Abstract
We studied transcription initiation in the mitochondria of higher plants, with particular respect to promoter structures. Conserved elements of these promoters have been successfully identified by in vitro transcription systems in different species, whereas the involved protein components are still unknown. Proteins binding to double-stranded oligonucleotides representing different parts of the pea (Pisum sativum) mitochondrial atp9 were analyzed by denaturation-renaturation chromatography and mobility-shift experiments. Two DNA-protein complexes were detected, which appeared to be sequence specific in competition experiments. Purification by hydroxyapatite, phosphocellulose, and reversed-phase high-pressure liquid chromatography separated two polypeptides with apparent molecular masses of 32 and 44 kD. Both proteins bound to conserved structures of the pea atp9 and the heterologous Oenothera berteriana atp1 promoters and to sequences just upstream. Possible functions of these proteins in mitochondrial promoter recognition are discussed.
In many genetic systems the level of expression of a gene is controlled by modulating the efficiency of transcription through changing the concentrations and activities of transcription initiation factors.
Proteins involved in transcription initiation in mitochondria have been purified and characterized from yeast (Saccharomyces cerevisiae) and animals (Tracy and Stern, 1995). In yeast two nuclear-encoded proteins are required for the initiation of transcription on a conserved nonanucleotide promoter, a core RNA polymerase of 145 kD (sc-mtRNAP), and a 43-kD specificity factor (sc-mtTFB; Kelly et al., 1986; Schinkel et al., 1987; Lisowsky and Michaelis, 1988; Shadel and Clayton, 1993). Whereas the RNA polymerase shows similarities to bacteriophage RNA polymerases (Masters et al., 1987), sc-mtTFB resembles bacterial ς factors (Jang and Jaehning, 1991). Transcriptional initiation is preceded by promoter-specific DNA binding of a complex of both proteins (Schinkel et al., 1988; Magus et al., 1994). A third factor potentially involved in the initiation process is sc-mtTFA, a 19-kD protein containing two HMG-like DNA-binding domains. sc-mtTFA has a moderately stimulating effect on transcriptional initiation in vitro, most likely due to its primary function of compacting and organizing yeast mtDNA (Diffley and Stillman, 1991; Fisher et al., 1991, 1992; Parisi et al., 1993).
In xl mitochondrial transcription is similarly initiated with the approximately 140-kD RNA polymerase xl-mtRNAP, requiring the dissociable factor xl-mtTFB, which is approximately 40 kD (Bogenhagen and Insdorf, 1988). Both proteins have been isolated in high purity, but their respective genes have not yet been cloned. The 30-kD xl-mtTFA, a protein with two HMG boxes, binds preferentially to the control region adjacent to the major promoters of xl-mtDNA and shows pronounced stimulation of transcription in vitro, similar to the homologous protein in yeast (Antoshechkin and Bogenhagen, 1995).
Mitochondrial transcription factors of the mtTFA family have also been purified and cloned from humans (h-mtTFA, 25 kD; Fisher and Clayton, 1988; Fisher et al., 1991; Parisi and Clayton, 1991) and mice (m-mtTFA, 23.5 kD; Larsson et al., 1996). Both proteins bind specifically to regions located upstream of the respective mitochondrial promoters and have a strong enhancing effect on the initiation of transcription (Larsson et al., 1996; Fisher et al., 1987, 1989). The h-mtRNA polymerase has recently been identified in mammalia as a phage-like, single-subunit enzyme that is similar to the respective yeast protein (Tiranti et al., 1997).
Investigation of the transcriptional initiation process in plant mitochondria has so far been focused on structural promoter studies by in vitro transcription analysis. Such in vitro systems have been established for wheat (Triticum aestivum; Hanic-Joyce and Gray, 1991), maize (Zea mays; Rapp and Stern, 1992), and pea (Pisum sativum; Binder et al., 1995) mitochondria. The architecture of the atp1 promoter in maize was analyzed by deletion analysis, linker-scanning analysis, and site-directed mutagenesis. These investigations identified a CRTA motif, conserved among monocot promoters, to represent the core promoter with an A-rich upstream domain enhancing initiation efficiency (Rapp and Stern, 1992; Rapp et al., 1993). A similar, although more extended nonanucleotide motif, 5′-(−7)CRTAAGAGA(+2)-3′, was found to surround transcription initiation sites in many promoters from dicot plants. Transcriptional studies with a pea mitochondrial lysate showed that the conserved nonanucleotide motif is recognized in homologous and heterologous in vitro transcription reactions, suggesting common transcriptional features among dicot plants. A 5′ deletional analysis of the atp9 promoter from pea mitochondria determined a minimal upstream region required for efficient in vitro initiation. The functional promoter structure extends up to nucleotide position −25 and consists of a nonanucleotide motif and, as in monocot plants, an upstream A(+T)-rich region (Binder et al., 1995).
The pea atp9 promoter structure now provides a template to search for DNA-binding proteins, which specifically recognize this region and thus could be involved in the transcription initiation process. As a purification strategy for such proteins we used the method of Fisher et al. (1991), a sequence of purification steps under denaturing and native conditions. Since this approach was successfully used for the purification of mtTFA proteins from the evolutionary distant organisms S. cerevisiae and humans (cell culture), we reasoned that this method might also represent a suitable strategy with which to purify a similar factor from plant mitochondria.
