Abstract
Toll-like receptor (TLR) signaling is one of the most important signaling cascades of the innate immune system of vertebrates. Studies in invertebrates have focused on the fruit fly Drosophila melanogaster and the nematode Caenorhabditis elegans, and there is little information regarding the evolutionary origin and ancestral function of TLR signaling. In Drosophila, members of the Toll-like receptor family are involved in both embryonic development and innate immunity. In C. elegans, a clear immune function of the TLR homolog TOL-1 is controversial and central components of vertebrate TLR signaling including the key adapter protein myeloid differentiation primary response gene 88 (MyD88) and the transcription factor NF-κB are not present. In basal metazoans such as the cnidarians Hydra magnipapillata and Nematostella vectensis, all components of the vertebrate TLR signaling cascade are present, but their role in immunity is unknown. Here, we use a MyD88 loss-of-function approach in Hydra to demonstrate that recognition of bacteria is an ancestral function of TLR signaling and that this process contributes to both host-mediated recolonization by commensal bacteria as well as to defense against bacterial pathogens.
Toll-like receptors (TLRs) are conserved throughout animal evolution but appear to serve different functions in different model organisms. TLRs are transmembrane receptors with extracellular leucin-rich repeat (LRR) motifs and an intracellular Toll/interleukin-1 receptor (TIR) domain. Upon stimulation of TLRs, the key adapter protein MyD88 associates with the cytosolic part of the TLR through a homophilic interaction of the TIR domains and then recruits the IL-1R–associated kinase (IRAK), which subsequently associates with the TNFR-associated factor (TRAF). TRAF recruits the TGF-β activated kinase 1 (TAK1). The kinase TAK1 induces a phosphorylation cascade finally leading to the nuclear translocation of the transcription factors NF-κB via the inhibitor of kappa B-kinase (IKK) signalosome or c-Jun via the c-Jun N-terminal kinase (JNK)/p38 branch of TLR signaling (1).
The Toll pathway was initially identified to be essential in early embryonic development in Drosophila (2). In addition to its crucial role in the establishment of the dorsal–ventral axis, Drosophila Toll was shown to be involved in muscle development (3) and heart formation (4). Later on, it was discovered that Toll signaling in Drosophila also contributes to defense reactions against bacteria as well as to antifungal defense by regulating, among others, the expression of the antifungal peptide drosomycin in adult flies (5, 6). Further immunity functions have been identified for Toll-7 (7) and Toll-8 (8). Studies in the mosquito Aedes aegypti also identified MyD88-dependent Toll signaling to mediate immune defenses against dengue viruses (9). One other invertebrate model system, the nematode Caenorhabditis elegans lacks central proteins of the canonical TLR-signaling cascade (10). Only one Toll homolog, termed TOL-1, was identified in C. elegans (10). The fact that TOL-1 mutants show strong developmental defects despite mutants for the putative signaling cascade displaying no developmental abnormalities led to the belief that TOL-1 in C. elegans might function as a cell–cell adhesion protein in neurons (10). Other reports state an additional involvement of TOL-1 in pathogen defense (11). In addition, a TIR-domain–containing protein called tir-1 is required for resistance to fungal and bacterial infection in C. elegans (12, 13).
Vertebrate homologs of Toll, the TLRs, are receptors of the immune system. Vertebrate TLRs are involved in eliminating pathogens and controlling commensal colonization (1, 14, 15) by recognizing conserved microbe-associated molecular patterns (MAMPs) including lipopolysaccharides, flagellin, and peptidoglycans (1, 16). Therefore, it was proposed that immune function of TLR signaling involving NF-κB and MyD88 has evolved within the bilaterians (17). However, recent genome projects in the nonbilaterians Hydra magnipapillata (18) and Nematostella vectensis (19) revealed the presence of TLRs, MyD88, and NF-κB (20, 21). Their role in bacterial recognition and innate immunity, however, remains to be shown (22). Cnidaria are a sister group to the Bilateria (19) and one of the earliest branches in the animal tree of life (Fig. 1A). The recent genome project of the cnidarian H. magnipapillata identified a conserved TLR-signaling cascade (21, 23) (Fig. 1B and Table S1), making Hydra a suitable model for addressing questions of the ancestral function of TLR signaling. Is the TLR pathway involved in the defense against bacterial pathogens or in maintaining specific host–microbe interactions? Does it affect the mechanisms and routes by which functionally diverse bacteria colonize their host? Is it involved in developmental processes such as axis formation? To gain insight into these questions, we performed MyD88 loss-of-function experiments in Hydra vulgaris [AEP strain (24)]. We used a combination of microarray-based gene expression screening and 16S rRNA-gene sequencing to detect changes in both the Hydra transcriptome and the associated microbiota. Further, we investigated the role of TLR signaling in pathogen defense against Pseudomonas aeruginosa. The patterns of differentially regulated host genes as well as changes in the bacterial colonization process and pathogen susceptibility in MyD88-knockdown polyps point to a role of TLR signaling in sensing of bacteria, be it associated commensals or pathogens. Thus, this functional analysis clearly identifies a role of TLR signaling in innate immunity in an animal at the base of metazoan evolution.
