Abstract
Rice (Oryza sativa L.) grain is a major dietary source of cadmium (Cd), which is toxic to humans, but no practical technique exists to substantially reduce Cd contamination. Carbon ion-beam irradiation produced three rice mutants with <0.05 mg Cd⋅kg−1 in the grain compared with a mean of 1.73 mg Cd⋅kg−1 in the parent, Koshihikari. We identified the gene responsible for reduced Cd uptake and developed a strategy for marker-assisted selection of low-Cd cultivars. Sequence analysis revealed that these mutants have different mutations of the same gene (OsNRAMP5), which encodes a natural resistance-associated macrophage protein. Functional analysis revealed that the defective transporter protein encoded by the mutant osnramp5 greatly decreases Cd uptake by roots, resulting in decreased Cd in the straw and grain. In addition, we developed DNA markers to facilitate marker-assisted selection of cultivars carrying osnramp5. When grown in Cd-contaminated paddy fields, the mutants have nearly undetectable Cd in their grains and exhibit no agriculturally or economically adverse traits. Because mutants produced by ion-beam radiation are not transgenic plants, they are likely to be accepted by consumers and thus represent a practical choice for rice production worldwide.
Cadmium (Cd), a contaminant that enters the food chain from multiple natural and industrial sources, is toxic to the kidneys, particularly to the proximal tubular cells, where it accumulates and leads to renal dysfunction (1). In Japan, itai-itai disease (renal osteomalacia), which is characterized by spinal and leg bone pain, is recognized as chronic toxicity caused by excess Cd in drinking water and crops (2). To reduce the risk of Cd poisoning, the Joint Food and Agriculture Organization/World Health Organization (FAO/WHO) Expert Committee on Food Additives established a provisional tolerable monthly Cd intake of 25 µg⋅kg−1 body weight (3), and the Codex Alimentarius Commission of the FAO/WHO established maximum Cd levels in food crops (4). The international maximum limit for rice is 0.4 mg Cd⋅kg−1 polished rice. Rice is a staple food for nearly half of the world’s population, and global production and consumption of rice increased approximately threefold from 1960 to 2011 (5). The demand for rice continues to grow, so it is necessary to produce low-Cd rice to reduce the potential risk that Cd poses to human health.
There are substantial genotypic differences in Cd accumulation in rice (6, 7), concentrations generally being higher in indica-type cultivars than in japonica-type cultivars. Genetic loci determining genotypic differences in Cd accumulation have been identified by quantitative trait locus (QTL) analysis of several mapping populations (8, 9). Recently, genes involved in Cd uptake by the root (10–13), root vacuole sequestration (14, 15), root xylem loading (16, 17), and phloem transport in the node (18) have been found in rice, so the physiological and molecular processes of Cd transport in rice are increasingly well understood (19). Although regulation of Cd transport by transgenic techniques may enable us to reduce Cd accumulation in rice grain, commercial transgenic rice is not currently acceptable in many countries, such as Japan. Many consumers fear eating food produced by transgenic plants.
Energetic heavy-ion beams have been recently used to generate mutants in higher plants because they induce mutations with high frequency at a relatively low dose (i.e., at which virtually all plants survive), and they induce a broad spectrum of phenotypes without affecting other plant characteristics (20, 21). Using this technique, unique varieties of some flowers and trees have been commercialized, but this has not yet occurred in crop plants. Mutants produced by ion-beam radiation are not transgenic, so they are more likely to be accepted by consumers.
In the present study, we report (i) nontransgenic rice mutants with nearly cadmium-free grain produced by irradiation with heavy-ion beams and (ii) the development of a DNA marker for further breeding based on the identification of the gene (OsNRAMP5) responsible for low Cd uptake. Field studies show that these mutants have nearly nondetectable levels of Cd in the grain, even when cultivated in paddy fields contaminated with high levels of Cd.
Results
Isolation of Low-Cd-Accumulating Rice Mutants.