MATERIALS AND METHODS
Preparation of ds-Oligonucleotides
ds-Oligonucleotides were prepared by annealing equi-molar amounts of two complementary single-stranded oligonucleotides in 50 mm Tris-HCl, pH 7.6, 10 mm MgCl2, 5 mm DTT, 1 mm spermidine, and 1 mm EDTA. Annealing reactions (50 μL) were incubated at 90°C for 2 min and slowly cooled to room temperature overnight. Oligonucleotides were obtained by annealing the following single-stranded oligonucleotides with the corresponding antisense oligonucleotides (not shown): P1 (PA-79: 5′-GAACTGCTTGCTTATGTGAGGTTCTTT-3′), P2 (PA-52: 5′-CCTCTCGCTTGTTCATCTTGTTTTGAG-3′), P3 (PA25: 5′-TACTCGACGAAATAATAGCATAAGAGA-3′), P4 (PA+3: 5′-AGATATTGGACATTGAGTCCACTTCG-3′), P5 (PA+30: 5′-ATATCACACCTATTTGAGTCGGGAGTT-3′), O1 (OA-79: 5′-GAGCACATCGAAATTTCCAATCCGGTT-3′), O2 (OA-52: 5′-CCAAGCCAGGTAAGCAAGTTCCT-TTTC-3′), O3 (OA-25: 5′-TAAAGAAAGTTGATAAATCAT-AAGAGA-3′), O4 (OA+3: 5′-AGCAAAGTCCCTAGTC-AAAGGTGGTTG-3′), O5 (OA+30: 5′-GGAAGTAGTACG-CCCGGTTCACAGGTT-3′), PtrnF (PIII: 5′-CTCTTGTC-TTCCGTCTTTTG-3′), and Pnad5 (Exc+: 5′-TACCTAAAC-CAATCATCATATCGAC-3′).
Radioactive Labeling of ds-Oligonucleotides
Thirteen picomoles of ds-oligonucleotides was 5′-labeled by T4 polynucleotide kinase (Boehringer Mannheim) with 70 μCi of [γ-32P]ATP (3000 Ci/mmol). Labeling reactions were incubated for 45 min at 37°C. Microspin S200 columns (Pharmacia) were used according to the manufacturer's instructions to remove unincorporated [γ-32P]ATP. ds-Oligonucleotide concentration and total radioactivity were determined by photometric measurement and scintillation counting, respectively. The specific activities of the probes varied between 800 and 2000 cpm/fmol.
Isolation of Pea Mitochondria
Pea (Pisum sativum L., var Lancet) seedlings were grown in the dark for 7 d. Mitochondria were isolated and purified as described previously (Binder and Brennicke, 1993).
Denaturation-Renaturation Chromatography of Mitochondrial Proteins
Purified pea mitochondria (approximately 2 g) were carefully resuspended in 30 mL of ice-cold buffer W (100 mm NaPO4, pH 6.8, 250 mm Suc, 15% glycerol, 1 mm DTT, and 1 mm PMSF) and centrifuged for 15 min at 27,000g. Mitochondria were then resuspended in 50 mL of boiling lysis buffer L (100 mm NaPO4, pH 6.8, 2% SDS, 20 mm DTT, and 1 mm PMSF). The mixture (approximately 40 mg of total protein) was boiled for 5 min until the lysate had cleared and was then diluted 10-fold with buffer D (100 mm NaPO4, pH 6.8, 1 mm DTT, and 1 mm PMSF).
Hydroxyapatite chromatography (Bio-Rad) was performed at room temperature on a 70-mL column equilibrated in buffer D adjusted to 0.1% SDS. The column was loaded with the diluted mitochondrial lysate and extensively washed with buffer D containing 0.1% SDS. Elution was performed with a linear gradient of 100 to 500 mm NaPO4, pH 6.8, in 0.1% SDS, 1 mm DTT, and 1 mm PMSF. Fifty fractions of 4 mL each were collected and assayed for DNA-binding activity by mobility-shift experiments. Hydroxyapatite fractions with DNA-binding activity were pooled, renatured by adding 2 volumes of renaturation buffer RII (20 mm Tris-HCl, pH 8.0, 0.2 mm EDTA, 15% glycerol, 4% Triton X-100, 2 mm DTT, and 0.5 mm PMSF), and incubated on ice for 20 min.
Chromatography on phosphocellulose (Sigma) was carried out at 4°C. Renatured hydroxyapatite fractions were loaded onto a 20-mL column equilibrated in phosphocellulose buffer (10 mm Tris-HCl, pH 8.0, 0.1 mm EDTA, 7.5% glycerol, 0.1% Triton X-100, 1 mm DTT, and 0.75 mm PMSF) containing 100 mm NaCl. Proteins were eluted with a linear gradient of 0.1 to 1.0 m NaCl in phosphocellulose buffer. Fifty 5-mL fractions were collected and assayed for DNA-binding activity.
Mobility-Shift Assays
Aliquots of hydroxyapatite fractions were renatured by adding 4 volumes of renaturation buffer RI (9 mm Tris-HCl, pH 8.0, 45 mm KCl, 0.9 mm EDTA, 44.5% glycerol, 2% Triton X-100, and 1 mm DTT) and incubating on ice for 20 min. Two microliters of the renatured protein fractions was mixed with 5 μL of hydroxyapatite shift mixture (20 mm Tris-HCl, pH 8.0, 20 mm MgCl2, 200 μg/mL BSA, and 100 mg/mL ds-poly[d{I-C}]), 1 μL of radiolabeled probe (approximately 15,000 cpm), and 2 μL of double-distilled water. In the phosphocellulose fractions, 2-μL aliquots were added to 7 μL of phosphocellulose shift mixture (14.3 mm Tris-HCl, pH 8.0, 14.3 mm MgCl2, 35.7% glycerol, 140 mg/mL BSA, 140 μg/mL ds-poly[d{I-C}]), and 1 μL of radiolabeled probe (approximately 15,000 cpm). Binding reactions were incubated at 25°C for 20 min, supplemented with 1 μL of loading solution (200 mm Tris-HCl, pH 8.0, 0.1% [w/v] bromphenol blue, 0.1% [w/v] xylenecyanole, and 50% glycerol) and electrophoresed on native 4% polyacrylamide gels (acrylamide:bisacrylamide, 30:1) in 10 mm Tris-HCl, pH 8.0, 5 mm sodium acetate, and 1.5 mm EDTA.
