Abstract
During oxidative stress in E. coli, the SufABCDSE stress response pathway mediates iron-sulfur (Fe-S) cluster biogenesis rather than the Isc pathway. To determine why the Suf pathway is favored under stress conditions, the stress response SufS-SufE sulfur transfer pathway and the basal housekeeping IscS-IscU pathway were directly compared. We found that SufS-SufE cysteine desulfurase activity is significantly higher than IscS-IscU at physiological cysteine concentrations and after exposure to H2O2. Mass spectrometry analysis demonstrated that IscS-IscU is more susceptible than SufS-SufE to oxidative modification by H2O2. These important results provide biochemical insight into the stress resistance of the Suf pathway.
INTRODUCTION
Iron-sulfur (Fe-S) clusters in metalloproteins carry out myriad cellular functions [1,2]. Fe-S cluster biogenesis requires proteins that donate sulfur and iron, pre-assemble clusters, and traffic Fe-S clusters to target metalloproteins [3–5]. Fe-S cluster biogenesis is sensitive to oxygen due to the proclivity of iron, sulfide, and protein sulfhydryl groups to be modified by oxygen or reactive oxygen species [6]. In Escherichia coli, the Isc system carries out Fe-S cluster assembly under normal conditions while the Suf pathway is required for Fe-S cluster biogenesis under oxidative stress conditions [7–10].
Both Isc and Suf use superficially similar mechanisms to mobilize sulfur for Fe-S cluster assembly. The homodimeric IscS and SufS cysteine desulfurase enzymes catalyze the pyridoxal-phosphate (PLP)-dependent removal of sulfur from L-cysteine substrate resulting in a protein-bound persulfide (R-S-SH) intermediate. This persulfide S0 species (also referred to as sulfane sulfur) is reduced and incorporated into the Fe-S cluster as sulfide (S2−) during assembly on a scaffold protein (IscU or the SufBC2D complex) [11–21]. Due to the reactivity of both the persulfide intermediate and active site sulfhydryl groups on the enzymes [22,23], oxidative stress has the potential to block the sulfur donation step of Fe-S cluster biogenesis. Genetic evidence has shown that the Isc system is not efficient at Fe-S cluster assembly under oxidative stress, raising the question of whether sulfur trafficking by the Suf pathway may be more resistant to disruption than the Isc system [24].
IscU and SufE are structural (but not sequence) homologues that each interact with their cognate cysteine desulfurase enzymes to accept S0 via a thiol exchange mechanism [17,19,20,25]. While IscU is a bona fide scaffold protein where the full Fe-S cluster can be assembled, SufE uses a single active site cysteine residue (C51) for accepting S0 and does not bind a nascent Fe-S cluster [20]. SufE then further traffics the S0 to SufB within the SufBC2D scaffold complex where the nascent cluster is assembled [21]. SufE enhances the cysteine desulfurase activity of SufS, although the exact mechanism of enhancement is unclear. SufBC2D further increases SufE-dependent enhancement of SufS via an unknown mechanism [20]. In contrast, IscU was recently shown to not enhance the desulfurase activity of IscS [26].
To determine if sulfur trafficking by the Suf pathway is more resistant to oxidative stress than the Isc pathway, we directly compared the oxidative stress resistance of the SufS-SufE sulfur transfer pathway to that of the E. coli IscS-IscU system. We discovered that SufS-SufE are more active than IscS-IscU at physiological concentrations of L-cysteine and that SufS-SufE activity is more resistant to H2O2 exposure than IscS-IscU. Furthermore, IscS and IscU are more sensitive to oxidative modification by H2O2 than SufS and SufE. The functional ramifications of these results for defining the relative roles of Isc and Suf are discussed.
MATERIALS AND METHODS
RESULTS
Kinetic Analysis of SufS Activity in the Presence of SufE
Native SufS, SufE, SufBC2D, IscS, and IscU proteins were purified to homogeneity and PLP cofactor occupancy was greater than 90% for IscS and SufS (Supplemental Figure 1). Using 2 mM L-cysteine with 2 mM DTT, SufS liberated 2.6 nmol of S2− min−1mg−1, which is 20 times lower than IscS (51.7 nmol of S2− min−1mg−1) (Supplemental Figure 2). Based on this low turnover number, under these reaction conditions SufS is hardly able to qualify as a bona fide enzyme. Previously, activities of 19 nmol of S2− min−1mg−1 for SufS and 380 nmol of S2− min−1mg−1 for IscS were measured using 12 mM cysteine and 50 mM DTT [14]. Under the same conditions used in the previous study, we observed activities of 7.9 nmol of S2− min−1mg−1 for SufS and 312.8 nmol of S2− min−1mg−1 for IscS (data not shown). Addition of 4 molar equivalents of SufE (adding 2 µM SufE to 0.5 µM SufS) increases SufS activity to 41.9 nmol of S2− min−1mg−1 so that it is comparable to IscS (Supplemental Figure 2). Further addition of 4 molar equivalents of the SufBC2D complex (2 µM SufBC2D complex) to SufS and SufE further enhanced SufS activity to 172.6 nmol of S2− min−1mg−1, making SufS a more efficient sulfur mobilization enzyme than IscS under these conditions (Supplemental Figure 2). In agreement with recently published reports, we found that IscU, the sulfur receptor for IscS, did not enhance IscS activity under these conditions (Supplemental Figure 2) [26].
SufS removes sulfur from L-cysteine and forms persulfide (S0) on the active site residue C364. The persulfide intermediate of E. coli SufS directly transfers the sulfur atom to residue C51 of SufE and SufS activity is enhanced specifically by SufE [19,20]. To further probe the SufS-SufE reaction, we performed kinetic analyses of E. coli SufS while varying both components, L-cysteine and SufE, using the methylene blue assay to quantify sulfide production [20]. This in vitro reaction requires a non-physiological reductant (such as DTT) to reduce persulfide (S0) to sulfide (S2−) on SufS and SufE thereby allowing the sulfide to react with DMPD. The concentration of cysteine was varied from 0 to 500 µM in the presence of 4 µM SufE (Figure 1A) while the concentration of SufE was varied from 0 to 15 µM SufE at a fixed 2 mM concentration of L-cysteine (Figure 1B). Under these conditions, SufS showed Michaelis-Menten enzyme kinetics for L-cysteine and SufE as its two substrates. The kinetic parameters are listed in Table 1. Previous studies of the Erwinia chrysanthemi SufS-SufE reported that the SufS-SufE Km for L-cysteine was 500 µM and the Vmax = 900 mU/mg, which are both higher than the values measured for E. coli SufS-SufE (Table 1) suggesting that the E. coli system has a higher affinity for the L-cysteine substrate but is a somewhat slower system [18]. We also found that SufE where C51 has been covalently blocked with iodoacetamide (SufEalk) was able to inhibit SufS activity in the presence of unalkylated SufE with a Ki of 0.19 µM (Supplemental Figure 3). This inhibition occurred regardless of the presence of the SufBC2D complex.
