Abstract
We previously developed a biological containment system using recombinant Salmonella Typhimurium strains that are attenuated yet capable of synthesizing protective antigens. The regulated delayed attenuation and programmed self-destructing features designed into these S. Typhimurium strains enable them to efficiently colonize host tissues and allow release of the bacterial cell contents after lysis. To turn such a recombinant attenuated Salmonella vaccine (RASV) strain into a universal DNA vaccine-delivery vehicle, our approach was to genetically modify RASV strains to display a hyperinvasive phenotype to maximize Salmonella host entry and host cell internalization, to enable Salmonella endosomal escape to release a DNA vaccine into the cytosol, and to decrease Salmonella-induced pyroptosis/apoptosis that allows the DNA vaccine time to traffic to the nucleus for efficient synthesis of encoded protective antigens. A DNA vaccine vector that encodes a domain that contributes to the arabinose-regulated lysis phenotype but has a eukaryotic promoter was constructed. The vector was then improved by insertion of multiple DNA nuclear-targeting sequences for efficient nuclear trafficking and gene expression, and by increasing nuclease resistance to protect the plasmid from host degradation. A DNA vaccine encoding influenza WSN virus HA antigen delivered by the RASV strain with the best genetic attributes induced complete protection to mice against a lethal influenza virus challenge. Adoption of these technological improvements will revolutionize means for effective delivery of DNA vaccines to stimulate mucosal, systemic, and cellular protective immunities, and lead to a paradigm shift in cost-effective control and prevention of a diversity of diseases.
Keywords: needle-free, vaccinology
Orally administered, live attenuated pathogens, such as Salmonella enterica, have been developed as homologous vaccines and as carriers of heterologous antigens because of their capacity for efficient mucosal antigen delivery to elicit mucosal (1), systemic (2, 3), and cellular immune responses (4) against the immunizing antigens (5, 6). However, biological containment systems are required to address the potential risk posed by the unintentional release of these genetically modified organisms into the environment, a subject of considerable concern. In previous studies, we devised and constructed a unique Salmonella Typhimurium biological containment and antigen delivery system designed to cause programmed cell lysis after colonization of host lymphoid tissues in vivo to deliver protective antigens (2–4). The system is composed of two parts (2). The first component is the S. Typhimurium strain, which features a deletion of asdA and arabinose-regulated expression of murA, two genes required for the synthesis of the peptidoglycan layer of the bacterial cell wall. The strain also contains additional mutations intended to enhance bacterial cell lysis and antigen delivery. The Δ(gmd-fcl)-26 mutation deletes genes encoding enzymes for GDP-fucose synthesis, thereby precluding the formation of colanic acid, a polysaccharide made in response to stress associated with cell wall damage. The ΔrelA1123 mutation uncouples cell wall-less death from dependence on protein synthesis to further ensure that the bacteria do not survive in vivo or after excretion and to allow for maximum antigen production when confronted with amino acid starvation resulting from a lack of aspartate semialdehyde synthesis because of the asdA mutation. The deletion of endA encoding the periplasmic endonuclease I enzyme, which was widely used in cloning strains to facilitate higher transformation frequencies, was also included to increase plasmid survival upon its release into the host cell. The second component is the plasmid, which allows for arabinose-regulated murA and asdA expression and C2-regulated synthesis of antisense asdA and murA mRNA transcribed from the P22 PR promoter. An arabinose-regulated c2 gene is present in the chromosome. Upon invasion of host tissues, which is an arabinose-free environment, transcription of asdA, murA, and c2 ceases and concentrations of their gene products decrease because of cell division. The drop in C2 activates PR driving synthesis of antisense mRNA to block translation of any residual asdA and murA mRNA. These concerted activities lead to bacterial cell lysis (2).
We also reported previously that the ΔaraBAD and ΔaraE mutations were included in the original lysis strains to create recombinant attenuated Salmonella vaccine (RASV) strains exhibiting a delayed lysis phenotype (6, 7). The ΔaraBAD denotes the deletion of structural genes for catabolism of arabinose, thereby preventing the use of arabinose retained in the cell cytoplasm at the time of immunization. The ΔaraE mutation, which deletes the gene for arabinose transport, enhances retention of arabinose by precluding its leakage from the cell. This inability to use arabinose prolongs time to lysis in vivo by one to two cell divisions, allowing increasing cell numbers and thus enhancing antigen delivery (6, 7). Vaccination with such RASVs resulted in induced antibody or cellular immune responses to a released bolus of pneumococcal, influenza, and mycobacterial antigens to induce protective immunity (2–4).
We developed means that enable S. Typhimurium to escape the endosome after invasion. Salmonella strains induce intestinal epithelial cells to take them up into a Salmonella-containing vacuole (SCV) (8) and then manipulate the intracellular trafficking of the vacuole to promote survival and replication of the pathogen. The sifA gene is a Salmonella pathogenicity island 2 (SPI-2)-encoded, type III secretion system (T3SS)-secreted effector protein that governs conversion of the SCV into filaments. Deletion of sifA releases Salmonella into the cytosol (9). We previously reported that an RASV-regulated delayed lysis strain harboring the sifA deletion mutation escaped from the endosome before lysis, allowing class I presentation of the influenza nucleoprotein (NP) antigen, resulting in induction of a desired protective cellular immune response against influenza virus (4).
An asdA deletion mutant of Shigella flexneri has been used to deliver a DNA vaccine in animals (10), but the immune responses were weak, presumably because the bacteria did not persist long enough to efficiently invade host tissues. Attenuated Salmonella have also been used for DNA vaccine delivery, although none of these strains was designed to undergo programmed lysis (10, 11). Therefore, DNA vaccine delivery may benefit from the regulated delayed lysis system, resulting in improved immune responses to the vectored antigens.
We have a long-standing interest in developing delivery platforms that could be used for rapid development of vaccines against viral pathogens. Influenza remains one of the most significant infectious diseases worldwide, averaging about 40,000 deaths and over 200,000 hospitalizations annually in the United States alone and up to 1.5 million deaths worldwide (12). Recent events such as the high virulence of the influenza A virus that infected people in Hong Kong in 1997 (13), the swine-origin H1N1 virus that spread worldwide (14), and the advent of laboratory plasmid-based reverse genetics systems to generate influenza A viruses, have highlighted the potential of influenza A virus as a bioterrorist weapon (15). Yearly epidemics and the more infrequent pandemics occur because of two separate mechanisms, both relating to the potential for changes in hemagglutinin (HA) and neuraminidase (NA). Yearly epidemics are caused by antigenic drift, which is the accumulation of mutations in the HA or NA genes in the currently circulating virus, such as it is able to avoid or overcome immunity generated by the original virus. The more infrequent pandemics can be caused by antigenic shift, the replacement of either HA or NA subtypes with novel ones, resulting in influenza subtypes to which the general population is totally naive. This situation presents humans with a novel antigenic experience that often results in the highest morbidity and mortality. Unfortunately, infection and recovery from influenza does not result in life-long immunity to all strains of influenza because influenza viruses frequently undergo antigenic changes. Increasing the speed of producing a matching vaccine is key in the context of response to an influenza epidemic. This study describes the development of a genetically modified RASV-improved DNA vaccine-delivery platform, which is vectoring the WSN HA protein to exploit the immune response to Salmonella infection yet to release a DNA vaccine vector that will solely specify synthesis of WSN HA antigen in the immunized host to induce a protective immune response against WSN virus challenge.
