Abstract
Tandem repeats (TRs) have extremely high mutation rates and are often considered to be neutrally evolving DNA. However, in coding regions, TR copy number mutations can significantly affect phenotype and may facilitate rapid adaptation to new environments. In several human genes, TR copy number mutations that expand polyglutamine (polyQ) tracts beyond a certain threshold cause incurable neurodegenerative diseases. PolyQ-containing proteins exist at a considerable frequency in eukaryotes, yet the phenotypic consequences of natural variation in polyQ tracts that are not associated with disease remain largely unknown. Here, we use Arabidopsis thaliana to dissect the phenotypic consequences of natural variation in the polyQ tract encoded by EARLY FLOWERING 3 (ELF3), a key developmental gene. Changing ELF3 polyQ tract length affected complex ELF3-dependent phenotypes in a striking and nonlinear manner. Some natural ELF3 polyQ variants phenocopied elf3 loss-of-function mutants in a common reference background, although they are functional in their native genetic backgrounds. To test the existence of background-specific modifiers, we compared the phenotypic effects of ELF3 polyQ variants between two divergent backgrounds, Col and Ws, and found dramatic differences. In fact, the Col-ELF3 allele, encoding the shortest known ELF3 polyQ tract, was haploinsufficient in Ws × Col F1 hybrids. Our data support a model in which variable polyQ tracts drive adaptation to internal genetic environments.
Keywords: circadian clock, hybrid incompatibility, microsatellite
In coding regions, tandem repeat (TR) copy number variation can have profound phenotypic effects (1). For example, TR copy number mutations that expand polyglutamine (polyQ) tracts past a threshold number of glutamines can cause incurable neurodegenerative diseases, such as Huntington disease and Spinocerebellar Ataxias (2, 3). PolyQ tract length correlates with onset and severity of polyQ expansion disorders, but for intermediate polyQ tracts this correlation is far weaker (4–8), suggesting that genetic and environmental modifiers exist (9–12). Despite their potential for pathogenicity, variable polyQ tracts occur frequently in eukaryotic proteins, many of them functioning in development and transcription (1, 13–15). Model organism studies have suggested that coding TRs are an important source of quantitative genetic variation that facilitates evolutionary adaptation (1, 16–19). For example, TR copy number variation in the yeast gene FLO1 correlates linearly with flocculation (20), a phenotype that is important for stress survival (17). As polyQ tracts often mediate protein interactions (2, 3, 21), polyQ-encoding TR copy number mutations could produce large and possibly adaptive phenotypic shifts.
To determine the phenotypic impact of naturally occurring polyQ variation (18, 22, 23) in a genetically tractable model, we focused on the gene EARLY FLOWERING 3 (ELF3), which encodes a polyQ tract that is highly variable across divergent Arabidopsis thaliana strains (accessions) (19, 24). ELF3 is a core component of the circadian clock and a potent repressor of flowering, and is considered a “hub protein” for its many interactions with various proteins (24–31). Consequently, elf3 loss-of-function mutants show pleiotropic phenotypes: they flower early, show poor circadian function, and grow long embryonic stems (hypocotyls) in light (25–27, 29, 30, 32). SNPs in ELF3 affect shade avoidance, a fitness-relevant plant trait (24, 33). ELF3 polyQ variation has been suggested to correlate with two parameters of the circadian clock: period and phase (19). The ELF3 polyQ tract may mediate ELF3 membership in protein complexes, although thus far no ELF3-binding protein is known to bind it (26, 28–30). We discovered that altering polyQ tract length has dramatic effects on ELF3-dependent phenotypes and that these effects are dependent on genetic background.
Results
ELF3-TR Variation Affects ELF3-Dependent Phenotypes.