Figure 1.
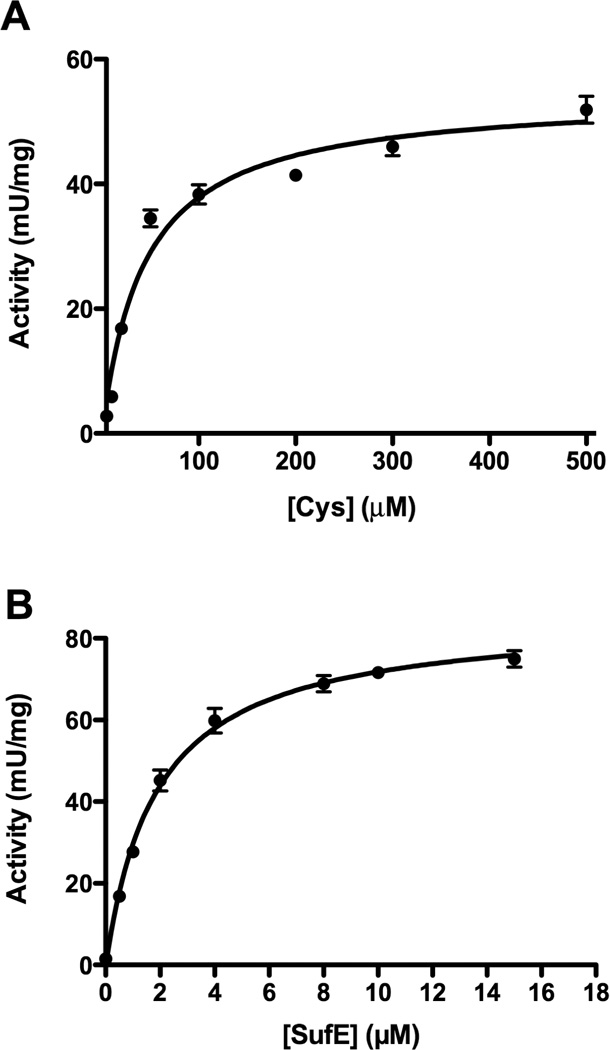
Kinetic analysis of SufS activity in response to varied substrate concentrations. The reactions contained (A) 0.5 µM SufS, 4 µM SufE, 2 mM DTT and 10 – 500 µM L-cysteine or (B) 0.5 µM SufS, 0 – 15 µM SufE, 2 mM DTT, and 2 mM L-cysteine. The lines are the best fits to the Michaelis – Menten equation obtained using GraphPad Prism.
Table 1.
Kinetic parameters of the SufS cysteine desulfurase.
| Cysteine dependenta | SufE dependentb | |
|---|---|---|
| Km (µM) | 43.5 ± 5.8 | 1.9 ± 0.1 |
| Vmax (mU/mg) | 54.3 ± 1.9 | 85.4 ± 1.8 |
| R2 | 0.95 | 0.99 |
Reaction conditions were: 0.5 µM SufS, 4 µM SufE, 2 mM DTT and 5 – 500 µM L-cysteine.
Reaction conditions were: 0.5 µM SufS, 0 – 15 µM SufE, 2 mM DTT, and 2 mM L-cysteine.
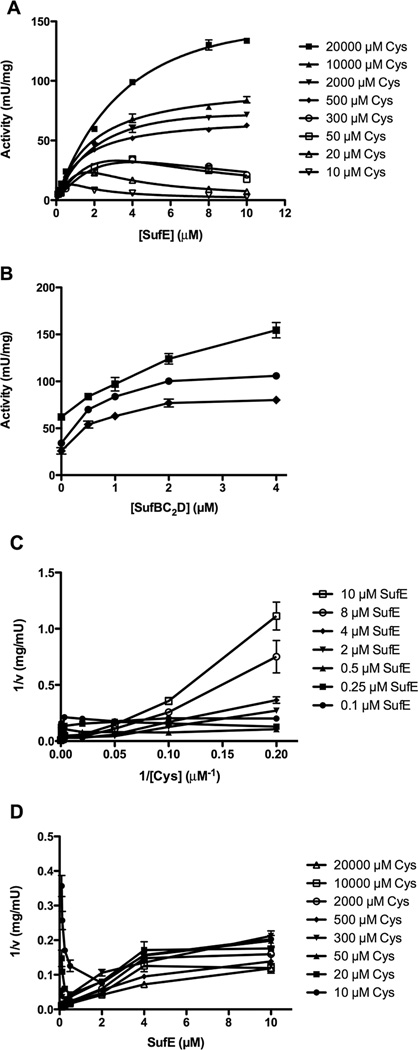
SufS Displays non-Michaelis-Menten Kinetics at Low but Physiological Cysteine Concentrations
SufS activity deviated from Michaelis-Menten enzyme kinetics when it was measured as a function of different concentrations of SufE but over a wider range of fixed L-cysteine levels (10 µM to 20 mM) (Figure 2A). At L-cysteine concentrations below 300 µM, increasing the concentration of SufE actually decreased sulfide formation by SufS (Figure 2A). As long as the L-cysteine concentration remained at 500 µM or higher the inhibition by SufE was not observed and SufS showed Michaelis-Menten kinetics (compare Figures 1B and 2A). Intracellular L-cysteine concentrations in E. coli are variable depending on growth conditions but can often be in the range of 100 – 200 µM [27], which is below the mM levels often used for in vitro cysteine desulfurase enzyme assays, so the deviation of SufS-SufE from Michaelis-Menten behavior under these conditions may be physiologically relevant.