Results
Construction and Characterization of an Improved DNA Vaccine Vector to Enhance Plasmid Nuclear Import and Increase the Resistance to Attack by Mammalian Nucleases.
We constructed a DNA vaccine vector pYA3650 (SI Results, Fig. S1A, and Table S1) that complemented the regulated delayed lysis strain (2–4). It is well known that nonviral gene-delivery systems are promising tools for gene therapy and DNA vaccination applications. Compared with viral-based systems, these systems possess several advantages, including excellent safety profiles, an essentially unlimited DNA carrying capacity, and so forth. However, gene expression from such plasmids in vivo remains much lower, largely because of the inability of the DNA to effectively translocate through the nuclear pore complexes. One of the major mechanisms of nonviral DNA vaccine vectors import into the nuclei in nondividing eukaryotic cells is dependent on DNA nuclear targeting sequences (DTS). The SV40 enhancer, which is known to bind to over 10 distinct transcription factors, is an excellent DTS (16). The minimum requirement for this function is a single copy of a 72-bp element of the SV40 enhancer, in combination with the CMV immediate-early gene enhancer/promoter (CMV E/P) (17). In addition, nuclease degradation of DNA vaccine vectors after delivery and during trafficking to the nucleus is another barrier that leads to inefficient DNA vaccination. Homopurine-rich tracts in the bovine growth hormone polyadenylation signal [BGH poly(A)] were identified as labile sequences (18), and replacement of BGH poly(A) with SV40 late poly(A) has improved resistance to attack from mammalian nucleases (19). To increase the efficiency of our DNA vaccine vector delivery system, we first inserted the SV 40 72-bp repeat enhancer, into the original DNA vaccine vector pYA3650 to serve as a DTS (I) to generate plasmid pYA3836 (Table S1). Then the BGH poly(A) in pYA3836 was replaced with the SV40 late poly(A) to obtain plasmid pYA4050 (Fig. S2A and Table S1). The enhanced green fluorescent protein (EGFP) genes were fused in frame with the Kozak sequence and inserted into vectors pYA4050 and pYA3650 down-stream of the CMV promoter to yield pYA4271 and pYA4272 (Table S1), respectively. This process allowed us to compare the levels of protein synthesis from the improved DNA vaccine vector pYA4050 to the original DNA vaccine vector pYA3650. The plasmid pYA4271 (pYA4050 encoding EGFP) allowed higher level synthesis of EGFP comparing to the plasmid pYA4272 (pYA3650 encoding EGFP) in human INT-407 intestinal cells (Fig. S3). However, mice immunized with RASV strain χ8888 harboring the DNA vaccine vector pYA4611 (pYA4050 encoding WSN HA) (Fig. S2B and Table S1) did not show significant protection from challenge with 100 times the LD50 of rWSN, although this vaccine strain induced moderate serum IgG responses to WSN HA in orally immunized mice (Fig. S2C).
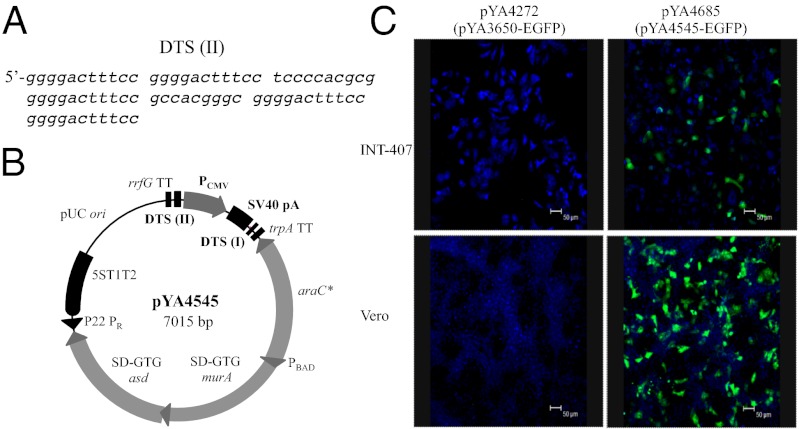
These results led us to search for more efficient methods to further improve our Salmonella DNA vaccine delivery system. Transcription factor NF-κB is found in almost all animal cell types. The binding affinity of NF-κB to their DNA-binding sites (κB sites) is high and the translocation of NF-κB-DNA complexes into the nucleus is rapid (minutes). Depending on their position relative to the encoding gene, the binding sites could also act as transcriptional enhancers that further increase gene-expression levels. Salmonella infection rapidly stimulates the synthesis of eukaryotic transcription factors, such as NF-κB and AP-2 (20). We postulated that the plasmid DNA with κB and AP-2 binding sites would allow newly synthesized NF-κB or AP-2, during Salmonella infection, to bind to the plasmid DNA in the cytoplasm and transport it to the nucleus through the protein nuclear import machinery. To test our hypothesis, we designed a cassette of synthetic DTS (II) (Fig. 1A), which is comprised of multiple binding sites for transcription factors NF-κB and AP-2. The DTS (II) was inserted into pYA4050 upstream of CMV E/P as an additional DNA nuclear targeting/enhancer sequence to yield the improved DNA vaccine vector pYA4545 (Fig. 1B). One additional beneficial feature of pYA4545 is that it possesses 24 putative immune enhancing CpG motifs that may contribute to enhancement of an innate immune response. Plasmid pYA4545 was fully sequenced. The EGFP gene was then fused in frame with the Kozak sequence and inserted into pYA4545 down-stream of the CMV promoter to yield pYA4685 (Table S1). High-level synthesis of EGFP, visualized by confocal microscopy, was found in INT-407 and Vero mammalian cells transfected with pYA4685 (pYA4545 encoding EGFP), but was not seen in the same cells transfected with a plasmid derivative of the original DNA vector pYA4272 (pYA3650 encoding EGFP) (Fig. 1C). These results (Fig. 1C and Fig. S3) indicated that DTS II (synthetic NF-κB and AP-2 binding sites) is potentially responsible for the further enhanced EGFP synthesis in pYA4685 transfected cells.