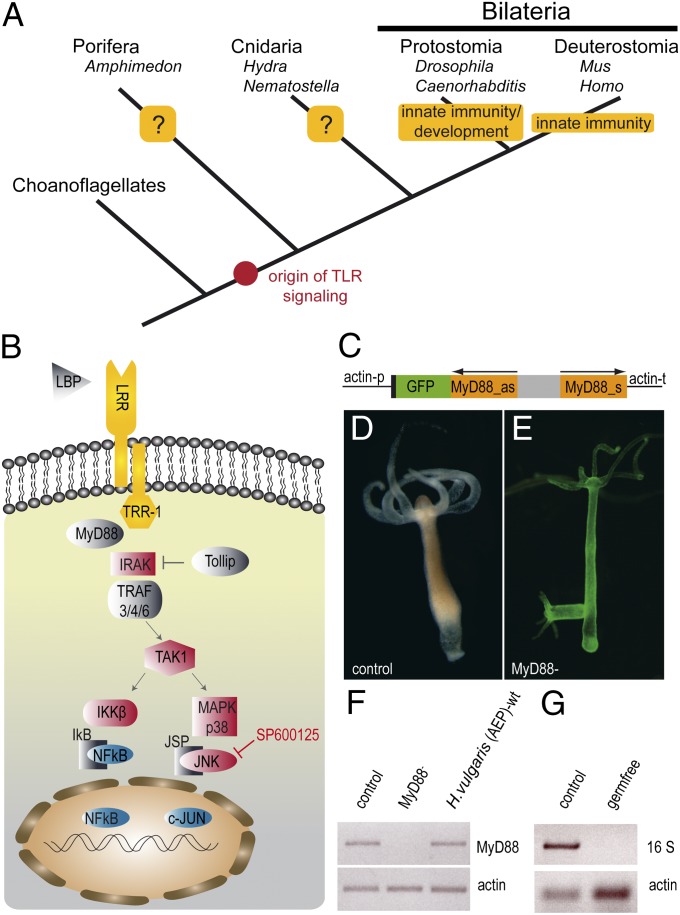
Fig. 1.
Interference with the conserved Hydra TLR-signaling pathway. (A) Function of TLR signaling during metazoan evolution. Cnidaria are the sistergroup to all bilateria and they diverged from the common eumetazoan ancestor ∼600–700 million years ago (19). (B) Schematic representation of the conserved TLR-signaling pathway in Hydra. Note that the functional Hydra TLR is assembled by two proteins (HyLRR and HyTRR) (23) and the exogenous JNK inhibitor SP600125 is shown. (C) MyD88–Hairpin construct for generation of transgenic Hydra (p, promoter; T, terminator, as, antisense; s, sense). (D) Live image of a MyD88-control polyp (control). (E) Live image of a MyD88-knockdown polyp (MyD88−) showing EGFP expression in the endodermal and the ectodermal cell lineage. (F) RT-PCR amplifying myd88 shows down-regulation in MyD88− polyps compared with control polyps and wild-type (WT) H. vulgaris (AEP). RT-PCR was normalized using the Hydra actin gene. (G) Absence of bacteria after antibiotic treatment was confirmed by PCR amplification of the bacterial 16S rRNA gene on genomic DNA normalized to Hydra actin.
Results
Generation of MyD88-Knockdown (MyD88−) H. vulgaris (AEP).
To analyze the function of TLR signaling in the basal metazoan Hydra, we generated a stable transgenic H. vulgaris (AEP) line with drastically reduced expression levels of the universal adapter protein MyD88 using a hairpin cassette containing the myd88 antisense and sense sequences fused to the reporter gene egfp (Fig. 1C). The transformation of two- to four-cell–stage embryos via microinjection resulted in the founder polyp MyD88-D3. This polyp showed mosaic distribution of GFP+ cells, which could be enriched or depleted in clones generated by budding, depending on where the bud formed. By several rounds of asexual proliferation, two stable lines were established. We termed them the MyD88-control (control) line, which contained no remaining GFP+ cells (Fig. 1D), and the MyD88-knockdown (MyD88−) line, which expressed the transgene in the endodermal and ectodermal cell lines (Fig. 1E). The resulting double-stranded (ds)RNA triggers the RNAi machinery (25–28), which leads to a decrease in the endogenous MyD88 transcript as shown by RT-PCR (Fig. 1F). Because both lines were generated from the same founder polyp by asexual reproduction, we were able to analyze the effects of a drastically decreased MyD88-expression level without the need to account for differences in genomic background. Neither line displayed any obvious developmental or behavioral abnormalities.
Absence of Bacteria as Well as MyD88 Deficiency Influence Central Parts of the TLR-Signaling Cascade.
To assess the transcriptional consequences of a MyD88 knockdown and identify potential downstream effector genes of the TLR-signaling cascade, we performed microarray analyses. Expression levels of both MyD88− polyps as well as germ-free polyps (Fig. 1G) were compared with control polyps. The MyD88− polyps combined with the germ-free polyps provided unique resources that allowed us to directly investigate the connection between TLR signaling and the regulation of associated bacterial diversity. Statistical analysis was carried out by ANOVA with Student–Newman–Keuls (SNK) post hoc tests and false discovery rate (FDR) correction. The microarray data independently validate the successful MyD88 knockdown. Contig 11552, encoding for MyD88, shows a 4.29-fold down-regulation (P < 0.001) in MyD88− polyps and is not differentially expressed in germ-free polyps (fold change 1.09, N.S.) (Table S1). To check for transcriptional changes of other putative members of the TLR cascade, the H. vulgaris (AEP) transcriptome (29) was screened for homologs of previously described members of the pathway. The majority of central cascade members are present in H. vulgaris (AEP) (Fig. 1A and Table S1). Various central components of the putative TLR cascade including members of the TRAF family of ubiquitin protein ligases, the kinase TAK1, MAP-kinase p38, and the JNK inhibitor JSP-1 show significantly decreased expression in germ-free and/or MyD88-deficient conditions (Table S1). We hypothesize therefore the existence of positive feedback loops of the putative effector transcription factors NF-κB and c-Jun on certain upstream pathway components, pointing toward a functional unity of these proteins in the bacterial sensing process in vivo.
Gene-Expression Profiling of MyD88-Knockdown and Germ-Free Polyps.
