Abstract
The pyrenoid is a subcellular microcompartment in which algae sequester the primary carboxylase, ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco). The pyrenoid is associated with a CO2-concentrating mechanism (CCM), which improves the operating efficiency of carbon assimilation and overcomes diffusive limitations in aquatic photosynthesis. Using the model alga Chlamydomonas reinhardtii, we show that pyrenoid formation, Rubisco aggregation, and CCM activity relate to discrete regions of the Rubisco small subunit (SSU). Specifically, pyrenoid occurrence was shown to be conditioned by the amino acid composition of two surface-exposed α-helices of the SSU: higher plant-like helices knock out the pyrenoid, whereas native algal helices establish a pyrenoid. We have also established that pyrenoid integrity was essential for the operation of an active CCM. With the algal CCM being functionally analogous to the terrestrial C4 pathway in higher plants, such insights may offer a route toward transforming algal and higher plant productivity for the future.
Keywords: algal photosynthesis, carbon fixation, chloroplast, protein engineering
The catalytic activities of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) are often considered to be flawed, inefficient, or at best confused (1, 2), and yet the resultant photosynthetic CO2 assimilation into three-carbon sugars (“C3” pathway) sustains life on earth. Served by a single active site, which evolved under anaerobic conditions, Rubisco carboxylase function became competitively inhibited by O2, a by-product of photosynthetic light reactions. Relatively high atmospheric CO2 concentrations, in equilibrium with the aquatic dissolved inorganic carbon (DIC) pool, continued to sustain Rubisco carboxylase activity through the gradually increasing O2 concentration, and beyond the great oxidation event at some 2.4 billion years ago (bya). However, between 0.6 and 1 bya, cyanobacterial photosynthesis was limited by inorganic carbon supply to the extent that some form of carbon-concentrating mechanism (CCM) may then have evolved (3). Subsequently, most aquatic eukaryotic lineages also developed a CCM to suppress oxygenase activity and the associated wasteful photorespiration, and overcome CO2 limitation due to slow diffusivity in water (4).
When plants conquered land, the role of stomata, and internalization of gas exchange surfaces to minimize liquid phase resistance, allowed CO2 diffusion to supply carboxylase activity of Rubisco. Despite subsequent variations in atmospheric CO2 concentrations, for 400 million years C3 terrestrial plants have dominated carbon storage in biomass and soils. Today, they sequester nearly one-third of anthropogenic CO2 emissions, as well as providing food and fuel for the burgeoning global population, via many staple crops. However, it is thought that CO2 starvation and a changing climate over the past 20–30 million years have been major selection pressures to develop other mechanisms of CO2 concentration by terrestrial plants, which tend to use a four-carbon organic acid currency to power a biochemical CCM in the C4 pathway and crassulacean acid metabolism (CAM). Today, in a warming world, as crop yields plateau (5) and the human population continues to grow, understanding the fundamental genetic and mechanistic properties of such biochemical CCM processes could be a prelude to their introduction into staple crops such as rice (6).
In parallel, we also need to learn from cyanobacteria and eukaryotic algae, which currently sequester an equivalent proportion of anthropogenic CO2 emissions. In aquatic photosynthetic microorganisms, CO2 assimilation is supported by a range of biophysical CCMs, which use transmembrane transporters and carbonic anhydrases to concentrate DIC intracellularly by up to 1,000-fold (7), with Rubisco packaged into an associated subcellular microcompartment. The confinement of Rubisco helps to limit CO2 back-diffusion (CCM “leakage”) inherently associated with the slow turnover rate of the enzyme. Understanding the molecular determinants of these biophysical CCM processes, which improve the operating efficiency of Rubisco, could also provide a route toward augmenting crop productivity for the future (8).
The cyanobacterial CCM sequesters Rubisco into carboxysomes, which are semipermeable microcompartments surrounded by a protein shell that resembles viral capsids (9). Nearly all eukaryotic algal lineages possess a functionally analogous structure, the pyrenoid (10), which is also found in one terrestrial plant group, the ancient hornworts (11). In transmission electron micrographs, pyrenoids appear as unstructured electron-dense compartments in the chloroplast stroma, usually traversed by several thylakoid membranes. Although pyrenoids seemingly lack a protein coat or membrane, a starch sheath is often deposited at the immediate periphery. The presence of a pyrenoid is associated with the occurrence of a CCM (12), although the importance of the pyrenoid in the functional operation of a CCM has hitherto not been demonstrated. The bundle sheath in C4 photosynthesis has a similar role in sequestering Rubisco and containing CO2 leakage (13). Based on C4 isotopic discrimination models (14) and species abundance in cyanobacterial and eukaryotic phytoplankton assemblages (15, 16), nearly one-half of the 105 petagrams of carbon fixed each year (17) is contingent on some form of CCM and Rubisco compartmentation. Whereas the C4 and CAM biochemical systems, and the cyanobacterial carboxysome, are increasingly well resolved structurally and functionally (9, 18), the algal pyrenoid remains without a precise molecular definition.
Chlamydomonas reinhardtii is a model unicellular green alga, both in terms of genome sequencing and associated genetic resources (19), and particularly because of an extensive program evaluating the impact of site-directed mutagenesis on Rubisco large subunit (LSU) and small subunit (SSU), giving insight into Rubisco kinetic properties (1). Chlamydomonas induces a CCM under atmospheric CO2 concentrations. When the CCM is induced, >90% of Rubisco is packaged into a single, large pyrenoid (20) (Fig. 1A), but the mechanisms controlling this mobilization are unknown. Chlamydomonas Rubisco mutants containing a hybrid enzyme composed of native LSU and foreign (higher plant) SSU display reduced levels of photosynthetic growth and systematically lack a pyrenoid (21), implying a relationship between SSU, pyrenoid, and a functional CCM. The work in this paper focuses on specific solvent-exposed Rubisco SSU regions, i.e., two external α-helices, based on the hypothesis that Rubisco aggregation within the pyrenoid could be mediated by extrinsic protein interactions. Using a site-directed mutagenesis approach, we show that these discrete SSU regions condition pyrenoid formation, and that confinement of Rubisco into the pyrenoid is coupled to the operation of an active CCM in Chlamydomonas. Such insights may well potentiate the introduction of algal CCM components into higher plants, for which the formation of a microcompartment containing Rubisco, within the chloroplast, may well be an essential prerequisite (8).
