Abstract
Transcription initiation by eukaryotic RNA polymerase (Pol) III relies on the TFIIE-related subcomplex C82/34/31. Here we combine cross-linking and hydroxyl radical probing to position the C82/34/31 subcomplex around the Pol III active center cleft. The extended winged helix (WH) domains 1 and 4 of C82 localize to the polymerase domains clamp head and clamp core, respectively, and the two WH domains of C34 span the polymerase cleft from the coiled-coil region of the clamp to the protrusion. The WH domains of C82 and C34 apparently cooperate with other mobile regions flanking the cleft during promoter DNA binding, opening, and loading. Together with published data, our results complete the subunit architecture of Pol III and indicate that all TFIIE-related components of eukaryotic and archaeal transcription systems adopt an evolutionarily conserved location in the upper part of the cleft that supports their functions in open promoter complex formation and stabilization.
RNA polymerase III (Pol III) is the largest eukaryotic RNA polymerase, composed of 17 subunits with a total molecular weight of ∼0.7 MDa (1). Pol III synthesizes certain small untranslated RNAs (e.g., tRNAs, 5S rRNA, U6 snRNA, and 7SL RNA) involved in RNA processing and translation and in protein translocation (2, 3). Human Pol III mutations have been implicated in a neurodegenerative disorder, hypomyelinating leukodystrophy (4–6).
The three eukaryotic RNA polymerases share a similar 12-subunit core as represented by the X-ray structure of yeast Pol II (1). In addition to the core, Pol III contains five specific subunits forming two subcomplexes, C37/53 and C82/34/31. The C37/53 subcomplex participates in promoter opening, transcription termination, and polymerase reinitiation (7–9). The C34 subunit of the C82/34/31 subcomplex plays a role in open complex formation and in recruiting Pol III to the preinitiation complex (PIC) through interaction with TFIIB-related factor 1 (Brf1) (10–13). The human RPC62/39/32 subcomplex, homologous to the yeast C82/34/31 complex, is dissociable and required for promoter-specific initiation (11).
The two Pol III-specific subcomplexes contain structural domains homologous to domains in the Pol II transcription factors TFIIF and TFIIE, including the TFIIF-related dimerization module in the C37/53 subcomplex and several TFIIE-related winged helix (WH) domains in subunits C82 and C34 (14–20). TFIIE is composed of two subunits, Tfa1 and Tfa2, in yeast, or TFIIEα and TFIIEβ in humans. Whereas Tfa1 bears an extended WH (eWH) domain in its N-terminal region, Tfa2 has two adjacent WH domains (21, 22). Two adjacent Tfa2-related WH domains are also present in the C34 subunit and its human homolog RPC39. Pol I contains the A49/34.5 subcomplex that features a TFIIF-like dimerization module and a tandem WH domain that contains two Tfa2-like WH folds in the C-terminal region of the A49 subunit (18). The crystal structure of the human C82 homolog RPC62 contains four Tfa1-like eWH domains (eWH1–4) that are structurally organized around a C-terminal coiled-coil stalk (20). A Tfa1-related eWH domain is also present in the archaeal transcription factor E (TFE) (22).
Recent cryo-EM studies on the overall structural organization of the yeast Pol III suggested locations of the Pol III-specific subcomplexes and their contacts with the 12-subunit polymerase core. The C37/53 dimerization module was positioned into the electron density adjacent to the lobe domain of the C128 subunit on one side of the polymerase cleft, similar to the localization of the TFIIF dimerization module and the A49/34.5 dimerization module on Pol II and Pol I, respectively (23–25). This structural arrangement was supported by site-specific photo-cross-linking and hydroxyl radical probing analyses that provided direct positional mapping for protein interactions (8).
The C82/34/31 subcomplex was proposed to occupy a large electron density region between the clamp and the stalk of the polymerase core, on the side of the cleft opposite the lobe (20, 24, 25). By fitting the crystal structure of hRPC62 into the yeast Pol III cryo-EM envelope, Lefèvre et al. proposed a model in which eWH2, eWH3, and the coiled-coil region are positioned on the clamp and eWH1 and eWH4 are exposed to solvent for single-stranded DNA binding (20). An alternative orientation was proposed by Fernández-Tornero et al. that placed eWH4 closer to the stalk of the polymerase core (26). Thus, it currently remains unclear how C82 and the C82/34/31 subcomplex are positioned on Pol III. To resolve this issue, protein–protein interactions between the C82/34/31 subcomplex and the Pol III core must be mapped.
Here we site-specifically inserted photo-cross-linking amino acids into C160 and C82 and identified protein regions involved in C82-polymerase core interactions by site-directed hydroxyl radical probing. Our data reveal that C82 is anchored on the Pol III clamp via its domains eWH1 and eWH4. Mass spectrometric (MS) analysis of peptides obtained by tryptic digestion of lysine–lysine cross-linked Pol III is consistent with these results, and additionally locates the coiled-coil stalk of C82 at the polymerase clamp–stalk junction near the ABC27 (Rpb5) and ABC23 (Rpb6) subunits of the Pol III core. Cross-linking-MS analysis also positions the two adjacent WH domains of C34 above the active site cleft. The resulting detailed interaction map completes the subunit architecture of Pol III and provides insights into the function of the C82/34/31 subcomplex during transcription initiation.
Results
C82 and C34 Reside on the Polymerase Clamp.