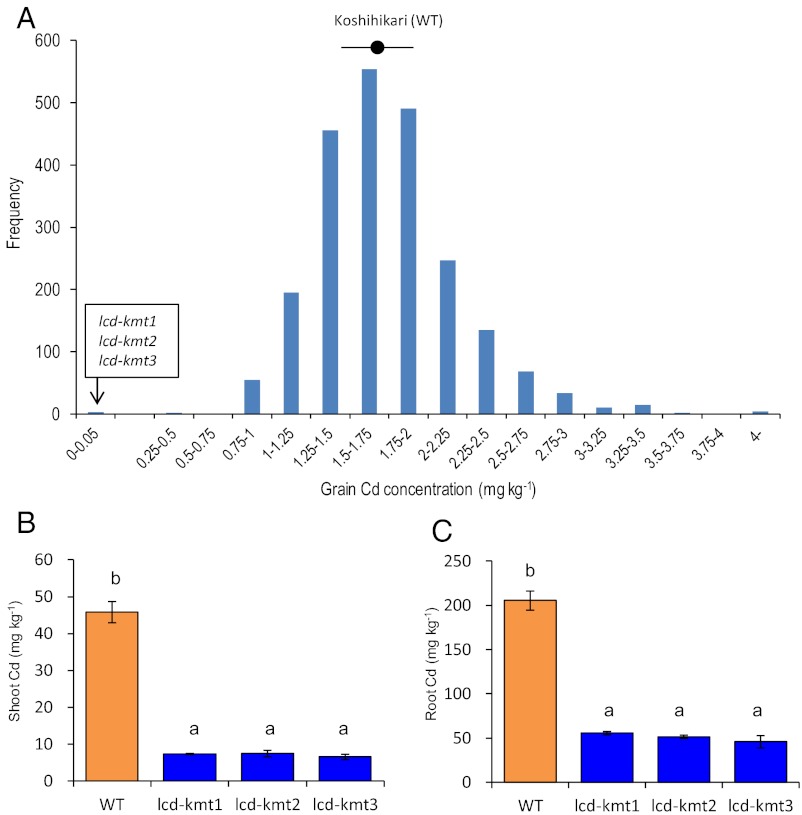
We irradiated seeds of the most popular Japanese temperate japonica rice cultivar, Koshihikari, with accelerated carbon ions. Three low-Cd mutants (lcd-kmt1, lcd-kmt2, and lcd-kmt3) were identified in initial screening for grain Cd concentration from among 2,592 M2 plants grown in Cd-polluted soil. The grain Cd concentration in the three mutants was <0.05 mg⋅kg−1, compared with an average of 1.73 mg⋅kg−1 in the WT Koshihikari parent (Fig. 1A). The root and shoot Cd concentrations were significantly lower in all M3 lcd-kmt mutants than in the WT (Fig. 1 B and C) when the seedlings were exposed to Cd in hydroponics. The concentrations of iron (Fe), zinc (Zn), and copper (Cu) in shoots and roots did not differ significantly between the lcd-kmt mutants and the WT (Table S1). However, the manganese (Mn) concentration in the shoots was significantly lower in the mutants (73.6–79.7 mg⋅kg−1) than in the WT (1,004 mg⋅kg−1). There was no difference in plant growth among the WT and lcd-kmt1 or lcd-kmt2 mutants, but the growth of lcd-kmt3 was reduced (Table S1) under the sufficient Mn level in hydroponics. These results suggest that Cd might be transported via the Mn pathway into the roots.
Fig. 1.
(A) Frequency distribution of grain Cd concentration in rice mutants (2,592 M2 plants) grown in pots filled with Cd-contaminated soil. The circle and range bar represent the mean and SD of grain Cd concentration in Koshihikari (288 plants). (B and C) Cd concentrations in the shoots and roots of WT Koshihikari and of three low-Cd Koshihikari mutants (lcd-kmt1, lcd-kmt2, and lcd-kmt3) grown in hydroponic culture containing 0.18 µM Cd. Bars labeled with different letters differ significantly (P < 0.05, ANOVA followed by Tukey's test).
Metal Concentrations in Grain and Agronomic Traits of Field Grown lcd-kmt Mutants.
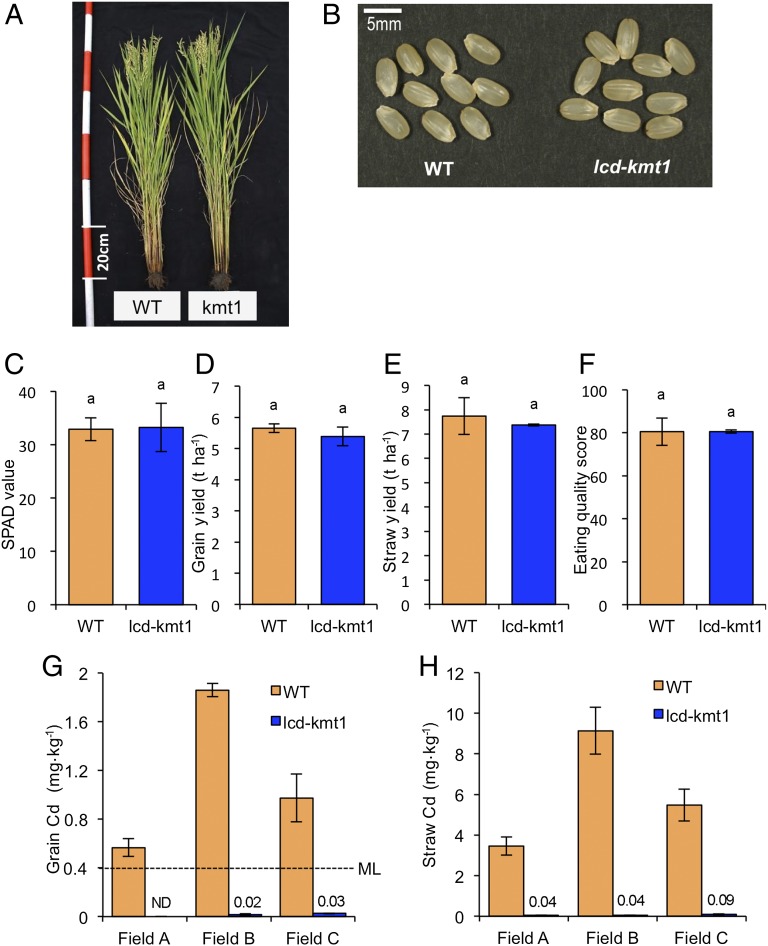
The M4 lcd-kmt mutants and WT were cultivated together in paddy fields to evaluate their metal concentrations and agronomic traits. There were no apparent differences in plant or grain morphologies between WT and lcd-kmt1 (Fig. 2 A and B) or between WT and lcd-kmt2 (Fig. S1 A and C). In addition, there were no significant differences in soil plant analysis development (SPAD) value for chlorophyll content (Fig. 2C), grain and straw yields (Fig. 2 D and E), or eating quality (Fig. 2F) between WT and lcd-kmt1. Similar results were found between WT and lcd-kmt2 (Table S2). This was in contrast to the lcd-kmt3 mutant, which had significantly earlier heading, smaller plant size, higher panicle number, and lower grain and straw yields than the WT (Fig. S1 B and C and Table S2). The concentration of Cd in the grain (unpolished rice) of lcd-kmt1 was extremely low, near the limit of detection (<0.01 mg⋅kg−1), whereas the Cd concentration in the WT grain exceeded the maximum limit of 0.4 mg⋅kg−1 (Fig. 2G). Indeed, the Cd concentration in lcd-kmt1 was <3% of that in the WT. The Cd concentration in the straw was also much lower in lcd-kmt1 than in the WT (Fig. 2H). Similar results were observed for Cd in grain and straw of lcd-kmt2 and lcd-kmt3 (Table S3).