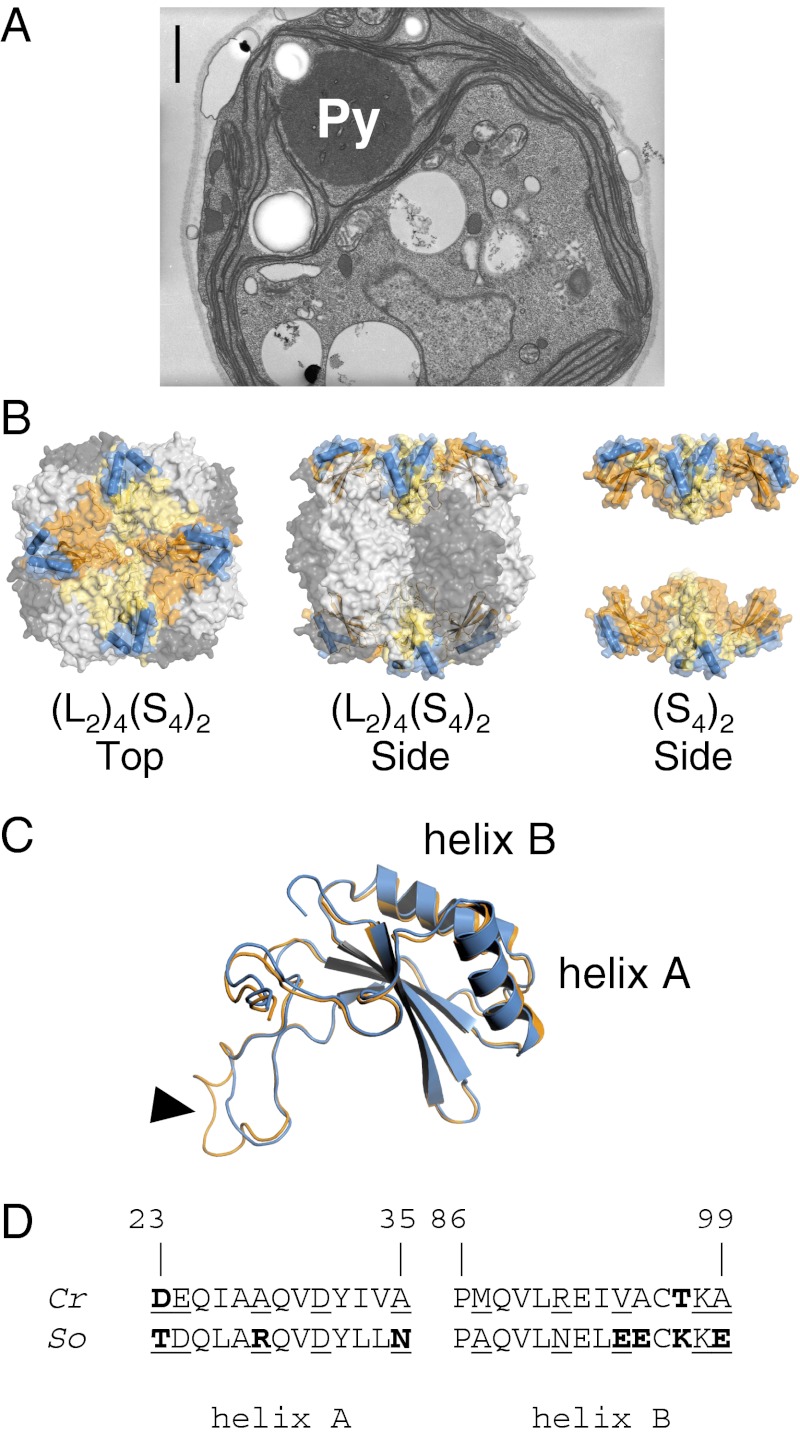
Fig. 1.
(A) Electron micrograph of a wild-type Chlamydomonas cell, grown under air-level CO2. Rubisco is packaged into a single conspicuous pyrenoid (Py). (Scale bar, 1 µm.) (B) Structure of the hexadecameric Rubisco form I of Chlamydomonas. The catalytic core is composed of four antiparallel LSU dimers (dark/light gray), stabilized by eight SSUs. Each SSU (orange/yellow) has two solvent-exposed α-helices (blue) that contribute to one-third of the surface-exposed SSU residues. (C) Despite 50% difference in amino acid composition, folding of Chlamydomonas (orange) and spinach (blue) SSU is highly conserved, as shown by the near-perfect overlay. The algal and higher plant SSUs differ primarily by the length of the βA–βB loop (arrowhead), which extends into the solvent channel of the holoenzyme. (D) Comparison of the amino acid composition of the Chlamydomonas reinhardtii (Cr) and Spinacia oleracea (So) SSU α-helices (22, 23). Numbering is relative to the Chlamydomonas sequence (24). The helices in spinach are distinctly more hydrophilic than in Chlamydomonas. Hydrophilic/polar residues differing between the two species are highlighted in bold; solvent-exposed residues are underlined.
Results
Land Plant Rubisco SSU with Substituted Algal α-Helices Sustains Growth in Vivo in Chlamydomonas Despite Compromised Rubisco Kinetics in Vitro.
Form I Rubisco, which is common to cyanobacteria, green algae, and land plants, is composed of a catalytic core of four LSU dimers, capped by four SSUs on both sides of a solvent channel (Fig. 1B). Based on the crystal structures (22, 23), and internal regions affecting Rubisco active site (24), we focused on the two solvent-exposed α-helices of the Rubisco SSU, to determine whether differences in Rubisco packaging between pyrenoidless higher plant hybrid enzymes, as represented by the spinach hybrid (21), and wild-type Chlamydomonas, could be mediated by extrinsic protein interactions. The two α-helices (A and B) contribute to one-third of the SSU surface-exposed residues (Fig. 1B). The spinach and Chlamydomonas SSUs are structurally very similar (Fig. 1C), and the two α-helices have an identical number of residues (24) but differ markedly in amino acid composition (Fig. 1D). Transformation vectors containing cDNA encoding for either spinach or Chlamydomonas SSU (21) were modified by PCR to encode for the following four chimeric SSU variants: spinach Rubisco small-subunit gene (RbcS) with individually substituted Chlamydomonas α-helix A or B sequences (yielding mutants “helix A,” with 7 amino acid mutations T23D–D24E–L26I–R28A–L33I–L34V–N35A, and “helix B,” also with 7 amino acid mutations A87M–N91R–L93I–E94V–E95A–K97T–E99A); spinach RbcS with substituted Chlamydomonas α-helix A and B sequences (mutant “helix AB,” combining all 14 mutations); and finally, a reciprocal construct, which substituted both spinach RbcS α-helix sequences into a wild-type Chlamydomonas RbcS (“reciprocal” mutant, mirroring the 14 mutations of “helix AB”). Transformation vectors were electroporated into a wallless, photosynthesis-deficient RbcS deletion strain (25). All Rubisco enzyme mutants thus retained the native Chlamydomonas LSU catalytic core but differed in the composition of their chimeric SSU, which combined algal and higher plant regions (Fig. S1A). Wild-type Chlamydomonas and original spinach hybrid enzymes were used as controls for the experiments (21).