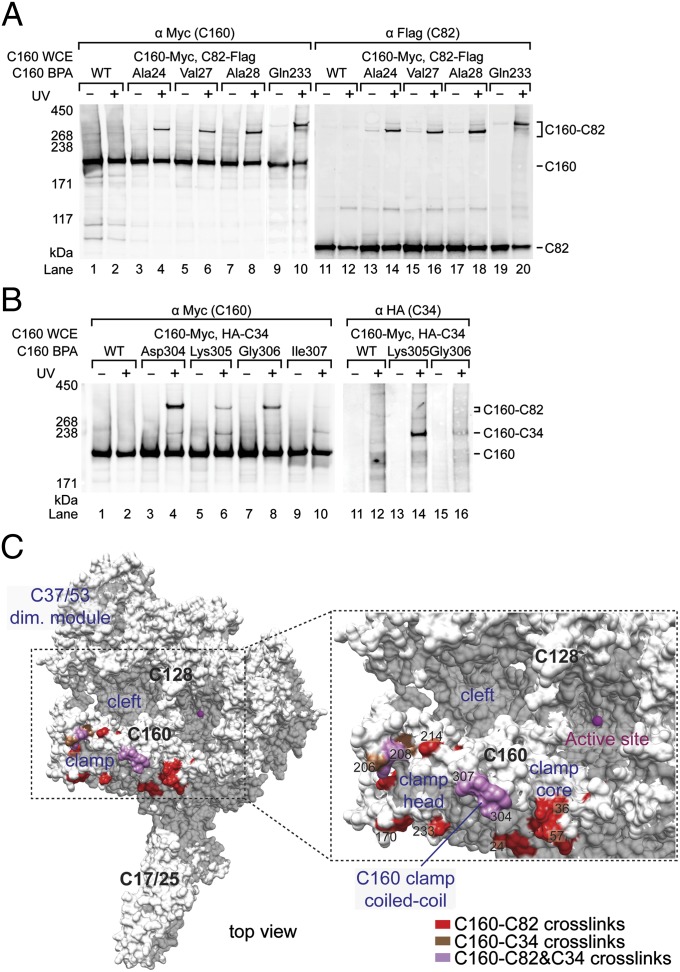
To elucidate protein interactions for the Pol III active center cleft, we replaced surface residues of the C160 clamp core and clamp head domains with the nonnatural amino acid photo-cross-linker p-benzoyl-L-phenylalanine (BPA). The sites were chosen at presumed interfaces of the Pol III core with the C82/34/31 subcomplex based on previous Pol III models (24, 25, 27). We conducted photo-cross-linking by using yeast whole-cell extracts containing C160-BPA derivatives in the immobilized template assay to probe potential protein targets within the Pol III PIC. C160-cross-linked polypeptides were found to have estimated molecular weights of 80 and 30 kDa, corresponding approximately to the sizes of C82 and C34 or C31. We epitope-tagged the possible candidates and confirmed the cross-linked polypeptides by identifying the epitope-containing cross-linked fusion in Western blot analysis, as described (8). Fig. 1A shows Western blot analysis revealing C160–C82 cross-links identified by antibodies against epitope-tagged C160 and C82 (anti-Myc for C160 and anti-Flag for C82).
Fig. 1.
C160 BPA photo-cross-linking indicates C82 and C34 reside on the C160 clamp domain. (A) Western blot of C82–C160 cross-links. Amino acid positions in C160 clamp replaced by BPA are indicated above the lanes. C160 and cross-linking bands were visualized by probing with anti-Myc antibody (C160-Myc, Left), and the cross-linking bands are confirmed to be C160–C82 fusion by probing Flag-tagged C82 (Right). C160 WCE, C160 whole-cell extract; UV + or −, with or without UV irradiation; WT, wild-type C160 with no BPA replacement. (B) C34-C160 cross-links. C160 and C34–C160 cross-linking bands were visualized by anti-Myc antibody (Left) and anti-HA antibody to reveal N-terminally HA-tagged C34 (Right). As indicated, these C160-BPA derivatives also generate simultaneous C82-C160 cross-links. (C) Positions in C82– and C34–C160 cross-links in polymerase clamp. The yeast Pol III core-C37/53 surface model is colored white. Magenta sphere: magnesium ion in the polymerase active site. As indicated, C160 residues yielding C82 and C34 cross-links are shown in red and brown, respectively. The residues cross-linking to both C82 and C34 are colored mauve.
This analysis reveals C82 cross-linking to C160 residues spanning from the upstream clamp core to the downstream clamp head (Fig. 1C; summarized in Table S1). Additionally, we found that four consecutive residues (Asp304 to Ile307) in the coiled-coil motif of the clamp core yielded a cross-link to C82 and another cross-link to C34 (Fig. 1 B and C). Cross-linking to C34 is also located from Ala206 to Pro209 in the clamp head, and His207 and Asn208 shows simultaneous cross-linking to C82 (Fig. 1C). Taken together, our cross-linking results indicate that the C82/34/31 subcomplex is mainly anchored on the clamp domain of C160 via its C82 subunit, and that C34 contributes to contacts along the rim of the clamp.
C82 Is Involved in a Protein Interaction Network.
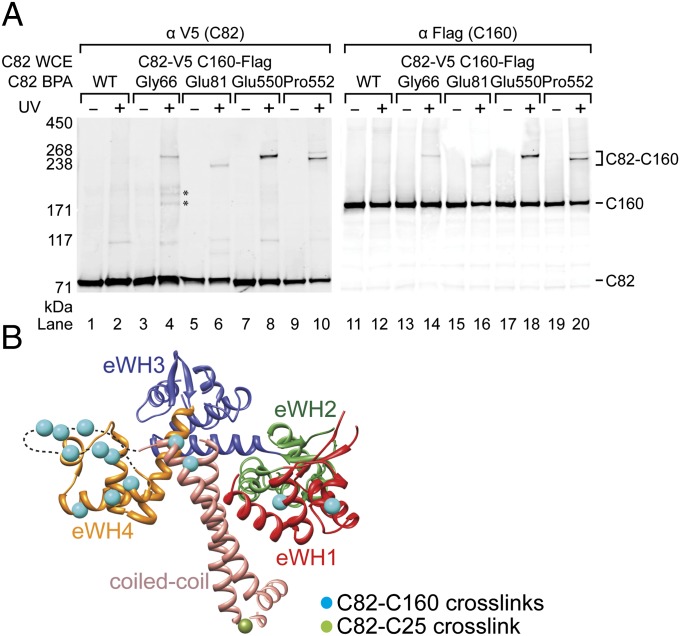
Our C160 cross-linking results indicate that C82 and C34 reside on the clamp domain but did not suffice to position the domains of these subunits on the Pol III core. We therefore conducted an extensive BPA cross-linking analysis for C82 that is summarized in Fig. S1A and Table S2. The data reveal C82–C31 and C82–C34 intrasubcomplex cross-links. Representative C82–C31 cross-links are shown in Fig. S1B. C31-cross-linked residues mostly lie within the C82 eWH1 and eWH2 domains, and one resides in its coiled-coil domain (Fig. S1A). By contrast, C34-cross-linked residues are scattered in eWH3 and eWH4, except for one in eWH2 (Fig. S1 A and C). This C82–C34 binding interface is consistent with the previously proposed C82–C34 interaction obtained by protein pull-down experiments (20). C82 shows distinct interaction surfaces for C31 and C34, which suggests a mainly linear connectivity for the three subunits, consistent with a previous mass spectrometric analysis (28).