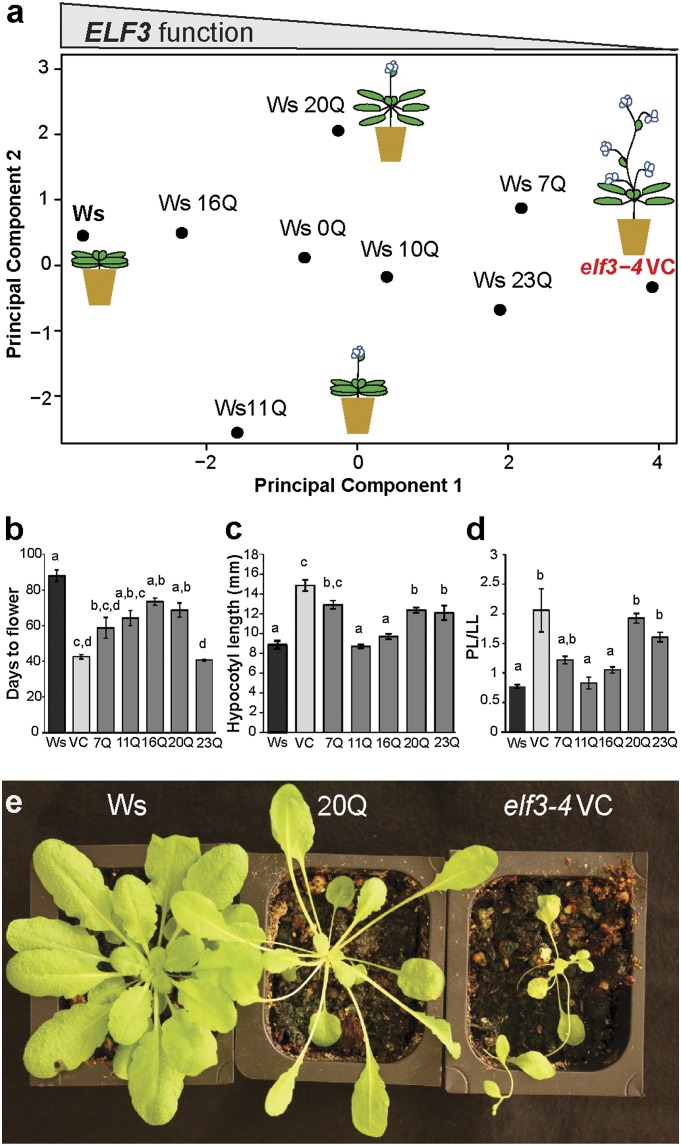
Among 181 natural A. thaliana accessions, the ELF3-TR encoded between 7 and 29Q (Fig. S1A and Table S1). For comparison, polyQ expansions over 20Q are associated with disease in the context of the SCA6 gene, although most other disease-associated polyQ expansions are longer (2, 19, 24). The most frequent ELF3-TR encoded 16Q, whereas the shortest TR (7Q) was found in the reference strain Col-0. We set out to test whether naturally occurring ELF3-TR alleles affect ELF3-dependent phenotypes, and whether they do so in a linear manner as suggested by association studies (19) and found for coding TR variation in other genes (16, 20). We generated expression-matched transgenic lines for most natural ELF3-TR alleles in the loss-of-function elf3-4 mutant (Ws background, Fig. S1C and Tables S2 and S3) (32) and measured their flowering time and circadian clock-related phenotypes (Fig. 1 and Fig. S2 A–G). ELF3-TR variation significantly affected ELF3-dependent phenotypes, but there was no evidence of a linear relationship. The different ELF3-TR alleles resulted in phenotypes ranging from nearly full complementation of elf3-4 to nearly phenocopying the loss-of-function mutant. We used principal components analysis (PCA) to describe the complex effects of ELF3-TR alleles on all tested ELF3-dependent phenotypes (Fig. 1A and Fig. S2 H–J). Principal component 1 (PC1) corresponds to general functionality of ELF3 in all measured phenotypes, with wild-type Ws and mutant elf3-4 defining the extremes. Separation along PC1 is driven by the tendency of plants with functional ELF3 to show short hypocotyls, late flowering, increased rosette leaf number, and short petioles (Fig. 1 B–D and Fig. S2). The endogenous ELF3-16Q allele complemented both the early-flowering and long-hypocotyl phenotypes of elf3-4 (Fig. 1 B–D and Fig. S2). In contrast, both the long ELF3-23Q and the short ELF3-7Q allele (endogenous TR alleles in Br-0/Bur-0 and Col-0, respectively) behaved similarly to the elf3-4 loss-of-function allele (Figs. 1 B–D and Fig. S2), although they are functional in their native backgrounds. Neither Col-0 nor Br-0 and Bur-0 show the phenotypic characteristics of elf3-mutants [early flowering (34), long hypocotyls (35), and long petioles (36)], suggesting that ELF3-TR alleles may interact with background-specific modifiers. ELF3-0Q, an artificial ELF3 allele lacking the TR, partially complemented elf3-4 (Fig. 1A and Fig. S2). Hence, the polyQ-encoding TR is not necessary for all ELF3 function, but changes in TR copy number are sufficient to enhance or ablate ELF3 function.
Fig. 1.