A number of photosynthetic competent colonies were recovered for all four constructs, showing that the SSU α-helix substitutions did not inhibit SSU folding or assembly of the holoenzyme. Randomly selected transformants were further analyzed with respect to occurrence of pyrenoid and CCM activity, via photosynthetic capacity, Rubisco abundance and kinetics in vitro, pyrenoid ultrastructure, Rubisco localization, and in vivo affinity for inorganic carbon.
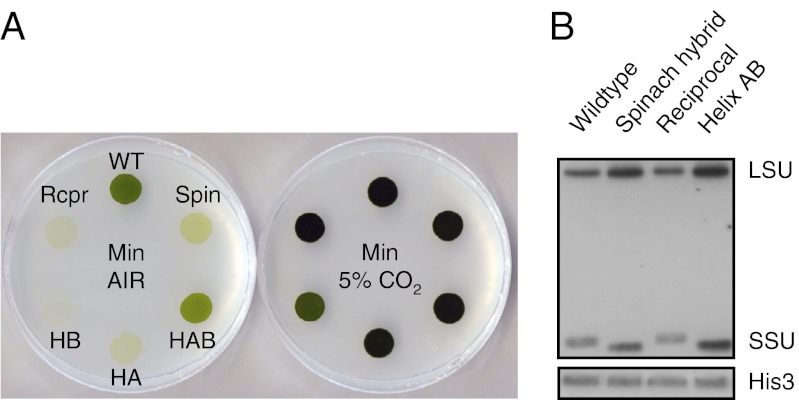
Photosynthetic growth phenotype was first examined via spot tests, revealing that under ambient CO2 (when the CCM is normally induced) all SSU mutants had reduced levels of growth compared with wild type (Fig. 2A, Left). Helix AB, however, had the least severe phenotype, and was also the only strain other than wild type that could be grown continuously in medium containing only air-equilibrated inorganic carbon. Maximum rate of photosynthesis of wild type, measured by O2 evolution, was 78 µmol O2⋅mg−1 chlorophyll⋅h−1. Helix AB had a photosynthetic rate on average seven times lower than wild type, spinach hybrid 16 times lower, and reciprocal 20 times lower (Table 1). These differences in photosynthetic capacity were not a consequence of reduced levels of Rubisco as shown by Western blot analysis (Fig. 2B) and by spot test under 5% CO2 (Fig. 2A, Right). Because growth was rescued under elevated CO2, we set out to determine whether the contrasting photosynthetic capacity at ambient CO2 was a consequence of compromised kinetic properties of Rubisco or a less efficient supply of carboxylating substrate to Rubisco via the CCM, or both.
Fig. 2.
(A) Comparison of photoautotrophic growth phenotype under CCM-induced (air-level CO2) and CCM-repressed conditions (5% CO2, vol/vol). Abbreviations: WT, wild type; Spin, spinach hybrid; HAB, helix AB; HA, helix A; HB, helix B; Rcpr, reciprocal. (B) Western blot analysis of total soluble proteins. The 53-kDa LSU and 15- to 16-kDa SSU bands are marked. The variation in SSU size reflects the shorter form in spinach (123 amino acids, versus 140 in Chlamydomonas). Helix AB has a spinach-like SSU (except for the α-helices), whereas reciprocal has a Chlamydomonas-like SSU (except, again, for the α-helices). Histone H3 served as a loading control. Corresponding Coomassie-stained gel in shown in Fig. S2.
Table 1.
Maximum photosynthetic rate and CCM efficacy
| Enzyme | Pmax*, µmol O2⋅mg−1 chl⋅h−1 | ∆13C*, ‰ | Leakage, % |
| Wild type | 78.1 ± 4.7 | 7.5 ± 0.8 | 18 |
| Helix AB | 10.6 ± 2.5 | 8.1 ± 2.3 | 25 |
| Spinach hybrid | 4.8 ± 1.2 | 18.9 ± 2.4 | 68 |
| Reciprocal | 3.9 ± 0.7 | 23.5 ± 4.7 | 86 |
*Values are the means ± 1 SD of three biological replicates.
When analyzing the kinetic properties of purified and activated Rubisco from helix AB, we found that the maximum rate of carboxylation (Vc) was six times lower than spinach hybrid Vc (21), and eight times lower than wild-type Vc (Table 2). Helix AB also had a significantly poorer capacity to discriminate between CO2 and O2 as shown by the lower specificity factor (Ω = 54, versus 65 for spinach hybrid and 61 for wild type). However, helix AB had a maximum photosynthetic rate more than twice the rate measured for spinach hybrid and was viable when continuously grown in minimal medium. These results suggested that a CCM was perhaps compensating for the kinetically impaired enzyme, whereas the CCM may have become inoperative in the spinach hybrid.
Table 2.
Kinetic properties of Rubisco purified from wild type and pyrenoid-restored Rubisco mutant (helix AB)
| Enzyme | Vc*, µmol⋅mg−1⋅h−1 | Kc*, µM CO2 | Ko*†, µM O2 | Vc/Kc‡ | Ko/Kc‡ | Vc/Vo‡ | Ω (VcKo/VoKc)‡ |
| Wild type | 110 ± 9 | 30 ± 2 | 523 ± 18 | 3.7 | 17 | 3.5 | 61 ± 2 |
| Helix AB | 13 ± 3 | 35 ± 3 | 299 ± 59 | 0.4 | 7 | 7.7 | 54 ± 2 |
Carboxylation (c) and oxygenation (o) kinetic properties of Rubisco purified from spinach hybrid have been determined by the authors before (mutant SSSO in ref. 21): Vc = 97, Kc = 34, Ko = 539, Vc/Kc = 2.9; Ko/Kc = 16; Vc/Vo = 4.1; Ω = 65. K (in micromolar concentration) is the Km and V (in micromolar hour−1 milligram−1) is the Vmax of purified and activated Rubisco.
