Neuroblastoma, an aggressive neural crest-derived malignancy of infants and young children, accounts for a large proportion of all pediatric cancer mortality, with overall survival less than 50% for high-risk disease. Current chemotherapy regimens are administered according to a risk stratification incorporating age, stage, histology, and amplification of the MYCN oncogene as primary factors for outcome (1). However, within the highest-risk group outcome is highly variable, and additional approaches are needed to identify those patients who would benefit from alternative therapies. In an article in PNAS entitled “A functional MYCN signature predicts outcome in neuroblastoma irrespective of MYCN amplification” (2), Valentijn et al. present an analysis distinguishing a subset of patients with high MYC protein levels but lacking MYCN gene amplification or high mRNA levels. They identify a set of MYCN-regulated genes in these patients regulating cell cycle, DNA repair, and apoptotic pathways and that are predictive of poor outcome.
The MYCN transcription factor was confirmed as an amplified oncogene more than two decades ago, and its function and role in neuroblastoma tumorigenesis has been intensely studied since then (3). Amplification of MYCN has proven to be a major negative prognostic factor distinguishing cases with very poor overall survival. However, neuroblastoma is a highly heterogeneous disease, and only approximately half of children with clinically high-risk disease and poor outcome display MYCN amplification (1). In addition, transgenic animal models of neuroblastoma suggest that amplification is not required for tumorigenesis. Rather, aberrant MYCN expression during neural crest development blocks differentiation of neural crest precursors favoring subsequent transformation and tumorigenesis (4). Valentijn et al. now define an MYCN-dependent transcriptional signature that identifies a subset of patients with high-risk disease whose tumors have elevated MYCN protein without MYCN amplification or elevated mRNA expression levels. By linking protein levels and direct transcriptional targets with clinical outcomes, this work further elucidates MYCN function in neuroblastoma. This also helps to explain some of the discrepancies between previous mRNA expression-based MYCN signatures studies that have yielded “nonoverlapping” prognostic signatures (5–7) and did not account for MYCN protein levels.
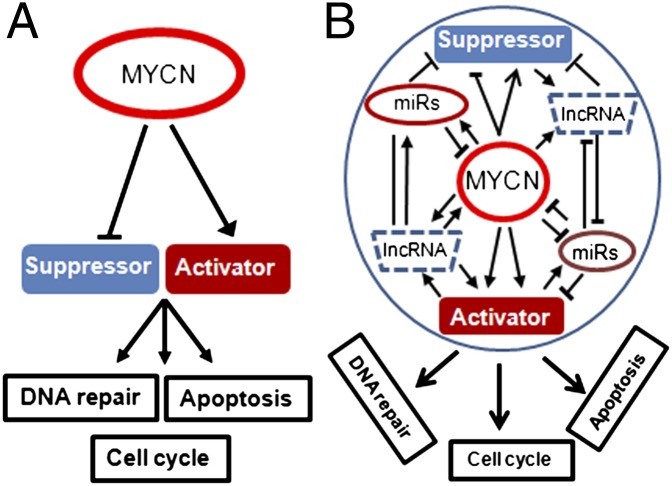
Despite decades of effort, the direct MYCN targets that account for its oncogenic functions remain enigmatic. Normally, MYCN is transiently expressed during embryogenesis and has essential roles in development (8). It typically regulates transcription through binding to E-box motifs in target gene promoters as an MYCN/myc-associated factor X (MAX) heterodimer. Aberrant expression of MYCN induces a plethora of phenotypic changes in neuroblastoma cell lines that are protumorigenic. These include increased proliferation and DNA repair, as well as inhibition of differentiation. However, MYCN also sensitizes neuroblastoma to genotoxic damage and lowers the apoptotic threshold of NB cells in culture (9). These conflicting phenotypes are highlighted at the molecular level by the findings that transcription of both p53 and its primary inhibitor, murine double minute (MDM2), is directly activated by MYCN (10, 11). Furthermore, it is clear that MYCN/DNA interactions can repress transcription of antioncogenic genes through both direct and indirect mechanisms, as recently demonstrated for high affinity nerve growth factor receptor TRKA and p75 neurotrophin receptor (p75NTR) (12) as well as MAP Kinase phosphatase 3 (MKP3) (13). Thus, the oncogenic influence of MYCN is an integration of direct and indirect transcriptional effects with diverse molecular functions (Fig. 1).