rpHPLC
rpHPLC was performed on a C4 column (Bioselect, Eurosil, Knauer, Berlin, Germany; 4.6 × 250 mm, medium pore size, 300 Å). Fifteen milliliters of pooled phosphocellulose fractions was loaded onto the column equilibrated in an aqueous solution containing 5% acetonitrile and 0.1% trifluoroacetic acid. Proteins were eluted by a linear gradient of 5 to 80% acetonitrile in 0.1% aqueous trifluoroacetic acid. One-hundred 1-mL fractions were collected. Fractions were lyophilized and the pellets were resuspended in 10 μL of buffer D adjusted to 100 mm NaPO4. After incubation for 30 min at room temperature, 2-μL aliquots were renatured as described for hydroxyapatite fractions and then tested for DNA-binding activity in mobility-shift experiments.
Recovery and Renaturation of Proteins after Purification by SDS-PAGE
SDS-polyacrylamide gel slices were covered with 350 μL of elution buffer (50 mm Tris-HCl, pH 8.0, 0.1 mm EDTA, 150 mm NaCl, 10% glycerol, and 1 mm DTT) and crushed with a glass pestle until fine gel slurries were obtained. After an overnight incubation at 4°C, the slurries were pipetted onto ultrafiltration tubes (Microcon 10, Amicon, Beverly, MA) equipped with prefilters (pore size, 0.2 μm). Samples were centrifuged at 8500g for 60 min to separate the eluted proteins from polyacrylamide debris and to concentrate the proteins. The final volume of the concentrated proteins was 5 to 15 μL. Aliquots with a volume of 0.5 μL were renatured by the addition of 2 μL of renaturation buffer RI and tested for DNA-binding activity. The remaining proteins were supplemented with 10 μL of sample buffer (125 mm Tris-HCl, pH 6.8, 2% SDS, 17% glycerol, 0.01% bromphenol blue [w/v], and 5% [v/v] β-mercapto-ethanol), resolved alongside a molecular weight marker (Amersham) on analytical 12.5% SDS-polyacrylamide gels (Laemmli, 1970), and detected by silver staining.
Determination of Relative Binding Activity
Mobility-shift reactions were performed with renatured rpHPLC fractions containing one of the two DNA-binding proteins (fraction no. 49, a 32-kD protein, or fraction no. 51, a 44-kD protein) and different ds-oligonucleotides (P1-P5 or O1-O5). The total radioactivity in each binding reaction was adjusted to 15,000 cpm. DNA-protein complexes were detected by autoradiography with preflashed radiographic films (Hyperfilm-MP, Amersham), and absolute signal intensities were determined by densitometric analysis using a scanner (model ED-8000, Epson, Torrance, CA) and imaging software (ImageQuant 2.1, Molecular Dynamics, Sunnyvale, CA). Absolute signal intensities were corrected by subtracting local background intensities and then multiplying with normalizing factors to compensate for differences resulting from the different specific activities of the ds-oligonucleotides in each experiment. Relative binding was compared by setting the highest normalized signal intensity determined in one experiment to 100%.
RESULTS
Purification of DNA-Binding Proteins from Pea Mitochondria by Denaturation-Renaturation Chromatography
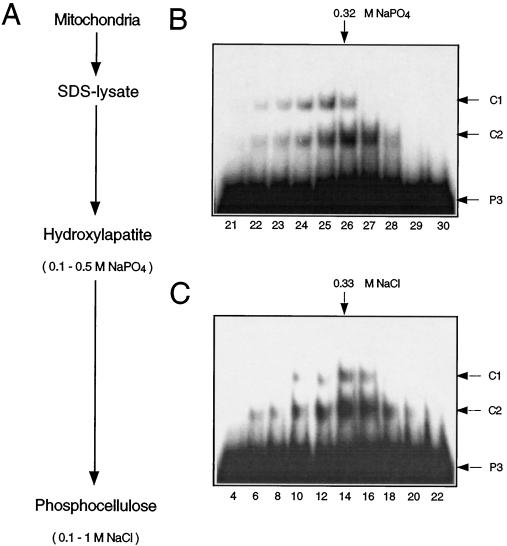
Proteins from pea mitochondria binding to ds-oligonucleotides representing a mitochondrial promoter were purified with the denaturation-renaturation chromatography procedure developed by Fisher et al. (1991; Fig. 1A).
Figure 1.
Purification of DNA-binding proteins from pea mitochondria. A, Purification scheme. Pea mitochondria were denatured by boiling in an SDS-containing buffer. The SDS-lysate was loaded onto a hydroxyapatite column and eluted with a linear NaPO4 gradient (0.1–0.5 m). Aliquots of the eluted fractions were renatured and tested for DNA-binding activity in mobility-shift experiments (B). Fractions with binding activity (nos. 22–28) were pooled, renatured, and loaded onto a phosphocellulose column. The column was eluted with a linear NaCl gradient (0.1–1 m), and fractions were assayed for DNA-binding activity by mobility-shift analysis (C). The approximate NaPO4 and NaCl concentrations of fractions with the highest binding activities are indicated at the upper margins. Arrows in the right margins indicate the positions of the unbound ds-oligonucleotide P3 and of the DNA-protein complexes C1 and C2. Fraction numbers are given at the bottom in B and C.