Concerning the interaction of C82 with the Pol III core, residues in eWH1, eWH4, the first insertion of eWH3, and the C-terminal coiled-coil stalk yield C82–C160 cross-links (Fig. 2A and Fig. S1A). By mapping the positions cross-linked to C160 on the yeast C82 homology model based on the human RPC62 structure, we reveal a protein interaction surface for C160 binding that comprised eWH1, eWH4, and the coiled-coil stalk of C82 (Fig. 2B). In addition to C160 and intrasubcomplex cross-links, Leu620-BPA near the tip of C82 coiled-coil showed simultaneous cross-links to C25 of the polymerase stalk and C53 of the C37/53 subcomplex (Fig. S1 A and D). This result is consistent with our previous data indicating that the C53 N terminus extends toward the polymerase stalk to allow for cross-linking to C82 and C25 (8).
Fig. 2.
C82 BPA photo-cross-linking. (A) Western blot of C82–C160 cross-linking. BPA-substituted residues in C82 are indicated above. C82 and cross-linking fusion bands are revealed by anti-V5 antibody (Left). The Flag-epitope tagged C160 in the cross-linking bands is revealed by anti-Flag antibody (Right). Asterisks indicate unidentified cross-linked polypeptides. (B) Amino acid positions cross-linked to C160 in the yeast C82 model. The yeast C82 homology model (displayed as the ribbon model of Cα trace) is generated by the Modeler program (42) using the human RPC62 structure (PDB 2XUB) (20) as the template. The dashed line indicates the missing residues (aa547–567 in C82) in the eWH4. C82 structural domains are colored in red (eWH1), green (eWH2), blue (eWH3), orange (eWH4), and salmon (coiled-coil). The color scheme is used for all following figures. Residues cross-linking to C160 and C25 are highlighted with cyan and olive spheres, respectively.
C82 eWH1 and eWH4 Reside on the Polymerase Clamp.
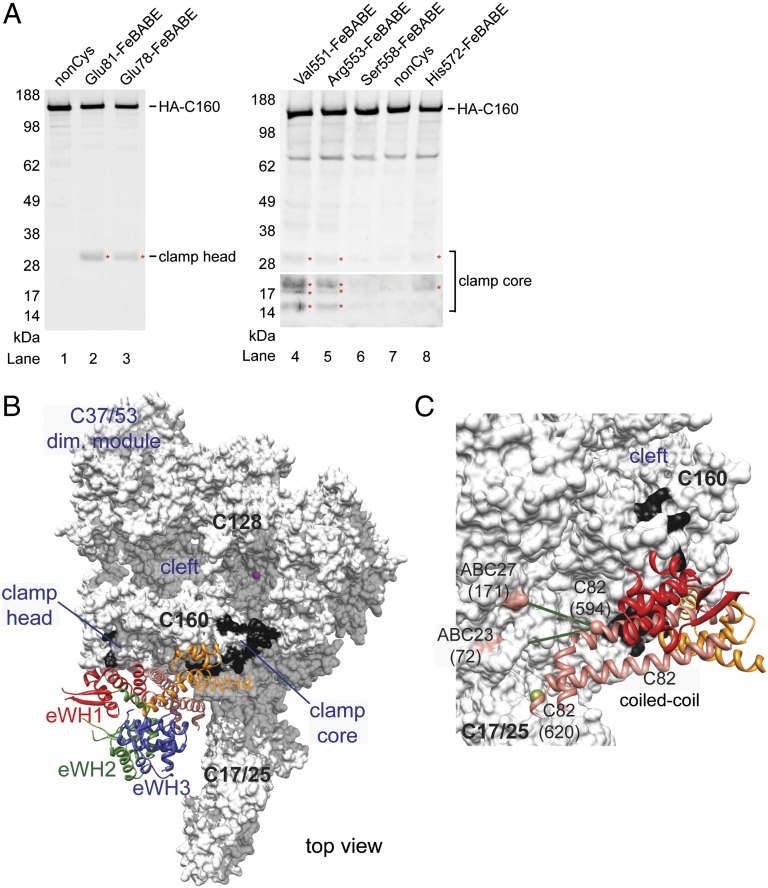
To more precisely localize C82 domains on the C160 clamp domain, we applied directed hydroxyl radical probing analysis with C82-FeBABE conjugates. We purified a series of recombinant C82 single-cysteine variants to tether the ⋅OH-generating FeBABE reagent at defined Cys positions. These C82-FeBABE conjugates were combined with purified recombinant C34 and C31 to reconstitute the C82/34/31 subcomplex (purified proteins are shown in Fig. S2A). The reconstituted subcomplex was subsequently titrated into a C82 mutant yeast whole-cell extract (Fig. S2B) for Pol III PIC formation with the immobilized template assay. All FeBABE-tethered C82 variants are shown to restore transcription activity (Fig. S2C). Fig. 3A shows hydroxyl radical cleavage of C160 from individual C82 variants with FeBABE tethered at Cys positions in the eWH1 and eWH4 domains. By mapping the cleavage sites to the Pol III core structural model, we found that eWH1 and eWH4 are respectively localized to the clamp head and clamp core regions (Fig. 3B; summarized in Table S3).