Figure 2.
Substrate inhibition of SufS by SufE at lower concentrations of L-cysteine. (A) The reactions contain 0.5 µM SufS, 0 – 10 µM SufE, 2 mM DTT, and 10 – 20,000 µM L-cysteine (see embedded legend). (B) The reactions contain 0.5 µM SufS, 50 µM Cysteine, 2 mM DTT, 4 µM (●) or 8 µM (◆) SufE with increasing concentrations of SufBC2D (0 – 4 µM). A control reaction with 2 mM Cysteine, 2 mM DTT, 0.5 µM SufS, and 8 µM SufE with increasing concentrations of SufBC2D (0 – 4 µM) is also shown (■). Double reciprocal plots of kinetic data. Activity of 0.5 µM SufS, 2 mM DTT, and (C) varied 10 – 20,000 µM L-cysteine at several fixed concentrations of SufE or (D) varied 0.1 – 10 µM SufE at several fixed concentrations of L-cysteine. See embedded legend for symbol explanations.
To test whether inhibition by SufE affects SufBC2D enhancement of SufS at lower cysteine concentrations, we assayed SufBC2D enhancement at 50 µM cysteine where SufE showed inhibition of SufS (Figure 2B). For comparison SufBC2D enhancement at 2 mM L-cysteine (where SufE inhibition does not occur) is also shown in Figure 2B. The enhancement normally provided by the SufBC2D complex diminished as the fixed concentration of SufE increased, in stark contrast to the SufBC2D-dependent enhancement seen at higher L-cysteine levels (Figure 2B). These results indicate that SufBC2D cannot reverse the SufE inhibition of SufS that is seen at low cysteine concentrations.
The double reciprocal transformations of the kinetic data clearly show the SufS deviation from Michaelis-Menten behavior at lower cysteine concentrations (Figure 2C and 2D). At low fixed SufE concentrations, parallel lines are observed when initial velocity as a function of L-cysteine is plotted (Figure 2C). As the fixed concentration of SufE becomes inhibiting (2 µM SufE and above), the slopes of the reciprocal plots increase and the lines begin to cross at high L-cysteine concentrations (approaching the 1/v axis) as the SufE concentration approaches the substrate inhibition Ki (Figure 2C). Similarly, when L-cysteine is fixed at concentrations below 500 µM and initial velocity is plotted as a function of SufE, we observed that as SufE concentration increases (approaching the 1/v axis), the initial velocity sharply decreases (turns sharply upward) (Figure 2D). The activity plot and double reciprocal plots are qualitatively similar to those of O-acetylserine sulfhydrylase, a PLP-dependent enzyme that reacts via a ping-pong mechanism with substrate inhibition [28]. We attempted to fit our data with the appropriate rate equation for this type of substrate inhibition [28]. Unfortunately the quality of the fit was insufficient to instill confidence in the values for the substrate inhibition constant and other kinetic constants (data not shown). This leaves open the question of whether SufE inhibition is due to substrate inhibition. Previously it was shown that E. coli SufS itself (even in the absence of SufE) deviates from Michaelis-Menten kinetics, which may explain the difficulty in fitting the rate equation described for other enzymes [14].
The SufS-SufE system is More Active at Physiological Cysteine Concentrations Than IscS and IscS-IscU
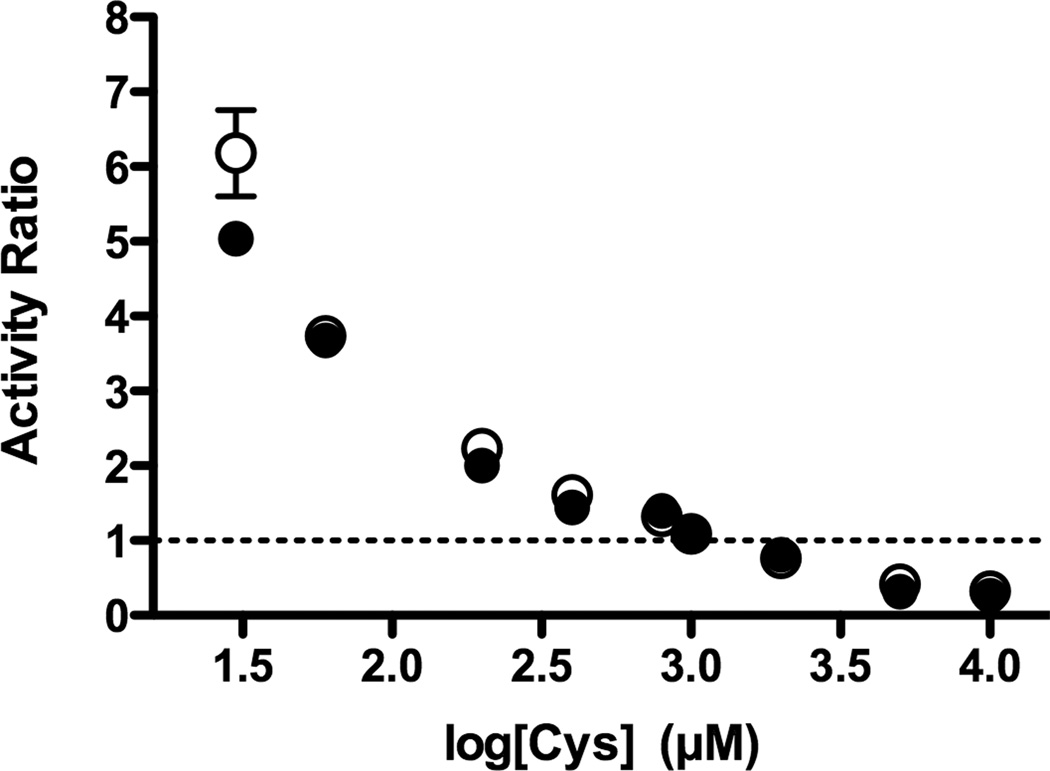
Next we directly compared the efficiency of the SufS-SufE sulfurtransferase system to that of the E. coli IscS and IscS-IscU proteins under the same conditions. The desulfurase activities of SufS-SufE, IscS alone, and IscS-IscU were measured at different concentrations of L-cysteine. A 1:3 molar ratio of SufS to SufE or IscS to IscU was used throughout. At a 1:3 molar ratio of SufS (0.5 µM) to SufE (1.5 µM), SufE does not show measurable inhibition of SufS activity over the range of L-cysteine concentrations used (30 µM – 10 mM). For ease of comparison, the activity of SufS-SufE at each L-cysteine concentration was divided by the activity of IscS alone or IscS-IscU measured under the same conditions and these activity ratios were plotted as a function of L-cysteine (Figure 3). For the activity ratios generated by these calculations, values greater than 1 indicate that SufS-SufE have a higher activity than IscS or IscS-IscU at those specific L-cysteine concentrations (Figure 3). This comparison reveals that the SufS-SufE system has higher cysteine desulfurase activity than IscS or the IscS-IscU system at physiological L-cysteine concentrations (up to 200 µM). At 30 µM L-cysteine SufS-SufE activity was 6-fold higher than IscS or the IscS-IscU system and remained at least 2-fold higher until the L-cysteine concentration exceeded 200 µM. Only at high L-cysteine concentrations above 1 mM did IscS or the IscS-IscU system begin to exceed SufS-SufE activity. These results also showed no activity difference between IscS alone compared to the IscS-IscU mixture over the range of L-cysteine tested (Figure 3).
