Abstract
Stability of the long noncoding-polyadenylated nuclear (PAN) RNA from Kaposi's sarcoma-associated herpesvirus is conferred by an expression and nuclear retention element (ENE). The ENE protects PAN RNA from a rapid deadenylation-dependent decay pathway via formation of a triple helix between the U-rich internal loop of the ENE and the 3′-poly(A) tail. Because viruses borrow molecular mechanisms from their hosts, we searched highly abundant human long-noncoding RNAs and identified putative ENE-like structures in metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) and multiple endocrine neoplasia-β (MENβ) RNAs. Unlike the PAN ENE, the U-rich internal loops of both predicted cellular ENEs are interrupted by G and C nucleotides and reside upstream of genomically encoded A-rich tracts. We confirmed the ability of MALAT1 and MENβ sequences containing the predicted ENE and A-rich tract to increase the levels of an intronless β-globin reporter RNA. UV thermal denaturation profiles at different pH values support formation of a triple-helical structure composed of multiple U•A-U base triples and a single C•G-C base triple. Additional analyses of the MALAT1 ENE revealed that robust stabilization activity requires an intact triple helix, strong stems at the duplex-triplex junctions, a G-C base pair flanking the triplex to mediate potential A-minor interactions, and the 3′-terminal A of the A-rich tract to form a blunt-ended triplex lacking unpaired nucleotides at the duplex-triplex junction. These examples of triple-helical, ENE-like structures in cellular noncoding RNAs, are unique.
Keywords: RNA stability, RNA tertiary interactions
Most of the mammalian genome is transcribed into noncoding RNA (ncRNA) (1). The functions of long ncRNAs (lncRNAs; >200 nt) are only beginning to be elucidated (2). Recent studies have revealed that lncRNA stability varies widely (<0.5 h to >48 h) and have proposed that transcript half-life is one factor that regulates biological function (3, 4), but the underlying mechanisms remain poorly understood.
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) and multiple endocrine neoplasia-β (MENβ) RNAs are highly abundant, nuclear-localized lncRNAs, the precursors of which contain tRNA-like structures known as mascRNA for MALAT1 and menRNA for MENβ, both of which are substrates for RNase P (5–9). The resulting 5′-cleavage products of MALAT1 and MENβ RNAs terminate with a highly conserved sequence containing two U-rich motifs and a genomically encoded A-rich tract at the 3′ ends (8, 9). In vitro decay assays have shown that mutations in the U-rich motifs and A-rich tract destabilize mouse MALAT1 RNA fragments, suggesting the structure of this region contributes to RNA stability despite the lack of a traditional poly(A) tail (8, 10).
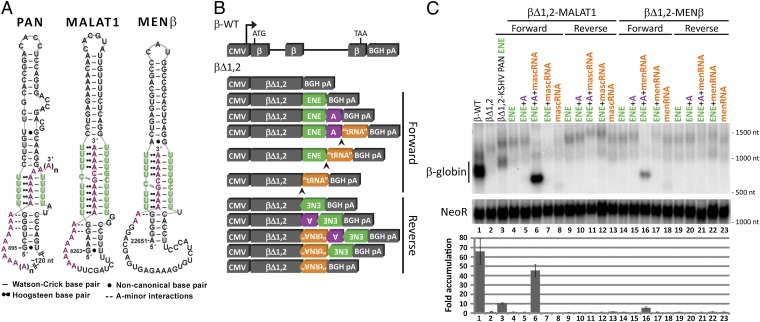
Previously, an expression and nuclear retention element (ENE) near the 3′ end of the lnc polyadenylated nuclear (PAN) RNA (∼500,000 copies per cell) produced by the Kaposi’s sarcoma-associated herpesvirus (KSHV) was identified and characterized (11, 12). The ENE (Fig. S1A) contains a U-rich internal loop that interacts with the 3′-poly(A) tail to protect PAN from a rapid nuclear deadenylation-dependent decay pathway (12–14). Crystallographic analysis of the KSHV PAN ENE core complexed with oligo(A9) revealed a triple helix composed of five consecutive U•A-U (where • denotes the Hoogsteen face and - denotes the Watson–Crick face) base triples extended by A-minor interactions with G-C base pairs flanking the U-rich loop (Fig. 1A) (15).
Fig. 1.
Models and stabilization activity of the predicted cellular ENE-like structures. (A) Schematic diagrams for the predicted ENE-like structures from MALAT1 (nucleotides 8263–8355) and MENβ (nucleotides 22651–22743) RNAs are based upon the known secondary and tertiary contacts of the PAN ENE (nucleotides 895–959) (14, 15). Nucleotides in the U-rich internal loop are in green and nucleotides in the poly(A) tail and A-rich tract are in purple. (B) Schematic diagrams of the β-globin plasmid constructs with different combinations of the predicted cellular ENE (green), A-rich tract (purple), and tRNA-like sequence (orange, representing mascRNA or menRNA) from MALAT1 or MENβ in either the forward or reverse orientation (see Fig. S2 for construct details). β-Globin expression is driven by the CMV promoter; the RNase P cleavage site is indicated by an arrowhead; and there is a bovine growth hormone polyadenylation signal (BGH pA) downstream of the 3′ UTR. (C) Northern blots (Upper) were probed for β-globin and Neomycin resistance (NeoR) RNAs, with the βΔ1,2-KSHV PAN ENE (βΔ1,2–79F in ref. 12) included as a control. RNA sizes are indicated to the right of the blot. Results were quantitated (Lower) by normalizing the β-globin signal to the NeoR signal, which served as a loading and transfection control. The empty βΔ1,2 reporter level was set at an arbitrary value of 1. Fold-accumulation is the average of at least three independent experiments; error bars represent SD.