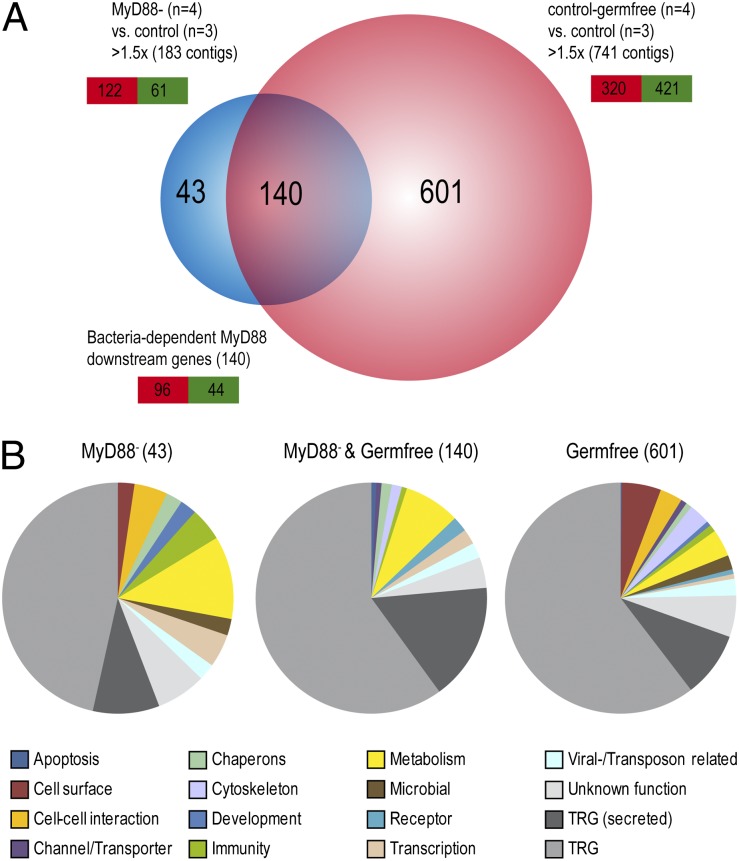
After identifying the conserved components of the TLR pathway in Hydra, we aimed to identify downstream target genes in an analysis not based on gene homologies. Therefore, only contigs exceeding a significant (P ≤ 0.05) fold-change threshold of 1.5 in at least one of the comparisons were considered for further analyses. This resulted in 183 differentially regulated (122 down-regulated, 61 up-regulated) contigs in MyD88− polyps (0.5% of total contigs) and in 741 differentially regulated contigs (320 down-regulated, 421 up-regulated) in germ-free polyps (2.0% of total contigs) (Fig. 2A). Interestingly, the signature of MyD88− polyps overlapped substantially (76.5%) with the germ-free signature (Fig. 2A). The overlapping signature of 140 contigs included a large proportion (>75%) of taxonomical restricted genes (TRGs), i.e., contigs of unknown function with no homolog detected in other species (Fig. 2B). A total of 21% of these TRGs lack a transmembrane domain but have a predicted signal peptide. This is notable because such secreted peptides might directly interact with associated bacteria. Annotated transcripts included metabolic genes such as carbonic anhydrases, protein-modifying enzymes like kinases and ubiquitinases, receptors, chaperones, viral/transposon-related genes such as transposases and reverse transcriptases, as well as transcription factors. Table 1 shows the fold changes of six representative genes. Additionally, we included three significantly (P < 0.05) differentially regulated genes of the germ-free vs. control comparison that had a significant fold change below the 1.5× threshold in the MyD88− samples (Table 1). These genes include previously described TLR-target genes such as a lectin, bcl-2, and alkaline phosphatase (30–32). The differential expression of these representative target genes was validated by qRT-PCR (Fig. S1). All analyzed contigs show a higher fold change in the absence of bacteria than in MyD88-deficient conditions (Table 1). The fact that expression of more than 75% of the MyD88-responsive transcripts is also altered in germ-free polyps (Fig. 2A) suggests that these MyD88-downstream genes are bacteria responsive.
Fig. 2.
Microarray analysis reveals differential gene expression due to MyD88 down-regulation and the absence of the associated microbiota. (A) Graphic representation of differentially regulated (≥1.5-fold change, P ≤ 0.05) contigs in MyD88− and germ-free compared with control polyps. Note the overlap between both experiments. Down-regulated contigs are highlighted in red, up-regulated contigs in green. (B) Categorization of differential contigs. Pie charts were separated in MyD88- but not bacterial-regulated contigs (Left), MyD88- as well as bacterial-regulated contigs (Center), and MyD88-independent bacterial-regulated contigs (Right). Contigs were assigned into self-chosen categories.
Table 1.
Differential expression of representative genes in MyD88− and germ-free polyps
| Annotation | MyD88− |
Germ-free |
|||
| Contig* | Fold change | P value | Fold change | P value | |
| Secreted peptide | 732 | 0.66 | 0.002 | 0.44 | 0.001 |
| Secreted EGF | 12837 | 0.57 | 0.006 | 0.50 | 0.005 |
| 24241 | 0.57 | 0.004 | 0.49 | 0.007 | |
| Secreted protein | 16151 | 0.63 | 0.000 | 0.54 | 0.004 |
| T-box | 19777 | 0.64 | 0.000 | 0.45 | 0.001 |
| Cadherin | 34924 | 0.54 | 0.001 | 0.30 | 0.001 |
| 14903 | 0.59 | 0.009 | 0.33 | 0.002 | |
| Secreted protein | 43476 | 0.62 | 0.026 | 0.40 | 0.002 |
| Gal-lectin | 1372 | 0.73 | 0.005 | 0.39 | 0.004 |
| bcl-2 | 7659 | 0.72 | 0.015 | 0.56 | 0.005 |
| Alkaline phosphatase | 45829 | 0.70 | 0.000 | 0.49 | 0.002 |
*Contigs are available at http://compagen.zoologie.uni-kiel.de/.
Notably, MyD88-dependent transcripts were not enriched for genes with known developmental functions (Fig. 2B). However, a developmental role of certain TRGs cannot be excluded. In comparison, the 601 transcripts that are regulated by the presence of the bacterial microbiota in a MyD88-independent way include a noticeable proportion of genes involved in cell-surface components and cell–cell interaction like mucins, lectins, and cadherins. Because the glycocalyx of epithelial cells is a habitat for Hydra-associated bacteria, it is possible that these colonizers induce changes in their own Hydra-cell–associated environment.