*Values are the means ± 1 SD of three biological replicates.
†The Michaelis–Menten constant is measured as the O2 inhibition constant for the carboxylation reaction (Kio) and is assumed to equal Ko (also noted KmO2).
‡Calculated values.
Algal Rubisco SSU α-Helices Are Sufficient and Necessary for Pyrenoid Formation.
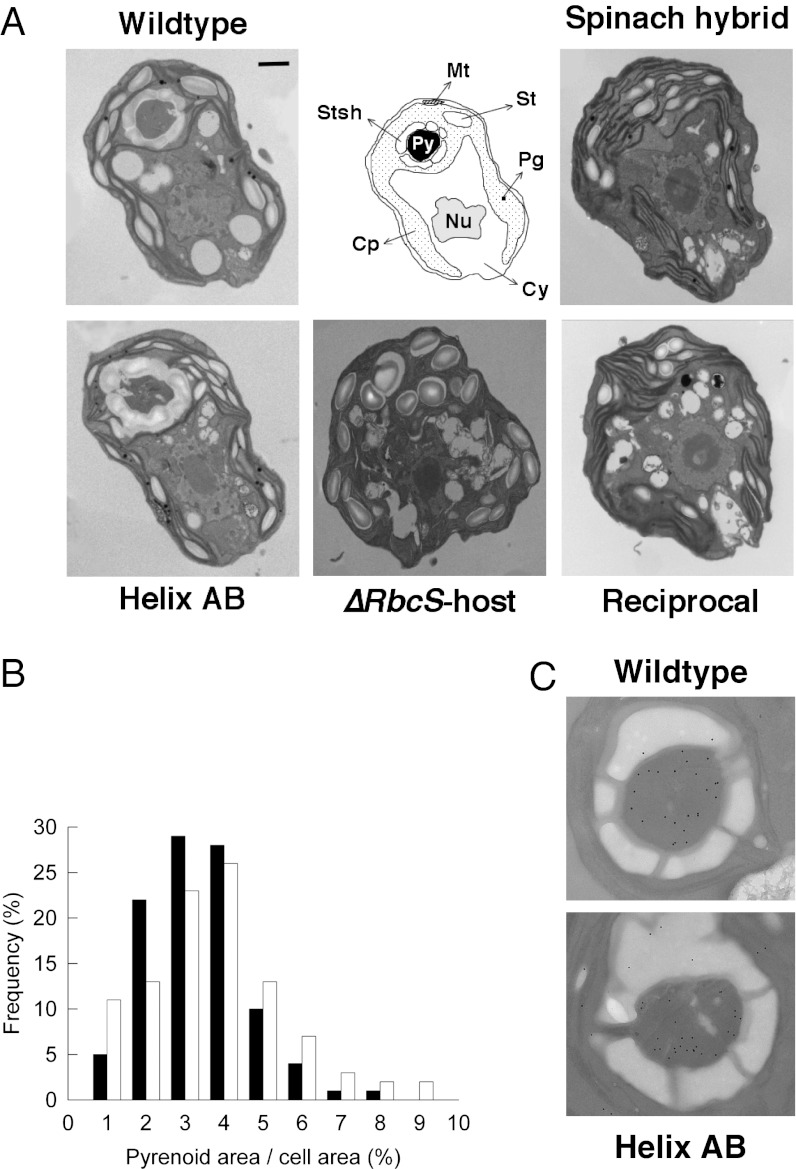
When examining the mutant strains with electron microscopy, we found that the pyrenoid, which was absent in the spinach hybrid (21), was restored in helix AB (Fig. 3A). This showed that the replacement of the spinach SSU α-helices with those from Chlamydomonas was sufficient to mobilize Rubisco into a pyrenoid. The subcellular location in the stroma, and relative size (Fig. 3B) of the restored pyrenoid was indistinguishable from wild type. Single α-helix replacements alone, in contrast, were insufficient to restore a pyrenoid (Fig. S3), suggesting that residues on both helices are essential for pyrenoid assembly. Proof that both Chlamydomonas SSU α-helices are an absolute requirement for pyrenoid integrity was shown by the reciprocal mutant: the pyrenoid was lost when both spinach SSU α-helices were introduced into an otherwise Chlamydomonas wild-type LSU+SSU holoenzyme (Fig. 3A). No electron-dense clustering of Rubisco was observed in any section of the reciprocal mutant, and the pyrenoidless phenotype was identical to the one previously observed for the spinach hybrid (Fig. 3A). These ultrastructural observations were validated by immunogold labeling of Rubisco (Fig. 3C). Labeling in wild type and helix AB was predominantly confined to the pyrenoid, with 84 and 76% of Rubisco packaged inside the microcompartment. These percentages were consistent with an induced CCM. In contrast, immunolocalization of Rubisco in the pyrenoid-negative lines (spinach hybrid and reciprocal) showed Rubisco was redistributed throughout the stroma, similar to higher plant chloroplasts. The subcellular distribution of Rubisco also had profound effects on the stacking of photosynthetic membranes, with the formation of grana-like structures in the two pyrenoid-lost mutants (see spinach hybrid and reciprocal in Fig. 3A and Fig. S4). Pyrenoid-positive strains, on the contrary, had a thylakoid arrangement characteristic of green algae (a few membranes appressed over longer intervals).
Fig. 3.