DNA-binding activity of eluted and renatured column fractions was assayed by mobility-shift analysis with a labeled ds-oligonucleotide (P3, 27 bp) covering the promoter core structure of the pea atp9 gene from −25 to +2 relative to the first transcribed nucleotide. This sequence was sufficient in vitro to support transcription initiation and did not require any further upstream or downstream sequences (Binder et al., 1995; S. Dombrowski and S. Binder, unpublished results). Two distinct DNA protein complexes, C1 and C2, were identified in fractions eluting at approximately 320 mm NaPO4 from the hydroxyapatite column (Fig. 1B). Fractions containing binding activity were pooled, renatured, and chromatographed on a phosphocellulose column. Pronounced C1 and C2 formation was observed in fractions eluting at approximately 330 mm NaCl (Fig. 1C).
Analysis of phosphocellulose fractions by SDS-PAGE did not allow a clear correlation between individual protein bands and the binding activities (data not shown), and additional purification steps were thus required to identify the DNA-binding proteins. Prior to further enrichment procedures, we investigated to what extent the observed complex formation is specific for the atp9 promoter sequence.
Analysis of DNA-Binding Specificity
Competition experiments with different ds-oligonucleo-tides offered a convenient approach for determining how specific proteins in complexes C1 and C2 recognize the atp9 promoter sequence. Binding reactions were performed with the 330 mm NaCl fraction from the phosphocellulose column (fraction no. 14) and labeled oligonucleotide P3, which covers the entire promoter (Fig. 2). A 10-fold molar excess of unlabeled P3 was found to be sufficient for almost complete suppression of the C1 and C2 signals (Fig. 2; lane 3). In contrast, the heterologous competitors PtrnF (20 bp) and Pnad5 (22 bp) only slightly reduced complex formation when present in 20- or even 40-fold molar excess (Fig. 2; lanes 4–7). The two competitor sequences are parts of mitochondrial genes and have no similarity to the promoter consensus. These results suggest that the DNA-binding proteins in C1 and C2 have, to some extent, sequence-specific binding properties.
Figure 2.
Competition experiments. Aliquots of the 330 mm NaCl phosphocellulose fraction were incubated with the labeled ds-oligonucleotide P3 in the presence of unlabeled P3 (lane 2) and the heterologous competitors PtrnF (20 bp, lanes 4 and 5) and Pnad5 (22 bp, lanes 6 and 7). The molar excesses in which the competing DNA molecules were added over P3 are indicated in the second line at the top. Binding reactions in the absence of mitochondrial proteins (lane 1) and in the absence of competitor (lane 2) served as negative and positive controls, respectively. A homologous competition experiment was performed to determine the molar excess of unlabeled P3 required for an almost complete suppression of the C1 and C2 signals (lane 3). Arrows at left indicate the positions of the DNA-protein complexes C1 and C2 and of the unbound ds-oligonucleotide P3.
Separation of the Two DNA-Binding Activities by rpHPLC
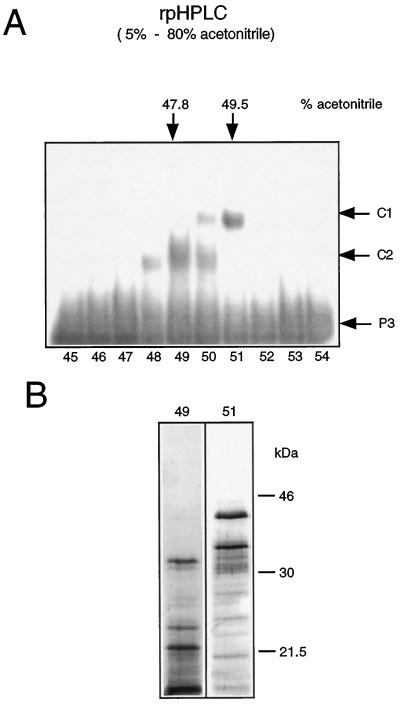
The denaturation-renaturation chromatography of pea mitochondrial proteins showed that the DNA-binding activity of the proteins in C1 and C2 can be recovered after denaturation in SDS. This observation allows the application of rpHPLC as a next purification step. rpHPLC has a high protein-separation capability but has to be carried out under partially denaturing conditions, since organic solvents are used for elution. Therefore, proteins purified by rpHPLC must be renatured before activity assays can be conducted.
Phosphocellulose fractions with DNA-binding activity (Fig. 1C, fraction nos. 6–20) were pooled and chromatographed on an rpHPLC C4 column. The two DNA-binding activities were efficiently separated by the rpHPLC (Fig. 3A). The highest C1 formation was observed with fraction no. 51, which was collected at 49.5% acetonitrile, whereas the peak of C2-binding activity was detected in fraction no. 49, which was collected at 47.8% acetonitrile.
Figure 3.
Separation of DNA-binding proteins by rpHPLC. A, Phosphocellulose fractions with binding activities (Fig. 1C, fraction nos. 6–20) were pooled and chromatographed on an rpHPLC C4 column. Fractions eluted with a linear acetonitrile gradient (5–80%) were tested after renaturation for DNA-binding activity by mobility-shift analysis using ds-oligonucleotide P3. Arrows in the right margin indicate the positions of the DNA-protein complexes C1 and C2 and of the unbound ds-oligonucleotide P3. Fraction numbers are given at the bottom. B, SDS-PAGE analysis of rpHPLC fraction nos. 49 and 51. Proteins were resolved on 12.5% SDS-polyacrylamide gels and visualized by silver staining. Molecular masses of marker proteins are given in kilodaltons.
The separation of DNA-binding activities confirms that C1 and C2 are formed by at least two different proteins rather than by dimeric and monomeric complexes of the same protein. SDS-PAGE analysis showed that the protein compositions in the fractions with peak binding activities are still too complex to allow assignment of the DNA-binding proteins (Fig. 3B).