Bioinformatic searches subsequently uncovered ENEs in RNAs produced by diverse viruses, suggesting a widespread mechanism to counteract host decay pathways (16). Because viruses borrow mechanisms from their hosts, we searched for ENE-like structures in cellular ncRNAs and identified candidate ENE-like structures in MALAT1 and MENβ ncRNAs. Here, we assess the RNA stabilization activity and characterize the triple-helical features of the putative ENE-like structures from the 3′ ends of mature MALAT1 and MENβ RNAs (Fig. 1A). These structures have U•A-U base triples flanked by two stems, one predicted to mediate A-minor interactions. The 3′ sides of the U-rich internal loops can form 11 or 9 consecutive Watson–Crick base pairs with the downstream A-rich tract for MALAT1 or MENβ RNA, respectively, and the 5′ sides are proposed to engage in Hoogsteen interactions. Interrupting the U•A-U triple-helical stack is a predicted C•G-C (or C•C-G) triple with an adjacent C-G (or G-C) base pair.
Results
Identification of RNA Sequences from MALAT1 and MENβ with Stabilization Activity.
To identify ENE-like structures in cellular ncRNAs, we: (i) performed a literature search for highly-abundant, nuclear-localized lncRNAs in human cells; (ii) visually inspected the 3′ region of each transcript for U-rich sequences upstream of a poly(A) tail or A-rich tract; and (iii) used Mfold to predict the secondary structure. MALAT1 and MENβ ncRNAs fulfilled these criteria. Both putative ENEs are predicted by Mfold to form a stem-loop structure with an asymmetric U-rich internal loop reminiscent of the KSHV PAN ENE (Fig. S1) (17).
Previously, we assessed the functionality of an ENE by its ability to increase the cellular abundance of an unstable, intronless β-globin (βΔ1,2) reporter (12, 16). To identify MALAT1 and MENβ sequences that confer stabilization activity, a series of βΔ1,2 reporters was created by inserting various combinations of the predicted ENE, A-rich tract, and tRNA-like region from either MALAT1 or MENβ into the 3′ UTR (Fig. 1B and Fig. S2).
Plasmids were transiently transfected into HEK293T cells and mRNA levels were measured via Northern blot analysis (Fig. 1C). Levels of the βΔ1,2 reporter RNAs increased 45- and 6-fold relative to the empty βΔ1,2 construct upon insertion of the putative ENE, A-rich tract, and tRNA-like region from MALAT1 and MENβ, respectively, in the forward orientation (Fig. 1C, lanes 6 and 16). Both transcripts appear to have been processed by RNase P because they are shorter and show a sharper band than the empty (polyadenylated) βΔ1,2 reporter RNA. Neither the reverse orientation nor any subset of inserted sequences significantly increased βΔ1,2 transcript levels. Specifically, (i) transcripts with the predicted ENE but without the A-rich tract did not accumulate, whether 3′-end processing proceeded via the canonical cleavage and polyadenylation pathway using the BGH pA signal or via RNase P cleavage by recognition of the mascRNA or menRNA structure [Fig. 1C, lanes 4 and 14 for poly(A) and lanes 7 and 17 for RNase P]; (ii) transcript accumulation did not occur when the ENE and A-rich tract were embedded internally within the transcript (Fig. 1C, lanes 5 and 15 vs. lanes 6 and 16); and (iii) the presence of a tRNA-like structure that could be processed by RNase P did not suffice for transcript accumulation (Fig. 1C, lanes 6–8 and 16–18). These findings argue that a functional ENE-like structure forms only when the predicted ENE and A-rich tract reside at the 3′ end of a transcript.
To ask whether direct interactions occur between the predicted MALAT1 and MENβ ENEs and the A-rich tracts, gel-shift assays were performed with 15mer oligonucleotides that mimic the corresponding A-rich tract for MALAT1 (5′-A9GCA4-3′) or MENβ (5′-CAG2A5GCA4-3′) (Fig. S3 A–C). When two Us on the 5′ sides of the U-rich internal loops were substituted with C in the MALAT1 (U8271C/U8275C) or MENβ (U22657C/U22661C) predicted ENEs, the A-rich oligonucleotide bound more weakly than to the WT ENE (Fig. S3). We conclude that the A-rich tract interacts directly with the U-rich internal loops of the proposed MALAT1 and MENβ ENEs, perhaps by forming triple helices.
Mutational Analyses Support Formation of RNA Triple Helices in Vivo.