ELF3-TR variation has nonlinear phenotypic effects. (A) PCA of developmental traits of all ELF3-TR copy number variants. A. thaliana images illustrate ELF3-TR effects on the traits days to flower and hypocotyl length under SD and LD, petiole-length/leaf-length ratio (PL/LL) under SD only, and rosette leaf number under LD only. The contributions of specific phenotypes to PCs are in Fig. S2J. Representative TR copy number alleles are shown from an analysis including all alleles (for all alleles see Fig. S2 H and I). (B) Days to flower under SD conditions for selected lines. n = 6 plants per transgenic line. (C) Hypocotyl length at 15 d under SD for selected lines. n = 20–30 seedlings per transgenic line. (D) PL/LL of the fourth leaf for selected lines. Data are from the same plants as in B. (E) Plants carrying the ELF3-20Q allele (Center) are specific hypomorphs under SD with the elongated petioles of the elf3-4 mutant (vector control, VC, Right) and a wild-type flowering phenotype (Ws, Left). ELF3-TR alleles are indicated with the number of Qs encoded, Ws is wild-type, VC is the elf3-4 vector control. Error bars are SEs of means. Genotypes labeled with different letters differed significantly in phenotype by Tukey-HSD test with α = 0.05. For all Ws-background phenotype data, see Fig. S2 A–G. Data are from multiple independently generated expression-matched (Fig. S1C) T3 and T4 lines for each TR copy number allele (Table S2). These experiments were repeated at least once with similar results. The tested ELF3-20Q lines contained unique insertions that did not affect genes with known function (Table S3).
PC2 separated ELF3-20Q and ELF3-11Q, which behaved as hypomorphs in certain phenotypes but not others (Fig. 1A). For example, ELF3-20Q plants had significantly longer hypocotyls than wild-type and its petioles phenocopied the extremely long petioles of the elf3-4 mutant (Fig. 1 C–E), but they did not differ from wild-type in flowering time (days to flower, Fig. 1B). The existence of both general and specific hypomorphs suggests that polyQ variation affects the multiple ELF3 functions separately.
As part of a protein complex, ELF3 affects expression of Phytochrome-interacting factor 5 (PIF5) and Pseudoresponse regulator 9 (PRR9) (28, 37, 38). PIF5 and PRR9 expression were strongly affected by ELF3 polyQ variation (Fig. S3). ELF3-16Q phenocopied wild-type PRR9 and PIF5 expression, and the hypomorphic ELF3-23Q phenocopied elf3-4 (28, 37, 38), mirroring their developmental phenotypes. Consistent with their divergence along PC2 (Fig. 1A), ELF3-11Q and ELF3-20Q differed in their effect on PRR9 expression, but not on PIF5 expression (Fig. S3 A and B), demonstrating that ELF3 polyQ variation differentially affects the regulation of downstream genes.
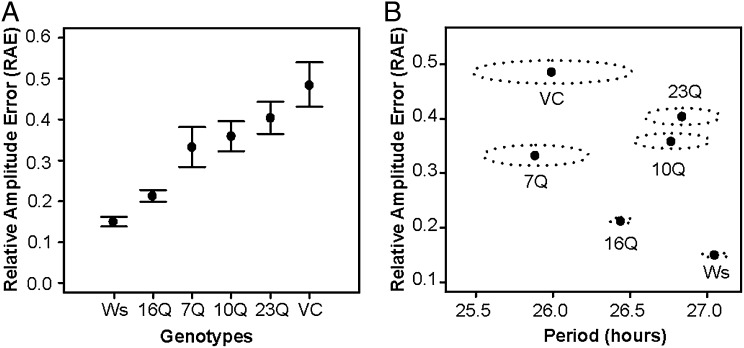
ELF3-TR Variation Modulates the Precision of the Circadian Clock.
To directly assess the role of ELF3 polyQ variation in the circadian clock, we used the CCR2::LUC reporter system (25, 39). We observed little difference in circadian period among wild-type Ws and tested ELF3-TR alleles (Fig. S4A), contradicting a previously observed association of TR copy number with period in natural accessions (19). However, we found that the relative amplitude error (RAE) of oscillation varies substantially across ELF3-TR genotypes (Fig. 2A and Fig. S4B). RAE measures the precision of a circadian period (40): high RAE values (>0.4) indicate poor oscillation and clock dysfunction (41). The endogenous Ws ELF3-16Q nearly complemented the elf3-4 RAE defect, whereas the TR alleles ELF3-7Q, ELF3-10Q, and ELF3-23Q showed higher RAE, approaching arrhythmic elf3-4 levels (Fig. 2 A and B), consistent with their hypomorphic performance in other ELF3 traits (close to elf3-4 in PC1) (Fig. 1A). Taken together, these results suggest that ELF3 polyQ tract length is a critical determinant of circadian clock precision—but not period length—in A. thaliana.