Fig. 2.
Agronomic traits of Koshihikari and lcd-kmt mutants grown in the field. (A) Plant morphologies of WT Koshihikari and lcd-kmt1. (B) Morphologies of unpolished rice grains. (C) Chlorophyll content in the flag leaf determined using a SPAD meter. (D) Grain yield. (E) Straw yield. (F) Eating quality scores evaluated using a taste analyzer; values >80 are considered “good quality.” No significant differences in agronomic traits or eating quality were observed between the WT and lcd-kmt1 (P > 0.05, ANOVA followed by Tukey's test). (G and H) Cd concentration of unpolished rice (G) and straw (H). Plants were grown in Cd-polluted paddy fields in three regions of Japan. Data are presented as means ± SD (n = 5). ND, not detected; ML, maximum allowed Cd concentration for rice (Codex Alimentarius Commission).
The Mn concentration in the grain of lcd-kmt mutants was approximately one-third that of the WT, and an even greater difference of nearly 30-fold was evident in the Mn concentration in the straw (Table S3). The concentrations of Cu, Fe, and Zn in grains of lcd-kmt1 and lcd-kmt2 were similar to those of the WT and slightly higher in lcd-kmt3 than in the WT. This was presumably because of the smaller size of lcd-kmt3 plants. There was no significant difference in Fe concentration in the straw between the WT and lcd-kmt mutants, whereas that of Zn was a little lower in lcd-kmt1 and lcd-kmt2.
Gene Identification.
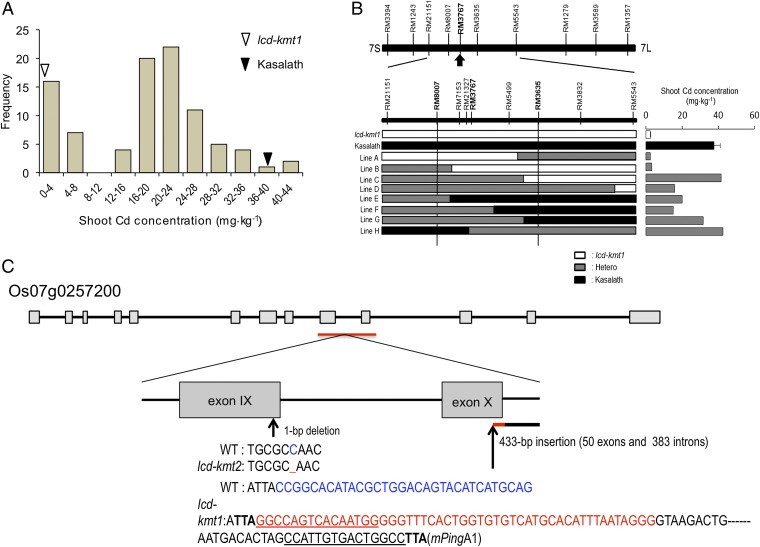
We developed an F2 population by crossing Kasalath, an indica-type rice cultivar, with lcd-kmt1 and then performed positional cloning of the gene(s) responsible for reduced Cd uptake by lcd-kmt1. Among the 92 F2 individuals, 22 plants were categorized as having a similarly low shoot Cd concentration to lcd-kmt1, whereas 70 plants showed a relatively high shoot Cd concentration (Fig. 3A). The segregation ratio was not significantly different from a 1:3 low:high ratio (χ2 = 0.058, P = 0.810), suggesting that the low-Cd trait of lcd-kmt1 is controlled by a single recessive gene. The gene locus associated with shoot and root Cd and Mn concentrations was localized on the short arm of chromosome 7 (Table S4). Linkage analysis showed that the gene was localized in the interval defined by the simple sequence repeat markers RM8007 and RM3635 (Fig. 3B). The maximum logarithm of odds values for all four traits were found at RM3767 (Table S4), which was located 9.07 Mbp from the distal end of the short arm of chromosome 7.