Subset of MyD88-Downstream Genes Is Regulated by JNK.
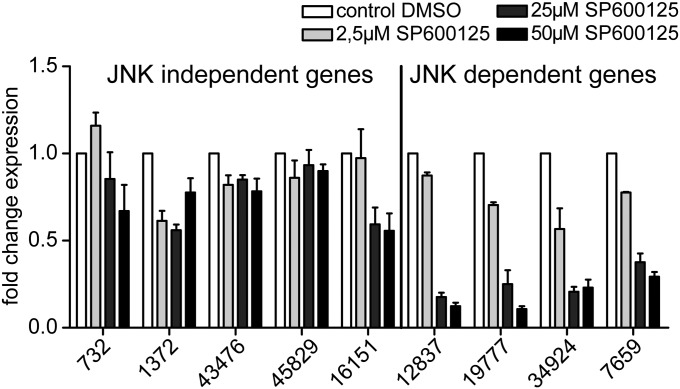
Upon stimulation by MAMPs, the TLR-signaling cascade in vertebrates can regulate the expression of target genes via two major downstream branches, leading to the nuclear translocation of the transcription factor NF-κB or c-Jun (Fig. 1A) (1). In Hydra, the expression levels of putative members of the JNK/p38 branch of TLR signaling leading to c-Jun translocation (TAK1, p38α, p38β, and JSP-1) are significantly affected by both the absence of bacteria as well as by silencing the MyD88 expression (Table S1). To directly analyze the role of JNK in TLR signaling, we examined the expression of the nine representative contigs (Table 1) in the presence of the JNK-specific inhibitor SP600125 (33, 34), which was shown to act specifically in the cnidarian Hydra (34). As shown in Fig. 3, four of nine candidate genes were negatively regulated by SP600125 in a concentration-dependent manner. This includes bcl-2 (contig 7659), a known NF-κB target gene in vertebrates (31). Thus, bcl-2 might be transcriptionally regulated by both NF-κB and c-Jun transcription factors. To exclude an unspecific, broad effect of the inhibitor SP600125, we conducted qRT-PCRs with a set of nine random genes. None of these genes were affected by SP600125 treatment (Fig. S2). Our results point toward a key role of MAP-kinase signaling in the MyD88-mediated perception of bacterial signals via TLRs in Hydra.
Fig. 3.
JNK phosphorylation is mediating the expression of several MyD88-downstream genes. Relative expression level of the candidate genes upon administration of the JNK inhibitor SP600125 (34), determined by qRT-PCR. Note that the expression of 12837, 19777, 34924, and 7659 is influenced by SP600125 in a concentration-dependent manner. cDNA amounts were equilibrated by elongation factor 1 α. The graphic shows means + SD (n = 3).
MyD88-Deficient Hydra Display Delayed Bacterial Recolonization upon Antibiotic Treatment.
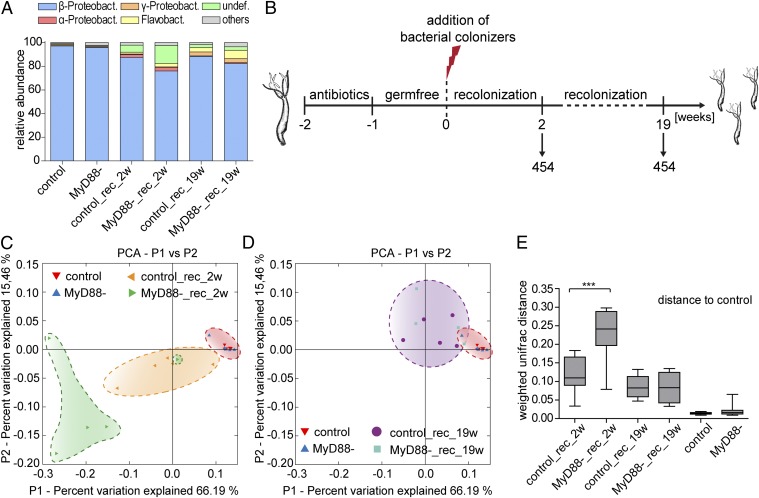
Next we examined whether a MyD88 knockdown affects the colonizing microbiota. We performed 454 pyrosequencing of the variable region 2 (V2) of the bacterial 16S rRNA gene, amplified from total DNA of control and MyD88− polyps after a 4-wk period of cocultivation. We found the microbiota of both control and MyD88− polyps to be dominated by β-proteobacteria (Fig. 4A and Fig. S3), mainly of the genus Curvibacter. On average, Curvibacter sp. accounts for 94% of the microbiota in control polyps and 92% in MyD88− polyps (Fig. 4A). This bacterium was previously cosequenced with the genome of H. magnipapillata (18). Rare fractions of the Hydra-associated bacterial community include α-proteobacteria, γ-proteobacteria, and flavobacteria (Fig. 4A).
Fig. 4.
454 sequencing of bacterial 16S rRNA reveals minor impact of MyD88 in bacterial colonization of Hydra polyps. (A) Bar charts representing bacterial communities of Hydra polyps on class level (means of five replicates). Rare bacterial taxa (<1% relative abundance) were grouped to the fraction “others.” (B) Experimental design. Germ-free MyD88− and control polyps were inoculated with potential bacterial colonizers derived from pond water, Hydra-culture supernatant, and Hydra-tissue homogenates. Single polyps were removed from clonally growing cultures 2 wk and 19 wk postinoculation and subjected to 454 sequencing of the microbiota. (C and D) Bacterial communities clustered using principle coordinate analysis of the weighted UniFrac distance matrix. Percent variation explained by the principle coordinates is indicated in the axes. (E) Weighted UniFrac differences calculated by pairwise comparisons of the bacterial profile to control polyps. Statistical analysis was carried out using two-tailed t test (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001).