Fig. 3.
Localization of C82 on the C160 clamp. (A) Directed hydroxyl radical cleavage of C160. Western blots of C160 cleavage from tethered FeBABE in C82 eWH1 and eWH4 are respectively shown in the left and right panels. C-terminally HA-tagged C160 fragments are revealed by anti-HA antibody. FeBABE positions in C82 are indicated above the lanes. Cleavage fragments in the lower-molecular-weight range are displayed with signals amplified (Bottom Right). Red asterisks mark identified cleavage fragments. Approximate locations of cleavage sites in C160 are indicated. (B) C82-Pol III core model. C82 (ribbon model) is manually docked as a rigid body on the Pol III core model (white surface model) based on directed hydroxyl radical cleavage and cross-linking-MS analyses. Hydroxyl radical cleavage regions in C160 clamp are highlighted in black covering 11 residues centered at the deduced cut site. (C) Cross-linked lysine pairs between C82 coiled-coil and subunits of the Pol III core. Green lines connect cross-linked lysine pairs from C82 (Lys594) to ABC27 (Lys171) and ABC23 (Lys72). The BPA substitution at C82 Leu620, highlighted as an olive sphere, cross-links to C25.
Cross-Linking-MS Analysis of Pol III.
Although BPA cross-linking and hydroxyl radical probing localized C82 eWH1/4 on the C160 clamp, additional structural restraints are required to further define the interface between C82 and the Pol III core for structural modeling. To this end, we used lysine–lysine cross-linking followed by MS identification of the cross-linked peptides as recently described for Pol I (23). A purified endogenous Pol III sample was subjected to cross-linking with disuccinimidyl suberate (DSS), a reagent that reacts with primary amines present in lysine side chain and protein N terminus. DSS cross-linking of Pol III was monitored by tryptic digestion and MS analysis. A complete list of cross-linked lysines and their corresponding peptide pairs is provided in Table S4.
The cross-linking identified linkage pairs that validate a previous model of the Pol III core that was based on the Pol II structure (27, 29). In addition, distance restraints position the Pol III-specific subcomplex C37/53 on the Pol III core, supporting the previously proposed model for the C37/53 dimerization module on the lobe domain of C128 (Fig. S3 A and B) (8, 24, 25, 29). Cross-links also reveal the location of the C-terminal region of C37 in the active center cleft (Fig. S3B) and are generally consistent with contacts between the N-terminal region of C53 and the coiled-coil domain of C82 and the C17/25 stalk subcomplex (Fig. S1D), supporting previous BPA cross-linking and FeBABE analyses (8).
Model for C82 Binding to the Pol III Core.
A schematic summary for C82 lysine–lysine cross-links is depicted in Fig. S4. We observed intramolecular cross-links to support the yeast C82 homology model based on the human RPC62 structure (29). In combination with the C82-FeBABE hydroxyl radical analysis (see above), we used intermolecular cross-links between C82 coiled-coil and the Pol III core subunits ABC27 (Rpb6) and ABC23 (Rpb5) to manually dock the C82 model on the Pol III core (Fig. 3 B and C). In the resulting model, the C82 domains eWH1 and eWH4 are located on the clamp head and clamp core in C160, respectively, whereas the C82 coiled-coil extends toward subunits ABC27 and ABC23. C82 coiled-coil is also positioned adjacent to the Pol III stalk, consistent with BPA cross-linking between the tip of C82 coiled-coil and C25 of the Pol III stalk (Fig. 3C). Further, the model satisfies all BPA photo-cross-linking results for C160 and C82 (Fig. S4 B and C).
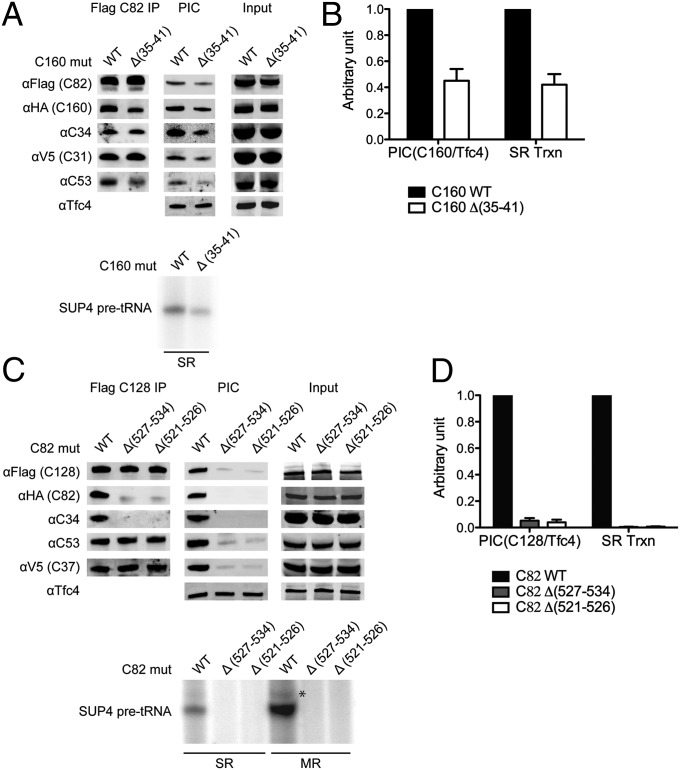
To validate our model of C82 on the Pol III core, we generated yeast strains containing mutations in the predicted C160 clamp–C82 interface and tested their in vivo and in vitro phenotypes. We obtained a deletion mutant C160 ∆ (35–41) that shows a slow-growth phenotype at elevated temperature but no significant change in the cellular C160 protein level (Fig. 4A, Input). In an immunoprecipitation assay by immobilizing C82, the C34 and C31 subunits of the subcomplex are coimmunoprecipitated, whereas the mutant C160 and another Pol III subunit C53 are partially dissociated (Fig. 4A, Flag C82 IP). By analyzing PIC formation on immobilized SUP4 tDNA, C160 ∆ (35–41) mutant showed ∼50% reduction in Pol III recruitment (Fig. 4 A, PIC and B). In agreement with the defect in Pol III recruitment, we observed a concomitant decrease in transcription initiation for the C160 mutant based on the single-round transcription analysis (Fig. 4 A Lower and B).