Fig. 3.
Positional cloning of the gene. (A) Frequency distribution for shoot Cd concentration of 92 F2 seedlings derived from a cross between lcd-kmt1 and WT Kasalath, an indica cultivar. White and black triangles represent the mean shoot Cd concentration of lcd-kmt1 and Kasalath, respectively. (B) Gene locus for low shoot Cd concentration on chromosome 7. Arrow indicates the peak logarithm of odds for the putative QTL gene. Graphical genotypes of F2 plants having recombination in the candidate region (Left) and their shoot Cd concentrations (Right) are shown. White, black, and gray bars indicate regions homozygous for the lcd-kmt1 allele, homozygous for the Kasalath allele, and heterozygous for the two alleles, respectively. (C) Structure of OsNRAMP5 (Os07g0257200) and the mutation sites in lcd-kmt1 and lcd-kmt2. Exons and introns are indicated by gray bars and black lines, respectively. The arrow below exon IX indicates the position of a 1-bp deletion in lcd-kmt2 relative to the corresponding sequence in WT Koshihikari. The arrow below exon X indicates the position of a 433-bp insertion in lcd-kmt1. The blue WT nucleotides have been replaced by the red nucleotides in lcd-kmt1. The bold TTA sequences indicate 3-bp target-site duplications, and underlines indicate 15-bp terminal inverted repeats.
We found two genes, OsNRAMP5 (Os07g0257200) and OsNRAMP1 (Os07g0258400), annotated as putative heavy-metal transporters around RM3767 in the Rice Annotation Project Database (http://rapdb.dna.affrc.go.jp/). The OsNRAMP1 cDNA sequence was unchanged in the lcd-kmt mutants relative to the WT. On the other hand, the cDNA and genomic DNA sequences of OsNRAMP5 revealed a single-nucleotide deletion in exon IX of lcd-kmt2 and a 433-bp insertion in the exon X of lcd-kmt1 (Fig. 3C). The latter replaced the terminal 32 bp in exon X of the WT with 50 bp in lcd-kmt1; the remaining 383 bp of the insertion in lcd-kmt1 is expected to be spliced out with intron X. The inserted DNA sequence was identical to the sequence of mPingA1, a member of a class of miniature inverted-repeat transposable elements in rice (22). An ∼227-kbp deletion that included all of OsNRAMP5 was found in lcd-kmt3 (Fig. S2). On the basis of these results, we propose naming the mutant genes as osnramp5-1 for lcd-kmt1 (DNA Data Bank of Japan accession no. AB690552), osnramp5-2 for lcd-kmt2 (AB690553), and osnramp5-3 for lcd-kmt3.
OsNRAMP5 (AB690551) from the WT Koshihikari is predicted to encode a 538-aa protein. The single base pair deletion in osnramp5-2 results in aberrant translation of 53 aa before a new stop codon at amino acid 358 (Fig. S3). On the basis of the cDNA and genome sequencing data of lcd-kmt1, it is likely that an 11-aa region of the WT was replaced with 17 aa at the terminal position of exon X, resulting in a 544-aa protein in the osnramp5-1.
Microarray analysis showed a 2.5-fold increase in OsNRAMP5 expression for lcd-kmt1 compared with the WT (Table S5). The expression of other OsNRAMP genes did not change substantially. Moreover, marked changes in the expression of genes possibly involved in metal transport, such as OsIRT, OsHMA, and OsLCT1, were not found in the mutant. Rather, genes involved in the photosynthetic process were up-regulated considerably, and Fe-deficiency inducible genes were down-regulated in the lcd-kmt1 mutant.