Fig. 1.
Evidence that indicates the improvement of DNA vaccine vector. (A) Nucleotide sequence of synthetic DTS (II). (B) Map of improved DNA vaccine vector pYA4545. Plasmid sequences include the rrfG, trpA, and 5S ribosomal RNA transcriptional terminators, the PBAD, P22 PR, and PCMV promoters, the araC gene, and start codon-modified murA and asdA genes, DTS (I), DTS (II), and SV40 late poly(A). (C) Laser confocal images of EGFP (green signals) synthesis by DNA vaccine vector pYA3650 (original) and pYA4545 (improved) in INT-407 (Upper) and Vero (Lower) cells. Cell nuclear DNA was stained with TO-PRO-3 (blue signals) (Scale bars, 20 μm).
Construction of an Improved DNA Vaccine Encoding HA of Influenza A/WSN/33 Virus.
We used influenza A WSN HA as a model antigen to evaluate the ability of our regulated lysis strain to deliver an antigen encoded by the improved DNA vaccine vector to host tissues. A DNA fragment encoding WSN HA antigen with Kozak sequence was inserted downstream of the CMV promoter in the improved DNA vaccine vector pYA4545 to obtain pYA4859 (Fig. S4A). The level of WSN HA synthesis coming from pYA4859 was detected using EGFP as a fusion tag that was significantly higher than synthesized by vector pYA4611 (pYA4050 encoding WSN HA).
Salmonella invasion into host cells results in bacteria residing in an endosomal compartment-SCV. The release of a DNA vaccine because of programmed lysis within the SCV would be unlikely to stimulate an immune response, because of difficulties that the DNA vaccine would encounter getting to the nucleus. As stated above, we therefore introduced a deletion of the sifA gene (ΔsifA26) (4) into strain χ8888 [∆asdA19::TT araC PBAD c2 TT ∆PmurA7::TT araC PBAD murA ∆araBAD1923 ∆araE25 ∆(gmd-fcl)-26 ∆relA1123 ∆endA2311] (3), which enabled the Salmonella strain to immediately escape upon invasion from the SCV and rapidly multiply in the cytosol. This yielded RASV strain χ9354 (χ8888 with ΔsifA26) (Table S1). We then introduced plasmid pYA4859 into χ9354 to yield χ9354(pYA4859). This vaccine strain induced low serum IgG responses to WSN HA in mice after oral immunization (see Fig. 4 below) and mice immunized with such a vaccine strain showed moderate protection from challenge with 100 LD50 of rWSN virus. These results forced us to seek additional means to further increase DNA vaccine delivery efficiency by improvement of the lysis strains.
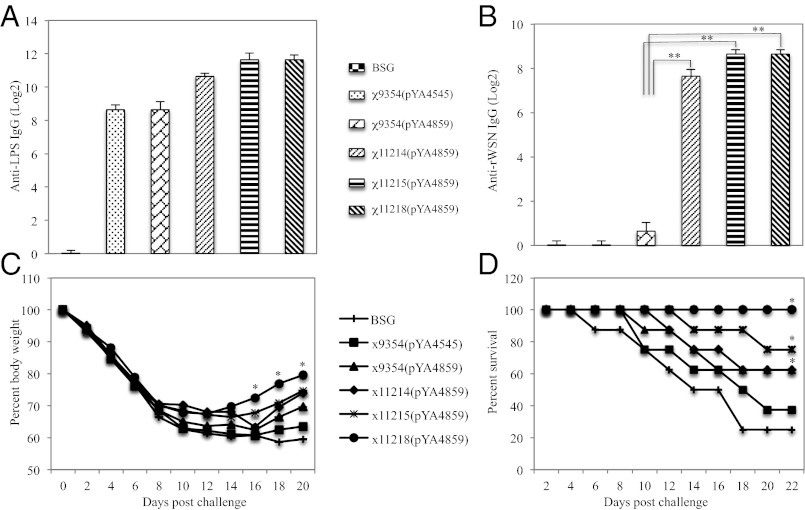
Fig. 4.
Immune responses in mice 5 wk after oral primary immunization and protection with RASV strains χ9354(pYA4545) (vector control), χ9354(pYA4859) (pYA4545 encoding WSN HA), χ11214(pYA4859) (∆PhilA::PtrcΔlacO888 hilA), χ11215(pYA4859) (∆tlpA, ∆sseL), and χ11218(pYA4859) (∆PhilA::PtrcΔlacO888 hilA, ∆tlpA, ∆sseL), or BSG. Induction of IgG titers against S. Typhimurium LPS (A) and inactivated WSN virus (B) detected by ELISA. Pooled serum samples (n = 8) from mice within a group were assayed and analyzed by ANOVA followed by Tukey’s range test, **P < 0.001. (C) Weight loss, *P < 0.01 and (D) percent survival of mice (n = 8) after an intranasal challenge with 100 LD50 of rWSN influenza virus at 8 wk PPI. Survival curve has a P < 0.0002 by log-rank (Mantel–Cox) test.
Retention of the Switch-On for Synthesis of Proteins Required for Invasiveness.
An early step in the establishment of S. Typhimurium murine infection is the penetration of the intestinal mucosa of the small intestine, mainly by bacteria invading M-cells overlying Peyer’s patches. The ability of Salmonella to invade intestinal cells of the host is also critical for efficient DNA vaccine delivery. The majority of genes responsible for the Salmonella invasive phenotype are encoded on SPI-1, and their transcription is controlled by the hilA transcriptional activator. The expression of hilA is regulated by environmental signals, including oxygen, osmolarity, pH, and growth phase, such that the presence of any one suboptimal condition results in repression of hilA expression and the invasive phenotype (21). On the other hand, although it was thought that Salmonella resides and proliferates within a membrane-bound vacuole in epithelial cells, it has recently been discovered that there are at least two transcriptionally distinct intracellular populations of replicating bacteria in epithelial cells: T3SS-2–induced intravacuolar bacteria and T3SS-1–induced flagellated bacteria that are invasion-primed, cytosolic, fast-replicating, moving freely within the cells, and well prepared to spread and invade new cells (22). Increase of the subpopulation of invasion-primed cytosolic Salmonella would greatly benefit DNA vaccine delivery.