Fig. 2.
ELF3-TR variation modulates the precision of the circadian clock. (A) RAE of CCR2::LUC circadian oscillation in seedlings with indicated ELF3-TR alleles. Bars represent 99% confidence intervals. (B) Mean values of circadian period and RAE (points) were measured in seedlings with indicated ELF3-TR alleles. Dotted ellipses represent SEMs for both period and RAE. Note that plants with high RAE have extremely unreliable estimates of circadian period. Bioluminescence rhythms from the CCR2::LUC reporter in ELF3-TR transgenic lines were used to measure circadian parameters under LL after 5 d of entrainment in 12-h light:12-h dark cycles. n = >100 seedlings for all genotypes. Aggregate data from four independent experiments are shown. See Fig. S4 for RAE and period data for all alleles.
ELF3-TR Variation Interacts with Genetic Background.
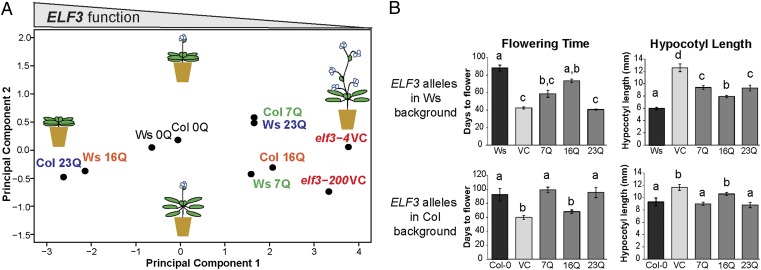
To test our hypothesis that ELF3-TR variation interacts with genetic background, we regenerated all ELF3-TR transgenic lines in the elf3-200 loss-of-function mutant with matched transgene expression (Col background; Fig. S1D and Table S4) (42). We used PCA to compare ELF3-TR effects between Ws and Col backgrounds (Fig. 3A and Fig. S5).
Fig. 3.
The phenotypic effects of ELF3-TR variation are strongly background-dependent. (A) PCA of developmental traits of all ELF3-TR alleles in Ws and Col genetic backgrounds. Shared color indicates a given ELF3-TR allele in both genetic backgrounds. A. thaliana images are as in Fig. 1A. The contributions of phenotypes to principal components are similar to Fig. 1A, except that PC2 is inverted (no effect on interpretation, loadings in Fig. S5C). Representative TR copy number alleles are shown from an analysis including all alleles (for all alleles see Fig. S5; for Col-background specific PCA, see Fig. S6). (B) Days to flower under SD and hypocotyl length under LD differ for particular TR alleles between Ws (Upper) and Col (Lower) backgrounds. ELF3-TR alleles are indicated with the number of Qs encoded, Ws and Col-0 are wild-type, elf3-4 and elf3-200 are respective vector controls (VC). Error bars represent SEM. Genotypes labeled with different letters differed significantly in phenotype by Tukey-HSD test with α = 0.05. For all Col-background phenotype data, see Fig. S6 A–G. Data are from multiple independently generated expression-matched (Fig. S1 C and D) T3 and T4 lines for each TR copy number allele (Table S4). These experiments were repeated at least once with similar results.
The Col-specific ELF3-7Q allele complemented elf3-200 in some traits, such as flowering time (in short days, SD) and hypocotyl length (in long days, LD), but not others (Fig. 3 A and B, and Figs. S5 and S6). This result may be because of the absence of the small 5′ intron from the ELF3 construct used in this study. However, there was still a dramatic spread of phenotypes: all longer ELF3-TR alleles (>20 Qs) nearly complemented elf3-200, delaying flowering and shortening hypocotyls, whereas few of the shorter alleles did (Fig. 3, and Figs. S5 and S6). Results were similar when the Col data were analyzed alone (Fig. S6). Thus, in contrast to our results in the Ws background, ELF3-TRs appeared to show a threshold effect for TR copy number in the Col background. We speculate that the intensive laboratory propagation of the Col-0 accession may have altered selection on the ELF3-TR, resulting in an extremely short “hypomorphic” allele, whereas under natural conditions a longer TR might be more functional.