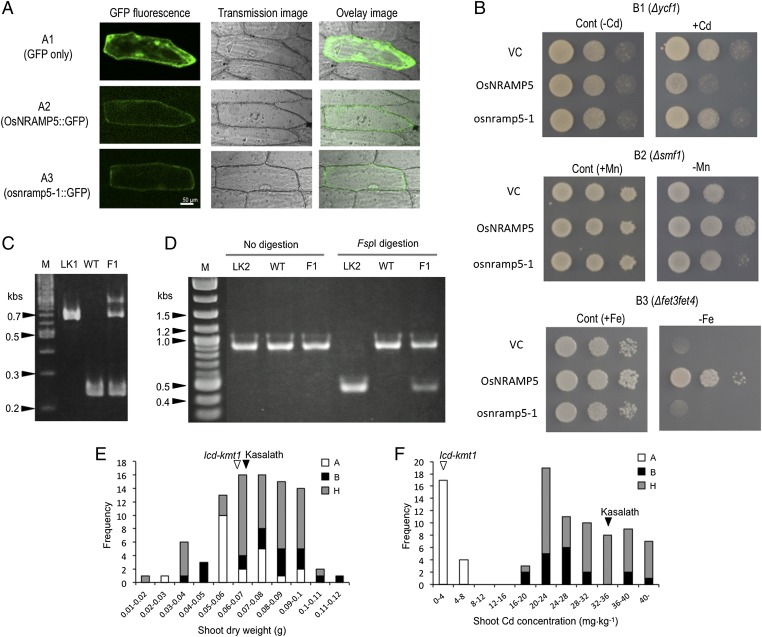
osnramp5-1 fused with GFP was observed at the periphery of the cells but not inside the cells (Fig. 4A), indicating the same localization as OsNRAMP5-GFP (10). This suggests that the mutation in osnramp5-1 did not alter the subcellular localization to the cell membrane. The growth of yeast cells expressing osnramp5-1 was not affected by the Cd treatment (Fig. 4B), although the growth of transformed mutant yeast cells expressing OsNRAMP5 was strongly impaired by Cd. This suggests that the osnramp5-1 could not transport Cd into yeast cells, whereas the WT OsNRAMP5 was able to do so. Furthermore, the mutant protein osnramp5-1 could not transport Mn and Fe, in addition to its inability to transport Cd.
Fig. 4.
(A) Subcellular localization of OsNRAMP5 and osnramp5-1 in transformed onion epidermal cells. (A, 1) GFP only; (A, 2) OsNRAMP5::GFP fusion protein; (A, 3) osnamp5-1::GFP fusion protein. (B) Growth of yeast cells transformed with the vector control (VC), OsNRAMP5, or osnramp5-1. Yeasts were spotted at three dilutions (optical densities at 600 nm of 0.1, 0.01, and 0.001, left to right). (B, 1) Growth of yeast Δycf1 (Cd-sensitive mutant) cells; (B, 2) growth of Δsmf1 (Mn-uptake mutant) yeast cells; (B, 3) growth of Δfet3fet4 (Fe-uptake mutant) yeast cells. Cont, control, with (+) or without (−) the specified metal. (C and D) DNA fragments of the genomic region containing the mutation amplified by PCR. (C) M, size marker; LK1, lcd-kmt1; WT, wild-type Koshihikari; F1, F1 progeny of lcd-kmt1 × Koshihikari. (D) M, size marker; LK2, lcd-kmt2; WT, wild-type Koshihikari; F1, F1 progeny of lcd-kmt2 × Koshihikari. Where indicated, amplified samples were digested with FspI before electrophoresis. (E) Frequency distribution for shoot dry weight of F2 plants derived from a cross between lcd-kmt1 and Kasalath. Using the developed marker (C), the 88 F2 plants were classified into three genotype classes: (A) those homozygous for the osnramp5-1 allele of lcd-kmt1, (B) those homozygous for the OsNRAMP5 allele of Kasalath, and (H) those that were heterozygous for the two alleles. (F) Frequency distribution for shoot Cd concentrations of F2 plants used in E.
Development of Genetic Markers for Breeding Low-Cd Rice.
DNA markers that detect polymorphism in the region of OsNRAMP5 would be useful for developing new cultivars with the low-Cd trait. Thus, we designed primer sets to amplify the mutated region. Different PCR fragment patterns could be readily detected between lcd-kmt1 and WT, because there is a 433-bp insertion in lcd-kmt1 (Fig. 4C). The F1 heterozygous genotype derived from lcd-kmt1 × WT appeared as two bands on the gel. In contrast, no differences in PCR fragment sizes were observed between the undigested PCR products of lcd-kmt2 and WT (Fig. 4D). Although these two alleles differ in length by only 1 bp, the mutation created a unique FspI site in lcd-kmt2. FspI digestion cut the PCR product of lcd-kmt2 (LK2) into two fragments of equal size, whereas the PCR product of WT was not cut by this enzyme. The alleles from both lcd-kmt2 and WT could be detected in the F1.
Using the developed genetic marker, we tested whether the mutant osnramp5-1 allele significantly decreases Cd accumulation in F2 plants derived from a cross between lcd-kmt1 and Kasalath. All F2 plants homozygous for the osnramp5-1 allele of lcd-kmt1 had significantly lower shoot Cd concentrations than those homozygous for the OsNRAMP5 allele of Kasalath and those that were heterozygous for the two alleles (Fig. 4F). This demonstrates that the allelic effect of osnramp5-1 contributes to decreased Cd in rice plants. In addition, the plants homozygous for lcd-kmt1 did not exhibit any significant decrease in their shoot dry weight (Fig. 4E), even if the genetic background was changed by crossing. These results indicate that introduction of the osnramp5-1 allele into the other cultivars might not affect plant growth under the Mn-sufficient conditions.