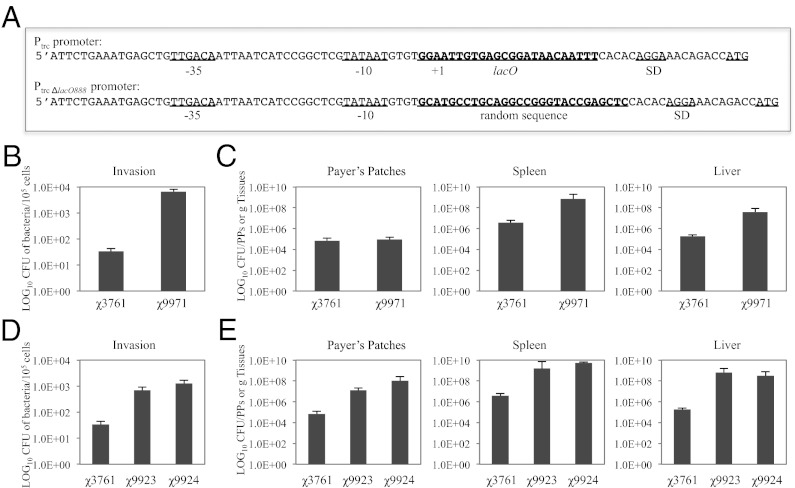
To construct Salmonella strains that constitutively exhibit the hyperinvasive phenotype, even after entering and traversing the epithelial monolayer lining the intestine, and subsequently increase the subpopulation of invasion-primed cytosolic Salmonella for efficient DNA vaccine delivery, we replaced the promoter of the hilA gene, encoding the HilA regulator, with a man-made promoter PtrcΔlacO888 (Fig. 2A), in which the operator lacO sequence was replaced with a random sequence as a spacer to enable constitutive synthesis of HilA even when the lacI gene is present and active in the host strain. The resulting derivative of the wild-type χ3761 is χ9971 (∆PhilA::PtrcΔlacO888 hilA) (Table S1). Strain χ9971 was able to invade and replicate in human intestinal INT-407 cells (multiplicity of infection 50:1) (Fig. 2B and Fig. S5A) and colonize mouse tissues in significantly greater numbers, specifically in spleen and liver, than the wild-type χ3761 (Fig. 2C) in orally immunized mice. We then inserted the ∆PhilA::PtrcΔlacO888 hilA deletion-insertion mutation into S. Typhimurium vaccine strain χ9354, which displays the regulated delayed lysis phenotype to facilitate efficient delivery of DNA vaccine. The resulting strain is χ11214 (χ9354 + ΔPhilA::Ptrc ΔlacO888 hilA) (Table S1).
Fig. 2.
Experiments detecting enhanced invasion and colonization of RASV strains by genetic modifications of hilA, tlpA, and sseL genes. (A) Nucleotide sequence of Ptrc promoter and synthetic promoter PtrcΔlacO888, in which the operator lacO sequence was replaced with a random sequence spacer. (B) Invasion of S. Typhimurium χ3761 (wild-type) and χ9971 (∆PhilA::PtrcΔlacO888 hilA) into INT-407 cells. Values are the mean ± SD of three experiments with triplicate wells. (C) Colonization of mice (three per group) with S. Typhimurium χ3761 (wild-type) and χ9971 (∆PhilA::PtrcΔlacO888 hilA) at day 6 following oral inoculation with 109 CFU bacteria. (D) Invasion of S. Typhimurium χ3761 (wild-type), χ9923 (ΔtlpA), and χ9924 (∆sseL) into INT-407 cells. Values are the mean ± SD of three experiments with triplicate wells. (E) Colonization of mice with S. Typhimurium χ3761 (wild-type), χ9923 (ΔtlpA), and χ9924 (∆sseL) at day 6 following oral inoculation with 109 CFU bacteria.
Engineering RASVs to Reduce Salmonella Induced Host Cell Pyroptosis/Apoptosis.
Salmonella strains induce host cell death during infection by several mechanisms and this is likely to diminish transcription of a DNA vaccine after trafficking to the nucleus (23, 24). Salmonella grown under conditions to express the SPI-1 T3SS activate the NLRC4 inflammasome in macrophages (25, 26), which activates caspase-1 leading to cell death by a process termed pyroptosis (23). In contrast, Salmonella-induced epithelial cell death, which has features of delayed classic apoptosis, depends on both the SPI-2 T3SS and the spv locus even though Salmonella invasion involves the SPI-1 T3SS (27).
Two major SPI-2 T3SS-secreted effectors that induce apoptosis have been identified: SpvB and SseL. The spv locus is required for the induction of apoptosis in human macrophages (28) and SseL, a Salmonella deubiquitinase, has been shown to be involved in macrophage cytotoxicity (29). In addition, SseL deubiquitinates IκBα, the major regulator of the classic NF-κB activation pathway. NF-κB activation is significantly increased after infection of macrophages with an SseL mutant, and this activity can be decreased to wild-type levels by complementation with a low copy plasmid synthesizing SseL. These results show that the action of SseL is to decrease NF-κB signaling, thereby decreasing the antiapoptotic and proinflammatory effects of this pathway (29) and support the idea that SseL may also act in concert with other effectors to decrease activation of innate immunity. We hypothesized that deletion of sseL from RASV strains would reduce Salmonella-induced apoptosis, enhance innate immune responses by enhancing NF-κB activation, and simultaneously enhance NF-κB-mediated nuclear targeting of the DNA vector.
In addition, Salmonella strains lacking the spv locus are dramatically attenuated (30) such that a spv mutation is not suitable as a means to reduce Salmonella-induced apoptosis. Consequently, we searched for other virulence regulator candidates as targets for mutation to reduce apoptosis. One interesting putative virulence regulator is the thermosensing gene regulator TlpA. TlpA has the ability to alter its DNA-binding according to variation in temperature, and concomitantly, its DNA regulatory characteristics functions (31, 32). Strong homologies to the operator sequence of tlpA are found in front of defined virulence genes, such as the spvABCD genes. Like the spv genes, tlpA is located on the large virulence-associated plasmid (33, 34) and is conserved in Salmonella carrying the virulence plasmid. Thus, the question is whether deletion of tlpA can reduce Salmonella-induced apoptosis to enhance DNA vaccine delivery.