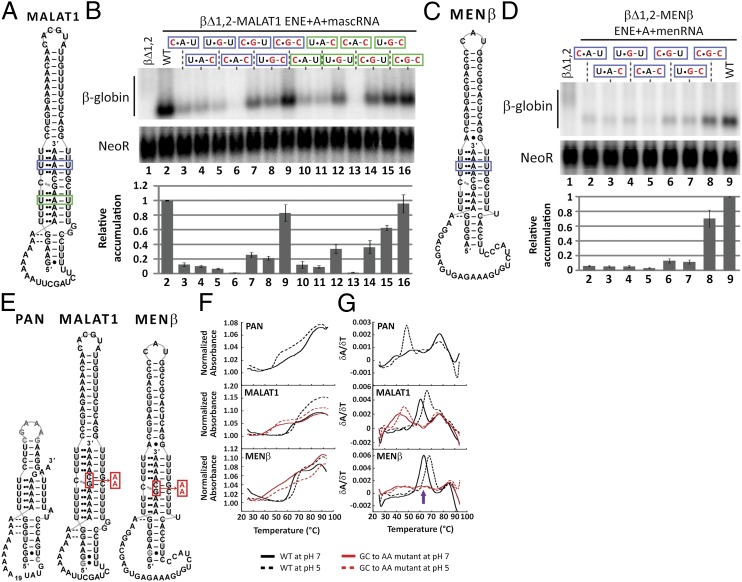
Stabilization by the KSHV ENE is conferred by a major groove RNA triple helix, in which the U-rich internal loop sequesters the 3′-poly(A) tail to form five consecutive U•A-U triples (13–15). To determine whether the predicted cellular ENEs engage their downstream A-rich tracts to form triple helices in vivo, we converted one putative U•A-U triple stepwise to an isosteric C•G-C triple in the βΔ1,2-MALAT1 and βΔ1,2-MENβ reporter constructs (Fig. 2 A and C, blue boxes). At the sites chosen in MALAT1 and MENβ, transcript levels decreased dramatically upon alteration of either one or two nucleotides of the predicted triplet (lanes 3–6 in Fig. 2B and lanes 2–5 in Fig. 2D). Single compensatory mutations (C•G-U and U•G-C), which should form a C•G Hoogsteen or G-C Watson–Crick base pair, respectively, slightly increased transcript levels (lanes 7 and 8 in Fig. 2B, lanes 6 and 7 in Fig. 2D). However, the double compensatory (C•G-C) mutants exhibited the highest transcript levels, with recoveries of ∼80% and 70% relative to WT for MALAT1 and MENβ, respectively (lane 9 in Fig. 2B; lane 8 in Fig. 2D). These results support formation of as many as four consecutive U•A-U base triples by both ENEs, as pictured in Fig. 2 A and C.
Fig. 2.
In vivo and in vitro biochemical evidence for an RNA triple-helical structure. (A and C) The same schematics as in Fig. 1A indicate the putative U•A-U base triples replaced with C•G-C in a blue (upper) or green (lower) box. (B and D) Northern blot analysis of β-globin and NeoR mRNAs (Upper) and quantitations (Lower) were performed as in Fig. 1C. Black nucleotides are WT sequence; mutated nucleotides are red. The WT βΔ1,2 reporter level was set at an arbitrary value of 1. Relative accumulation is the average of at least three independent experiments; error bars represent SD. (E) Schematic diagrams show the RNAs used in UV thermal denaturation experiments. The alignment of the PAN ENE core with A32GA2 is arbitrary. The red box and arrow denote the GC dinucleotide that was substituted with AA in MALAT1 ENE+A and MENβ ENE+A. (F) Plots of normalized absorbance at 260 nm versus temperature and (G) first derivative plots (δA/δT) versus temperature are shown for the PAN ENE core + A32GA2, the MALAT1 ENE+A WT and GC-to-AA, and the MENβ ENE+A WT and GC-to-AA mutant. Melting profiles of WT sequences are in black and the GC-to-AA mutant RNAs are in red. A solid line denotes pH 7; a dashed line denotes pH 5. Note that the MENβ ENE+A GC-to-AA mutant exhibits three transitions or peaks. The peak at 55–70 °C (purple arrow) shifted by several degrees in different experiments and once split into two defined peaks, making it difficult to explain its origin.
Unlike the viral ENEs identified so far, the U-rich internal loops of both cellular ENEs have intervening C and G nucleotides (Fig. 1A and Fig. S1). Therefore, we also engineered mutations into the lower predicted U•A-U triple helix in the βΔ1,2-MALAT1 ENE+A+mascRNA reporter. When a single U•A-U base triple was stepwise replaced with C•G-C (Fig. 2A, green box), the single- and double-nucleotide mutations decreased transcript levels, whereas full conversion to a C•G-C triple restored transcript levels to ∼100% of WT (Fig. 2B, lanes 10–16). Thus, the ENE-like structure of MALAT1 (and presumably MENβ) appears to form stabilizing U•A-U triple helices in vivo on both sides of the C and G nucleotides in the U-rich loop.
Thermal Denaturation Provides Evidence for a C•G-C Base Triple.
The proposed ENE-like structures of MALAT1 and MENβ RNA could potentially include a C•G-C base triple (Fig. 1A). We undertook UV thermal denaturation studies, a useful diagnostic tool because the melting profiles of DNA and RNA triplexes exhibit two distinct transitions: the first represents denaturation of the Hoogsteen face and the second denaturation of the Watson–Crick face (18, 19). Formation of C•G-C triples is pH-sensitive because the Hoogsteen face is stabilized by protonation of cytosine’s N3 position at lower pH (18, 20). Thus, comparison of RNA melting profiles at pH 5 and pH 7 can potentially reveal whether the Hoogsteen transition is pH dependent, suggesting the presence of a C•G-C triple (18, 20).
Melting profiles were determined for the five (three WT and two mutant) RNAs depicted in Fig. 2E. For MALAT1 and MENβ RNAs, the WT sequences extend from the predicted ENE to the end of the A-rich tract, but the mutants replace GC in the A-rich tract with AA to disrupt the putative triple helix. We also examined the KSHV PAN ENE core, which does not contain the GC interruption (15), with a 39-nt A-rich tract added to form an intramolecular RNA triple helix. We first performed gel-shift assays to confirm that the predicted MALAT1 and MENβ ENEs bound several orders of magnitude more weakly to oligo(A15) than to an A-rich oligonucleotide with GC (Figs. S3 B and C and S4).