To investigate whether TLR signaling plays a role in the microbiota assembly process, we generated germ-free control and germ-free MyD88− polyps by antibiotic treatment and reinfected them with a complex pool of potential colonizing bacteria retrieved from the supernatant of a nongerm-free culture, pond water, and Hydra tissue homogenates (Fig. 4B). Individual polyps were examined for their microbiota by 454 pyrosequencing 2 wk and 19 wk postbacterial inoculation. Principle coordinate analysis (PCoA) of the weighted UniFrac metric revealed that after 2 wk, bacterial communities of MyD88− polyps (MyD88−_rec_2w) display a relatively high separation from the preantibiotic control state, whereas control samples (control_rec_2w) cluster in close proximity to the preantibiotic state (red cloud) (Fig. 4 C and E). After 19 wk of recolonization (Fig. 4 D and E), this initial difference between the bacterial communities of control and MyD88− polyps disappeared. Taken together, these results demonstrate a role of MyD88-mediated TLR signaling in the reestablishment of Hydra-associated bacterial communities by promoting bacterial recolonization. However, MyD88 does not appear to be essential for maintaining bacterial homeostasis in adult polyps of H. vulgaris (AEP).
TLR Signaling in Pathogen Defense in Hydra.
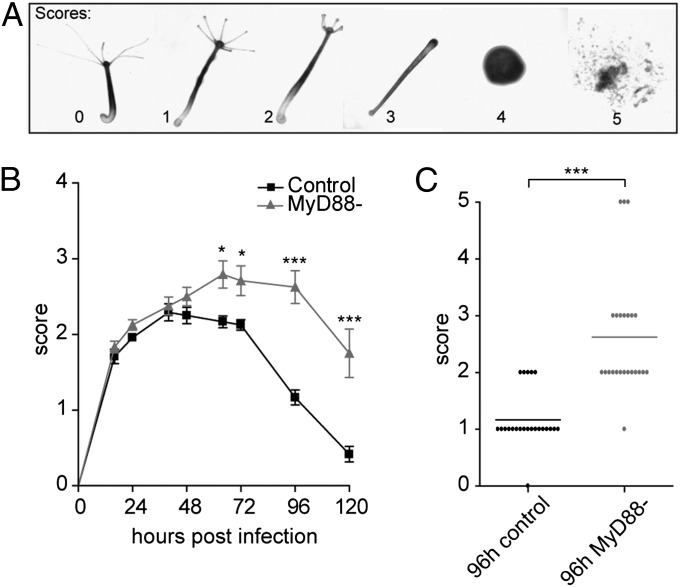
P. aeruginosa is a pathogenic bacteria that can infect a broad range of host organisms including plants, nematodes, insects, and mice (35). To examine whether silencing of MyD88 affects pathogen defense in Hydra, we used the isolate of P. aeruginosa, strain UCBPP-PA14 (PA14; http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/www.tax.cgi?id=208963) (36) to infect MyD88− and control polyps. Single polyps (n = 24 each) were cultured in medium containing 1.8 × 108 PA14 cells per milliliter. Polyps were screened daily and the impact of the pathogenic bacterium was scored according to the disease phenotypes presented in Fig. 5A. Twenty-four hours postinfection, MyD88− as well as control polyps suffered from severe tentacle shortening (mean score ∼2) (Fig. 5B). Forty-eight hours postinfection, the disease state was characterized by a complete loss of tentacles in a subset of polyps both in control (7/24 polyps with score 3) and MyD88− (10/24). Whereas the control polyps started to recover, disease severity increased in MyD88− polyps in the following 2 d (Fig. 5B), culminating in the complete lysis of 3 MyD88− polyps after 96 h (Fig. 5C). Taken together, the PA14 infection assay shows a significantly higher pathogen susceptibility of MyD88− polyps compared with control polyps.
Fig. 5.
MyD88− and control polyps show differential susceptibility to infection by P. aeruginosa (P.a.) (A) Phenotypic scores of the Hydra infection model. Disease always starts with swelling of the tentacle tips (score 1), followed by subsequent shortening (score 2), and loss (score 3) of tentacles. Score 4 indicates the loss of body shape with maintenance of an intact epithelium. Score 5 is characterized by tissue lysis. (B) Temporal profile of P.a.14 infection in Hydra. Polyps were incubated in 1 mL Hydra medium containing 1.8 × 108 cfu P.a.14. Values are plotted as mean + SEM (n = 24). (C) Detailed representation of the time point 96 h postinfection from B. Each dot represents one polyp; horizontal line shows the mean. Note that three polyps died in the MyD88-knockdown group, whereas the maximum score observed in the control group was 2 (tentacle shortening). Statistical significance was tested by two-tailed Mann–Whitney test (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001).
Discussion
Bacterial Sensing Is an Ancient Function of TLR Signaling.
In vertebrates, most TLRs act as pattern recognition receptors (PRRs) and are involved in the elimination of pathogens and controlling commensal colonization (1, 14, 15). In contrast, the role of Drosophila Toll receptors is less uniform. Their temporal and spatial expression patterns during embryonic development point toward roles in developmental processes by either inducing signaling cascades or acting as cell-adhesion molecules (37). However, a role in development is clearly proven only for Toll (2). In addition, Toll (5, 6) as well as Toll-7 (7) and Toll-8 (8) act in immune defense against fungi, bacteria, or viruses. This functional duality prompted us to search for the ancestral role of TLR signaling in a more basal metazoan (22).