Discussion
By using ion-beam mutagenesis, we succeeded first in producing nontransgenic rice mutants that accumulate very low Cd in grain of <3% that in Koshihikari, the most popular Japanese temperate japonica rice cultivar. Physiological studies in hydroponic culture demonstrated that decreased Cd uptake by roots leads to low levels of Cd in the shoot and grain of these mutants (Fig. 1). Our QTL analysis suggests that a Fe and Cd transporter gene, OsNRAMP1, on chromosome 7 was the most likely candidate gene, but the OsNRAMP1 cDNA sequence was unchanged in the lcd-kmt mutants. Microarray analysis showed that the expressions of three other genes, OsIRT1, OsIRT2, and OsHMA3, previously related to Cd transport in rice, did not differ between the WT and lcd-kmt1 mutant (Table S5). Instead, we found that the three mutant lines each had a different mutation [i.e., a transposon (mPingA1) insertion, a single-base pair deletion, and a large deletion], in the same gene OsNRAMP5, which is located near OsNRAMP1 (Fig. 3C and Fig. S2). A mPingA1 was probably activated by the ion beams (23), then transposed into a preferred insertion site (TTA) in an exon of OsNRAMP5 (22). It has been reported recently that OsNRAMP5 is involved in Mn, Fe, and Cd transport in rice roots (10, 13). Interestingly, in our previous study (10) the RNAi-induced silencing of OsNRAMP5 in rice promoted Cd translocation to shoots, although root Cd uptake was decreased. In these OsNRANP5-RNAi plants, the expression of OsNRANP5 was suppressed but the expressions of several Fe deficiency-inducible genes were up-regulated. In contrast, the expression of osnramp5-1 present in the lcd-kmt1 plant was increased but the expressions of Fe deficiency-inducible genes were down-regulated (Table S5). Therefore, the differential pattern in root-to-shoot Cd translocation between the RNAi-plants and lcd-kmt mutants could be partly explained by the different expression of Fe deficiency-inducible genes.
Although the osnramp5 mutant gene was expressed in the roots, the mutant transporter proteins failed to mediate uptake of Cd, Mn, and Fe in yeast (Fig. 4B), indicating loss of function of these metal transporters in the cell membrane. A highly conserved consensus transport motif (CTM) between transmembrane domains 8 and 9 was transformed into a hydrophobic segment in osnramp5-1 and was truncated in osnramp5-2 (Fig. S4). The CTM in NRAMP (natural resistance-associated macrophage protein) has been implicated in the interaction with ATP-coupling subunits and to be important for metal transport by these proteins (24). Within the CTM motif, the Gly-347 residue (based on position in OsNRAMP5) is absolutely conserved in all members of the NRAMP family. This residue could be especially important for metal transport activity because in mammalian NRAMP2, a mutant in which glycine is substituted with valine, lost NRAMP2 function in yeast (25). The Gly-347 residue is absent from the osnramp5 mutant proteins (Fig. S3). Therefore, such changes might affect Cd and Mn transport via the cell membrane in the roots.
The lcd-kmt1 and lcd-kmt2 mutants did not exhibit significant negative effects on plant or grain morphology, eating quality, grain yield, or straw yield (Fig. 2 and Table S2), indicating that a transposon insertion or a single base pair deletion on OsNRAMP5 does not negatively affect agronomic traits. In contrast, lcd-kmt3 had earlier heading and smaller plant size than the WT, presumably because of the large DNA deletions in this mutant line (Fig. S2 and Table S2). These results indicate that lcd-kmt1 and lcd-kmt2 can be used directly in breeding programs.
Field trials showed that the lcd-kmt mutants have nearly undetectable Cd concentrations in their grain and staw (Fig. 2 G and H). Although root Cd concentrations were not measured in field conditions, root Cd uptake by lcd-kmt mutants is presumed to be substantially lower than in WT. If a small amount of Cd enters the root cells via other cell membrane metal transporters such as OsIRT1 (11) and OsNRAMP1 (12), a kind of “firewall” system might sequester Cd in the root vacuoles via a functional OsHMA3 transporter (14, 15). Therefore, a defective gene (osnramp5) working together with a functional gene (OsHMA3) may be responsible for the drastic decrease in grain and straw Cd concentrations in the lcd-kmt mutants.