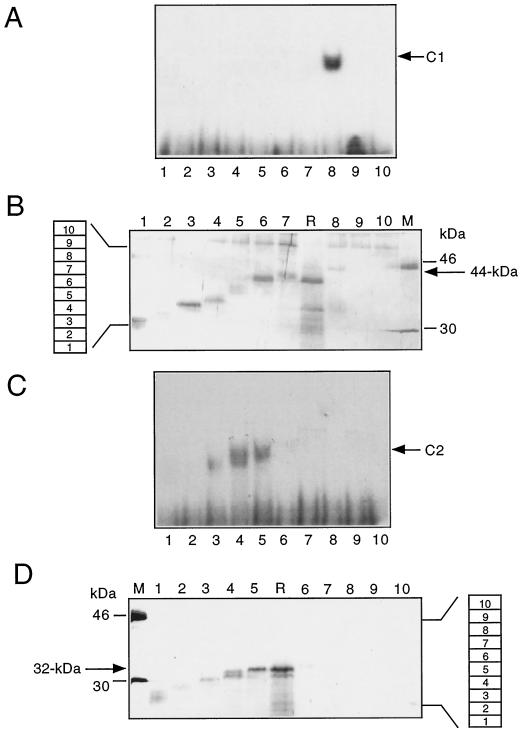
Size Determination of the Two DNA-Binding Proteins
The apparent molecular masses of the proteins in complexes C1 and C2 were estimated by gel electrophoresis. rpHPLC fractions with peak binding activities (Fig. 3A, fractions 49 and 51) were resolved by electrophoresis on preparative SDS-polyacrylamide gels, which were subsequently cut into 3-mm slices. Proteins from individual gel slices were recovered by elution, and aliquots were tested for DNA-binding activity after renaturation (Fig. 4, A and C). The remaining proteins were again loaded onto analytical SDS-polyacrylamide gels (Fig. 4, B and D) and co-electrophoresed with a molecular size marker (lanes M) and with aliquots of the original rpHPLC fractions, which served as reference samples (lanes R). Apparent sizes of proteins with DNA-binding activity could thus be estimated by comparing protein bands present on the silver-stained gels with shift signals in the corresponding lanes of the mobility-shift assays.
Figure 4.
Size determination of the purified proteins by gel-dissection experiments. rpHPLC fraction nos. 49 and 51 were electrophoresed on two separate, preparative 12.5% SDS-polyacryl-amide gels (not shown). Each gel was cut into 10 slices, as indicated by the numbered boxes at the left of B and at the right of D. A and C, Proteins were eluted from these gel slices and aliquots of the eluted proteins were tested for DNA binding in mobility-shift assays. Arrows indicate the signals of DNA-protein complexes C1 and C2. Numbers of the polyacrylamide slices are given at the bottom. B and D, The remaining proteins were electrophoresed on analytical 12.5% SDS-polyacrylamide gels alongside an aliquot of rpHPLC fraction no. 51 or no. 49 (lanes R in B and D). Proteins were detected by silver staining. Numbers at the top correspond to the gel slice numbers. Molecular masses of marker proteins are given in kilodaltons (lanes M).
Analysis of rpHPLC fraction no. 51 (Fig. 3A) identified a DNA-binding protein with an apparent molecular mass of approximately 44 kD (Fig. 4, A and B, lanes 8). A polypeptide migrating at 50 kD is present in all lanes in which eluted proteins were resolved in the analytical protein gel (Fig. 4B, lanes 1–10). This protein is absent from both the reference and marker lanes (lanes R and M) and is therefore very likely a contaminant that was introduced during elution or renaturation. Other more diffuse bands in lane 8 are most likely degradation products of the 44-kD protein, since they are too small to represent intact proteins recovered from gel slice no. 8.
The analysis of rpHPLC fraction no. 49 (Fig. 3A) identified a DNA-binding activity from gel slice nos. 3 to 5, containing proteins with apparent molecular masses of 30 to 32 kD and forming complex C2 with P3 (Fig. 4, C and D, lanes 5). Signal structures in the autoradiogram of the mobility-shift gel (Fig. 4C, lanes 3 and 4) possibly identify more than one complex-forming protein with similar sizes in the range of 30 to 32 kD (Fig. 4D). The resolution of complex C2 achieved in the mobility-shift analyses of different mitochondrial fractions did not show whether one or more complexes was formed by different proteins in this size range (Figs. 1, B and C, and 4C). Since there was at least one protein participating, we consider complex C2 as being derived from one protein of 32 kD, without precluding the involvement of others. Further specific investigation will be required to resolve the question of additional DNA-binding polypeptides of this size range.
DNA-Binding Specificity of the Two Proteins to Different Parts of Two Mitochondrial-Promoter Regions
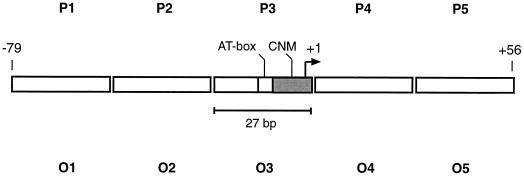
The competition experiments suggested that the detected proteins from pea mitochondria show sequence-specific binding to the ds-oligonucleotides, representing a plant mitochondrial promoter region (Fig. 2). To gather more experimental information about these proteins and their potential for recognizing the promoter sequences, we investigated binding to different parts of the pea atp9 and the heterologous Oenothera berteriana atp1 promoter regions and their sequence vicinities. The latter promoter was selected because it has been recognized and correctly activated in an in vitro transcription assay in pea (Binder et al., 1995).