The cnidarian Hydra possesses a bona fide TLR-signaling cascade (Fig. 1A) (21), making it a suitable model for investigating the ancestral function of the TLR pathway. Together with previous results showing that the HyLRR-2 receptor in Hydra is able to bind flagellin in vitro (23), the in vivo results of this study suggest a role for TLR signaling in bacterial recognition in the cnidarian Hydra. Notably, MyD88 loss of function, as well as the absence of commensal bacteria in germ-free polyps, causes significant and overlapping changes in the transcriptome (Fig. 2). A subset of differential genes is regulated by the JNK/p38 branch of TLR signaling (Fig. 3). This shows that TLR signaling in Hydra does not act unidirectionally via the transcription factor NF-κB but is also linked to MAP kinases. Notably, a p38 MAPK cascade was identified as a key component of the C. elegans immune response and is also present in Monosiga brevicollis, a single-cell eukaryote that is most closely related to metazoans (22). Because JNK and p38 in Hydra are also activated by many other stimuli, including G-protein–coupled receptors (38) and the wnt pathway (34), innate immune reactions appear to be controlled by a complex signal transduction network.
MyD88 Promotes Reestablishment of Bacterial Homeostasis After Disturbance.
Many studies show that microbes have direct beneficial effects on the host (39–42). This is supported by the fact that a dysregulation of host–bacteria homeostasis seems to be involved in the occurrence of disorders, like chronic inflammatory bowel diseases (43, 44). Hydra polyps are colonized by species-specific bacterial communities (45), which are largely determined by the host rather than by the environment (45, 46). After verifying that TLR signaling in Hydra is clearly involved in the recognition of bacteria, we therefore asked whether this active cross-talk is involved in the maintenance of host–bacterial homeostasis. Our results show no impact of impaired TLR signaling on the composition of the bacterial microbiota in healthy animals. However, after active disturbance of the bacterial community by antibiotic treatment or upon bacterial infection with P. aeruginosa, MyD88-mediated TLR signaling promotes a reestablishment of bacterial homeostasis. This is concordant with several mechanisms to restrict active immune signaling to disturbance, such as pathogen defense while being hyporesponsive to the healthy commensal microbiota (47). These mechanisms include (i) a decreased apical surface expression of TLRs, (ii) spatial segregation of host cells and commensal bacteria by mucus layers to limit detection of invasive bacteria that crossed the epithelial barrier, and (iii) active bacterial suppression of host immunity, e.g., via the induction of the TLR-signaling suppressor Tollip (Fig. 1B) by commensal bacteria (47, 48).
MyD88-Dependent Target Genes Include Taxonomically Restricted and Conserved Genes.
Interestingly, around 75% of differential contigs could not be assigned to functional categories due to the lack of BLAST hits (Fig. 2). This fraction of TRGs (49) is largely overrepresented compared with the overall fraction of TRGs in the whole transcriptome. This large number might indicate that the TLR-dependent response toward commensal bacteria is by and large taxon specific. Furthermore, 21% of these TRGs possess a predicted signal peptide. Because these secreted peptides may contribute to the properties of Hydra’s epithelial environment, they might also affect the colonizing microbiota. We have previously shown (50) that antimicrobial peptides, apart from their role in defense against pathogenic bacteria, also have regulatory functions in host–bacterial homeostasis. This adds support to our previously proposed hypothesis (49) that taxonomically restricted host defense molecules facilitate the disarming of taxon-specific microbial attackers, and at the same time shape the colonizing microbiota. Nevertheless, a small proportion of MyD88-target genes is conserved throughout the animal kingdom. These genes include previously described vertebrate MyD88-dependent NFκB-target genes such as lectins or bcl-2 (30, 31). In addition, alkaline phosphatase in zebrafish also responds to LPS through a mechanism that involves MyD88. It is required to detoxify LPS and prevent intestinal inflammation in response to the resident microbiota (32). We propose that the differentially expressed alkaline phosphatase in Hydra (Table 1) plays a similar role.
Conclusion
Based on our results we propose that the TLR/MyD88 pathway is an ancestral immune signaling pathway predating the evolution of TLR-dependent immune signaling pathways at the origin of metazoan evolution. Recognizing and managing the bacterial communities typically present at epithelial surfaces throughout the animal kingdom likely contributed to its evolution and maintenance.
Materials and Methods
Experimental details are provided in SI Materials and Methods.
Animal Culture.
Experiments were carried out using H. vulgaris (AEP). All animals were cultured under constant, identical environmental conditions including culture medium, food, and temperature according to standard procedures. All animal experiments were conducted in accordance with the German law concerning animal experimentation.
Generation of Transgenic H. vulgaris (AEP) Expressing an EGFP–MyD88–Hairpin Fusion Construct in Endodermal and Ectodermal Cells.
For generation of H. vulgaris (AEP) egfp:myd88–hairpin transgenics, a hairpin construct was designed. By selecting for EGFP expression, polyps with full endodermal and ectodermal expression of EGFP (MyD88 knockdown) were generated.
Generation of Germ-Free Hydra.
Polyps were incubated for 1 wk in an antibiotic solution containing 50 μg/mL each of ampicillin, rifampicin, streptomycin, and neomycin with daily exchange of the solution.
Custom-Made H. vulgaris (AEP) Microarray.
The microarray is based on a full transcriptome of H. vulgaris (AEP) sequenced by 454 technology (29). This results in a microarray platform having 45,220 oligos of 60 nucleotides in length, resembling 37,063 unique contigs.
RNA Isolation and Microarray Gene Expression Experiments.
Total RNA was isolated from 15 polyps using the TRIzol Plus protocol (Invitrogen). Labeled cDNAs were hybridized to custom-made Agilent Hydra (AEP) gene expression microarray slides (4 × 44K).
Microarray Data Extraction, Filtering, and Analysis.
Raw microarray image files were processed and quality checked by Agilent’s Feature Extraction 10.7 Image Analysis software. Statistical analysis was conducted by ANOVA with SNK post hoc test and FDR correction for multiple comparisons.
qRT-PCR.
qRT-PCR was conducted in biological triplicates (n = 3), using the GoTaq qPCR Master Mix (Promega) and a 7300 Real-Time PCR system (ABI).