The melting profiles of all five RNA constructs at pH 7 were biphasic, consistent with denaturation of a triple-helical structure (Fig. 2F). Melting temperatures (TM) for the two major transitions were extracted by locating the two largest maxima from a first derivative versus temperature plot (Fig. 2G); these values are presented in Table 1. For the three WT RNA constructs (PAN ENE core + A32GA2, MALAT1 ENE+A and MENβ ENE+A), the TM for the first transition (TM,1) was 56–63 °C and the second melting transition (TM,2) was 77–84 °C. Small differences in TM likely arise from the varying GC content of the three RNA structures (Fig. 1A). The GC-to-AA mutants for MALAT1 ENE+A and MENβ ENE+A have a TM,2 similar to WT; however, TM,1 is 16 °C and 21 °C lower than their WT counterparts (Table 1). Although the GC-to-AA mutation weakens the triple-helical portion of the structures, the flanking stem loops are as stable as WT, consistent with our proposed model (Fig. 1A). Additional support for assigning the first transition to melting of the Hoogsteen faces of triple helices came from analyzing the same bimolecular constructs used in the gel-shift assays. Melting profiles were monophasic for the KSHV PAN ENE core and the putative MALAT1 and MENβ ENEs alone, but the curves were biphasic when these same RNAs were folded in the presence of an A-rich oligonucleotide to form triplexes (Fig. S5). These results argue that an RNA triple helix forms in both unimolecular and bimolecular systems.
Table 1.
RNA melting temperatures (TM) at pH 5 and 7
| RNA with predicted or known ENE* | pH 7 |
pH 5 |
||
| TM,1† | TM,2 | TM,1 | TM,2 | |
| KSHV PAN ENE core + A32GA2 | 56 ± 1 | 76.8 ± 0.6 | 49.0 ± 0.7 | 77 ± 1 |
| MALAT1 ENE+A WT | 60.9 ± 0.2 | 76.6 ± 0.6 | 66.2 ± 0.3 | 75 ± 1 |
| MALAT1 ENE+A GC to AA | 45 ± 1 | 76.5 ± 0.4 | 46.7 ± 0.6 | 75.7 ± 0.3 |
| MENβ ENE+A WT | 63.3 ± 0.5 | 83.9 ± 0.7 | 69.0 ± 0.9 | 86.3 ± 0.3 |
| MENβ ENE+A GC to AA | 41.9 ± 0.5 | 83 ± 1 | 46.3 ± 0.4 | 85.6 ± 0.6 |
*Schematics of RNA structures are shown in Fig. 2E
†Melting temperatures are reported in degrees Celsius as the average of three independent measurements ± SD.
At pH 5, the melting profiles for the five RNAs (Fig. 2E) were biphasic, but the TM—mainly TM,1—were shifted compared with pH 7 (Fig. 2F and Table 1). TM,2 at pH 5 is within 1 °C of the values determined at pH 7 except for the MENβ ENE+A WT and GC-to-AA mutant, which increased by ∼2.5 °C at pH 5. The MENβ structure may be more stable at pH 5 because of formation of a noncanonical A•C base pair in the upper stem. For KSHV PAN ENE, TM,1 is 7 °C lower at pH 5 than pH 7, suggesting destabilization of the five U•A-U triples, consistent with previous studies of pure T•A-T or U•A-U triplets (20, 21). In contrast, the predicted WT MALAT1 and MENβ ENE+A RNAs exhibit TM,1s that are 5–6 °C greater at pH 5 than pH 7. Like WT, both mutant ENE+A RNAs have a higher TM,1 at pH 5, consistent with stabilization by a protonated cytosine. These pH effects support our structural model, suggesting formation of a C•G-C base triple within the triple helix of the cellular ENEs (Fig. 1A).
To examine the GC interruption using in vivo stabilization assays, the possible C•G-C or C•C-G triple was stepwise replaced with U•A-U in the βΔ1,2-MALAT1 ENE+A+mascRNA reporter (Fig. S6). The mutant with U•A-U replacing C•G-C accumulates to higher βΔ1,2 transcript levels (∼60% of WT) than that with U•A-U replacing C•C-G (∼20% of WT) (Fig. S6B, lanes 4 and 6). This result supports formation of C•G-C rather than C•C-G in vivo, although the incomplete rescue is suggestive of additional tertiary or protein interactions.
Nucleotide Content and Alignment of the U-Rich Loop and A-Rich Tract Are Important for Robust Stabilization.
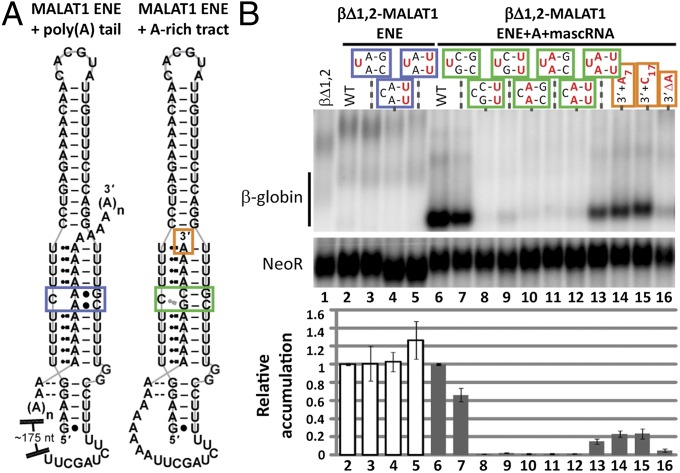
In the β-globin reporter assay, the βΔ1,2 transcripts with the MALAT1 or MENβ ENE insert alone did not accumulate (Fig. 1C, lanes 4 and 14), likely because the 3′-poly(A) tail of the reporter does not contain the GC nucleotides important for strong interaction with the ENE-like structures (Fig. 2 E–G, and Figs. S4 and S6). To test this explanation, the U-rich internal loop and A-rich tract were incrementally converted to all U and A nucleotides, respectively, in the βΔ1,2-MALAT1 ENE and βΔ1,2-MALAT1 ENE+A+mascRNA reporters. The βΔ1,2 transcripts with the mutant MALAT1 ENE, which would be predicted to interact with the poly(A) tail as the KSHV PAN ENE does, exhibited no substantial increase in accumulation relative to WT, including the all-U loop mutant (Fig. 3B, lanes 2–5). Increasing the U content in the internal loop of the βΔ1,2-MALAT1 ENE+A+mascRNA reporter decreased transcript levels (Fig. 3B, lanes 7–9) and increasing the A content of the A-rich tract did not restore stabilization until the U loop and A tract contained all U and A nucleotides, respectively (Fig. 3B, lanes 10–13). For the all-U/all-A mutant, the rescue was only ∼15% relative to WT, similar to the reporter level observed with one copy of the KSHV PAN ENE (Fig. 1C, lane 3 and Fig. 3B, lane 13). We conclude that G and C nucleotides in the U-rich loop and A-rich tract of the proposed MALAT1 ENE-like structure are important for robust stabilization activity.