Fig. 4.
Association of the C82/34/31 subcomplex with the Pol III core. (A) Western blot analyses of coimmunoprecipitation and Pol III PIC formation with yeast whole-cell extract containing a C160 mutation. Coimmunoprecipitation was conducted with anti-Flag agarose to immobilize Flag-epitope-tagged C82 (Flag C82 IP; Upper Left). Pol III PIC formation assay was conducted with the immobilized SUP4 tDNA (Upper Middle). The level of a TFIIIC subunit Tfc4 in the PIC is also shown. As indicated, whole-cell extracts containing wild-type (WT) or internal deletion Δ (35–41) C160 were used. Single-round in vitro transcription of the isolated Pol III PIC was assayed (Lower). (B) Immunoblot and in vitro transcription signals were quantified from three independent experiments with WT signals set to 1. Error bars indicate SD. (C) Western blot analyses of coimmunoprecipitation and Pol III PIC formation with yeast whole-cell extracts containing C82 mutations. Similar to the analyses in A, whole-cell extracts containing WT or the indicated mutations in C82 eWH4 were used in coimmunoprecipitation, PIC formation, and in vitro transcription analyses. Different from the analysis in A, immune pull-down was conducted by immobilizing Flag-epitope-tagged C128, and both single-round (SR) and multiple-round (MR) transcription were assayed (Lower). An asterisk indicates the read-through transcripts. (D) The results for PIC formation and SR transcription from C are quantified and plotted with WT signals set to 1. Errors bars indicate SD from three independent experiments.
Further mutagenesis study on C82 allowed us to isolate two internal deletion mutations in C82 eWH4 that confer temperature-sensitive growth defect. Coimmunoprecipitation analysis by immobilizing the Pol III core subunit C128 indicates that these mutations lead to dissociation of C82 and C34 from the Pol III core (Fig. 4C). This association defect severely compromises Pol III recruitment and causes complete loss of in vitro transcription activity (Fig. 4 C and D), in agreement with a role for the human C82/34/31 homolog in transcription initiation (11). Taken together, our mutational and functional analyses support the proposed model for C82 binding to the Pol III core.
C34 Bridges the Active Center Cleft.
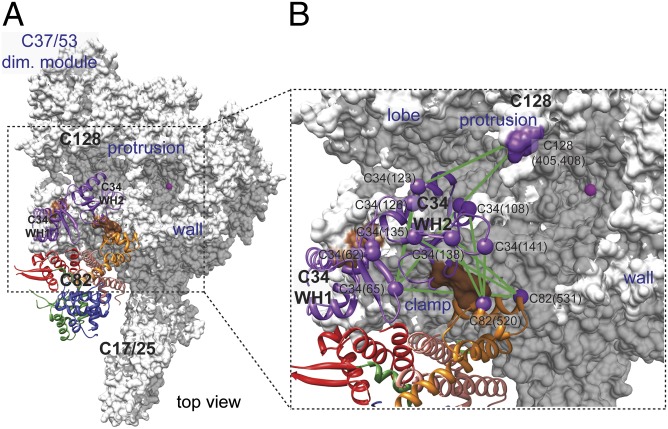
In addition to C82 positioning, we used lysine–lysine cross-linked peptide pairs to model the positions for C34 WH domains. Homology models for C34 WH1 and WH2 were generated based on the WH structures in mouse and human RPC39, respectively (Protein Data Bank accession numbers 2DK8 and 2DK5). Based on a total of eight intersubunit cross-links, we docked the C34 WH2 model above the Pol III active center cleft to contact C128 protrusion, C160 clamp coiled-coil, and the eWH4 domain of the previously modeled C82 (Fig. 5). With the positioning for WH2, the adjacent WH1 is localized on the rim of the C160 clamp head based on five intrasubunit cross-links with WH2 and a single intersubunit cross-link with C82 eWH4. The positions for WH1 and WH2 are consistent with the localization of C34 on the polymerase clamp based on the BPA cross-linking results for C160 (Figs. 5 and 1C). Further, several cross-link pairs are observed between the C34 WH domains and the C37 C-terminal region, supporting the previously described location for the C37 C-terminal region near the Pol III active site (8) (Fig. S3C).
Fig. 5.
Model of C34 WH domains in Pol III based on lysine–lysine cross-links. Model of C34 and C82 on the Pol III core is shown on the left. C34 WH1 and WH2 ribbon models are colored purple. In the Pol III cleft view on the right, green lines connect intrasubunit and intersubunit cross-linked lysines (purple spheres with labels indicating subunit names and residue numbers). Brown patches on C160 clamp indicate the BPA-substituted residues in C160 that cross-linked to C34.