SP600125 JNK Inhibitor Treatment.
For the treatment with SP600125 (A.G. Scientific), polyps (25 each) were incubated at a density of 1 polyp per milliliter in SP600125 diluted in 0.1% DMSO/Hydra medium in the dark for 24 h at 18 °C (34).
DNA Extraction and Sequencing of 16S rRNA Genes.
For total DNA extraction, single polyps were subjected to the DNeasy Blood and Tissue kit (Qiagen). For sequencing of the bacterial 16s rRNA genes, the variable region 2 (V2) was amplified using the universal forward primer V2_B_Pyro_27F and the barcoded reverse primer V2_A_338R. A sample of each library was run on an Agilent Bioanalyzer before emulsion PCR and sequencing as recommended by Roche.
16S rRNA 454 Analysis.
16S rRNA amplicon sequence analysis was conducted using the Qiime 1.3 package (51). Fig. S4 shows the alpha-diversity curves for all samples ensuring sufficient sequencing depth.
P. aeruginosa Infection.
Experiments were carried out using the P. aeruginosa strain PA14. Single MyD88-knockdown and -control polyps (n = 24 each) were incubated in single wells of 24-well plates and incubated in 1 mL of the PA14 solution with an optical density (OD600) of 0.1 containing 1.8 × 108 cells per milliliter. Polyps were screened daily and scored by following the criteria shown in Fig. 5.
Supplementary Material
Acknowledgments
We thank Doris Willoweit-Ohl and Jörg Wittlieb for their expert technical assistance, Elke Blohm-Sievers and Diethard Tautz for enabling microarray analysis in Hydra, Heinke Buhtz for her technical support in the 454 sequencing process, Gunther Jansen and Hinrich Schulenburg for providing the Pseudomonas aeruginosa PA14 strain and for their support in the infection experiments. This work was funded by the Deutsche Forschungsgemeinschaft (DFG) through Grant Bo 848/17-1 and by grants from the DFG Cluster of Excellence programs “The Future Ocean” and “Inflammation at Interfaces.”
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE32383); and the 16S 454 data are deposited at the metagenome analysis server MG-RAST (Project ID 1719).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213110109/-/DCSupplemental.
References
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124(4):783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Anderson KV, Jürgens G, Nüsslein-Volhard C. Establishment of dorsal-ventral polarity in the Drosophila embryo: Genetic studies on the role of the Toll gene product. Cell. 1985;42(3):779–789. doi: 10.1016/0092-8674(85)90274-0. [DOI] [PubMed] [Google Scholar]
- 3.Halfon MS, Keshishian H. The Toll pathway is required in the epidermis for muscle development in the Drosophila embryo. Dev Biol. 1998;199(1):164–174. doi: 10.1006/dbio.1998.8915. [DOI] [PubMed] [Google Scholar]
- 4.Wang J, et al. Expression, regulation, and requirement of the toll transmembrane protein during dorsal vessel formation in Drosophila melanogaster. Mol Cell Biol. 2005;25(10):4200–4210. doi: 10.1128/MCB.25.10.4200-4210.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86(6):973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 6.Rosetto M, Engström Y, Baldari CT, Telford JL, Hultmark D. Signals from the IL-1 receptor homolog, Toll, can activate an immune response in a Drosophila hemocyte cell line. Biochem Biophys Res Commun. 1995;209(1):111–116. doi: 10.1006/bbrc.1995.1477. [DOI] [PubMed] [Google Scholar]
- 7.Nakamoto M, et al. Virus recognition by Toll-7 activates antiviral autophagy in Drosophila. Immunity. 2012;36(4):658–667. doi: 10.1016/j.immuni.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akhouayri I, Turc C, Royet J, Charroux B. Toll-8/Tollo negatively regulates antimicrobial response in the Drosophila respiratory epithelium. PLoS Pathog. 2011;7(10):e1002319. doi: 10.1371/journal.ppat.1002319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xi Z, Ramirez JL, Dimopoulos G. The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog. 2008;4(7):e1000098. doi: 10.1371/journal.ppat.1000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pujol N, et al. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr Biol. 2001;11(11):809–821. doi: 10.1016/s0960-9822(01)00241-x. [DOI] [PubMed] [Google Scholar]
- 11.Tenor JL, Aballay A. A conserved Toll-like receptor is required for Caenorhabditis elegans innate immunity. EMBO Rep. 2008;9(1):103–109. doi: 10.1038/sj.embor.7401104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Couillault C, et al. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat Immunol. 2004;5(5):488–494. doi: 10.1038/ni1060. [DOI] [PubMed] [Google Scholar]
- 13.Liberati NT, et al. Requirement for a conserved Toll/interleukin-1 resistance domain protein in the Caenorhabditis elegans immune response. Proc Natl Acad Sci USA. 2004;101(17):6593–6598. doi: 10.1073/pnas.0308625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wen L, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455(7216):1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Round JL, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332(6032):974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pasare C, Medzhitov R. Toll-like receptors: Linking innate and adaptive immunity. Adv Exp Med Biol. 2005;560:11–18. doi: 10.1007/0-387-24180-9_2. [DOI] [PubMed] [Google Scholar]
- 17.Kim DH, Ausubel FM. Evolutionary perspectives on innate immunity from the study of Caenorhabditis elegans. Curr Opin Immunol. 2005;17(1):4–10. doi: 10.1016/j.coi.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 18.Chapman JA, et al. The dynamic genome of Hydra. Nature. 2010;464(7288):592–596. doi: 10.1038/nature08830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Putnam NH, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317(5834):86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 20.Lange C, et al. Defining the origins of the NOD-like receptor system at the base of animal evolution. Mol Biol Evol. 2011;28(5):1687–1702. doi: 10.1093/molbev/msq349. [DOI] [PubMed] [Google Scholar]