Fig. 3.
Functional significance of the intervening C and G nucleotides in the U-rich loop and A-rich tract. (A) A schematic diagram is shown for the ENE-like structures predicted to form by the putative MALAT1 ENE with a 3′-poly(A) tail (nucleotides 8263–8336) or its adjacent A-rich tract (nucleotodes 8263–8355). The blue, green, and orange boxes outline the nucleotides targeted for mutagenesis. (B) Northern blot analysis of β-globin and NeoR mRNAs (Upper) and quantitation (Lower) were performed as in Fig. 1C. Black denotes WT sequence; mutated nucleotides are red. The plus (+) and delta (Δ) symbols represent a nucleotide insertion and deletion, respectively. The WT βΔ1,2 reporter levels were set at an arbitrary value of 1 for the βΔ1,2-MALAT1 ENE WT (white bar, lane 2) and the βΔ1,2-MALAT1 ENE+A+mascRNA WT (gray bar, lane 6). Each mutant was normalized relative to its WT counterpart. Relative accumulation is the average of at least three independent experiments; error bars represent SD.
The intervening GC nucleotides might function to align the A-rich tract specifically with the U-rich loop to create a blunt triple-helical terminus (Fig. 1A). In their absence, overhanging A nucleotides at the 3′ end could create a substrate for a 3′ exonuclease to initiate decay (22). To test this idea, A7 and C17 were inserted between the A-rich tract and mascRNA of the βΔ1,2-MALAT1 ENE+A+mascRNA construct (Fig. 3A). Transcript levels were ∼20% of WT, comparable to the all-U/all-A mutant (Fig. 3B, lanes 13–15). When we tested the effect of shortening the A-rich tract by deleting one A (Fig. 3A, orange box), transcript levels dropped ∼21-fold relative to WT (Fig. 3B, lane 16). These results argue that the intervening G and C nucleotides in the U-rich internal loop contribute to transcript abundance by generating a precisely-aligned, triple-helical structure with a blunt end.
Cellular ENEs Require Structures Other than a Triple Helix for Stabilization Activity.
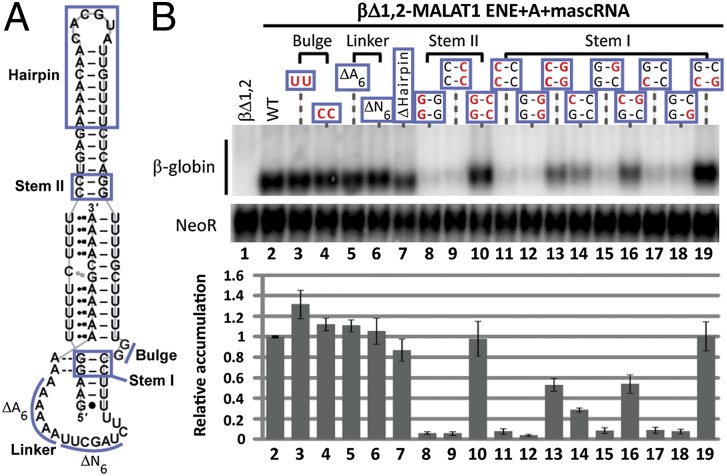
Mutational analysis of the KSHV PAN ENE identified additional features that are functionally important for stabilizing activity (14). Starting with the βΔ1,2-MALAT1 ENE+A+mascRNA reporter construct, we mutated five additional predicted structural features (Fig. 4A: bulge, linker, hairpin, and stems I and II) and assayed stabilization activity. The predicted GG bulge adjacent to the 3′ side of the U-rich loop was substituted with either UU or CC, slightly increasing levels compared with WT (Fig. 4B, lanes 3 and 4) and suggesting that these nucleotides do not form the G•U base pairs predicted by Mfold (Fig. S1B). Deletions within the predicted single-stranded linker that connects the U-rich loop to the A tract [AAAAAA (ΔA6) or UAGCUU (ΔN6)] yielded levels similar to WT, suggesting no important role for these nucleotides (Fig. 4B, lanes 5 and 6). The hairpin that extends stem II can likewise be deleted and replaced with a GCAA tetraloop, producing βΔ1,2 transcript levels ∼80% relative to WT (Fig. 4B, lane 7).
Fig. 4.