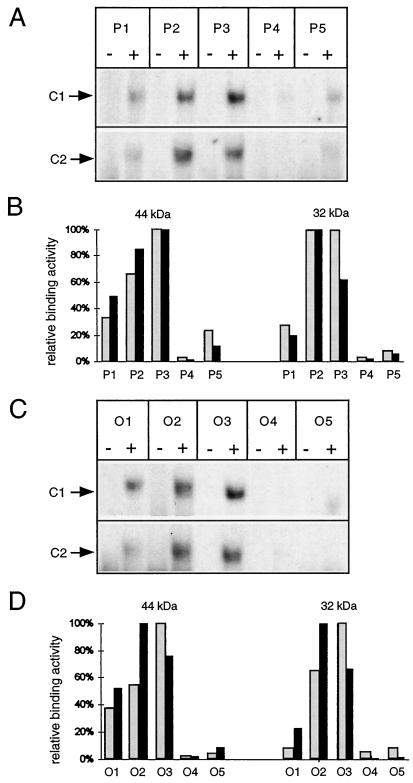
Two series of ds-oligonucleotides, P1 to P5 and O1 to O5, represent 27-bp sections flanking the respective transcriptional start sites (Fig. 5). Binding reactions of the labeled ds-oligonucleotides to the rpHPLC-purified proteins were analyzed on 4% native polyacrylamide gels, and complex formation was compared by densitometric evaluation of mobility-shift experiments. Two independent sets of experiments (Fig. 6) reveal the strongest shift signals and thus preferential binding of both proteins, with ds-oligonucleotides representing the core promoter (P3/O3) and the respective upstream oligonucleotides (P2/O2). Minor binding is observed with P1 and O1 (covering regions further upstream), and almost no binding to the downstream ds-oligonucleotides (P4/O4 and P5/O5) is observed. This study suggests that the identified proteins have similar binding properties, binding preferentially to DNA regions containing the conserved promoter elements (P3/O3) and the nonconserved, upstream sequences from both plant species (P2/O2).
Figure 5.
Schematic representation of ds-oligonucleotides used in the investigation of protein binding in different sections of two promoter regions. Two series of ds-oligonucleotides were used to investigate protein binding in different sections of the pea atp9 (P1-P5) and the O. berteriana atp1 (O1-O5) promoter regions. Each ds-oligonucleotide represents a different section of one promoter region and was used as the DNA substrate in mobility-shift experiments (Fig. 6). The contiguous sequences of the two ds-oligonucleotide arrays correspond to the complete sequences of the atp9 and atp1 promoter regions from nucleotide position −79 to +56, respectively. Both promoter regions contain conserved nonanucleotide motifs (CNM) and AT-rich regions (AT-box). Numbering of the nucleotide positions refers to the transcriptional starting point (nucleotide position +1), represented by a bent arrow.
Figure 6.
DNA-binding specificity in different sections of the pea atp9 and the O. berteriana atp1 promoter regions. A and C, Autoradiograms of mobility-shift experiments. rpHPLC fractions containing either the 32-kD protein (fraction no. 49) or the 44-kD protein (fraction no. 51) were used to assay C2 and C1 formation. Complexes are indicated by arrows. The ds-oligonucleotides used in the individual reactions are given at the top. Binding reactions were carried out in the presence (+) or absence (−) of mitochondrial proteins. B and D, Quantitative evaluation of two sets of mobility-shift experiments (autoradiograms in A and C representing one set). Relative binding affinities of the 44- and 32-kD proteins to the individual ds-oligonucleotides were calculated from normalized C1 and C2 signal intensities (see Methods). Gray and black bars represent the relative binding affinities derived from the first and the second data sets, respectively.
DISCUSSION
DNA-Binding Proteins from Pea Mitochondria
The denaturation-renaturation method developed by Fisher et al. (1991) and rpHPLC were used to purify proteins that preferentially bind to mitochondrial promoter sequences from higher plants. This approach identified 32- and 44-kD proteins from pea mitochondria. The 32-kD proteins behave as distinct proteins in the SDS-PAGE analysis, whereas their ds-oligonucleotide complexes cannot be clearly distinguished by their gel migration; therefore they may represent variant protein forms. The DNA-binding activities of these proteins are recovered after denaturation and elute at similar conditions during chromatography on hydroxyapatite, phosphocellulose, and rpHPLC (Figs. 1 and 3).
Comparison of the Pea Proteins with mtDNA-Binding Proteins from Other Organisms
The denaturation-renaturation purification procedure used here to enrich pea mitochondrial proteins was originally developed to purify mtTFA proteins from yeast and animal mitochondria. It therefore appears reasonable to compare characteristics of the proteins identified in pea with those of sc- and h-mtTFA for similarities and differences in biochemical properties.
The first similarity is their survival through the harsh denaturing treatment and their regaining DNA-binding activity through renaturation in the presence of Triton X-100. Furthermore, these two pea mitochondrial proteins co-elute at approximately 320 mm NaPO4 from the hydroxyapatite column, comparable to the NaPO4 concentrations required to elute sc-mtTFA (approximately 330 mm) and h-mtTFA (approximately 300 mm) from this matrix. However, in phosphocellulose chromatography, the pea proteins elute at approximately 330 mm NaCl, whereas significantly higher salt concentrations are required to recover h-mtTFA (580 mm) and sc-mtTFA (700 mm NaCl; Fisher et al., 1991). No DNA-binding activity is detected with pea mitochondrial proteins mobilized at comparably high NaCl concentrations. The pea 32- and 44-kD proteins thus show purification characteristics quite different from those of the sc-mtTFA and h-mtTFA polypeptides.
The 44-kD protein has a molecular mass similar to xl-mtTFB proteins (approximately 40 kD; Bogenhagen and Insdorf, 1988; Antoshechkin and Bogenhagen, 1995) and sc-mtTFB (43 kD; Schinkel et al., 1987). Although the sc-mtTFB activity can also be recovered after denaturing SDS-PAGE (Schinkel et al., 1987), it seems unlikely that the 44-kD pea polypeptide represents a plant mtTFB homolog. In contrast to mtTFB, which binds to promoters only in a binary complex with the respective mitochondrial RNA polymerase, the 44-kD protein from pea shows significant binding to promoter regions, and binding does not depend on the presence of other proteins (such as the mtRNA polymerase, which is more than 100 kD; Figs. 2 and 6).