(A) Representative electron micrographs of the wallless Chlamydomonas RbcS deletion mutant T60-3 (∆RbcS-host), complemented with Chlamydomonas (wild type), spinach (spinach hybrid), or Chlamydomonas/spinach chimeric RbcS constructs (helix AB and reciprocal). When the two SSU helices are encoded by the native Chlamydomonas sequence, a pyrenoid is formed (as in wild type and helix AB), irrespective of the nature of the rest of the SSU. However, when the two SSU helices are encoded by the spinach sequence, Rubisco does not aggregate into a pyrenoid, even when the rest of the SSU is Chlamydomonas-like (as in reciprocal). A phenotype of thylakoid hyperstacking was generally observed in the pyrenoidless strains (spinach hybrid and reciprocal; Fig. S4). Abbreviations: Cy, cytosol; Nu, nucleus; Cp, chloroplast; Py, pyrenoid; Stsh, starch sheath; Mt, mitochondrion; St, stromal starch grain; Pg, plastoglobule. (Scale bar, 500 nm.) (B) Proportional cross-sectional area of the pyrenoid in wild type (black columns) and helix AB (white columns). The area occupied by the “restored” pyrenoid in helix AB is not statistically different from wild type (unpaired t test, P = 0.143). (C) Detail of representative wild-type and helix AB pyrenoids probed with an antibody against wheat Rubisco. The density of pyrenoid labeling relative to the rest of the chloroplast was similar in both strains (unpaired t test, P = 0.077). Note that the starch sheath is interrupted by gaps allowing the passage of thylakoids.
Pyrenoid Is Essential for the Expression of a CCM in Chlamydomonas.
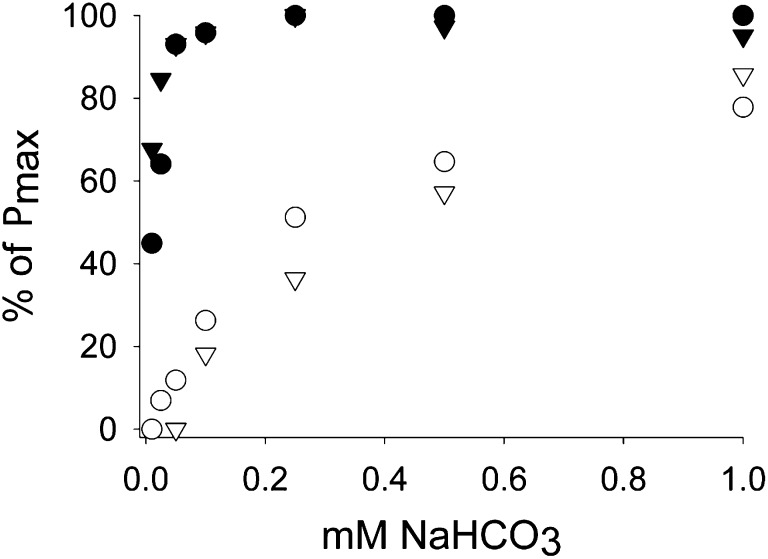
Pyrenoid occurrence was also directly related to CCM expression. The activity of a CCM in wild type and helix AB was confirmed by measuring real-time carbon isotope discrimination (Δ13C). This measure was used to derive the extent of CCM efficiency or leakage (26, 27), which is low (typically <10%) for a fully functional CCM in Chlamydomonas and high (theoretically up to 100%) for photosynthesis reliant solely on CO2 diffusion. The helix AB transformants, with their restored pyrenoid, had a CCM activity similar to wild type when grown under CCM-induced conditions (Table 1). Both spinach hybrid and reciprocal, in contrast, displayed a “leaky” phenotype, consistent with photosynthesis supplied by CO2 diffusion, with no CCM and without a pyrenoid. Relative affinity for DIC in vivo, as measured by oxygen evolution (Fig. 4), was consistent with these observations. The combined Δ13C and DIC affinity data suggest that the pyrenoid and CCM were coupled, and had helped to restore growth when associated with a kinetically compromised Rubisco in helix AB mutants. These important observations prove that the pyrenoid is a functional component of the algal CCM, mediated by the Rubisco SSU α-helices, even when the enzyme is catalytically impaired.
Fig. 4.
O2 evolution of DIC-starved cells, incubated with increasing concentrations of sodium bicarbonate (0.005–1 mM) in an illuminated oxygen electrode chamber. To enable a direct comparison between the different strains, data were expressed in fraction of maximum photosynthetic rate. Absolute maximum rates are given in Table 1. Pyrenoid-positive strains (wild type and helix AB) have a photosynthesis saturating at low levels of NaHCO3, indicating the presence of a CCM. Legend: wild type (●), spinach hybrid (○), helix AB (▼), and reciprocal (▽).
Discussion
With the simple expedient of substituting discrete regions of the Rubisco SSU, we have knocked out and restored both the pyrenoid and the CCM in Chlamydomonas, providing conclusive evidence for the functional importance of the Rubisco SSU in pyrenoid formation and that the algal pyrenoid is essential for the efficacy of the CCM. The SSU had long been suspected to play a role in stabilizing the catalytic LSU octamer, and specific SSU residues at the LSU interface influence maximal catalysis. In a previous study (28), substitution of Tyr-32 with Ala produced catalytic properties quite similar to those of the helix AB enzyme. Perhaps differences in the identity of the neighboring residues (Ile-33 versus Leu-33) influence the interaction of Tyr-32 with the large subunit to bring about these negative effects on catalysis. Our work suggests additional roles for the SSU in aquatic photosynthesizers equipped with a CCM. The amino acid composition of the Chlamydomonas SSU α-helices is distinctly more hydrophobic compared with higher plant SSU α-helices (Fig. 1D), suggesting that helix-mediated packaging of Rubisco into pyrenoids may be driven by hydrophobic interactions.
A parsimonious explanation for pyrenoid formation could therefore be a spontaneous aggregation of Rubisco by exclusion of water molecules (via transient hydrophobic interactions), whereas more hydrophilic SSU helices, as in the spinach hybrid and reciprocal, might interfere with this mechanism. In higher plants, it has been shown that thylakoid stacking releases more stromal volume (∼17%) for the free diffusion of macromolecules, including Rubisco (29). This is consistent with our observation that the helix substitutions appear to have a pleiotropic effect on the degree of thylakoid stacking: when the pyrenoid is lost (i.e., when Rubisco is redistributed to the stroma), mutant cells also display a hyperstacking phenotype. A less parsimonious explanation would involve an active mechanism through some sort of yet unidentified linker protein that interacts with the SSU helices.