Fig. 1.
The MYCN oncogenic functions include altering DNA repair, p53 activities, and cell cycle regulation to drive neuroblastoma tumorigenesis. (A) Through interaction with gene promoters, MYCN directly regulates the transcription of oncogenic activators and tumor suppressors. (B) The addition of microRNA (miRs) and lncRNAs transcripts now place MYCN at the center of a complex regulatory network of direct and indirect effects complicating interpretation of transcriptional profiles.
The incorporation of microRNA and other noncoding RNAs into the scheme of MYCN-mediated oncogenesis adds an additional layer of complexity to the concept of an MYCN-prognostic signature. MYCN directly activates microRNAs that promote tumorigenesis and an aggressive phenotype (reviewed in ref. 14). These include miR-17-92 and miR-9, which promote survival and metastasis of neuroblastoma. It also likely inhibits expression of microRNAs such as miR-34a, which acts as a tumor suppressor by directly targeting MYCN and antiapoptotic factors such as B-cell lymphoma 2 (BCL2) (15). Because a single microRNA may target hundreds of downstream genes and can inhibit translation as well as transcription, and because MYCN regulates a large number of microRNAs, deciphering the oncogenic influence of MYCN on both the overall transcriptome and proteome of a cell becomes highly complex. This underscores the need to combine clinical outcome data and gene expression studies with protein data to uncover key critical molecular targets and pathways for therapeutic intervention. In addition, current high-throughput DNA and RNA sequencing methods and improved chromatin immunoprecipitation protocols now permit the comprehensive analysis of the MYCN transcriptome in cancer. Such studies are revealing interactions with recently discovered classes of noncoding RNAs, such as lncRNAs (long intergenic noncoding RNAs), with important functions in normal development and cancer (16, 17). Thus, we must constantly redefine and expand the components of an “MYCN-dependent prognostic signature” as our understanding of transcription regulation in normal and malignant cells progresses.
A consistent observation is that prognostic gene expression signatures for neuroblastoma, generated with diverse methodologies in different clinical cohorts, often identify noncoincident gene sets (5–7). The work by Valetijn et al. (2) highlights several issues with these data. First, gene and protein expression are not equivalent. Posttranscriptional mechanisms are well described that dramatically alter mRNA and protein stability. MYCN stabilization through LIN28b-mediated repression of Let-7 microRNAs was recently shown to contribute to neuroblastoma development (18). Second, oncogenes often act through both transcriptional activation and repression, which may be particularly important regarding inhibition of tumor suppressor microRNAs driving differentiation (19). Third, although the 157 gene signatures they delineate and several previous neuroblastoma gene signatures share few common genes, gene ontology analysis reveals that these signatures do converge on the common themes of altered cell cycle, DNA repair, and differentiation pathways.
Major intensification of chemotherapy for neuroblastoma over the past decade has only modestly improved survival yet markedly increased short- and long-term side toxicities (20). Our current ability to rapidly obtain comprehensive genome-wide data sets defining gene expression has generated a plethora of data regarding the oncogenic functions of MYCN and transcriptional targets. However, as demonstrated by Valentijn et al. (2), protein levels often do not correlate with gene expression. Furthermore, noncoding RNA functions have added multiple new layers to the complexity of both transcriptional and translational regulation. Going forward, analyzing, interpreting, and translating this wealth of information into effective molecularly targeted therapy remains an urgent challenge for molecular biologists and pediatric oncologists.
Footnotes
The author declares no conflict of interest.
See companion article on page 19190.