Intersubunit cross-linked peptide pairs within the C82/34/31 subcomplex further suggest the positions of the C34 C-terminal region and C31 on C82. The C34 C-terminal region contacts C82 eWH3, and C31 is positioned on the surface formed by C82 eWH1, eWH2, and the coiled-coil stalk (Fig. S5 A and B). The C31 position is also supported by cross-links with the C160 clamp head, the C17/25 stalk, and ABC27 (Fig. S5A). Furthermore, the distinct location of C34 and C31 and no observable cross-links between C34 and C31 are in perfect agreement with the linear connectivity for the C82/34/31 subcomplex indicated by our C82 BPA-cross-linking results and a previous MS study (28). Taken together, the multiple experimental approaches have enabled us to arrive at the complete domain architecture for the 17-subunit Pol III enzyme.
Discussion
Here we present the complete subunit domain architecture of Pol III, the largest eukaryotic RNA polymerase, derived from an integrative approach that combines nonnatural amino acid photo-cross-linking, directed hydroxyl-radical probing, MS analysis of lysine–lysine chemically cross-linked peptides, and molecular modeling based on X-ray crystallographic structures. This work identified subunit–subunit interfaces within Pol III, and in particular revealed the location of subcomplex C82/34/31 around the Pol III clamp and cleft. The C82 domains eWH1 and eWH4 are localized on the clamp head and clamp core, respectively, whereas the C82 C-terminal coiled-coil domain binds between the polymerase clamp and stalk and reaches near the subunits ABC27 and ABC23. This model differs from previously proposed relative orientations of the C82 human homolog structure on Pol III based on electron microscopic data alone (20, 24, 25). The C34 domain WH2 resides between the C160 coiled-coil motif on one side of the polymerase cleft and the C128 protrusion domain on the other side, whereas the C34 domain WH1 localizes to the clamp head. C31 is positioned adjacent to the clamp head and contacts C82 eWH1, eWH2, and its coiled-coil domain, ABC27, and the C17/25 stalk. The subunit domain model of Pol III derived here is generally consistent with the molecular envelope of the Pol III EM structure (24, 25) (Fig. S6 A and B).
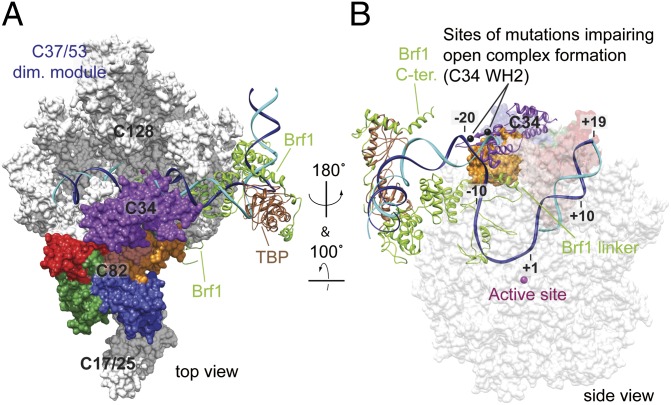
To derive implications for the function of C82 and C34 in Pol III initiation, we extended the Pol III model to a model of the Pol III open promoter complex (Fig. 6). The open promoter complex model additionally contains the TATA box-binding protein TBP and the TFIIB-related factor Brf1 based on previous analysis of the related Pol II open complex (37). This model is consistent with previously reported protein–DNA cross-linking results, in which C34 was cross-linked to the farthest upstream position at −21 with respect to the transcription start +1, and C82 was cross-linked both upstream at positions −8/−7 and downstream at +11 (Fig. 6B) (13, 38). The C34 location correlates well with its double-stranded DNA binding ability and its functional roles in DNA melting (12, 20). With their positions flanking the DNA bubble of the Pol III open complex model (Fig. 6B), the C34 WH domains potentially contribute their DNA binding residues to function in initial strand separation and/or subsequent bubble stabilization. In accordance with this proposed mechanism, two amino acid residues in the WH2 domain of C34 that were previously reported to be involved in DNA binding and open complex formation (12, 20) may interact with the upstream edge of the transcription bubble (Fig. 6B). Our data and published results thus converge on a model that C34 is positioned over the active center cleft to stabilize the DNA bubble and aid in open complex formation. This model relies on C34 mobility, because promoter DNA must first be loaded into the cleft during the transition from the closed to the open complex, and this requires transient displacement of C34 from the location observed here. Weak evidence to support C34 mobility comes from the slightly different locations for C34 domains in Pol III and Pol III-DNA models derived by EM (20, 24, 25) but merits further exploration.
Fig. 6.
Pol III architecture and open promoter complex. (A) Model for the Pol III open promoter complex. The model of Pol III-Brf1-TBP-DNA open promoter complex was built based on the Pol II-TFIIB-TBP open complex (37) and the Brf1-TBP-DNA structure (43). Template and nontemplate DNA strands are in blue and cyan, respectively. The Pol III core-C37/53 model is displayed as the white surface model. Ribbon models for Brf1 and TBP are colored yellow-green and brown, respectively. The C82 surface model is colored according to the domain color scheme as above, and both C34 WH surface models are colored purple. (B) Protein–DNA organization in the Pol III active center. The Pol III open promoter complex in A is displayed with different orientation to view the Pol III active center. The Pol III core is semitransparent and C34 is changed to the ribbon model. Lys135 and Lys138 in C34 WH2 are highlighted as black spheres. These two amino acids are required for DNA binding and open complex formation (12, 20).
C82 and C34 may cooperate with the TFIIB-related factor Brf1 to stimulate open complex formation. In our Pol III model, C82 eWH4 and C34 WH domains are in the vicinity of the polymerase clamp coiled-coil (Fig. 6A and Fig. S6C). In Pol II, the clamp coiled-coil interacts with a region in TFIIB, the B-linker, which was also implicated in DNA opening (37, 39). During Pol III initiation, Brf1 likely contacts C82 and C34, because C34 was shown to bind Brf1 during Pol III recruitment (10, 40). Situated at the upstream edge of the DNA bubble (Fig. 6B), C82, C34, and Brf1 may provide essential protein–DNA interactions for initial spontaneous strand separation, similar to the function of σ2/3 in the bacterial system (30, 37, 39, 41). In particular, the C34 WH domains and C82 eWH4 could stabilize the emerging nontemplate DNA strand, whereas the template DNA strand slips into the cleft and binds to its floor (31, 32, 37). On the downstream side of the DNA bubble, C82 eWH1 could contact downstream DNA (Fig. 6A). Further, the C37 C-terminal domain and another transcription factor, Bdp1, may also bind the DNA bubble (8, 33), because they localize near the Pol III active center (Fig. S3C).