In an intriguing parallel, aggregation of Rubisco inside the cyanobacterial carboxysome was also shown to involve the SSU (30, 31). A Rubisco-organizing protein, with several SSU-like repeats, links adjacent Rubisco units in a lattice-like arrangement. Additionally, the linkers also interact with a carboxysomal carbonic anhydrase, to form a bicarbonate dehydration complex that supplies the active sites of Rubisco with CO2. It is not unreasonable to suggest that the pyrenoid may comprise an analogous molecular scaffold, considering that other enzymes of the Calvin–Benson–Bassham (CBB) cycle are also reliant on small intrinsically unstructured proteins to form transient supramolecular complexes (32).
Whether the interaction between Rubisco holoenzymes is mediated by some additional protein-linker system or by specific individual residues remains to be investigated experimentally. Nearly all reported pyrenoids appear amorphous, with no lattice-like arrangement of Rubisco. The partial redistribution of Rubisco when Chlamydomonas is grown under elevated CO2 (33), and need for access to allow organic CBB cycle substrate and product exchanges, also suggest considerable plasticity in pyrenoid organization. In addition to Rubisco, the catalytic chaperone Rubisco activase and photosystem I enriched thylakoid membranes (34) are present in the pyrenoid. Specific components of the CCM are also localized within the pyrenoid, most significantly a thylakoid luminal carbonic anhydrase, which has been suggested to facilitate CO2 conversion close to the active sites of Rubisco, with the CCM apparently using the intrathylakoid membranes as a conduit for CO2 delivery (35).
Our observations also make a major contribution toward coupling pyrenoid occurrence and CCM activity in Chlamydomonas. Although certain molecular components associated with the CCM have been identified (reviewed in refs. 36 and 37), the link between Rubisco molecular structure, aggregation, and pyrenoid function had not previously been proven. In evolutionary terms, there is an inverse relationship between Rubisco specificity and the activity of a CCM in cyanobacteria, chlorophyte algae, and higher plants (38). C3 plants, and green algae without a CCM (39) or reliant on CO2 diffusion, have a Rubisco with high specificity for CO2 (Ω ∼ 80–100). Cyanobacteria, backed by a powerful CCM, have a Rubisco with low selectivity (Ω ∼ 50). Chlamydomonas has an intermediate specificity (Ω ∼ 60), and growth by the helix AB mutant cells, with compromised kinetic properties (Ω ∼ 54), was largely rescued by the restored CCM and pyrenoid. If Rubisco has evolved from a fast, low-affinity carboxylase to a slow, high-affinity carboxylase, it would be interesting to determine the extent that the operation of a CCM, and provision of elevated CO2 via the pyrenoid, has modified kinetic properties.
Results presented here provide a major insight into the molecular organization of the algal pyrenoid and expression of the CCM. If a pyrenoid could be expressed heterologously, as has been demonstrated for the carboxysome (40), this could offer significant opportunities to promote CCM activity and algal productivity in bioenergy systems. In addition, the occurrence of a pyrenoid and CCM in one group of terrestrial plants, the hornworts, together with our fundamental observations on aggregation of Rubisco, pyrenoid formation, and CCM activity, perhaps offer a route by which elements of an algal CCM could be used to promote higher plant and crop productivity in the future.
Materials and Methods
Strains and Culture Conditions.
All strains used in this work have the deletion mutant ΔRbcS-T60-3, which has no functional copy of the genes encoding for a Rubisco SSU (25), as common genetic background. All strains are maintained at 25 °C in the dark on medium containing 10 mM acetate and 1.5% Bacto agar (wt/vol) (41). For biochemical analysis, cells were grown as described in ref. 21. For physiological analysis, cells were grown as described in ref. 27.
Plasmids and Host Transformation.
A 4.7-kb EcoRI restriction fragment, containing a codon-optimized spinach RbcS cDNA with the native algal chloroplast signaling peptide, under the control of the Chlamydomonas RbcS1 UTRs and introns 1 and 2 (21), was modified by overlap PCR (42) to substitute the coding sequences of the SSU α-helices with those of Chlamydomonas, first singly and then in combination. The reverse construct (substituting the algal helices with the spinach helices) was undertaken on a vector that contained the native Chlamydomonas RbcS1, flanked by the same regulatory sequences as above. Mutated cDNA sequences are given in Fig. S1B. T-60 host cells were transformed via electroporation and selected for restored photosynthesis on minimal medium (without acetate) in the light (80 µmol photons·m−2·s−1) (21).
Photosynthesis Assay.
Spot tests were used to screen the strains by plating 5 × 105 dark-grown cells on minimal medium in the light (80 µmol photons·m−2·s−1) at 25 °C under ambient CO2 or 5% CO2. Photosynthetic rate was determined from the rate of O2 evolution of 1.5 × 107 internal DIC pool-depleted cell, under 250 µmol photons·m−2·s−1 in response to increasing concentrations of NaHCO3 using an oxygen electrode (Rank Brothers). Cells were prepared according to Badger et al. (43). The light intensity was validated as nonphotoinhibitory by analysis of light response curves measured with a PAM chlorophyll fluorometer (Walz).
Western Blot.
Total soluble proteins were isolated by sonication on ice for 3 min in extraction buffer (50 mM bicine, pH 8.0, 10 mM NaHCO3, 10 mM MgCl2, 1 mM DTT), solubilized in SDS-loading buffer [0.1 M Tris·HCl, pH 7.8, 10% (vol/vol) glycerol, 2% (wt/vol) SDS, 25 mM DTT, 0.1% BPB], and analyzed by SDS/PAGE on 12% acrylamide gel. Denatured proteins were transferred electrophoretically to nitrocellulose membrane and visualized with rabbit primary antibody against wheat Rubisco (44), and anti-rabbit IgG-HRP conjugate and ECL+ Western Blotting detection system (Perkin-Elmer). Histone H3 (ab1791; Abcam) and Coomassie blue staining were used as loading control (30 µg total protein/lane).
Rubisco Kinetics Assay.
Rubisco holoenzymes were isolated from total soluble protein extracts by ultracentrifugation on 10–30% continuous sucrose gradient (45). Carboxylation (c) and oxygenation (o) kinetic constants (where K is the Km and V is the Vmax) of purified and activated Rubisco were assayed by measuring the rate of incorporation of acid stable NaH14CO3 (K in micromolar concentration; V in micromolar hour−1 milligram−1) (45). Enzymes from the reciprocal and single-helix mutants were not purified, and the spinach hybrid had been assayed before (21).
Isotopic Discrimination.