Figure 3.
Direct activity comparison of the SufS-SufE and IscS-IscU sulfur transfer systems. SufSSufE activity was divided by IscS activity (closed circles ●) or the IscS-IscU activity (open circles ○) and the ratios were plotted as a function of the L-cysteine concentration in the reaction. The reactions contain 0.5 µM SufS or IscS, 1.5 µM SufE or IscU, and 0.03 – 10 mM L-cysteine and DTT.
IscS-IscU Activity is More Sensitive to H2O2 Exposure Than SufS-SufE
The Suf pathway is activated to build Fe-S clusters during oxidative stress in E. coli and deletion of the suf operon causes disruption of Fe-S cluster biosynthesis by oxidative stress [7–9,29,30]. In contrast, the Isc system is unable to carry out Fe-S cluster assembly in vivo upon exposure to reactive oxygen species like H2O2 [24]. Active site cysteine residues and persulfide intermediates in sulfur trafficking may react with oxidants like H2O2 depending on their exact pKa values [22,23]. To test if the SufS-SufE or IscS-IscU sulfur trafficking pathways are maintained under oxidative stress, we compared their relative in vitro H2O2 sensitivity. It is difficult to test for H2O2 sensitivity in the present of DTT due to the propensity for DTT itself to react with and consume H2O2 and the ability of DTT to reverse some H2O2-mediated thiol oxidation products, such as sulfenic acid [31,32]. Therefore the desulfurase reactions were carried out in the presence of H2O2 but in the absence of DTT under anaerobic conditions (Figure 4). Since the SufE and IscU sulfur acceptors may not be as efficiently recycled in the absence of DTT (see above), they were used in a 10:1 excess over SufS and IscS. The concentration of L-cysteine was increased to 2 mM to ensure adequate activity could be measured in the presence of H2O2. Interestingly, in the absence of DTT, excess IscU was now able to enhance IscS desulfurase activity by 1.5 fold (Figure 4A). This result suggests that if DTT is present it will normally outcompete IscU to release persulfide from IscS and explains why IscU enhancement is not usually observed in the unmodified assay where DTT is present.
Figure 4.
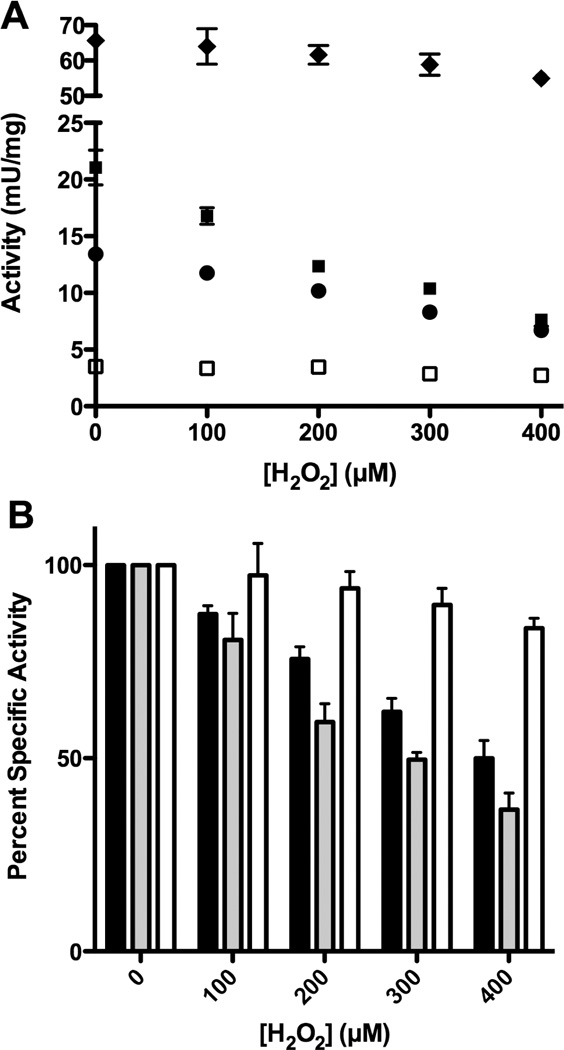
The sensitivity of SufS-SufE and IscS-IscU to H2O2 during the cysteine desulfurase reaction. 1 µM SufS or IscS and (where indicated) 10 µM SufE or IscU were mixed for 5 min. 2 mM L-cysteine was added to initiate the reaction followed immediately by 0 – 400 µM H2O2. After 30 minutes the reaction was quenched by heating at 95 °C for 5 minutes, followed by the addition of 2 mM DTT to reduce and release sulfide for measurement as described in Supplementary Materials Methods. All steps were carried out anaerobically. (A) Desulfurase activity of SufS (□), IscS (●), IscS-IscU (■) and SufS-SufE (◆). (B) Percent activity of IscS (black bar), IscS-IscU (light grey bar), and SufS-SufE (white bar) compared to their activity without H2O2.