Our results suggest an evolutionary conservation of the mechanisms used by TFIIE-related components in the three eukaryotic and the archaeal transcription initiation machineries. In Pol II, TFIIE has been mapped to the coiled-coil motif and the clamp head of Rpb1 (34, 35). In Pol I, the TFIIE-related A49 tandem winged-helix domain occupies a position above the active center cleft similar to C34 (23). The archaeal TFIIE-like factor TFE was also localized near the clamp coiled-coil (36). Thus, all TFIIE-related components share the location within the upper active center cleft and a function in promoting formation and stabilization of the transcription bubble.
Materials and Methods
Detailed descriptions are available in SI Materials and Methods for plasmids and yeast strains, BPA photo-cross-linking, preparation of proteins, in vitro transcription, hydroxyl radical cleavage, and cross-linking-MS analysis. Immobilized DNA templates containing either SUP4 tRNA or U6 snRNA genes were prepared as previously described (8). The Pol III core-C37/53 model was built as previously described (8).
Supplementary Material
Acknowledgments
We thank Cheng-Feng Lo and Jin-Cheng Lee for assistance with cloning and development of purification strategies for C82, C34, and C31. This work was supported by National Science Council Grant 100-2311-B-001-013-MY3 and a Career Development Award (to H.-T.C.) from Academia Sinica. P.C. was supported by Deutsche Forschungsgemeinschaft Grant SFB646, TR5, Nanosystems Initiative Munich, the BioImaging Network, a European Research Council Advanced Grant, and a Jung-Stiftung grant. F.H. is supported by a European Molecular Biology Organization long-term fellowship and by European Commission Grant FP7-PEOPLE-IEF.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211665109/-/DCSupplemental.
References
- 1.Cramer P, et al. Structure of eukaryotic RNA polymerases. Annu Rev Biophys. 2008;37:337–352. doi: 10.1146/annurev.biophys.37.032807.130008. [DOI] [PubMed] [Google Scholar]
- 2.Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23(12):614–622. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Schramm L, Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16(20):2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- 4.Bernard G, et al. Mutations of POLR3A encoding a catalytic subunit of RNA polymerase Pol III cause a recessive hypomyelinating leukodystrophy. Am J Hum Genet. 2011;89(3):415–423. doi: 10.1016/j.ajhg.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tétreault M, et al. Recessive mutations in POLR3B, encoding the second largest subunit of Pol III, cause a rare hypomyelinating leukodystrophy. Am J Hum Genet. 2011;89(5):652–655. doi: 10.1016/j.ajhg.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saitsu H, et al. Mutations in POLR3A and POLR3B encoding RNA Polymerase III subunits cause an autosomal-recessive hypomyelinating leukoencephalopathy. Am J Hum Genet. 2011;89(5):644–651. doi: 10.1016/j.ajhg.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kassavetis GA, Prakash P, Shim E. The C53/C37 subcomplex of RNA polymerase III lies near the active site and participates in promoter opening. J Biol Chem. 2010;285(4):2695–2706. doi: 10.1074/jbc.M109.074013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu CC, Lin YC, Chen HT. The TFIIF-like Rpc37/53 dimer lies at the center of a protein network to connect TFIIIC, Bdp1, and the RNA polymerase III active center. Mol Cell Biol. 2011;31(13):2715–2728. doi: 10.1128/MCB.05151-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landrieux E, et al. A subcomplex of RNA polymerase III subunits involved in transcription termination and reinitiation. EMBO J. 2006;25(1):118–128. doi: 10.1038/sj.emboj.7600915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Werner M, Chaussivert N, Willis IM, Sentenac A. Interaction between a complex of RNA polymerase III subunits and the 70-kDa component of transcription factor IIIB. J Biol Chem. 1993;268(28):20721–20724. [PubMed] [Google Scholar]
- 11.Wang Z, Roeder RG. Three human RNA polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes Dev. 1997;11(10):1315–1326. doi: 10.1101/gad.11.10.1315. [DOI] [PubMed] [Google Scholar]
- 12.Brun I, Sentenac A, Werner M. Dual role of the C34 subunit of RNA polymerase III in transcription initiation. EMBO J. 1997;16(18):5730–5741. doi: 10.1093/emboj/16.18.5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartholomew B, Durkovich D, Kassavetis GA, Geiduschek EP. Orientation and topography of RNA polymerase III in transcription complexes. Mol Cell Biol. 1993;13(2):942–952. doi: 10.1128/mcb.13.2.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carter R, Drouin G. The increase in the number of subunits in eukaryotic RNA polymerase III relative to RNA polymerase II is due to the permanent recruitment of general transcription factors. Mol Biol Evol. 2010;27(5):1035–1043. doi: 10.1093/molbev/msp316. [DOI] [PubMed] [Google Scholar]
- 15.Knutson BA, Hahn S. Yeast Rrn7 and human TAF1B are TFIIB-related RNA polymerase I general transcription factors. Science. 2011;333(6049):1637–1640. doi: 10.1126/science.1207699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naidu S, Friedrich JK, Russell J, Zomerdijk JC. TAF1B is a TFIIB-like component of the basal transcription machinery for RNA polymerase I. Science. 2011;333(6049):1640–1642. doi: 10.1126/science.1207656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Werner F, Grohmann D. Evolution of multisubunit RNA polymerases in the three domains of life. Nat Rev Microbiol. 2011;9(2):85–98. doi: 10.1038/nrmicro2507. [DOI] [PubMed] [Google Scholar]