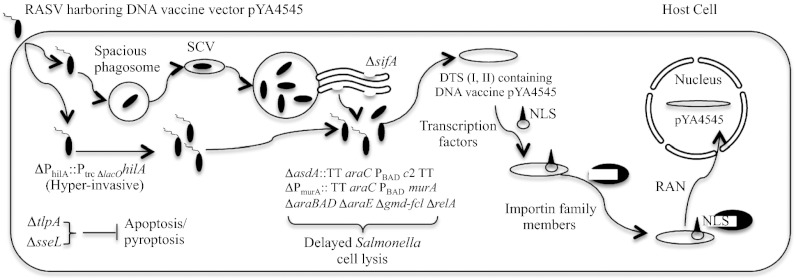
To explore the impact of deletions in the sseL and tlpA genes on Salmonella-induced apoptosis, S. Typhimurium strains χ9923 (ΔtlpA181) and χ9924 (ΔsseL116) were constructed (Table S1). Elimination of synthesis of TlpA (χ9923) and SseL (χ9924) reduce apoptosis in mice infected with Salmonella. Representative H&E images show apoptotic bodies (Fig. S6, arrows) in the lower villus/crypt regions of the ileum in BALB/c mice infected with wild-type S. Typhimurium UK-1 strain compared with wild-type strains harboring a tlpA gene mutation or a sseL gene mutation, or control uninfected mice (Fig. S6). Furthermore, significantly higher numbers of bacteria were recovered from χ9923 (ΔtlpA181)- and χ9924 (ΔsseL116)-infected INT-407 cells (multiplicity of infection 50:1) than those from cells infected by the wild-type strain (Fig. 2D and Fig. S5B). In addition, strains χ9923 (ΔtlpA181) and χ9924 (ΔsseL116) colonized mouse tissues in significantly greater numbers than their wild-type parent strain χ3761 at day 6 postinfection (Fig. 2E). We then constructed the final RASV strain χ11218 (χ9354 harboring ΔPhilA::Ptrc ΔlacO888 hilA, ΔtlpA181, and ΔsseL116) (Fig. S4B). Improved DNA vaccine vector pYA4545 was then introduced into these RASV strains described above for further evaluation. The effects of the genetic attributes and DTS (II) on the process of DNA vaccine delivery by RASV strains is illustrated in Fig. 3. Briefly, RASV strains with enhanced invasiveness, that are inhibited in inducing apoptosis/pyroptosis, escape from the endosome to lyse in the cytosol to deliver a DNA vaccine engineered for maximal survival and targeting to the nucleus. Newly synthesized transcription factors, which contain nuclear localization sequences and DNA binding proteins, bind to the plasmid DNA in the cytoplasm to create protein–DNA complexes. Thus, the complex is recognized by the protein import machinery through the nuclear localization sequences and is thereby targeted to the nucleus.
Fig. 3.
Diagram of model illustrating the effects of genetic attributes and DTS (II) on the process of DNA vaccine delivery by RASV strains displaying delayed regulated lysis phenotype. NLSs, nuclear localization sequences; Ran, Ras-related nuclear protein. Details are outlined in the text.
Evaluation of Antibody Responses and Protection Against Viral Challenge.
To evaluate the efficacy of improved DNA vaccine vector delivery by RASV strains harboring hyperinvasive or reduced Salmonella-induced apoptosis phenotypes, mice were orally immunized with RASV strains χ9354(pYA4859), χ11214(pYA4859) (ΔPhilA::Ptrc ΔlacO888 hilA), χ11215(pYA4859) (ΔtlpA181, ΔsseL116), χ11218(pYA4859) (ΔPhilA::Ptrc ΔlacO888 hilA, ΔtlpA181, ΔsseL116), vector control χ9354(pYA4545) (Table S1), or PBS containing 0.01% gelatin (BSG), and boosted at weeks 1, 4, and 7 postprimary immunization (PPI) with the same strains. All mice survived, and no signs of disease were observed during the entire experimental period. The antibody responses to Salmonella LPS and to influenza antigen HA in the sera of immunized mice were measured at week 5 PPI by ELISA. The total IgG antibody titers elicited against LPS by vaccine strains χ11214(pYA4859), χ11215(pYA4859), and χ11218(pYA4859) are significantly higher than those induced by strain χ9354(pYA4859) and the vector controls (Fig. 4A), reflecting better invasion and colonization by the improved RASV strains. HA-specific IgG was detected in the sera from mice immunized with χ9354(pYA4859), χ11214(pYA4859), χ11215(pYA4859), and χ11218(pYA4859) (Fig. 4B) but not in sera from mice immunized with the χ9354(pYA4545) (vector control). However, the IgG response to HA antigen was at a much lower level in mice immunized with RASV strain χ9354(pYA4859) than the IgG responses induced in mice immunized with χ11214(pYA4859), χ11215(pYA4859), and χ11218(pYA4859).
Immunized mice were intranasally infected with 100 LD50 of rWSN influenza virus. Mice infected with the rWSN influenza strain showed ruffled fur, hunched posture, trembling, and a continuous weight loss as signs of infection from the second day after challenge that progressed with time. Mice immunized with χ11218(pYA4859) recovered from influenza infection earlier as indicated by the alleviation of symptoms by 10 d after challenge, compared with mice immunized with χ11214(pYA4859) and χ11215(pYA4859), which recovered by 16 d; mice immunized with χ9354(pYA4859) and the vector-immunized control groups continued to become sicker. This result is also evident by weight-recovery data of mice immunized with χ11218(pYA4859) (ΔPhilA::Ptrc ΔlacO888 hilA, ΔtlpA181, ΔsseL116), compared with mice immunized with χ11215(pYA4859) (ΔtlpA181, ΔsseL116), χ11214(pYA4859) (ΔPhilA::Ptrc ΔlacO888 hilA), χ9354(pYA4859), or with vector control χ9354(pYA4545) or BSG (Fig. 4C). Mice immunized with strain χ11218(pYA4859) survived, whereas mice immunized with χ11215(pYA4859), χ11214(pYA4859), χ9354(pYA4859), vector control χ9354(pYA4545), or BSG commenced dying 6 d after challenge. All mice immunized orally with χ11218(pYA4859) were protected (100%) against 100 LD50 of rWSN virus challenge compared with 75% survivors in the group immunized with χ11215(pYA4859), 62% survivors in the group immunized with χ11214(pYA4859) or χ9354(pYA4859), 37% survivors in the group immunized with vector control χ9354(pYA4545), and 25% survivors in the group immunized with BSG (Fig. 4D). The survival curves of groups χ11218(pYA4859), χ11215(pYA4859), χ11214(pYA4859), are significantly different from χ9354(pYA4859), vector control χ9354(pYA4545), and BSG (Fig. 4D), although unexpectedly in this experiment some mice survived that should have died.
Discussion
Current means for delivery of DNA vaccines by intramuscular injection or by use of gene guns do not result in delivery of the vaccine to mucosal as well as to a diversity of internal lymphoid tissues important in yielding a high-level sustained (i.e., memory) protective immunity to pathogens. When using invasive recombinant bacterial vectors for delivery, it is necessary that viral antigens requiring glycosylation and posttranslational modifications be synthesized within cells of the immunized host and not by the attenuated bacteria.