Using this modified assay, we found that as the H2O2 concentration increased from 0 to 400 µM, sulfide production by IscS and IscS-IscU decreased by 50% or more (Figure 4). In contrast, sulfide production by SufS-SufE only decreased by about 10 – 15%. The percent decrease in IscS-IscU activity was greater than the percent decrease in the activity of IscS alone, suggesting that IscU enhancement of IscS is largely abolished in response to H2O2, possibly due to oxidative damage to IscU (Figure 4B). Furthermore, total sulfide production by SufS-SufE was always from 3 – 9 fold higher than IscS or IscS-IscU throughout the entire range of H2O2 concentrations used (Figure 4A). Together these results demonstrate that SufS-SufE sulfide production is more resistant to oxidative stress exposure than sulfide production by IscS or IscS-IscU.
Oxidation of IscS-IscU and SufS-SufE Residues After H2O2 exposure
The decrease in IscS and IscS-IscU activity in response to H2O2 suggests that important active site residues or reaction intermediates are damaged by oxidative stress. To map the sites of oxidation in the Isc and Suf sulfur transfer proteins, anaerobic cysteine desulfurase reactions were carried out in the presence of H2O2 as described above (in the absence of DTT) except that the reactions were quenched and trapped by the addition of tricholoroacetic acid (TCA) rather than by heating. TCA-trapped samples were alkylated, trypsinized, and analyzed by LC-MS without any further reduction steps as described in Supplementary Materials. Oxidation of the active site Cys residues C328 from IscS, C51 from SufE, and Cys 364 from SufS as well as conserved C63 and C106 in IscU were confirmed by MS/MS analysis of those peptides. The different oxidative modifications detected for the active site Cys residues or their reaction intermediates are summarized in Supplemental Table 2. Using this protocol, stable sulfenic acid modifications were not observed but the more stable sulfinic and sulfonic acid oxidation products were detected. The m/z peak areas for each modified peptide were separately quantified (see Supplementary Materials). For ease of comparison, the signal intensity for the oxidized forms of each specific Cys-containing peptide were pooled and divided by the total signal intensity for all forms of that Cys-containing peptide (Figure 5). These values can be used for relative comparisons between samples.
Figure 5.
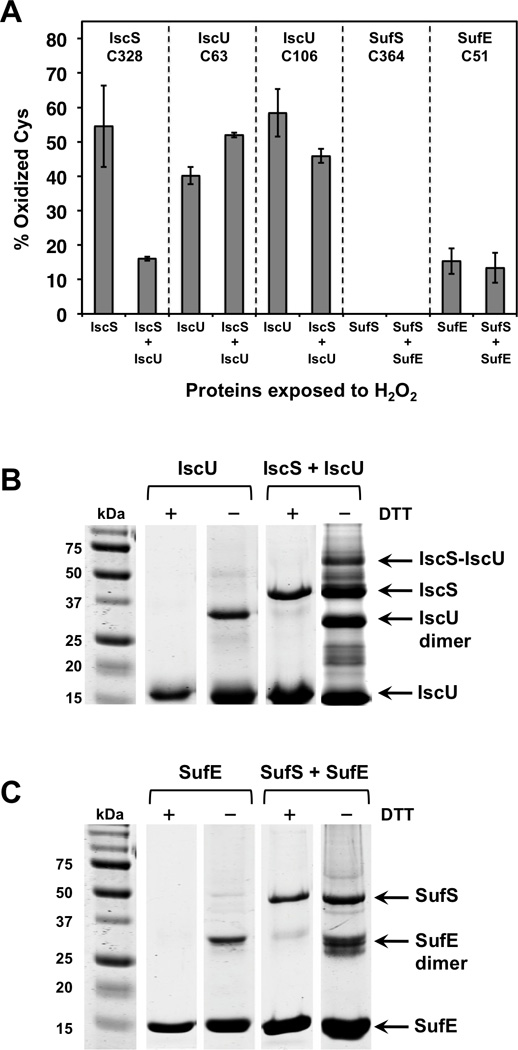
(A) Percent oxidation of active site Cys residues in the IscS, IscU, SufS, and SufE proteins after H2O2 exposure during the cysteine desulfurase reaction. (B) and (C) Reducing and non-reducing 12% SDS-PAGE gel separation of H2O2 treated proteins. The proteins were treated the same way as the samples for mass spectrometry analysis (see text). Proteins were precipitated with 10% TCA and dissolved in 1 X SDS loading buffer with or without DTT. Samples were heated at 95 °C for 10 min before loading on the gel. (B) IscU and IscS-IscU gel separation. (C) SufE and SufS-SufE gel separation.
For IscS treated with 400 µM H2O2, peptides with oxidative modification to the active site C328 accounted for 54% of the total signal intensity (Figure 5A), in rough agreement with the decrease in IscS activity observed under the same conditions (Figure 4). IscS C328 was more protected when IscU was added since the oxidized forms of C328 only represented 16% of the total signal intensity in that sample. In contrast, the total oxidation of IscU C63 by 400 µM H2O2 was 40% for IscU alone and 52 % for IscU in the presence of IscS. IscU C106 was fairly similar with 58% oxidation for IscU alone and 46% for IscU in the presence of IscS (Figure 5A). These results show that IscS is sensitive to oxidation by H2O2 during the desulfurase reaction cycle. While IscU seems to help prevent direct oxidation of IscS C328, probably by binding to and protecting IscS, we did detect disulfide bond formation between IscS-IscU (see below), which was not directly quantified by MS. IscU itself is oxidized by H2O2 even if IscS is present. Oxidized IscU can no longer enhance IscS activity and may also actively decrease IscS activity by acting as an inhibitor that competes with undamaged IscU for access to IscS. The percent oxidation of IscU C63 and C106 correlates with the percent decrease in IscS-IscU activity upon exposure to 400 µM H2O2 (Figure 4).
In contrast to IscS, oxidized forms of the peptides containing the SufS active site C364 were not detected after H2O2 exposure under these conditions, indicating that residue has intrinsic resistance to oxidative damage (Figure 5A). Peptides with oxidative modification to the SufE active site C51 were observed but only accounted for 15% of total signal in the absence of SufS and 13% in the presence of SufS. The generally lower levels of Cys oxidation in the SufS and SufE proteins correlate with their higher activity in the presence of H2O2 (Figure 4).