Surprisingly, there were no differences in the leaf chlorophyll contents between WT and lcd-kmt mutants (Fig. 2C and Table S2), even though the shoot (straw) Mn concentration of the lcd-kmt mutants was markedly lower than that of the WT (Table S3), and several genes involved in the photosystem were up-regulated significantly (Table S5). Being adapted to the reducing conditions in paddy soils, rice accumulates high Mn in shoots of up to 2,000 mg⋅kg−1, an order of magnitude higher than that in soybean shoots (26), without damage (27). One T-DNA insertion line and RNAi lines of OsNRANP5 in rice exhibited severe growth inhibition, although the Mn concentration in the straw was 100–200 mg⋅kg−1 in soil culture (13). In contrast, the lcd-kmt mutants did not show any adverse growth with <100 mg⋅kg−1 Mn in straw. Additionally, the F2 plants harboring the osnramp5-1 allele did not exhibit a significant decrease in shoot dry weight, even when their genetic background was changed by crossing. Our results indicate that rice may require less Mn for normal growth than is typically present in the shoot, and the introduction of osnramp5-1 allele into other rice cultivars might not induce the growth inhibition under the Mn-sufficient conditions. Further investigation is needed on the effects of osnramp5 alleles on plant growth and various agronomic traits under low-Mn conditions. The mutant osnramp5 alleles did not significantly reduce Fe in grain and straw of the lcd-kmt mutants, indicating that other Fe transporters, such as OsIRT1 and OsIRT2, are more important than a functional OsNRAMP5 for Fe transport.
We have developed DNA markers that can be used to introduce the mutant nramp5 alleles into various cultivars including indica type by means of marker-assisted selection. Indeed, breeding programs have been launched to transfer the low-Cd trait into other popular cultivars in Japan. These mutant alleles would also reduce the Cd concentration in rice straw being fed to livestock, thereby greatly reducing bioaccumulation of Cd in meat. We therefore believe that our findings provide an important tool for reducing the Cd levels in rice and that the risk of Cd exposure via the food chain will be greatly reduced.
Materials and Methods
Seeds of rice (Oryza sativa L. cv. Koshihikari) were irradiated with 320 MeV carbon ions (12C6+) at a dose of 40 Gy. Three low-Cd mutants (lcd-kmt1, lcd-kmt2, and lcd-kmt3) were identified according to the grain Cd concentration of 2,592 M2 plants determined by inductively coupled plasma mass spectroscopy (ICP-MS). Positional cloning was conducted to identify the gene loci responsible for reduced Cd uptake by lcd-kmt1. The cDNA and genomic DNA of OsNRAMP5 in the WT and lcd-kmt mutants were amplified by PCR and sequenced. Molecular methods were applied to observe the gene location in the rice plants and to analyze gene function. DNA markers were developed to assist breeding of low-Cd rice based on the sequences in the mutation regions for lcd-kmt1 or lcd-kmt2. The cDNA sequences determined in this study have been submitted using the SAKURA nucleotide sequence data submission system through the Web server at the DNA Data Bank of Japan (DDBJ; http://sakura.ddbj.nig.ac.jp/) and are deposited in the DDBJ database under accession nos. AB690551 (OsNRAMP5), AB690552 (osnramp5-1), and AB690553 (osnramp5-2). Further details on the procedures used are available in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank S. Mori, H. Hirochika, M. Yano, Y. Nagamura, K. Naito, and P. Blamey for helpful suggestions on this study; and K. Abe, H. Yamaguchi, T. Ara, T. Ibaraki, and T. Honma for help during our field experiments. This work was supported by the Program for the Promotion of Basic Research Activities for Innovative Biosciences (S.I., N.K.N., and H.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The cDNA sequences reported in this paper have been deposited in the DNA Data Bank of Japan, http://ddbj.nig.ac.jp [accession nos. AB690551 (OsNRAMP5), AB690552 (osnramp5-1), and AB690553 (osnramp5-2)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211132109/-/DCSupplemental.
References
- 1.European Food Safety Authority Scientific opinion on tolerable weekly intake for cadmium. EFSA J. 2011;9(2):1975. [Google Scholar]
- 2.Tsuchiya K. Epidemiological studies on cadmium in the environment in Japan: Etiology of itai-itai disease. Fed Proc. 1976;35(12):2412–2418. [PubMed] [Google Scholar]
- 3. Food and Agriculture Organization/World Health Organization (2010) Joint FAO/WHO Expert Committee on Food Additives, Seventy-Third Meeting, Geneva, 8–17 June 2010. Summary and Conclusions. Available at: www.who.int/foodsafety/publications/chem/summary73.pdf. Accessed June 30, 2012.
- 4. Codex Alimentarius (2008) CODEX STAN 193-1995, Codex general standard for contaminants and toxins in foods and feed. Available at: www.codexalimentarius.net/download/standards/17/CXS_193e.pdf. Accessed June 30, 2012.
- 5. International Rice Research Institute (2011). World Production and Consumption of Domestic Milled Rice. Available at: http://ricestat.irri.org/vis/wrs_quickCharts.php. Accessed June 30, 2012.