Attenuated strains of pathogenic and nonpathogenic bacteria have been widely explored for potential as DNA vaccine delivery systems. However, delivery of DNA vaccines encoding viral antigens by attenuated invasive bacteria (Shigella, Salmonella, and Listeria), first described in the 1990s (10, 35), has not been very successful. In 2008, after 5 y of effort, we described a means for the regulated delayed lysis in vivo of an RASV strain to deliver a bolus of recombinant protective antigen and to confer complete biological containment with no vaccine cells persisting in vivo or surviving if excreted (2). However, this vector was unsuitable to deliver DNA vaccines for several reasons. First, Salmonella internalization is the first step of DNA vaccine delivery by RASVs. The frequency of host cell homing by Salmonella is too low. Second, Salmonella induces apoptosis/pyroptosis such that the nucleus of infected cells has a diminished ability to support transcription of introduced DNA vaccines. Third, Salmonella enters into an endosome such that bacterial lysis would not facilitate a DNA vaccine reaching the nucleus for transcription.
We report here that we have now solved these and other problems by constructing a RASV with enhanced invasiveness that is inhibited in inducing apoptosis/pyroptosis and that escapes from the endosome to lyse in the cytosol for delivery of a DNA vaccine engineered for maximal survival and targeting to the nucleus. Using the original lysis vector pYA3650 (Fig. S1A and Fig. S7) as backbone, we developed a much improved DNA vaccine vector with enhanced transmission to the nucleus for gene expression. Using this system to deliver the WSN HA antigen successfully induced protective immunity to influenza virus challenge in mice immunized with the RASV strains described above. To reduce the doses needed to induce protective immunity, these Salmonella host-vector systems are being improved by including mutations, such as ΔasdA27::TT araC PBAD c2 TT (36), ΔPmurA25::TT araC PBAD murA (4), and Δ(araC PBAD)-5::P22 PR araBAD, to enhance lysis. The resulting RASV vaccine strain induced protective immunity to influenza virus challenge in mice with one booster. In addition, the regulated delayed lysis phenotype as described above results in the release of lipid A endotoxin that is inflammatory via interaction with Toll-like receptor-4 and MD2 (37); this can also enhance induction of pyroptosis/apoptosis (38). To preclude this, we generated and fully evaluated a deletion-insertion mutation that contains the lpxE gene from Francisella tularensis that has been codon-optimized for high-level expression in Salmonella results to produce 1-dephosphoryl lipid A, which is totally nontoxic, and yet should be a safe adjuvant for recruitment of innate immunity (39, 40).
Conventional approaches to influenza vaccination, using a live attenuated or an inactivated virus vaccine, are effective at inducing neutralizing antibodies but must be reformulated each year as new antigenic variant strains arise. The live attenuated virus vaccine also elicits cellular immunity, although it does not appear to provide additional protection in humans (41). Perhaps of greater importance is the generation of more broadly reactive vaccines. The Matrix protein 2 (M2) sequence is less variable than HA and NA sequences and the immune responses recognizing these epitopes appear to be able to reduce or prevent infection with a wider range of influenza virus strains (42). In addition, influenza NP could potentially provide broad-based protection: T-cell epitopes in NP are well defined and both CD8+ and CD4+ T cells play an important role in protection afforded by NP (43).
Using recently developed recombinant Salmonella vectors, we have developed a unique means to enhance protective immunity against influenza challenge in mice by inducing cellular immunity to NP (4). We have also developed recombinant Salmonella vectors to deliver M2e fused to the woodchuck hepatitis virus core (3). Our RASV HA DNA vaccine delivery system can be redesigned with new HA antigens in 2 wk and manufactured in billions of doses at a low cost within several months from the time a decision is made to change antigenic components. We expect that a combination of these three different vaccines would afford a better protection than conferred by vaccination with any of the three individual vaccines.
By combining the developments described above, our influenza vaccine is designed to induce mucosal immunity and specific neutralizing antibody immunity against HA (encoded by DNA vaccine vectors) and broad-based immunity (antibodies to M2e and CTL responses to conserved NP) capable of at least partially controlling heterologous influenza strains. Ultimately, we will develop a S. Typhi vector for use in human trials and work out formulations of vaccines as thermostable lyophilized preparations for needle-free delivery to an optimal mucosal tissue. If successful, our efforts would ensure development of preventive influenza vaccines that can induce broad cross-protective responses and that can be administered as soon as an epidemic or bioterrorist attack is declared or even before. Successes will also lead to other applications for safe, efficacious delivery of recombinant vaccines or therapeutic reagents against other important pathogens that cause significant morbidity or mortality.
Materials and Methods
Construction of Improved DNA Vaccine Vector.
We first inserted the Simian virus 40 (SV40) 72-bp repeat enhancer into pYA3650 using primers SphI-SV40 second and PflMI-araC-SV40 (Table S2) to create plasmid pYA3836 (Table S1). Then, the BGH poly(A) in pYA3650 was replaced with the SV40 late poly(A) using primers XhoI-SV40 polyA and SphI-SV40 polyA (Table S2); the resulting plasmid was pYA4050 (Table S1). Next, we designed a synthetic DTS (II) (Table S2) based on conserved mammalian NF-κB and AP-2 binding sites. A DTS (II) fragment was inserted into pYA4050 using primers PmlI-DTS up-1, DTS up-2, DTS down-1, and KpnI-DTS down-2 (Table S2) to yield the improved DNA vaccine vector pYA4545 (Fig. 1B). In addition, EGFP genes were fused in frame with the Kozak sequence and inserted into vectors pYA3650 and pYA4545 downstream of the CMV promoter to yield pYA4272 and pYA4685 using primers KpnI-EGFP and XhoI-EGFP (Table S2), respectively.
Construction of Hyperinvasive RASV Strains.
To construct the hyper-invasive Salmonella strains, we designed a promoter PtrcΔlacO888 (Table S2). PtrcΔlacO888 was built into a suicide vector using primers HindIII-Ptrc 888 up-1, Ptrc 888 up-2, Ptrc 888 down-1, and BamHI- Ptrc 888 down-2. The resulting suicide vector was pYA4641 (Table S1). Wild-type strain χ3761 was conjugated with Escherichia coli strain χ7213 harboring suicide vector pYA4641 to generate mutant strain χ9971.
Construction of RASV Strains to Reduce Host Cell Pyroptosis/Apoptosis.
Two S. Typhimurium mutant strains χ9923 and χ9924 were constructed using suicide vectors pYA4620 and pYA4621. Then mutations ΔtlpA181 or/and ΔsseL116 were introduced into S. Typhimurium lysis strain χ9354 by conjugation, to achieve strains χ11215 and χ11218.
Immunization of Mice and Sample Collection.