Comparing TR allele effects between the two backgrounds revealed striking differences. For example, the ELF3-23Q allele was a general hypomorph in the Ws background (elf3-4), whereas it produced highly functional ELF3 in the Col background (elf3-200) (Fig. 3). In turn, the ELF3-16Q allele produced highly functional ELF3 in the Ws background (elf3-4), but was a general hypomorph in the Col background (elf3-200). The consistent performance of the artificial ELF3-0Q allele across backgrounds suggests that the background effect is TR-dependent (Fig. 3A and Fig. S5). Collectively, our results suggest that ELF3-TR alleles interact with background-specific modifiers.
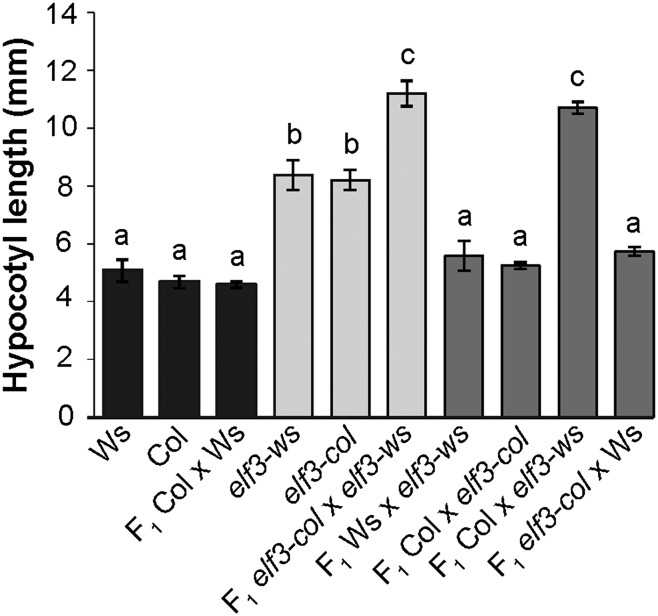
Col ELF3 Allele Is Haploinsufficient in Col × Ws Hybrids.
To address whether Ws and Col-specific background effects are sufficient for altered hybrid phenotypes, we generated F1 populations between wild-type and elf3-null plants in the Ws and Col backgrounds and measured ELF3 function by assessing hypocotyl length. Ws × Col F1 hybrids resembled their wild-type parents (Fig. 4). F1 hybrids containing both loss-of-function alleles had significantly longer hypocotyls than either parent (Fig. 4). Both ELF3 alleles were haplosufficient in F1 crosses within their native backgrounds, as expected for recessive mutants (Fig. 4). In stark contrast, we observe that ELF3-Col, but not ELF3-Ws, phenocopied the extreme hypocotyl length of the double loss-of-function mutant (Fig. 4). Consistent with the results from our transgenic lines, our F1 hybrid data suggest that full ELF3 function depends on a permissive genetic background.
Fig. 4.
ELF3-Col is haploinsufficient in a hybrid Col × Ws genetic background. Hypocotyl length under SD was measured in seedlings from parental and F1 lines. elf3-ws is the elf3-4 loss-of-function mutant in the Ws background; elf3-col is the elf3-200 loss-of-function mutant in the Col background. Reciprocal crosses for each F1 showed similar results. n = 15–20 for each genotype, except for F1Ws x elf3-ws (n = 5). Error bars are SEM. Genotypes labeled with different letters differed significantly by Tukey-HSD test with α = 0.05.
Discussion
Our results demonstrate that natural ELF3 polyQ variation that is not associated with disease has dramatic phenotypic consequences, and that these consequences depend on genetic background. For ELF3, in at least the Ws background, the relationship between TR copy number and phenotype does not follow a linear or threshold pattern as observed for other coding TR and polyQ disorders (1, 2, 16, 17, 20). Studies correlating TR variation with phenotype often apply linear models, treating TR copy number as a quantitative variable (19, 22, 23). Our data show that this approach is not appropriate for all TRs.
Instead, ELF3-TR alleles seem “matched” to specific genetic backgrounds in which they are functional, whereas they are incompatible with other backgrounds. The haploinsufficiency of the elf3-col allele in Ws × Col hybrids supports this interpretation. In contrast, the ELF3-Ws allele is haplosufficient in hybrids, indicating that the ELF3-Col × Ws incompatibility is asymmetric. This observation agrees with Orr’s assertion that incompatibility between recently diverged populations is usually asymmetrical, because it tends to arise from the derived allele (i.e., ELF3-Col) (43). Variable TRs, and the ELF3-TR in particular, have been previously suggested as agents of adaptation to new external environments (1, 16, 17, 20, 24, 44). Our results suggest that polyQ-encoding TRs are also agents of coadaptation within genomes.