References
- 1.Cohn SL, et al. INRG Task Force The International Neuroblastoma Risk Group (INRG) classification system: An INRG Task Force report. J Clin Oncol. 2009;27(2):289–297. doi: 10.1200/JCO.2008.16.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valentijn LJ, et al. Functional MYCN signature predicts outcome of neuroblastoma irrespective of MYCN amplification. Proc Natl Acad Sci USA. 2012;109:19190–19195. doi: 10.1073/pnas.1208215109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brodeur GM, Seeger RC, Schwab M, Varmus HE, Bishop JM. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science. 1984;224(4653):1121–1124. doi: 10.1126/science.6719137. [DOI] [PubMed] [Google Scholar]
- 4.Hansford LM, et al. Mechanisms of embryonal tumor initiation: Distinct roles for MycN expression and MYCN amplification. Proc Natl Acad Sci USA. 2004;101(34):12664–12669. doi: 10.1073/pnas.0401083101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Preter K, et al. Accurate outcome prediction in neuroblastoma across independent data sets using a multigene signature. Clin Cancer Res. 2010;16(5):1532–1541. doi: 10.1158/1078-0432.CCR-09-2607. [DOI] [PubMed] [Google Scholar]
- 6.Ohira M, et al. A review of DNA microarray analysis of human neuroblastomas. Cancer Lett. 2005;228(1-2):5–11. doi: 10.1016/j.canlet.2005.01.054. [DOI] [PubMed] [Google Scholar]
- 7.Vermeulen J, et al. Predicting outcomes for children with neuroblastoma using a multigene-expression signature: A retrospective SIOPEN/COG/GPOH study. Lancet Oncol. 2009;10(7):663–671. doi: 10.1016/S1470-2045(09)70154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charron J, et al. Embryonic lethality in mice homozygous for a targeted disruption of the N-myc gene. Genes Dev. 1992;6(12A):2248–2257. doi: 10.1101/gad.6.12a.2248. [DOI] [PubMed] [Google Scholar]
- 9.Gamble LD, Kees UR, Tweddle DA, Lunec J. MYCN sensitizes neuroblastoma to the MDM2-p53 antagonists Nutlin-3 and MI-63. Oncogene. 2012;31(6):752–763. doi: 10.1038/onc.2011.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L, et al. p53 is a direct transcriptional target of MYCN in neuroblastoma. Cancer Res. 2010;70(4):1377–1388. doi: 10.1158/0008-5472.CAN-09-2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slack A, et al. The p53 regulatory gene MDM2 is a direct transcriptional target of MYCN in neuroblastoma. Proc Natl Acad Sci USA. 2005;102(3):731–736. doi: 10.1073/pnas.0405495102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iraci N, et al. A SP1/MIZ1/MYCN repression complex recruits HDAC1 at the TRKA and p75NTR promoters and affects neuroblastoma malignancy by inhibiting the cell response to NGF. Cancer Res. 2011;71(2):404–412. doi: 10.1158/0008-5472.CAN-10-2627. [DOI] [PubMed] [Google Scholar]
- 13.Marshall GM, et al. SIRT1 promotes N-Myc oncogenesis through a positive feedback loop involving the effects of MKP3 and ERK on N-Myc protein stability. PLoS Genet. 2011;7(6):e1002135. doi: 10.1371/journal.pgen.1002135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buechner J, Einvik C. N-myc and noncoding RNAs in neuroblastoma. Mol Cancer Res. 2012;10(10):1243–1253. doi: 10.1158/1541-7786.MCR-12-0244. [DOI] [PubMed] [Google Scholar]
- 15.Cole KA, et al. A functional screen identifies miR-34a as a candidate neuroblastoma tumor suppressor gene. Mol Cancer Res. 2008;6(5):735–742. doi: 10.1158/1541-7786.MCR-07-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niland CN, Merry CR, Khalil AM. Emerging roles for long non-coding RNAs in cancer and neurological disorders. Front Genet. 2012;3:25. doi: 10.3389/fgene.2012.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brunner AL, et al. Transcriptional profiling of lncRNAs and novel transcribed regions across a diverse panel of archived human cancers. Genome Biol. 2012;13(8):R75. doi: 10.1186/gb-2012-13-8-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molenaar JJ, et al. LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat Genet. 2012 doi: 10.1038/ng.2436. [DOI] [PubMed] [Google Scholar]
- 19.Shohet JM, et al. A genome-wide search for promoters that respond to increased MYCN reveals both new oncogenic and tumor suppressor microRNAs associated with aggressive neuroblastoma. Cancer Res. 2011;71(11):3841–3851. doi: 10.1158/0008-5472.CAN-10-4391. [DOI] [PubMed] [Google Scholar]
- 20.Perwein T, et al. Survival and late effects in children with stage 4 neuroblastoma. Pediatr Blood Cancer. 2011;57(4):629–635. doi: 10.1002/pbc.23036. [DOI] [PubMed] [Google Scholar]