Disulfide bond formation is another potential consequence of H2O2 oxidation of Cys thiolates. The MS analysis was conducted without a reduction step (to allow detection of oxidized sulfane sulfur species) and may not adequately detect disulfide-bonded fragments, which tend to poorly ionize. Therefore, we also analyzed each oxidized sample qualitatively for the formation of mixed disulfides. After 400 µM H2O2 treatment, TCA-trapped samples were resuspended and separated by SDS-PAGE under both reducing (+DTT) and non-reducing (−DTT) conditions (Figure 5B, 5C). Regardless of H2O2 treatment, no high molecular weight species were detected for SufS and IscS alone and each protein migrated at its monomer molecular weight irrespective of DTT addition (data not shown). However, both SufE and IscU form disulfide bonded homodimers that are clearly delineated in the non-reducing gel (Figure 5B, 5C). Quantification of the intensity of the gel bands indicates that the relative level of SufE homodimer is fairly constant at about 18% of the total protein regardless of the addition of H2O2 (data not shown and Figure 5C). In contrast, the relative amount of IscU homodimer increases from 12% to 28% of total IscU protein upon exposure to H2O2. In the samples containing both IscS and IscU, the IscU homodimer increased to 39% of total IscU protein and we also observed the appearance of a new higher molecular weight species that runs at the expected size for a covalent IscS-IscU heterodimer. The IscS-IscU heterodimer band was excised from the gel and analyzed by mass spectrometry. The mass spectrometry results showed the presence of an H2O2-induced disulfide or polysulfide bond between IscS C328 and IscU C63, which is consistent with previous studies [33]. A disulfide bonded SufS-SufE heterodimer was not observed under our experimental conditions although small amounts of such a species have been seen for 35S-labeled SufS-SufE analyzed on a non-reducing gel [20]. Based on these results it appears that upon exposure to H2O2, both IscU and the IscS-IscU complex have a greater propensity to form covalently linked dimers compared to SufE and the SufS-SufE complex, providing an additional mechanism by which IscS activity may be inhibited by H2O2 exposure. While we did not absolutely quantify the total levels of IscU-IscU and IscS-IscU disulfide linked dimers, their formation would likely contribute to the decrease in IscS-IscU activity observed upon H2O2 treatment.
DISCUSSION
Substrate Inhibition of SufS by SufE May be a Physiological Adaptation
Using label transfer assays and surface plasmon resonance measurements we previously showed that SufS-SufE interact in the absence of L-cysteine with a KD of 0.36 µM [21]. Furthermore, previous yeast two-hybrid experiments indicate that the SufS C364S mutant, which cannot form a persulfide intermediate, interacts as well with SufE as the wild type SufS [34]. These published studies confirm that SufE interacts strongly with SufS regardless of SufS persulfide state, which is consistent with the potential substrate inhibition we observe at lower L-cysteine levels. Substrate inhibition by SufE could be a mechanism to limit SufS activity when cellular L-cysteine pools drop below a critical threshold. Measurable inhibition by SufE begins to occur if L-cysteine levels drop below 500 µM and if the ratio of SufE:SufS simultaneously increases beyond 4:1(which in the experiment is 2 µM SufE to 0.5 µM SufS). Depending on the exact in vivo ratio of SufE:SufS, which has not currently been measured, substrate inhibition may occur in vivo. Further experiments are necessary to fully explore this enzymatic behavior and its physiological relevance.
SufS-SufE Provide a More Robust Sulfur Transfer System Than the Isc Pathway
We found that SufS-SufE has higher cysteine desulfurase activity than IscS or IscS-IscU at physiological L-cysteine concentrations (200 µM and below), especially if the SufE:SufS ratio is maintained at 3:1 or lower. The higher activity of SufS-SufE at lower cysteine concentrations may be physiologically important for its oxidative stress resistance. Cysteine biosynthetic genes are upregulated under oxidative stress possibly to replenish free cysteine used for glutathione biosynthesis or replacement of oxidized protein thiols [35,36]. There is also evidence that L-cysteine is actively exported to the periplasm during oxidative stress to protect that sub-cellular compartment [37]. Since SufS-SufE has a higher desulfurase activity than IscS-IscU at lower cysteine concentrations the Suf system may be better able to maintain Fe-S cluster biosynthesis under conditions where L-cysteine availability decreases.
We observed a pronounced activity difference between the Isc and Suf sulfur trafficking proteins when they were exposed to H2O2 during the cysteine desulfurase reaction cycle. Under these conditions, IscS and IscS-IscU activity was inhibited while SufS-SufE activity was largely resistant to the H2O2 stress. Similar results were obtained when the resting proteins were exposed to H2O2 stress prior to initiating the desulfurase reaction (Supplemental Figure 4). MS analysis of the proteins shows that during enzyme turnover the active site Cys residues of IscS and IscU are sensitive to oxidation, forming dead-end sulfinic and sulfonic acid species as well as mixed disulfide heterocomplexes. In contrast, active site C364 of SufS remained unmodified throughout the stress. In addition, MS analysis revealed that the highly reactive S0 persulfide intermediates on IscS, IscU, and SufE, could also react with H2O2 to form cysteine-S-sulfonate derivatives (Supplemental Table 2). This is not surprising given that persulfides tend to have lower pKa values than thiols, making them an “activated” form of sulfur that could readily react with oxidants. Indeed in some organisms a cysteinyl persulfide is the substrate for enzymatic sulfur-oxidation rather than elemental sulfur (S8) and is oxidized to a cysteine-S-sulfonate derivative as part of the reaction cycle [38–40]. The relative stress resistance of the SufS-SufE system indicates that the active site Cys thiolates and persulfide intermediates for this sulfur transfer pathway are partially protected from reactive oxygen species compared to IscS-IscU.
In summary, the results above show that the SufS-SufE and SufS-SufE-SufBC2D sulfur transfer partners maintain higher desulfurase activity upon exposure to oxidative stress than the analogous IscS and IscS-IscU systems. The robust activity of SufS-SufE at physiological cysteine concentrations, coupled with the resistance of SufS-SufE activity to oxidative stress, indicate that the E. coli Suf pathway is well-suited to carry out Fe-S cluster biogenesis when it is induced under stress conditions.
Supplementary Material
Highlights.