Discrimination against the stable carbon isotope (Δ13C) was measured using a mass spectrometer, downstream of an open gas cuvettete system, after cold trapping of air passing over photosynthetic active Chlamydomonas, into a vacuum line and water vapor scrubbing, as described previously (27). Leakage was calculated from Δ13C, fractionation during carboxylation (27‰), discrimination during dissolution of CO2 (1.1‰), and liquid phase diffusion (0.7‰), using the authors’ model (27).
Microscopy.
Samples for electron microscopy were fixed as described in ref. 21. The same blocks that provided thin sections for ultrastructural analysis were used for immunogold labeling (using Ni instead of Cu grids). Embedded material was etched to unmask epitopes and remove oxidizing agents, before incubation with the primary (1:5,000 dilution of a wheat Rubisco antibody) and secondary antibodies. The fraction of Rubisco in the pyrenoid was calculated for 100 randomly selected pyrenoid-positive cells. Gold particle density in the pyrenoid was multiplied by the pyrenoid area (average, 0.78 µm2 for wild type and 0.98 µm2 for helix AB) and then expressed as a percentage of the total particles in the chloroplast (stroma plus pyrenoid; the average area of the stroma was 12.7 µm2 for wild type and 13.9 µm2 for helix AB). Cytosolic gold particles density served as a measure of nonspecific labeling and was deducted from the pyrenoidal and stromal densities.
Supplementary Material
Acknowledgments
This research was supported by Biotechnology and Biological Sciences Research Council Grant BB/I024518/1 (“Combining Algal and Plant Photosynthesis”) and Department of Energy Grant DE-FG02-00ER15044.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1210993109/-/DCSupplemental.
References
- 1.Spreitzer RJ, Salvucci ME. Rubisco: Structure, regulatory interactions, and possibilities for a better enzyme. Annu Rev Plant Biol. 2002;53:449–475. doi: 10.1146/annurev.arplant.53.100301.135233. [DOI] [PubMed] [Google Scholar]
- 2.Tcherkez GGB, Farquhar GD, Andrews TJ. Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc Natl Acad Sci USA. 2006;103(19):7246–7251. doi: 10.1073/pnas.0600605103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riding R. Cyanobacterial calcification, carbon dioxide concentrating mechanisms, and Proterozoic-Cambrian changes in atmospheric composition. Geobiology. 2006;4(4):299–316. [Google Scholar]
- 4.Raven JA. The role of marine biota in the evolution of terrestrial biota: Gases and genes. Biogeochemistry. 1997;39(2):139–164. [Google Scholar]
- 5.Zhu XG, Long SP, Ort DR. Improving photosynthetic efficiency for greater yield. Annu Rev Plant Biol. 2010;61:235–261. doi: 10.1146/annurev-arplant-042809-112206. [DOI] [PubMed] [Google Scholar]
- 6.Covshoff S, Hibberd JM. Integrating C4 photosynthesis into C3 crops to increase yield potential. Curr Opin Biotechnol. 2012;23(2):209–214. doi: 10.1016/j.copbio.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 7.Badger MR, et al. The diversity and coevolution of Rubisco, plastids, pyrenoids, and chloroplast-based CO2-concentrating mechanisms in algae. Can J Bot. 1998;76(6):1052–1071. [Google Scholar]
- 8.Price GD, Badger MR, von Caemmerer S. The prospect of using cyanobacterial bicarbonate transporters to improve leaf photosynthesis in C3 crop plants. Plant Physiol. 2011;155(1):20–26. doi: 10.1104/pp.110.164681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM. Protein-based organelles in bacteria: Carboxysomes and related microcompartments. Nat Rev Microbiol. 2008;6(9):681–691. doi: 10.1038/nrmicro1913. [DOI] [PubMed] [Google Scholar]
- 10.Lee RE. Phycology. 4th Ed. Cambridge, UK: Cambridge Univ Press; 2008. [Google Scholar]
- 11.Smith EC, Griffiths H. A pyrenoid-based carbon-concentrating mechanism is present in terrestrial bryophytes of the class Anthocerotae. Planta. 1996;200(2):203–212. [Google Scholar]
- 12.Giordano M, Beardall J, Raven JA. CO2 concentrating mechanisms in algae: Mechanisms, environmental modulation, and evolution. Annu Rev Plant Biol. 2005;56:99–131. doi: 10.1146/annurev.arplant.56.032604.144052. [DOI] [PubMed] [Google Scholar]
- 13.Kromdijk J, et al. Bundle sheath leakiness and light limitation during C4 leaf and canopy CO2 uptake. Plant Physiol. 2008;148(4):2144–2155. doi: 10.1104/pp.108.129890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lloyd J, Farquhar GD. 13C discrimination during CO2 assimilation by the terrestrial biosphere. Oecologia. 1994;99(3):201–215. doi: 10.1007/BF00627732. [DOI] [PubMed] [Google Scholar]
- 15.Zwirglmaier K, et al. Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ Microbiol. 2008;10(1):147–161. doi: 10.1111/j.1462-2920.2007.01440.x. [DOI] [PubMed] [Google Scholar]
- 16.Granum E, Raven JA, Leegood RC. How do marine diatoms fix 10 billion tonnes of inorganic carbon per year? Can J Bot. 2005;83(7):898–908. [Google Scholar]
- 17.Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. Primary production of the biosphere: Integrating terrestrial and oceanic components. Science. 1998;281(5374):237–240. doi: 10.1126/science.281.5374.237. [DOI] [PubMed] [Google Scholar]
- 18.Majeran W, et al. Structural and metabolic transitions of C4 leaf development and differentiation defined by microscopy and quantitative proteomics in maize. Plant Cell. 2010;22(11):3509–3542. doi: 10.1105/tpc.110.079764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merchant SS, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318(5848):245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borkhsenious ON, Mason CB, Moroney JV. The intracellular localization of ribulose-1,5-bisphosphate carboxylase/oxygenase in Chlamydomonas reinhardtii. Plant Physiol. 1998;116(4):1585–1591. doi: 10.1104/pp.116.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Genkov T, Meyer M, Griffiths H, Spreitzer RJ. Functional hybrid rubisco enzymes with plant small subunits and algal large subunits: Engineered rbcS cDNA for expression in Chlamydomonas. J Biol Chem. 2010;285(26):19833–19841. doi: 10.1074/jbc.M110.124230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor TC, Backlund A, Bjorhall K, Spreitzer RJ, Andersson I. First crystal structure of Rubisco from a green alga, Chlamydomonas reinhardtii. J Biol Chem. 2001;276(51):48159–48164. doi: 10.1074/jbc.M107765200. [DOI] [PubMed] [Google Scholar]