- 6.Arao T, Ishikawa S. Genotypic differences in cadmium concentration and distribution of soybean and rice. Jpn Agric Res Q. 2006;40:21–30. [Google Scholar]
- 7.Uraguchi S, et al. Root-to-shoot Cd translocation via the xylem is the major process determining shoot and grain cadmium accumulation in rice. J Exp Bot. 2009;60(9):2677–2688. doi: 10.1093/jxb/erp119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishikawa S, et al. A major quantitative trait locus for increasing cadmium-specific concentration in rice grain is located on the short arm of chromosome 7. J Exp Bot. 2010;61(3):923–934. doi: 10.1093/jxb/erp360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishikawa S, Ae N, Yano M. Chromosomal regions with quantitative trait loci controlling cadmium concentration in brown rice (Oryza sativa) New Phytol. 2005;168(2):345–350. doi: 10.1111/j.1469-8137.2005.01516.x. [DOI] [PubMed] [Google Scholar]
- 10.Ishimaru Y, et al. Characterizing the role of rice NRAMP5 in manganese, iron and cadmium transport. Sci Rep. 2012;2:286. doi: 10.1038/srep00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakanishi H, Ogawa I, Ishimaru Y, Mori S, Nishizawa NK. Iron deficiency enhances cadmium uptake and translocation mediated by the Fe2+ transporters OsIRT1 and OsIRT2 in rice. Soil Sci Plant Nutr. 2006;52:464–469. [Google Scholar]
- 12.Takahashi R, et al. The OsNRAMP1 iron transporter is involved in Cd accumulation in rice. J Exp Bot. 2011;62(14):4843–4850. doi: 10.1093/jxb/err136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sasaki A, Yamaji N, Yokosho K, Ma JF. Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice. Plant Cell. 2012;24(5):2155–2167. doi: 10.1105/tpc.112.096925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyadate H, et al. OsHMA3, a P1B-type of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles. New Phytol. 2011;189(1):190–199. doi: 10.1111/j.1469-8137.2010.03459.x. [DOI] [PubMed] [Google Scholar]
- 15.Ueno D, et al. Gene limiting cadmium accumulation in rice. Proc Natl Acad Sci USA. 2010;107(38):16500–16505. doi: 10.1073/pnas.1005396107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Satoh-Nagasawa N, et al. Mutations in rice (Oryza sativa) heavy metal ATPase 2 (OsHMA2) restrict the translocation of zinc and cadmium. Plant Cell Physiol. 2012;53(1):213–224. doi: 10.1093/pcp/pcr166. [DOI] [PubMed] [Google Scholar]
- 17.Takahishi R, et al. The OsHMA2 transporter is involved in root-to-shoot translocation of Zn and Cd in rice. Plant Cell Environ. 2012;35(11):1948–1957. doi: 10.1111/j.1365-3040.2012.02527.x. [DOI] [PubMed] [Google Scholar]
- 18.Uraguchi S, et al. Low-affinity cation transporter (OsLCT1) regulates cadmium transport into rice grains. Proc Natl Acad Sci USA. 2011;108(52):20959–20964. doi: 10.1073/pnas.1116531109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uraguchi S, Fujiwara T. Cadmium transport and tolerance in rice: Perspectives for reducing grain cadmium accumulation. Rice. 2012;5:5. doi: 10.1186/1939-8433-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kazama Y, et al. Characterization of highly efficient heavy-ion mutagenesis in Arabidopsis thaliana. BMC Plant Biol. 2011;11:161. doi: 10.1186/1471-2229-11-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka A, Shikazono N, Hase Y. Studies on biological effects of ion beams on lethality, molecular nature of mutation, mutation rate, and spectrum of mutation phenotype for mutation breeding in higher plants. J Radiat Res (Tokyo) 2010;51(3):223–233. doi: 10.1269/jrr.09143. [DOI] [PubMed] [Google Scholar]
- 22.Kikuchi K, Terauchi K, Wada M, Hirano HY. The plant MITE mPing is mobilized in anther culture. Nature. 2003;421(6919):167–170. doi: 10.1038/nature01218. [DOI] [PubMed] [Google Scholar]
- 23.Maekawa M, Hase Y, Shikazono N, Tanaka A. Induction of somatic instability in stable yellow leaf mutant of rice by ion beam irradiation. Nucl Instrum Meth B. 2003;206:579–585. [Google Scholar]
- 24.Curie C, Alonso JM, Le Jean M, Ecker JR, Briat JF. Involvement of NRAMP1 from Arabidopsis thaliana in iron transport. Biochem J. 2000;347(Pt 3):749–755. [PMC free article] [PubMed] [Google Scholar]
- 25.Pinner E, Gruenheid S, Raymond M, Gros P. Functional complementation of the yeast divalent cation transporter family SMF by NRAMP2, a member of the mammalian natural resistance-associated macrophage protein family. J Biol Chem. 1997;272(46):28933–28938. doi: 10.1074/jbc.272.46.28933. [DOI] [PubMed] [Google Scholar]
- 26.Ishikawa S, Ae N, Sugiyama M, Murakami M, Arao T. Genotypic variation in shoot cadmium concentration in rice and soybean in soils with different levels of cadmium contamination. Soil Sci Plant Nutr. 2005;51:101–108. [Google Scholar]
- 27.Lidon FC, Barreiro MG, Ramalho JC. Manganese accumulation in rice: Implications for photosynthetic functioning. J Plant Physiol. 2004;161(11):1235–1244. doi: 10.1016/j.jplph.2004.02.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.