We speculate that the observed background effects arise from background-specific polymorphisms in genes encoding physically interacting proteins (26, 28–30). TRs have a far higher mutation rate than nonrepeated regions (10−4 per site per generation for TR vs. 10−8 for SNPs) (45, 46) and, as we show, their expansion or contraction can have dramatic phenotypic impact. ELF3’s partner proteins may have acquired compensatory mutations to accommodate new ELF3-TR variants and vice versa. Alternative explanations for the background effects are compensatory mutations in ELF3 (intragenic suppressors), or ELF3 interactions that are unique to a given background. Intragenic variation and protein modification can play an important role in polyQ-mediated phenotypes (47, 48). At least for the ELF3-Col allele, however, our F1 data are not consistent with intragenic suppressors.
Consistent with polyQ-mediated background effects, in at least one case a modifier mutation has been shown to delay onset of Huntington disease (11). Hypothetically, population genetic approaches could identify incompatible alleles that may contribute to variable disease onset in patients with polyQ expansions and to ELF3-dependent background effects in A. thaliana. However, the great diversity of TR alleles compared with SNP alleles and the small number of individuals carrying specific TR alleles render a population genetics approach infeasible. Extensive genetic mapping or other experimental approaches will be needed to identify the determinants of ELF3-TR–dependent background effects.
As TRs are rapidly evolving, we speculate that polyQ-mediated incompatibilities and the resulting fitness loss in hybrids and their offspring may contribute to disruption of gene flow between closely related populations. This speciation mechanism would be of particular importance for organisms with many polyQ-encoding TRs, thousands of offspring, and an inbreeding lifestyle. Even in humans, however, about 1% of proteins contain polyQ tracts (13, 14, 45). Our results identify TR copy number variation, and in particular polyQ variation, as a phenotypically important class of genetic variation that warrants genome-wide assessment in model organisms, crops, and humans alike.
Materials and Methods
Information on the genotypes and experimental growth conditions, recombinant DNA techniques and generation of ELF3 transgenic plants, and RNA extraction, processing and quantitative real-time PCR used in this study is provided in SI Materials and Methods. See Tables S5–S7 for primer information.
Developmental Phenotype Assays.
Phenotypic assays were performed with expression-matched, homozygous T3 or T4 transgenic plants (Fig. S1 C and D and Tables S2–S4). For hypocotyl length and flowering-time experiments, plants were grown and measured in a pseudorandomized design under SD and LD conditions. More details are provided in SI Materials and Methods.
Luciferase Imaging and Period Analysis.
Luciferase assays were performed with lines harboring the CCR2::LUC reporter. Further details are provided in SI Materials and Methods.
Principal Components Analysis.
PCA on transformed phenotype data was performed using the R function prcomp (R Foundation for Statistical Computing, 2011, http://www.r-project.org/). More details are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank D. Nusinow for providing the ELF3 promoter, M. Nordborg for Arabidopsis thaliana accessions, and S. Fields for critical comments on the manuscript. This work was supported by National Human Genome Research Institute Interdisciplinary Training in Genome Sciences Grants T32HG000035-16 (to S.F.U.) and 2T32HG35-16 (to M.O.P.); Deutsche Forschungsgemeinschaft Grant DA1061/4-1 (to S.J.D.); and Royalty Research Fund Grant RRF4365 (to C.Q.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
1Deceased October 7, 2006.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211021109/-/DCSupplemental.