The molecular masses of the identified pea proteins also suggest that neither of them resembles a mitochondrial RNA polymerase, which is 112 kD in Chenopodium album (Weihe et al., 1997) and is most likely of a similar size in mitochondria of plants in general, including pea (Cermakian et al., 1996).
Recognition of Protein-Binding Sites in Plant Mitochondrial Promoter Regions
The binding studies with the two series of ds-oligonucleotides (Fig. 6) revealed that the formation of the respective DNA-protein complexes is a nonrandom, sequence-dependent process. The strongest protein binding was observed at two sites upstream of the transcription start sites in both the pea atp9 and the O. berteriana promoter regions. The proximal, preferred protein-binding sites contain the two important sequence elements identified in plant mitochondrial promoters. The conserved nonanucleotide motif and the AT-box are covered by the ds-oligonucleotides P3 and O3 (Fig. 5). Consequently, one or both elements may be recognized by proteins binding immediately upstream of the respective transcription start site. However, the more distal, second protein-binding sites contained in P2 and O2 do not share any extended sequence similarities, either with each other or with P3 and O3. Therefore, sequence elements important for this second binding site cannot easily be correlated with a conserved consensus sequence.
In mitochondrial light-strand promoters of human and mouse, h-mtTFA and m-mtTFA specifically recognize such “hidden” elements. Here, binding sites were identified by DNase I footprinting and further analyzed by methylation interference assays. The DNase I footprints of both proteins extend approximately between nucleotides −10 and −40 relative to the respective transcription start site. Although these two upstream regions show only very limited sequence similarities, several of the directly contacted nucleotides were found conserved between the two species. Heterologous footprint experiments and in vitro transcription reactions revealed that these cryptic, conserved features are sufficient for specific mtTFA binding and transcriptional stimulation (Fisher et al., 1987, 1989).
In plant mitochondrial promoters, similar investigations are required to map binding sites more precisely and to identify critical protein-nucleotide contacts. Such studies will reveal to what extent the conserved nonanucleotide motif and/or the AT-box play a role in the specific binding of the 32- and 44-kD proteins. The 5′ deletional analysis of the pea atp9 promoter suggests that engagement of the protein-binding site in the conserved promoter region, the P3-section, is, at least in vitro, sufficient for the effective initiation of transcription (Binder et al., 1995). Therefore, protein binding to additional upstream site(s) in the P2- or even in the P1-section may have a more enhancing effect on the transcription initiation process.
Are the Identified Pea Mitochondrial Proteins Connected with the Initiation of Transcription in Plant Mitochondria?
The promoter-binding properties of the 32- and the 44-kD proteins tentatively support a preferential connection of these polypeptides to the transcription initiation process in pea mitochondria.
In addition, enhanced transcription initiation activity is observed upon the addition of a fraction containing the two promoter-binding proteins to an in vitro transcription reaction (data not shown). However, this stimulating mitochondrial fraction (330 mm NaCl phosphocellulose fraction; fraction 14 in Fig. 1) is far too complex and has too many other proteins to allow a connection with the 32- and 44-kD proteins. Enhancing effects were observed in analogous in vitro transcription experiments when h-mtTFA-containing protein fractions were incubated with partially purified human mtRNA polymerase (Fisher and Clayton, 1988) or when xl-mtTFA was added to mixtures of xl-mtTFB and xl-mtRNAP (Bogenhagen and Insdorf, 1988).
The increased transcript amount could alternatively be explained by an improved RNA stability. Preliminary investigations showed that the slow degradation rate of T7 RNA polymerase transcripts of the pea atp9 region in mitochondrial lysate preparations used for in vitro transcription is not influenced by the addition of the 330 mm NaCl phosphocellulose fraction (data not shown). Future work should analyze the observed transcriptional effect in more detail, which should elucidate the putative stimulatory function of each DNA-binding protein separately.
ACKNOWLEDGMENTS
We are very grateful to Dr. Thomas Lisowsky for helpful hints concerning the denaturation-renaturation chromatography and to Dr. Richard Reinhard for generously providing excellent support and rpHPLC facilities. We also thank Dr. Hans Peter Braun and Dr. Lutz Grohmann for many suggestions about protein-purification techniques.
Abbreviations:
- ds
double-stranded
- h
human
- HMG protein
high-mobility group protein
- m
mouse
- mtRNAP
mitochondrial RNA polymerase
- mtTFA or mtTFB
mitochondrial transcription factor A or B
- rpHPLC
reversed-phase HPLC
- sc
Saccharomyces cerevisiae
- xl
Xenopus laevis
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft, a Landesforschungsschwerpunkt Baden-Württemberg, the Human Frontiers Science Program, and the Fonds der Chemischen Industrie.