We characterize the kinetics of the SufS-SufE sulfur transfer system
We directly compare SufS-SufE to the IscS-IscU system from E. coli
SufS-SufE are more active at physiological L-cysteine levels than IscS-IscU
SufS-SufE activity is more resistant to oxidative stress than IscS-IscU
ACKNOWLEDGMENTS
This study was supported by U.S. National Institutes of Health Grant GM 81706 to F.W.O. The authors would like to thank Dr. Patricia Dos Santos for insightful discussions and Jennifer R. Bethard, manager of the Mass Spectrometry Facility at the Medical University of South Carolina, for expert assistance in the mass spectrometry analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Materials Available.
REFERENCES
- 1.Meyer J. Iron-sulfur protein folds, iron-sulfur chemistry, and evolution. J Biol Inorg Chem. 2008;13:157–170. doi: 10.1007/s00775-007-0318-7. [DOI] [PubMed] [Google Scholar]
- 2.Sheftel A, Stehling O, Lill R. Iron-sulfur proteins in health and disease. Trends Endocrinol Metab. 2010;21:302–314. doi: 10.1016/j.tem.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 3.Shepard EM, Boyd ES, Broderick JB, Peters JW. Biosynthesis of complex iron-sulfur enzymes. Curr Opin Chem Biol. 2011;15:319–327. doi: 10.1016/j.cbpa.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 4.Xu XM, Moller SG. Iron-sulfur clusters: biogenesis, molecular mechanisms, and their functional significance. Antioxid Redox Signal. 2011;15:271–307. doi: 10.1089/ars.2010.3259. [DOI] [PubMed] [Google Scholar]
- 5.Rouault TA. Biogenesis of iron-sulfur clusters in mammalian cells: new insights and relevance to human disease. Dis Model Mech. 2012;5:155–164. doi: 10.1242/dmm.009019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imlay JA. Iron-sulphur clusters and the problem with oxygen. Mol Microbiol. 2006;59:1073–1082. doi: 10.1111/j.1365-2958.2006.05028.x. [DOI] [PubMed] [Google Scholar]
- 7.Nachin L, El Hassouni M, Loiseau L, Expert D, Barras F. SoxR-dependent response to oxidative stress and virulence of Erwinia chrysanthemi: the key role of SufC, an orphan ABC ATPase. Mol Microbiol. 2001;39:960–972. doi: 10.1046/j.1365-2958.2001.02288.x. [DOI] [PubMed] [Google Scholar]
- 8.Nachin L, Loiseau L, Expert D, Barras F. SufC: an unorthodox cytoplasmic ABC/ATPase required for [Fe-S] biogenesis under oxidative stress. Embo J. 2003;22:427–437. doi: 10.1093/emboj/cdg061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Outten FW, Djaman O, Storz G. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol Microbiol. 2004;52:861–872. doi: 10.1111/j.1365-2958.2004.04025.x. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi Y, Tokumoto U. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J Biol Chem. 2002;277:28380–28383. doi: 10.1074/jbc.C200365200. [DOI] [PubMed] [Google Scholar]
- 11.Kambampati R, Lauhon CT. IscS is a sulfurtransferase for the in vitro biosynthesis of 4-thiouridine in Escherichia coli tRNA. Biochemistry. 1999;38:16561–16568. doi: 10.1021/bi991119r. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi Y, Nakamura M. Functional assignment of the ORF2-iscS-iscU-iscA-hscB-hscA-fdx-ORF3 gene cluster involved in the assembly of Fe-S clusters in Escherichia coli. J Biochem (Tokyo) 1999;126:917–926. doi: 10.1093/oxfordjournals.jbchem.a022535. [DOI] [PubMed] [Google Scholar]
- 13.Agar JN, Zheng L, Cash VL, Dean DR, Johnson MK. Role of the IscU Protein in Iron-Sulfur Cluster Biosynthesis: IscS-mediated Assembly of a 2Fe-2S Cluster in IscU. J Am Chem Soc. 2000;122:2136–2137. [Google Scholar]
- 14.Mihara H, Kurihara T, Yoshimura T, Esaki N. Kinetic and mutational studies of three NifS homologs from Escherichia coli: mechanistic difference between L-cysteine desulfurase and L-selenocysteine lyase reactions. J Biochem (Tokyo) 2000;127:559–567. doi: 10.1093/oxfordjournals.jbchem.a022641. [DOI] [PubMed] [Google Scholar]
- 15.Schwartz CJ, Djaman O, Imlay JA, Kiley PJ. The cysteine desulfurase, IscS, has a major role in in vivo Fe-S cluster formation in Escherichia coli. Proc Natl Acad Sci U S A. 2000;97:9009–9014. doi: 10.1073/pnas.160261497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith AD, Agar JN, Johnson KA, Frazzon J, Amster IJ, Dean DR, Johnson MK. Sulfur transfer from IscS to IscU: the first step in iron-sulfur cluster biosynthesis. J Am Chem Soc. 2001;123:11103–11104. doi: 10.1021/ja016757n. [DOI] [PubMed] [Google Scholar]
- 17.Urbina HD, Silberg JJ, Hoff KG, Vickery LE. Transfer of sulfur from IscS to IscU during Fe/S cluster assembly. J Biol Chem. 2001;276:44521–44526. doi: 10.1074/jbc.M106907200. [DOI] [PubMed] [Google Scholar]
- 18.Loiseau L, Ollagnier-de-Choudens S, Nachin L, Fontecave M, Barras F. Biogenesis of Fe-S cluster by the bacterial Suf system: SufS and SufE form a new type of cysteine desulfurase. J Biol Chem. 2003;278:38352–38359. doi: 10.1074/jbc.M305953200. [DOI] [PubMed] [Google Scholar]
- 19.Ollagnier-de-Choudens S, Lascoux D, Loiseau L, Barras F, Forest E, Fontecave M. Mechanistic studies of the SufS-SufE cysteine desulfurase: evidence for sulfur transfer from SufS to SufE. FEBS Lett. 2003;555:263–267. doi: 10.1016/s0014-5793(03)01244-4. [DOI] [PubMed] [Google Scholar]
- 20.Outten FW, Wood MJ, Munoz FM, Storz G. The SufE protein and the SufBCD complex enhance SufS cysteine desulfurase activity as part of a sulfur transfer pathway for Fe-S cluster assembly in Escherichia coli. J Biol Chem. 2003;278:45713–45719. doi: 10.1074/jbc.M308004200. [DOI] [PubMed] [Google Scholar]