Other structural features contribute to stabilization. (A) The same MALAT1 schematic as in Fig. 1A has blue lines designating mutated regions. (B) Northern blot analysis of β-globin and NeoR RNAs (Upper) and quantitations (Lower) were performed as in Fig. 1C. WT nucleotides are black; mutated nucleotides are red. The delta (Δ) symbol represents a nucleotide deletion. In Δhairpin, a GCAA tetraloop was substituted to maintain a predicted stem-loop structure. The WT βΔ1,2 reporter level was set at an arbitrary value of 1. Relative accumulation is the average of at least three independent experiments; error bars represent SD.
In contrast, creating double C-C and G-G mismatches in stems I and II of the predicted ENE dramatically reduced βΔ1,2-MALAT1 ENE+A+mascRNA transcript levels, with compensatory mutations restoring stabilization activity to ∼50% and ∼100%, respectively (Fig. 4B, lanes 8–13). Partial rescue by the stem I compensatory mutant indicates the existence of other tertiary interactions, such as the A-minor interactions documented for the KSHV ENE (14, 15). Introducing a C-C or G-G mismatch for either of the stem I G-C base pairs (upper and lower) decreased reporter levels, but compensatory mutants restored activity to ∼50% and ∼100% of WT for the upper and lower base pair, respectively (Fig. 4B, lanes 14–19). These structure–function relationships suggest that stabilization activity of the MALAT1 ENE depends on highly structured stems near the duplex-triplex junctions and at least one strong A-minor interaction with the loop-proximal G-C base pair (14, 15).
Discussion
Although RNA triple helices were discovered more than 50 y ago, their in vivo existence and functional roles have remained elusive. The only cellularly encoded example of a eukaryotic RNA triple helix with three or more consecutive base triples is in the telomerase RNA (20, 23–26). Structural studies of the KSHV PAN ENE revealed that a triple helix forms when the U-rich internal loop of the ENE engages the 3′-poly(A) tail of PAN RNA; this structure effectively counteracts host nuclear RNA decay pathways (12–15). Here, we have exploited our understanding of the KSHV PAN ENE to discover triple-helical, ENE-like structures in two cellular lncRNAs: MALAT1 and MENβ.
We showed that sequences harboring the predicted ENE, A-rich tract, and tRNA-like structure from MALAT1 and MENβ RNAs confer stabilization activity on an intronless β-globin reporter transcript (Fig. 1 B and C) that had previously been used to validate five viral ENEs (12, 16). Formation of ENE-like structures was subsequently confirmed by mutational analyses. When predicted U•A-U base triples were mutated stepwise, the greatest recovery of stabilization activity was observed for the triple mutants with C•G-C (Fig. 2 A–D). Thermal denaturation studies on the predicted ENE-containing RNAs from MALAT1 and MENβ revealed two major melting transitions, a signature of DNA and RNA triplex structures (18, 19), for both intrastrand (Fig. 2F) and interstrand (Fig. S5) RNA interactions. The TM,1 is higher at pH 5 than at pH 7, consistent with the presence of a C•G-C triplet in the MALAT1 and MENβ ENE+A structures (Fig. 1A). This predicted C•G-C triplet can be replaced with U•A-U in the βΔ1,2-MALAT1 ENE+A+mascRNA reporter yielding 60% stabilization activity (Fig. S6). Collectively, our in vivo and in vitro results are consistent with the predicted ENEs and A-rich tracts of MALAT1 and MENβ forming triple-helical structures.
The U-rich internal loops of the viral (12, 16) and cellular ENEs characterized so far differ in both the number and identity of nucleotides (Fig. 1A). Based on our mutational analyses using the βΔ1,2-MALAT1 ENE+A+mascRNA reporter, the unique C and G nucleotides of the cellular ENEs likely fix the alignment of the A-rich tract with the U-rich loop so that the 3′-terminal A of the A-rich tract forms a blunt-ended triplex (SI Discussion). The presence of unpaired terminal nucleotides, as in mutants with overhangs (+A7 or +C17) or a nucleotide deletion (ΔA) at the 3′ end, decreases levels of the βΔ1,2 reporter (Fig. 3). The lack of robust stabilization activity conferred by the βΔ1,2-MALAT1 ENE all-U mutant (Fig. 3B, lane 5) indicates that a strong ENE-like structure cannot form with a 3′ tail that is purely poly(A) in vivo. Varying βΔ1,2 levels (1.1- to 9.2-fold differences in accumulation) have been observed for one copy of different viral ENEs tested in the β-globin reporter assay (12, 16). Differences could arise from improper ENE folding because of the sequence context or the distance between the ENE and 3′-poly(A) tail. Such factors have not been tested systematically, although increasing the U content of the U-rich loop unexpectedly decreases stabilization activity of the KSHV PAN ENE (14).
Overall, features critical for robust stabilization of the βΔ1,2-MALAT1 ENE+A+mascRNA reporter RNA were similar to those established for the KSHV PAN ENE (14): strong duplex-triplex junctions and the ability to form at least one A-minor interaction with the uppermost G-C base pair of stem I (Fig. 4). Structures or sequences that are dispensable for stabilization include the predicted apical hairpin, single-stranded linker, and bulged nucleotides as defined in Fig. 4A. Although the mutational analysis was performed only on the putative MALAT1 ENE-like structure, we predict a similar overall architecture for the MENβ ENE, given its high similarity (Fig. 1A). However, it is surprising that the stabilization observed for the βΔ1,2-MALAT1 ENE+A+mascRNA reporter is ∼sevenfold greater than the βΔ1,2-MENβ ENE+A+menRNA reporter when both have similar U-rich loops and A-rich tracts (Fig. 1 B and C). The proposed MENβ ENE-like structure may be less stable because of fewer U•A-U base triples in the lower triplex, fewer bulged nucleotides, fewer A-minor interactions in stem I, a weaker noncanonical A•G base pair in stem II, a longer single-stranded linker, an unknown trans-regulatory protein, or an unknown factor related to the β-globin reporter assay (Fig. 1A).