S. Typhimurium DNA vaccine strains were individually grown as described above. Groups of eight 7-wk-old female BALB/c mice were orally vaccinated at week zero with the following dose: 1.5 × 109 CFU χ8888(pYA4050) (vector control) and 1.2 × 109 CFU χ8888(pYA4611) (pYA4050 encoding WSN HA) or 1.4 × 109 CFU χ9354(pYA4545) (vector control), 1.2 × 109 CFU χ9354(pYA4859) (pYA4545 encoding WSN HA), 1.1 × 109 CFU χ11214(pYA4859), 1.3 × 109 CFU χ11215(pYA4859), and 1.0 × 109 CFU χ11218(pYA4859). Booster immunizations were given to all immunization groups three times later at weeks 1, 4, and 7 PPI. The immunized mice were monitored for 8 wk for any evidence of illness by observing them daily for evidence of diarrhea, ruffled (ungroomed) fur, or irritability. None of these symptoms of infection was observed in any of the mice. Blood collected at week 5 PPI by cheek-pouch bleeding was monitored for the presence of antibodies against HA or S. Typhimurium LPS by ELISA.
Virus Challenge.
Groups of mice were lightly anesthetized with 0.05 mL/20 g body weight of a ketamine mixture administered intramuscularly. Sedated mice were intranasally infected with 100 LD50 of purified rWSN virus in a total volume of 30 μL (15 μL per nostril for all experiments). An aliquot of the virus used for challenge was back-titrated on Madin–Darby canine kidney cells to ascertain the exact dose given to mice. The challenged mice were inspected daily for signs of infection such as ruffled fur, hunched posture, and weighed on alternate days until 21 d to monitor the progression of infection. Percent weight loss was calculated for individual mice in each group by comparing their daily weight to the prechallenge weight of the animal. Mortality was used as end-point in the challenge studies; however, mice that succumbed to infection or had to be euthanized were promptly removed.
Statistical Analysis.
Differences in antibody titers were determined using ANOVA and statistically different means (P < 0.05) were further separated using Tukey’s range test.
Supplementary Material
Acknowledgments
We thank Dr. Qingke Kong for construction of suicide vectors pYA4620 and pYA4621; Drs. Shamaila Ashraf and Xiangmin Zhang for technical support on virus propagation and challenge; Dr. Andrew Pekosz for providing the influenza virus strains; and the WM Keck Bioimaging Facility at Arizona State University. This study is funded by US Department of Agriculture Grant 03-35204-13748 and National Institutes of Health Grants R01 AI065779, R01 AI056289, R01 AI093348, and R21CA152456.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217554109/-/DCSupplemental.
References
- 1.Xin W, Li Y, Mo H, Roland KL, Curtiss R., III PspA family fusion proteins delivered by attenuated Salmonella enterica serovar Typhimurium extend and enhance protection against Streptococcus pneumoniae. Infect Immun. 2009;77(10):4518–4528. doi: 10.1128/IAI.00486-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kong W, et al. Regulated programmed lysis of recombinant Salmonella in host tissues to release protective antigens and confer biological containment. Proc Natl Acad Sci USA. 2008;105(27):9361–9366. doi: 10.1073/pnas.0803801105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ameiss K, et al. Delivery of woodchuck hepatitis virus-like particle presented influenza M2e by recombinant attenuated Salmonella displaying a delayed lysis phenotype. Vaccine. 2010;28(41):6704–6713. doi: 10.1016/j.vaccine.2010.07.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashraf S, Kong W, Wang S, Yang J, Curtiss R., III Protective cellular responses elicited by vaccination with influenza nucleoprotein delivered by a live recombinant attenuated Salmonella vaccine. Vaccine. 2011;29(23):3990–4002. doi: 10.1016/j.vaccine.2011.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Germanier R, Füer E. Isolation and characterization of Gal E mutant Ty 21a of Salmonella typhi: A candidate strain for a live, oral typhoid vaccine. J Infect Dis. 1975;131(5):553–558. doi: 10.1093/infdis/131.5.553. [DOI] [PubMed] [Google Scholar]
- 6.Curtiss R, III, et al. Salmonella enterica serovar Typhimurium strains with regulated delayed attenuation in vivo. Infect Immun. 2009;77(3):1071–1082. doi: 10.1128/IAI.00693-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curtiss R, III, et al. New technologies in using recombinant attenuated Salmonella vaccine vectors. Crit Rev Immunol. 2010;30(3):255–270. doi: 10.1615/critrevimmunol.v30.i3.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galán JE. Salmonella interactions with host cells: Type III secretion at work. Annu Rev Cell Dev Biol. 2001;17:53–86. doi: 10.1146/annurev.cellbio.17.1.53. [DOI] [PubMed] [Google Scholar]
- 9.Brumell JH, Goosney DL, Finlay BB. SifA, a type III secreted effector of Salmonella typhimurium, directs Salmonella-induced filament (Sif) formation along microtubules. Traffic. 2002;3(6):407–415. doi: 10.1034/j.1600-0854.2002.30604.x. [DOI] [PubMed] [Google Scholar]
- 10.Sizemore DR, Branstrom AA, Sadoff JC. Attenuated bacteria as a DNA delivery vehicle for DNA-mediated immunization. Vaccine. 1997;15(8):804–807. doi: 10.1016/s0264-410x(96)00252-6. [DOI] [PubMed] [Google Scholar]
- 11.Darji A, et al. Oral somatic transgene vaccination using attenuated S. Typhimurium. Cell. 1997;91(6):765–775. doi: 10.1016/s0092-8674(00)80465-1. [DOI] [PubMed] [Google Scholar]
- 12.Thompson WW, et al. Influenza-associated hospitalizations in the United States. JAMA. 2004;292(11):1333–1340. doi: 10.1001/jama.292.11.1333. [DOI] [PubMed] [Google Scholar]
- 13.Anonymous Centers for Disease Control and Prevention (CDC) Update: Isolation of avian influenza A(H5N1) viruses from humans—Hong Kong, 1997–1998. MMWR Morb Mortal Wkly Rep. 1998;46(52–53):1245–1247. [PubMed] [Google Scholar]
- 14. WHO (2010) Situation updates–Pandemic (H1N1) 2009. Available at www.who.int/csr/disease/swineflu/updates/en/. Accessed October 29, 2012.