- 21.Layer G, Gaddam SA, Ayala-Castro CN, Ollagnier-de Choudens S, Lascoux D, Fontecave M, Outten FW. SufE transfers sulfur from SufS to SufB for iron-sulfur cluster assembly. J Biol Chem. 2007;282:13342–13350. doi: 10.1074/jbc.M608555200. [DOI] [PubMed] [Google Scholar]
- 22.Allison WS. Formation and Reactions of Sulfenic Acids in Proteins. Accounts of Chemical Research. 1976;9:293–299. [Google Scholar]
- 23.Everett SA, Wardman P. Perthiols as antioxidants: radical-scavenging and prooxidative mechanisms. Methods Enzymol. 1995;251:55–69. doi: 10.1016/0076-6879(95)51110-5. [DOI] [PubMed] [Google Scholar]
- 24.Jang S, Imlay JA. Hydrogen peroxide inactivates the Escherichia coli Isc iron-sulphur assembly system, and OxyR induces the Suf system to compensate. Mol Microbiol. 2010;78:1448–1467. doi: 10.1111/j.1365-2958.2010.07418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldsmith-Fischman S, et al. The SufE sulfur-acceptor protein contains a conserved core structure that mediates interdomain interactions in a variety of redox protein complexes. J Mol Biol. 2004;344:549–565. doi: 10.1016/j.jmb.2004.08.074. [DOI] [PubMed] [Google Scholar]
- 26.Iannuzzi C, Adinolfi S, Howes BD, Garcia-Serres R, Clemancey M, Latour JM, Smulevich G, Pastore A. The role of CyaY in iron sulfur cluster assembly on the E. coli IscU scaffold protein. PLoS One. 2011;6:e21992. doi: 10.1371/journal.pone.0021992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park S, Imlay JA. High levels of intracellular cysteine promote oxidative DNA damage by driving the fenton reaction. J Bacteriol. 2003;185:1942–1950. doi: 10.1128/JB.185.6.1942-1950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cook PF, Wedding RT. A reaction mechanism from steady state kinetic studies for O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2. J Biol Chem. 1976;251:2023–2029. [PubMed] [Google Scholar]
- 29.Lee JH, Yeo WS, Roe JH. Induction of the sufA operon encoding Fe-S assembly proteins by superoxide generators and hydrogen peroxide: involvement of OxyR, IHF and an unidentified oxidant-responsive factor. Mol Microbiol. 2004;51:1745–1755. doi: 10.1111/j.1365-2958.2003.03946.x. [DOI] [PubMed] [Google Scholar]
- 30.Tokumoto U, Kitamura S, Fukuyama K, Takahashi Y. Interchangeability and distinct properties of bacterial Fe-S cluster assembly systems: functional replacement of the isc and suf operons in Escherichia coli with the nifSU-like operon from Helicobacter pylori. J Biochem (Tokyo) 2004;136:199–209. doi: 10.1093/jb/mvh104. [DOI] [PubMed] [Google Scholar]
- 31.Winterbourn CC, Metodiewa D. Reactivity of biologically important thiol compounds with superoxide and hydrogen peroxide. Free Radical Biology and Medicine. 1999;27:322–328. doi: 10.1016/s0891-5849(99)00051-9. [DOI] [PubMed] [Google Scholar]
- 32.Zhang N, Schuchmann HP, Vonsonntag C. The Reaction of Superoxide Radical-Anion with Dithiothreitol - a Chain Process. Journal of Physical Chemistry. 1991;95:4718–4722. [Google Scholar]
- 33.Kato S, Mihara H, Kurihara T, Takahashi Y, Tokumoto U, Yoshimura T, Esaki N. Cys-328 of IscS and Cys-63 of IscU are the sites of disulfide bridge formation in a covalently bound IscS/IscU complex: implications for the mechanism of iron-sulfur cluster assembly. Proc Natl Acad Sci U S A. 2002;99:5948–5952. doi: 10.1073/pnas.082123599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sendra M, Ollagnier de Choudens S, Lascoux D, Sanakis Y, Fontecave M. The SUF iron-sulfur cluster biosynthetic machinery: sulfur transfer from the SUFS-SUFE complex to SUFA. FEBS Lett. 2007;581:1362–1368. doi: 10.1016/j.febslet.2007.02.058. [DOI] [PubMed] [Google Scholar]
- 35.Zheng M, Wang X, Templeton LJ, Smulski DR, LaRossa RA, Storz G. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J Bacteriol. 2001;183:4562–4570. doi: 10.1128/JB.183.15.4562-4570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blanchard JL, Wholey WY, Conlon EM, Pomposiello PJ. Rapid changes in gene expression dynamics in response to superoxide reveal SoxRS-dependent and independent transcriptional networks. PLoS One. 2007;2:e1186. doi: 10.1371/journal.pone.0001186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohtsu I, Wiriyathanawudhiwong N, Morigasaki S, Nakatani T, Kadokura H, Takagi H. The L-cysteine/L-cystine shuttle system provides reducing equivalents to the periplasm in Escherichia coli. J Biol Chem. 2010;285:17479–17487. doi: 10.1074/jbc.M109.081356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sauve V, Bruno S, Berks BC, Hemmings AM. The SoxYZ complex carries sulfur cycle intermediates on a peptide swinging arm. J Biol Chem. 2007;282:23194–23204. doi: 10.1074/jbc.M701602200. [DOI] [PubMed] [Google Scholar]
- 39.Rohwerder T, Sand W. The sulfane sulfur of persulfides is the actual substrate of the sulfur-oxidizing enzymes from Acidithiobacillus and Acidiphilium spp. Microbiology. 2003;149:1699–1710. doi: 10.1099/mic.0.26212-0. [DOI] [PubMed] [Google Scholar]
- 40.Quentmeier A, Friedrich CG. The cysteine residue of the SoxY protein as the active site of protein-bound sulfur oxidation of Paracoccus pantotrophus GB17. FEBS Lett. 2001;503:168–172. doi: 10.1016/s0014-5793(01)02727-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.