [PubMed] [Google Scholar]
- 53. Xie L. S., Qin W., Fan J. M., Huang J., Xie X. S., Li Z. (2010) The role of C1GALT1C1 in lipopolysaccharide-induced IgA1 aberrant O-glycosylation in IgA nephropathy. Clin. Invest. Med. 33, E5–E13 [DOI] [PubMed] [Google Scholar]
- 54. Yamada K., Kobayashi N., Ikeda T., Suzuki Y., Tsuge T., Horikoshi S., Emancipator S. N., Tomino Y. (2010) Down-regulation of core 1 β1,3-galactosyltransferase and Cosmc by Th2 cytokine alters O-glycosylation of IgA1. Nephrol. Dial. Transplant. 25, 3890–3897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Qin W., Zhong X., Fan J. M., Liu X. R., Li Z., Ma X. Y. (2011) [Effect of methylation modification on the expression of Cosmc gene in peripheral B lymphocyte of IgA nephropathy patients]. Sichuan Da Xue Xue Bao Yi Xue Ban 42, 762–765 [PubMed] [Google Scholar]