- 18.Geiger SR, et al. RNA polymerase I contains a TFIIF-related DNA-binding subcomplex. Mol Cell. 2010;39(4):583–594. doi: 10.1016/j.molcel.2010.07.028. [DOI] [PubMed] [Google Scholar]
- 19.Vannini A, Cramer P. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol Cell. 2012;45(4):439–446. doi: 10.1016/j.molcel.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 20.Lefèvre S, et al. Structure-function analysis of hRPC62 provides insights into RNA polymerase III transcription initiation. Nat Struct Mol Biol. 2011;18(3):352–358. doi: 10.1038/nsmb.1996. [DOI] [PubMed] [Google Scholar]
- 21.Luo J, Fishburn J, Hahn S, Ranish J. An integrated chemical cross-linking and mass spectrometry approach to study protein complex architecture and function. Mol Cell Proteomics. 2012;11(2):M111.008318. doi: 10.1074/mcp.M111.008318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meinhart A, Blobel J, Cramer P. An extended winged helix domain in general transcription factor E/IIE alpha. J Biol Chem. 2003;278(48):48267–48274. doi: 10.1074/jbc.M307874200. [DOI] [PubMed] [Google Scholar]
- 23.Jennebach S, Herzog F, Aebersold R, Cramer P. Crosslinking-MS analysis reveals RNA polymerase I domain architecture and basis of rRNA cleavage. Nucleic Acids Res. 2012;40(12):5591–5601. doi: 10.1093/nar/gks220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vannini A, et al. Molecular basis of RNA polymerase III transcription repression by Maf1. Cell. 2010;143(1):59–70. doi: 10.1016/j.cell.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 25.Fernández-Tornero C, et al. Conformational flexibility of RNA polymerase III during transcriptional elongation. EMBO J. 2010;29(22):3762–3772. doi: 10.1038/emboj.2010.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernández-Tornero C, Böttcher B, Rashid UJ, Müller CW. Analyzing RNA polymerase III by electron cryomicroscopy. RNA Biol. 2011;8(5):760–765. doi: 10.4161/rna.8.5.16021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jasiak AJ, Armache KJ, Martens B, Jansen RP, Cramer P. Structural biology of RNA polymerase III: Subcomplex C17/25 X-ray structure and 11 subunit enzyme model. Mol Cell. 2006;23(1):71–81. doi: 10.1016/j.molcel.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 28.Lane LA, et al. Mass spectrometry reveals stable modules in holo and apo RNA polymerases I and III. Structure. 2011;19(1):90–100. doi: 10.1016/j.str.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 29.Jennebach S. 2011. RNA polymerase I domain architecture and basis of rRNA cleavage. PhD dissertation (Ludwig-Maximilians-Universität München, Munich) [Google Scholar]
- 30.Feklistov A, Darst SA. Structural basis for promoter-10 element recognition by the bacterial RNA polymerase σ subunit. Cell. 2011;147(6):1257–1269. doi: 10.1016/j.cell.2011.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naji S, Bertero MG, Spitalny P, Cramer P, Thomm M. Structure-function analysis of the RNA polymerase cleft loops elucidates initial transcription, DNA unwinding and RNA displacement. Nucleic Acids Res. 2008;36(2):676–687. doi: 10.1093/nar/gkm1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheung AC, Sainsbury S, Cramer P. Structural basis of initial RNA polymerase II transcription. EMBO J. 2011;30(23):4755–4763. doi: 10.1038/emboj.2011.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kassavetis GA, Letts GA, Geiduschek EP. The RNA polymerase III transcription initiation factor TFIIIB participates in two steps of promoter opening. EMBO J. 2001;20(11):2823–2834. doi: 10.1093/emboj/20.11.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen HT, Warfield L, Hahn S. The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nat Struct Mol Biol. 2007;14(8):696–703. doi: 10.1038/nsmb1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grünberg S, Warfield L, Hahn S. Architecture of the RNA polymerase II preinitiation complex and mechanism of ATP-dependent promoter opening. Nat Struct Mol Biol. 2012;19(8):788–796. doi: 10.1038/nsmb.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grohmann D, et al. The initiation factor TFE and the elongation factor Spt4/5 compete for the RNAP clamp during transcription initiation and elongation. Mol Cell. 2011;43(2):263–274. doi: 10.1016/j.molcel.2011.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kostrewa D, et al. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature. 2009;462(7271):323–330. doi: 10.1038/nature08548. [DOI] [PubMed] [Google Scholar]
- 38.Kassavetis GA, Han S, Naji S, Geiduschek EP. The role of transcription initiation factor IIIB subunits in promoter opening probed by photochemical cross-linking. J Biol Chem. 2003;278(20):17912–17917. doi: 10.1074/jbc.M300743200. [DOI] [PubMed] [Google Scholar]
- 39.Liu X, Bushnell DA, Wang D, Calero G, Kornberg RD. Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science. 2010;327(5962):206–209. doi: 10.1126/science.1182015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khoo B, Brophy B, Jackson SP. Conserved functional domains of the RNA polymerase III general transcription factor BRF. Genes Dev. 1994;8(23):2879–2890. doi: 10.1101/gad.8.23.2879. [DOI] [PubMed] [Google Scholar]
- 41.Artsimovitch I, et al. Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins. Cell. 2005;122(3):351–363. doi: 10.1016/j.cell.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 42.Eswar N, Eramian D, Webb B, Shen MY, Sali A. Protein structure modeling with MODELLER. Methods Mol Biol. 2008;426:145–159. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
- 43.Juo ZS, Kassavetis GA, Wang J, Geiduschek EP, Sigler PB. Crystal structure of a transcription factor IIIB core interface ternary complex. Nature. 2003;422(6931):534–539. doi: 10.1038/nature01534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.