The half-life values reported for mature MALAT1 and MENβ RNAs vary widely: 3–16.5 h for MALAT1 and 1–8 h for MENβ (3, 9, 27). For nuclear RNA accumulation, formation of the predicted ENE-like structures at the 3′ end is only one feature that may be critical (5). Mouse knockout studies show MALAT1 and MENβ RNAs are not essential under normal conditions (28–31), despite the lack of paraspeckles in MENβ knockout mice (ref. 32 and references therein). MALAT1 has been implicated in pre-mRNA splicing via modulation of SR proteins and in activating a cell-growth control gene program (6, 29, 33–35). Because the ENEs discovered so far reside in lncRNAs with nonoverlapping biological functions, all ncRNAs should be included when searching for additional cellular ENE-like structures. Similarly, structural variations, such as noncanonical base triples, should be considered because the MALAT1 and MENβ ENEs contain C and G nucleotides that apparently form a C•G-C triple. Ultimately, solving the 3D structures of the MALAT1 or MENβ ENEs will illuminate the perturbations occasioned by inclusion of C and G nucleotides and inform future searches for structures critical to stabilization of RNA 3′ ends.
Materials and Methods
Plasmids and Mutagenesis.
The β-WT (with introns) and βΔ1,2 plasmids have a pcDNA3 backbone and were previously described (Fig. S2) (12, 36). The βΔ1,2 constructs containing the human MALAT1 ENE (nucleotides 8254–8336), MALAT1 ENE+A-rich tract (nucleotides 8254–8355), MALAT1 ENE+A-rich tract+mascRNA (nucleotides 8254–8424), mascRNA (nucleotides 8356–8424), MENβ ENE (nucleotides 22643–22716), MENβ ENE+A-rich tract (nucleotides 22643–22743), MENβ ENE+A-rich tract+menRNA (nucleotides 22643–22812), and menRNA (nucleotides 22744–22812) fragments were generated by PCR using human genomic DNA as a template. PCR fragments were subsequently inserted into the ApaI site of βΔ1,2, which is 137 nt upstream of the BGH pA signal sequence of AATAAAA, using standard molecular biology techniques (12). Nucleotide numbering corresponds to the sequences listed for human MALAT1 and human MENβ under accession NR_002819.2 and GQ859162.1, respectively. The sequence numbering for menRNA is a continuation from GQ859162.1. The βΔ1,2-MALAT1 ENE+mascRNA (Δnucleotides 8337–8355) and βΔ1,2-MENβ ENE+menRNA (Δnucleotides 22717–22743) constructs were created using site-directed mutagenesis per the manufacturer’s protocol (QuikChange). Nucleotide changes at the locations specified in the figures were created using site-directed mutagenesis. For the UV melting studies, the MALAT1 ENE+A-rich tract (nucleotides 8263–8355) and MENβ ENE+A-rich tract (nucleotides 22649–22743) sequences begin with GG to enhance transcription and were cloned into the EcoRI and Nhe1 sites in the pHDV plasmid [a gift from Graeme Conn (Emory University, Atlanta, GA) (37)]. The previously described PAN ENE core-pHDV plasmid (15) had the sequence 5′-GTATA32GA2-3′ inserted between the PAN ENE core and HDV ribozyme sequences.
β-Globin Reporter Assays, RNA Preparation, and Gel-Shift Assays.
β-Globin reporter assays (12, 16), RNA preparation for gel-shift and thermal denaturation assays (15), and gel-shift assays (15) were performed as previously described; modifications are noted in SI Materials and Methods.
Thermal Denaturation Assays.
RNA samples (∼0.25 μM) were prepared in a degassed buffer solution (25 mM sodium cacodylate pH 5 or 7, 50 mM KCl, and 1 mM MgCl2), heated at 95 °C for 3 min, snap-cooled on ice for 10 min, and allowed to equilibrate at room temperature for 1.5 h before being transferred to a stoppered 1-cm quartz cuvettete. For the bimolecular system, RNA samples contained a 1:1 molar ratio of ENE:A-rich oligo. Absorbance and temperature data were collected using a Chirascan CD spectrometer (Applied Photophysics) equipped with a temperature-controlled turret. Absorbance was monitored at 260 nm as the temperature increased at a rate of ∼0.4 °C/min from 25 to 94 °C and collected at 0.5 °C intervals. Individual melting profiles were analyzed using Origin software (OriginLab) to subtract buffer background, to take the first derivative of absorbance with respect to temperature (δA/δT), and to smooth the data over a 4 °C window using the Savitzky–Golay method. TM values were determined from the peak maxima of the first derivative versus temperature plot. TM values are an average of three independent measurements and error represents SD. The SD was ≤1 °C, supporting the reproducibility of the measurements.
Supplementary Material
Acknowledgments
We thank M.-D. Shu for βΔ1,2-MALAT1 ENE, forward and reverse plasmids, R. Mitton-Fry and S. DeGregorio for pHDV plasmids with WT MALAT1 ENE+A and MENβ ENE+A, and O. Itsathiphaisarn for PAN ENE core-pHDV plasmid with 5′-GTATA32GA2-3′ insert. This work was supported by Grant GM26154 from the National Institutes of Health, and Postdoctoral Fellowship Grant 122267-PF-12-077-01-RMC (to J.A.B.) from the American Cancer Society. J.A.S. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217338109/-/DCSupplemental.