References
- 1.Gemayel R, Vinces MD, Legendre M, Verstrepen KJ. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu Rev Genet. 2010;44:445–477. doi: 10.1146/annurev-genet-072610-155046. [DOI] [PubMed] [Google Scholar]
- 2.Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: Mechanisms and common principles. Nat Rev Genet. 2005;6(10):743–755. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- 3.Orr HT. Polyglutamine neurodegeneration: Expanded glutamines enhance native functions. Curr Opin Genet Dev. 2012;22(3):251–255. doi: 10.1016/j.gde.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrew SE, et al. The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington’s disease. Nat Genet. 1993;4(4):398–403. doi: 10.1038/ng0893-398. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt BJ, Greenberg CR, Allingham-Hawkins DJ, Spriggs EL. Expression of X-linked bulbospinal muscular atrophy (Kennedy disease) in two homozygous women. Neurology. 2002;59(5):770–772. doi: 10.1212/wnl.59.5.770. [DOI] [PubMed] [Google Scholar]
- 6.Schöls L, Bauer P, Schmidt T, Schulte T, Riess O. Autosomal dominant cerebellar ataxias: Clinical features, genetics, and pathogenesis. Lancet Neurol. 2004;3(5):291–304. doi: 10.1016/S1474-4422(04)00737-9. [DOI] [PubMed] [Google Scholar]
- 7.van de Warrenburg BP, et al. Spinocerebellar ataxias in the Netherlands: Prevalence and age at onset variance analysis. Neurology. 2002;58(5):702–708. doi: 10.1212/wnl.58.5.702. [DOI] [PubMed] [Google Scholar]
- 8.Zühlke C, Dalski A, Schwinger E, Finckh U. Spinocerebellar ataxia type 17: Report of a family with reduced penetrance of an unstable Gln49 TBP allele, haplotype analysis supporting a founder effect for unstable alleles and comparative analysis of SCA17 genotypes. BMC Med Genet. 2005;6:27. doi: 10.1186/1471-2350-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fryer JD, et al. Exercise and genetic rescue of SCA1 via the transcriptional repressor Capicua. Science. 2011;334(6056):690–693. doi: 10.1126/science.1212673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gehrking KM, et al. Partial loss of Tip60 slows mid-stage neurodegeneration in a spinocerebellar ataxia type 1 (SCA1) mouse model. Hum Mol Genet. 2011;20(11):2204–2212. doi: 10.1093/hmg/ddr108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Metzger S, et al. Huntingtin-associated protein-1 is a modifier of the age-at-onset of Huntington’s disease. Hum Mol Genet. 2008;17(8):1137–1146. doi: 10.1093/hmg/ddn003. [DOI] [PubMed] [Google Scholar]
- 12.Zijlstra MP, et al. Levels of DNAJB family members (HSP40) correlate with disease onset in patients with spinocerebellar ataxia type 3. Eur J Neurosci. 2010;32(5):760–770. doi: 10.1111/j.1460-9568.2010.07352.x. [DOI] [PubMed] [Google Scholar]
- 13.Faux NG, et al. Functional insights from the distribution and role of homopeptide repeat-containing proteins. Genome Res. 2005;15(4):537–551. doi: 10.1101/gr.3096505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karlin S, Brocchieri L, Bergman A, Mrazek J, Gentles AJ. Amino acid runs in eukaryotic proteomes and disease associations. Proc Natl Acad Sci USA. 2002;99(1):333–338. doi: 10.1073/pnas.012608599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Molla M, Delcher A, Sunyaev S, Cantor C, Kasif S. Triplet repeat length bias and variation in the human transcriptome. Proc Natl Acad Sci USA. 2009;106(40):17095–17100. doi: 10.1073/pnas.0907112106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sawyer LA, et al. Natural variation in a Drosophila clock gene and temperature compensation. Science. 1997;278(5346):2117–2120. doi: 10.1126/science.278.5346.2117. [DOI] [PubMed] [Google Scholar]
- 17.Smukalla S, et al. FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell. 2008;135(4):726–737. doi: 10.1016/j.cell.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michael TP, et al. Simple sequence repeats provide a substrate for phenotypic variation in the Neurospora crassa circadian clock. PLoS ONE. 2007;2(8):e795. doi: 10.1371/journal.pone.0000795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tajima T, Oda A, Nakagawa M, Kamada H, Mizoguchi T. Natural variation of polyglutamine repeats of a circadian clock gene ELF3 in Arabidopsis. Plant Biotechnol. 2007;24:237–240. [Google Scholar]
- 20.Verstrepen KJ, Jansen A, Lewitter F, Fink GR. Intragenic tandem repeats generate functional variability. Nat Genet. 2005;37(9):986–990. doi: 10.1038/ng1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stott K, Blackburn JM, Butler PJ, Perutz M. Incorporation of glutamine repeats makes protein oligomerize: Implications for neurodegenerative diseases. Proc Natl Acad Sci USA. 1995;92(14):6509–6513. doi: 10.1073/pnas.92.14.6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindqvist C, Laakkonen L, Albert VA. Polyglutamine variation in a flowering time protein correlates with island age in a Hawaiian plant radiation. BMC Evol Biol. 2007;7:105. doi: 10.1186/1471-2148-7-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Malley KG, Ford MJ, Hard JJ. Clock polymorphism in Pacific salmon: Evidence for variable selection along a latitudinal gradient. Proc Biol Sci. 2010;277(1701):3703–3714. doi: 10.1098/rspb.2010.0762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiménez-Gómez JM, Wallace AD, Maloof JN. Network analysis identifies ELF3 as a QTL for the shade avoidance response in Arabidopsis. PLoS Genet. 