- 15.Krug RM. The potential use of influenza virus as an agent for bioterrorism. Antiviral Res. 2003;57(1–2):147–150. doi: 10.1016/s0166-3542(02)00207-3. [DOI] [PubMed] [Google Scholar]
- 16.Young JL, Benoit JN, Dean DA. Effect of a DNA nuclear targeting sequence on gene transfer and expression of plasmids in the intact vasculature. Gene Ther. 2003;10(17):1465–1470. doi: 10.1038/sj.gt.3302021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Firak TA, Subramanian KN. Minimal transcriptional enhancer of simian virus 40 is a 74-base-pair sequence that has interacting domains. Mol Cell Biol. 1986;6(11):3667–3676. doi: 10.1128/mcb.6.11.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ribeiro SC, Monteiro GA, Prazeres DM. The role of polyadenylation signal secondary structures on the resistance of plasmid vectors to nucleases. J Gene Med. 2004;6(5):565–573. doi: 10.1002/jgm.536. [DOI] [PubMed] [Google Scholar]
- 19.Carswell S, Alwine JC. Efficiency of utilization of the simian virus 40 late polyadenylation site: Effects of upstream sequences. Mol Cell Biol. 1989;9(10):4248–4258. doi: 10.1128/mcb.9.10.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vitiello M, et al. Interleukin-8 production by THP-1 cells stimulated by Salmonella enterica serovar Typhimurium porins is mediated by AP-1, NF-kappaB and MAPK pathways. Cytokine. 2004;27(1):15–24. doi: 10.1016/j.cyto.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 21.Bajaj V, Hwang C, Lee CA. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol Microbiol. 1995;18(4):715–727. doi: 10.1111/j.1365-2958.1995.mmi_18040715.x. [DOI] [PubMed] [Google Scholar]
- 22.Knodler LA, et al. Dissemination of invasive Salmonella via bacterial-induced extrusion of mucosal epithelia. Proc Natl Acad Sci USA. 2010;107(41):17733–17738. doi: 10.1073/pnas.1006098107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fink SL, Cookson BT. Pyroptosis and host cell death responses during Salmonella infection. Cell Microbiol. 2007;9(11):2562–2570. doi: 10.1111/j.1462-5822.2007.01036.x. [DOI] [PubMed] [Google Scholar]
- 24.van der Velden AW, Lindgren SW, Worley MJ, Heffron F. Salmonella pathogenicity island 1-independent induction of apoptosis in infected macrophages by Salmonella enterica serotype Typhimurium. Infect Immun. 2000;68(10):5702–5709. doi: 10.1128/iai.68.10.5702-5709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franchi L, et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in Salmonella-infected macrophages. Nat Immunol. 2006;7(6):576–582. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- 26.Miao EA, et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol. 2006;7(6):569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 27.Paesold G, Guiney DG, Eckmann L, Kagnoff MF. Genes in the Salmonella pathogenicity island 2 and the Salmonella virulence plasmid are essential for Salmonella-induced apoptosis in intestinal epithelial cells. Cell Microbiol. 2002;4(11):771–781. doi: 10.1046/j.1462-5822.2002.00233.x. [DOI] [PubMed] [Google Scholar]
- 28.Libby SJ, Lesnick M, Hasegawa P, Weidenhammer E, Guiney DG. The Salmonella virulence plasmid spv genes are required for cytopathology in human monocyte-derived macrophages. Cell Microbiol. 2000;2(1):49–58. doi: 10.1046/j.1462-5822.2000.00030.x. [DOI] [PubMed] [Google Scholar]
- 29.Rytkönen A, et al. SseL, a Salmonella deubiquitinase required for macrophage killing and virulence. Proc Natl Acad Sci USA. 2007;104(9):3502–3507. doi: 10.1073/pnas.0610095104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gulig PA, et al. Molecular analysis of spv virulence genes of the Salmonella virulence plasmids. Mol Microbiol. 1993;7(6):825–830. doi: 10.1111/j.1365-2958.1993.tb01172.x. [DOI] [PubMed] [Google Scholar]
- 31.Hurme R, Berndt KD, Normark SJ, Rhen M. A proteinaceous gene regulatory thermometer in Salmonella. Cell. 1997;90(1):55–64. doi: 10.1016/s0092-8674(00)80313-x. [DOI] [PubMed] [Google Scholar]
- 32.Ahmer BM, Tran M, Heffron F. The virulence plasmid of Salmonella typhimurium is self-transmissible. J Bacteriol. 1999;181(4):1364–1368. doi: 10.1128/jb.181.4.1364-1368.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoertt BE, Ou J, Kopecko DJ, Baron LS, Warren RL. Novel virulence properties of the Salmonella typhimurium virulence-associated plasmid: Immune suppression and stimulation of splenomegaly. Plasmid. 1989;21(1):48–58. doi: 10.1016/0147-619x(89)90086-3. [DOI] [PubMed] [Google Scholar]
- 34.Elsinghorst EA, Baron LS, Kopecko DJ. Penetration of human intestinal epithelial cells by Salmonella: Molecular cloning and expression of Salmonella typhi invasion determinants in Escherichia coli. Proc Natl Acad Sci USA. 1989;86(13):5173–5177. doi: 10.1073/pnas.86.13.5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sizemore DR, Branstrom AA, Sadoff JC. Attenuated Shigella as a DNA delivery vehicle for DNA-mediated immunization. Science. 1995;270(5234):299–302. doi: 10.1126/science.270.5234.299. [DOI] [PubMed] [Google Scholar]
- 36.Li Y, et al. Evaluation of new generation Salmonella enterica serovar Typhimurium vaccines with regulated delayed attenuation to induce immune responses against PspA. Proc Natl Acad Sci USA. 2009;106(2):593–598. doi: 10.1073/pnas.0811697106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poltorak A, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science. 1998;282(5396):2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 38.Bannerman DD, Goldblum SE. Mechanisms of bacterial lipopolysaccharide-induced endothelial apoptosis. Am J Physiol Lung Cell Mol Physiol. 2003;284(6):L899–L914. doi: 10.1152/ajplung.00338.2002. [DOI] [PubMed] [Google Scholar]
- 39.Alderson MR, McGowan P, Baldridge JR, Probst P. TLR4 agonists as immunomodulatory agents. J Endotoxin Res. 2006;12(5):313–319. doi: 10.1179/096805106X118753. [DOI] [PubMed] [Google Scholar]
- 40.Kong Q, et al. Salmonella synthesizing 1-dephosphorylated [corrected] lipopolysaccharide exhibits low endotoxic activity while retaining its immunogenicity. J Immunol. 2011;187(1):412–423. doi: 10.4049/jimmunol.1100339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harper SA, Fukuda K, Cox NJ, Bridges CB. Advisory Committee on Immunization Practices Using live, attenuated influenza vaccine for prevention and control of influenza: supplemental recommendations of the Advisory Committee on Immunization Practices (ACIP) MMWR Recomm Rep. 2003;52(RR-13):1–8. [PubMed] [Google Scholar]
- 42.Fiers W, et al. M2e-based universal influenza A vaccine. Vaccine. 2009;27(45):6280–6283. doi: 10.1016/j.vaccine.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 43.Ulmer JB, et al. Protective CD4+ and CD8+ T cells against influenza virus induced by vaccination with nucleoprotein DNA. J Virol. 1998;72(7):5648–5653. doi: 10.1128/jvi.72.7.5648-5653.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.