References
- 1.Carninci P. Tagging mammalian transcription complexity. Trends Genet. 2006;22(9):501–510. doi: 10.1016/j.tig.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark MB, et al. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22(5):885–898. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tani H, et al. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res. 2012;22(5):947–956. doi: 10.1101/gr.130559.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hutchinson JN, et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernard D, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29(18):3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stadler P. Evolution of the long non-coding RNAs MALAT1 and MEN. In: Ferreira C, Miyano S, Stadler P, editors. Advances in Bioinformatics and Computational Biology. Berlin, Heidelberg: Springer; 2010. pp. 1–12. [Google Scholar]
- 8.Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135(5):919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sunwoo H, et al. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19(3):347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guhaniyogi J, Brewer G. Regulation of mRNA stability in mammalian cells. Gene. 2001;265(1–2):11–23. doi: 10.1016/s0378-1119(01)00350-x. [DOI] [PubMed] [Google Scholar]
- 11.Sun R, Lin SF, Gradoville L, Miller G. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc Natl Acad Sci USA. 1996;93(21):11883–11888. doi: 10.1073/pnas.93.21.11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conrad NK, Steitz JA. A Kaposi’s sarcoma virus RNA element that increases the nuclear abundance of intronless transcripts. EMBO J. 2005;24(10):1831–1841. doi: 10.1038/sj.emboj.7600662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conrad NK, Mili S, Marshall EL, Shu MD, Steitz JA. Identification of a rapid mammalian deadenylation-dependent decay pathway and its inhibition by a viral RNA element. Mol Cell. 2006;24(6):943–953. doi: 10.1016/j.molcel.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 14.Conrad NK, Shu MD, Uyhazi KE, Steitz JA. Mutational analysis of a viral RNA element that counteracts rapid RNA decay by interaction with the polyadenylate tail. Proc Natl Acad Sci USA. 2007;104(25):10412–10417. doi: 10.1073/pnas.0704187104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitton-Fry RM, DeGregorio SJ, Wang J, Steitz TA, Steitz JA. Poly(A) tail recognition by a viral RNA element through assembly of a triple helix. Science. 2010;330(6008):1244–1247. doi: 10.1126/science.1195858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tycowski KT, Shu MD, Borah S, Shi M, Steitz JA. Conservation of a triple-helix-forming RNA stability element in noncoding and genomic RNAs of diverse viruses. Cell Rep. 2012;2(1):26–32. doi: 10.1016/j.celrep.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plum GE, Park YW, Singleton SF, Dervan PB, Breslauer KJ. Thermodynamic characterization of the stability and the melting behavior of a DNA triplex: A spectroscopic and calorimetric study. Proc Natl Acad Sci USA. 1990;87(23):9436–9440. doi: 10.1073/pnas.87.23.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoyne PR, Gacy AM, McMurray CT, Maher LJ., 3rd Stabilities of intrastrand pyrimidine motif DNA and RNA triple helices. Nucleic Acids Res. 2000;28(3):770–775. doi: 10.1093/nar/28.3.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Theimer CA, Blois CA, Feigon J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol Cell. 2005;17(5):671–682. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 21.James PL, Brown T, Fox KR. Thermodynamic and kinetic stability of intermolecular triple helices containing different proportions of C+*GC and T*AT triplets. Nucleic Acids Res. 2003;31(19):5598–5606. doi: 10.1093/nar/gkg782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolin SL, Sim S, Chen X. Nuclear noncoding RNA surveillance: Is the end in sight? Trends Genet. 2012;28(7):306–313. doi: 10.1016/j.tig.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Felsenfeld G, Davies DR, Rich A. Formation of a 3-stranded polynucleotide molecule. J Am Chem Soc. 1957;79(8):2023–2024. [Google Scholar]
- 24.Felsenfeld G, Rich A. Studies on the formation of two- and three-stranded polyribonucleotides. Biochim Biophys Acta. 1957;26(3):457–468. doi: 10.1016/0006-3002(57)90091-4. [DOI] [PubMed] [Google Scholar]
- 25.Shefer K, et al. A triple helix within a pseudoknot is a conserved and essential element of telomerase RNA. Mol Cell Biol. 2007;27(6):2130–2143. doi: 10.1128/MCB.01826-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiao F, Cech TR. Triple-helix structure in telomerase RNA contributes to catalysis. Nat Struct Mol Biol. 2008;15(6):634–640. doi: 10.1038/nsmb.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedel CC, Dölken L, Ruzsics Z, Koszinowski UH, Zimmer R. Conserved principles of mammalian transcriptional regulation revealed by RNA half-life. Nucleic Acids Res. 2009;37(17):e115. doi: 10.1093/nar/gkp542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakagawa S, Naganuma T, Shioi G, Hirose T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J Cell Biol. 2011;193(1):31–39. doi: 10.1083/jcb.201011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakagawa S, et al. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012;18(8):1487–1499. doi: 10.1261/rna.033217.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang B, et al. The lncRNA MALAT1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2(1):111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eißmann M, et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012;9(8) doi: 10.4161/rna.21089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakagawa S, Hirose T. Paraspeckle nuclear bodies-useful uselessness? Cell Mol Life Sci. 2012;69(18):3027–3036. doi: 10.1007/s00018-012-0973-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tripathi V, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39(6):925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang L, et al. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147(4):773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyagawa R, et al. Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA. 2012;18(4):738–751. doi: 10.1261/rna.028639.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lykke-Andersen J, Shu MD, Steitz JA. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103(7):1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- 37.Walker SC, Avis JM, Conn GL. General plasmids for producing RNA in vitro transcripts with homogeneous ends. Nucleic Acids Res. 2003;31(15):e82. doi: 10.1093/nar/gng082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.