2010;6(9):e1001100. doi: 10.1371/journal.pgen.1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Covington MF, et al. ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell. 2001;13(6):1305–1315. doi: 10.1105/tpc.13.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu XL, Covington MF, Fankhauser C, Chory J, Wagner DR. ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell. 2001;13(6):1293–1304. doi: 10.1105/tpc.13.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McWatters HG, Bastow RM, Hall A, Millar AJ. The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature. 2000;408(6813):716–720. doi: 10.1038/35047079. [DOI] [PubMed] [Google Scholar]
- 28.Nusinow DA, et al. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475(7356):398–402. doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshida R, et al. Possible role of early flowering 3 (ELF3) in clock-dependent floral regulation by short vegetative phase (SVP) in Arabidopsis thaliana. New Phytol. 2009;182(4):838–850. doi: 10.1111/j.1469-8137.2009.02809.x. [DOI] [PubMed] [Google Scholar]
- 30.Yu J-W, et al. COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol Cell. 2008;32(5):617–630. doi: 10.1016/j.molcel.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zagotta MT, Shannon S, Jacobs C, Meeks-Wagner R. Early-flowering Mutants of Arabidopsis thaliana. Aust J Plant Physiol. 1992;19(4):411–418. [Google Scholar]
- 32.Zagotta MT, et al. The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J. 1996;10(4):691–702. doi: 10.1046/j.1365-313x.1996.10040691.x. [DOI] [PubMed] [Google Scholar]
- 33.Coluccio MP, Sanchez SE, Kasulin L, Yanovsky MJ, Botto JF. Genetic mapping of natural variation in a shade avoidance response: ELF3 is the candidate gene for a QTL in hypocotyl growth regulation. J Exp Bot. 2011;62(1):167–176. doi: 10.1093/jxb/erq253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shindo C, et al. Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol. 2005;138(2):1163–1173. doi: 10.1104/pp.105.061309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maloof JN, et al. Natural variation in light sensitivity of Arabidopsis. Nat Genet. 2001;29(4):441–446. doi: 10.1038/ng777. [DOI] [PubMed] [Google Scholar]
- 36.Pérez-Pérez JM, Serrano-Cartagena J, Micol JL. Genetic analysis of natural variations in the architecture of Arabidopsis thaliana vegetative leaves. Genetics. 2002;162(2):893–915. doi: 10.1093/genetics/162.2.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kolmos E, et al. A reduced-function allele reveals that EARLY FLOWERING3 repressive action on the circadian clock is modulated by phytochrome signals in Arabidopsis. Plant Cell. 2011;23(9):3230–3246. doi: 10.1105/tpc.111.088195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dixon LE, et al. Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr Biol. 2011;21(2):120–125. doi: 10.1016/j.cub.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doyle MR, et al. The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature. 2002;419(6902):74–77. doi: 10.1038/nature00954. [DOI] [PubMed] [Google Scholar]
- 40.Plautz JD, et al. Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms. 1997;12(3):204–217. doi: 10.1177/074873049701200302. [DOI] [PubMed] [Google Scholar]
- 41.Izumo M, Sato TR, Straume M, Johnson CH. Quantitative analyses of circadian gene expression in mammalian cell cultures. PLOS Comput Biol. 2006;2(10):e136. doi: 10.1371/journal.pcbi.0020136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosso MG, et al. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol. 2003;53(1-2):247–259. doi: 10.1023/B:PLAN.0000009297.37235.4a. [DOI] [PubMed] [Google Scholar]
- 43.Orr HA. The population genetics of speciation: The evolution of hybrid incompatibilities. Genetics. 1995;139(4):1805–1813. doi: 10.1093/genetics/139.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sureshkumar S, et al. A genetic defect caused by a triplet repeat expansion in Arabidopsis thaliana. Science. 2009;323(5917):1060–1063. doi: 10.1126/science.1164014. [DOI] [PubMed] [Google Scholar]
- 45.Legendre M, Pochet N, Pak T, Verstrepen KJ. Sequence-based estimation of minisatellite and microsatellite repeat variability. Genome Res. 2007;17(12):1787–1796. doi: 10.1101/gr.6554007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ossowski S, et al. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science. 2010;327(5961):92–94. doi: 10.1126/science.1180677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duennwald ML, Jagadish S, Muchowski PJ, Lindquist S. Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc Natl Acad Sci USA. 2006;103(29):11045–11050. doi: 10.1073/pnas.0604547103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kratter IH, Finkbeiner S. PolyQ disease: Too many Qs, too much function? Neuron. 2010;67(6):897–899. doi: 10.1016/j.neuron.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.