LITERATURE CITED
- Antoshechkin I, Bogenhagen DF. Distinct roles for two purified factors in transcription of Xenopus mitochondrial DNA. Mol Cell Biol. 1995;15:7032–7042. doi: 10.1128/mcb.15.12.7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder S, Brennicke A. Transcription initiation sites in mitochondria of Oenotheraberteriana. J Biol Chem. 1993;268:7849–7855. [PubMed] [Google Scholar]
- Binder S, Hatzack F, Brennicke A. A novel pea mitochondrial in vitro transcription system recognizes homologous and heterologous mRNA and tRNA promoters. J Biol Chem. 1995;270:22182–22189. doi: 10.1074/jbc.270.38.22182. [DOI] [PubMed] [Google Scholar]
- Bogenhagen DF, Insdorf NF. Purification of Xenopuslaevis mitochondrial RNA polymerase and identification of a dissociable factor required for specific transcription. Mol Cell Biol. 1988;8:2910–2916. doi: 10.1128/mcb.8.7.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermakian N, Tatsuya MI, Cedergren R, Gray MW. Sequences homologous to yeast mitochondrial and bacteriophage T3 and T7 RNA polymerases are widespread throughout the eukaryotic lineage. Nucleic Acids Res. 1996;24:648–654. doi: 10.1093/nar/24.4.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley JFX, Stillman B. A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc Natl Acad Sci USA. 1991;88:7864–7868. doi: 10.1073/pnas.88.17.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RP, Clayton DA. Purification and characterization of human mitochondrial transcription factor 1. Mol Cell Biol. 1988;8:3496–3509. doi: 10.1128/mcb.8.8.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RP, Lisowsky T, Breen GAM, Clayton DA. A rapid, efficient method for purifying DNA-binding proteins. J Biol Chem. 1991;266:9153–9160. [PubMed] [Google Scholar]
- Fisher RP, Lisowsky T, Parisi MA, Clayton DA. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J Biol Chem. 1992;267:3358–3367. [PubMed] [Google Scholar]
- Fisher RP, Parisi MA, Clayton DA. Flexible recognition of rapidly evolving promoter sequences by mitochondrial transcription factor 1. Genes Dev. 1989;3:2202–2217. doi: 10.1101/gad.3.12b.2202. [DOI] [PubMed] [Google Scholar]
- Fisher RP, Topper JN, Clayton DA. Promoter selection in human mitochondria involves binding of a transcription factor to orientation-independent upstream regulatory elements. Cell. 1987;50:247–258. doi: 10.1016/0092-8674(87)90220-0. [DOI] [PubMed] [Google Scholar]
- Hanic-Joyce PJ, Gray MW. Accurate transcription of a plant mitochondrial gene in vitro. Mol Cell Biol. 1991;11:1035–2039. doi: 10.1128/mcb.11.4.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang SH, Jaehning JA. The yeast mitochondrial RNA polymerase specificity factor, MTF1, is similar to bacterial sigma factors. J Biol Chem. 1991;266:22671–22677. [PubMed] [Google Scholar]
- Kelly JL, Greenleaf AL, Lehman IR. Isolation of the nuclear gene encoding a subunit of the yeast mitochondrial RNA polymerase. J Biol Chem. 1986;261:10348–10351. [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larsson N-G, Garman JD, Oldfors A, Barsh GS, Clayton DA. A single mouse gene encodes the mitochondrial transcription factor A and a testis-specific nuclear HMG-box protein. Nature Genet. 1996;13:296–302. doi: 10.1038/ng0796-296. [DOI] [PubMed] [Google Scholar]
- Lisowsky T, Michaelis G. A nuclear gene essential for mitochondrial replication suppresses a defect of mitochondrial transcription. Mol Gen Genet. 1988;214:218–223. doi: 10.1007/BF00337714. [DOI] [PubMed] [Google Scholar]
- Magus DA, Jang SH, Jaehning JA. Release of the yeast mitochondrial RNA polymerase specificity factor from transcription complexes. J Biol Chem. 1994;269:26568–26574. [PubMed] [Google Scholar]
- Masters BS, Stohl LL, Clayton DA. Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell. 1987;51:89–99. doi: 10.1016/0092-8674(87)90013-4. [DOI] [PubMed] [Google Scholar]
- Parisi MA, Clayton DA. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science. 1991;252:965–969. doi: 10.1126/science.2035027. [DOI] [PubMed] [Google Scholar]
- Parisi MA, Xu B, Clayton DA. A human mitochondrial transcriptional activator can functionally replace a yeast mitochondrial HMG-box protein both in vivo and in vitro. Mol Cell Biol. 1993;13:1951–1961. doi: 10.1128/mcb.13.3.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp WD, Lupold DS, Mack S, Stern DB. Architecture of the maize mitochondrial atp1 promoter as determined by linker-scanning and point mutagenesis. Mol Cell Biol. 1993;13:7232–7238. doi: 10.1128/mcb.13.12.7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp WD, Stern DB. A conserved 11 nucleotide sequence contains an essential promoter element of the maize mitochondrial atp1 gene. EMBO J. 1992;11:1065–1073. doi: 10.1002/j.1460-2075.1992.tb05145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinkel AH, Groot-Koerkamp MJA, Teunissen AWRH, Tabak HF. Mitochondrial RNA polymerase of Saccharomycescerevisiae: composition and mechanism of promoter recognition. EMBO J. 1988;7:3255–3262. doi: 10.1002/j.1460-2075.1988.tb03192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinkel AH, Groot-Koerkamp MJA, Touw EPW, Tabak HF. Specificity factor of yeast mitochondrial RNA polymerase. Purification and interaction with core RNA polymerase. J Biol Chem. 1987;262:12785–12791. [PubMed] [Google Scholar]
- Shadel GS, Clayton DA. Mitochondrial transcription initiation. J Biol Chem. 1993;268:16083–16086. [PubMed] [Google Scholar]
- Tiranti V, Savoia A, Forti F, D'Apolito M-F, Centra M, Rocchi M, Zeviani M. Identification of the gene encoding the human mitochondrial RNA polymerase (h-mtRPOL) by cyberscreening of the Expressed Sequence Tags database. Hum Mol Genet. 1997;6:615–625. doi: 10.1093/hmg/6.4.615. [DOI] [PubMed] [Google Scholar]
- Tracy RL, Stern DB. Mitochondrial transcription initiation: promoter structures and RNA polymerases. Curr Genet. 1995;28:205–216. doi: 10.1007/BF00309779. [DOI] [PubMed] [Google Scholar]
- Weihe A, Hedtke B, Börner T. Cloning and characterization of a cDNA encoding a bacteriophage-type RNA polymerase from the higher plant Chenopodiumalbum. Nucleic Acids Res. 1997;25:2319–2325. doi: 10.1093/nar/25.12.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]