- 21.Miller DJ, et al. The innate immune repertoire in cnidaria—ancestral complexity and stochastic gene loss. Genome Biol. 2007;8(4):R59. doi: 10.1186/gb-2007-8-4-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irazoqui JE, Urbach JM, Ausubel FM. Evolution of host innate defence: Insights from Caenorhabditis elegans and primitive invertebrates. Nat Rev Immunol. 2010;10(1):47–58. doi: 10.1038/nri2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bosch TC, et al. Uncovering the evolutionary history of innate immunity: The simple metazoan Hydra uses epithelial cells for host defence. Dev Comp Immunol. 2009;33(4):559–569. doi: 10.1016/j.dci.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 24.Wittlieb J, Khalturin K, Lohmann JU, Anton-Erxleben F, Bosch TC. Transgenic Hydra allow in vivo tracking of individual stem cells during morphogenesis. Proc Natl Acad Sci USA. 2006;103(16):6208–6211. doi: 10.1073/pnas.0510163103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 26.McManus MT, Sharp PA. Gene silencing in mammals by small interfering RNAs. Nat Rev Genet. 2002;3(10):737–747. doi: 10.1038/nrg908. [DOI] [PubMed] [Google Scholar]
- 27.Zamore PD. Ancient pathways programmed by small RNAs. Science. 2002;296(5571):1265–1269. doi: 10.1126/science.1072457. [DOI] [PubMed] [Google Scholar]
- 28.Kennerdell JR, Carthew RW. Heritable gene silencing in Drosophila using double-stranded RNA. Nat Biotechnol. 2000;18(8):896–898. doi: 10.1038/78531. [DOI] [PubMed] [Google Scholar]
- 29.Hemmrich G, et al. Molecular signatures of the three stem cell lineages in Hydra and the emergence of stem cell function at the base of multicellularity. Mol Biol Evol. 2012 doi: 10.1093/molbev/mss134. 10.1093/molbev/mss13. [DOI] [PubMed] [Google Scholar]
- 30.Hsu DK, Hammes SR, Kuwabara I, Greene WC, Liu FT. Human T lymphotropic virus-I infection of human T lymphocytes induces expression of the beta-galactoside-binding lectin, galectin-3. Am J Pathol. 1996;148(5):1661–1670. [PMC free article] [PubMed] [Google Scholar]
- 31.Catz SD, Johnson JL. Transcriptional regulation of bcl-2 by nuclear factor kappa B and its significance in prostate cancer. Oncogene. 2001;20(50):7342–7351. doi: 10.1038/sj.onc.1204926. [DOI] [PubMed] [Google Scholar]
- 32.Bates JM, Akerlund J, Mittge E, Guillemin K. Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe. 2007;2(6):371–382. doi: 10.1016/j.chom.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bennett BL, et al. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc Natl Acad Sci USA. 2001;98(24):13681–13686. doi: 10.1073/pnas.251194298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Philipp I, et al. Wnt/beta-catenin and noncanonical Wnt signaling interact in tissue evagination in the simple eumetazoan Hydra. Proc Natl Acad Sci USA. 2009;106(11):4290–4295. doi: 10.1073/pnas.0812847106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rahme LG, et al. Plants and animals share functionally common bacterial virulence factors. Proc Natl Acad Sci USA. 2000;97(16):8815–8821. doi: 10.1073/pnas.97.16.8815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rahme LG, et al. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268(5219):1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 37.Kambris Z, Hoffmann JA, Imler JL, Capovilla M. Tissue and stage-specific expression of the Tolls in Drosophila embryos. Gene Expr Patterns. 2002;2(3-4):311–317. doi: 10.1016/s1567-133x(02)00020-0. [DOI] [PubMed] [Google Scholar]
- 38.Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298(5600):1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 39.Douglas AE, Minto LB, Wilkinson TL. Quantifying nutrient production by the microbial symbionts in an aphid. J Exp Biol. 2001;204(Pt 2):349–358. doi: 10.1242/jeb.204.2.349. [DOI] [PubMed] [Google Scholar]
- 40.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122(1):107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 41.Nyholm SV, McFall-Ngai MJ. The winnowing: Establishing the squid-vibrio symbiosis. Nat Rev Microbiol. 2004;2(8):632–642. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- 42.Rawls JF, Samuel BS, Gordon JI. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci USA. 2004;101(13):4596–4601. doi: 10.1073/pnas.0400706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.French N, Pettersson S. Microbe-host interactions in the alimentary tract: The gateway to understanding inflammatory bowel disease. Gut. 2000;47(2):162–163. doi: 10.1136/gut.47.2.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ott SJ, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53(5):685–693. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fraune S, Bosch TC. Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc Natl Acad Sci USA. 2007;104(32):13146–13151. doi: 10.1073/pnas.0703375104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fraune S, Abe Y, Bosch TC. Disturbing epithelial homeostasis in the metazoan Hydra leads to drastic changes in associated microbiota. Environ Microbiol. 2009;11(9):2361–2369. doi: 10.1111/j.1462-2920.2009.01963.x. [DOI] [PubMed] [Google Scholar]
- 47.Cario E, Podolsky DK. Intestinal epithelial TOLLerance versus inTOLLerance of commensals. Mol Immunol. 2005;42(8):887–893. doi: 10.1016/j.molimm.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 48.Otte JM, Cario E, Podolsky DK. Mechanisms of cross hyporesponsiveness to Toll-like receptor bacterial ligands in intestinal epithelial cells. Gastroenterology. 2004;126(4):1054–1070. doi: 10.1053/j.gastro.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 49.Khalturin K, Hemmrich G, Fraune S, Augustin R, Bosch TC. More than just orphans: Are taxonomically-restricted genes important in evolution? Trends Genet. 2009;25(9):404–413. doi: 10.1016/j.tig.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 50.Fraune S, Augustin R, Bosch TC. Embryo protection in contemporary immunology: Why bacteria matter. Commun Integr Biol. 2011;4(4):369–372. doi: 10.4161/cib.4.4.15159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.