- 23.Andersson I. Large structures at high resolution: The 1.6 Å crystal structure of spinach ribulose-1,5-bisphosphate carboxylase/oxygenase complexed with 2-carboxyarabinitol bisphosphate. J Mol Biol. 1996;259(1):160–174. doi: 10.1006/jmbi.1996.0310. [DOI] [PubMed] [Google Scholar]
- 24.Spreitzer RJ. Role of the small subunit in ribulose-1,5-bisphosphate carboxylase/oxygenase. Arch Biochem Biophys. 2003;414(2):141–149. doi: 10.1016/s0003-9861(03)00171-1. [DOI] [PubMed] [Google Scholar]
- 25.Khrebtukova I, Spreitzer RJ. Elimination of the Chlamydomonas gene family that encodes the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase. Proc Natl Acad Sci USA. 1996;93(24):13689–13693. doi: 10.1073/pnas.93.24.13689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berry JA. Studies of mechanisms affecting the fractionation of carbon isotopes in photosynthesis. In: Rundel PW, Ehleringer JR, Nagy KA, editors. Ecological Studies. Vol 68. New York: Springer; 1989. pp. 82–94. [Google Scholar]
- 27.Meyer M, Seibt U, Griffiths H. To concentrate or ventilate? Carbon acquisition, isotope discrimination and physiological ecology of early land plant life forms. Philos Trans R Soc Lond B Biol Sci. 2008;363(1504):2767–2778. doi: 10.1098/rstb.2008.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Genkov T, Spreitzer RJ. Highly conserved small subunit residues influence rubisco large subunit catalysis. J Biol Chem. 2009;284(44):30105–30112. doi: 10.1074/jbc.M109.044081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chow WS, Kim EH, Horton P, Anderson JM. Granal stacking of thylakoid membranes in higher plant chloroplasts: The physicochemical forces at work and the functional consequences that ensue. Photochem Photobiol Sci. 2005;4(12):1081–1090. doi: 10.1039/b507310n. [DOI] [PubMed] [Google Scholar]
- 30.Cot SSW, So AKC, Espie GS. A multiprotein bicarbonate dehydration complex essential to carboxysome function in cyanobacteria. J Bacteriol. 2008;190(3):936–945. doi: 10.1128/JB.01283-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Long BM, Rae BD, Badger MR, Price GD. Over-expression of the β-carboxysomal CcmM protein in Synechococcus PCC7942 reveals a tight co-regulation of carboxysomal carbonic anhydrase (CcaA) and M58 content. Photosynth Res. 2011;109(1-3):33–45. doi: 10.1007/s11120-011-9659-8. [DOI] [PubMed] [Google Scholar]
- 32.Graciet E, et al. The small protein CP12: A protein linker for supramolecular complexes. Biochemistry. 2003;42(27):8163–8170. doi: 10.1021/bi034474x. [DOI] [PubMed] [Google Scholar]
- 33.Ramazanov Z, et al. The induction of the CO2-concentrating mechanism is correlated with the formation of the starch sheath around the pyrenoid of Chlamydomonas reinhardtii. Planta. 1994;195(2):210–216. [Google Scholar]
- 34.McKay RML, Gibbs SP. Composition and function of pyrenoids: Cytochemical and immunocytochemical approaches. Can J Bot. 1991;69(5):1040–1052. [Google Scholar]
- 35.Raven JA. CO2-concentrating mechanisms: A direct role for thylakoid lumen acidification? Plant Cell Environ. 1997;20(2):147–154. [Google Scholar]
- 36.Moroney JV, et al. The carbonic anhydrase isoforms of Chlamydomonas reinhardtii: Intracellular location, expression, and physiological roles. Photosynth Res. 2011;109(1-3):133–149. doi: 10.1007/s11120-011-9635-3. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, Duanmu D, Spalding MH. Carbon dioxide concentrating mechanism in Chlamydomonas reinhardtii: Inorganic carbon transport and CO2 recapture. Photosynth Res. 2011;109(1-3):115–122. doi: 10.1007/s11120-011-9643-3. [DOI] [PubMed] [Google Scholar]
- 38.Whitney SM, Houtz RL, Alonso H. Advancing our understanding and capacity to engineer nature’s CO2-sequestering enzyme, Rubisco. Plant Physiol. 2011;155(1):27–35. doi: 10.1104/pp.110.164814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palmqvist K, Sültemeyer D, Baldet P, Andrews TJ, Badger MR. Characterisation of inorganic carbon fluxes, carbonic anhydrase(s) and ribulose-1.5-biposphate carboxylase-oxygenase in the green unicellular alga Coccomyxa. Planta. 1995;197(2):352–361. [Google Scholar]
- 40.Bonacci W, et al. Modularity of a carbon-fixing protein organelle. Proc Natl Acad Sci USA. 2012;109(2):478–483. doi: 10.1073/pnas.1108557109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spreitzer RJ, Mets L. Photosynthesis-deficient mutants of Chlamydomonas reinhardtii with associated light-sensitive phenotypes. Plant Physiol. 1981;67(3):565–569. doi: 10.1104/pp.67.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Higuchi R, Krummel B, Saiki RK. A general method of in vitro preparation and specific mutagenesis of DNA fragments: Study of protein and DNA interactions. Nucleic Acids Res. 1988;16(15):7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Badger MR, Kaplan A, Berry JA. Internal inorganic carbon pool of Chlamydomonas reinhardtii. Plant Physiol. 1980;66(3):407–413. doi: 10.1104/pp.66.3.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Howe CJ, et al. Location and nucleotide sequence of the gene for the proton-translocating subunit of wheat chloroplast ATP synthase. Proc Natl Acad Sci USA. 1982;79(22):6903–6907. doi: 10.1073/pnas.79.22.6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spreitzer RJ, Jordan DB, Ogren WL. Biochemical and genetic analysis of an RuBP carboxylase/oxygenase-deficient mutant and revertants of Chlamydomonas reinhardtii. FEBS Lett. 1982;148(1):117–121. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.